Significance
Many current vaccines seek to elicit protective antibody responses along precise evolutionary pathways, and a fundamental understanding of genetic, structural, and biophysical antibody development features will accelerate guided vaccine strategies. Here we determine the structural and biophysical features associated with improvement or fitness of an antibody targeting the HIV fusion peptide, a known vulnerable site on HIV-1. We show that around 1% of possible antibody mutations can improve virus recognition. Beneficial mutations were often clustered far from the binding interface and altered protein structure and dynamics, leading to reduced antibody thermal stability and higher binding affinity to the HIV envelope trimer. These data quantify the genetic and biophysical landscape for improvement of a vaccine-elicted antibody and provide a rational framework to guide HIV vaccines.
Keywords: broadly neutralizing antibodies, HIV-1 vaccines, fusion peptide, somatic hypermutation, yeast display
Abstract
Vaccine-based elicitation of broadly neutralizing antibodies holds great promise for preventing HIV-1 transmission. However, the key biophysical markers of improved antibody recognition remain uncertain in the diverse landscape of potential antibody mutation pathways, and a more complete understanding of anti–HIV-1 fusion peptide (FP) antibody development will accelerate rational vaccine designs. Here we survey the mutational landscape of the vaccine-elicited anti-FP antibody, vFP16.02, to determine the genetic, structural, and functional features associated with antibody improvement or fitness. Using site-saturation mutagenesis and yeast display functional screening, we found that 1% of possible single mutations improved HIV-1 envelope trimer (Env) affinity, but generally comprised rare somatic hypermutations that may not arise frequently in vivo. We observed that many single mutations in the vFP16.02 Fab could enhance affinity >1,000-fold against soluble FP, although affinity improvements against the HIV-1 trimer were more measured and rare. The most potent variants enhanced affinity to both soluble FP and Env, had mutations concentrated in antibody framework regions, and achieved up to 37% neutralization breadth compared to 28% neutralization of the template antibody. Altered heavy- and light-chain interface angles and conformational dynamics, as well as reduced Fab thermal stability, were associated with improved HIV-1 neutralization breadth and potency. We also observed parallel sets of mutations that enhanced viral neutralization through similar structural mechanisms. These data provide a quantitative understanding of the mutational landscape for vaccine-elicited FP-directed broadly neutralizing antibody and demonstrate that numerous antigen-distal framework mutations can improve antibody function by enhancing affinity simultaneously toward HIV-1 Env and FP.
The tremendous circulating sequence diversity of HIV-1 and its capacity to evade host immunity pose unique challenges for vaccine design (reviewed in refs. 1, 2). Broadly neutralizing antibodies (bNAbs) identified from HIV-1 patients target conserved vulnerable epitopes on the HIV-1 envelope protein (Env) to prevent HIV-1 infection, and HIV-1 bNAb elicitation has become a major goal for HIV-1 vaccine design (3, 4). Several HIV-1 vulnerable epitopes have been described (5) including the following: the V1V2 apex (6, 7), the CD4-binding site (8–10), the membrane-proximal external region (11, 12), the glycan-V3 region (also known as N332 glycan supersite) (10, 13), the highly glycosylated region at the center of the silent face on the gp120 subunit (14), and the fusion peptide (FP), which is required for viral entry (15–17). Several complementary approaches seek to develop immunogens that elicit broadly neutralizing HIV-1 antibodies, with promising results (4, 18, 19). However, vaccine-elicited HIV-1 antibodies are often either less potent or less broad than many of the bNAbs identified from human patients. There is a pressing need to better understand bNAb developmental pathways and outline the genetic and structural antibody features that can provide enhanced neutralization breadth and potency for HIV-1 vaccines.
Clinical data show that broadly neutralizing serum antibodies develop naturally in around 20% of individuals with chronic HIV-1 infection (20, 21) and that a smaller number of individuals show highly potent neutralization (22, 23). bNAbs develop via the accumulation of somatic hypermutations (SHM) and affinity maturation following their initial B-cell selection and expansion. While HIV-1–infected individuals may have high titers against HIV-1 antigens, the rarity of bNAbs suggests that the mutations acquired during bNAb development are correspondingly rare and/or that the mutational pathways to effective bNAb development are explored only in the context of chronic HIV-1 antigen exposure over years of viral infection (24). Evidence that high viral load is correlated with higher HIV-1 serum breadth and potency also suggests that larger numbers of sampled antibody maturation pathways are correlated with broad HIV-1 neutralization (20). A major question for HIV-1 vaccine design is thus how to quickly and effectively induce B-cell maturation to bNAbs from a small number of controlled immunizations.
HIV-1 bNAbs are often highly somatically mutated (25–28), and reverting these mutations to antibody germline sequences results in drastic reductions of neutralization breadth and potency (9, 29–31). Complementarity determining regions (CDRs) are known to be mutational hotspots and are often in direct contact with antigen to enable recognition. Antibody framework region (FR) mutations can also modulate neutralization breadth and potency by altering the structural orientation, particularly at heavy:light interface alignments, and by altering the intrinsic flexibility of the paratope (16, 32–35). Not all observed bNAb somatic mutations are required for high neutralization breadth and potency (36), and a better understanding of the critical mutations and antibody structural features (both naturally elicited and vaccine-induced) to improve neutralization breadth and potency would enhance our understanding of bNAb development and guide efficient HIV-1 vaccine strategies (16, 17, 37–41).
Among various immunization approaches reported for eliciting broadly neutralizing antibodies (16–19, 38–42), one strategy targets the HIV-1 FP epitope and has elicited 59 and 31% HIV-1 neutralization breadth in rhesus macaques and mice, respectively, against a broad panel of 208 HIV-1 strains (16, 17). Priming the immune response with soluble FP followed by three Env trimer boosts elicited antibodies with FP-targeted trimer recognition and broad HIV-1 neutralization; however, the structural mechanisms that enable effective anti-FP antibody maturation and pathway selection are not fully understood. Several key unanswered questions include the following: 1) What are the structural features of FP recognition vs. trimer recognition that may appropriately guide antibody development?; 2) What critical parameters control virus neutralization of FP-targeting bNAbs?; and 3) How do beneficial mutations fit into the entire landscape of possible mutational pathways, and what fraction of possible mutations provide increased affinity, neutralization breadth, and potency?
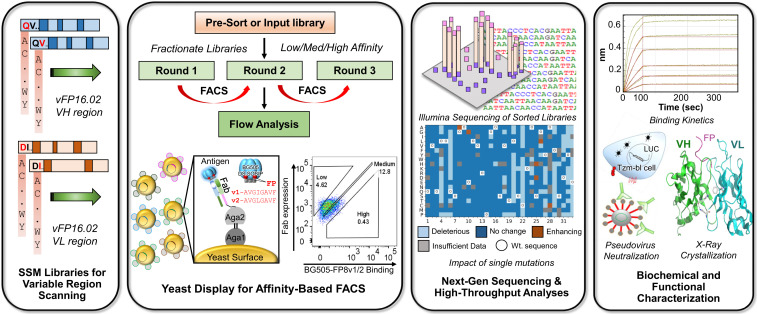
To address these questions, we characterized the genetic and functional fitness landscape of an anti-FP bNAb and used biophysical and structural techniques to follow the mechanisms of antibody improvement. We implemented these screening strategies using the anti-FP bNAb vFP16.02, a vaccine-elicited antibody that neutralizes ∼30% of HIV-1 viral isolates (16). We applied yeast display and fluorescence-activated cell sorting (FACS) coupled with next-generation sequencing (NGS) to identify single mutations that enhanced binding or fitness to multiple HIV-1 BG505 Env trimer variants (Fig. 1). We mapped possible single mutations by their functional impacts and identified a panel of mutations with enhanced binding affinity and neutralization. Structural analyses of a subset of these mutations provided insights into the mechanisms of enhanced neutralization. These data confirm that several parallel mutational pathways exist for HIV-1 bNAb improvement and underscore the importance of improved Env trimer affinity for enhancing neutralization potency and breadth in HIV-1 vaccine designs.
Fig. 1.
Experimental workflow for comprehensive functional analysis of all possible single mutations in an HIV-1 bNAb. SSM libraries were designed for VH and VL regions of the anti–HIV-1 FP bNAb vFP16.02. SSM libraries were cloned into yeast Fab surface display libraries and screened by FACS to determine the functional impact of each possible single mutation. Single-mutation display libraries were sorted for their affinity against a BG505 DS-SOSIP HIV-1 Env trimer with two different FP sequences. Sorted yeast libraries were prepped for NGS to enable quantitative variant tracking across three screening rounds; bioinformatic analyses of NGS data were used to interpret the functional impact of each possible amino acid mutation. Selected variants were characterized for neutralization activity and affinity against soluble FP and against HIV-1 Env. Structural analyses were performed to understand the mechanistic basis of anti–HIV-1 FP bNAb improvement.
Results
High-Throughput Functional Analysis of the vFP16.02 Mutational Landscape.
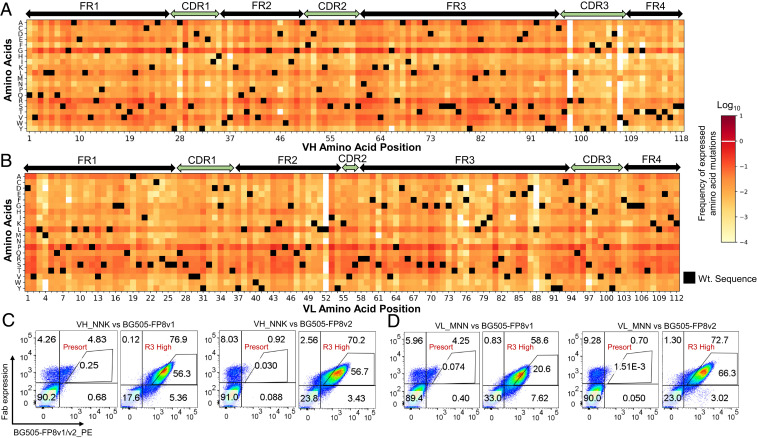
We evaluated the functional impact of all single amino acid (AA) mutations over the entire heavy chain variable region (VH) and light chain variable region (VL) of vFP16.02 using site-saturation mutagenesis (SSM) for deep mutational scanning (43, 44) (Fig. 1). We performed SSM on vFP16.02 VH and VL genes and cloned the resulting libraries into a yeast surface display (45–47) while ensuring an >250-fold theoretical library coverage based on the number of DNA codon variants included in the library throughout all cloning steps (32 codons for each amino acid, see Materials and Methods and SI Appendix, Table S1). On an amino acid level we observed >95% coverage in the presort libraries for both SSM libraries generated (Dataset S2). Antibody Fab libraries were expressed on the yeast surface using a galactose-inducible bidirectional promoter for functional evaluation. Heat maps in Fig. 2 A and B show the frequency of single mutations in yeast libraries after FACS enrichment of populations with fully assembled surface Fab display (hereafter referred to as the VL+ library) (SI Appendix, Fig. S1A).
Fig. 2.
vFP16.02 single mutation library generation and yeast display functional screening. Heat maps for the Fab-expressing VL+ SSM libraries of (A) the vFP16.02 VH region, and (B) the VL region, that were confirmed to express as Fabs on yeast surface (SI Appendix, Fig. S1A). (C) VH_NNK libraries sorted for high affinity against BG505-FP8v1 and BG505-FP8v2 showed enhanced binding affinity after three rounds of sorting by FACS. Presort (input) libraries were compared to the Round 3 sorted libraries to evaluate the degree of affinity enrichment. (D) VL_MNN libraries were similarly sorted against BG505-FP8v1 and BG505-FP8v2 and showed enhanced binding affinity against both Env variants after three rounds of sorting. In addition to sorting for high-affinity populations as shown here, we also sorted each SSM library to identify medium-affinity and low-affinity clonal variants in parallel (Fig. 1 and SI Appendix, Fig. S1).
We next applied FACS to fractionate antibody variant libraries into low-, medium-, and high-affinity populations (47, 48). We leveraged NGS analysis and quantitative data mining to track antibody sequences across different sort groups and to assess the functional impact of each mutation in comprehensive library-scale experiments. We assessed binding affinity against two BG505 DS-SOSIP.664 HIV-1 Env trimers (49, 50): one trimer carried the most prevalent 8-mer FP (FP8) sequence (BG505-FP8v1: AVGIGAVF), and another trimer presented the second-most-common FP8 (BG505-FP8v2: AVGLGAVF) (15).
Yeast libraries were sorted into affinity gates that compared the intensities of surface Fab expression with antigen binding (47, 48) (Fig. 1). Yeast-carrying mutations detrimental to Env binding were enriched by sorting with a low-affinity gate. Likewise, yeast with mutations that had no effect or a similar affinity as the template monoclonal vFP16.02 Fab were sorted into a medium-affinity gate, and finally, a population with enhanced binding affinity was sorted and enriched using a high-affinity gate. After three rounds of FACS screening, the high-affinity groups showed phenotypic binding improvements, where ∼60% of the sorted high-affinity population for most libraries against both antigens fell within the original Round 1 high-affinity gate; the expected phenotypic changes were also observed in medium- and low-affinity groups (Fig. 2 C and D and SI Appendix, Fig. S1). The changes that we observed in sorted yeast populations across screening rounds suggested that our sort strategies had successfully binned mutational variants according to their functional affinity characteristics.
Single Mutations Enable Improvements in HIV-1 Affinity and Neutralization.
We performed NGS analysis of sorted yeast libraries to quantitatively track AA mutations driving the phenotypic differences that were observed in sorted populations. We performed Illumina 2 × 300-bp MiSeq NGS of plasmids isolated from each sorted population to track the enrichment of single-mutation variants throughout screening rounds. We applied an enrichment ratio (ER) formula (Materials and Methods) to quantify the distribution of each vFP16.02 variant among sorted populations (47, 51, 52) (Fig. 3A and SI Appendix, Fig. S2). Each mutation was assigned to an affinity group (deleterious/no-change/affinity enhancing) in which it had the highest ER (SI Appendix, SI Methods).
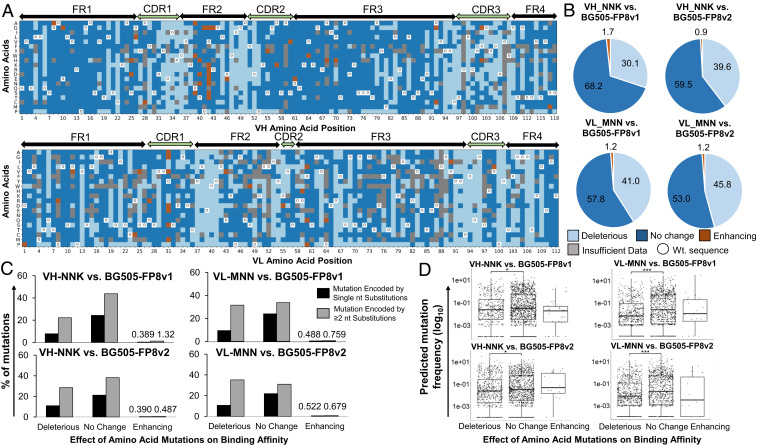
Fig. 3.
Comprehensive single-mutation landscape analysis of the vFP16.02 VH and VL regions. (A) Heat map displaying the functional impact of single mutations against BG505-FP8v1 antigen. Heat maps against BG505-FP8v2 are provided in SI Appendix, Fig. S2. (B) Pie charts quantifying the impact of single-mutation VH_NNK and VL_MNN libraries against BG505-FP8v1 and -v2 antigens. (C) Minimum number of DNA nucleotide substitution mutations required for each single-amino-acid mutation from germline, grouped by functional mutation impacts. (D) Probability of every possible mutation arising in a murine repertoire using a gene-specific substitution matrix model, with grouping by amino acid mutation functional impact. A two-sample Kolmogorov–Smirnov test was used to judge the statistical significance: *P < 0.05; ***P < 0.0001.
Around 1.3% of single mutations showed a beneficial effect on HIV-1 Env-binding affinity, whereas 59.6% had a binding affinity highly similar to the template vFP16.02 gene, and the remaining 39.1% of single mutations were detrimental to HIV-1 Env binding (Fig. 3B and SI Appendix, Table S2). Our large-scale affinity analysis showed that beneficial vFP16.02 mutations clustered in antibody FRs, especially FR2, for both VH and VL. We observed very few mutations in CDRs, suggesting that the vFP16.02 paratope had a more limited potential for affinity-enhancing mutations than FR residues, possibly a result of prior in vivo CDR optimization (16).
To evaluate the rarity of different single mutations among the population, we examined the minimum number of DNA base mutations needed to achieve high-affinity AA substitutions. We observed that most high-affinity mutations required two DNA base substitutions, which are considered to be more rare mutational events than single mutations (Fig. 3C). In total, 35/99 (35.4%) affinity-enhancing mutations required only a single DNA mutation, whereas 64/99 (64.6%) of affinity-enhancing AA mutations required a double-nucleotide mutation. To further characterize the relative rarity of SHMs grouped by their binding affinity impact, we compared the relative affinity performance of each mutation (enhancing, deleterious, or no change) to its probability of occurring in the murine repertoire using a gene-specific substitution matrix model (53). Probability analyses revealed that affinity-enhancing mutations identified for binding against the autologous BG505-FP8v1 trimer [one immunogen used in the original vFP16.02 discovery study (16)] were skewed toward more rare mutations (Fig. 3D). We observed that deleterious mutations generally comprised more rare mutations than the subset that did not substantially change binding affinity, and these differences were statistically significant (VH_NNK vs. BG505-FP8v1, P = 0.0116; VL_MNN vs. BG505-FP8v1, P = 9.02 × 10−11; VH_NNK vs. BG505-FP8v2, P = 0.0022, Fig. 3D).
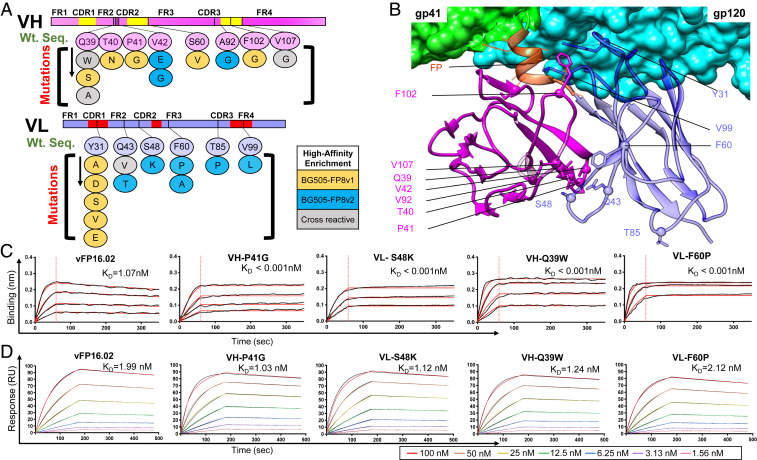
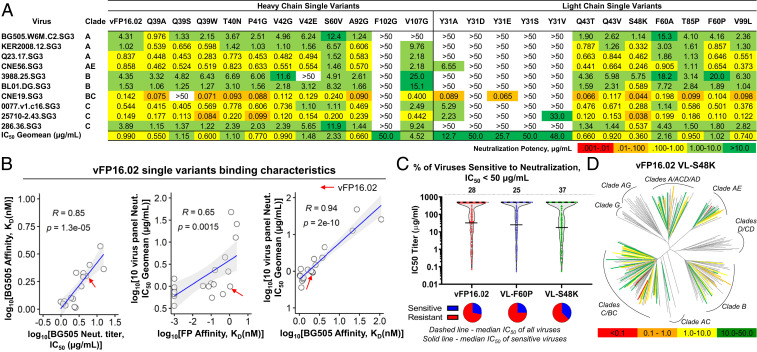
From our NGS data mining, we selected a panel of 23 single AA variants against BG505-FP8v1 and/or BG505-FP8v2 for IgG expression and characterization (11 VH mutations and 12 VL mutations; SI Appendix, Table S3). These beneficial mutations clustered around the FR2 and the VH:VL interface, but other locations were also represented (Fig. 4 A and B and SI Appendix, Figs. S3 and S4). We assessed binding affinities for each single variant against soluble FP8v1 peptide and against the BG505-FP8v1 trimer (Fig. 4 C and D and SI Appendix, Figs. S5 and S6). Surprisingly, we observed a ≥1,000-fold affinity improvement for several VH and VL single AA variants against the soluble FP8v1 peptide (Fig. 4C and SI Appendix, Fig. S5). Binding affinity improvements against trimer antigens were more measured and increased two- to three- fold with several variants (Fig. 4D and SI Appendix, Fig. S6 and Table S3). In addition to soluble antibody-based validation of high-affinity variants, we also selected a total of 24 variants from medium- and low-affinity gates for confirmation as yeast colonies by FACS. Of the 23 single-mutation Fab variants that expressed on yeast, 21/23 showed the expected binding characteristics based on our low- or medium-affinity predictions (SI Appendix, Fig. S7), To better understand how affinity differences for soluble FP vs. HIV-1 trimers influenced HIV-1 protection, we proceeded with antibody neutralization assays and variant structural characterization.
Fig. 4.
Selected single variants showed moderately enhanced affinity against the autologous BG505 trimer and orders of magnitude enhanced affinity against soluble FP. (A) Single mutational variants selected after FACS screening and bioinformatic analysis with locations highlighted within the antibody variable region. Twenty-three single-amino-acid variants against BG505-FP8v1/v2 were selected (11 VH and 12 VL mutations). Some mutations were identified in library screening against both BG505-FP8 v1 and v2. (B) Structural mapping of enriched single mutations onto a cryo-electron microscopy structure of vFP16.02 in complex with a BG505.SOSIP trimer (PDB ID 6CDO) (16). (C) Bio-layer interferometry response curves for key variants binding against soluble FP showing over 1,000-fold enhanced binding affinity (SI Appendix, Fig. S5). (D) Surface plasmon resonance response curves for binding against BG505-FP8v1 trimer for key single mutation variants (SI Appendix, Fig. S6).
Single-Mutation Variants Improve Neutralization Breadth and Potency.
We assessed mutational variants for HIV-1 neutralization breadth and potency against a panel of 12 viral isolates comprising 8 viruses with FP8v1 (AVGIGAVF), 2 viruses with FP8v2 (AVGLGAVF), and 1 virus each with FP8 sequences AVGIGAMI and AVGIGAMF (Fig. 5A and SI Appendix, Table S4). Neutralization potencies against the BG505 virus showed improvement for several single variants with improved binding affinity toward BG505-FP8v1 trimer (Fig. 5 A and B and SI Appendix, Table S3). The most potent single AA mutant, VL-S48K, showed three-fold improved potency in a 10-virus panel (geomean IC50 = 0.36 µg/mL compared to vFP16.02 geomean IC50 = 0.99 µg/mL) and an almost four-fold improved BG505 neutralization IC50 (1.14 vs. 4.31 µg/mL, Fig. 5A and SI Appendix, Tables S3 and S4, with maximum IC50 defined as 50 µg/mL). Interestingly, we observed that dramatic increases in soluble FP8v1 affinity (≥1,000-fold, SI Appendix, Table S3 and Fig. S5) did not provide commensurate enhancements of BG505 virus neutralization potency. One variant (VH-V42E) showed that three orders of magnitude increased affinity to the soluble fusion peptide, but resulted in a 50% decrease in neutralization potency (SI Appendix, Table S3 and Fig. S5A).
Fig. 5.
High-affinity single mutants showed improved HIV-1 neutralization breadth and potency that was closely correlated to BG505 trimer affinity. (A) Single variants analyzed for pseudovirus IC50 neutralization on a 10-virus panel comprising n = 8 FP8v1 and n = 2 FP8v2 viral isolates. (B) Relationships between BG505-FP8v1 IC50 neutralization titer and BG505-FP8v1 affinity (KD, Left); and relationship between IC50 neutralization titer on a 10-virus panel and fusion peptide affinity (Middle) and BG505_FP8v1 affinity (Right). (SI Appendix, Fig. S6). Pearson correlation coefficients are provided in the panels; red arrows indicate wild-type vFP16.02. Shaded areas indicate 95% confidence limits. (C) Neutralization breadth and potency assessed on a 208-viral isolate panel for wild-type vFP16.02, VL-S48K, and VL-F60P with a maximum IC50 of 50 µg/mL. (D) Neutralization dendrogram showing the diversity of 208 viral strains neutralized by VL-S48K with branches colored according to neutralization potency and nonneutralized viruses shown in gray. Also see a dendrogram for VL-F60P for comparison in SI Appendix, Fig. S8.
After aggregating data from the panel of expressed antibodies, it became clear that improved affinity against the BG505-FP8v1 trimer was closely correlated with improved BG505 neutralization potency (R = 0.85, P = 1.3 × 10−5, Fig. 5B), suggesting that trimer affinity is a potential driving factor in FP-targeted HIV-1 neutralization potency. Neutralization breadth also increased with BG505 trimer affinity (Fig. 5B), indicating that enhancing bNAb affinity against a single HIV-1 trimer variant (as performed in our screening assays) can improve potency against other diverse viruses. Enhanced soluble FP affinity was correlated with (but not sufficient for) enhanced HIV-1 neutralization breadth and potency (SI Appendix, Fig. S5 B, Middle). We compared antibody-binding kinetics against BG505-FP8v1 (on-rate [Kon] and off-rate [Koff]) with neutralization potencies (SI Appendix, Fig. S8). We noted a statistically significant positive correlation between BG505-FP8v1 Koff and BG505-FP8v1 neutralization titer, with a Pearson’s R value of 0.84 (P = 2.2 × 10−5); however, the correlation for Koff was not as strong as the correlation for overall affinity, KD (R = 0.85, P = 1.3 × 10−5, Fig. 5B). A similar trend in BG505 Koff and 10-virus panel neutralization IC50 geomean was observed (SI Appendix, Fig. S8 D and E). Together, these data suggest that trimer affinity was the most important driving factor in HIV-1 neutralization among the variants of this FP-targeting bNAb lineage.
To better understand the structural mechanisms of improved anti–HIV-1 affinity and neutralization, we selected two variants with different functional profiles for in-depth neutralization assays and structural study. First, we chose VL-S48K, which showed the largest observed improvements in soluble FP affinity, trimer affinity, and neutralization, and also VL-F60P, which showed strong enhancements in soluble FP affinity, but not in trimer binding affinity or neutralization. On a 208-virus panel, we observed a 33% enhanced neutralization breadth for VL-S48K compared to vFP16.02 (neutralizing 77 vs. 58 isolates at 50 µg/mL, Fig. 5 C and D and Dataset S1). In contrast, VL-F60P neutralized 25% of viruses (52 isolates) in the large panel, providing additional support for the hypothesis that BG505 SOSIP trimer affinity is directly correlated with neutralization breadth (despite the enhanced soluble FP8v1 affinity for VL-F60P) (SI Appendix, Fig. S9). Interestingly, large panel neutralization data show that VL-S48K neutralized two viruses carrying FP8v2 sequences which are not neutralized by vFP16.02, although with neutralization IC50 > 300 µg/mL (T250-4, IC50 = 314 µg/mL; and T278-50, IC50 = 470 µg/mL, Dataset S1). Thus, our mutational scanning revealed that a single mutation can broaden the specificity of vFP16.02 to neutralize viruses carrying FP8v2, while at the same time enhance potency toward FP8v1 strains.
Single Mutations Influence Local Structural Dynamics to Improve HIV-1 Neutralization.
We sought to define the structural basis of improved antibody neutralization breadth and potency in the vFP16.02 lineage. First, we performed X-ray cocrystallization of the two single variants with contrasting functional features (VL-S48K and VL-F60P) with their soluble fusion peptide antigen and compared those structures to the previously reported structure of the vFP16.02 antibody (Fig. 6A). We observed minor conformational changes in the CDR-H1 and -H2 of both single variants. The variable regions of both structures aligned closely to the unmutated vFP16.02 structure, with an overall C-alpha rmsd < 0.5 Å. The structural analysis, however, revealed the formation of a new hydrogen bond in VL-S48K between VH-W108 and the serine→lysine mutation at position 48, implying that interactions at the VH:VL interface could alter FP and Env trimer-binding characteristics.
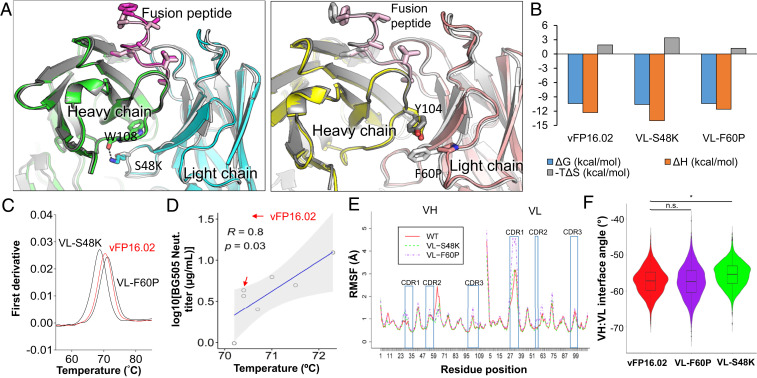
Fig. 6.
Structural basis of enhanced HIV-1 fusion peptide recognition. (A) Ribbon representation of an X-ray crystal structure of VL-S48K (Left) and VL-F60P (Right) in complex with FP8v1 and structurally superposed on wild-type vFP16.02 (shown in gray). (B) Thermodynamic binding interaction parameters obtained by isothermal titration calorimetry for VL-S48K, VL-F60P, and vFP16.02 in complex with the BG505-FP8v1.DS.SOSIP trimer. (C) nanoDSF measurements of melting temperature (Tm) for single vFP16.02 variants. VL-S48K and VL-F60P had opposing effects on the Tm of vFP16.02 (shown in red for comparison). (D) Correlation between Tm and BG505 neutralization titers for single-mutation neutralizing variants that were predicted to bind to BG505-FP8v1.DS.SOSIP. Shaded area indicates 95% confidence limits. (E) Average RMSF plot for three independent molecular dynamics simulation performed for vFP16.02, VL-S48K, and VL-F60P. (F) VH:VL interface angle distribution throughout molecular dynamics simulations, calculated using ABangle. VL-S48K showed a statistically significant change in interface angle with respect to vFP16.02 and VL-F60P with *P < 0.0001 using Mann–Whitney U test. vFP16.02 and VL-F60P interface angles were not statistically significant. n.s., not significant.
Next we explored VL-S48K and VL-F60P binding thermodynamics against the BG505-FP8v1 DS.SOSIP trimer using isothermal calorimetry (ITC) (Fig. 6B). In comparison to vFP16.02, VL-S48K (which had improved soluble FP affinity, HIV-1 trimer affinity, and neutralization potency) showed increased favorable enthalpy and a higher entropy change of binding to the BG505-FP8v1 trimer. In contrast, VL-F60P (which improved soluble FP affinity, but did not improve trimer affinity nor neutralization potency) exhibited decreases in favorable enthalpy and a lower entropy change of binding to the HIV-1 trimer. Binding of VL-F60P was not associated with improved trimer interactions; in fact, the magnitudes of both enthalpy and entropy changes were reduced, indicating less conformational restriction upon binding. Based on these ITC data, the VL-S48K mutation enables improved trimer surface interactions (larger negative enthalpy), albeit partially compensated by restriction (more unfavorably entropy) and suggestive that VL-S48K may permit additional flexibility or conformational sampling by the unbound antibody.
In light of these observed entropy changes, we measured the melting temperature of single variants using differential scanning fluorimetry to determine how single mutations can influence protein stability. We observed slight changes in the thermal stabilities of many single variants compared to vFP16.02 (SI Appendix, Fig. S10 and Table S3). Interestingly, we observed a close correlation between thermal stabilities of single variants and BG505 autologous neutralization for the variants that showed neutralization activity on a 10-virus panel (i.e., neutralization IC50 <50 µg/mL) (R = 0.80, P = 0.03, Fig. 6 C and D), suggesting that antibodies with slightly lower thermal stabilities may have better neutralization potencies. Similar features have been reported for other HIV-1 bNAbs (54), and these trends in thermal stability agreed with changes in entropy observed in ITC (Fig. 6B). We did not notice any correlations between soluble FP8v1 affinity and Tm (SI Appendix, Fig. S11).
To better understand possible conformational changes of key single mutation variants, we performed three independent molecular dynamics simulations for vFP16.02, VL-S48K, and VL-F60P, for a total of 2 µs. The root mean square fluctuation (RMSF), which monitors residue movement over the course of simulation, was 2.62 Å (SD = 1.56 Å) for the VL-F60P CDR-L1, whereas in the vFP16.02 and VL-S48K variants the CDR-L1 RMSFs were 1.71 Å (SD = 0.93 Å) and 1.79 Å (SD = 1.06 Å), respectively (Fig. 6E). Simulation trajectories revealed that both mutations appeared to increase local CDR-H1 RMSF and decrease local RMSF in FR-H3, which may contribute to the greatly enhanced soluble FP8v1 peptide affinity (Fig. 6E). In contrast, VL-F60P greatly increased CDR-L1 RMSF, suggesting that the lower CDR-L1 loop flexibility for the VL-S48K variant could assist in forming stable interactions with the HIV-1 Trimer Env surface (55). We also observed a change of around 2° in the VH:VL interface angle for VL-S48K compared to vFP16.02 and VL-F60P, and these changes were statistically significant (P < 0.0001, Fig. 6F). Taken together, these data show that single FR mutations in antigen distal regions modulated positional plasticity near the FP paratope to enhance trimer-binding affinities, resulting in the observed affinity, breadth, and potency improvements.
Multimutation Combinations Provide Minor Additive Effects for Neutralization.
We rationally designed 12 combination variants to determine possible synergistic effects from single-mutation combinations (SI Appendix, Table S5). Variants were designed by selecting mutations from different areas of the variable regions and also by combining mutations at various distances from the FP-binding pocket. We observed some minor additive effects, with neutralization IC50 potency against BG505 improved by seven-fold (SI Appendix, Tables S6 and S7). Combination variants did not enhance neutralization breadth on a 10-virus panel substantially beyond the improvements provided by single variants separately, showing that multiple parallel single mutations may often present similar or nonsynergistic structurally beneficial effects.
Discussion
Elicitation of bNAbs against HIV-1 Env is a major focus of HIV-1 vaccine development, and enhanced understanding of the mechanisms of bNAb recognition will inform HIV-1 vaccine designs. Here we performed a comprehensive single-mutation functional analysis of an anti-FP antibody to understand possible pathways for antibody improvement. We characterized the genetic mechanisms shaping the affinity maturation landscape, analyzed the probability of finding these mutations in vivo, and revealed the detailed structural mechanisms that led to affinity and HIV-1 neutralization improvements in this anti-FP bNAb lineage.
Our comprehensive mutational analyses suggest that multiple, parallel pathways exist for achieving higher neutralization breadth and potency via single-AA mutations. Deep mutational scanning and functional screening revealed that around 39% of the single mutations were detrimental to binding, 60% of mutations were neutral to changes in affinity, and around 1% of possible AA mutations showed affinity improvement. We identified multiple key structural changes, including the formation of a new hydrogen bond at the VH:VL interface and changes to conformational flexibility, as structural features of increased antibody breadth and potency against the HIV-1 FP epitope.
We observed a cluster of enhancing mutations around the framework region at the VH:VL interface; beneficial FR mutations have also been observed in prior studies (33–35, 56–61). Here we showed that these FR mutations can provide an enormous enhancement in affinity against soluble HIV-1 FP (in some cases, more than three orders of magnitude), but only a subset of FR mutations simultaneously conferred enhanced Env affinity for improved HIV-1 neutralization breadth and potency. Here we observed altered CDR loop flexibility as a result of FR mutations; other studies of antibodies targeting HIV-1 vulnerable epitopes also revealed some FR mutations that increased paratope flexibility (28). We did not identify new beneficial mutations making direct contact with the soluble FP8 antigen, suggesting that the FP-binding paratope may have been effectively optimized for vFP16.02 in vivo. We observed some yeast display selection for variants with increased thermal stability, including VH-Y31A, and prior studies have noted that thermal stability is correlated with enhanced protein expression and antibody specificity (61–64). Rare AA mutations can show altered biophysical properties that reduce thermal stability, and the link between decreased thermal stability and neutralization is supported by other bNAb studies that have examined the role of framework mutations to modulate antibody function (33, 63).
The most improved single mutant that we identified (VL-S48K) showed a 33% enhancement in neutralization breadth against a broad panel of 208 viral isolates and three-fold improvement in binding affinity to the BG505-FP8v1 trimer. VL-S48 is located in the FR2 region, distal to the FP-binding pocket, and appears to modulate antibody affinity by enabling a better fit for FP inside the binding pocket while also maintaining appropriately stable interactions with the surrounding BG505 Env trimer epitope. We show that many single mutations can enhance binding affinity to a soluble FP8 peptide, but may not provide substantial gains against the fully assembled HIV-1 trimer (SI Appendix, Table S3). A major challenge for FP-based vaccine development is thus finding ways to identify and elicit these “dual-purpose” and often rare mutations like VL-S48K that enhance affinity to both the soluble FP sequence and the complete HIV-1 trimer. Our comprehensive screening platform outlines a way to identify and track such dual-purpose mutations. Incorporation and discovery of rare mutations can also be used for improving the breadth and potency of therapeutic candidates.
Our HIV-1–binding kinetic assays and neutralization data reveal a direct correlation between BG505 neutralization potency and BG505 affinity (and also between HIV-1 neutralization breadth and BG505 affinity), suggesting that trimer affinity drives effective HIV-1 neutralization for FP-binding antibodies. These data lend further support to vaccination protocols that prime with FP peptide immunogens and boost with fully assembled trimers to enhance affinity against HIV-1 vulnerable epitopes (17). The observation that BG505 trimer affinity is also correlated with neutralization breadth suggests that, for FP-targeting antibodies, enhanced affinity for a particular trimer can lead to enhanced affinity against other trimers as well, reducing the breadth of trimer exposure that may be required for broad HIV-1 vaccines. We used ERs to effectively track functional mutations across sort rounds via NGS, although we mostly relied on binning clones into their highest prevalence affinity gate, and we observed that ERs did not correlate directly with the degree of antibody affinity improvement (SI Appendix, Fig. S12). Future studies will apply various experimental (47, 48, 65–67) and computational (51, 66, 68) techniques to optimize HIV-1 antibody:antigen affinity predictions via yeast display. Many of the high-affinity mutations that we report here required a minimum of two DNA base substitutions (Fig. 3D and SI Appendix, Figs. S13 and S14). Several studies have shown that rare mutations are important for improved HIV-1 neutralization breadth (9, 36, 69, 70). Acquiring rare mutations in vivo may require strong selection for FR mutations during affinity maturation and B-cell clonal expansion, potentially via carefully selected HIV-1 trimer vaccine boost antigens.
In summary, here we determined the comprehensive functional mutational landscape of HIV-1 bNAbs, revealing striking features of affinity, breadth, and neutralization. We performed biophysical and structural characterization of key mutations, which revealed the existence of multiple parallel pathways to improve HIV bNAbs. These data highlight the genetic and structural features for an antibody to acquire improved HIV-1 neutralization breadth and suggest that strategies to improve the HIV-1 trimer affinity of vaccine-elicited anti-FP antibodies offer a promising approach toward an effective HIV-1 vaccine.
Materials and Methods
Site Saturation Mutagenesis Library Construction for vFP16.02.
vFP16.02 VH and VL genes were cloned into the yeast display vector pCT_VHVL-Kappa1 (47) using multifragment infusion (Takara Bio).
The VH and VL SSM libraries were made separately using a one-pot single-site saturation mutagenesis protocol, which entails sequential nicking, degradation, and PCR generation of a mutated top strand of the template plasmid, followed by replication of the bottom strand incorporating the copied mutation (44). A detailed description of the yeast display vector system, SSM library generation, and yeast plasmid transformation is provided in SI Appendix, SI Methods.
Affinity Screening of Yeast Surface Display Fab Libraries.
Transformed yeast libraries were grown in SGCAA medium (20 g/L galactose, 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 5.4 g/L Na2HPO4, 8.6 g/L NaH2PO4.H2O; SGCAA from TEKnova) supplemented with 2 g/L dextrose (SGDCAA) for 36 h to induce Fab expression. We used the HIV BG505 SOSIP Env trimer with two different versions of fusion peptide: AVGIGAVF (BG505-FP8v1, the native FP8 sequence) and AVGIGALF (BG505-FP8v2). HIV-1 trimer antigens were biotinylated for screening and conjugated with streptavidin-PE (SA-PE) at 1:1 molar ratio (9) (Thermo Fisher Scientific). For affinity sorting, libraries were washed and stained with anti-FLAG FITC to label surface Fab expression (ANTI-FLAG M2-FITC, Sigma-Aldrich), along with SA-PE–conjugated HIV-1 trimers at 70 nM for BG505-FP8v1 and 100 nM for BG505-FP8v2, separately. After sorting, samples were collected and grown in low-pH SDCAA (20 g/L dextrose, 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 10.4 g/L trisodium citrate, and 7.4 g/L citric acid monohydrate). For the first round of sorting where the yeast population was separated into low, medium, and high affinity, at least 3 × 107 yeast cells were stained in a three-way sort for low, medium, and high binding affinities, and VL + Fab-expressing populations were collected. Two additional rounds of enrichment for low, medium, and high affinity were performed.
MiSeq Preparation and Bioinformatic Analysis.
Plasmids were isolated from sorted yeast as described previously (67). Briefly, VH and VL regions were amplified from the yeast display vector using Kapa Hifi HotStart ReadyMix (Kapa Biosystems, Roche) (47). An additional round of PCR was done to add barcodes and adaptors for Illumina 2 × 300 sequencing.
Raw fastq Illumina sequences were quality-filtered for a quality score of 30 over 90% of the raw reads using Fastxtoolkit (v0.0.14) (http://hannonlab.cshl.edu/fastx_toolkit/). Quality-filtered NGS libraries were processed as previously described to obtain in-frame antibody AA sequences (47, 71) (Dataset S2). We calculated the ER of each single-amino-acid mutation in the sorted libraries to track the change in mutation prevalence across each round of sorting, calculated as:
| [1] |
See SI Appendix, SI Methods, for additional details regarding bioinformatic analysis.
Antibody Expression and Characterization.
Antibodies were expressed and purified as described previously (16). Briefly, VH and VL plasmids (1:1 DNA ratio) were cotransfected into Expi293F cells using Turbo293 transfection reagent (Speed Biosystems) following manufacturer’s protocols. See SI Appendix, SI Methods, for a detailed description of antibody expression, purification, and affinity assays.
Neutralization Assays.
10-virus panel.
A single round of entry neutralization assays using TZM-bl target cells was performed to assess monoclonal antibody neutralization as described (72). Briefly, the monoclonal antibodies were tested via five-fold serial dilutions starting at up to 500 μg/mL. Monoclonal antibodies were mixed with the virus stocks in a total volume of 50 μL and incubated at 37 °C for 1 h. We then added 20 μL of TZM-bl cells (0.5 million/mL) to the mixture and incubated it at 37 °C overnight. On day 2, 130 μL complete Dulbecco’s modified Eagle medium was added, and cells were lysed on day 3 and assessed for luciferase activity measured in relative light units. The 50 and 80% inhibitory concentrations (IC50 and IC80) were determined using a hill-slope regression analysis as described (72).
208-virus panel.
To assess monoclonal antibody neutralization on a panel of 208 HIV-1 Env-pseudotyped viruses, automated 384-well microneutralization assays were performed as described previously (73).
X-ray Crystallization and Structural Analysis.
FP8v1 (AVGIGAVF) was dissolved in 100% dimethylsulfoxide at a 100 mg/mL concentration and then mixed with Fabs in a 10:1 molar ratio to reach a final protein complex concentration of 10 mg/mL. Fab:FP8 complexes were screened for crystallization using 576 conditions from Hampton screen, Precipitant Synergy screen, and Qiagen Wizard screen with a mosquito robot delivering 100 nL of reservoir solution mixing with 100 nL of protein solution per drop. Further details on X-ray crystallization and structural analysis are provided in SI Appendix, SI Methods.
Isothermal Titration Calorimetry.
The binding of Fab fragments of vFP16.02 and the variants VL-S48K and VL-F60P to the BG505-FP8v1 DS.SOSIP trimer was studied thermodynamically by ITC at 25 °C using a VP-ITC microcalorimeter from Malvern Instruments. BG505-FP8v1 and the antibody fragments were prepared in phosphate-buffered saline (PBS), pH 7.4, and the solutions were thoroughly degassed prior to the experiments. In each titration, the solution containing the antibody fragment was added stepwise in 10-μL aliquots to the stirred calorimetric cell (cell volume ∼1.4 mL) containing BG505-FP8v1 at 0.7 to 0.8 µM. The concentration of Fab in the syringe was 25 to 30 µM. Analysis of ITC data is in SI Appendix, SI Methods
Differential Scanning Fluorimetry.
Melting temperatures of the antigen-binding fragments were determined by nano differential scanning fluorimetry (nanoDSF) on a Prometheus NT.48 instrument (NanoTemper Technologies). A total of 10 μL of protein samples at a concentration between 0.5 and 1.0 mg/mL in PBS were loaded into capillaries and placed on the sample holder. A temperature gradient from 40 to 90 °C was scanned at 1 °C/min, and the intrinsic fluorescence intensity ratio (350 nm:330 nm) was recorded. The data analysis was performed using Prometheus NT.48 control software.
Molecular Dynamics Simulation.
Three molecular dynamics (MD) simulations were carried out in order to characterize the vFP16.02 wild-type, VL-S48K, and VL-F60P Fabs under physiological conditions. The X-ray structure 6CDO (16) was used as initial atomistic model. VL-S48K and VL-F60P mutants were generated using PyMOL software (the PyMOL Molecular Graphics System, Version 2.0, Schrödinger) MD simulations were performed using NAMD2.13 engine (74) with the CHARMM36 force field (75, 76); additional details are provided in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Francisco Martinez Becerra and Jennifer Hackett for help with FACS experiments and NGS, respectively; Krisha McKee and Matias Fernando Gutierrez Gonzalez for help with data analysis; and Tim Whitehead for experimental advice and guidance. This work was supported by the University of Kansas Departments of Pharmaceutical Chemistry and Chemical Engineering; by NIH Grants 1DP5OD023118, R21AI143407, R01AI141452, P20GM103638, and P20GM113117; and by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH. It was also supported by National Institute of Allergy and Infectious Diseases, the National Cancer Institute (HHSN261200800001E to A.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011653118/-/DCSupplemental.
Data Availability
Raw sequence and structural data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA678717 and in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under Protein Data Bank (PDB) IDs 6WWC and 6WX2, respectively.
Change History
July 26, 2021: Figure 4 has been updated; please see accompanying Correction for details.
References
- 1.Gaschen B., et al., Diversity considerations in HIV-1 vaccine selection. Science 296, 2354–2360 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Rey F. A., Lok S.-M., Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 172, 1319–1334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes B. F., Burton D. R., Mascola J. R., Multiple roles for HIV broadly neutralizing antibodies. Sci. Transl. Med. 11, eaaz2686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong P. D., Mascola J. R., HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Chuang G.-Y., et al., Structural survey of broadly neutralizing antibodies targeting the HIV-1 Env trimer delineates epitope categories and characteristics of recognition. Structure 27, 196–206.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLellan J. S., et al., Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker L. M.et al.; Protocol G Principal Investigators , Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J., et al., Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou T., et al., Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou T.et al.; NISC Comparative Sequencing Program , Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guenaga J., Wyatt R. T., Structure-guided alterations of the gp41-directed HIV-1 broadly neutralizing antibody 2F5 reveal new properties regarding its neutralizing function. PLoS Pathog. 8, e1002806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., et al., Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longo N. S., et al., Multiple antibody lineages in one donor target the glycan-V3 supersite of the HIV-1 envelope glycoprotein and display a preference for quaternary binding. J. Virol. 90, 10574–10586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T.et al.; NISC Comparative Sequencing Program , A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity 48, 500–513.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong R., et al., Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu K., et al., Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong R.et al.; NISC Comparative Sequencing Program , Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell 178, 567–584.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders K. O., et al., Targeted selection of HIV-specific antibody mutations by engineering B cell maturation. Science 366, eaay7199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steichen J. M., et al., A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 366, eaax4380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray E. S.et al.; CAPRISA002 Study Team , The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85, 4828–4840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hraber P., et al., Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simek M. D., et al., Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83, 7337–7348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landais E., Moore P. L., Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology 15, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao H.-X.et al.; NISC Comparative Sequencing Program , Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheid J. F., et al., Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Xiao X., Chen W., Feng Y., Dimitrov D. S., Maturation pathways of cross-reactive HIV-1 neutralizing antibodies. Viruses 1, 802–817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corti D., et al., Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5, e8805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheid J. F., et al., Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mouquet H., et al., Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467, 591–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X.et al.; NISC Comparative Sequencing Program , Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao X., et al., Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: Implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390, 404–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong P. D., Mascola J. R., Human antibodies that neutralize HIV-1: Identification, structures, and B cell ontogenies. Immunity 37, 412–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein F., et al., Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ovchinnikov V., Louveau J. E., Barton J. P., Karplus M., Chakraborty A. K., Role of framework mutations and antibody flexibility in the evolution of broadly neutralizing antibodies. eLife 7, e33038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomlinson I. M., et al., The imprint of somatic hypermutation on the repertoire of human germline V genes. J. Mol. Biol. 256, 813–817 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Georgiev I. S., et al., Antibodies VRC01 and 10E8 neutralize HIV-1 with high breadth and potency even with Ig-framework regions substantially reverted to germline. J. Immunol. 192, 1100–1106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burton D. R., Advancing an HIV vaccine; advancing vaccinology. Nat. Rev. Immunol. 19, 77–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dosenovic P., et al., Immunization for HIV-1 broadly neutralizing antibodies in human Ig Knockin mice. Cell 161, 1505–1515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan H., et al., Glycan masking focuses immune responses to the HIV-1 CD4-binding site and enhances elicitation of VRC01-class precursor antibodies. Immunity 49, 301–311.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jardine J. G., et al., HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian M., et al., Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell 166, 1471–1484.e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jardine J., et al., Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fowler D. M., Fields S., Deep mutational scanning: A new style of protein science. Nat. Methods 11, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrenbeck E. E., et al., Plasmid-based one-pot saturation mutagenesis. Nat. Methods 13, 928–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boder E. T., Wittrup K. D., Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Chao G., et al., Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Wang B., et al., Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat. Biotechnol. 36, 152–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reich L. L., Dutta S., Keating A. E., SORTCERY: A high-throughput method to affinity rank peptide ligands. J. Mol. Biol. 427, 2135–2150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon Y. D., et al., Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ou L.et al.; VRC Production Program , Preclinical development of a fusion peptide conjugate as an HIV vaccine immunogen. Sci. Rep. 10, 3032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koenig P., et al., Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc. Natl. Acad. Sci. U.S.A. 114, E486–E495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mason D. M., et al., High-throughput antibody engineering in mammalian cells by CRISPR/Cas9-mediated homology-directed mutagenesis. Nucleic Acids Res. 46, 7436–7449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheng Z.et al.; NISC Comparative Sequencing Program , Gene-specific substitution profiles describe the types and frequencies of amino acid changes during antibody somatic hypermutation. Front. Immunol. 8, 537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finton K. A. K., et al., Ontogeny of recognition specificity and functionality for the broadly neutralizing anti-HIV antibody 4E10. PLoS Pathog. 10, e1004403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dingens A. S., et al., Complete functional mapping of infection- and vaccine-elicited antibodies against the fusion peptide of HIV. PLoS Pathog. 14, e1007159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khalifa M. B., et al., Effects on interaction kinetics of mutations at the VH-VL interface of Fabs depend on the structural context. J. Mol. Recognit. 13, 127–139 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Kumagai I., Nishimiya Y., Kondo H., Tsumoto K., Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10. J. Biol. Chem. 278, 24929–24936 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Masuda K., et al., The role of interface framework residues in determining antibody V(H)/V(L) interaction strength and antigen-binding affinity. FEBS J. 273, 2184–2194 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Nakanishi T., Tsumoto K., Yokota A., Kondo H., Kumagai I., Critical contribution of VH-VL interaction to reshaping of an antibody: The case of humanization of anti-lysozyme antibody, HyHEL-10. Protein Sci. 17, 261–270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi H., Tamura H., Shimba N., Shimada I., Arata Y., Role of the domain-domain interaction in the construction of the antigen combining site. A comparative study by 1H-15N shift correlation NMR spectroscopy of the Fv and Fab fragments of anti-dansyl mouse monoclonal antibody. J. Mol. Biol. 243, 494–503 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Warszawski S., et al., Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLOS Comput. Biol. 15, e1007207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimitrov J. D., Kaveri S. V., Lacroix-Desmazes S., Thermodynamic stability contributes to immunoglobulin specificity. Trends Biochem. Sci. 39, 221–226 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Julian M. C., Li L., Garde S., Wilen R., Tessier P. M., Efficient affinity maturation of antibody variable domains requires co-selection of compensatory mutations to maintain thermodynamic stability. Sci. Rep. 7, 45259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kowalski J. M., Parekh R. N., Mao J., Wittrup K. D., Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J. Biol. Chem. 273, 19453–19458 (1998). [DOI] [PubMed] [Google Scholar]
- 65.Adams R. M., Mora T., Walczak A. M., Kinney J. B., Measuring the sequence-affinity landscape of antibodies with massively parallel titration curves. eLife 5, e23156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalsky C. A., et al., Rapid fine conformational epitope mapping using comprehensive mutagenesis and deep sequencing. J. Biol. Chem. 290, 26457–26470 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medina-Cucurella A. V., Whitehead T. A., Characterizing protein-protein interactions using deep sequencing coupled to yeast surface display. Methods Mol. Biol. 1764, 101–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Blarcom T., et al., Precise and efficient antibody epitope determination through library design, yeast display and next-generation sequencing. J. Mol. Biol. 427, 1513–1534 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Wiehe K., et al., Functional relevance of improbable antibody mutations for HIV broadly neutralizing antibody development. Cell Host Microbe 23, 759–765.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Umotoy J.et al.; IAVI Protocol C Investigators; IAVI African HIV Research Network , Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity 51, 141–154.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DeKosky B. J., et al., Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. U.S.A. 113, E2636–E2645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X., et al., Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sarzotti-Kelsoe M., et al., Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips J. C., et al., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Best R. B., et al., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guvench O., et al., CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 7, 3162–3180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence and structural data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA678717 and in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under Protein Data Bank (PDB) IDs 6WWC and 6WX2, respectively.