Significance
The DREB1/CBF transcription factors function as master switches for cold stress adaptation in Arabidopsis. Cold stress strongly induced the DREB1 genes, and elucidation of the mechanisms of DREB1 induction is important to clarify the cold stress responses in plants. We revealed that the clock-related MYB proteins RVE4 and RVE8 were quickly transferred from the cytoplasm to the nucleus in response to cold stress and functioned as direct transcriptional activators of DREB1 expression. The other clock-related MYB proteins, CCA1 and LHY, suppressed DREB1 expression under unstressed conditions and were rapidly degraded under cold stress, which suggests that these factors indirectly regulate cold-inducible DREB1 expression. We concluded that posttranslational regulation of the clock-related transcription factors triggered cold-inducible gene expression.
Keywords: cold stress responses, Arabidopsis, cold-inducible DREB1/CBF expression, circadian clock-related transcription factors, posttranslational regulation
Abstract
Cold stress is an adverse environmental condition that affects plant growth, development, and crop productivity. Under cold stress conditions, the expression of numerous genes that function in the stress response and tolerance is induced in various plant species, and the dehydration-responsive element (DRE) binding protein 1/C-repeat binding factor (DREB1/CBF) transcription factors function as master switches for cold-inducible gene expression. Cold stress strongly induces these DREB1 genes. Therefore, it is important to elucidate the mechanisms of DREB1 expression in response to cold stress to clarify the perception and response of cold stress in plants. Previous studies indicated that the central oscillator components of the circadian clock, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), are involved in cold-inducible DREB1 expression, but the underlying mechanisms are not clear. We revealed that the clock-related MYB proteins REVEILLE4/LHY-CCA1-Like1 (RVE4/LCL1) and RVE8/LCL5 are quickly and reversibly transferred from the cytoplasm to the nucleus under cold stress conditions and function as direct transcriptional activators of DREB1 expression. We found that CCA1 and LHY suppressed the expression of DREB1s under unstressed conditions and were rapidly degraded specifically in response to cold stress, which suggests that they act as transcriptional repressors and indirectly regulate the cold-inducible expression of DREB1s. We concluded that posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression. Our findings clarify the complex relationship between the plant circadian clock and the regulatory mechanisms of cold-inducible gene expression.
Plants, as sessile organisms, have evolved to develop resilient systems to adapt to and survive in extreme temperature conditions at the molecular, cellular, and physiological levels. In response to cold stress, a large number of genes have been reported to be induced in diverse plant species (1, 2). The products of these genes function to enhance freezing stress tolerance and to regulate gene expression and signal transduction under cold stress conditions. The dehydration-responsive element/C-repeat (DRE/CRT) with a conserved core motif of A/GCCGAC has been identified as a cis-acting element that activates gene expression in response to both cold and dehydration stresses in plants (3, 4). Three Arabidopsis (Arabidopsis thaliana) transcription factors, DRE binding protein 1A/C-repeat binding factor 3 (DREB1A/CBF3), DREB1B/CBF1, and DREB1C/CBF2, bind to the DRE and act as master switches in cold-inducible gene expression (5, 6). Induction of the DREB1 genes triggers the cold-responsive transcriptional cascade followed by the expression of many cold-inducible genes encoding proteins that function in cold stress response and tolerance (2, 7–9). Thus, elucidation of the mechanisms of DREB1 expression in response to cold stress is important to clarify the perception and response of cold stress in plants.
Many transcription factors have been shown to regulate the cold-inducible expression of DREB1s (10). The MYC-like transcription factor INDUCER OF CBF EXPRESSION 1 (ICE1) was first identified as a transcription factor in the cold-inducible expression of DREB1A using the ice1-1 mutant (11). However, recent data have demonstrated that DREB1A repression in ice1-1 is achieved by DNA methylation-mediated gene silencing triggered by an inserted T-DNA, not by genetic regulation, and that ICE1 is not involved in DREB1 expression (10, 12, 13). Among six CALMODULIN-BINDING TRANSCRIPTION ACTIVATORs (CAMTAs) (14), CAMTA3 and CAMTA5 were shown to activate the expression of DREB1B and DREB1C (15, 16). The expression of many cold-inducible genes, including DREB1A and DREB1C, is associated with circadian regulation, and these genes are strongly induced in daytime and weakly induced at night (17, 18). CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and its close homolog, LATE ELONGATED HYPOCOTYL (LHY), which are key components of circadian oscillators (19, 20), function as important positive regulators in the cold-responsive expression of DREB1A and DREB1C (15, 21). Detailed analyses of these transcription factors revealed that plants recognize cold stress as two different signals, rapid and gradual temperature decreases, and that each of the three DREB1 genes is differentially induced in response to these two stress signals through two different signal transduction pathways. CAMTA3 and CAMTA5 respond to a rapid temperature decrease, strongly inducing the expression of DREB1B and DREB1C. In contrast, CCA1 and LHY induce the expression of DREB1A and DREB1C in response to both rapid and gradual temperature decreases (15). The presence of the two different signaling pathways leading to the expression of DREB1s has allowed plants to efficiently acquire freezing tolerance in response to a gradual temperature decrease during season changes and a sudden temperature drop during the night and abnormal weather (15).
Therefore, the regulatory factors involved in the cold-inducible expression of DREB1A and DREB1C associated with circadian regulation are likely CCA1 and LHY (15). However, even in cca1 lhy double mutants, their cold-inducible expression persisted, exhibiting a circadian rhythm (15), which indicates that other clock-related factors may be involved in their expression. CCA1 and LHY are suppressors of the expression of several core clock genes, such as TIMING OF CAB EXPRESSION 1 (TOC1)/PSEUDO-RESPONSE REGULATOR 1 (PRR1), PRR5, PRR7, and PRR9 at normal temperature (22–24). The molecular mechanisms by which circadian clock-related factors, including CCA1 and LHY, activate cold stress-specific expression of DREB1s have not yet been clarified.
We analyzed the mechanisms of the cold-inducible expression of DREB1A and DREB1C that are associated with circadian regulation. We found that the clock-related MYB transcription factors REVEILLE4/LHY-CCA1-Like1 (RVE4/LCL1) and RVE8/LCL5 directly activate this cold-inducible expression, and CCA1 and LHY indirectly regulate the cold-inducible expression of DREB1s as transcriptional repressors. Notably, we revealed that the activities of these transcription factors are posttranslationally regulated in response to cold stress.
Results
Evening Element Is Required for the Cold Induction of DREB1A.
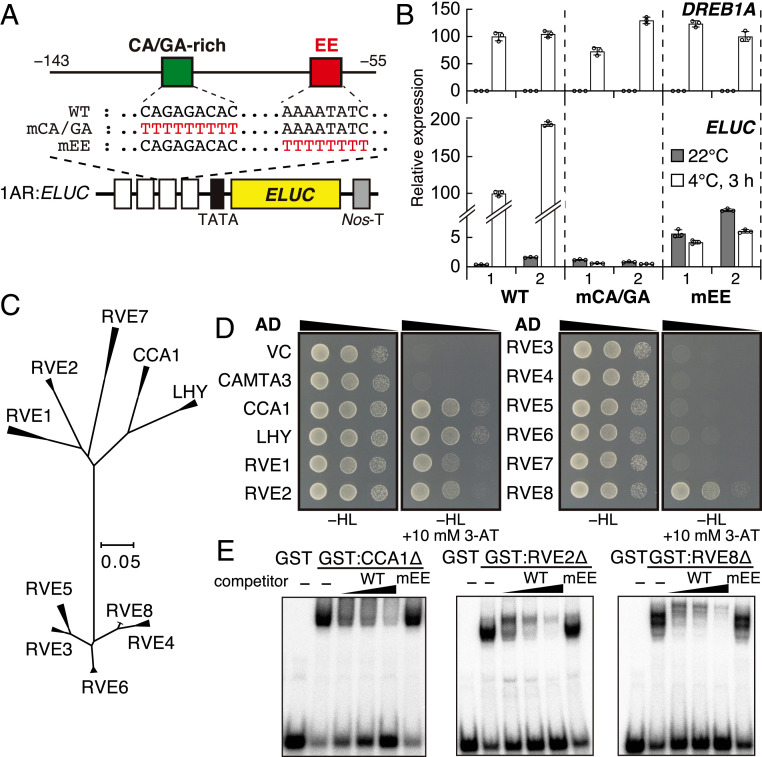
Previously, we revealed that a DREB1A promoter region (1AR:ELUC, −143 to −55 bp from the transcription start site) contains sufficient cis-acting elements for the cold-inducible expression of DREB1A (10). To identify the cis-acting elements in 1AR, we compared the promoter sequences of DREB1A orthologs in Brassicaceae species with those in A. thaliana. Four Brassicaceae species (Arabidopsis lyrata, Capsella rubella, Brassica rapa, and Boechera stricta) have three DREB1 genes with a tandem arrangement similar to that of A. thaliana (SI Appendix, Fig. S1). We therefore compared the DREB1A promoter sequences in the five Brassicaceae species (SI Appendix, Fig. S2A). In their 1AR homologous regions, a CA/GA-enriched sequence (CA/GA-rich) was conserved, except in the AlDREB1A promoter, and the Evening Element (EE) was completely conserved in all these DREB1A promoters. The EE is a major cis-acting element in the promoters of circadian-controlled genes and is known to be a CCA1/LHY binding site (22, 23, 25). We also aligned the DREB1B and DREB1C promoters in the five Brassicaceae species and found an EE in all the DREB1C promoters but not in the DREB1B promoters (SI Appendix, Fig. S2 B and C). We generated transgenic Arabidopsis plants expressing the emerald Luc (ELUC) reporter gene driven by the mutated 1AR fragments (Fig. 1A). The transgenic plants were treated at 4 °C for 3 h from 2 h after dawn (LL2). In the plants carrying the CA/GA-rich mutated 1AR or EE mutated 1AR (1ARmEE), cold induction of ELUC was completely lost (Fig. 1B). Additionally, in the 1ARmEE plants, the ELUC expression levels at 22 °C were mildly increased compared with those of the wild-type (WT), implying that CA/GA-rich and EE positively regulate DREB1A expression under cold stress conditions, and EE also negatively regulates its expression under unstressed conditions.
Fig. 1.
Mutation analysis of the 1AR element in the DREB1A promoter and identification of transcription factors that bind to the Evening Element (EE) of 1AR. (A) Schematic diagram of the 1AR:ELUC reporter construct. The emerald luciferase (ELUC) reporter gene driven by four tandem repeats of a DREB1A promoter fragment (−143 to −55; 1AR) and its minimal promoter (TATA) is expressed in transgenic Arabidopsis. (B) Expression of the ELUC reporter gene in response to cold stress. Two representative lines of each construct are shown. The bars refer to the mean ± SD of three biological replicates. (C) Phylogenetic tree of the CCA1/LHY/RVE transcription factors in Brassicaceae species. The alignment is shown in SI Appendix, Fig. S3. (D) Yeast one-hybrid assays between the 1AR element and CCA1/LHY/RVE transcription factors. For the reporter, four tandemly repeated 1AR fragments were cloned in a pHisi-1 vector. (E) Electrophoretic mobility shift assays of CCA1, RVE2, and RVE8.
Since we found that EE is an essential cis-acting element for DREB1A induction and the EE binding proteins CCA1 and LHY have been shown to be involved in the cold-inducible expression of DREB1A (15, 21), we focused on RVE family proteins, including CCA1/LHY, as candidate regulators of DREB1A induction (Fig. 1C and SI Appendix, Fig. S3). To screen the RVE proteins that bind to 1AR, we performed yeast one-hybrid assays (Y1H). Among eight RVE proteins (26–28) (RVE1 to RVE8; Fig. 1C and SI Appendix, Fig. S3), RVE1, RVE2, and RVE8 interacted with 1AR in addition to CCA1/LHY (Fig. 1D). We performed electrophoretic mobility shift assays and found that GST-fusion proteins of CCA1, RVE2, and RVE8 fragments physiologically interacted with 1AR. The addition of unlabeled 1AR caused competition for the binding of the probes, but the addition of a 1AR with a mutated EE did not, indicating that these factors can directly recognize the EE (Fig. 1E).
RVE4 and RVE8 Activate Cold-Inducible DREB1A Expression.
We next generated transgenic plants overexpressing the sGFP fusion of the RVE family genes encoding EE binding proteins and analyzed the expression levels of DREB1A under cold stress conditions (SI Appendix, Fig. S4). Although RVE4 did not interact with 1AR in Y1H, we overexpressed this gene because it has high homology with RVE8 (Fig. 1C). The cold stress treatment was initiated at three time points, 2 h (LL2), 6 h (LL6), and 14 h (LL14), after the start of the light period by gradual cooling to 4 °C, and the plants were incubated for 3 h. In the vector control (VC) plants, DREB1A expression was strongly induced by cold stress from LL2 and LL6 but weakly induced by cold stress from LL14. In the plants overexpressing CCA1-sGFP, LHY-sGFP, RVE4-sGFP, and RVE8-sGFP, DREB1A expression was similarly induced by the cold stress from LL2 and LL6 and more strongly induced by that from LL14 compared with that in the VC plant. In contrast, its induction by LL2 and LL6 was comparable but was significantly decreased in the RVE1-sGFP- and RVE2-sGFP-overexpressing plants compared with the VC plants. These results suggest that not only CCA1/LHY but also the other RVE homologs could regulate cold-inducible DREB1A expression.
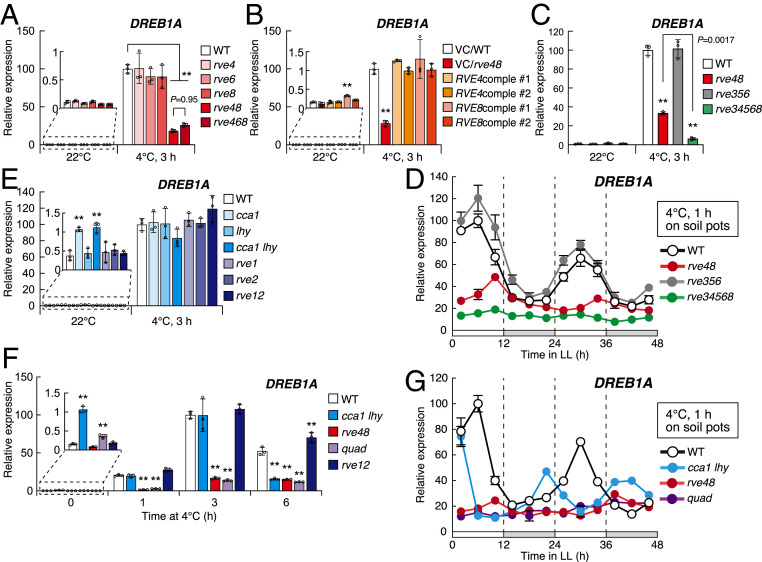
We then analyzed DREB1A expression using plants with single and multiple mutations of the RVE4, RVE6, and RVE8 genes (Fig. 2A and SI Appendix, Fig. S5). In all the single mutants, the DREB1A expression levels were similar to those in the WT plants under cold stress conditions at 4 °C for 3 h from LL2. In contrast, the expression was substantially decreased in the rve4 rve8 mutant (rve48), and this reduction was complemented by the expression of RVE4 or RVE8 in the mutant plants (Fig. 2B). Moreover, the decreased level of DREB1A expression was similar in the rve4 rve6 rve8 (rve468) triple mutants compared with that in rve48 (Fig. 2A). Since DREB1A was still induced by cold stress at low levels in the rve48 mutants, we generated rve3 rve4 rve5 rve6 rve8 (rve34568) quintuple mutant plants and analyzed DREB1A expression. In the quintuple mutant, DREB1A expression was further reduced to a slight level under cold stress conditions, but the DREB1A expression level in the rve3 rve5 rve6 (rve356) mutant was similar to that in the WT plants (Fig. 2C). These results suggest that originally, RVE4 and RVE8 activate DREB1A expression under cold stress conditions in the plants. RVE3, RVE5, and RVE6 showed only partial effects on the expression of DREB1A when the activities of RVE4 and RVE8 were lost. To compare the circadian rhythms of cold-inducible expression of the DREB1A genes in the rve48, rve356, and rve34568 mutant plants, we measured the expression levels of these genes after cold stress treatments for 1 h every 4 h during 2 d after the transition from light/dark cycles to free-running conditions using these mutant plants (Fig. 2D). In rve48, DREB1A expression was almost constant at low levels, which were similar to those in the subjective night, and DREB1A expression was constant at more reduced levels in rve34568. However, the expression pattern of DREB1A in rve356 was similar to that in the WT plants, which also indicated that RVE4 and RVE8 are the major transcriptional activators of DREB1A under cold stress conditions.
Fig. 2.
Cold-inducible DREB1A expression in the mutant plants of CCA1/LHY/RVE genes. (A–C) Cold-inducible expression of DREB1A in the mutant plants of RVEs and the complementation lines of rve4 rve8. Two-week-old seedlings of the mutant plants of RVEs (A and C) and the complementation lines (B) grown on agar plates were gradually chilled at 4 °C for 3 h from 2 h after dawn (LL2). As complementation lines, the GFP fusion genes of the genomic fragments, including the promoter regions of RVE4 and RVE8, were expressed in rve4 rve8 (RVE4comple and RVE8comple, respectively). Two representative lines of each construct are shown. (D) Effects of the circadian clock on the cold-inducible DREB1A expression in the rve multiple mutant plants under cold stress conditions. Two-week-old seedlings grown in soil pots under a 12-h light/12-h dark cycle were transferred to free-running conditions under continuous light from dawn. Cold treatments were initiated every 4 h from 2 h to 46 h after the beginning of the free-running conditions. At each time point, the seedlings were rapidly cooled at 4 °C for 1 h. (E) Cold-inducible expression of DREB1A in the mutant plants of CCA1/LHY and RVE1/RVE2. The expression levels of DREB1A before and after the cold stress treatment from LL2 (22 °C and 4 °C, 3 h) were measured. (F) Time course analyses of DREB1A expression were performed using the double and quadruple (quad: cca1 lhy rve48) mutant plants of CCA1/LHY and RVEs. Two-week-old seedlings grown on agar plates were gradually chilled at 4 °C for 1 h, 3 h, and 6 h from 2 h after dawn (LL2). (G) Effects of the circadian clock on the cold-inducible DREB1A expression in the double and quadruple mutant plants of CCA1/LHY and RVE4/RVE8. Experimental conditions were the same as in D. The bars refer to the mean ± SDs in three biological replicates. The letters above the bars indicate significant differences (**P < 0.01 as analyzed using one-way ANOVA followed by a Tukey’s post hoc test) in the gene expression levels at each time point.
We then analyzed DREB1A expression using plants with single and double mutations of the CCA1, LHY, RVE1, and RVE2 genes. In the single, cca1 lhy and rve1 rve2 (rve12) double mutant plants, significant changes in DREB1A expression were not detected under cold stress conditions (Fig. 2E). We further generated a quadruple mutant plant of CCA1/LHY and RVE4/RVE8 (cca1 lhy rve4 rve8; quad) by crossing with these double mutant plants (Col background) and examined the time course of DREB1A expression using the quadruple and double mutant plants (Fig. 2F). In cca1 lhy, the expression levels under unstressed conditions were clearly increased compared with those in the WT, while those under cold stress conditions for 1 h and 3 h were similar to those in the WT. In contrast, the expression levels of DREB1A were clearly decreased in rve48 and quad but were similar in rve12 to those in the WT after both 1-h and 3-h cold stress treatments. However, under cold stress conditions for 6 h, DREB1A expression was decreased in cca1 lhy as well as in rve48 and quad and increased in rve12. All these results suggest that CCA1 and LHY function as negative regulators under unstressed conditions and positive regulators under prolonged cold stress conditions. Moreover, RVE1 and RVE2 function as negative regulators under prolonged cold stress conditions. To examine the circadian rhythms of cold-inducible expression levels of the three DREB1 genes in rve48, cca1 lhy, and quad, we analyzed the expression levels of these genes after cold stress treatments for 1 h every 4 h during 2 d after the transition from light/dark cycles to free-running conditions (Fig. 2G and SI Appendix, Fig. S6). In rve48 and quad, the expression of DREB1A and DREB1C was almost constant at low levels, which were similar to those in the subjective night, although shifted, but obvious rhythmic patterns of these genes were observed in cca1 lhy. The expression levels of DREB1B were not strongly decreased in any of the mutants. These results indicated that RVE4 and RVE8 primarily act as transcriptional activators of cold-inducible expression of DREB1A and DREB1C associated with circadian control.
RVE4 and RVE8 Regulate the Expression of DREB1-Downstream Genes and Freezing Tolerance.
We then analyzed the expression of the DREB1-downstream genes RD29A and COR15A in the mutant plants (SI Appendix, Fig. S7A). The cold-inducible expression levels of these genes were decreased in the rve48 and quad plants treated for 6 to 24 h at 4 °C compared with those in the WT plants. We conducted freezing tolerance tests in the morning using the mutants (SI Appendix, Fig. S7 B and C). The cca1 lhy plants showed a freezing-sensitive phenotype, as previously shown (21). The rve48 and quad plants were sensitive to freezing stress. In contrast, the rve12 plants revealed increased freezing tolerance compared with the WT plants. These results suggest that RVE4/RVE8 and CCA1/LHY function as positive regulators and RVE1/RVE2 function as negative regulators in freezing tolerance, mediating the regulation of the cold-inducible expression of genes downstream of DREB1.
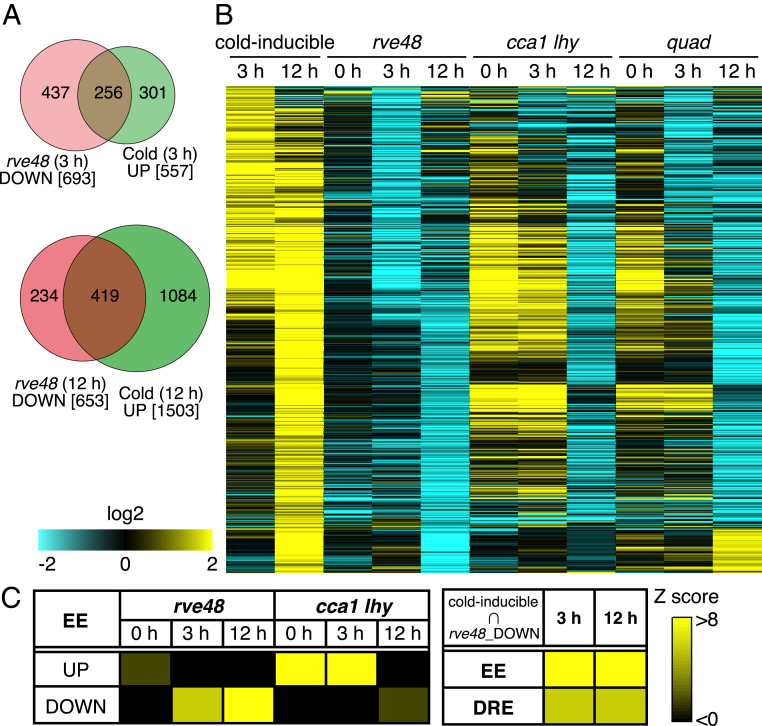
To comprehensively identify downstream genes of RVE4/RVE8 in the cold stress response, we further performed RNA sequence analyses using the mutant and WT plants under normal growth conditions at LL2 (0 h) and cold stress conditions at 4 °C for 3 h and 12 h (Fig. 3 and SI Appendix, Table S1 and Dataset S1). In the WT plants, 557 and 1,503 genes were induced by the cold stress treatments for 3 h and 12 h, respectively. In rve48, 758 (0 h), 693 (3 h), and 653 (12 h) genes were down-regulated compared with those in the WT at each time point. Many genes were also down-regulated in cca1 lhy (0 h: 1,273, 3 h: 1,206, 12 h: 1,246) and quad (0 h: 529, 3 h: 955, 12 h: 884) compared with those in the WT. Among the down-regulated genes in rve48, 256 genes (∼37%; 3 h) and 419 genes (∼64%; 12 h) were cold inducible in the WT at each time point (Fig. 3A). Most of them were different from the RVE8-induced genes under normal growth conditions reported previously (29). On the other hand, many of the cold-inducible genes at 3 h were up-regulated, and those at 12 h were similarly down-regulated in cca1 lhy (Fig. 3B). Moreover, many cold-inducible genes were up-regulated in cca1 lhy under normal growth conditions (0 h). The differentially expressed genes in quad reflected the results in both rve48 and cca1 lhy (Fig. 3B). We investigated the presence of the EE in the promoters of the differentially expressed genes in each mutant plant and revealed the high frequency of EEs in the promoter regions of the down-regulated genes in rve48 and the up-regulated genes in cca1 lhy. The EE and DRE were enriched in the promoter regions of genes that were cold inducible and down-regulated in the rve48, which suggests that RVE4 and RVE8 directly regulate the expression of many cold-inducible genes, including DREB1A and DREB1C (Fig. 3C).
Fig. 3.
Transcriptome analysis of the mutant plants of CCA1, LHY, RVE4, and RVE8. (A) Venn diagram between the cold-inducible genes and the down-regulated genes in the rve4 rve8 double mutant plant. (B) Hierarchical clustering of the genes that were induced by cold stress and the down-regulated genes in the double and quadruple (quad) mutant plants of CCA1, LHY, RVE4, and RVE8. In the heatmaps, yellow and cyan show up-regulated and down-regulated genes, respectively. (C) Enrichment of cis-acting elements in the promoters of the differentially expressed genes (DEGs) in the mutants and the cold-inducible genes that were down-regulated in the rve48 mutant. Frequency of the cis-acting elements in the promoters (upstream 1 kb) in the DEGs was compared with the random-selected genes.
RVE4 and RVE8 Accumulate in Nuclei under Cold Stress.
To elucidate the mechanisms by which CCA1/LHY/RVE4/RVE8 regulate DREB1 expression, we analyzed the time course of their expression (SI Appendix, Fig. S8). Under normal growth conditions at 22 °C, these genes were highly expressed in the morning, and the expression levels gradually decreased, as previously reported (27, 28). Under cold stress conditions, high expression levels of CCA1 and RVE4 were maintained, and those of LHY and RVE8 gradually decreased, but their expression levels were higher than those under normal growth conditions except for those of the 24-h cold treatment. Because the expression of these genes was maintained at high levels but not induced by cold stress, and even with overexpression, DREB1A expression under normal growth conditions was not increased (SI Appendix, Fig. S4 B and D), we assumed that these genes were controlled at the posttranscriptional level to regulate cold stress-inducible DREB1 expression.
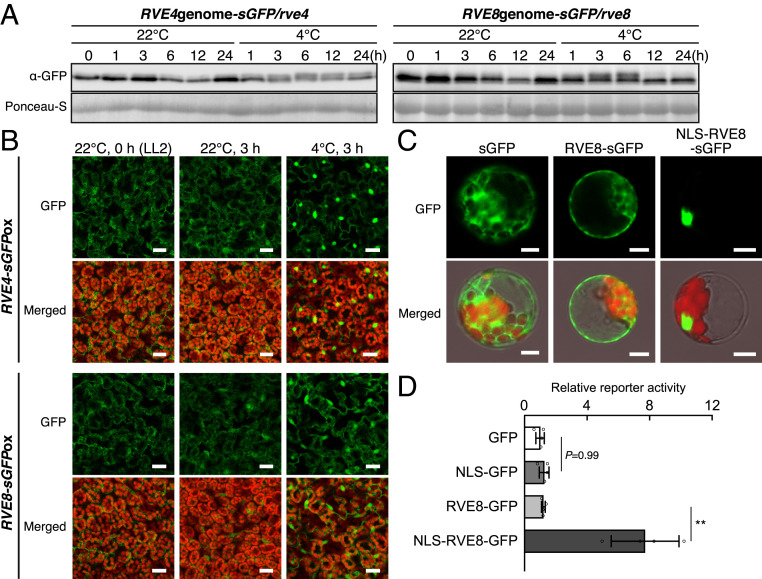
We examined the protein levels of RVE4 and RVE8 under normal growth and cold stress conditions. We expressed the sGFP fusion genes of RVE4 and RVE8 driven by their own promoters in each single mutant plant. Immunoblot analyses indicated that RVE4-sGFP and RVE8-sGFP accumulated more in the daytime than at subjective night in the transgenic plants under normal growth conditions (Fig. 4A). Under cold stress conditions, the levels of these two proteins were almost constant, and a clear shifted band was detected under cold stress conditions at some time points, suggesting that these proteins might undergo a modification in response to cold stress. We then observed the subcellular localization of these two proteins. GFP fluorescence of RVE4 was mainly observed in the cytoplasm under normal growth conditions and was detected in the nuclei under cold stress for 3 h (SI Appendix, Fig. S9A). In contrast, the fluorescence of RVE8 could not be clearly observed in these plants, probably due to low expression levels.
Fig. 4.
Protein accumulation levels and subcellular localization of RVE4 and RVE8 in response to cold stress. (A) Immunoblot analyses using transgenic Arabidopsis plants expressing RVE4-sGFP and RVE8-sGFP driven by their own promoters. The GFP fusion genes of the genomic fragment, including the promoter region of RVE4 or RVE8, were expressed in each single mutant. Two-week-old seedlings grown on agar plates were gradually chilled at 4 °C up to 24 h from LL2. (B) GFP fluorescence of RVE4-sGFP and RVE8-sGFP in 2-wk-old transgenic Arabidopsis seedlings overexpressing RVE4-sGFP and RVE8-sGFP driven by the CaMV 35S promoter. (Scale bars, 20 μm.) (C) GFP fluorescence of RVE8-sGFP in the transient expression assay in Arabidopsis protoplasts. GFP fusion genes of RVE8 and a nuclear localization signal (NLS) of SV40 driven by the CaMV 35S promoter were expressed. (Scale bars, 20 μm.) (D) Reporter activity in the transient expression assay in Arabidopsis protoplasts. The ELUC gene driven by four tandemly repeated 1AR elements (the same as Fig. 1A) was used as a reporter. The constructs in C were used as effectors. The bars refer to the mean ± SD of four biological replicates. The asterisks indicate significant differences (**P < 0.01 as analyzed using one-way ANOVA followed by a Tukey’s post hoc test) between two effectors.
To clarify the subcellular localization of RVE4 and RVE8, we examined their localization by using transgenic plants overexpressing the sGFP fusion genes of RVE4 and RVE8, in which DREB1A expression was analyzed, as shown in SI Appendix, Fig. S4D. These proteins were detected primarily in the cytoplasm and weakly in the nuclei under unstressed conditions in the morning (LL2) and during the subjective night (LL14) (SI Appendix, Figs. S4B and S9B). Under cold stress conditions for 3 h, these proteins were strongly observed in the nuclei. Their clear nuclear localization was detected within 15 min after the seedlings were transferred from 22 °C to 4 °C (Fig. 4B and SI Appendix, Fig. S9C). In turn, when the plants were transferred back from 4 °C to 22 °C, nuclear-localized RVE4 and RVE8 were reduced within 10 min (SI Appendix, Fig. S9D). Obvious nuclear localization of both proteins was observed in response to a temperature decrease below 10 °C (SI Appendix, Fig. S9E). We then fused RVE8-sGFP to the nuclear localization signal peptide of SV40 and expressed the fusion protein with the ELUC reporter gene driven by the 1AR fragment in Arabidopsis protoplasts. The fusion protein was localized in the nuclei of the protoplasts at 22 °C (Fig. 4C), and the reporter activity was increased by the fusion protein compared with RVE8-sGFP (Fig. 4D), suggesting that activation of RVE8 is regulated by its localization.
We further investigated the subcellular localization of RVE4 and RVE8 in response to other stimuli, heat stress (37 °C), high light stress (1,000 μE), high salinity stress (250 mM NaCl), and abscisic acid (ABA) (50 μM) treatment, in the plants overexpressing RVE4 and RVE8. These two proteins were also localized in the nuclei under heat stress, although they did not respond to other stresses (SI Appendix, Fig. S10A). However, heat stress did not induce DREB1A expression (SI Appendix, Fig. S10B), suggesting that the nuclear localization of RVE4 and RVE8 was important but not sufficient for DREB1A induction. The RVE4/RVE8-sGFP proteins driven by their own promoter showed no shifted bands under heat stress (SI Appendix, Fig. S10C).
CCA1 and LHY Are Degraded in Response to Cold Stresses.
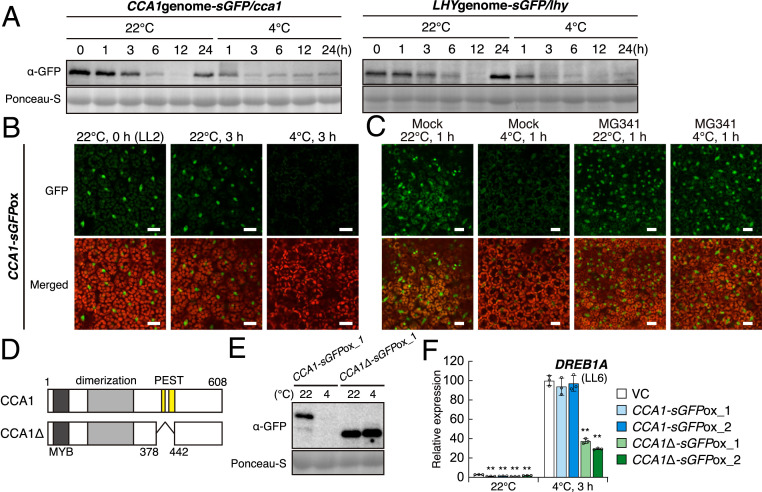
We then analyzed the protein levels of CCA1 and LHY in transgenic Arabidopsis plants expressing the sGFP fusion gene of CCA1 or LHY driven by their own promoters in each single mutant plant. Immunoblot analyses indicated that CCA1-sGFP and LHY-sGFP highly accumulated in the morning at 22 °C and gradually decreased by the subjective night, which reflected their gene expression. Under cold stress conditions, their levels decreased more quickly than those under the normal temperature until 3 h and remained at very low levels (Fig. 5A). The GFP fluorescent signals of these proteins in the transgenic plants also decreased quickly under cold stress conditions, similar to the results of the immunoblot analyses (SI Appendix, Fig. S11A).
Fig. 5.
Protein accumulation levels of CCA1 and LHY in response to cold stress. (A) Immunoblot analyses using transgenic Arabidopsis plants expressing CCA1-sGFP and LHY-sGFP driven by their own promoters. sGFP-fused genomic fragments, including the promoter regions of CCA1 and LHY, were expressed in each single mutant. Two-week-old seedlings grown on agar plates were gradually chilled at 4 °C up to 24 h from LL2. (B) GFP fluorescence of CCA1-sGFP in transgenic Arabidopsis plants. Two-week-old seedlings overexpressing CCA1-sGFP driven by the CaMV 35S promoter were used for GFP observation. (Scale bars, 20 μm.) (C) Effect of a 26S proteasome inhibitor on the GFP fluorescence of CCA1-sGFP in response to cold stress. (Scale bars, 20 μm.) (D) Schematic structure of CCA1 and CCA1Δ. Dark gray, light gray, and yellow bars indicate the MYB DNA binding domain, dimerization domain, and PEST sequences, respectively. The truncated fragment of CCA1 (Δ379 to 441) was named CCA1Δ. (E) Immunoblot analyses using transgenic Arabidopsis plants expressing CCA1-sGFP or CCA1Δ-sGFP driven by the CaMV 35S promoter. Two-week-old seedlings grown on agar plates were gradually chilled at 4 °C for 3 h from LL2. (F) The gene expression of DREB1A in transgenic Arabidopsis plants overexpressing CCA1Δ-sGFP. The CCA1-sGFP-overexpressing plants are the same lines as shown in SI Appendix, Fig. S4. The bars refer to the mean ± SD in three biological replicates. The asterisks indicate significant differences (**P < 0.01 as analyzed using one-way ANOVA followed by a Tukey’s post hoc test) compared to the expression levels in the VC plants at each time point.
To confirm the degradation of CCA1 and LHY under cold stress conditions, we examined the subcellular localization of the CCA1 and LHY proteins using transgenic plants overexpressing these genes, as shown in SI Appendix, Fig. S4B. CCA1-sGFP and LHY-sGFP were detected in the nuclei at 22 °C, but their fluorescence almost disappeared after 3 h under cold stress conditions (Fig. 5B and SI Appendix, Fig. S11B). However, when MG341, a 26S proteasome inhibitor, was pretreated, fluorescence signals were observed even under cold stress conditions (Fig. 5C and SI Appendix, Fig. S12A). Because two potential PEST sequences (397 to 409 and 422 to 443; CCA1, 415 to 431 and 466 to 486; LHY) were found in CCA1 and LHY, we generated transgenic plants overexpressing truncated CCA1 (CCA1Δ; Δ378 to 442; Fig. 5D and SI Appendix, Fig. S12B). The CCA1Δ-overexpressing plants as well as the CCA1-overexpressing plants exhibited longer hypocotyls and petioles than the control plants, suggesting that CCA1Δ maintains the function of CCA1 under unstressed conditions (SI Appendix, Fig. S12C). In these transgenic plants, CCA1Δ did not degrade even under cold stress conditions (Fig. 5E), and the DREB1A expression levels in the CCA1Δ-overexpressing plants under cold stress were lower than those in the VC plants, while overexpression of CCA1 did not affect its expression (Fig. 5F). We also analyzed the stability of CCA1 and LHY under heat stress, but these proteins were stable at 37 °C for 3 h as well as under other stresses, such as high light, high salinity, and ABA treatment (SI Appendix, Fig. S13), suggesting that not only accumulation of RVE4 and RVE8 in the nuclei but also degradation of CCA1 and LHY may be necessary for induction of DREB1A under cold stress conditions.
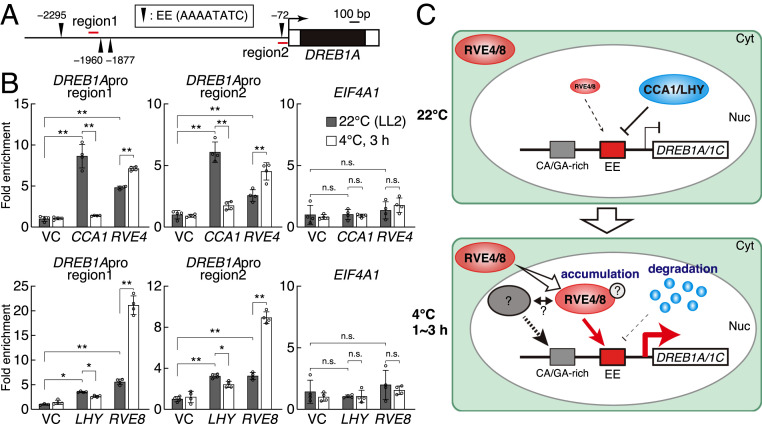
We further conducted chromatin immunoprecipitation (ChIP)-qPCR using transgenic plants expressing CCA1/LHY/RVE4/RVE8-sGFP driven by their own promoters (Fig. 6 A and B). All proteins clearly bound to the 1AR, including the EE and an upstream region near the other three EEs in the DREB1A promoter under unstressed conditions in the morning (LL2). In response to cold stress from LL2, the binding of RVE4 and RVE8 significantly increased, and CCA1 and LHY binding was reduced (Fig. 6C). During the subjective night (LL14), which showed low levels of cold-responsive DREB1A induction, RVE4 and RVE8 significantly bound to the upstream region, but not to the 1AR region, under the cold stress conditions (SI Appendix, Fig. S14). These results support the hypothesis that CCA1/LHY negatively regulates the expression of DREB1A under normal growth conditions, and RVE4/RVE8 positively regulates its expression under cold stress conditions. The EE in the 1AR was considered a major cis-acting element that regulates the cold-inducible expression of DREB1A activation by RVE4 and RVE8.
Fig. 6.
ChIP assays of the CCA1, LHY, and RVE proteins and schematic model of the expression of DREB1A and DREB1C. (A) Schematic structure of the DREB1A promoter. Black arrowhead indicates the EE (AAAATATC). Enrichment of two regions around EEs (1 and 2; red lines) was measured in ChIP-qPCR analyses. (B) ChIP-qPCR analyses using transgenic Arabidopsis plants expressing CCA1-sGFP, LHY-sGFP, and RVE-sGFP driven by their own promoters. The transgenic Arabidopsis plants used in Figs. 4A and 5A were analyzed. The cold stress treatment was started 2 h after dawn (LL2). The bars refer to the mean ± SD in three or four replicates. The asterisks and "n.s." indicate significant (*P < 0.05, **P < 0.01) and no differences (P > 0.05), respectively, as analyzed using one-way ANOVA followed by a Tukey’s post hoc test) between two samples. (C) Schematic model illustrating the expression of DREB1A and DREB1C under normal growth and cold stress conditions. Under normal growth conditions, CCA1 and LHY suppress DREB1 expression. Under cold stress conditions, RVE4/RVE8 accumulate in the nuclei, and CCA1 and LHY are degraded. High levels of RVE4 and RVE8 induce the expression of DREB1A and DREB1C through the cis-acting element EE.
Discussion
Cold stress-responsive gene expression of DREB1A and DREB1C is associated with circadian regulation (15). We revealed that EE is an essential cis-acting element for cold-responsive DREB1A expression using transgenic plants harboring the ELUC reporter gene (Fig. 1), and since the EE was also completely conserved in all the DREB1C promoters among the Brassicaceae species (SI Appendix, Fig. S2C), EE is also considered to be an essential cis-acting element for cold-responsive DREB1C expression. We revealed that the clock-related MYB transcription factors, including CCA1, LHY, and RVEs, could bind to the EE in their promoters (Fig. 1 D and E). Furthermore, among these MYB proteins, RVE4 and RVE8 were shown to be transcriptional activators for the cold-inducible expression of DREB1A and DREB1C because the expression of DREB1A was increased in the RVE4- and RVE8-overexpressing plants, and the circadian rhythms of the cold-inducible expression of DREB1A and DREB1C were lost almost completely in the rve48 double mutant plants (Fig. 2 and SI Appendix, Figs. S4 and S6). In the rve34568 quintuple mutant plants, the levels of cold-inducible DREBA expression were further reduced compared with those in rve48 but not changed in the rve356 mutant compared with that in the WT plants (Fig. 1D). Considering that effects of RVE3, RVE5, and RVE6 on the DREB1A expression were only observed in the plants lacking the RVE4 and RVE8 genes, RVE4 and RVE8 are the major transcription factors that activate the cold stress-responsive expression of DREB1A and DREB1C. RVE4 and RVE8, together with RVE6, have been reported to be transcriptional activators that directly control the expression of clock genes repressed by CCA1 and LHY with EEs in their promoters (24, 29). However, we demonstrated that the RVE4 and RVE8 transcription factors directly regulate different target genes with EE in their promoters in response to cold stress.
Detailed subcellular localization analyses of the RVE4 and RVE8 proteins suggested a mechanism of the target gene alteration caused by temperature changes. We found that the RVE4 and RVE8 proteins were quickly and reversibly transferred from the cytoplasm to the nucleus in response to a temperature decrease below 10 °C (Fig. 4 and SI Appendix, Fig. S9). The expression of DREB1s is also induced in response to a temperature decrease below 10 °C (15). This consistency of the temperature indicates that cold stress-responsive translocation of RVE4 and RVE8 into the nuclei could play an important role in the induction of DREB1 expression. However, their translocation from the cytoplasm to the nucleus was observed not only under cold stress conditions but also under heat stress conditions (SI Appendix, Fig. S10). In general, plant circadian clocks have temperature compensation and maintain robust rhythms over a wide temperature range from ∼12 °C to 27 °C (30). Since RVE4 and RVE8 translocate to the nucleus in response to both cold and heat stresses, loss of temperature compensation of the circadian clock is considered to be closely related to their translocation into the nucleus. Recently, RVE4 and RVE8 were reported to activate the expression of HSF-independent heat-inducible genes (31). However, as DREB1A expression did not respond to heat stress (SI Appendix, Fig. S10B), accumulation of RVE4 and RVE8 in the nucleus may be necessary but not sufficient for its induction. In our immunoblot analysis, we detected shifted bands of RVE8 and RVE4 under cold stress conditions but not under heat stress conditions (SI Appendix, Fig. S10C), which suggests that posttranslational modification of these proteins is required to trigger the expression of DREB1s.
Previous studies have reported that CCA1/LHY are positive regulators of the cold-responsive expression of DREB1s (15, 21). Our experiments using CCA1/LHY-overexpressing plants also showed increased expression of DREB1A under cold stress conditions (SI Appendix, Fig. S4B). However, GFP fluorescence and immunoblot analyses showed that both the CCA1 and LHY proteins were degraded under cold stress conditions (Fig. 5), suggesting that they were not direct activators of DREB1 expression. CCA1 and LHY are repressors for the expression of some core circadian clock genes with EEs in their promoters (23). These proteins were detected at low levels under cold stress conditions (Fig. 5A) and may indirectly and positively affect DREB1 expression via suppression of the expression of genes for factors that inhibit DREB1 expression. Because RVE1 and RVE2 are downstream genes of CCA1 and LHY under normal growth conditions (27, 32), CCA1 and LHY may regulate DREB1A expression via regulation of RVE1 and RVE2, even under cold stress conditions. RVE1 and RVE2 likely function as suppressors of DREB1A expression because the expression of DREB1A was suppressed in the RVE1- and RVE2-overexpressing plants and was increased in the rve1 rve2 mutants under cold stress conditions (Fig. 2F and SI Appendix, Fig. S4F). Therefore, CCA1 and LHY may positively regulate the expression of DREB1A and DREB1C via suppressing of REV1 and RVE2 expression under cold stress conditions. Detailed analyses of the expression of RVE1 and RVE2 under cold stress conditions using the cca1 lhy mutant likely reveal a relationship between CCA1/LHY and RVE1/RVE2, and the roles of CCA1/LHY should be elucidated in the expression of DREB1A and DREB1C under cold stress conditions.
However, under nonstressed normal growth conditions (22 °C), DREB1A expression was decreased in the CCA1-sGFP-overexpressing plants and was increased in the cca1 and cca1 lhy mutant plants (Fig. 2 E and F and SI Appendix, Fig. S4B). These data suggest that CCA1 and LHY (mainly CCA1) suppress DREB1A expression under normal growth conditions. Under normal growth conditions, CCA1 and LHY are abundantly present in the nucleus (Fig. 5A) and may suppress the leakage of DREB1A expression by directly binding to its promoter. These CCA1 and LHY proteins were rapidly degraded under cold stress conditions, whereas they are abundantly present in the nucleus under heat stress conditions (SI Appendix, Fig. S13). Since CCA1 and LHY are thought to function as repressors even under heat stress conditions, nuclear-translocated RVE4 and RVE8 may not function as activators due to competition for their binding activity. In transgenic plants expressing CCA1Δ, which was stabilized by removing the proteolytic signal (Fig. 5E), the cold-inducible expression of DREB1A was suppressed compared with that in the VC plants, unlike the transgenic plants expressing intact CCA1 (Fig. 5F). High levels of RVE4 and RVE8 and low levels of CCA1 and LHY in the nucleus may be essential for the induction of DREB1A and DREB1C through the cis-acting element EE under cold-stress conditions. Moreover, transcription factors other than EE binding proteins, such as CA/GA-rich binding proteins, may be necessary for the induction of DREB1A and DREB1C under cold stress conditions. We speculate that these unknown factors probably form complexes with these circadian clock-related factors under cold stress conditions to specifically express cold-inducible genes (Fig. 6C).
Materials and Methods
Plant Materials.
The Arabidopsis seeds of rve1 (SALK_025754), rve2 (SALK_074896), rve3 (SALK_001480), rve4 (SALK_051728), rve5 (SAIL_769_A09), rve6 (SAIL_548_F12), and rve8 (SALK_053482) were obtained from the Arabidopsis Biological Resource Center (ABRC). The Col-0 backgrounded cca1, lhy, and cca1 lhy seeds were kindly provided by Takafumi Yamashino, Nagoya University, Nagoya, Japan. For the transgenic Arabidopsis plants, the coding sequences or genomic fragments including the promoter regions of each gene were amplified and cloned into restriction enzyme sites of the pGH_35S:sGFP vector (33). Oligomers for genotyping and cloning are shown in SI Appendix, Table S2.
Growth and Cold Stress Treatment.
The plants were grown on germination medium agar plates or in peat moss soil pots at 22 ± 1 °C under a 12-h light/12-h dark cycle at a photon flux density of 50 ± 10 μmol m−2 s−1 of white light. For gradual cold stress treatment, 14-d-old whole seedlings on agar plates were chilled in a 4 °C cold chamber. For rapid cold stress treatment, 16-d-old seedlings in soil pots were directly transferred to a 4 °C cold chamber.
RT-qPCR.
RT-qPCR was performed as previously described (10). The total RNA extraction from plants was conducted with RNAiso Plus (TaKaRa Bio, Inc.). cDNA was synthesized from plant total RNA by utilizing a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT-qPCR was performed using the QuantStudio 3 Real-Time PCR system and software (ver. 1.2; Applied Biosystems). Power SYBR Green Master Mix (Applied Biosystems) was used for amplification. IPP2 was used as the quantitative control for the template. Oligomers for qPCR are shown in SI Appendix, Table S2.
Sequence Alignment and Phylogenetic Tree.
The promoter sequences of DREB1s and the protein sequences of CCA1/LHY/RVEs in Brassicaceae species were obtained from the Phytozome database (34). Alignment of the promoter sequences was constructed by using mVISTA (35, 36). The protein sequences were aligned and clustered by using ClustalX 2.1 (37). The alignments were manually adjusted by using the GeneDoc 2.6 program (www.nrbsc.org/gfx/genedoc/). The phylogeny of the selected sequences was constructed by MEGA6 (38). The distances between branches were calculated by the neighbor-joining method based on 1,000 bootstrap samples.
Yeast One-Hybrid Assay.
Yeast transformation was conducted according to the Yeast Protocols Handbook (Clontech). For the reporter, four tandemly repeated 1AR fragments were cloned into a pHisi-1 vector. They were integrated into the genomic DNA of a yeast YM4271 strain. For the effector, the coding sequence of each gene was cloned into a pGADT7 vector. The transformed yeast (OD600 = 1, 0.1, and 0.01) was spotted on SD/−His, Leu (control) and SD/−His, Leu + 10 mM 3-AT (selective) agar plates.
Electrophoretic Mobility Shift Assay.
Expression and purification of the recombinant GST fusion proteins and electrophoresis mobility shift assays were performed as previously reported (18) with minor modifications. Oligomers for the probe and competitors are shown in SI Appendix, Table S2. For probe binding, 2 pmol of the probe fragment was mixed with 100 ng of the recombinant proteins and incubated for 15 min at room temperature. The reaction mixtures were resolved by electrophoresis through a 6% polyacrylamide gel in 0.5× Tris-borate-ethylenediaminetetra-acetic acid buffer at 150 V for 90 min. For competition, 0.02 μmol, 0.2 μmol, and 2 μmol of the unlabeled DNA fragment mixed with the recombinant proteins were incubated for 5 min at room temperature before probe binding.
Freezing Tolerance Test.
Freezing treatments were performed as previously described with minor modifications (10, 39). Ten-day-old whole seedlings on agar plates were chilled in the −2 °C cold chamber for 2 h. After the generation of ice nuclei, the temperature was decreased by −1 °C/h until −9 °C and then stored at 4 °C overnight. After recovery at 22 °C for 10 d, the seedlings that generated new leaves were counted for survival.
RNA Sequencing.
From 1 μg of the total RNA, mRNA was purified by using a NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). cDNA libraries were constructed by using the NEBNext Ultra II RNA Library Kit for Illumina (New England Biolabs). The adaptors were provided in the NEBNext Multiple Oligos for Illumina (New England Biolabs). The libraries were sequenced by NextSEq. 500 (Illumina). The single ends of the libraries were sequenced for 86 nt using the Illumina Genome Analyzer IIx. The reads were mapped to the Arabidopsis reference sequence (TAIR10). A heatmap was generated by using Gene Cluster 3.0 (40) with correlation (uncentered) as the similarity metric and single linkage as the clustering method. Java TreeView (41) was used to visualize the results. The RNA-sequencing data were deposited in the DNA Data Bank of Japan under accession number DRA01211.
Immunoblot Analysis.
Protein extraction from seedlings, SDS-polyacrylamide gel electrophoresis, and immunoblotting were performed as previously described (42). A polyclonal anti-GFP antibody (1:4,000 dilution) (43) and a horseradish peroxidase-conjugated anti-rabbit IgG antibody (1:4,000 dilution; Pierce) were used to detect the sGFP-fused proteins. Signals were detected with the ECL Select Western Blotting Detection Reagent (GE Healthcare) and an Image Quant LAS-4000 system (GE Healthcare). Following chemiluminescence detection, the membrane was stained with a solution of 0.1% (wt/vol) Ponceau S (Sigma-Aldrich), and 5% (vol/vol) acetic acid to ensure protein loading.
GFP Observation.
An LSM Exciter confocal laser microscope (Zeiss) was used to detect GFP fluorescence. For cold stress treatment, 14-d-old whole seedlings on agar plates were transferred to chilled water in a 4 °C cold chamber.
Transient Expression Assays with Arabidopsis Mesophyll Protoplasts.
Transient transformation of Arabidopsis mesophyll protoplasts was performed as previously described (44). The mesophyll protoplasts were isolated from the WT plants grown in soil pots for 5 wk. A GUS gene driven by the CaMV 35S promoter in a pBI221 vector (TaKaRa Bio, Inc.) was used as an internal control.
ChIP.
ChIP was conducted as previously described (45) with minor modifications. To 0.5 to 1 g of the seedlings, 5 mL of the extraction buffer including 0.5% formaldehyde was added and incubated for 10 min. After the cross-linking was stopped by addition of a glycine solution, the nuclei-enriched fraction was isolated. The resuspended chromatin solution was sonicated six times for 10 s each with an ultrasonic disrupter UD-211 (TOMY SEIKO). The polyclonal anti-GFP antibody (43) was used for IP. The immunoprecipitated DNA was purified with NucleoSpin Gel and PCR Clean-Up (TaKaRa Bio, Inc.) with Buffer NTB (TaKaRa Bio, Inc.). The oligomers for qPCR are shown in SI Appendix, Table S2.
Accession Numbers.
Sequences of genes described in this article can be found in The Arabidopsis Information Resource (https://www.arabidopsis.org/) under the following accession numbers: CCA1 (AT2G46830), LHY (AT1G01060), RVE1 (AT5G17300), RVE2 (AT5G37260), RVE3 (AT1G01520), RVE4 (AT5G02840), RVE5 (AT4G01280), RVE6 (AT5G52660), RVE7 (AT1G18330), RVE8 (AT3G09600), DREB1A/CBF3 (AT4G25480), DREB1B/CBF1 (AT4G25490), DREB1C/CBF2 (AT4G25470), RD29A (AT5G52310), and COR15A (AT2G42540).
Supplementary Material
Acknowledgments
We thank Yuriko Tanaka, Tomomi Shinagawa, and Ayumi Furuta for the excellent technical assistance provided and Etsuko Toma for the skilled editorial assistance provided. We also thank Dr. Takafumi Yamashino (Nagoya University) for kindly providing the cca1 lhy mutant seeds. This work was supported by Grants-in-Aid for Scientific Research for Young Scientists (B) (no. 17K15413 to S.K.) and for Scientific Research (A) (no. 18H03996 to K.Y.-S.) and by grants for Basic Science Research Projects from the Sumitomo Foundation (190666 to S.K.) and for Scientific Research on Innovative Areas (no. 15H05960 to K.Y.-S.) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021048118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Thomashow M. F., Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi-Shinozaki K., Shinozaki K., Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Baker S. S., Wilhelm K. S., Thomashow M. F., The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi-Shinozaki K., Shinozaki K., A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F., Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Liu Q., et al., Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomashow M. F., Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fowler S., Thomashow M. F., Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maruyama K., et al., Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Kidokoro S., et al., DREB1A/CBF3 is repressed by transgene-induced DNA methylation in the Arabidopsis ice1-1 mutant. Plant Cell 32, 1035–1048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinnusamy V., et al., ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomashow M. F., Torii K. U., SCREAMing twist on the role of ICE1 in freezing tolerance. Plant Cell 32, 816–819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J. S., Kidokoro S., Shinozaki K., Yamaguchi-Shinozaki K., DNA demethylase ROS1 prevents inheritable DREB1A/CBF3 repression by transgene-induced promoter methylation in the Arabidopsis ice1-1 mutant. Plant Mol. Biol. 104, 575–582 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H., A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Kidokoro S., et al., Different cold-signalling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29, 760–774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty C. J., Van Buskirk H. A., Myers S. J., Thomashow M. F., Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikkelsen M. D., Thomashow M. F., A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60, 328–339 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Kidokoro S., et al., The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151, 2046–2057 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alabadí D., Yanovsky M. J., Más P., Harmer S. L., Kay S. A., Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12, 757–761 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Nohales M. A., Kay S. A., Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 23, 1061–1069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong M. A., Farré E. M., Thomashow M. F., Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7241–7246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagel D. H., et al., Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E4802–E4810 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamioka M., et al., Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28, 696–711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalit-Kaneh A., Kumimoto R. W., Filkov V., Harmer S. L., Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc. Natl. Acad. Sci. U.S.A. 115, 7147–7152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael T. P., McClung C. R., Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 130, 627–638 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carré I. A., Kim J. Y., MYB transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 53, 1551–1557 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Rawat R., et al., REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 16883–16888 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawat R., et al., REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7, e1001350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu P. Y., Devisetty U. K., Harmer S. L., Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2, e00473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould P. D., et al., The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18, 1177–1187 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B., Gao Z., Liu X., Sun D., Tang W., Transcriptional profiling reveals a time-of-day-specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis. Plant Cell 31, 2353–2369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., et al., Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51, 512–525 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Fujita Y., et al., Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Goodstein D. M., et al., Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubchak I., et al., Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10, 1304–1306 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frazer K. A., Pachter L., Poliakov A., Rubin E. M., Dubchak I., VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin M. A., et al., Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidokoro S., et al., Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 81, 505–518 (2015). [DOI] [PubMed] [Google Scholar]
- 40.de Hoon M. J., Imoto S., Nolan J., Miyano S., Open source clustering software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Saldanha A. J., Java Treeview–Extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Ohama N., et al., The transcriptional cascade in the heat stress response of Arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell 28, 181–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka H., et al., Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70, 599–613 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Yoo S. D., Cho Y. H., Sheen J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Gendrel A. V., Lippman Z., Martienssen R., Colot V., Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.