Abstract
The current rate of species extinction is rapidly approaching unprecedented highs, and life on Earth presently faces a sixth mass extinction event driven by anthropogenic activity, climate change, and ecological collapse. The field of conservation genetics aims at preserving species by using their levels of genetic diversity, usually measured as neutral genome-wide diversity, as a barometer for evaluating population health and extinction risk. A fundamental assumption is that higher levels of genetic diversity lead to an increase in fitness and long-term survival of a species. Here, we argue against the perceived importance of neutral genetic diversity for the conservation of wild populations and species. We demonstrate that no simple general relationship exists between neutral genetic diversity and the risk of species extinction. Instead, a better understanding of the properties of functional genetic diversity, demographic history, and ecological relationships is necessary for developing and implementing effective conservation genetic strategies.
Keywords: conservation genetics, adaptive potential, inbreeding depression, genetic load, species extinction
Are Species with Little Genetic Diversity Endangered?
Climate change caused by human activity is currently responsible for widespread ecological disruption and habitat destruction, with an ensuing unprecedented rate of species loss known as the Anthropocene Mass Extinction (1–4). This catastrophic scenario poses a serious threat to the future of life and human survival on Earth that has sparked a global sense of emergency about the need to preserve the diversity of life on the planet. However, this emergency has also fostered the development and implementation of imperfect and often simplistic conservation strategies with potentially detrimental consequences for the preservation of life on Earth (5).
A fundamental conception underlying many of these strategies is the importance attributed to intraspecific genetic diversity (Box 1), measured at markers scattered across the genome, for assessing the extinction risk of species facing rapid environmental change and habitat destruction (6–8). Specifically, low genetic diversity is often interpreted as an indicator of inbreeding depression and increased genetic drift. Moreover, low genetic diversity is related to reduced individual life span and health, along with a depleted capacity for population growth (9). In contrast, high levels of genetic diversity are often seen as key to promoting population survival and guaranteeing the adaptive potential of natural populations in the face of rapidly changing environmental pressures (10). These principles are reflected in strategies such as genetic rescue, where the genetic diversity of a threatened or endangered population is increased by facilitating gene flow from a population with high levels of diversity (11).
Box 1.
Neutral genetic diversity and effective population size
The amount of neutral genetic variation that segregates in a population is influenced by two opposing factors: mutation and genetic drift. Mutation leads to an increase in genetic diversity, whereas drift tends to reduce variation. In mutation–drift equilibrium, the amount of genetic diversity results from a balance between the rate of incorporation of novel mutations and the rate at which variation is lost through drift. A variety of population genetic measures has been developed as an attempt to summarize the levels of genetic diversity. A classic and often-used measure is nucleotide diversity (12), or π, which is calculated as the average number of nucleotide differences per site between pairs of sequences in a sample.
Neutral genetic diversity is proportional to the mutation rate (µ) and population size (N), reflecting the balance between mutation and drift. This can most directly be seen when considering the lineages of a pair of sequences backward in time (i.e., their genetic ancestry). Let us first consider a simple scenario of a diploid population with N individuals evolving under a Wright–Fisher (WF) model with discrete generations, constant population size, and random mating. The probability of any two lineages coalescing (i.e., finding a common ancestor) in the previous generation is 1/(2N), and the expected time to coalescence is the inverse of this rate, 2N generations (13). If mutations occur along a lineage at a rate of µ mutations per generation, then a pair of lineages accumulate on average 2µ(2N) = 4Nµ mutations over this time span. Accordingly, under the assumption of an infinite sites model, it follows that E[π] = 4Nµ, i.e., both the mutation rate and the population size determine the average number of pairwise differences. It is possible to relax many of the assumptions of the WF model (e.g., differences in numbers of breeding males and females, age-structured populations, partial self-fertilization) by simply exchanging the census population size N with the coalescent effective population size NeC (13). If the mutation rate is known, then NeC can be estimated empirically from genetic data by dividing the nucleotide diversity by 4µ.
However, the assumption of long-term constant population size can hardly be applied to natural populations, which frequently undergo expansions and contractions. While a population size can still be estimated from neutral diversity, this “diversity” Ne is determined by demographic events occurring in the history of a population, including ancient bottlenecks and migration events. Importantly, the estimated Ne offers little information about the immediate adaptive potential of a population. The emergence and fate of beneficial mutations in a population undergoing environmental change depend not on the long-term historical demography of the population, but instead on the shorter time frame of when the adaptive mutations establish in the population (14) (see also Fig. 4). Accordingly, rather than interpreting the levels of genetic diversity within a population, conservation genetics studies might benefit from estimating the contemporary effective population size. For example, the variance effective population size (NeV) measures the fluctuation and dispersion of allele frequencies across generations and can be directly estimated from temporal data for a specified period of time (15). The distinction between long-term, short-term, and contemporary estimates of Ne (16) is particularly relevant as vulnerable natural populations usually undergo rapid population decline, a scenario that can lead to significant differences between different estimates of Ne. Various estimators of contemporary effective population size and their strengths, weaknesses, and assumptions are reviewed in Luikart et al. (17).
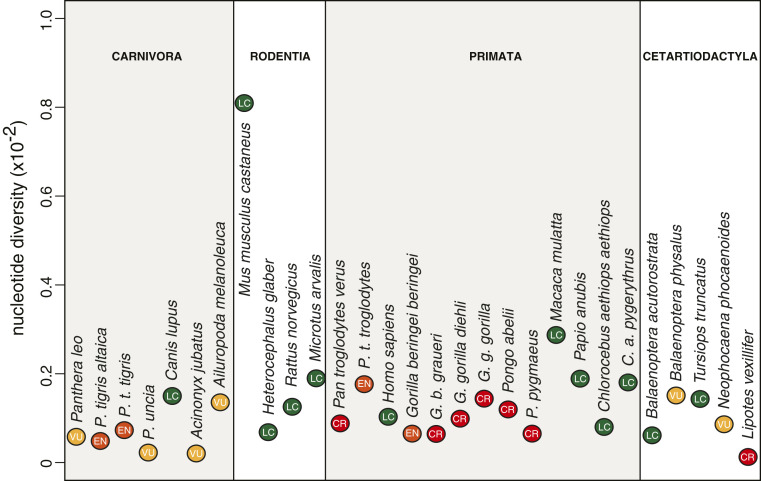
Nonetheless, supporting empirical evidence for the existence of a causal relationship between genetic diversity and population viability or adaptive potential is weak. If genetic diversity is indeed a major factor affecting the health and survival of populations in the wild, then one would expect endangered species to show, on average, lower levels of diversity. However, the International Union for Conservation of Nature (IUCN) Red List status is only a poor predictor of a species’ genome-wide nucleotide diversity (18–21) (Fig. 1). It has been previously argued that this lack of correlation reflects a deficient classification and that genome-wide patterns of neutral diversity should be incorporated into IUCN’s listing criteria to more accurately assess the likelihood of future extinction and the associated need for conservation (20, 22, 23). However, a more parsimonious explanation is that the lack of correlation is due to genetic diversity having only a minor predictive role for the viability or extinction risk of a species. Furthermore, the increased rate of genetic drift in small populations inevitably leads to low levels of neutral genetic diversity over time (Box 1). Hence, it is expected that some endangered species show low levels of neutral genetic diversity as a result of small population numbers and high levels of genetic drift, but this does not imply that genetic factors, such as those listed in Table 1, are causally responsible for decreasing population sizes or have an impact on population health and survival.
Fig. 1.
Genome-wide nucleotide diversity is a poor predictor of IUCN’s Red List status. Mammalian nucleotide diversity estimates were taken from Robinson et al. (42). Some critically endangered (CR) species, such as the Yangtze river dolphin or baiji (Lipotes vexillifer), show extremely low levels of nucleotide diversity. However, low levels of diversity are not necessarily due to recent anthropogenic pressures [the observed low genetic diversity in the baiji is due to a population bottleneck during the last glacial maximum ∼20 kya (114)]. Other endangered (EN) and critically endangered species, such as the Sumatran orangutan (Pongo abelii), show levels of diversity that fall well within the range of least concern (LC) species.
Table 1.
Classification of genetic extinction models
| Extinction model | Trigger | Genetic mechanism leading to reduced fitness | Selected references |
| Inbreeding depression | Breeding of related individuals | Recessive deleterious mutations become homozygous due to inbreeding (although other mechanisms have been proposed). | Charlesworth and Willis (28); Hedrick and Garcia-Dorado (117) |
| Mutational meltdown | Ineffective selection due to small/reduced population size | Many slightly deleterious mutations become fixed due to strong genetic drift in small populations. | Lynch et al. (118); Agrawal and Whitlock (44) |
| Maladaptation to environment | Changing environment [but see Brady et al. (119)] | Organisms are maladapted to the environment. | Tallmon et al. (120); Gomulkiewicz and Holdt (121) |
In fact, genetic factors are clearly not the primary drivers of the current unprecedented high rate of species extinction. Nongenetic factors such as overkill, habitat destruction and fragmentation, the introduction of invasive species, rapid climate change, or pollution, reduce the growth rates of species and cause their populations to decline (24). Nonetheless, genetic aspects such as inbreeding depression or lack of adaptive diversity can certainly contribute to the final extinction of an already diminished and inbred population (25, 26).
Conservation genetic practice rests on the assumption that measured levels of genetic diversity provide a direct indicator of the degree to which genetic factors contribute to the risk of extinction, and that increasing or protecting genetic diversity in small populations can mitigate this risk (e.g., ref. 27). The scope of our paper is to critically examine such claims in the light of recent population genetic insights as well as empirical results that were made possible by the eruption of genomic datasets that became available over the last decade. We propose that neutral genetic diversity has only very limited relevance for conservation genetics. Instead, individual genomes harbor functionally important, genomically localized variation that can severely impact individual fitness and should, therefore, guide conservation efforts. Population genetic models suggest that the relationship between neutral diversity, functional diversity, and population viability, can be unintuitive and rely on several unknown parameters. Thus, we argue for the necessity of mapping adaptive genetic variation in the genome, and for a better understanding of the determinants of genetic load and its consequences. Importantly, however, the conservation of species in their natural habitats or ecosystems can ultimately be successful only when the primary nongenetic causes for population decline are mitigated as well.
Inbreeding Depression and Hybrid Vigor
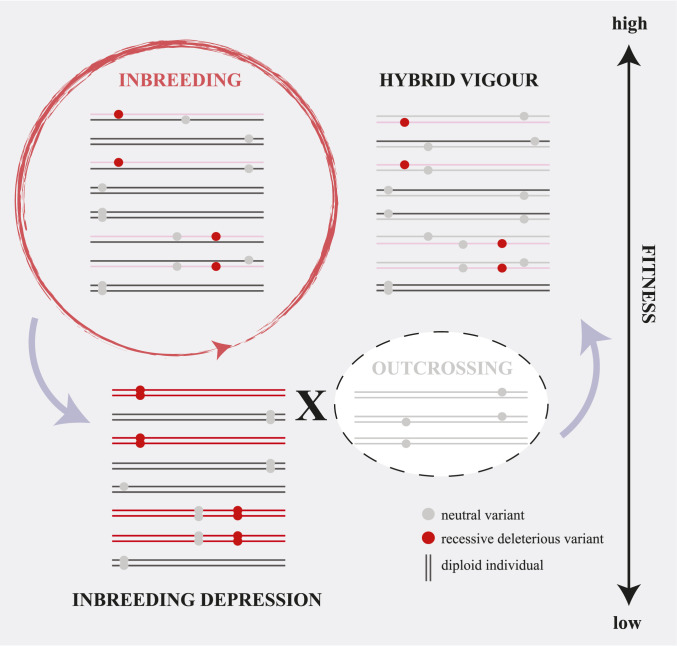
Two processes often cited in the conservation genetics literature to link the amount of neutral genetic diversity to individual fitness are “inbreeding depression” and “hybrid vigor.” The impact each of these factors has on fitness arises from the exposure or masking of recessive deleterious variants (28). In the case of inbreeding depression, the reduction in fitness of individuals is caused by mating between related individuals as a result, for example, of habitat fragmentation (29). Such consanguineous mating leads to an increase in genomic segments of identity by descent harboring recessive deleterious variants that become exposed in homozygosity in some individuals (Fig. 2), and in extreme cases can lead to the extinction of populations or species (Table 1). An inverse process occurs in the case of outbreeding, where a fitness increase in the population can be observed as a result of hybrid vigor, as recessive deleterious mutations become masked in heterozygous states in hybrid individuals (30) (Fig. 2).
Fig. 2.
Inbreeding depression and hybrid vigor. Inbreeding leads to an increase in homozygous genotypes. Recessive deleterious variants that become homozygous lead to a reduction in fitness in affected individuals (i.e., inbreeding depression). Outcrossing with individuals from a different population reduces homozygous genotypes and thus increases individual fitness again (hybrid vigor).
Although often conflated in conservation genetic literature, inbreeding depression is not mechanistically linked to low levels of genetic diversity. Inbreeding depression can happen in a population of any size and is probably worse in large, more diverse populations (31). Conversely, there are species with strikingly low levels of diversity but no signs of inbreeding [e.g., the brown hyena (32)]. A recent study by Kyriazis et al. (33) has proposed that using a population with high levels of genetic diversity for genetic rescue can in fact be ineffective for preventing the extinction of small populations. By leveraging population genetic simulations on ecological models, the authors demonstrated that this occurs via the introduction of a large number of recessive deleterious variants whose combined effects are potentially more harmful than the increase in fitness associated with hybrid vigor. Moreover, they showed that outcrossing with a population with low genetic diversity can introduce deleterious variants that had drifted to fixation due to small population sizes (33). Thus, according to their simulations, a population with intermediate levels of genetic diversity is most effective for rescuing a small endangered population.
Considering more complex relationships between genetic variants and fitness, such as epistasis and gene–environment interactions, makes the relation between inbreeding, outcrossing, and fitness even less predictable. Facilitated outcrossing and migration can lead to a species-wide reduction in diversity and prevent local adaptation, leading to a further long-term negative effect on species survival (34). Furthermore, as populations diverge and ultimately evolve into distinct species, hybrids between them gradually become inviable and infertile. Thus, even though outcrossing might prevent inbreeding depression, it can also lead to reduced fitness because of noncompatible mutations within hybrid individuals. Recombination in the hybrid population can disrupt parental gene combinations (“coadapted gene complexes”) and thus expose incompatibilities that lead to a decline in fitness over several generations (35). Experimental studies that examine hybrid fitness are often limited to the F1 and F2 generations and thus do not take this potential decline in fitness due to outcrossing depression in later generations into account (36). Although outcrossing depression between diverged populations can be predicted to some degree from sequence divergence (37–39), we are only beginning to fully understand how hybrid vigor over time transforms into hybrid inviability as a function of genetic drift and the individual and epistatic fitness effects of mutations (40). A better understanding of population genetic mechanisms will facilitate an evaluation of the relative risks of inbreeding and outcrossing depression. However, it is clear that a simple relation with neutral genetic diversity is not warranted.
Low Genetic Diversity and Mutation Load
Besides the effects of inbreeding on fitness, it is also often assumed that low genetic diversity indicates that selection is ineffective and that random genetic drift dominates allele frequency dynamics. As a consequence, deleterious mutations are not effectively removed and/or increase in frequency, resulting in reduced mean fitness of the individuals in a population (i.e., mutation load; see SI Appendix, Text S1). Such a reduction in fitness can lead to reduced population size and a higher risk of stochastic demographic extinction. Moreover, in very small populations, this increase in drift leads to the fixation of slightly deleterious mutations and, in a vicious circle, to further reduced population size, even less effective selection, and eventually, to the extinction of the population (a process often referred to as “mutational meltdown”; see Table 1). Although we do not discount that, over long periods of time, a lack of effective selection and the accumulation of deleterious mutations will most certainly impact the genomic and physiological integrity of an organism, it is not exactly clear when reduced neutral genetic diversity is indicating problematic levels of genetic drift, or when population size becomes too small to prevent a decline in population viability. Experiments in Drosophila suggest that the species can survive for many generations with population sizes as small as 25 individuals without showing any signs of reduced fitness compared to outbred wild populations (41). Furthermore, certain species, such as the San Nicolas island fox (42), the vaquita porpoise (43), or the brown hyena (32), have a near absence of genetic variation, even though no fitness reduction or any apparent genetically linked diseases have been observed. From a theoretical perspective, it is possible for a species to persist even with high levels of deleterious mutation load (44). Population persistence crucially depends on the specific conditions of how and when selection acts (SI Appendix, Text S1), and a better understanding of the ecological relevance of mutation load is thus necessary to predict its effect on the extinction risk of a population (44). Furthermore, the purging of partially recessive deleterious mutations might be another important factor to explain the long-term survival and apparent lack of inbreeding depression in some small populations (45).
The recent availability of genomic datasets has made it possible to directly estimate and compare the mutation load across different species. Contrary to expectations commonly assumed in conservation genetics, species with small population census size seem to contain less (additive) mutation load than species with larger population sizes (46). Notwithstanding, measuring differences in mutation load between diverged species is challenging and potentially prone to bioinformatic biases and measurement errors (47) (SI Appendix, Text S1).
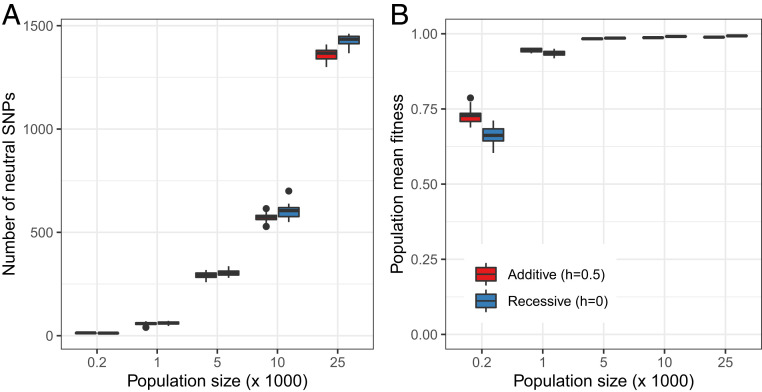
A more tractable question is how demographic differences between populations of the same species have affected the distribution of genetic diversity and load. In this case, only mutations that segregate in the ancestral population of the species have to be considered, and the alignment of genomes is less error-prone than when comparing genomes from different species (47, 48). Results point to an existing but weak relation between neutral diversity and mutation load in bottlenecked populations (49, 50). For example, in humans, the Out-of-Africa bottleneck ∼50,000 y ago has strongly reduced neutral genetic diversity in non-African human populations. However, even in one of the most extremely bottlenecked human populations, the Greenlandic Inuit, the estimated additive mutation load shows, at most, only a slight increase compared to African populations (51). Although the Inuit carry deleterious mutations at higher frequencies than other populations, they also carry fewer deleterious variants overall. This is consistent with population genetics theory that suggests that the number of deleterious variants per individual is only modestly affected by a reduction in population size since many deleterious mutations are lost during the bottleneck (47, 49, 52). More generally, a significant increase in additive mutation load is expected only under severe and extended reductions in population size that exceed that of the human Out-of-Africa bottleneck (49) and lead to a reduction in population mean fitness as a result of an increase in the frequency of slightly deleterious mutations through drift (Fig. 3). Mutation load due to strongly recessive mutations can show a more immediate response to changes in population size or surges of inbreeding (47, 51). However, similar to additive load, the long-term equilibrium value hardly depends on population size and thus cannot be predicted from neutral diversity (Fig. 3).
Fig. 3.
Forward in time simulations over a wide range of population sizes show mutation load is only modestly affected by Ne. The forward in time population genetic simulations with SLiM 3 (115) assumed a constant population size, ranging from 200 to 25,000 individuals. We simulated a total of 1,000 evenly distributed exons over a genetic length of 10 cM, with a nonsynonymous to synonymous ratio of 2:1, and a mutation rate of 1e-5 per exon. The selection coefficient of the nonsynonymous mutations was sampled from a gamma distribution as estimated in Huber et al. (98), and synonymous mutations were assumed to be neutral. We considered either additive (h = 0.5; red) or recessive (h = 0; blue) deleterious mutations. We simulated 100,000 generations and recorded genetic diversity and mean fitness of individuals in the population at the end of each run. (A) The number of segregating synonymous sites is proportional to the population size, as expected from neutral theory. (B) Above a long-term population size of 5,000, the population mean fitness is close to the maximum, and independent of population size. Only with a fairly small population size of fewer than 1,000 individuals over prolonged periods of time does fitness become severely reduced due to the fixation of slightly deleterious mutations. This result, i.e., that only a few hundred individuals are necessary to prevent a decline in population fitness and viability, holds true under more realistic simulations of mammalian genomic parameters (33), and when extending the model to allow for beneficial mutations and epistasis (116).
Neutral Diversity Does Not Predict Adaptive Potential
Another key concept in the conservation genetic literature is that of adaptive (or evolutionary) potential, which can be defined as the ability of populations to respond to shifts in environmental and selective pressures by means of phenotypic and/or molecular changes (53) and thus prevent extinction due to maladaptation (Table 1). Accordingly, it is assumed that populations with higher genetic diversity have more adaptive potential because of higher levels of standing genetic variation, which makes them more robust to changing environmental conditions and, thus, more suitable for conservation efforts (54). However, evidence from surveying genetic diversity of wild populations is showing that such a relationship might not always exist. A study evaluated the effects of introducing steelhead trout (Oncorhynchus mykiss), a species known to hatch in rivers and live in the ocean before returning to freshwater for spawning, from California into Lake Michigan in the 1880s (55). Upon its introduction to Lake Michigan, steelhead trout began to use the lake as a surrogate ocean and, despite this remarkable shift in environmental conditions and a strong population bottleneck, showed distinct signatures of local adaptation (55). Similarly, an experimental study in brook trout (Salvelinus fontinalis) suggests that low genetic diversity does not prevent or predict transplantation success into fishless ponds over a wide gradient of ecological variables (56). As another example, dramatically bottlenecked northern European populations of Arabidopsis lyrata and Arabidopsis thaliana show strong genomic footprints of adaptation (57, 58), and experimental evidence suggests that their strongly reduced genetic diversity has not affected their ability to adapt to local environmental conditions (59–61). To investigate the determinants of the rate of genetic adaptation more generally, population genomic studies have analyzed genome-wide polymorphism and divergence data from many different species and concluded that low-diversity taxa do not seem to accumulate adaptive substitutions at a substantially lower rate than high-diversity taxa (62, 63), although within certain groups such a relationship might exist (63, 64).
These results question the existence of a simple general relationship between the levels of genome-wide genetic diversity observed in a population and its potential to genetically adapt to changing environmental conditions. Instead, the nature of genetic diversity segregating at particular loci seems to be substantially more important (65, 66), and selection acting on these variants makes their distribution and evolutionary trajectories potentially quite distinct from neutral variants. As stated by Lewontin in 1974, “The question was never really how much genetic variation is there but rather what is the nature of genetic variation for fitness in a population” (67). Genetic diversity at few selected loci can be maintained for millions of years in species or populations via balancing selection (68, 69), including overdominance or heterozygote advantage, temporal/spatial variation in selective pressures, negative frequency-dependent selection, and pleiotropy (70). In fact, balancing selection might constitute a widespread and important mechanism enabling rapid adaptation (71). The classical example of balancing selection is the major histocompatibility complex (MHC) region, which is responsible for antigen presentation and immune response in vertebrate species (70). It has been shown that rare MHC alleles can confer increased resistance when parasites are introduced into a host population where they were previously not present (72). Shifts in parasite composition can lead to adaptive changes in the frequency of MHC alleles, as observed in sticklebacks (73), birds (74), and frogs (75). Such shifts in parasite composition can be a response to climatic changes (76) and thus might become increasingly relevant for future conservation efforts.
Notably, MHC diversity is not related to effective population size when comparing central chimpanzees and humans (77), but is strongly correlated with environmental factors, such as pathogen load, in the latter (78). Similarly, in a study involving invasive cane toads (Rhinella marina) in Australia, the levels of genetic diversity at loci involved in resistance to heat and dehydration are either weakly or not at all correlated with effective population size, even in the case of severe bottlenecks (79). Thus, genetic variation that might become relevant in future climatic conditions is not necessarily largest in populations with the most genome-wide genetic diversity. However, the current paradigm in conservation genetics would misleadingly suggest that the population with the most neutral diversity has the highest “adaptive potential” regarding future climatic change. Consequently, a recent study on bottlenose dolphin populations (Tursiops aduncus) has argued for an evaluation of the levels of diversity contained within the MHC region, rather genome-wide patterns of neutral genetic diversity, for conservation purposes (80).
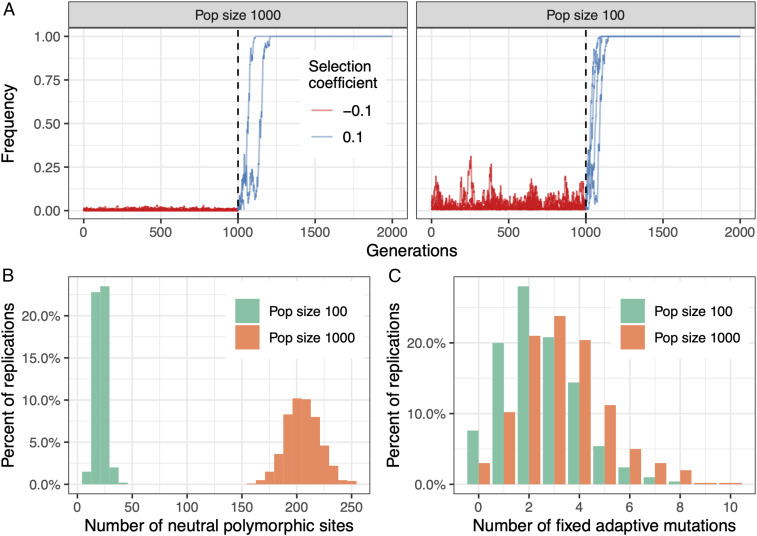
Balancing selection is not the only mechanism distorting the relation between neutral diversity and adaptive potential. If adaptation involves preexisting mutations that shift from effectively deleterious to beneficial as the environment changes, then adaptation can be almost unaffected by population size since the frequency of segregating deleterious mutations is on average higher in small populations due to less effective selection (Fig. 4). Evidence from experimental evolution and empirical population genetic studies indicates that beneficial mutations are typically deleterious in the absence of selective pressures (81), suggesting that such dynamics likely occur in nature. Similarly, quantitative genetic models show that stabilizing selection and pleiotropy can strongly reduce the correlation between neutral diversity and additive variation of a quantitative trait (54). Importantly, neutral genetic diversity is determined by demographic events occurring in the history of a population, including ancient bottlenecks and migration events (Box 1), whereas the emergence and fate of beneficial mutations after environmental change depend on the population size immediately after the change (14, 82) (Fig. 4). Hence, in order to predict the adaptive potential of populations, it is necessary to estimate the immediate effective population size, the effect size and rate of mutations that contribute to adaptation, and the mutational target size (14, 83), all of which are parameters that are difficult to assess (81).
Fig. 4.
Adaptive potential can be fairly unrelated to neutral genetic diversity. In this example, adaptation involves preexisting mutations that shift from deleterious (s = −0.1) to beneficial (s = 0.1) to mimic the response of two different populations to a shift in environmental conditions. Here, selection is strong enough such that deleterious mutations are effectively selected and thus unlikely to fix even in the smaller population, which is a fundamental assumption of this model. (A) The allele frequency trajectories of selected mutations are plotted as a function of time (note that the trajectories of multiple mutations are overlapping). Before the change in environmental conditions at generation 1,000 (dashed vertical line), the two populations had vastly different population sizes of 1,000 (Left) and 100 individuals (Right), respectively. In the larger population, where selection is more effective, deleterious mutations segregate at lower frequencies (red lines, Left) compared to the smaller population (red lines, Right). After the change in environmental conditions, both populations are composed of 100 individuals, and segregating deleterious mutations become beneficial (blue lines). (B) Neutral genetic diversity right after the change in environmental conditions is, as expected, 10 times larger in the historically larger population. (C) However, because the fixation probability increases with allele frequency and deleterious mutations segregate at higher frequencies in smaller populations, the number of fixed adaptive mutations after the change in environmental conditions has a very similar distribution in both populations, whereby the effects of historical population size are much less apparent. The plots in B and C summarize results of 500 replicates.
Finally, it is often argued that genetic rescue increases the genetic diversity and, therefore, the adaptive potential of low-diversity populations. This implies that an increase in fitness is not predominantly caused by heterosis, but rather that adaptive mutations are transferred from the large to the small population, i.e., there is adaptive variation segregating at high enough frequencies in the source (larger) population. Such an assumption depends, as above, on the mutation rate, the mutational target size, and the effective population size (14, 81, 83), but also on past environmental conditions that might have shaped adaptive diversity in the source population. This is nicely exemplified by adaptive introgression that resulted from a series of admixture events between archaic and modern humans (84–92). Even though Neanderthals and Denisovans had very low levels of genetic diversity and small long-term effective population sizes, the two groups were exposed to harsh climatic and biotic conditions for thousands of generations and thus were able to accumulate adaptive genetic variants that were then introgressed into modern human populations and facilitated adaptation to high altitude (86), pathogens (88), and cold climatic conditions (89). Accordingly, the choice to concentrate conservation efforts on populations with higher levels of genetic diversity might only be effective in a very restricted set of scenarios (93, 94).
The Future of Conservation Genetics in the Face of Mass Extinction
We provide several empirical and theoretical arguments that challenge the commonly assumed simplistic relation between genome-wide patterns of neutral genetic diversity and population persistence or adaptive potential. We argue that such a relation remains speculative and should thus be excluded from conservation strategies. Instead, we suggest alternative approaches that focus on gaining a better understanding of the genetic basis of deleterious and beneficial variation.
In general, a better knowledge of crucial population genetic parameters, including “dominance” and “epistasis,” will allow us to better predict the effects of reduced population size, inbreeding, and outcrossing, on fitness and adaptive potential (40, 47, 48, 54, 95–98). For example, better knowledge of the distribution of dominance coefficients in natural populations (e.g., 96) will lead to an improved understanding of the relevance of purging for inbreeding load in small populations (99) and the dynamics of genetic rescue (33, 100). Moreover, more detailed knowledge of epistasis (e.g., refs. 40 and 101) will lead to a better prediction of the time course of outbreeding depression in hybrid populations (35). Improving the knowledge of population genetic processes affecting natural populations could, in turn, lead to more effective conservation strategies. For example, it would be possible to predict in which species and under which demographic histories genetic rescue can be most successful (33), which individuals or populations to use when implementing genetic rescue, or when to expect a possibly undesirable replacement of native genomes by the introduced immigrant alleles (100). While the estimation of these population genetic parameters remains quite challenging, recent work has shown novel ways of inference using model-based approaches (40, 96, 101–103). Likewise, bioinformatic and comparative genomic methods can provide useful information on the location and strength of deleterious variation (104), but their reliance on assumptions should be carefully evaluated (SI Appendix, Text S1).
Furthermore, we argue for the need to better understand functional genetic variation that is relevant for future environmental and climatic change. To investigate relevant variation, at least three conditions have to be satisfied. First, the environmental variables negatively affecting a species (e.g., drought) have to be known, as well as their predicted geographical and/or temporal expansion in the future. Second, tolerance or robustness to the changing environments needs to be variable within the species and exhibit a significant genetic basis (e.g., some populations are genetically more drought resistant than others). Third, it must be possible to map the corresponding genetic variants and to predict which individuals will be most tolerant or robust to the detrimental environment. Such an approach could be used to select individuals for managed translocations and selective breeding or to protect the most promising populations of a species given predictable changes in climatic and environmental variables (105). Recent empirical and experimental studies illustrate the exciting potential for mapping adaptive variation and incorporating genomics to study and predict traits of conservation importance (106–108). However, we want to emphasize that complex gene–environment interactions and genetic incompatibilities between source and recipient populations can greatly reduce the benefit of genetic mixing (109), and that there is no guarantee that such facilitated evolutionary strategies are effective enough to counteract the drastic climatic and ecological challenges of the Anthropocene. Furthermore, in many instances, such genomic approaches might be too laborious and impractical compared to strategies that rather focus on alleviating nongenetic threats to the population (24). Finally, we agree that the best way to preserve biodiversity is to restore and conserve natural ecosystems, as has been previously proposed (56). The protection or reestablishment of a functioning ecosystem might provide a more effective alternative to treating single species as isolated, independently evolving entities (110–113).
Supplementary Material
Acknowledgments
We thank Chris Kyriazis, Athanasios Kousathanas, Alexander Salis, Joshua Schmidt, and two anonymous reviewers for helpful comments on the manuscript. J.C.T. and C.D.H. are supported by the Australian Research Council through Discovery Grants IN180100017 (J.C.T.) and DP190103606 (C.D.H.), and an Australian Research Council Discovery Early Career Researcher Award Fellowship DE180100883 (C.D.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015096118/-/DCSupplemental.
Data Availability
There are no data underlying this work.
References
- 1.Pimm S. L., et al., The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Ceballos G., et al., Accelerated modern human-induced species losses: Entering the sixth mass extinction. Sci. Adv. 1, e1400253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceballos G., Ehrlich P. R., Dirzo R., Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. U.S.A. 114, E6089–E6096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceballos G., Ehrlich P. R., The misunderstood sixth mass extinction. Science 360, 1080–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Knight A. T., et al., Knowing but not doing: Selecting priority conservation areas and the research-implementation gap. Conserv. Biol. 22, 610–617 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Frankham R., Briscoe D. A., Ballou J. D., Introduction to Conservation Genetics (Cambridge University Press, 2002). [Google Scholar]
- 7.Reed D. H., Frankham R., Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237 (2003). [Google Scholar]
- 8.Frankham R., Genetics and extinction. Biol. Conserv. 126, 131–140 (2005). [Google Scholar]
- 9.Spielman D., Brook B. W., Frankham R., Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U.S.A. 101, 15261–15264 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markert J. A., et al., Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 10, 205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weeks A. R., Stoklosa J., Hoffmann A. A., Conservation of genetic uniqueness of populations may increase extinction likelihood of endangered species: The case of Australian mammals. Front. Zool. 13, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M., Li W. H., Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakeley J., Coalescent Theory: An Introduction (Roberts Publishers, 2009). [Google Scholar]
- 14.Messer P. W., Petrov D. A., Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28, 659–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waples R. S., A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121, 379–391 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., Estimation of effective population sizes from data on genetic markers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1395–1409 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luikart G., Ryman N., Tallmon D. A., Schwartz M. K., Allendorf F. W., Estimation of census and effective population sizes: The increasing usefulness of DNA-based approaches. Conserv. Genet. 11, 355–373 (2010). [Google Scholar]
- 18.Doyle J. M., Hacking C. C., Willoughby J. R., Sundaram M., Andrew DeWoody J., Mammalian genetic diversity as a function of habitat, body size, trophic class, and conservation status. J. Mammal. 96, 564–572 (2015). [Google Scholar]
- 19.Brüniche-Olsen A., Kellner K. F., Anderson C. J., Andrew DeWoody J., Runs of homozygosity have utility in mammalian conservation and evolutionary studies. Conserv. Genet. 19, 1295–1307 (2018). [Google Scholar]
- 20.Brüniche-Olsen A., Kellner K. F., DeWoody J. A., Island area, body size and demographic history shape genomic diversity in Darwin’s finches and related tanagers. Mol. Ecol. 28, 4914–4925 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Grossen C., Guillaume F., Keller L. F., Croll D., Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nat. Commun. 11, 1001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankham R., Bradshaw C. J. A., Brook B. W., Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 170, 56–63 (2014). [Google Scholar]
- 23.Willoughby J. R., et al., The reduction of genetic diversity in threatened vertebrates and new recommendations regarding IUCN conservation rankings. Biol. Conserv. 191, 495–503 (2015). [Google Scholar]
- 24.Foden W. B., et al., Climate change vulnerability assessment of species. Wiley Interdiscip. Rev. Clim. Change 10, e551 (2019). [Google Scholar]
- 25.Hedrick P. W., Lacy R. C., Allendorf F. W., Soule M. E., Directions in conservation biology: Comments on Caughley. Conserv. Biol. 10, 1312–1320 (1996). [Google Scholar]
- 26.Caughley G., Directions in conservation biology. J. Anim. Ecol. 63, 215 (1994). [Google Scholar]
- 27.Ralls K., Sunnucks P., Lacy R. C., Frankham R., Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 251, 108784 (2020). [Google Scholar]
- 28.Charlesworth D., Willis J. H., The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Schlaepfer D. R., Braschler B., Rusterholz H.-P., Baur B., Genetic effects of anthropogenic habitat fragmentation on remnant animal and plant populations: A meta-analysis. Ecosphere 9, e02488 (2018). [Google Scholar]
- 30.Kim B. Y., Huber C. D., Lohmueller K. E., Deleterious variation shapes the genomic landscape of introgression. PLoS Genet. 14, e1007741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson J. A., et al., Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci. Adv. 5, eaau0757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westbury M. V., et al., Extended and continuous decline in effective population size results in low genomic diversity in the world’s rarest hyena species, the brown hyena. Mol. Biol. Evol. 35, 1225–1237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyriazis C. C., Wayne R. K., Lohmueller K. E., Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett., 10.1002/evl3.209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolodny O., et al., Reconsidering the management paradigm of fragmented populations. 10.1101/649129 (24 May 2019). [DOI] [Google Scholar]
- 35.Edmands S., Timmerman C. C., Modeling factors affecting the severity of outbreeding depression. Conserv. Biol. 17, 883–892 (2003). [Google Scholar]
- 36.Bell D. A., et al., The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol. 34, 1070–1079 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Coyne J. A., Orr H. A., Patterns of speciation in Drosophila. Evolution 43, 362–381 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Yukilevich R., Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 66, 1430–1446 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Roux C., et al., Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 14, e2000234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dagilis A. J., Kirkpatrick M., Bolnick D. I., The evolution of hybrid fitness during speciation. PLoS Genet. 15, e1008125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilligan D. M., Woodworth L. M., Montgomery M. E., Briscoe D. A., Frankham R., Is mutation accumulation a threat to the survival of endangered populations? Conserv. Biol. 11, 1235–1241 (1997). [Google Scholar]
- 42.Robinson J. A., et al., Genomic flatlining in the endangered Island Fox. Curr. Biol. 26, 1183–1189 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Morin P. A., et al., Reference genome and demographic history of the most endangered marine mammal, the vaquita. Mol. Ecol. Resour., 10.1111/1755-0998.13284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal A. F., Whitlock M. C., Mutation load: The fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst. 43, 115–135 (2012). [Google Scholar]
- 45.Robinson J. A., Brown C., Kim B. Y., Lohmueller K. E., Wayne R. K., Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr. Biol. 28, 3487–3494.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Valk T., van der Valk T., de Manuel M., Marques-Bonet T., Guschanski K., Estimates of genetic load in small populations suggest extensive purging of deleterious alleles. bioRxiv [Preprint] (2019). 10.1101/696831 (Accessed 12 February 2021). [DOI]
- 47.Simons Y. B., Sella G., The impact of recent population history on the deleterious mutation load in humans and close evolutionary relatives. Curr. Opin. Genet. Dev. 41, 150–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henn B. M., Botigué L. R., Bustamante C. D., Clark A. G., Gravel S., Estimating the mutation load in human genomes. Nat. Rev. Genet. 16, 333–343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons Y. B., Turchin M. C., Pritchard J. K., Sella G., The deleterious mutation load is insensitive to recent population history. Nat. Genet. 46, 220–224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keinan A., Mullikin J. C., Patterson N., Reich D., Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 39, 1251–1255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen C. T., et al., The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: Insights from the Greenlandic Inuit. Genetics 205, 787–801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Do R., et al., No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 47, 126–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann A. A., Sgrò C. M., Kristensen T. N., Revisiting adaptive potential, population size, and conservation. Trends Ecol. Evol. 32, 506–517 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Willi Y., Van Buskirk J., Hoffmann A. A., Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433–458 (2006). [Google Scholar]
- 55.Willoughby J. R., Harder A. M., Tennessen J. A., Scribner K. T., Christie M. R., Rapid genetic adaptation to a novel environment despite a genome-wide reduction in genetic diversity. Mol. Ecol. 27, 4041–4051 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Yates M. C., Bowles E., Fraser D. J., Small population size and low genomic diversity have no effect on fitness in experimental translocations of a wild fish. Proc. Biol. Sci. 286, 20191989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huber C. D., Nordborg M., Hermisson J., Hellmann I., Keeping it local: Evidence for positive selection in Swedish Arabidopsis thaliana. Mol. Biol. Evol. 31, 3026–3039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takou M., et al., Maintenance of adaptive dynamics and no detectable load in a range-edge out-crossing plant population. Mol. Biol. Evol., 10.1093/molbev/msaa322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leinonen P. H., et al., Local adaptation in European populations of Arabidopsis lyrata (Brassicaceae). Am. J. Bot. 96, 1129–1137 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Agren J., Schemske D. W., Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 194, 1112–1122 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Hämälä T., Mattila T. M., Savolainen O., Local adaptation and ecological differentiation under selection, migration, and drift in Arabidopsis lyrata. Evolution, 10.1111/evo.13502 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Galtier N., Adaptive protein evolution in animals and the effective population size hypothesis. PLoS Genet. 12, e1005774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rousselle M., et al., Is adaptation limited by mutation? A timescale-dependent effect of genetic diversity on the adaptive substitution rate in animals. PLoS Genet. 16, e1008668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gossmann T. I., et al., Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol. Biol. Evol. 27, 1822–1832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reed D. H., Frankham R., How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55, 1095–1103 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Hoelzel A. R., Rus Hoelzel A., Bruford M. W., Fleischer R. C., Conservation of adaptive potential and functional diversity. Conserv. Genet. 20, 1–5 (2019). [Google Scholar]
- 67.Lewontin R. C., The Genetic Basis of Evolutionary Change (Columbia University Press, 1974). [Google Scholar]
- 68.Leffler E. M., et al., Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 339, 1578–1582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teixeira J. C., et al., Long-term balancing selection in LAD1 maintains a missense trans-species polymorphism in humans, chimpanzees, and bonobos. Mol. Biol. Evol. 32, 1186–1196 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Key F. M., Teixeira J. C., de Filippo C., Andrés A. M., Advantageous diversity maintained by balancing selection in humans. Curr. Opin. Genet. Dev. 29, 45–51 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Stern D. B., Lee C. E., Lee C. E., Evolutionary origins of genomic adaptations in an invasive copepod. Nat. Ecol. Evol. 4, 1084–1094 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Phillips K. P., et al., Immunogenetic novelty confers a selective advantage in host-pathogen coevolution. Proc. Natl. Acad. Sci. U.S.A. 115, 1552–1557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eizaguirre C., Lenz T. L., Kalbe M., Milinski M., Rapid and adaptive evolution of MHC genes under parasite selection in experimental vertebrate populations. Nat. Commun. 3, 621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biedrzycka A., et al., Blood parasites shape extreme major histocompatibility complex diversity in a migratory passerine. Mol. Ecol. 27, 2594–2603 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Savage A. E., Zamudio K. R., Adaptive tolerance to a pathogenic fungus drives major histocompatibility complex evolution in natural amphibian populations. Proc. Biol. Sci. 283, 20153115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Polley L., Hoberg E., Kutz S., Climate change, parasites and shifting boundaries. Acta Vet. Scand. 52, S1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maibach V., Hans J. B., Hvilsom C., Marques-Bonet T., Vigilant L., MHC class I diversity in chimpanzees and bonobos. Immunogenetics 69, 661–676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manczinger M., et al., Pathogen diversity drives the evolution of generalist MHC-II alleles in human populations. PLoS Biol. 17, e3000131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Selechnik D., et al., Increased adaptive variation despite reduced overall genetic diversity in a rapidly adapting invader. Front. Genet. 10, 1221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manlik O., et al., Is MHC diversity a better marker for conservation than neutral genetic diversity? A case study of two contrasting dolphin populations. Ecol. Evol. 9, 6986–6998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jensen J. D., On the unfounded enthusiasm for soft selective sweeps. Nat. Commun. 5, 5281 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Barton N., Understanding adaptation in large populations. PLoS Genet. 6, e1000987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kardos M., Luikart G., The genetic architecture of fitness drives population viability during rapid environmental change. Am. Nat., 10.1086/713469 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Mendez F. L., Watkins J. C., Hammer M. F., A haplotype at STAT2 Introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am. J. Hum. Genet. 91, 265–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mendez F. L., Watkins J. C., Hammer M. F., Global genetic variation at OAS1 provides evidence of archaic admixture in Melanesian populations. Mol. Biol. Evol. 29, 1513–1520 (2012). [DOI] [PubMed] [Google Scholar]
- 86.Huerta-Sánchez E., et al., Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E., Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dannemann M., Andrés A. M., Kelso J., Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human Toll-like receptors. Am. J. Hum. Genet. 98, 22–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Racimo F., et al., Archaic adaptive introgression in TBX15/WARS2. Mol. Biol. Evol. 34, 509–524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Racimo F., Marnetto D., Huerta-Sánchez E., Signatures of archaic adaptive introgression in present-day human populations. Mol. Biol. Evol. 34, 296–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Enard D., Petrov D. A., Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell 175, 360–371.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rees J. S., Castellano S., Andrés A. M., The genomics of human local adaptation. Trends Genet. 36, 415–428 (2020). [DOI] [PubMed] [Google Scholar]
- 93.Garcia de Leaniz C., et al., A critical review of adaptive genetic variation in atlantic salmon: Implications for conservation. Biol. Rev. Camb. Philos. Soc. 82, 173–211 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Hohenlohe P. A., et al., Conserving adaptive potential: Lessons from Tasmanian devils and their transmissible cancer. Conserv. Genet. 20, 81–87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henn B. M., et al., Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. U.S.A. 113, E440–E449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huber C. D., Durvasula A., Hancock A. M., Lohmueller K. E., Gene expression drives the evolution of dominance. Nat. Commun. 9, 2750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mackay T. F., Moore J. H., Why epistasis is important for tackling complex human disease genetics. Genome Med. 6, 124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huber C. D., Kim B. Y., Marsden C. D., Lohmueller K. E., Determining the factors driving selective effects of new nonsynonymous mutations. Proc. Natl. Acad. Sci. U.S.A. 114, 4465–4470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Caballero A., Bravo I., Wang J., Inbreeding load and purging: Implications for the short-term survival and the conservation management of small populations. Heredity 118, 177–185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harris K., Zhang Y., Nielsen R., Genetic rescue and the maintenance of native ancestry. Conserv. Genet. 20, 59–64 (2019). [Google Scholar]
- 101.Sohail M.et al.; Genome of the Netherlands Consortium; Alzheimer’s Disease Neuroimaging Initiative , Negative selection in humans and fruit flies involves synergistic epistasis. Science 356, 539–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Balick D. J., Do R., Cassa C. A., Reich D., Sunyaev S. R., Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet. 11, e1005436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Marsden C. D., et al., Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. U.S.A. 113, 152–157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Oosterhout C., Mutation load is the spectre of species conservation. Nat. Ecol. Evol. 4, 1004–1006 (2020). [DOI] [PubMed] [Google Scholar]
- 105.van Oppen M. J. H., Oliver J. K., Putnam H. M., Gates R. D., Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carvalho C. S., et al., Combining genotype, phenotype, and environmental data to delineate site‐adjusted provenance strategies for ecological restoration. Mol. Ecol. Resour. 21, 44–58 (2020). [DOI] [PubMed] [Google Scholar]
- 107.Fuller Z. L., et al., Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science 369, eaba4674 (2020). [DOI] [PubMed] [Google Scholar]
- 108.Selmoni O., Rochat E., Lecellier G., Berteaux-Lecellier V., Joost S., Seascape genomics as a new tool to empower coral reef conservation strategies: An example on north-western Pacific Acropora digitifera. Evol. Appl. 13, 1923–1938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoffmann A. A., Miller A. D., Weeks A. R., Genetic mixing for population management: From genetic rescue to provenancing. Evol. Appl., 10.1111/eva.13154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Berger J., Stacey P. B., Bellis L., Johnson M. P., A mammalian predator–prey imbalance: Grizzly bear and wolf extinction affect avian Neotropical migrants. Ecol. Appl. 11, 947–960 (2001). [Google Scholar]
- 111.Beschta R. L., Ripple W. J., Large predators and trophic cascades in terrestrial ecosystems of the western United States. Biol. Conserv. 142, 2401–2414 (2009). [Google Scholar]
- 112.Seddon P. J., Griffiths C. J., Soorae P. S., Armstrong D. P., Reversing defaunation: Restoring species in a changing world. Science 345, 406–412 (2014). [DOI] [PubMed] [Google Scholar]
- 113.Calderón K., et al., Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME J. 11, 272–283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou X., et al., Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat. Commun. 4, 2708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haller B. C., Messer P. W., SLiM 3: Forward genetic simulations beyond the Wright-Fisher model. Mol. Biol. Evol. 36, 632–637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schultz S. T., Lynch M., Mutation and extinction: The role of variable mutational effects, synergistic epistasis, beneficial mutations, and degree of outcrossing. Evolution 51, 1363–1371 (1997). [DOI] [PubMed] [Google Scholar]
- 117.Hedrick P. W., Garcia-Dorado A., Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol. Evol. 31, 940–952 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Lynch M., Conery J., Bürger R., Mutational meltdowns in sexual populations. Evolution 49, 1067–1080 (1995). [DOI] [PubMed] [Google Scholar]
- 119.Brady S. P., et al., Causes of maladaptation. Evol. Appl. 12, 1229–1242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tallmon D. A., Luikart G., Waples R. S., The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 19, 489–496 (2004). [DOI] [PubMed] [Google Scholar]
- 121.Gomulkiewicz R., Holt R. D., When does evolution by natural selection prevent extinction? Evolution 49, 201–207 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no data underlying this work.