Significance
The insect respiratory system consists of tubular tracheae that transport oxygen to the organs. We show that, in the insect pest Riptortus pedestris, the establishment of an essential symbiosis in the gut with the aerobic bacterial species Burkholderia insecticola triggers the development of an extensive tracheal network enveloping the gut. Genetically blocking the trachea formation prevents this gut symbiosis. We further discovered that the reactive oxygen species-generating enzyme Duox is crucial for the formation and stabilization of tracheae by forming protein cross-links in the tracheal matrix. Reactive oxygen species generated by Duox can be scavenged with antioxidants such as N-acetylcysteine, and feeding insects with this compound prevents tracheal formation and symbiosis, suggesting that antioxidants can be used as novel insecticides.
Keywords: symbiosis, Duox, Burkholderia, Riptortus pedestris, trachea
Abstract
Most animals harbor a gut microbiota that consists of potentially pathogenic, commensal, and mutualistic microorganisms. Dual oxidase (Duox) is a well described enzyme involved in gut mucosal immunity by the production of reactive oxygen species (ROS) that antagonizes pathogenic bacteria and maintains gut homeostasis in insects. However, despite its nonspecific harmful activity on microorganisms, little is known about the role of Duox in the maintenance of mutualistic gut symbionts. Here we show that, in the bean bug Riptortus pedestris, Duox-dependent ROS did not directly contribute to epithelial immunity in the midgut in response to its mutualistic gut symbiont, Burkholderia insecticola. Instead, we found that the expression of Duox is tracheae-specific and its down-regulation by RNAi results in the loss of dityrosine cross-links in the tracheal protein matrix and a collapse of the respiratory system. We further demonstrated that the establishment of symbiosis is a strong oxygen sink triggering the formation of an extensive network of tracheae enveloping the midgut symbiotic organ as well as other organs, and that tracheal breakdown by Duox RNAi provokes a disruption of the gut symbiosis. Down-regulation of the hypoxia-responsive transcription factor Sima or the regulators of tracheae formation Trachealess and Branchless produces similar phenotypes. Thus, in addition to known roles in immunity and in the formation of dityrosine networks in diverse extracellular matrices, Duox is also a crucial enzyme for tracheal integrity, which is crucial to sustain mutualistic symbionts and gut homeostasis. We expect that this is a conserved function in insects.
Many animals possess a gut microbiota that is composed of pathogenic, commensal, and mutualistic microorganisms (1, 2). In insects, gut symbiotic microorganisms play a broad range of physiological roles in host metabolism and immunity through digestion of food, supplementation of essential nutrients that are scarce in food, degradation of phytotoxins and pesticides, and prevention of pathogen invasion by stimulation of the host immune system, production of antibiotics, or competition for limited nutrients and colonization sites (1, 3–6). Keeping symbiotic populations in the gut poses specific challenges: the hosts should specifically sustain the beneficial microbes while, at the same time and place, they need to winnow out pathogens and parasites acquired through feeding. To maintain gut homeostasis, some insects have evolved a symbiont sorting organ or filter in the gut (7, 8) or develop a specific gut compartment called the crop or crypt to harbor beneficial microbes (9). Specific physiological conditions in the gut lumen, such as pH, osmotic pressure, and oxygen concentration probably also contribute to create the favorable conditions for gut symbiosis. Except for gut symbionts in wood-feeding insects and honeybees, most insects’ gut symbionts are aerobic bacteria belonging to the Proteobacteria (10, 11), suggesting that oxygen supplementation is critical to maintain quality and quantity of the gut symbiont population. However, it is poorly understood how the oxygen supply to the gut is established and how it affects the gut symbiosis.
In addition, studies in Drosophila have revealed that gut mucosal immunity plays an important role to eliminate unfavorable bacteria and to keep gut homeostasis (12, 13). Ingested bacteria, especially pathogens that come in contact with the gut epithelia, activate immune responses such as the production of antimicrobial peptides (AMPs) and reactive oxygen species (ROS) (14, 15). The activated gut immunity maintains the gut homeostasis by eliminating the invading environmental microbes, which may be harmful to the host. In particular, bactericidal ROS produced by the membrane-bound enzyme dual oxidase (Duox) is thought to antagonize bacterial growth and play a major role in gut mucosal immunity (12, 13, 16–20). Duox generates ROS via its C-terminal NADPH oxidase domain that produces H2O2 in the gut lumen and its N-terminal peroxidase-homology domain that converts the H2O2 into highly bactericidal hypochlorous acid. In Drosophila fed with a gut pathogen, silencing of Duox led to an increased pathogen load in the gut and the death of the insect (12). It was proposed that the Duox- and ROS-dependent immunity in the gut is dominant over AMP production that serves as a failsafe system in case of infection with ROS-resistant microbes (21).
In addition to the direct immune function, Duox-mediated ROS also contributes to gut homeostasis in insects by stabilizing the peritrophic membrane of the midgut through the formation of covalent dityrosine bonds between matrix proteins (22, 23). The peritrophic matrix of insects forms a layer composed of chitin and glycoproteins that lines the midgut lumen. This layer protects the midgut epithelium from abrasive food particles and microbes. In the mosquito Anopheles gambiae, Duox-RNA interference (RNAi) leads to the disruption of a dityrosine network (DTN), which cross-links proteins in the peritrophic membrane, and a loss of the matrix’s barrier function (23). Consequently, gut microbes gain direct contact with gut epithelial cells and activate chronic AMP expression, which provokes in turn a dramatic reduction of the bacterial midgut population. A similar function of Duox was reported in the tick Ixodes scapularis (24). Thus, Duox can interact in the insect gut with ingested microbes in two ways, by producing bactericidal ROS or by generating a physical barrier. In contrast to these well-described functions in mucosal immunity, little is known about the impact of the Duox enzyme on mutualistic bacteria.
The bean bug Riptortus pedestris (Fig. 1A), a notorious pest of legume crops, has a particular symbiotic organ located in the posterior midgut (25–27) (Fig. 1B). This region, called M4, is composed of several hundred crypts organized in two parallel rows. The lumen of these crypts is entirely filled with a specific gut symbiont of the genus Burkholderia. This is a nutritional symbiosis based on recycling by the gut symbiont of insect metabolic waste into high-value metabolites used by the host (28). This interaction is essential for optimal development and reproduction of the bean bug host (26). Every new generation of R. pedestris acquires the symbiont from the environmental soil by oral intake during feeding and drinking. A narrow channel located in the midgut, at the entrance of the M4, as well as microbe–microbe competition in the gut crypts, sorts out the specific partner from the ingested soil microbiota, resulting in the specific colonization of the symbiotic organ (7, 29). The few hundreds of bacterial cells that are taken up by the symbiotic organ during infection multiply in a few days to a population of tens of millions that completely occupy the luminal space of all the crypts in the M4 (30). The crypts control this very large symbiont population using immune-related AMPs as well as M4-specific symbiotic peptides (28, 31–33).
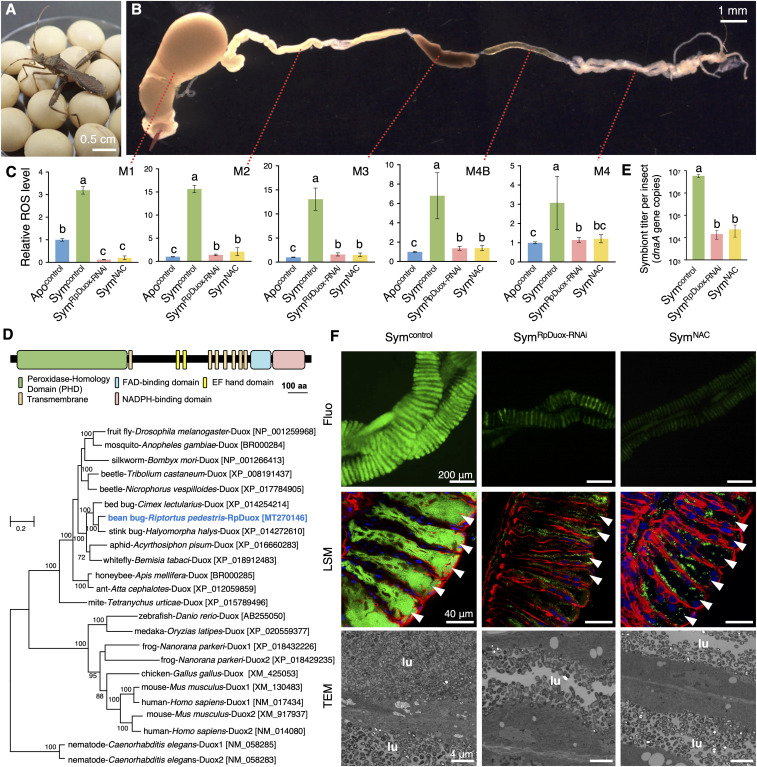
Fig. 1.
RpDuox-mediated ROS is important for gut symbiosis. (A) An adult male R. pedestris feeding on soybeans. (B) The dissected midgut of R. pedestris. (C) Relative ROS level in the different midgut regions of Apo (Apocontrol), Sym (Symcontrol), RpDuox-RNAi (SymRpDuox-RNAi), and NAC-fed (SymNAC) insects. All values were normalized by the ROS level of Apo insects. (D) The predicted domains of the RpDuox protein (Top). Maximum-likelihood phylogeny based on amino acid sequences (Bottom). Numbers in parentheses indicate accession numbers. Bootstrap values (>60%) are shown near each node. (E) The number of symbionts colonizing the M4. Data shown in C and E are mean ± SD of n = 9 and n = 6 insects, respectively. Different letters indicate statistically significant differences (P < 0.05). The statistical significance of differences between samples was analyzed by a Kruskal–Wallis test with Bonferroni correction. (F) Microscopy images of the M4: epifluorescence microscopy (first row), laser scanning confocal microscopy (second row; green, symbionts; blue, host nuclei; red, host cytoskeleton; white arrowheads, midgut crypts), and transmission electron microscopy (third row; lu, luminal region of the midgut crypt).
Given the cardinal role of ROS produced by Duox in insect gut immunity and the fact that the Burkholderia symbionts stay in direct contact with the crypt epithelial cells (34), we postulated in this study that, besides the AMPs, Duox and ROS are also involved in the management of the symbiosis in the midgut crypts of R. pedestris. However, our findings did not support an antimicrobial immune function of Duox, but revealed an unanticipated pivotal role for the enzyme in the formation of a tracheal network enveloping the symbiotic organ, which is in turn essential for supplying sufficient oxygen for respiration to the symbiotic organ and the aerobic gut symbiont.
Results
Gut Symbionts Induce the Generation of R. pedestris Duox-Dependent ROS in the R. pedestris Midgut.
To determine if the gut symbiont Burkholderia insecticola induces ROS in the symbiotic organ of R. pedestris, we measured the relative ROS level in the lysate of dissected gut compartments of apo-symbiotic and symbiotic insects (Apo and Sym insects, respectively) using the fluorescent dye 2′,7′-dichlorofluorescein diacetate. An approximately threefold higher level of ROS was detected in the symbiotic organ of Sym insects compared to that of Apo insects, confirming that the colonization of the crypts by the B. insecticola symbiont stimulates ROS production (Fig. 1C). Moreover, the ROS level in the digestive tract of the midgut, the M1, M2, and M3 anterior regions (Fig. 1B), equally increased after symbiont colonization, indicating that the colonization by B. insecticola induces systemic high ROS levels throughout the whole midgut regions even if the symbiont resides only in the posterior symbiotic midgut region M4 (Fig. 1 B and C). To demonstrate whether Duox generates the symbiont-mediated ROS, the homologous gene of R. pedestris (RpDuox) was down-regulated by RNAi (SI Appendix, Fig. S1A). The RpDuox transcript was identified based on homology with the Drosophila melanogaster protein in a R. pedestris transcriptome dataset generated from the infected symbiotic organ. We found one RpDuox copy in R. pedestris, displaying 74.7% homology, and the encoded protein had all the functional domains of typical Duox proteins from insects to humans (22) (Fig. 1D). RpDuox-RNAi reduced the ROS level in the symbiotic organ, as well as in the other midgut regions of Sym insects, to a similar level as Apo insects (Fig. 1C), demonstrating that induction of ROS by symbiont colonization is due to Duox activity. Similarly, the feeding of Sym insects with the antioxidant compound N-acetylcysteine (NAC) (35), added to the drinking water, also decreased the ROS levels of the midgut regions to similar concentrations as in Apo insects (Fig. 1C). Taken together, these results demonstrate that the infection of B. insecticola induces production of ROS in the entire midgut region via the RpDuox enzyme.
RpDuox-Produced ROS Is Important for the Gut Symbiosis but Not as a Microbicidal Molecule.
The production of infection-associated ROS is compatible with a mucosal immune response directed against the gut symbiont in the symbiotic organ. To test this hypothesis, we estimated the population size of symbionts in the M4 of Sym, RpDuox-RNAi (SymRpDuox-RNAi), and NAC-fed (SymNAC-fed) insects by qPCR. In Drosophila, silencing of Duox led to a reduced microbicidal ROS production and a concomitant increased total number of gut microbes (12). However, contrary to Drosophila, the titer of the Burkholderia symbiont in the M4 of R. pedestris was decreased more than 100 times in RpDuox-RNAi insects compared to Sym insects (Fig. 1E). Also, feeding of the antioxidant NAC to Sym insects reduced the number of gut symbionts to similarly low levels (Fig. 1E). Microscopy analysis confirmed that RpDuox-RNAi and NAC feeding caused the collapse of the symbiosis (Fig. 1F). Laser scanning microscopy and transmission electron microscopy (TEM) images showed that, in the symbiotic organ of RpDuox-RNAi and NAC-fed insects, gut symbionts were present close to the epithelial surface of the midgut crypts, but few symbionts were observed in the interior part of the crypt lumen (Fig. 1F). These results indicate that RpDuox plays a pivotal role in controlling gut symbiosis, but by a mechanism that is unrelated to a bactericidal action of ROS against the symbiont.
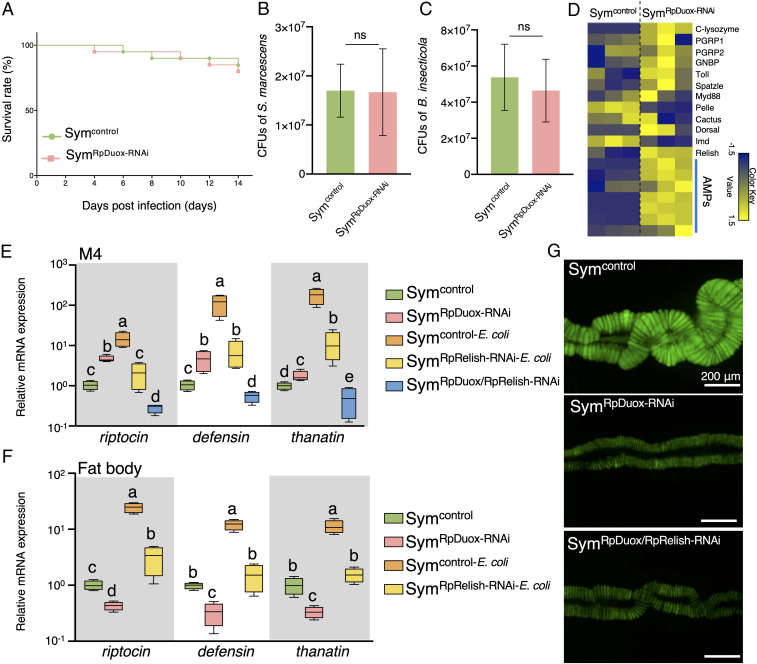
We next investigated the possible relation of RpDuox activity and bacterial infection and multiplication in the anterior digestive regions of the midgut. Although the ROS level was significantly decreased in M3 by RpDuox-RNAi (Fig. 1C), we found that the survival rate of R. pedestris after oral infection with the entomopathogen Serratia marcescens was not affected (Fig. 2A), suggesting that removing Duox-mediated ROS production did not render the insects more susceptible to the pathogen. Accordingly, the number of S. marcescens cells in the M3 region of the midgut was not significantly changed after RpDuox RNAi (Fig. 2B). Moreover, the number of B. insecticola cells that were transiently present in the M3 region after symbiont ingestion was also not altered in RpDuox-RNAi insects (Fig. 2C). Together, these results demonstrate that Duox-mediated ROS generation is not important to regulate the bacterial titer of pathogens or symbionts in the nonsymbiotic midgut regions.
Fig. 2.
RpDuox function is not related to gut mucosal immunity in the bean bug. (A) The survival rate of Sym (Symcontrol) and RpDuox-RNAi (SymRpDuox-RNAi) insects after oral infection with 107 cells per milliliter of S. marcescens (n = 20 insects). (B) Number of S. marcescens colonies from the M3 region of third-instar nymph Symcontrol and SymRpDuox-RNAi insects. (C) The number of Burkholderia symbionts in the M3 of the third-instar nymph Symcontrol and SymRpDuox-RNAi insects. CFU, colony-forming units. (D) Expression profile from RNA sequencing data of immunity-related genes in the M4 of Symcontrol and SymRpDuox-RNAi insects. Each column is a replicate experiment. (E) The expression level of AMPs in the M4 of Sym insects without immune stimulation (Symcontrol), Symcontrol with septic injury by Escherichia coli injection in the hemolymph (Symcontrol-E. coli), RpRelish-RNAi with E. coli injection (SymRpRelish-RNAi-E.coli), RpDuox-RNAi without bacterial injection (SymRpDuox-RNAi), and double RpDuox/RpRelish-RNAi insects without bacterial injection (SymRpDuox/RpRelish-RNAi). (F) The expression level of AMPs in the fat body. Data shown in B and C are from n = 5 insects, and those in E and F are from n = 4 insects. Different letters indicate statistically significant differences (P < 0.05). The statistical significance was analyzed by the Mann–Whitney U test (B and C) or Kruskal–Wallis test with Bonferroni correction (E and F). Error bars indicate SDs; ns, not statistically significant. (G) Effect of RpDuox/RpRelish double RNAi on symbiosis. Green color indicates the GFP signal from gut-colonizing Burkholderia symbionts.
Immune Stimulation by RpDuox-RNAi Is Not Responsible for Symbiosis Collapse.
In addition to its bactericidal activity, Duox-mediated ROS is also known to contribute to the construction of the midgut peritrophic membrane, as shown in A. gambiae (23). Knockdown of duox in A. gambiae leads to a decreased barrier function of the peritrophic matrix and enhanced immune stimulation and AMP expression by gut microbes coming in close contact with epithelial cells. However, in contrast with the majority of insects, most hemipteran insects, including R. pedestris, do not possess a peritrophic membrane in the midgut (36). Nevertheless, the gene-expression profile comparison between the symbiotic organs of Sym and RpDuox-RNAi insects showed that expression of immunity-related genes, especially genes encoding AMPs (37), was increased in the midgut of RpDuox-RNAi insects (Fig. 2 D and E), but not in the fat body (Fig. 2F). It should be noted, however, that this up-regulated AMP gene expression in the midgut by RpDuox-RNAi was significantly lower than in immune-stimulated insects by septic injury (Fig. 2 E and F). Because induced immunity in the symbiotic organ could antagonize the gut symbiont and thus, in principle, be at the origin of the symbiosis defect in RpDuox-RNAi insects, we aimed at knocking down the expression of the AMP genes in the bean bug. In the brown-winged green bug Plautia stali, a stinkbug related to R. pedestris in the infraorder Pentatomomorpha but belonging to a different family, the NF-κB transcription factor Relish of the Imd pathway is an essential regulator of AMP gene expression (38). RpRelish-RNAi in R. pedestris reduced the expression of all three tested AMP genes in the symbiotic organ as well as the fat body, indicating that, also in R. pedestris, Relish is a critical regulator of the expression of the AMP immune effectors (Fig. 2 E and F). After simultaneously silencing both RpRelish and RpDuox, the AMP expression was not up-regulated anymore in the symbiotic organ (Fig. 2E), showing that the induced AMP expression in the M4 by knockdown of RpDuox is dependent on the canonical Imd immune pathway. Importantly, the green fluorescent protein (GFP) signal of Burkholderia gut symbionts in double RpDuox/RpRelish-RNAi insects was not restored to control levels but remained similar to single RpDuox-RNAi insects (Fig. 2G and SI Appendix, Fig. S2). Therefore, we can conclude that the up-regulation of the immune response does not directly contribute to disrupting the symbiotic relationship in RpDuox-RNAi insects.
Symbiont Colonization Triggers Tracheal Development, and RpDuox-Mediated ROS Stabilizes the Respiratory Network by Forming a Dityrosine Network.
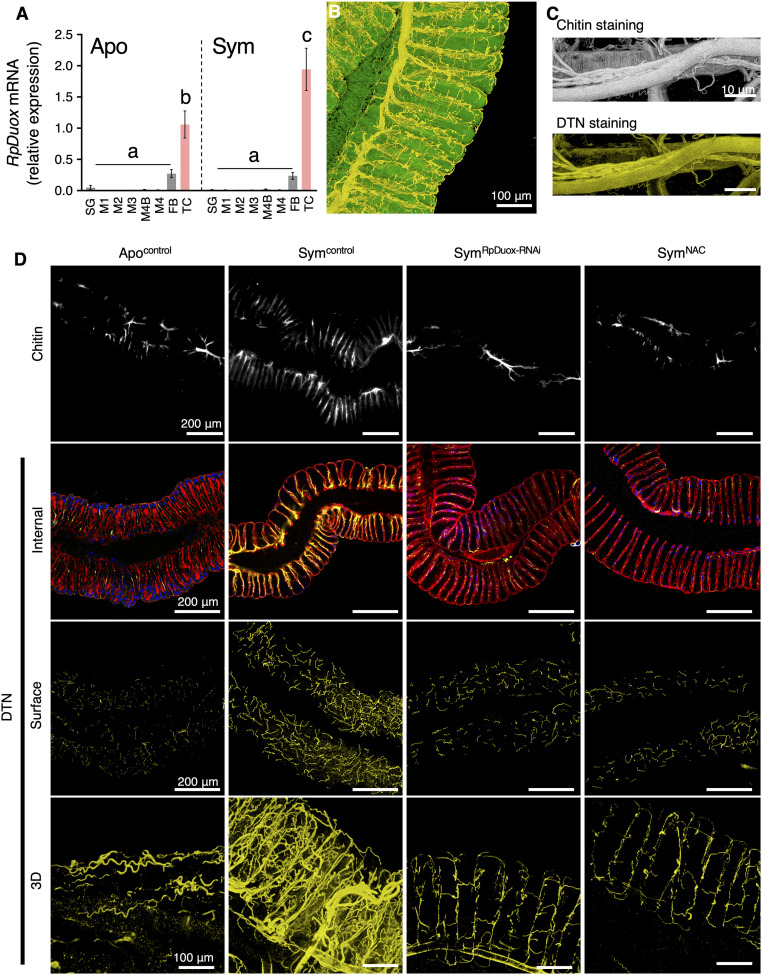
Thus, contrary to our initial expectation, the role of RpDuox and the ROS produced by it in the R. pedestris–Burkholderia symbiosis is not related to mucosal gut immunity. To find a clue leading to the specific function of RpDuox in the symbiosis, we measured its tissue-specific expression by RT-qPCR. Surprisingly, the expression of the RpDuox gene in Apo and Sym insects was incomparably high in the tracheae relative to all other tested tissues, suggesting that the RpDuox-mediated generation of ROS may have an important function in the respiratory system for maintaining gut symbiosis (Fig. 3A). Although the expression level of RpDuox in Sym insects was slightly higher than in Apo insects, the gene was also very strongly expressed in the tracheae of the latter insects, indicating that RpDuox is constitutively expressed in the tracheal network (Fig. 3A). The insect tracheal system consists of a tubular epithelial network containing a luminal chitin and protein matrix (23, 39–41). High RpDuox expression levels in the tracheae could be in agreement with the previously documented function of Duox-mediated ROS in shaping extracellular matrices by forming dityrosine cross-links between proteins and thereby creating a DTN (22). Immunostaining of a dissected midgut by a dityrosine-specific antibody indeed revealed the presence of a DTN enveloping the symbiotic organ of R. pedestris that likely corresponds to a respiratory network composed of tracheae and tracheoles (Fig. 3B). In addition to protein, chitin is another major constituent of the tracheal matrix (41). Chitin staining with calcofluor-white Fluorescent Brightener 28 revealed a tracheal network around the symbiotic organ that colocalized with the network revealed with the dityrosine-specific antibody (Fig. 3C), confirming that the DTN highlights tracheae and that dityrosines are abundant in them. Interestingly, both chitin and dityrosine staining revealed that the tracheal network enveloping the symbiotic organ and the tracheoles connecting inside the midgut crypt tissue developed much stronger in Sym insects compared to Apo insects (Fig. 3D). This morphological response suggests that colonization of the crypts by the large number of the Burkholderia gut symbiont is an oxygen-requiring process that induces the development of the respiratory network to supply enough oxygen to the symbiotic organ. Moreover, the tracheal network, especially the ramified tracheoles, did develop in response to the colonization of the gut symbiont Burkholderia, not only around the crypt-bearing M4 but also in the other midgut regions as well as the fat body (SI Appendix, Fig. S3 A and B), indicating that the activity of the symbiotic organ provokes a systemic oxygen stress in the whole insect body. This is furthermore in agreement with the ROS accumulation in all midgut regions (Fig. 1C). Accordingly, the higher ROS levels in the midgut of Sym insects compared to Apo insects resulted from the strongly developed attached tracheal network, since their careful removal from the symbiotic organ significantly reduced the detected ROS level (SI Appendix, Fig. S1B). Strikingly, the DTN as well as the chitin mesh in the symbiotic organ of Sym insects was strongly reduced by RpDuox-RNAi or feeding with the antioxidant NAC (Fig. 3D). Thus, RpDuox is indispensable for the establishment of the symbiosis-induced tracheal network in R. pedestris, and the increased ROS level in the midgut produced by RpDuox after symbiont infection is associated with the development of a tracheal network along the different midgut regions. The RNAi inactivation of RpDuox also abolished the formation of the enhanced tracheal network in the digestive, anterior midgut regions M1, M2, and M3 (SI Appendix, Fig. S3A) as well as the fat body (SI Appendix, Fig. S3B). These observations are in agreement with the suppression of RpDuox expression in the entire insect body, as can be expected from the double-stranded RNA (dsRNA) injection procedure to induce gene silencing.
Fig. 3.
Symbiont colonization triggers tracheal development, and RpDuox-mediated ROS stabilizes it by forming a DTN. (A) The relative expression of RpDuox in different tissues of Apo and Sym insects. Data shown are mean ± SD (n = 4 insects). Different letters indicate statistically significant differences (P < 0.05). The statistical significance of differences between samples was analyzed by a Kruskal–Wallis test with Bonferroni correction. (B) A 3D image of the tracheal network in the M4 of a Symcontrol insect. (C) Staining of the chitin and DTN of tracheae detached from the M4. (D) The tracheal network in the M4 of Apo (Apocontrol), Sym (Symcontrol), RpDuox-RNAi (SymRpDuox-RNAi), and NAC-fed (SymNAC) insects. The tracheal network, stained with Fluorescent Brightener 28 (calcofluor-white) to visualize chitin (first row), DTN in the internal section of the M4 (second row), DTN located on the surface of the M4 (third row), and 3D reconstitution of DTN of the tracheal network (fourth row). The colors in B–D are as follows: white, chitin; yellow, DTN; red, host cytoskeleton; blue, host nuclei; green, GFP signal derived from gut-colonizing symbionts.
Destabilization of the Respiratory Network by RpDuox-RNAi Causes Hypoxia in the Midgut.
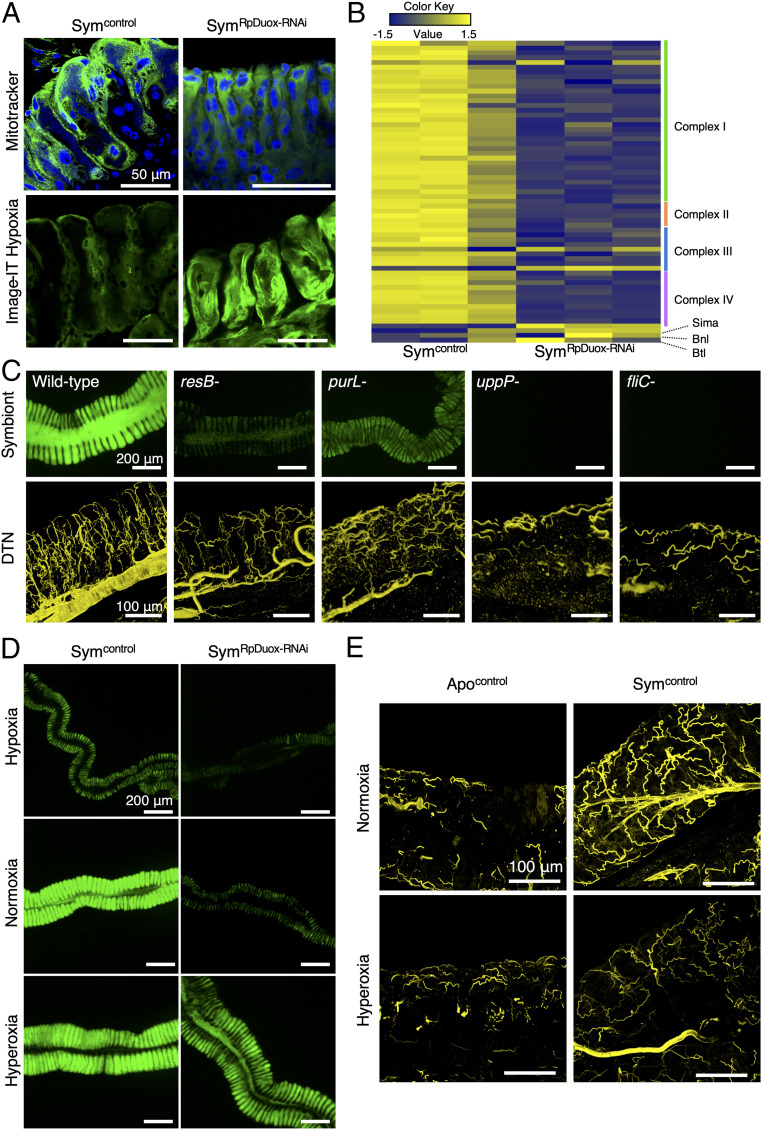
Based on these findings, we made the hypothesis that the collapse of the respiratory system by RpDuox-RNAi will cause a hypoxic environment in the symbiotic organ. We first measured in the crypt epithelial cells the activity of the mitochondria using MitoTracker dye. In Sym insects, a high number of active mitochondria was detected in the epithelial cells of the crypts of the symbiotic organ, which was drastically decreased after RpDuox-RNAi (Fig. 4A). Correspondingly, the expression of genes encoding the mitochondrial respiratory chain was down-regulated in RpDuox-RNAi insects (Fig. 4B), confirming that the symbiotic organ entered into a hypoxic condition after RpDuox-RNAi. Moreover, staining the symbiotic organ with Image-IT dye, which is a real-time hypoxia indicator, revealed a high fluorescence in the epithelia of the midgut crypts after RpDuox-RNAi compared with control Sym insects, further supporting that the disorganization of the respiratory network by RpDuox silencing causes hypoxia of the symbiotic organ (Fig. 4A). Of note, our finding that AMP-encoding genes are up-regulated in the hypoxic M4 of RpDuox-RNAi insects in a Relish-dependent manner (Fig. 2 D and E) mirrors previous findings in Drosophila, demonstrating that a hypoxic environment activates the expression of AMPs via the Toll and IMD pathways (42, 43).
Fig. 4.
Destabilization of the respiratory network by RpDuox-RNAi causes hypoxia in the midgut. (A) Active mitochondria in Sym (Symcontrol) and RpDuox-RNAi (SymRpDuox-RNAi) insects stained by MitoTracker, a membrane potential fluorescence dye, which stains active mitochondria in live cells. Green, MitoTracker; blue, host nuclei (Top). Hypoxia in the M4 revealed by staining with Image-IT hypoxia fluorescence dye, a real-time oxygen detector (green; Bottom). (B) Heat map of relative expression levels of genes related to the host respiratory chain, Sima, Branchless (bnl), and Breathless (btl), in Symcontrol and SymRpDuox-RNAi insects. Each column is a replicate experiment. (C) Colonization level of the wild type, resB-, purL-, uppP-, and fliC-deficient B. insecticola mutants in the symbiotic organ (Top). The DTN of the tracheal network from the symbiotic organs infected with wild type and Burkholderia mutants (Lower). Yellow color indicates the DTN signal of the tracheal network. (D) Observation by epifluorescence microscopy of the midgut of Symcontrol and SymRpDuox-RNAi insects reared in hypoxia (6 to 12% O2), normoxia (21% O2), and hyperoxia (39 to 42% O2) environments. Green is the GFP signal derived from Burkholderia gut symbionts. (E) Tracheal network in the M4 of Apocontrol and Symcontrol insects reared under normoxia and hyperoxia conditions.
Hypoxia could reduce the metabolic activity (43) of the crypt epithelial cells and their capacity to feed the bacterial symbiont, thereby provoking the collapse of the symbiosis. Some insect genes related to glycolysis and most genes of the tricarboxylic acid cycle were down-regulated in the M4 after RpDuox-RNAi, indicating that the metabolic activity of the crypt epithelial cells was indeed lower in RpDuox-RNAi insects than in Sym insects (SI Appendix, Fig. S4A). Furthermore, hypoxia is expected to also directly affect the proliferation of the Burkholderia symbiont. Species of the genus Burkholderia are obligate aerobic bacteria, although they can tolerate microaerobic and anaerobic conditions (44–46). Accordingly, the Burkholderia symbiont grew normally in normoxia (21% O2) but did not grow well in hypoxia (6 to 12% O2) and anoxia (<0.1% O2; SI Appendix, Fig. S4B). However, the bacteria incubated first in hypoxia or anoxia grew again when transferred to normoxia and cultured for an additional 48 h (SI Appendix, Fig. S4B), suggesting that B. insecticola needs enough oxygen for proliferation in vitro as well as in the midgut crypts. Furthermore, a B. insecticola mutant in the resB gene, essential for cytochrome c maturation and respiration, cannot properly colonize the M4 crypts (Fig. 4C). A reduced flux of oxygen and nutrients into the lumen of the crypts of RpDuox-RNAi insects could create a rapid depletion of these molecules, resulting in a gradient from the surface of the crypts toward their interior. Such an oxygen and/or nutrient gradient could explain the presence of a relatively high number of symbionts close to the epithelial surface of the midgut crypts and a lack of them in the interior part of the crypt lumen, as observed by TEM (Fig. 1F). Thus, the symbiosis defect in the RpDuox-RNAi insects is likely due to a lack of sufficient oxygen in the symbiotic organ that can sustain the respiratory activity of the symbiont.
Conversely, the high bacterial respiration in the M4 is the probable trigger of the extensive tracheal network in Sym insects. B. insecticola mutants in fliC, encoding the flagellin protein, and uppP, involved in cell wall synthesis, cannot colonize the M4 (7, 47), while a mutant in purL, involved in purine biosynthesis, can only weakly colonize the crypts (48) (Fig. 4C). The formation of tracheae was not, or less, stimulated in insects infected with these mutants (Fig. 4C), confirming that the mere presence of Burkholderia in the midgut is not sufficient and that bacterial proliferation in the M4 crypts is required to trigger tracheal development.
Increased Oxygen Supply Compensates for the Lack of Tracheal Development and Restores Symbiosis in RpDuox-RNAi Insects.
Next, we confirmed the crucial role of oxygen in gut symbiosis by rearing Sym and RpDuox-RNAi insects in hypoxia (6 to 12% O2), normoxia (21% O2), and hyperoxia (39 to 42% O2) conditions (SI Appendix, Fig. S4C). The GFP signal of gut symbionts was diminished in Sym insects reared in a low-oxygen environment (Fig. 4D). In agreement with the microscopy observation, qPCR confirmed the decreased number of Burkholderia in the M4 crypts (SI Appendix, Fig. S4D). In contrast, when Sym insects were reared in hyperoxia, the number of gut-colonizing symbionts was not significantly increased, suggesting that the oxygen supply to maintain symbiosis is sufficient in normoxia (Fig. 4D and SI Appendix, Fig. S4D).
Contrary to Sym insects reared in normoxia, those reared in hyperoxia did not develop much of an enhanced tracheal network in the symbiotic organ compared to Apo insects (Fig. 4E). These results indicate that the available oxygen in the hyperoxia condition is sufficient to support symbiosis without the need for more tracheae and that the high oxygen consumption by symbiont colonization in the midgut is the trigger of the enhanced tracheal development in Sym insects in normoxia.
As described above, RpDuox-RNAi insects harbor only few symbiotic bacteria under normoxia, and, when they were reared in hypoxia, their symbiont titer was decreased even further (Fig. 4D and SI Appendix, Fig. S4D). However, the M4 crypt population of gut symbionts in RpDuox-RNAi insects considerably recovered in a hyperoxia environment (Fig. 4D and SI Appendix, Fig. S4D). These results demonstrate that the availability of sufficient oxygen in the symbiotic organ is critical for a successful symbiosis and that a deficient oxygen supply is the major factor underpinning the symbiotic defect of RpDuox-RNAi insects.
Silencing of Regulators of Tracheal Development Simulates the Collapse of Symbiosis.
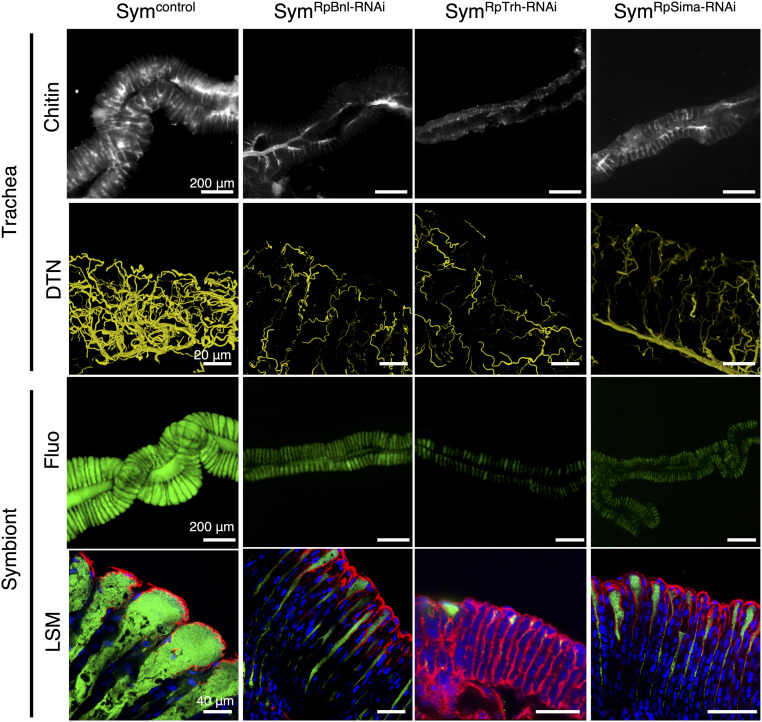
To confirm the importance of oxygen distribution by the respiratory network in gut symbiosis, we silenced branchless (RpBnl) and trachealess (RpTrh), two known genes important for the tracheal development in Drosophila (49–51), as well as RpSima, encoding the hypoxia-inducible factor (HIF)-1α, which is a central regulator of the transcriptional response to hypoxia (52) and controls the expression of Bnl and breathless (Btl), another tracheal regulator (53). Thus, Sima, Trh, Bnl, and Btl constitute a regulatory module that controls tracheal development under low oxygen. The RpTrh-RNAi insects showed a high mortality (SI Appendix, Fig. S5B), but the alive insects and those silenced for the other two genes had an underdeveloped respiratory network in the M4 (Fig. 5), in agreement with the expected role of RpBnl and RpTrh and further showing that oxygen-depletion signaling via Sima controls the tracheal development in the symbiotic organ. Crucially, the defect in the tracheal development in these RNAi insects is accompanied by a failure to establish the symbiosis with a full infection of the crypts, as revealed by the strongly decreased GFP signal derived from gut symbionts in the symbiotic organ (Fig. 5 and SI Appendix, Fig. S5A). Thus, the down-regulation of these three regulators of the respiratory system affected symbiosis in a very similar way as RpDuox-RNAi. The enhanced expression of RpSima, RpBnl, and RpBtl in the crypts of the RpDuox-RNAi insects (Fig. 4B) is in agreement with Sima still being active after RpDuox RNAi and thus with RpDuox acting downstream of the Sima/Btl/Trh/Bnl regulatory module.
Fig. 5.
Oxygen supply to the symbiotic organ via the tracheal network is important for gut symbiosis. Chitin staining (first row), 3D reconstruction of the DTN (second row), colonization by Burkholderia (third row), and crypt morphology (fourth row) in the M4 of Sym (Symcontrol), RpBnl-RNAi (SymRpBnl-RNAi), RpTrh-RNAi (SymRpTrh-RNAi-), and RpSima-RNAi (SymRpSima-RNAi) insects. Images were obtained by epifluorescence microscopy in the first and third rows and by laser scanning confocal microscopy in the second and fourth rows. White, chitin; yellow, DTN; green, gut symbiont; blue, host nuclei; red, host cytoskeleton.
Discussion
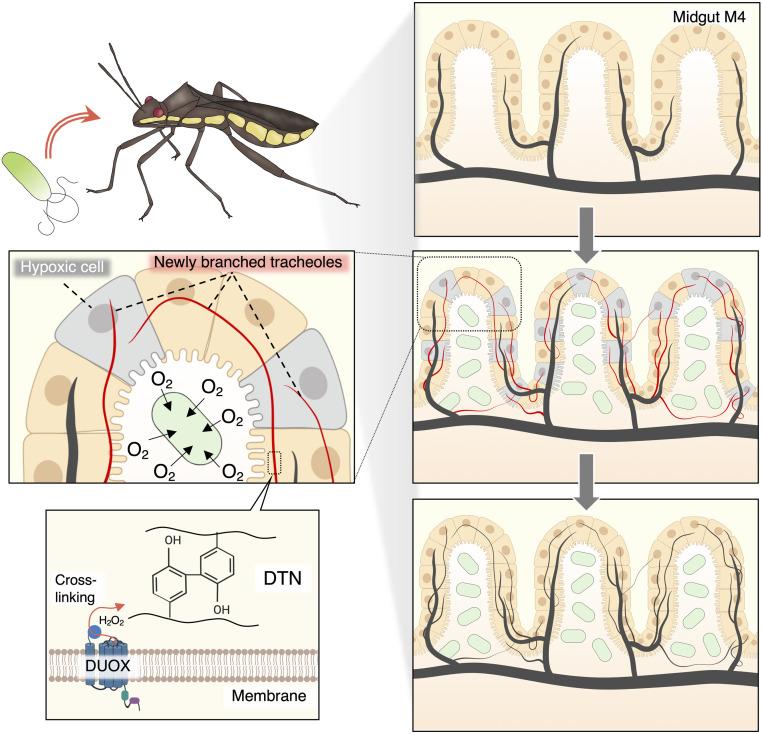
Our results demonstrate that the large population of the Burkholderia symbiont present in the body of the bean bug stimulates the sprouting of tracheal branches toward the symbiont-infected M4 crypts. We highlight a function of the Duox enzyme in this process, catalyzing the formation of a newly discovered tracheal DTN, which is essential for the stabilization of the respiratory network (Fig. 6). The tracheal system consists of a tubular epithelial network containing an extracellular luminal chitin and protein matrix (39, 40). Chitin is an essential component of this matrix, providing rigidity to the tracheal tubes, and mutants in chitin synthesis genes form misshapen tracheal tubes in Drosophila (41, 54). The matrix also contains proteins, including the zona pellucida domain proteins Piopio (Pio) and Dumpy (Dp), chitin-binding and remodeling proteins, and other unidentified protein components. They are thought to provide, together with the chitin scaffold, a structural network in the luminal space, regulating tube length, width, branching, and integrity (55–57). The requirement for Duox in the formation of new tracheae indicates that dityrosine cross-linking these matrix proteins is crucial for their structural function. Similar as in R. pedestris, we found that the expression of Duox is much higher in the tracheae than in the midgut in the phylogenetically diverse insects Gryllus bimaculatus (Orthoptera), Bombyx mori (Lepidoptera), Tribolium castaneum (Coleoptera), and D. melanogaster (Diptera; SI Appendix, Fig. S6). This conserved expression pattern suggests that the role of Duox in tracheae is common in the insects.
Fig. 6.
RpDuox mediates symbiosis and tracheal network stability. Tracheal network in the M4 before symbiont infection (Top Right). Development of the tracheal network is stimulated to supply oxygen to the M4 when colonized by the symbiont, which consumes high levels of oxygen (Middle Left and Right; red, newly ramified tracheoles; green, gut symbiont; gray, epithelial cells experiencing reduced oxygen levels and attracting growing tracheoles). The branched tracheoles are stabilized by RpDuox (Bottom Right). RpDuox stabilizes the DTN of the tracheal cuticle layer, which is critical for structurally maintaining trachea (Bottom Left). The developmental order is in the direction of the arrows.
The pivotal role of Duox-mediated ROS in the formation of the tracheal cuticle proposed here adds to a number of other structures composed of extracellular matrices requiring Duox for DTN formation. These structures include the earlier-mentioned peritrophic matrix of the gut in the mosquito A. gambiae as well as in the tick I. scapularis (23, 24), the eggshell of the kissing bug (58), the cuticle structure of wings in Drosophila (59, 60), the collagenous extracellular cuticle matrix that forms the outer covering of nematodes (61), and the hardened extracellular matrix of sea urchin eggs after fertilization (62). Therefore, the importance of Duox in forming DTNs as part of the biogenesis of extracellular cuticle structures is conserved and not limited to insects. This function could be the ancestral and primary function of Duox enzymes.
The formation of dityrosine cross-links in cuticle requires a NADPH oxidase to generate H2O2 and a heme peroxidase to catalyze the formation of a covalent bond between two neighboring tyrosine residues via tyrosyl radical formation and the reduction of H2O2. Potentially, both these functions are provided by Duox via its C-terminal NADPH oxidase domain and N-terminal peroxidase homology domain, respectively. In Caenorhabditis elegans, Duox is a self-contained catalyst that mediates tyrosine cross-linking independently (63). However, in mosquito, fly, and ticks, the peroxidase domain is only partially functional in DTN formation, and an additional secreted heme peroxidase protein is required for the formation of the dityrosine cross-links (23, 24, 59). The weak peroxidase activity of Duox in these organisms is believed to result from a mutation in a glutamic acid residue in the N-terminal peroxidase domain involved in heme binding (24). In the case of the R. pedestris RpDuox protein, the same point mutation is present, suggesting that the functionality of RpDuox may be similar to the tick, fly, and mosquito Duox enzymes. Therefore, we predict that the RpDuox enzyme works in concert with an additional, unidentified heme peroxidase to form the dityrosine bridges in the matrix proteins.
Often, the production of bactericidal ROS is presented as the principal function of Duox in insect physiology (14, 15, 22). In Drosophila, knockdown of Duox or Mesh, a positive regulator of Duox expression, leads to an increase of the gut microbiome load or to the persistence in the gut after oral infection of the entomopathogenic bacterium Erwinia carotovora ssp. carotovora strain Ecc15 (20, 64). A similar role in direct killing of gut microbes was proposed in C. elegans (19). In striking contrast, we did not observe, after Duox-RNAi in the bean bug, an increase of the pathogen S. marcescens in the M3 digestive region of the midgut or of the symbiont in M3 or M4 crypts (Figs. 1E and 2 B and C). On the contrary, we noticed a strongly reduced symbiotic population in M4 (Fig. 1 E and F). In the mosquito A. gambiae and the tick I. scapularis, silencing Duox or the gene encoding the Duox partner peroxidase decreased the microbial load in the midgut rather than increasing it (23, 24). Together, these results indicate that the direct bactericidal activity of Duox as proposed originally in Drosophila is not universal in the insects and other arthropods. In light of this conclusion, we propose that the exclusive emphasis on the role of Duox in the production of antimicrobial ROS in the gut of Drosophila (14, 15, 22) should be revisited. It is likely that Drosophila Duox is also involved in the synthesis of the peritrophic matrix in the midgut and contributes through this activity as well to gut immunity. In agreement with this hypothesis, a mutation of the Drosophila dcy gene, encoding the drosocrystallin chitin-binding protein, results in a thinner but not completely abolished peritrophic matrix, which renders the flies more sensitive to gut infections with entomopathogenic bacteria (65, 66). This phenotype is reminiscent of the Duox RNAi flies, although the latter were more severely affected in immune response (12). Transglutaminase is another enzyme that affects protein–protein cross-linking and permeability in the peritrophic matrix of Drosophila, as well as the tolerance of the flies to the gut microbiota (67). Thus, Duox could have a dual contribution to the gut immune homeostasis in the case of Drosophila. On the contrary, the load of aerobic bacteria in the Drosophila midgut is much lower than in the R. pedestris symbiotic organ (68). Therefore, Duox-mediated development of tracheae enveloping the midgut could possibly have a lesser importance for sustaining the microbiota or permitting pathogen development in Drosophila.
Nutritional cues via insulin-like and neuropeptide signaling give rise to changes in the tracheal architecture in the Drosophila midgut (69). Conversely, trachea-derived morphogens control midgut development by stimulating intestinal stem cells (70). Therefore, we can speculate that bacterial or host signals warning for enhanced nutritional requirements, are the triggers for the formation of the respiratory network enveloping the crypts, while, simultaneously, the growing tracheae produce morphogens that allow the swelling and opening of the crypts needed for bacterial colonization. On the other hand, hypoxia is the major known regulator of tracheal plasticity in Drosophila (53, 71). Therefore, low oxygen levels created by the metabolic activity of the proliferating symbionts and the crypt epithelial cells that nurture the symbiont could be the direct cause of tracheae genesis (Fig. 6). In agreement with the view of the symbiotic organ as an oxygen sink, we find no symbiosis-stimulated development of tracheae in insects reared in high oxygen levels (Fig. 4E) or when the insects were infected with symbiont mutants that have weak or no crypt colonization capacity (Fig. 4C). Moreover, Sima, Trh, and Bnl are required for tracheae development in a similar way as Duox. The bnl gene in Drosophila encodes a fibroblast growth factor homolog whose expression is highly increased in oxygen-starved nontracheal cells. Bnl exerts chemoattraction of tracheal cells via binding to the trachea-specific receptor Btl that controls the ramification of tracheae toward the Bnl-producing cells (49, 71). The transcription factor Trh is a tracheal cell identity regulator, controlling trachea-specific gene expression, including btl (50, 51). The HIF-1α homolog Sima in Drosophila is a central regulator of the transcriptional response to hypoxia. The Sima transcription factor is actively degraded in normoxia, dependent on the oxygen sensor Fatiga, but is stable and active in hypoxia (52, 72). Active Sima in hypoxia up-regulates the expression of btl in tracheal cells and bnl in nontracheal cells, thereby stimulating the development of the respiratory network (53). Thus, in R. pedestris, oxygen depletion in the symbiotic organ triggered by symbiont colonization is potentially enough as a signal to stabilize Sima in both the crypt epithelia and the local tracheal cells. Sima subsequently activates the transcription of, respectively, RpBnl and RpBtl, the latter also necessitating the tracheae-specific transcription factor Trh. This signaling cascade controls the expression of the genes required for the development of the tracheal network around the crypts, which most likely also includes RpDuox. Thus, RpDuox gene expression and protein activity, being an inherent part of tracheal development, is indirectly induced by the establishment of the symbiosis via the enhanced trachea formation.
In a remarkable analogous process to the tracheal development in the colonized midgut of R. pedestris, colonization of the mammalian small intestine by microbiota promotes angiogenesis and the development of the blood vessel network around the gut epithelia. This enhanced vascularization increases oxygenation of the villi, which have a modified morphology after colonization and an increased nutrient absorption activity (73–75). Thus, from insects to mammals, colonization of the gut by symbionts can create a local oxygen sink, triggering an analogous postembryonic developmental response.
Besides the midgut crypts of R. pedestris and related stinkbugs, other types of symbiotic organs in other insects, such as bacteriomes (9, 76), are equally enveloped by a tracheal network, presumably to provide oxygen to the respiring symbiotic bacteria (77, 78). These symbioses might therefore depend on Duox activity as well. Feeding Drosophila and C. elegans with different types of antioxidant chemicals inhibits Duox-produced ROS (19, 64). Here, we show that feeding R. pedestris with the antioxidant NAC is sufficient to prevent the accumulation of ROS produced by Duox and the sprouting of the tracheae, and this blocks the establishment of symbiosis. Since many insects obligatorily depend on their symbioses, triggering their collapse by the specific inhibition of the respiratory network with antioxidants could be a new route to fight insect pests.
Materials and Methods
Insect Rearing and Infection with Symbiont.
The R. pedestris strain used in this study was originally collected from a soybean field in Tsukuba, Ibaraki, Japan, and maintained in the laboratory for over 10 y. The bean bugs were reared in Petri dishes (90 mm in diameter and 20 mm high) under a long-day regimen (16 h light and 8 h dark) at 25 °C and fed with soybean seeds and distilled water containing 0.05% ascorbic acid (DWA). To infect insects with symbionts, B. insecticola (RPE75), GFP-expressing B. insecticola (RPE225), or mutant Burkholderia strains (SI Appendix, Table S1) were orally administered to second-instar nymphs by supplying the bacteria at a density of 107 cells per milliliter to the DWA. To feed NAC, DWA with 10 mg/mL of NAC was supplied to the bean bugs. T. castaneum was a gift of Ryo Futahashi (National Institute of Advanced Industrial Science and Technology Tsukuba Center, Tsukuba, Japan). G. bimaculatus and B. mori were purchased from a pet shop in Sapporo, Japan.
Detection of ROS Levels.
Fourth-instar nymphs of bean bugs were dissected in phosphate-buffered saline (PBS), pH 7.4, using fine forceps under a dissection microscope (S8APO and MZ FZ III; Leica), and each part of their midguts was transferred to 1.5-mL microcentrifuge tubes and homogenized by a pestle. The homogenized midgut samples were centrifuged at 15,000 rpm for 1 min to remove tissue debris, and the supernatant was transferred to the 96-well black polystyrene microplate and incubated with 50 μM 2′,7′-dichlorofluorescein diacetate (Thermo Fisher Scientific) for 20 min in the dark. The ROS level in each sample was measured by a microplate reader (Infinite F200 PRO; Tecan) using an excitation wavelength of 488 nm and an emission wavelength of 535 nm.
Synthesis of Double-Stranded RNA and RNAi.
Gene sequences of Duox, RpRelish, RpTrh, RpBnl, and RpSima were identified by an RNA-sequencing analysis of symbiotic insects and BLAST identification using the Drosophila proteins as search term (accession nos. MT270146–MT270150). Fragments of target genes for RNAi knockdown were obtained from R. pedestris cDNA. The fifth-instar nymphs of bean bugs were dissected in PBS, and total RNA was extracted from the M4 using RNAiso Plus (Takara Bio) and the RNeasy Mini Kit (Qiagen). The cDNA was synthesized from 1 μg of extracted total RNA by PrimeScript RT Reagent Kit with gDNA Erase (Takara Bio).
To synthesize dsRNA for RNAi, ∼500-bp fragments of the target gene were amplified from cDNA, with primers listed in SI Appendix, Table S2. The PCR products were cloned in the pT7Blue T-vector and transformed into Escherichia coli DH5ɑ cells. Clones were verified by sequencing. Next, the target gene was amplified by PCR from these plasmids using specific upstream and downstream primers, each extended with the T7 promotor sequences (SI Appendix, Table S2), to create a template for RNA polymerization. The dsRNA was synthesized from these PCR products by the MEGAscript RNAi kit (Thermo Fisher Scientific).
To silence a target gene, 100 ng of the corresponding dsRNA was injected into the hemolymph between the torso and hind leg of the symbiont-infected second-instar nymph of bean bugs after 2 d of symbiont infection using a glass capillary and a FemtoJet microinjector (Eppendorf). For triggering a septic injury, 107 E. coli DH5ɑ cells were similarly injected in the hemolymph. The phenotypic alteration in the insects was observed 1 wk after RNAi. The detailed timeline of the experiment is described in SI Appendix, Fig. S7.
Estimation of Infection Levels of Symbiotic Organs by qPCR.
To measure the number of symbionts present in the M4, DNA was extracted from the dissected M4 by the QIAamp DNA Mini Kit (Qiagen), and real-time qPCR was performed using primers BSdnaA-F and BSdnaA-R (SI Appendix, Table S2), which target the dnaA gene of the Burkholderia symbiont, the KAPA SYBR FAST qPCR Master Mix Kit (KAPA Biosystems), and the Roche LightCycler 96 System (Roche). The number of gut symbionts was calculated based on a standard curve for the dnaA gene containing 10, 102, 103, 104, 105, 106, and 107 copies per reaction of the target PCR product.
Gene Expression Determination by RT-qPCR.
To measure the mRNA expression level of the target genes, total RNA was extracted from the salivary gland, the midgut regions, the fat body, or the tracheae of the Apo and Sym bean bugs and reverse-transcribed to cDNA as described above. The real-time qPCR for target genes was performed as described above using primers listed in SI Appendix, Table S2. The efficiency of the RNAi process on target genes and change of gene-expression patterns of immune genes was measured 3 d after RNAi (SI Appendix, Fig. S7). The mRNA levels were normalized by the elongation factor 1-alpha (EF1α) housekeeping gene.
Microscopy Analyses.
To analyze the infection level of control, RNAi, or NAC-treated insects, the bean bugs were infected with GFP-expressing Burkholderia symbionts and injected with dsRNA or fed with NAC (SI Appendix, Fig. S7). After 1 wk, the M4 organs of fourth-instar bean bugs were dissected in PBS, transferred to a glass-bottom dish (Matsunami), and covered with a cover glass. The GFP signal derived from the M4-colonizing gut symbionts was observed with an epifluorescence microscope (DMI4000B; Leica).
Alternatively, to observe the colonization of the M4 by the gut symbiont with confocal microscopy, the bean bugs harboring GFP-expressing symbionts were dissected in PBS and the midgut was transferred to a 2-mL microcentrifuge tube. The gut samples were fixed with 4% paraformaldehyde (pH 7.4) for 10 min at 25 °C. The fixed samples were washed three times by PBS and permeabilized by PBS with 0.1% Triton X-100 (PBST) for 10 min at 25 °C. After permeabilization, the gut samples were washed again three times by PBS and stained with 4′6-diamidino-2-phenylindole (DAPI) and Alexa Fluor 647 Phalloidin for 30 min at 25 °C to stain, respectively, the nucleus and cytoskeleton of host cells. Then, the samples were transferred to a glass-bottom dish, mounted with ProLong Gold Antifade Mountant (Thermo Fisher Scientific), and observed under a laser scanning confocal microscope (TCS SP8; Leica).
Dityrosine networks were revealed by immunostaining. The whole gut of the fourth instar of control or RNAi bean bugs was dissected in PBS and fixed with 4% paraformaldehyde. The fixed midgut was permeabilized and incubated with blocking buffer (1% BSA in PBST) for 30 min at 25 °C. Then, the gut was stained with 1:400 dilution of anti-dityrosine monoclonal antibody (MyBioScience) overnight at 4 °C. Subsequently, the midgut was washed three times by PBS, each for 10 min, and then stained with 1:500 dilution of goat anti-mouse immunoglobulin G heavy and light chains conjugated with Alexa Fluor 555 (Abcam) for 1 h at 25 °C in the dark. The midgut was washed three times by PBS, each for 10 min, and then incubated with DAPI and phalloidin. The midgut was transferred to a glass-bottom dish, mounted, and observed with a laser scanning confocal microscope. The three-dimensional (3D) image of the DTN was reconstructed from z-stacked images using Leica LAS X software.
To observe the tracheal network by chitin fluorescent labeling, dissected midguts were stained with the Fluorescent Brightener 28 (Sigma-Aldrich) at a final concentration of 0.1 mg/mL for 5 min in the dark. Then, the midgut was washed three times by PBS, transferred to a glass-bottom dish, and observed with a confocal or epifluorescence microscope.
To analyze the hypoxic condition of the midgut, dissected M4 was stained with 10 μM Image-iT Green Hypoxia Reagent (Invitrogen) for 30 min at 25 °C and observed with a confocal microscope. To label active mitochondria, dissected midguts were incubated with 100 nM MitoTracker Green FM (Invitrogen) for 30 min at 25 °C and observed with a confocal microscope.
For TEM observations, the bean bugs were dissected with fine forceps in fixative solution (0.1 M sodium phosphate buffer containing 2.5% glutaraldehyde, pH 7.4). The isolated M4 was prefixed in the fixative solution at 4 °C overnight and postfixed in 2% osmium tetroxide at 4 °C for 1 h. After a series of dehydration steps by ethanol, the gut was embedded in Epon812 resin (TAAB). Ultrathin sections were made by using an ultramicrotome (EM UC7; Leica), mounted on a copper mesh, stained with uranyl acetate and lead citrate, and observed under a transmission electron microscope (H-7600; Hitachi).
Colony-Forming Unit Assay.
Cultured S. marcescens (107 cells per milliliter) were orally administrated to the third instar of Sym and RpDuox-RNAi bean bugs. The midgut M3 regions were dissected 3 d after infection and homogenized with a pestle in 100 µL of PBS. After the serial dilution of these M3 lysates, 10 µL of the dilutions was spotted on a Luria broth agar plate with 30 µg/mL of rifampicin, and plates were incubated at 30 °C for 1 d to obtain bacterial growth. To measure the symbiont titer in the M3 of Sym and RpDuox-RNAi bean bugs, dsRNA of RpDuox was injected to second-instar nymphs of Apo insects, and then newly molted third-instar nymphs were infected with cultured Burkholderia symbionts (107 cells per milliliter) and the M3 was dissected 24 h after infection. The dissected M3 was homogenized, and dilution series were spotted on a yeast glucose (YG) agar plate with 30 µg/mL of rifampicin, which was incubated for 2 d at 30 °C.
Bacterial Growth and Insect Rearing in Hypoxia and Hyperoxia Conditions.
To culture the Burkholderia symbiont in anaerobic or microaerophilic conditions, agar plates were placed in a 2.5-L W-zip standing pouch jar (Mitsubishi Gas Chemical) containing, respectively, an Anaero Pack-Anaero (O2 <0.1%) or an Anaero Pack-MicroAero (6% < O2 < 12%; Mitsubishi Gas Chemical). Plates were incubated for 48 h at 30 °C and subsequently for a further 48 h at 30 °C in normoxia condition.
To rear insects in a hypoxia environment, Petri dishes containing second-instar nymphs were placed in a 2.5-L rectangular jar containing an Anaero Pack-MicroAero. To create the hyperoxia condition, Petri dishes containing second-instar nymphs were placed in a culture jar, and oxygen was injected daily via a valve in order to maintain 39 to 42% oxygen as shown in SI Appendix, Fig. S4C. The oxygen content (as a percentage) in the jar was monitored by an oxygen indicator (OXY-M; JICKO). Additional methods are described in SI Appendix, Supplementary Text.
Supplementary Material
Acknowledgments
We thank H. Ooi (National Institute of Advanced Industrial Science and Technology [AIST]) for insect rearing; X.-Y. Meng and M. Kanno (AIST) for help with electron microscopy; and Y. Matsuura (The University of the Ryukyus), K. Takeshita (Akita Prefecture University), and S. Kanie (AIST) for critical comments on the manuscript. This study was supported by Japan Society for the Promotion of Science (JSPS) Research Fellowships for Young Scientists to S.J. (201911493) and T.O. (20170267 and 19J01106); by Ministry of Education, Culture, Sports, Science and Technology KAKENHI to Y.K. (18KK0211 and 20H03303); and by a JSPS-CNRS Bilateral Open Partnership Joint Research Project and the CNRS International Research Project “Ménage à Trois” to Y.K. and P.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020922118/-/DCSupplemental.
Data Availability
RNA-seq data of the M4 of Symcontrol and SymRpDuox-RNAi; gene sequences of RpDuox, RpBnl, RpTrh, RpSima, and RpRelish; and sequences of Duox of G. bimaculatus, B. mori, and T. castaneum data have been deposited in the National Center for Biotechnology Information (BioProject PRJDB9456) GenBank database (DRA accession numbers DRR215696-DRR215701; MT270146-MT270150; MT270151, NW_004582026, and NC_007418).
References
- 1.Moran N. A., Ochman H., Hammer T. J., Evolutionary and ecological consequences of gut microbial communities. Annu. Rev. Ecol. Evol. Syst. 50, 451–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley R. E., et al., Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douglas A. E., Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh H., Tago K., Hayatsu M., Kikuchi Y., Detoxifying symbiosis: Microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 35, 434–454 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Vogel H., et al., The digestive and defensive basis of carcass utilization by the burying beetle and its microbiota. Nat. Commun. 8, 15186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillon R. J., Dillon V. M., The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 49, 71–92 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Ohbayashi T., et al., Insect’s intestinal organ for symbiont sorting. Proc. Natl. Acad. Sci. U.S.A. 112, E5179–E5188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanan M. C., Rodrigues P. A. P., Agellon A., Jansma P., Wheeler D. E., A bacterial filter protects and structures the gut microbiome of an insect. ISME J. 10, 1866–1876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchner P., Endosymbiosis of Animals with Plant Microorganisms (Interscience Publishers, New York, 1965). [Google Scholar]
- 10.Jang S., Kikuchi Y., Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 41, 33–39 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Yun J. H., et al., Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ha E. M., Oh C. T., Bae Y. S., Lee W. J., A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Lee K. A., et al., Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Buchon N., Silverman N., Cherry S., Immunity in Drosophila melanogaster–From microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14, 796–810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuraishi T., Hori A., Kurata S., Host-microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 4, 375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha E. M., et al., Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat. Immunol. 10, 949–957 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Ha E. M., et al., Regulation of DUOX by the Galphaq-phospholipase Cbeta-Ca2+ pathway in Drosophila gut immunity. Dev. Cell 16, 386–397 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Bae Y. S., Choi M. K., Lee W. J., Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 31, 278–287 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Chávez V., Mohri-Shiomi A., Garsin D. A., Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect. Immun. 77, 4983–4989 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao X., et al., A Mesh-Duox pathway regulates homeostasis in the insect gut. Nat. Microbiol. 2, 17020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu J. H., et al., An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 25, 3693–3701 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S. H., Lee W. J., Role of DUOX in gut inflammation: Lessons from Drosophila model of gut-microbiota interactions. Front. Cell. Infect. Microbiol. 3, 116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S., Molina-Cruz A., Gupta L., Rodrigues J., Barillas-Mury C., A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 327, 1644–1648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Smith A. A., Williams M. S., Pal U., A dityrosine network mediated by dual oxidase and peroxidase influences the persistence of Lyme disease pathogens within the vector. J. Biol. Chem. 289, 12813–12822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohbayashi T., Mergaert P., Kikuchi Y., Host-symbiont specificity in insects: Underpinning mechanisms and evolution. Adv. Insect Phys. 58, 27–62 (2020). [Google Scholar]
- 26.Takeshita K., Kikuchi Y., Riptortus pedestris and Burkholderia symbiont: An ideal model system for insect-microbe symbiotic associations. Res. Microbiol. 168, 175–187 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Kaltenpoth M., Flórez L. V., Versatile and dynamic symbioses between insects and Burkholderia bacteria. Annu. Rev. Entomol. 65, 145–170 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Ohbayashi T., et al., Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J. 13, 1469–1483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh H., et al., Host-symbiont specificity determined by microbe-microbe competition in an insect gut. Proc. Natl. Acad. Sci. U.S.A. 116, 22673–22682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi Y., Ohbayashi T., Jang S., Mergaert P., Burkholderia insecticola triggers midgut closure in the bean bug Riptortus pedestris to prevent secondary bacterial infections of midgut crypts. ISME J. 14, 1627–1638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Futahashi R., et al., Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. PLoS One 8, e64557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mergaert P., Kikuchi Y., Shigenobu S., Nowack E. C. M., Metabolic integration of bacterial endosymbionts through antimicrobial peptides. Trends Microbiol. 25, 703–712 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Park K. E., et al., The roles of antimicrobial peptide, rip-thanatin, in the midgut of Riptortus pedestris. Dev. Comp. Immunol. 78, 83–90 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Kikuchi Y., Meng X.-Y., Fukatsu T., Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71, 4035–4043 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aruoma O. I., Halliwell B., Hoey B. M., Butler J., The antioxidant action of N-acetylcysteine: Its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radic. Biol. Med. 6, 593–597 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Goodchild A. J. P., Studies on the fucntional anatomy of the intestines of heteroptera. J. Zool. 141, 851–910 (1963). [Google Scholar]
- 37.Kim J. K., et al., Insect gut symbiont susceptibility to host antimicrobial peptides caused by alteration of the bacterial cell envelope. J. Biol. Chem. 290, 21042–21053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishide Y., et al., Functional crosstalk across IMD and Toll pathways: Insight into the evolution of incomplete immune cascades. Proc. Biol. Sci. 286, 20182207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi S., Kondo T., Development and function of the Drosophila tracheal system. Genetics 209, 367–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schottenfeld J., Song Y., Ghabrial A. S., Tube continued: Morphogenesis of the Drosophila tracheal system. Curr. Opin. Cell Biol. 22, 633–639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devine W. P., et al., Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 17014–17019 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandarra D., Biddlestone J., Mudie S., Muller H. A., Rocha S., Hypoxia activates IKK-NF-κB and the immune response in Drosophila melanogaster. Biosci. Rep. 34, e00127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou D., et al., Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: Hairy as a metabolic switch. PLoS Genet. 4, e1000221 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamad M. A., et al., Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob. Agents Chemother. 55, 3313–3323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sass A. M., et al., The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. ISME J. 7, 1568–1581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pessi G., et al., Response of Burkholderia cenocepacia H111 to micro-oxia. PLoS One 8, e72939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J. K., et al., Bacterial cell wall synthesis gene uppP is required for Burkholderia colonization of the Stinkbug Gut. Appl. Environ. Microbiol. 79, 4879–4886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J. K., et al., Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J. 8, 552–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland D., Samakovlis C., Krasnow M. A., Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87, 1091–1101 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Chung S., Chavez C., Andrew D. J., Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis. Dev. Biol. 360, 160–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilk R., Weizman I., Shilo B. Z., Trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 10, 93–102 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Lavista-Llanos S., et al., Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell. Biol. 22, 6842–6853 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Centanin L., et al., Cell autonomy of HIF effects in Drosophila: Tracheal cells sense hypoxia and induce terminal branch sprouting. Dev. Cell 14, 547–558 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Tonning A., et al., A transient luminal chitinous matrix is required to model epithelial tube diameter in the Drosophila trachea. Dev. Cell 9, 423–430 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Luschnig S., Bätz T., Armbruster K., Krasnow M. A., Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 16, 186–194 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Wang S., et al., Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 16, 180–185 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Jaźwińska A., Ribeiro C., Affolter M., Epithelial tube morphogenesis during Drosophila tracheal development requires Piopio, a luminal ZP protein. Nat. Cell Biol. 5, 895–901 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Dias F. A., et al., Ovarian dual oxidase (Duox) activity is essential for insect eggshell hardening and waterproofing. J. Biol. Chem. 288, 35058–35067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hurd T. R., Liang F. X., Lehmann R., Curly encodes dual oxidase, which acts with heme peroxidase curly Su to shape the adult Drosophila wing. PLoS Genet. 11, e1005625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anh N. T. T., Nishitani M., Harada S., Yamaguchi M., Kamei K., Essential role of Duox in stabilization of Drosophila wing. J. Biol. Chem. 286, 33244–33251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edens W. A., et al., Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J. Cell Biol. 154, 879–891 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong J. L., Wessel G. M., Extracellular matrix modifications at fertilization: Regulation of dityrosine crosslinking by transamidation. Development 136, 1835–1847 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meitzler J. L., Brandman R., Ortiz de Montellano P. R., Perturbed heme binding is responsible for the blistering phenotype associated with mutations in the Caenorhabditis elegans dual oxidase 1 (DUOX1) peroxidase domain. J. Biol. Chem. 285, 40991–41000 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ha E. M., et al., An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell 8, 125–132 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Kuraishi T., Binggeli O., Opota O., Buchon N., Lemaitre B., Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 108, 15966–15971 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hori A., Kurata S., Kuraishi T., Unexpected role of the IMD pathway in Drosophila gut defense against Staphylococcus aureus. Biochem. Biophys. Res. Commun. 495, 395–400 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Shibata T., et al., Transglutaminase-catalyzed protein-protein cross-linking suppresses the activity of the NF-κB-like transcription factor relish. Sci. Signal. 6, ra61 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Ren C., Webster P., Finkel S. E., Tower J., Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6, 144–152 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Linneweber G. A., et al., Neuronal control of metabolism through nutrient-dependent modulation of tracheal branching. Cell 156, 69–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z., Zhang Y., Han L., Shi L., Lin X., Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev. Cell 24, 133–143 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Jarecki J., Johnson E., Krasnow M. A., Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell 99, 211–220 (1999). [DOI] [PubMed] [Google Scholar]
- 72.Bruick R. K., McKnight S. L., A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Sommer F., Bäckhed F., The gut microbiota–Masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Reinhardt C., et al., Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 483, 627–631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stappenbeck T. S., Hooper L. V., Gordon J. I., Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U.S.A. 99, 15451–15455 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumann P., Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59, 155–189 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Łukasik P., et al., Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc. Natl. Acad. Sci. U.S.A. 115, E226–E235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weiss B., Kaltenpoth M., Bacteriome-localized intracellular symbionts in pollen-feeding beetles of the genus Dasytes (Coleoptera, Dasytidae). Front. Microbiol. 7, 1486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data of the M4 of Symcontrol and SymRpDuox-RNAi; gene sequences of RpDuox, RpBnl, RpTrh, RpSima, and RpRelish; and sequences of Duox of G. bimaculatus, B. mori, and T. castaneum data have been deposited in the National Center for Biotechnology Information (BioProject PRJDB9456) GenBank database (DRA accession numbers DRR215696-DRR215701; MT270146-MT270150; MT270151, NW_004582026, and NC_007418).