Significance
The human respiratory syncytial virus (RSV) is a significant cause of lower respiratory tract infections in the young and the elderly and a substantial burden to global human health. Currently, there are no specific and effective treatments for RSV infections. Here we report the first X-ray crystal structure of RSV nonstructural protein 2 (NS2), which revealed a unique fold. The combined biochemical and structural analyses of the NS2 structure identified a region that binds to and inhibits ubiquitination of an inactive form of RIG-I and MDA5, preventing downstream signaling and type I interferon production. Our study provides structural insight into the mechanism of how NS2 antagonizes IFN response to RSV infection and defines a target for antiviral development.
Keywords: RSV, nonstructural protein, NS2, IFN antagonist
Abstract
Human respiratory syncytial virus (RSV) nonstructural protein 2 (NS2) inhibits host interferon (IFN) responses stimulated by RSV infection by targeting early steps in the IFN-signaling pathway. But the molecular mechanisms related to how NS2 regulates these processes remain incompletely understood. To address this gap, here we solved the X-ray crystal structure of NS2. This structure revealed a unique fold that is distinct from other known viral IFN antagonists, including RSV NS1. We also show that NS2 directly interacts with an inactive conformation of the RIG-I–like receptors (RLRs) RIG-I and MDA5. NS2 binding prevents RLR ubiquitination, a process critical for prolonged activation of downstream signaling. Structural analysis, including by hydrogen-deuterium exchange coupled to mass spectrometry, revealed that the N terminus of NS2 is essential for binding to the RIG-I caspase activation and recruitment domains. N-terminal mutations significantly diminish RIG-I interactions and result in increased IFNβ messenger RNA levels. Collectively, our studies uncover a previously unappreciated regulatory mechanism by which NS2 further modulates host responses and define an approach for targeting host responses.
The human respiratory syncytial virus (RSV) is a major cause of lower respiratory tract infections and hospitalizations of young children, the elderly, and immunocompromised individuals worldwide (1, 2). RSV infection results in immune dysregulation and can exacerbate existing conditions, including asthma and chronic obstructive pulmonary disease. Furthermore, there are high rates of reinfection, as prior exposure to RSV does not lead to long-lasting immunity. Despite the significant burden of RSV on global health, there are limited prophylactic and therapeutic treatment options available. Our incomplete understanding of the viral components and of host responses that are elicited during RSV infection is a significant limitation.
RSV is a nonsegmented, negative-strand RNA virus and a member of the Pneumoviridae family (3). The RSV genome encodes two multifunctional nonstructural (NS) proteins, NS1 and NS2. Homologs or orthologs of NS1 and NS2 are lacking in other Mononegavirales members, including the related human metapneumovirus; therefore, the significance of these proteins to RNA viruses is unique and unlikely to be informed by studies of other viruses. Both NS1 and NS2 are implicated as interferon (IFN) antagonists and are known to play key roles in modulating host responses during early stages of infection. Recombinant viruses lacking NS1 or NS2 are attenuated in cell culture; infection with NS1/NS2-deleted viruses results in increased IFNβ messenger RNA (mRNA) levels compared with infection with wild-type (WT) RSV-infected cells, supporting the idea that NS1 and NS2 controls the host response (4–17). It was also noted that recombinant viruses lacking the NS2 gene display slower growth kinetics, suggesting that NS2 may play a role in replication of the WT virus due to its role in immune evasion (18). Infection of these NS2 deletion recombinant viruses in airway epithelial cells resulted in increased IFNβ mRNA levels that were not as high compared with a virus with a NS1 gene deletion or NS1/NS2 gene deletions (8, 16–18), indicating that NS2 may work in concert with NS1 to enhance the effect of NS1. NS2 does not have sequence homology to any known protein (19, 20). This underscores the need to further characterize NS2 to understand its functionally distinct role(s) in facilitating the viral pathogenesis of RSV, which remains poorly defined.
The retinoic acid-inducible gene I (RIG-I)–like receptors (RLRs) are host pattern recognition receptors that include RIG-I and melanoma differentiation-associated gene 5 (MDA5) and are known targets of RSV NS1 and NS2 (21–25). RIG-I and MDA5 each contain a cytoplasmic DEXD/H box helicase domain and can detect viral infection by recognizing pathogen-associated molecular patterns (PAMPs), such as viral replication by-products (26, 27). Both RIG-I and MDA5 contain two N-terminal caspase activation and recruitment domains (CARDs) that are linked to the helicase domain and followed by a C-terminal domain (CTD) (28, 29). In the absence of any PAMPs, RIG-I adopts an autoinhibited conformation, where the CARDs are sequestered from interactions with other CARD-containing molecules (30–33). Binding of viral RNAs to the helicase/CTD relieves RIG-I autoinhibition, releasing the N-terminal CARDs. MDA5 likely exists in an equilibrium between the autoinhibited and extended conformation, where the N-terminal CARDs assemble into oligomers to form filaments (34, 35). Important for the complete activation of RIG-I and MDA5 is the dephosphorylation of CARDs as well as ubiquitination by several E3 ubiquitin ligases, including TRIM25, Riplet, TRIM4, and MEX3C (36–40). Activation of RIG-I and MDA5 then allows for binding to the mitochondrial antiviral signaling protein (MAVS; also known as Cardif/VISA/IPS-1) (41–44). MAVS activation leads to downstream signaling and activation of transcription factors including IRF3, IRF7, and NF-κB. Phosphorylation of these transcription factors is critical for them to translocate into the nucleus and stimulate IFN α/β production, which can then trigger antiviral IFN-stimulated gene (ISG) expression.
Early studies suggested that RIG-I plays a predominant role in detecting RSV during the early phases of infection in airway epithelial cells to mount a rapid antiviral response (45–47). Small interfering RNA knockdown of RIG-I inhibited NF-κB and IRF3 activation post infection with RSV (45, 47). RSV infection also appeared to be restricted in RIG-I knockout cells, resulting in reduced expression of several IRF-3–dependent ISGs when compared with MDA5 knockout mouse embryonic fibroblast cells, which displayed delayed expression (46). However, subsequent studies demonstrated that MDA5 may have a more significant role in response to RSV infections than previously appreciated. Both MDA5 and RIG-I expression were found to be up-regulated in the nasopharyngeal washes from RSV-infected infants (48). MDA5 directly drives NF-κB activation in response to RSV infection that is RIG-I independent (47) and is required to prevent premature degradation of IRF3 (49). Furthermore, MDA5 and MAVS were found to colocalize with RSV genomic RNA and nucleoprotein in inclusion bodies to attenuate IFN signaling (50). Collectively, these studies suggest that MDA5 may play a previously underappreciated role during RSV infection.
NS2 interferes with RLR signaling by targeting molecules in the IFN-signaling pathway that are distinct steps in the pathway from NS1. Overexpression of NS2 inhibits RIG-I–mediated IRF3 activation when stimulated with a 5′ppp RNA (11). Coimmunoprecipitation (coIP) studies suggest that NS2 binds to RIG-I through interactions with the N-terminal CARDs (11). NS2 expression also decreases STAT2 expression levels when transfected into human tracheobronchial epithelial cells (51). RNA interference of NS2 expression in cells infected with RSV resulted in a loss of STAT2 inhibition. Furthermore, a ΔNS2 deletion virus lost the ability to suppress STAT2 expression or PKR when compared with WT or a ΔNS1 deletion virus (51), which may be associated with a role for NS2 to modulate ubiquitination of cellular proteins like STAT2 and to facilitate degradation (52–54). Altogether, these observations suggest that NS2 is capable of performing multiple functions as a viral antagonist of type I IFN signaling independent of NS1. While much insight has been gained through these studies, the molecular mechanisms for how NS2 mediates these functions remain undefined, including the lack of an atomic structure for RSV NS2. Lack of these insights limits our understanding of the unique role that NS2 plays in facilitating RSV pathogenesis and our ability to develop and treat RSV infections that are targeted to RSV nonstructural proteins.
To address this gap, we solved the X-ray crystal structure of RSV NS2, which revealed a unique fold that is distinct from other known IFN antagonists, including RSV NS1. Using the structure as a basis, we conducted a series of studies aimed at defining the molecular basis for NS2-mediated immune antagonism. We demonstrate that RSV NS2 directly interacts with both inactive RIG-I and MDA5 and that the binding of NS2 prevents RLR ubiquitination, activation, and downstream signaling. Our biochemical and structural analyses identified NS2 residues that are important for binding to RIG-I, which were subsequently validated by functional studies. Together, our results provide insights into the mechanism by which RSV NS2 modulates host pattern recognition receptors and define a molecular framework that defines a role of NS2 in host IFN antagonism.
Results
The RSV NS2 X-Ray Crystal Structure Represents a Unique Fold.
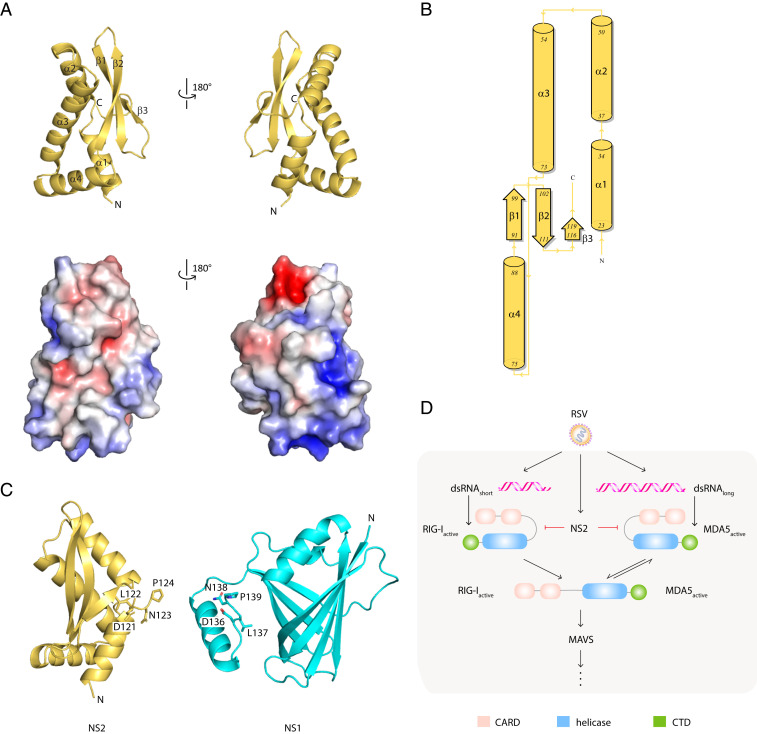
RSV NS2 is a viral antagonist of IFN signaling. Like RSV NS1, NS2 targets multiple host factors to limit host responses during infection. Recent studies, including those from our group, initiated structure–function analyses to define the immunoregulatory functions of NS1 (55). RSV NS2 is transcribed from a region near the 5′UTR and expressed at high levels in the host cytoplasm. NS2 is also thought to be multifunctional. However, unlike NS1, little is known about NS2, leaving a large gap in our understanding of how these two small nonstructural proteins facilitate immune modulation, viral replication, and viral pathogenesis. To gain additional insight, we generated crystals of selenomethionine-labeled NS2 (SI Appendix, Fig. S1A) and solved the X-ray crystal structure of NS2 by single-wavelength anomalous diffraction at 2.8 Ǻ resolution (Fig. 1A and SI Appendix, Table S1). The asymmetric unit contains three nearly identical molecules in the asymmetric unit (molecule A, molecule B, molecule C) (SI Appendix, Fig. S1B). While the solvent-accessible surface area is large enough to suggest homo-oligomerization of NS2 within the asymmetric unit, solution studies, including size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS), do not support NS2-NS2 interactions (SI Appendix, Fig. S1C).
Fig. 1.
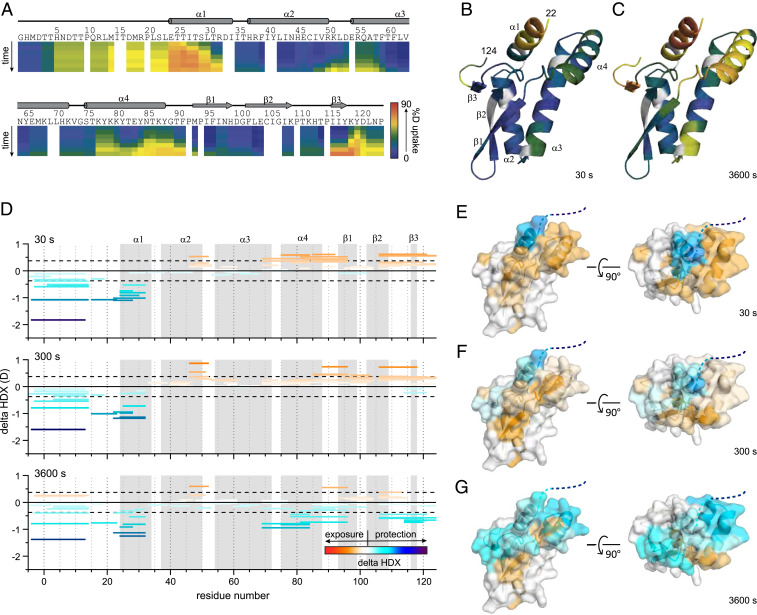
RSV NS2 adopts a unique mixed alpha–beta fold. (A) Cartoon representation of the X-ray crystal structure of NS2 (Top). Surface electrostatic representation of NS2 (Bottom). Red, white, and blue represent negative, neutral, and positive electrostatic potential, respectively (−5 to +5 kBT e−1). Orientation in Bottom is the same as in Top. (B) Wiring diagram of the secondary structural elements in NS2. Figure was generated using PDBSum. (C) Structural comparison of RSV NS2 (Left) and RSV NS1 (Right, PDB 5VJ2). The terminal four residues, DNLP, are highlighted in stick representation. (D) RLR function in IFNβ signaling. RIG-I and MDA5 contain two tandem caspase activation and recruitment domains (CARDs, pink) at the N terminus followed by a DEXD-box helicase domain (blue) and a CTD (green). RIG-I and MDA5 exists in an autoinhibited conformation that is relieved upon binding to immunostimulatory RNAs, exposing the CARDs for interaction with other CARD-containing proteins, including the mitochondrial-associated MAVS protein.
The X-ray structure of NS2 is a single globular domain with a mixed α/β fold (Fig. 1 A and B). In the structure, the N terminus contains a cluster of four alpha helices followed by a three-stranded, antiparallel beta sheet. Missing electron density for the N-terminal 22 residues and the C-terminal 7 residues indicates that these regions may be disordered in this X-ray crystal structure. The C-terminal proline residue (P124) appears to N-cap the helix α2, which may function to further stabilize the unique NS2 fold (Fig. 1C). Analysis of the electrostatic surface of NS2 shows two charged pockets composed of basic residues, including K68, K72, K77, K79, and K80, in helices α3 and α4 (Fig. 1 A and B).
Based on sequence alignments, no known functional motifs were identified from comparisons with the NS2 structure. Comparison of the NS2 structure with other existing protein structures using the DALI server (56) revealed no significant structural homology. The top-scoring hit is a yeast ubiquitin carboxyl-terminal hydrolase (Protein Data Bank [PDB] ID: 4W4U; Z score: 5.6; rmsd: 2.8). Although there is some similarity in the secondary structural elements that comprise the structure, there is no significant alignment to the NS2 structure that would suggest structural homology or functional correlations. Structural comparison of NS2 with our previously solved structure of RSV NS1 (PDB 5VJ2) showed no similarity to each other, with the exception of the four identical residues located at the C terminus of each molecule (amino acids 121-DLNP-124) (Fig. 1C). NS2 and NS1 have overlapping functions, but they lack sequence similarity and, as revealed through our comparisons here, these two proteins also lack structural homology. These findings further suggest that NS1 and NS2 may target different proteins in the host IFN induction and response pathways through distinct mechanisms to achieve synergistic regulation of host responses.
NS2 Binds to Inactive RIG-I and MDA5 and Prevents Activation.
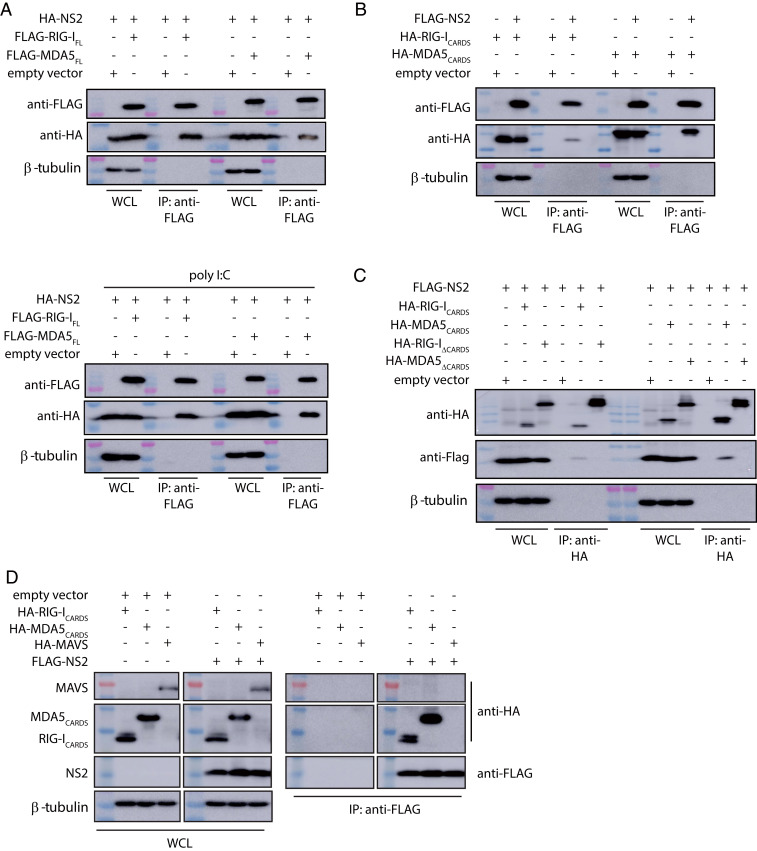
Early studies suggested that NS2 inhibits host signaling early in the IFN induction pathway (Fig. 1D) (25, 57); furthermore, these studies suggested that RIG-I, but not MDA5, was important for RSV-mediated IFN inhibition (11, 27, 46). Subsequent studies, however, suggested a greater role for MDA5 in response to RSV infections (11, 45, 48–50). To further probe these interactions, ectopically expressed FLAG-tagged full-length RIG-I (RIG-IFL) or FLAG-tagged full-length MDA5 (MDA5FL) were coexpressed with hemagglutinin (HA)-tagged NS2 in HEK 293T cells. Coimmunoprecipitation assays were performed using anti-FLAG resin. We found that NS2 interacted with both RIG-IFL and MDA5FL (Fig. 2A and SI Appendix, Fig. S1D). Because RIG-I and MDA5 undergo conformational changes and are activated upon binding to viral RNA PAMPs (Fig. 1D), co-IP assays were also performed on transfected HEK 293T cells that were treated with polyI:C (27), a synthetic double-stranded RNA mimic. NS2 interacts with RIG-IFL and was similar in the presence of polyI:C; NS2 interactions with MDA5FL appear to be slightly enhanced with polyI:C (Fig. 2 A, Bottom, and SI Appendix, Fig. S1D). To further validate which domains of RIG-I and MDA5 are required for NS2 interaction, FLAG-NS2 was coexpressed with either the HA-RLR CARD domains, which form protein–protein interactions with other CARD-containing proteins, or an HA-tagged–tethered RLR helicase/C-terminal domains (ΔCARDs) in HEK 293T cells. The co-IP results demonstrate that NS2 binds to RIG-ICARDs, consistent with previous studies (11), and to MDA5CARDs (Fig. 2B and SI Appendix, Fig. S1D). We did not observe an NS2 interaction with either RIG-I or MDA5 helicase/CTD domains (Fig. 2C and SI Appendix, Fig. S1D).
Fig. 2.
RSV NS2 coimmunoprecipitates with RIG-I and MDA5. (A) Empty expression plasmid (empty vector), FLAG-full-length RIG-I (FLAG-RIG-IFL) or FLAG- full-length MDA5 (FLAG-MDA5FL) were cotransfected with HA-tagged NS2 expression plasmid in HEK 293T cells in the absence (Top) or presence (Bottom) of poly I:C transfection. (B) Cells were cotransfected with empty vector, HA-RIG-ICARDs, or HA-MDA5CARDs with FLAG-NS2. (C) Cells were cotransfected with empty vector, HA-RIG-ICARDs, HA-MDA5CARDs, HA-RIG-I helicase/CTD (RIG-IΔCARDs), or HA-MDA5 helicase/CTD (MDA5ΔCARDs) with FLAG-NS2. (D) Cells were cotransfected with HA-RIG-ICARDS, HA-MDA5CARDs, or HA-MAVS with empty vector or FLAG-NS2. Western blots of IP and whole-cell lysates (WCL), including β-tubulin, are shown. Shown are representative results from experiments repeated in triplicate.
In contrast to RLR CARDs, MAVS, a mitochondrial-associated protein that also contains a single CARD domain at the N terminus, did not show binding. To test NS2 binding to the MAVS CARD, HEK 293T cells were cotransfected with HA-RLRs-CARDs, HA-MAVS, and FLAG-NS2. The coIP assays were performed 24 h post transfection. Our results reveal that NS2 does not interact with MAVS; in contrast, NS2 can readily bind to RIG-ICARDs and MDA5CARDs (Fig. 2D and SI Appendix, Fig. S1D).
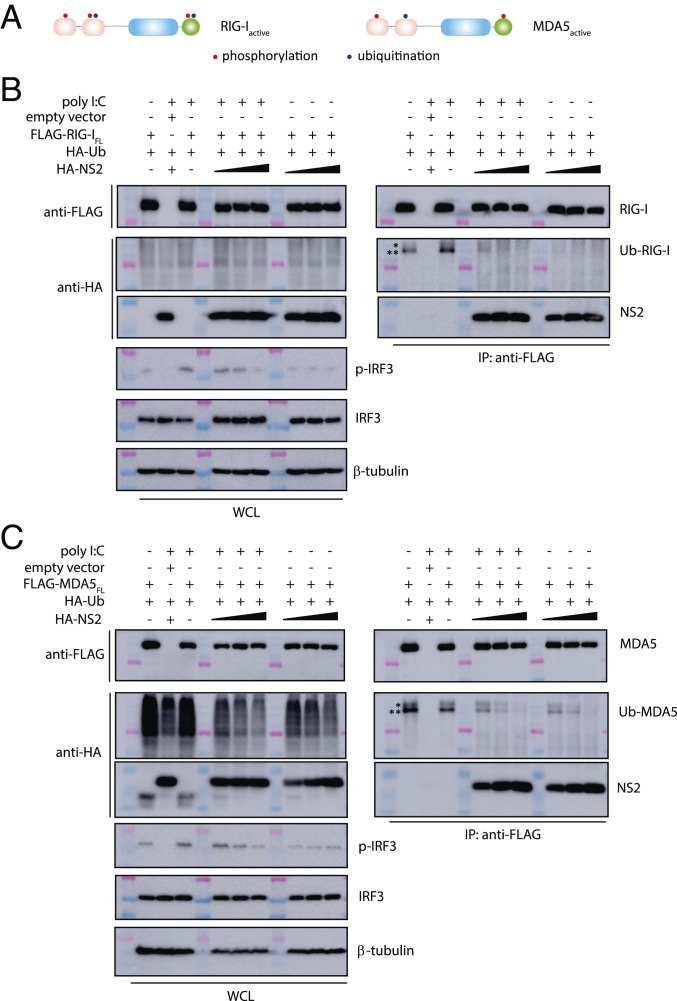
In addition to RNA binding, activation of RLRs is enhanced by their dephosphorylation and ubiquitination. These posttranslational modifications are thought to occur prior to interaction with MAVS and subsequent downstream signaling (Figs. 1D and 3A). To identify which RLR state NS2 binds to, HEK 293T cells were cotransfected with FLAG-tagged RIG-IFL or MDA5FL and HA-tagged ubiquitin after treatment with polyI:C. FLAG coIP results show that both RIG-IFL (Fig. 3 B, Right) and MDA5FL (Fig. 3 C, Right) are ubiquitinated. To test if NS2 binding prevents ubiquitination of RLRs, we repeated the same experiment in the presence of increasing concentrations of HA-tagged NS2. Expression of NS2 inhibited ubiquitination of RIG-IFL and MDA5FL in the absence or in the presence of polyI:C and in a concentration-dependent manner. These results are consistent with NS2 binding to an inactive state of the RLRs.
Fig. 3.
RSV NS2 inhibits RIG-I and MDA5 ubiquitination. (A) Simplified schematic of posttranslational modifications of RIG-I and MDA5. Domains of RIG-I and MDA5 that are phosphorylated (red) or ubiquitinated (blue) are indicated by a single dot. FLAG-coIPs (Right) of FLAG- (B) RIG-IFL or (C) MDA5FL with HA-ubiquitin and increasing concentrations of HA-NS2 (1, 2, and 4 μg) and in the absence or presence of polyI:C transfection. Band corresponding to Ub-RIG-I and Ub-MDA5 is denoted by a double asterisk (**). Unidentified ubiquitinated band is denoted by a single asterisk (*). Immunoblotting of phosphorylated IRF3 (p-IRF3), IRF3, and β-tubulin is shown below. Western blots of WCL are shown on the Left in B and C. Shown are representative results from experiments repeated in triplicate.
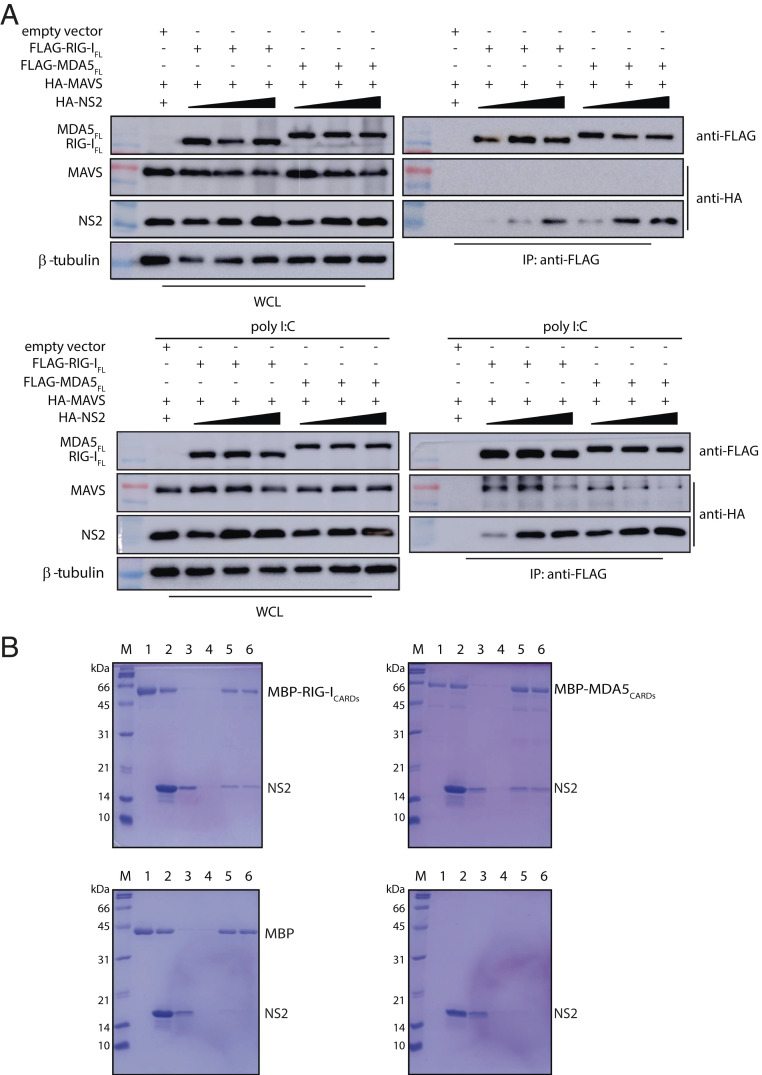
We next tested how the presence of NS2 affects RLR interaction with the MAVS protein, which is immediately downstream of RLR binding, with nonself RNA (Fig. 1D). HEK 293T cells were cotransfected with either FLAG-RIG-IFL or FLAG-MDA5FL, HA-MAVS, along with increasing concentrations of HA-tagged NS2. Without polyI:C, coIP results show that MAVS does not interact with RIG-IFL or MDA5FL in the presence of NS2 (Fig. 4A). When the cells are treated with polyI:C prior to performing the co-IP, we find that both RIG-IFL and MDA5FL now bind to MAVS, but that these interactions are diminished in the presence of increasing concentrations of NS2 (Fig. 4 A, Bottom). These results are consistent with previous studies demonstrating that RNA binding activates a conformational change in the RLRs, where RNA binding releases the CARDs to bind to MAVS (30–35). Our results here further support that NS2 binds to an inactive form of the RLRs, suggesting that NS2 binding likely prevents RLR activation.
Fig. 4.
RSV NS2 inhibits the interaction between RLRs and MAVS. (A) The coIP between FLAG-RIG-IFL or FLAG-MDA5FL with HA-tagged MAVS and increasing concentrations of HA-NS2 (1, 2, and 4 μg) in the absence (Top) or presence (Bottom) of poly I:C. WCL, whole-cell lysate. (B) In vitro pull-down assays between NS2 and RLR CARDs. Recombinantly expressed and purified MBP-RIG-ICARDs (Top Left), MBP-MDA5CARDs (Top Right), or MBP tag alone (Bottom Left) were immobilized on amylose resin prior to incubation with NS2. NS2 alone was also incubated with amylose resin (Bottom Right) as a negative control. Lanes correspond to the following: M, marker; 1, bound beads; 2, input; 3, wash 1; 4, wash 4; 5, final beads; 6, elution. Shown are representative results repeated in triplicate.
Since RLR signaling leads to phosphorylation and subsequent nuclear translocation of IRF3, we also examined the effect of NS2 binding to RIG-I or MDA5 on the phosphorylation of IRF3. Resulting data reveal that the overall levels of endogenous IRF3 remain relatively constant in the absence or presence of RIG-IFL or MDA5FL, and with or without polyI:C treatment (Fig. 3 A and B, Bottom Left). When we look at activation of IRF3 using an antibody that detects phosphorylated IRF3, we observe an NS2 concentration-dependent decrease in IRF3 phosphorylation in the presence of polyI:C and coexpressed with ubiquitin. Without polyI:C, the level of IRF3 phosphorylation is low and unchanged as NS2 expression level increases with both RIG-IFL or MDA5FL (Fig. 3 A and B, Bottom Left). Collectively, these results suggest that NS2 interacts with both RIG-I and MDA5 through the CARD domains in a manner that is independent of RNA binding and RLR activation.
NS2 Binds Directly to RLR CARDs.
To confirm that RSV NS2 directly binds to RIG-I and MDA5, we performed in vitro pull-down assays using recombinantly expressed and purified NS2, maltose-binding protein (MBP)-tagged RIG-ICARDs, and MBP-MDA5CARDs. MBP-RIG-ICARDs or MBP-MDA5CARDs were immobilized on amylose resin prior to incubation with NS2. MBP-RIG-ICARDs (Fig. 4 B, Top Left) and MBP-MDA5CARDs (Fig. 4 B, Top Right) both pull down untagged NS2, demonstrating a direct interaction between RLR CARDs and NS2. In contrast, NS2 does not bind to the MBP protein alone, an important negative control (Fig. 4 B, Bottom Left) or the amylose resin (Fig. 4 B, Bottom Right). These results support our co-IP results and show that the CARDs are important for NS2 binding to RLRs.
NS2 Is Conformationally Dynamic.
Next, we used hydrogen deuterium exchange coupled to mass spectrometry (HDX-MS) to characterize structural differences in hydrogen bonding and solvent accessibility of NS2 upon binding to RIG-I CARDs. HDX-MS, a well-known reporter of protein dynamics, conformational changes, and secondary structure, identifies changes in solvent accessibility or hydrogen bonding specifically for backbone amide groups. Digestion of MBP-NS2 with fungal protease XIII in solution coupled with online pepsin produced 69 unique peptides and 99% coverage of NS2 (SI Appendix, Fig. S2A). No statistically significant differences in deuterium uptake were observed for the protein with or without the MBP-tag (SI Appendix, Fig. S2B, black and blue, respectively).
The HDX-MS kinetic characteristics of MBP-NS2 are consistent with our crystal structure (Fig. 1) for short HDX times; at longer incubation times, increased HDX occurs (Fig. 5 A–C and SI Appendix, Fig. S3). The region covering the first 22 residues, which was missing in the X-ray crystal structure owing to the lack of electron density, and most of the residues in the N-terminal helix α1 undergo fast exchange within 30 s. This indicates a high degree of flexibility and/or lack of hydrogen bonding for the first 22 residues and suggests that much of helix α1 is more conformationally dynamic than expected for an α-helix, where most amides are involved in hydrogen bonding. After 3,600 s of D2O exchange, a high degree of deuterium uptake was also observed for the loop between helix α2 and α3, the loop connecting to strand β1 and strand β2 to the C terminus (Fig. 5 A–C), also consistent with a flexible structure lacking strong hydrogen bonding; interestingly, we also observed a high degree of deuterium uptake after 3,600 s for helix α4, suggesting that helix α4 is also conformationally dynamic.
Fig. 5.
HDX-MS of RSV NS2 reveals regions that are dynamic and protected upon binding to RIG-I CARDs. (A) Heatmap showing the extent of HDX for free NS2 for incubation times of 10, 30, 300, 900, 3,600, and 14,400 s. The heat maps were smoothed using “light smoothing” with the HDExaminer software. Secondary structures determined in the crystal structure are shown above. Deuterium uptake extents mapped onto the NS2 structure are shown for (B) 30 s and (C) 3,600 s, respectively. (D) Peptide-level difference maps showing absolute differences in HDX for all identified peptides after 30, 300, and 3,600 s of HDX (Top to Bottom). Each peptide is shown as a bar covering the peptide sequence. Global significance limits (black dash) indicate the minimum HDX difference to be statistically significant (P < 0.01). Increases (warm) and decreases (cool) in HDX are indicated by the gradient and correspond to the structures on the right (E–G). Secondary structures from A are indicated in gray. Representative peptide-level increases (warm colors) and decreases (cool colors) in HDX as a function of binding RIG-I CARDs mapped onto the crystal structure of NS2 after (E) 30 s, (F) 300 s, and (G) 3,600 s of exchange, respectively. Structures on the Left of E, F, and G are shown in the same orientation as those in B and C structures on the Right are rotated 90° along the y axis.
A Conformationally Dynamic Surface of NS2 Accommodates Binding with RIG-I CARDs.
Using HDX-MS assays, we further probed changes in backbone amide deuterium uptake for the NS2/RIG-ICARDS protein complex (SI Appendix, Fig. S4A). Many peptides covering NS2 show differences in HDX for NS2 alone compared with the NS2/RIG-I CARDs complex (Fig. 5D and SI Appendix, Fig. S4B). Both increases and decreases in deuterium incorporation are primarily localized near the N terminus and proximal structures (Fig. 5 E–G and SI Appendix, Fig. S4B). Decreased HDX is consistently observed for the N-terminal 32 residues. The surface surrounding these residues (i.e., helix α4 and the C-terminal 15 residues) exhibit, upon binding, several behaviors including consistently increased HDX and an interesting “crossing” or composite behavior, where increased HDX at shorter incubation times is followed by decreased HDX at longer incubation times. Presumably, the observed increases in HDX occur in select regions where flexibility increases upon binding. Once some amides saturate with deuterium, other NS2 regions that show protection dominate the composite HDX and exhibit a slower increase in HDX at longer times. For example, at early time points for peptides covering helix α4 and nearby loops, we observe a clear increase in HDX of ∼0.6 Da occurs. At later time points, we observed converged HDX and maintained exposure (covering α4 and its C-terminal loop), or we observed binding-induced decreased HDX (covering α4 and its N-terminal loop). Together, this composite behavior reveals both an increase in conformational flexibility and binding-induced protection by RIG-I CARDs for those peptide regions. Interestingly, these structures cover the conformationally dynamic regions of the unbound state of NS2, also including the C-terminal 15 residues (Fig. 5 A–C). Finally, increased HDX occurs for β1, β2, and the loop between α2 and α3, suggesting changes in conformation or dynamics occurring at a region remote from the binding site. No statistically significant differences in HDX are observed for any other regions of NS2 (i.e., for helices α2 and α3) (SI Appendix, Fig. S5). While we observed some protection of the C-terminal end and linker of the MBP tag for the NS2/RIG-I CARDs complex (SI Appendix, Fig. S2B, red), we find no direct binding between the MBP-tag and RIG-I CARDs. This protection is consistent with RIG-I CARDs binding the N-terminal region of NS2, inducing nonspecific protection of the proximal MBP tag, and the protection identifies a continuous RIG-I–binding surface on the NS2 α1 helix. The complexities observed in our HDX analysis may represent the conformational flexibility of RSV NS2 and widescale RIG-I-binding–induced increases in flexibility.
NS2 Disrupts RLR-Mediated Signaling.
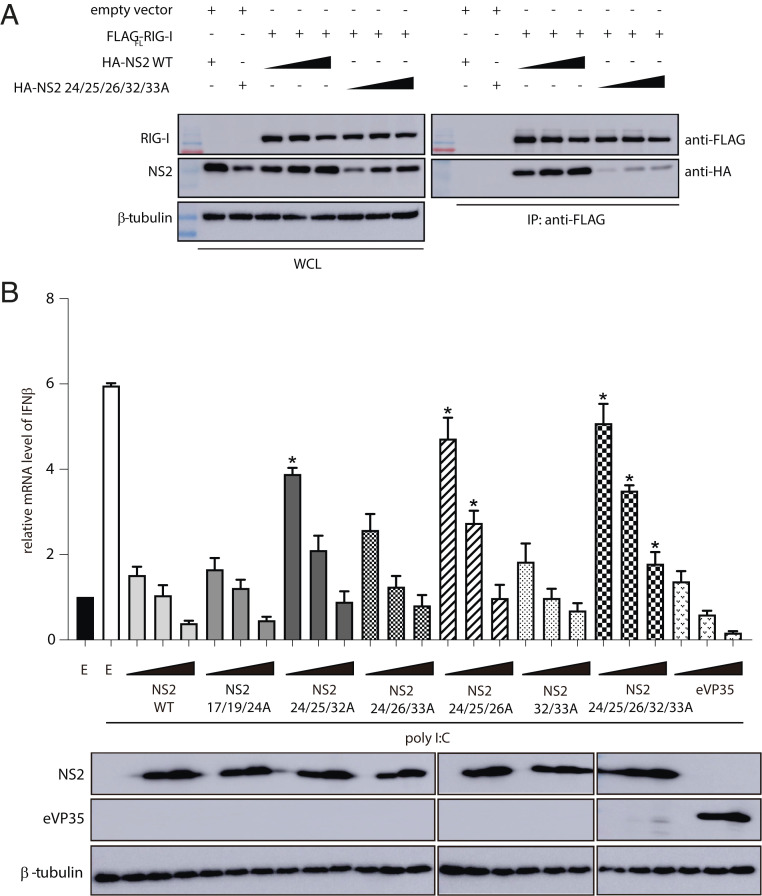
To assess the impact of NS2 binding on RLR-mediated signaling and to validate our structural findings, we introduced several alanine mutations in the NS2 N terminus, a dynamic region of RSV NS2 revealed by HDX-MS. We tested the ability of these NS2 mutants to bind RIG-I. HEK 293T cells were cotransfected with FLAG-RIG-IFL and increasing concentrations of HA-tagged NS2 WT or HA-tagged NS2 mutant 24/25/26/32/33A. We find that mutation of NS2 residues in helix α1 significantly reduces binding to RIG-I compared with NS2 WT (Fig. 6A), whereas alanine mutations of a similar number of residues in other regions, including in the NS2 α3-α4 helix (68/72/77/79/80) that shows increased HDX protection in the presence of RIG-I CARDs, do not impact interactions with RIG-I (SI Appendix, Fig. S6A). To test inhibition of IFNβ activation, we measured the IFNβ mRNA levels when stimulated by polyI:C by qRT-PCR. Transfection of WT NS2 showed that NS2 decreased the IFNβ mRNA levels compared with an empty vector control to levels that are comparable to RSV NS1 and Ebola virus VP35, a potent IFN antagonist (Fig. 6B and SI Appendix, Fig. S6B) (58–60). We found that mutation of NS2 residues in helix α1, which shows the greatest levels of HDX in the absence of RIG-I CARDs and increased protection in the presence of RIG-I CARDs, showed reduced suppression of IFNβ mRNA levels (Fig. 6B). NS2 mutants 24/25/32A and NS2 24/26/33A show some increased levels of IFNβ mRNA whereas NS2 mutants 24/25/26A and 24/25/26/32/33A show the highest levels of IFNβ mRNA. Mutations in the region N-terminal to helix α1 (17/19/24A) do not impact IFNβ mRNA levels (Fig. 6B). While the NS2 α3-α4 helix is not directly involved in RIG-I binding (SI Appendix, Fig. S6A), mutation of the charged residues in the NS2 α3-α4 helix (68/72/77/79/80A) shows partial suppression of IFNβ mRNA levels (SI Appendix, Fig. S6B), suggesting an alternative impact of this more conformationally dynamic region. Our combined results support a model in which NS2 antagonizes IFN signaling by binding to RIG-I and MDA5, preventing their activation and signaling functions, through interactions with the CARD region.
Fig. 6.
Mutation of RSV NS2 N terminus results in a loss of inhibition of IFNβ mRNA levels. (A) Immunoblot of Flag-RIG-I coIPs in the presence of increasing concentrations of NS2 WT or NS2 E24A/T25A/T26A/R32A/D33A (Right). WCL (Left), whole-cell lysate. Shown are representative results from experiments repeated in triplicate. (B) qRT-PCR of IFNβ mRNA levels of HEK 293T cells 8 h post transfection with increasing concentrations (0.2, 1, and 5 μg) of NS2 WT, NS2 N-terminal mutants, or eVP35 (control IFN antagonist) indicated and treated with polyI:C. The data represent the mean ± SEM of three independent experiments. Unpaired t test; *P < 0.05 mutants compared with NS2 WT at the respective plasmid concentrations. Western blots of protein expression are shown below.
Discussion
The cytoplasmic RLRs, RIG-I, and MDA5 serve critical functions in detecting initial infection and in stimulating host innate responses. Many RNA viruses, including RSV, have developed strategies to circumvent RLR-mediated antiviral responses by directly targeting RIG-I or MDA5. For example, influenza A virus NS1, SARS-CoV M protein, human metapneumovirus G protein, enterovirus 71 3C protein, New World arenaviruses Z protein, and severe fever with thrombocytopenia syndrome virus nonstructural proteins have been reported to suppress RIG-I signaling by directly interacting with RIG-I (61–66). Some viruses bind MDA5, including the paramyxovirus V protein (67, 68), while others, like West Nile virus NS1, target both RIG-I and MDA5 (69). Here we define key molecular mechanisms that allow RSV NS2 to antagonize RIG-I and MDA5.
RSV NS1 has been well characterized for its central role in facilitating host immune evasion by targeting various points in the IFNβ-signaling pathway (8, 10, 12, 14, 16, 17, 19, 70–72) and for inhibiting gene transcription in the nucleus (55, 73). However, less is known about RSV NS2 at a molecular and structural level, partly due to the notion that NS2 plays a reduced and perhaps redundant function to NS1. Here we show the crystal structure of NS2, which, until now, remained elusive to the field. Comparison with other known viral IFN antagonists reveals a unique arrangement of NS2 secondary structural elements that are conformationally dynamic. Further comparison with RSV NS1 shows a lack of any significant structural homology. The significance of the last four residues remains unknown as these residues do not comprise a recognizable motif. Our results suggest that NS2 targets different molecules from NS1 and likely utilizes distinct mechanisms to mediate IFN antagonism. Consistent with this model, we demonstrate that RSV NS2 is important for interactions that inhibit RIG-I activity. We confirmed that NS2 binds to RIG-I through interactions with the N-terminal CARDs, as was previously shown (11). Further structural analysis of NS2 revealed that these interactions are mediated through the N terminus of NS2, including the α1 helix, which becomes protected from HDX in the presence of RIG-I CARDs. Mutagenesis of helix α1 results in reduced RIG-I binding as well as the ability to inhibit IFN mRNA levels.
Earlier studies established a critical role for RIG-I in sensing RSV infection and in activating host innate immune responses, but more recent studies suggest that MDA5 likely has a more critical role in triggering responses that limit infection of RSV and closely related viruses (49, 50, 74–78). MDA5 is important for sustaining IRF3 activation in cells infected with RSV (49). RSV infection also stimulates the relocalization of MDA5 to inclusion bodies, which may serve to attenuate IFN responses (50). Here, our results define a molecular mechanism, one that unifies data previously observed for both RIG-I and for MDA5 by revealing that RSV NS2 can bind to both proteins. Importantly, our data support a model for NS2 binding to MDA5 through interactions with the CARDs, an interaction that had not been characterized previously. This interaction is specific to RLR and NS2 as NS2 does not bind to the MAVS CARD. Our data also revealed that NS2 likely interacts with the inactive RNA-free forms of RIG-I and MDA5 because NS2 binding occurs even in the absence of RNA PAMPs. NS2 binding prevents ubiquitination of RLRs, a posttranslational switch that is important for the sustained activation of the RLRs (23, 30, 79). We propose that loss of ubiquitination and MAVS binding results in the loss of sustained downstream signaling, which dampens the impact of RLR activation, thus preventing the RLR-MAVS axis from stimulating the phosphorylation of IRF3. Together, our studies support a model where RSV NS2 mediates inhibition of both RIG-I and MDA5 to effectively restrict host immune responses to RSV infection.
Our findings here further support the notion that RNA viruses, particularly those with negative-sense genomes with limited coding capacity, appear to use a multipronged strategy to evade host immunity. Our previous studies unexpectedly revealed that RSV NS1 is a structural paralog of the RSV matrix protein (55). In contrast, results here reveal that the NS2 structure bears no similarity to any other known proteins, including other viral immune antagonists or host proteins, supporting the unique and diverse functions of RSV nonstructural proteins that target unique and overlapping host targets. While we do not experimentally test this hypothesis, the current data support a model where RSV NS1 and NS2 may work together synergistically to suppress innate responses. NS1 and NS2 are RSV-specific proteins, not present in other virus genomes from the same family, and have remained enigmatic, in part due to their uniqueness. Our recent study of NS1 (55) together with results from the current study on NS2 fill a critical gap in our knowledge. Data from this current study also provide additional opportunities for targeted antiviral design and will facilitate studies to guide potential future development of attenuated vaccines for protection against RSV.
Materials and Methods
Cells and Plasmids.
The human embryonic kidney cell line 293T (American Type Culture Collection, CCL-3216) was maintained in complete Dulbecco’s modified Eagle’s medium (ThemoFisher, 11965) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich, F4135). The cells were incubated at 37 °C with 5% CO2.
The coding sequence for RSV NS2 (NCBI accession no. AY904041) (12) was synthesized in a pCDNA 3.1 vector (ThermoFisher) and then subcloned into mammalian expression plasmid pCAGGS with an N terminus FLAG tag and an HA tag. Mutations of NS2 were generated using the overlap PCR method and verified by sequencing. The full-length sequences of human RIG-I (1 to 226) and MDA5 (1 to 276) were used as templates to subclone different constructs into pCAGGS with an N terminus FLAG tag and HA tag, respectively.
Cloning, Protein Expression, and Purification.
Open reading frames corresponding to proteins of interest were subcloned and expressed in Escherichia coli (Novagen) by using methods previously described (55). Briefly, vectors encoding for MBP-tagged NS2 or RLR CARDs were transformed into E. coli and grown at 37 °C prior to induction with isopropyl β-d-1-thiogalactopyranoside at 18 °C for 14 h. Cells were harvested and lysed using an EmulsiFlex-C5 Homogenizer (Avestin). Lysate was clarified by centrifugation at 47,000 × g for 40 min at 4 °C. NS2 was purified using a series of chromatographic columns, including affinity and ion exchange, prior to Tev protease (New England Biolabs) cleavage of the MBP tag and final purification on a size exclusion column. Sample purity was monitored and assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels.
Antibodies and Reagents.
Monoclonal mouse anti-FLAG M2 antibody (F3165), monoclonal mouse anti-HA antibody (H3663), and monoclonal mouse anti–β-Tubulin antibody (T8328) were purchased from Sigma-Aldrich. Monoclonal rabbit anti–Phospho-IRF-3 (Ser396) antibody (29047) was purchased from Cell Signaling Technology. Monoclonal mouse anti–IRF-3 antibody (sc-33641) was purchased from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)-labeled goat anti-mouse (115-035-003) and goat anti-rabbit (111-035-003) secondary antibodies were obtained from Jackson ImmunoResearch. Low-molecular-weight polyI:C was purchased from InvivoGen.
Transfection.
HEK 293T cells grown to 80% confluence in six-well cell culture plates were transfected with targeted plasmids by using Lipofectamine 2000 transfection reagent (ThemoFisher, 11668019). Briefly, 2 μg of plasmid or polyI:C and 2 μL of Lipofectamine 2000 were diluted in 150 μL of serum-free OptiMEM medium (ThemoFisher, 31985070), respectively. After a 5-min incubation, Lipofectamine 2000 OptiMEM was directly pipetted into the medium containing the diluted plasmid and incubated at room temperature for 25 min. Then the DNA–lipid complex was added to the cell culture medium, and the cells were further incubated for 24 h at 37 °C. The gene expression efficiency and the interaction between targeted genes were evaluated by coimmunoprecipitation and immunoblotting.
Coimmunoprecipitation Assays.
Twenty-four hours post transfection with the indicated plasmids, HEK 293T cells were lysed in Nonidet P-40 lysis buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Nonidet P-40, 0.2 mM ethylenediaminetetraacetic acid [EDTA], 2 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 10% glycerol) containing protease inhibitors. Anti-HA affinity gel (Sigma-Aldrich, E6779) or anti-Flag M2 magnetic beads (Sigma-Aldrich, M8823) were incubated with lysates for 1 h at 4 °C, and the resin was then washed three times with cell lysis buffer and eluted at room temperature for 30 min using either HA peptide (Sigma-Aldrich, I2149) or Flag peptide (Sigma-Aldrich, F3290). Precipitates and whole-cell lysates were analyzed by SDS/PAGE and immunoblotting.
Immunoblotting.
After transfection, cells were washed with cold phosphate-buffered saline (PBS) and incubated on ice with Nonidet P-40 lysis buffer supplemented with protease inhibitors for 10 min. Cell lysates were then clarified by centrifugation at 16,873 × g for 20 min at 4 °C. Equal amounts of protein samples were diluted in 5× SDS/PAGE loading buffer and boiled for 5 min. Proteins were separated by SDS/PAGE and transferred onto polyvinylidene fluoride membranes (Sigma-Aldrich, IPVH00010). After blocking with PBS containing 5% nonfat milk powder and 0.05% Tween 20 (Sigma-Aldrich, P1397) for 2 h at room temperature, the membrane was incubated with specific primary antibodies overnight at 4 °C and then with the corresponding HRP-conjugated secondary antibodies at room temperature for 2 h at appropriate dilutions. The protein bands were visualized using a chemiluminescent HRP substrate (Sigma-Aldrich, WBKLS0500). Images of protein blots were obtained from an Amersham Imager 600 (GE Healthcare).
In Vitro Pull-Down Assays.
Amylose resin (New England Biolabs, E8022) was pre-equilibrated with buffer (10 mM MES [pH 7], 150 mM NaCl, 5 mM 2-mercaptoethanol) before adding the purified MBP-tagged proteins. Then the resin containing bait protein was incubated at 4 °C for 10 min, followed by three washes and subsequent resuspension in the buffer. The targeted prey proteins were applied to the resin and allowed to incubate at 4 °C for 20 min, and the resin was then washed five times and finally eluted at room temperature for 20 min in the above buffer containing 1% maltose (Sigma-Aldrich, M5895). Samples were taken at each step and visualized by Coomassie blue staining of SDS/PAGE.
HDX-MS Experiments.
All chemical reagents used in HDX-MS experiments were purchased from Sigma-Aldrich unless otherwise noted. Purified MBP-NS2 and copurified MBP-NS2/RIG-ICARDS were buffer-exchanged into 10 mM MES (pH 7), 150 mM NaCl, 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl). HDX was initiated by the addition of the same buffer solution except in pD 7.0 D2O (Cambridge Isotope Laboratories) to afford a final composition of 90/10% vol/vol D2O/H2O. After 10, 30, 60, 300, 900, 3,600, and 14,400 s of HDX at 4 °C, the reaction was quenched by 60% dilution with 3 M Urea and 1 mg protease from Aspergillus satoi type XIII in 1 x PBS, pH 2.2; the mixture was incubated at 37 °C for 3 min, and the samples were flash-frozen in N2(l). The thawed samples were injected onto a custom-built liquid chromatography assembly incorporating on-line digestion with a custom-packed pepsin column (2 mm × 20 mm), 3 min trapping and desalting on a ZORBAX Eclipse XDB C8 column (2.1 mm × 15 mm; Agilent) using water/0.1% trifluoroacetic acid and analytical separation using a 10.5-min gradient from 4 to 40% acetonitrile/0.1% formic acid on a Hypersil Gold C18 column (2.1 mm × 50 mm, Thermo Fisher). All valves, tubes, and columns (except for the pepsin column, which lost activity at low temperature) were submerged in an ice-water bath to minimize back-exchange. The peptides were then introduced via electrospray ionization into an LTQ fourier transform-ion cyclotron resonance (LTQ FT-ICR) mass spectrometer (Thermo Fisher). All experiments were conducted in duplicate unless otherwise indicated. Data analysis of the continuous HDX results was carried out with HDExaminer (version 2.5.1, Sierra Analytics). Peptide-level difference Woods' plots were drawn using absolute differences in deuterium uptake vs. the peptide coverage. Global significance limits were calculated using a two-tailed t test value (P < 0.01), and the SEM was calculated from the pooled SD of each state (80, 81).
HDX-MS Mapping Analysis.
Peptide mapping of the digests was done to provide a list of peptides that could be followed during HDX-MS data acquisition and analysis. Product-ion tandem mass spectrometry (MS/MS) spectra were collected in a data-dependent mode, in which the six most abundant ions from each scan were selected for MS/MS analysis. For the mapping of MBP-NS2, the MS/MS files from two parallel runs were analyzed using Byonic and Byologic (Protein Metrics). Searches were also carried out against a reverse sequence to discard ambiguous assignments.
X-Ray Crystallography.
Crystallization trials of purified NS2 and selenomethionine (SeMet)-labeled NS2 were initiated using commercially available screens (Hampton Research). Initial hits were obtained in 0.4 M K-Na-tartrate at a protein concentration of 5 mg/mL at room temperature. Conditions were optimized using in-house reagents to obtain diffraction quality crystals of ∼150 × 20 µm in size. Crystals were harvested by soaking in cryoprotectant composed of 0.8 M K-Na-Tartrate with 25% glycerol and vitrified in liquid nitrogen. Initial phases were obtained by single-wavelength anomalous diffraction of a SeMet NS2 crystal. Electron density was improved by density modification using Parrot (82) and automated model building using Buccaneer (83) to 3 Å. The NS2 structure was solved using MOLREP (84) of a native NS2 crystal to 2.8 Å. X-ray diffraction datasets for both crystals were collected at the Advanced Photon Source, Structural Biology Center at Sector 19-ID. HKL3000 was used to process datasets (85). Computational corrections for absorption in a crystal and imprecise calculations of the Lorentz factor resulting from a minor misalignment of the goniostat were applied (86, 87). Anisotropic diffraction was corrected to adjust the error model and to compensate for a radiation-induced increase of nonisomorphism within the crystal (88–90). The crystal data were merged to 2.8 Å resolution, but diffraction was highly anisotropic with diffraction in c direction reaching only ∼3.2 Å (<I>/<σ(I)>∼2 at 3.2 Å and ∼0.1 at 2.8 Å). Indexing, integration, and scaling indicated P6x symmetry. The space group ambiguity was resolved by analyzing systematic absences, and the choice between 62 and 64 was resolved during experimental phasing. The data-processing statistics are presented in SI Appendix, Table S1.
The native structure was solved with molecular replacement method using MOLREP (82) with Se-Met–based model as a search model. The solution was refined with REFMAC (91, 92) and then rebuilt and refined with REFMAC with NCS restraints. The final rounds of refinement for both structural models were performed by iteratively applying REFMAC refinement and manual rebuilding and corrections in COOT (93, 94). We also used TLS restraints in refinement (95) with the TLSMD server providing the best combination of TLS bodies (96). The results of refinement are summarized in SI Appendix, Table S1. The model quality was validated with Molprobity (97, 98).
SEC-MALS.
SEC-MALS experiments were performed using a DAWN-HELEOS II detector (Wyatt Technologies) coupled to a Superdex SD200 column (GE Healthcare) in buffer containing 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 7), 150 mM NaCl, and 2 mM TCEP. Samples (2 mg/mL) were injected and raw data were analyzed using ASTRA 6 software (Wyatt Technologies) to determine the weight averaged molecular mass. Protein concentrations were determined using the refractive index measured by an Optilab T-rEX (Wyatt Technologies) and a dn/dc = 0.185 mL3⋅g−1.
qRT-PCR of IFNβ mRNA.
Real-time qRT-PCR was used in the study. For measurement of targeted gene expression, total RNA was isolated using RNeasy Mini Kit (Qiagen, 74104) according to the manufacturer’s protocol. The following primer sets were used for the human IFNβ and GAPDH constant regions: IFNβ, CTAACTGCAACCTTTCGAAGC and GGAAAGAGCTGTAGTGGA‐GAAG; GAPDH, CGCAGAGCCTCGAGGAGAAG and ACAGGAGGACTTTGG‐GAACGAC. Quantitative RT-PCR was performed using the Power SYBR Green PCR Master Mix (Thermo Fishier Scientific, 4367659) on a StepOnePlus Real-Time PCR System (Thermo Fishier Scientific). Relative changes in mRNA levels of genes were assessed using the 2–ΔΔCT method and normalized to the housekeeping gene GAPDH.
Supplementary Material
Acknowledgments
We thank members of the D.W.L., G.K.A., and M.L.G. groups for assistance and support. This work was supported by NIH Grants R01AI107056 and R01AI40758 (to D.W.L.), Grant P01AI120943 (to G.K.A., D.W.L., and C.F.B.), and Grants R01AI114654, U19AI109945, and U19AI109664 (to C.F.B.). Mass spectrometry was supported by NIH Grants P41GM103422 and R24GM136766 (to M.L.G.). S.C. was funded in part by an American Heart Association Postdoctoral Fellowship (15POST25140009). D.W.L. is also funded partially by Children’s Discovery Institute Grant PDII2018702. X-ray data collection was performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, under US Department of Energy, Office of Biological and Environmental Research Contract DE-AC02-06CH11357.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020587118/-/DCSupplemental.
Data Availability
The atomic coordinates have been deposited in the Protein Data Bank, https://www.rcsb.org/ (PDB ID: 7LDK).
References
- 1.Collins P. L., Graham B. S., Viral and host factors in human respiratory syncytial virus pathogenesis. J. Virol. 82, 2040–2055 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths C., Drews S. J., Marchant D. J., Respiratory syncytial virus: Infection, detection, and new options for prevention and treatment. Clin. Microbiol. Rev. 30, 277–319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rima B.et al.; Ictv Report Consortium , ICTV virus taxonomy profile: Pneumoviridae. J. Gen. Virol. 98, 2912–2913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H., et al., Evaluation of recombinant respiratory syncytial virus gene deletion mutants in African green monkeys for their potential as live attenuated vaccine candidates. Vaccine 21, 3647–3652 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Le Nouën C., et al., Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc. Natl. Acad. Sci. U.S.A. 111, 13169–13174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng J., Lee S., Hotard A. L., Moore M. L., Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. MBio 5, e01704-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlender J., Bossert B., Buchholz U., Conzelmann K. K., Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74, 8234–8242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teng M. N., et al., Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74, 9317–9321 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehead S. S., et al., Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73, 3438–3442 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyapalle S., et al., Respiratory syncytial virus NS1 protein colocalizes with mitochondrial antiviral signaling protein MAVS following infection. PLoS One 7, e29386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling Z., Tran K. C., Teng M. N., Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 83, 3734–3742 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo M. S., Brazas R. M., Holtzman M. J., Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79, 9315–9319 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munir S., et al., Respiratory syncytial virus interferon antagonist NS1 protein suppresses and skews the human T lymphocyte response. PLoS Pathog. 7, e1001336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munir S., et al., Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J. Virol. 82, 8780–8796 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren J., et al., A novel mechanism for the inhibition of interferon regulatory factor-3-dependent gene expression by human respiratory syncytial virus NS1 protein. J. Gen. Virol. 92, 2153–2159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spann K. M., Tran K. C., Chi B., Rabin R. L., Collins P. L., Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J. Virol. 78, 4363–4369 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spann K. M., Tran K. C., Collins P. L., Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J. Virol. 79, 5353–5362 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng M. N., Collins P. L., Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J. Virol. 72, 5707–5716 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swedan S., Andrews J., Majumdar T., Musiyenko A., Barik S., Multiple functional domains and complexes of the two nonstructural proteins of human respiratory syncytial virus contribute to interferon suppression and cellular location. J. Virol. 85, 10090–10100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitko V., et al., Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-kappaB-dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 81, 1786–1795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H., Takahasi K., Fujita T., RIG-I-like receptors: Cytoplasmic sensors for non-self RNA. Immunol. Rev. 243, 91–98 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Leung D. W., Amarasinghe G. K., Structural insights into RNA recognition and activation of RIG-I-like receptors. Curr. Opin. Struct. Biol. 22, 297–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehwinkel J., Gack M. U., RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu B., Hur S., How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 12, 91–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brisse M., Ly H., Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front. Immunol. 10, 1586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goubau D., Deddouche S., Reis e Sousa C., Cytosolic sensing of viruses. Immunity 38, 855–869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato H., et al., Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Rehwinkel J., Reis e Sousa C., RIGorous detection: Exposing virus through RNA sensing. Science 327, 284–286 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Yoneyama M., Fujita T., RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227, 54–65 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Jiang F., et al., Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalinski E., et al., Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Luo D., et al., Structural insights into RNA recognition by RIG-I. Cell 147, 409–422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickey T. H., Song B., Pyle A. M., RNA binding activates RIG-I by releasing an autorepressed signaling domain. Sci. Adv. 5, eaax3641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu B., et al., Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Peisley A., et al., Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. U.S.A. 108, 21010–21015 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cadena C., et al., Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177, 1187–1200.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gack M. U., et al., TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Wies E., et al., Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38, 437–449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuniyoshi K., et al., Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity. Proc. Natl. Acad. Sci. U.S.A. 111, 5646–5651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan J., Li Q., Mao A. P., Hu M. M., Shu H. B., TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 6, 154–163 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Kawai T., et al., IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Meylan E., et al., Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Seth R. B., Sun L., Ea C. K., Chen Z. J., Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Xu L. G., et al., VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19, 727–740 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Liu P., et al., Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 81, 1401–1411 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loo Y. M., et al., Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82, 335–345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoboua F., Martel A., Duval A., Mukawera E., Grandvaux N., Respiratory syncytial virus-mediated NF-kappa B p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKK beta. J. Virol. 84, 7267–7277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scagnolari C., et al., Gene expression of nucleic acid-sensing pattern recognition receptors in children hospitalized for respiratory syncytial virus-associated acute bronchiolitis. Clin. Vaccine Immunol. 16, 816–823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grandvaux N., et al., Sustained activation of interferon regulatory factor 3 during infection by paramyxoviruses requires MDA5. J. Innate Immun. 6, 650–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lifland A. W., et al., Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 86, 8245–8258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramaswamy M., et al., Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344, 328–339 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Whelan J. N., Tran K. C., van Rossum D. B., Teng M. N., Identification of respiratory syncytial virus nonstructural protein 2 residues essential for exploitation of the host ubiquitin system and inhibition of innate immune responses. J. Virol. 90, 6453–6463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goswami R., et al., Viral degradasome hijacks mitochondria to suppress innate immunity. Cell Res. 23, 1025–1042 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swedan S., Musiyenko A., Barik S., Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J. Virol. 83, 9682–9693 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chatterjee S., et al., Structural basis for human respiratory syncytial virus NS1-mediated modulation of host responses. Nat. Microbiol. 2, 17101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holm L., Rosenstrom P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan Y. K., Gack M. U., Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 14, 360–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basler C. F., et al., The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U.S.A. 97, 12289–12294 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cárdenas W. B., et al., Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80, 5168–5178 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leung D. W., et al., Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 17, 165–172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bao X., et al., Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 4, e1000077 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan L., Briese T., Lipkin W. I., Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J. Virol. 84, 1785–1791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gack M. U., et al., Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5, 439–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei X., et al., The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J. Virol. 84, 8051–8061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santiago F. W., et al., Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J. Virol. 88, 4572–4585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siu K. L., et al., Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 284, 16202–16209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Motz C., et al., Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science 339, 690–693 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez K. R., Horvath C. M., Amino acid requirements for MDA5 and LGP2 recognition by paramyxovirus V proteins: A single arginine distinguishes MDA5 from RIG-I. J. Virol. 87, 2974–2978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H. L., et al., West Nile virus NS1 antagonizes interferon beta production by targeting RIG-I and MDA5. J. Virol. 91, e02396-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ban J., et al., Human respiratory syncytial virus NS 1 targets TRIM25 to suppress RIG-I ubiquitination and subsequent RIG-I-mediated antiviral signaling. Viruses 10, 716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu W., et al., The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J. Virol. 86, 7777–7789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y., Yang L., Wang H., Zhang G., Sun X., Respiratory syncytial virus non-structural protein 1 facilitates virus replication through miR-29a-mediated inhibition of interferon-α receptor. Biochem. Biophys. Res. Commun. 478, 1436–1441 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Tan Y. R., Peng D., Chen C. M., Qin X. Q., Nonstructural protein-1 of respiratory syncytial virus regulates HOX gene expression through interacting with histone. Mol. Biol. Rep. 40, 675–679 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Baños-Lara Mdel. R., Ghosh A., Guerrero-Plata A., Critical role of MDA5 in the interferon response induced by human metapneumovirus infection in dendritic cells and in vivo. J. Virol. 87, 1242–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gitlin L., et al., Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6, e1000734 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ikegame S., et al., Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 84, 372–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim W. K., et al., Deficiency of melanoma differentiation-associated protein 5 results in exacerbated chronic postviral lung inflammation. Am. J. Respir. Crit. Care Med. 189, 437–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yount J. S., Gitlin L., Moran T. M., López C. B., MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J. Immunol. 180, 4910–4918 (2008). [DOI] [PubMed] [Google Scholar]
- 79.Jiang X., et al., Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Houde D., Berkowitz S. A., Engen J. R., The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J. Pharm. Sci. 100, 2071–2086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weis D. D., Comment on Houde, D.; Berkowitz, S. A.; Engen, J. R., The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J. Pharm. Sci. 2011, 100, 2071–2086. J. Pharm. Sci. 108, 807–810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vagin A., Teplyakov A., Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Cowtan K., The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Cowtan K., Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solution: From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 87.Borek D., Minor W., Otwinowski Z., Measurement errors and their consequences in protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 59, 2031–2038 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Otwinowski Z., Borek D., Majewski W., Minor W., Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Borek D., Cymborowski M., Machius M., Minor W., Otwinowski Z., Diffraction data analysis in the presence of radiation damage. Acta Crystallogr. D Biol. Crystallogr. 66, 426–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borek D., Dauter Z., Otwinowski Z., Identification of patterns in diffraction intensities affected by radiation exposure. J. Synchrotron Radiat. 20, 37–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 92.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J., Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 (1999). [DOI] [PubMed] [Google Scholar]
- 93.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 94.Debreczeni J. E., Emsley P., Handling ligands with coot. Acta Crystallogr. D Biol. Crystallogr. 68, 425–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Winn M. D., Murshudov G. N., Papiz M. Z., Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Painter J., Merritt E. A., Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006). [DOI] [PubMed] [Google Scholar]
- 97.Davis I. W., et al., MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C., MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited in the Protein Data Bank, https://www.rcsb.org/ (PDB ID: 7LDK).