Significance
Mutations that activate LRRK2 protein kinase cause Parkinson's disease. LRRK2 phosphorylates a subset of Rab GTPases, in particular Rab8 and Rab10. We show here details related to the mechanism by which Rab10 phosphorylation blocks initiation of cilia formation, fundamental information related to how pathogenic LRRK2 interferes with normal cell physiology.
Keywords: LRRK2 kinase, Parkinson’s disease, Rab GTPase, primary cilia
Abstract
Mutations that activate LRRK2 protein kinase cause Parkinson's disease. We showed previously that Rab10 phosphorylation by LRRK2 enhances its binding to RILPL1, and together, these proteins block cilia formation in a variety of cell types, including patient derived iPS cells. We have used live-cell fluorescence microscopy to identify, more precisely, the effect of LRRK2 kinase activity on both the formation of cilia triggered by serum starvation and the loss of cilia seen upon serum readdition. LRRK2 activity decreases the overall probability of ciliation without changing the rates of cilia formation in R1441C LRRK2 MEF cells. Cilia loss in these cells is accompanied by ciliary decapitation, and kinase activity does not change the timing or frequency of decapitation or the rate of cilia loss but increases the percent of cilia that are lost upon serum addition. LRRK2 activity, or overexpression of RILPL1 protein, blocks release of CP110 from the mother centriole, a step normally required for early ciliogenesis; LRRK2 blockade of CP110 uncapping requires Rab10 and RILPL1 proteins and is due to failure to recruit TTBK2, a kinase needed for CP110 release. In contrast, deciliation probability does not change in cells lacking Rab10 or RILPL1 and relies on a distinct LRRK2 pathway. These experiments provide critical detail to our understanding of the cellular consequences of pathogenic LRRK2 mutation and indicate that LRRK2 blocks ciliogenesis upstream of TTBK2 and enhances the deciliation process in response to serum addition.
Activating mutations in Leucine rich repeat kinase 2 (LRRK2) kinase are responsible for inherited Parkinson’s disease. Pathogenic LRRK2 phosphorylates a subset of Rab GTPases (1, 2) that are master regulators of protein transport through the secretory and endocytic pathways (3, 4). Upon phosphorylation, these Rab GTPases bind to a new set of effector proteins (1), triggering likely diverse physiological events.
Together with the laboratories of Dario Alessi and Matthias Mann, we have shown that LRRK2 kinase activity blocks primary cilia formation in cultured cells (1) and in specific areas of mouse brains carrying pathogenic LRRK2 mutations (5). Inhibition of cilia formation requires Rab10 and a protein, RILPL1 (5), that binds with strong preference to phosphorylated Rab10 protein (1). This process is regulated by PPM1H phosphatase that is highly specific for pRab8A and pRab10 proteins (6). LRRK2-mediated Rab phosphorylation of Rab proteins has also been reported to alter centrosomal cohesion (7). Rab phosphorylation appears to be part of the normal process by which cilia formation is regulated, both in wild-type (WT) cells and even more so in cells expressing hyperactive mutant LRRK2 kinase, as depletion of PPM1H phosphatase in wild-type cells sufficient to block cilia formation (6).
Cilia growth and resorption are closely linked to the cell cycle. Upon serum starvation, removal of CP110 from the mother centriole is a key step in the initiation of cilia formation (8). Tau tubulin kinase 2 (TTBK2), a protein mutated in the inherited movement disorder spinocerebellar ataxia type 11, promotes the removal of CP110 and recruitment of IFT proteins to build the ciliary axoneme (8–10). TTBK2 is recruited selectively to the mother centriole by binding to CEP164 (11, 12). Huang et al. (13) showed that TTBK2 triggers ciliogenesis by phosphorylating MPP9, leading to MPP9 degradation and subsequent release of CP110/CEP97. TTBK2 also binds and phosphorylates CEP83 (14), further facilitating CP110 release and cilia formation.
We have carried out live-cell microscopy experiments to investigate the mechanisms by which LRRK2 activity inhibits cilia formation in cell culture and find that the blockade is early, upstream of TTBK2 recruitment to the mother centriole.
Results
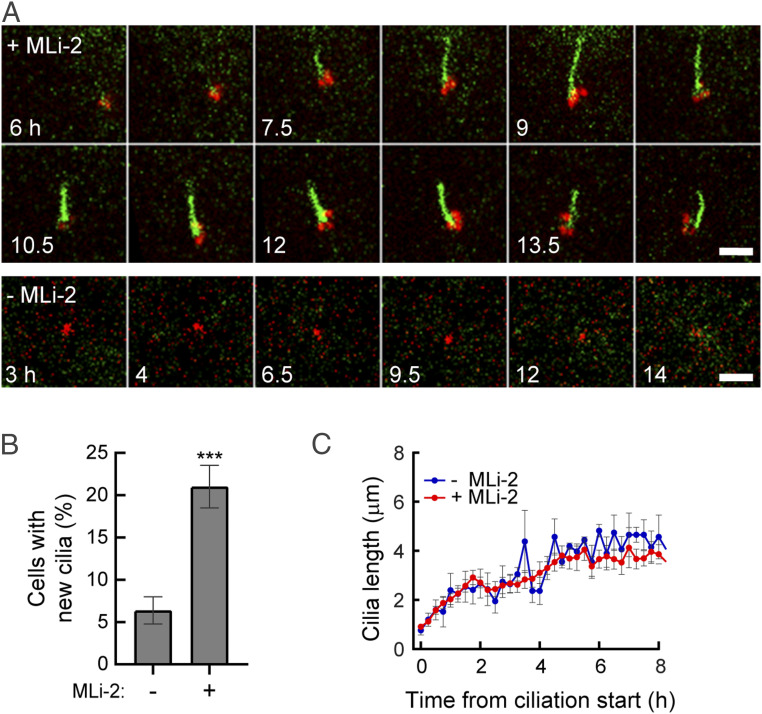
To better characterize the step(s) of cilia formation or disassembly altered by LRRK2 mediated Rab10 phosphorylation, we carried out live-cell video microscopy of knock-in mouse embryonic fibroblast (MEF) cells expressing pathogenic R1441C LRRK2 at endogenous levels; these cells were engineered to stably express the cilia-localized G protein coupled receptor, Somatostatin Receptor 3 (SSTR3) tagged with green fluorescent protein (GFP) (15) and a pericentrin-AKAP-450 centrosomal targeting domain [PACT, (16)] tagged with monomeric kusabira-orange 2 (mKO2) (red in Fig. 1A). To monitor cilia formation, cells were transferred to a serum-free medium and filmed for 8 h, with or without a LRRK2 specific inhibitor, MLi-2, with images captured every 15 min (Movies S1 and S2). We tracked cilia using SSTR3 instead of the common marker Arl13b since exogenous expression of Arl13b elongated primary cilia and altered their stability.
Fig. 1.
Pathogenic LRRK2 activity decreases the probability of cilia formation. (A) Time-lapse images of R1441C MEF cells stably expressing SSTR3-GFP (green) and mKO2-PACT (red). After 2 h of serum starvation ± MLi-2, cells were imaged every 15 min. In this and subsequent figures, intermediate time images were removed from the figure to conserve space. (Scale bar, 5 µm.) (B) Percentage of cells that generated primary cilia ± MLi-2 over 8 h of imaging. Values represent the mean ± SEM from three independent experiments, each with >20 cells. Significance was determined by the unpaired t test; ***P = 0.0026. (C) Rate of cilia growth. Lengths of cilia were measured as SSTR3+ cilia appeared (t = 0). Values represent the mean ± SEM from 3 to 7 (−MLi-2) and 9 to 21 (+MLi-2) cilia (cilia numbers were different at different time points over 8 h).
Addition of the LRRK2 inhibitor MLi-2 significantly increased the number of cells that generated a cilium under these low confluency conditions (22% versus 7%), as expected (Fig. 1 A and B). Focusing only on cells that grew cilia, the rate of cilia growth was indistinguishable in the presence or absence of LRRK2 inhibitor, plateauing after about 3 h of serum starvation (Fig. 1C). If a cell ciliated, the rate of growth was the same, independent of LRRK2 activity. Thus, pathogenic LRRK2 influences the probability that a cell will initiate cilia growth upon serum starvation.
To monitor cilia loss from LRRK2 mutant cells, cilia formation was first initiated by serum starvation (∼24 h), and then cells were transferred to serum containing medium ± MLi-2 (Fig. 2 A and C and Movie S3). This protocol enabled us to monitor the properties of the smaller proportion of cilia formed despite the presence of mutant LRRK2 activity (Fig. 2B); the process by which these cilia were lost could then be followed with or without active kinase. As previously reported, deciliation in MEFs includes “ectocytosis” or decapitation from the tips of cilia (17–19); yellow arrowheads in Fig. 2C).
Fig. 2.
Cilia loss upon serum readdition is enhanced by pathogenic LRRK2 activity (A) Timeline of this imaging experiment. (B) Probability of cilia loss during imaging is shown. Values represent the mean ± SEM from three independent experiments each containing >7 cells. Significance was determined by the unpaired t test; *P = 0.029. (C) Time-lapse images of R1441C LRRK2 MEF cells stably expressing SSTR3-GFP. Cells were serum starved for 24 h and cilia loss was then triggered by serum readdition ± MLi-2. After 2 h, cells were imaged every 15 min. Arrowheads indicate decapitated vesicles. (Scale bar, 5 µm.) (D) Individual bars indicate single cilia. Heights of the bars (red, +MLi-2 and blue, −MLi-2) represent the time at which a given cilium disappeared. Horizontal black bars indicate decapitation events. 27 MLi-2+ cilia and 11 MLi-2− cilia remained; 11 MLi-2+ cilia and 18 MLi-2− cilia disappeared during the imaging period. (E) Percentage of cilia that displayed decapitation events. Significance was determined by the unpaired t test; P = 0.94. (F) Average timing of events. Times of first decapitation for each cilium (P = 0.80), all decapitations (P = 0.45), and cilia loss (P = 0.85) are shown. Values represent the mean ± SEM. Significance was determined by the unpaired t test. (G) Rates of cilia loss. Lengths of cilia that disappeared during imaging were measured. Values represent the mean ± SEM from 3 to 18 (−MLi-2) and 3 to 11 (+MLi-2) cilia.
Each bar in Fig. 2D represents the lifetime of an individual cilium, with horizontal lines indicating the timing of decapitation events. The left-most bars in each set (red or blue) indicate cilia that were stable over the imaging time frame; the adjacent bars of decreasing height represent cilia with the indicated, shorter lifetimes (<8 h). Note that in the absence of MLi-2 (blue bars), there were fewer stable cilia and proportionately more cilia loss; the percent of cilia displaying decapitation was similar between treated and untreated cells (Fig. 2E). As quantified in Fig. 2F, we detected no significant difference in terms of the times at which a cilium displayed its first decapitation event ± MLi-2. In addition, the total number of subsequent decapitation events and their timing were similar, with or without LRRK2 activity (Fig. 2F). Thus, LRRK2 does not appear to influence the overall timing or pathway of decapitation. Nevertheless, LRRK2 activity increased the proportion of cells that lost their cilia over an 8 h observation window (Fig. 2 B and C). For those cells that lost cilia, the rate of loss was similar ± MLi-2 (Fig. 2G; >9 cells averaged for each curve). What differed was the overall probability of deciliation within the first 8 h of serum addition, which was 64% without inhibitor and 29% with inhibitor (Fig. 2B). LRRK2 activity led to a larger proportion of cells losing their cilia, as if kinase activity facilitated events that trigger cilia loss upon serum addition.
In control experiments, cilia loss was monitored in LRRK2 R1441C MEF cells compared with MEF cells harboring WT LRRK2 (SI Appendix, Fig. S1A). After 24 h of serum starvation, WT MEF cells had more cilia than R1441C LRRK2 cells as reported previously (1, 5). Ten hours after serum readdition, MLi-2 treated R1441C LRRK2 MEF cells had a similar number of cilia as WT cells; without MLi-2, cilia numbers were significantly reduced. Importantly, the behavior of MLi-2 treated R1441C MEF cells matched that of wild-type MEF cells. In addition, cilia loss probability for WT cells was unaltered by the presence of MLi-2 (SI Appendix, Fig. S1B). These data indicate that enhanced cilia loss is induced by hyperactive mutant LRRK2.
LRRK2 reduces cilia number via Rab10 phosphorylation and strong binding of pRab10 to RILPL1; both Rab10 and RILPL1 are essential for LRRK2’s effects (1, 5). We therefore examined whether Rab10 or RILPL1 are needed for LRRK2-enhanced cilia loss. Rab10 and RILPL1 were depleted from R1441C LRRK2 MEF cells by short hairpin RNA (shRNA) encoding lentiviruses (SI Appendix, Fig. S1C). These cells were serum starved for 24 h in the absence of MLi-2 to induce cilia formation (Fig. 3 A and B). Serum was then added, and cells were cultured further ± MLi-2; deciliation was monitored after 4, 7, or 10 h (Fig. 3 A and C). Consistent with data presented in Fig. 2, MLi-2 treatment stabilized cilia in control R1441C LRRK2 MEF cells even after 7 or 10 h incubation. Importantly, the ability of mutant LRRK2 activity to enhance cilia loss did not require Rab10 or RILPL1 (Fig. 3 C and D). Thus, LRRK2 activity enhances cilia loss after serum readdition, by a Rab10- and RILPL1-independent pathway.
Fig. 3.
Rab10 and RILPL1 do not contribute to LRRK2-mediated, enhanced cilia loss upon serum readdition. (A) Timeline of this imaging experiment. (B) Percentage of cells with primary cilia at time = 0. Values represent the mean ± SEM from four independent experiments, each containing >40 cells. Significance was determined by repeated measures one-way ANOVA with Dunnett post hoc test (control versus shRab10: P = 0.047, control versus shRILPL1: P = 0.159). (C and D) Percentage of remaining cilia compared with time = 0 (100%) after serum readdition. Line plot (C) and detailed bar graph for each time point (D) are shown. Significance was determined by repeated measures one-way ANOVA followed by Tukey’s multiple comparisons test (at 4 h, control +MLi-2 versus shRab10 [P = 0.037]; at 7 h, control +MLi-2 versus control −MLi-2 [P = 0.009], control +MLi+2 versus shRab10 [P = 0.024]; at 10 h, control +MLi-2 versus control −MLi-2 [P = 0.037], control +MLi-2 versus shRab10 [P = 0.022], and control +MLi+2 versus shRILPL1 [P = 0.029]).
Cilia dynamics are tightly linked to the cell cycle. Cilia are formed during G0/G1 phase and resorbed before mitosis begins (20). We next tested whether LRRK2 hyperactivity accelerated cell cycle reentry to enhance cilia loss after serum stimulation. After 24 h serum starvation to trigger cilia formation, cells were treated with serum to initiate cilia loss ± MLi-2 (for 8 h). Cell cycle status was then determined using propidium iodide staining flow cytometry (SI Appendix, Fig. S2 A and B). Eight hours after serum readdition, at a point in time when enhanced cilia loss was clearly detected (Fig. 2 D and F), most cells were still in G1/G0 phase and had not yet progressed further in the cell cycle, with or without MLi-2 treatment. Thus, enhanced cilia loss is not caused by altered kinetics of reentry into the cell cycle.
MLi-2 washout restores LRRK2 activity very rapidly, with pRab10 levels increasing within 30 min and cilia loss detected within hours (SI Appendix, Fig. S3 A and B). We next examined the deciliation process for cilia that were formed without kinase activity (+MLi-2) during serum starvation, followed by observation with or without LRRK2 inhibition in the continued absence of serum (SI Appendix, Fig. S4 A and B and Movies S4 and S5). This protocol examines the intrinsic stability of cilia after LRRK2 reactivation by MLi-2 washout. LRRK2 activity had no influence on the percent of cells with persistent cilia (SI Appendix, Fig. S4C), the percent of cells displaying decapitation (SI Appendix, Fig. S4D), the stability of cilia length (SI Appendix, Fig. S4E), the time of first decapitation, or the overall frequency of decapitations (SI Appendix, Fig. S4F). These experiments confirm that the loss of cilia seen in the presence of pathogenic LRRK2 activity relates to the process by which serum addition initiates cilia loss.
We also compared cilia loss rates for cilia formed in the presence (SI Appendix, Fig. S4G open bars) or absence (SI Appendix, Fig. S4G filled bars) of pathogenic LRRK2 activity. Under these conditions, we did not detect differences in enhanced cilia loss probability (SI Appendix, Fig. S4G) or timing of decapitation or deciliation (SI Appendix, Fig. S4H).
Similar findings were obtained if cilia from R1441C LRRK2 MEFs were generated with or without LRRK2 activity in the continued absence of serum (SI Appendix, Fig. S5 and Movie S6). Cilia that formed despite the presence of pathogenic kinase activity were just as persistent as those formed in the presence of kinase inhibitor over an 8 h observation period (SI Appendix, Fig. S5 A–C). Again, cells showed a similar fraction of total cilia displaying decapitation (SI Appendix, Fig. S5D) and comparable timing of first and total decapitation events (SI Appendix, Fig. S5E). Altogether, these data show that pathogenic LRRK2 influences cilia formation propensity and sensitivity to serum triggering the cilia resorption and loss process.
Mechanism of Ciliation Blockade.
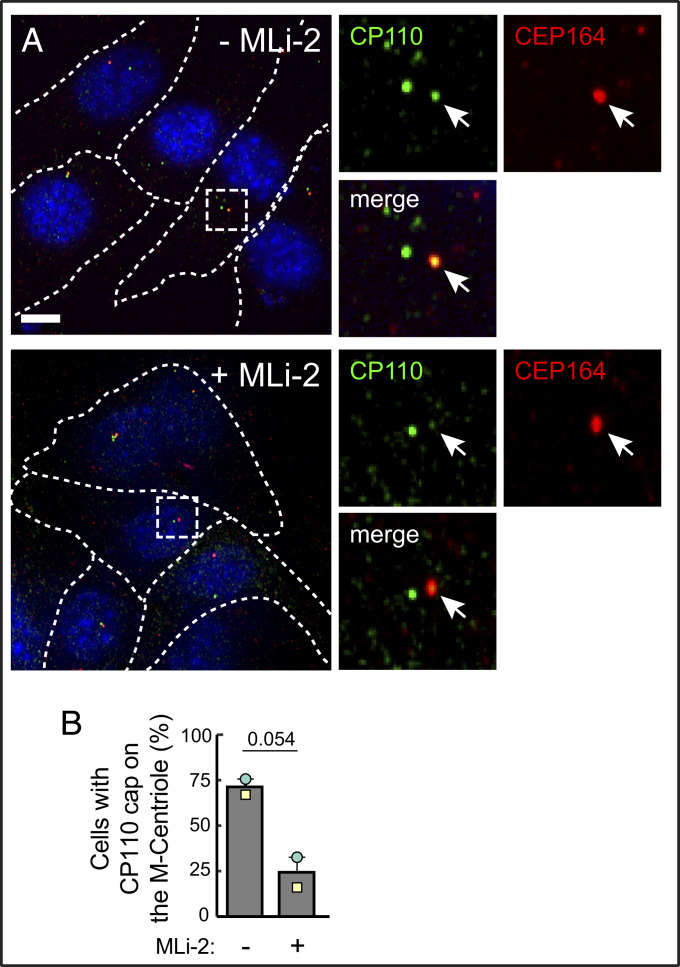
Removal of CP110 from the mother centriole is a key step in the initiation of cilia formation (8, 21). CP110 is recruited to the distal end of the centriole by binding to CEP97; the CP110-CEP97 complex is recruited to centrioles by MPP9, and together they suppress microtubule assembly (13). We therefore explored CP110 status at the mother centriole to check its relationship to the decreased probability of ciliogenesis in cells expressing pathogenic LRRK2.
Fig. 4 shows that in the presence of pathogenic LRRK2, upon overnight serum starvation conditions that reveal a ciliogenesis defect, both mother and daughter centrioles were capped with CP110 protein in ∼75% of cells. Treatment with MLi-2 to inhibit LRRK2 kinase reversed this block, enabling CP110 release from the mother centriole in 77% of cells within 24 h of treatment.
Fig. 4.
Pathogenic LRRK2 activity blocks release of CP110 from the mother centriole. (A) R1441C LRRK2 MEF cells were starved for 24 h ± MLi-2. Cells were then fixed with cold methanol and stained with rabbit anti-CP110 (green) and mouse anti-CEP164 (red) antibodies. Smaller images at right are enlarged regions boxed in the larger images at left. (B) Percent of cells harboring a CP110 cap at the mother (M) centriole, determined by CEP164 staining. Values represent the mean and SEM of two independent experiments (n > 30 in each experiment). Significance was determined by the paired t test, P = 0.054. (Scale bars, 10 µm.)
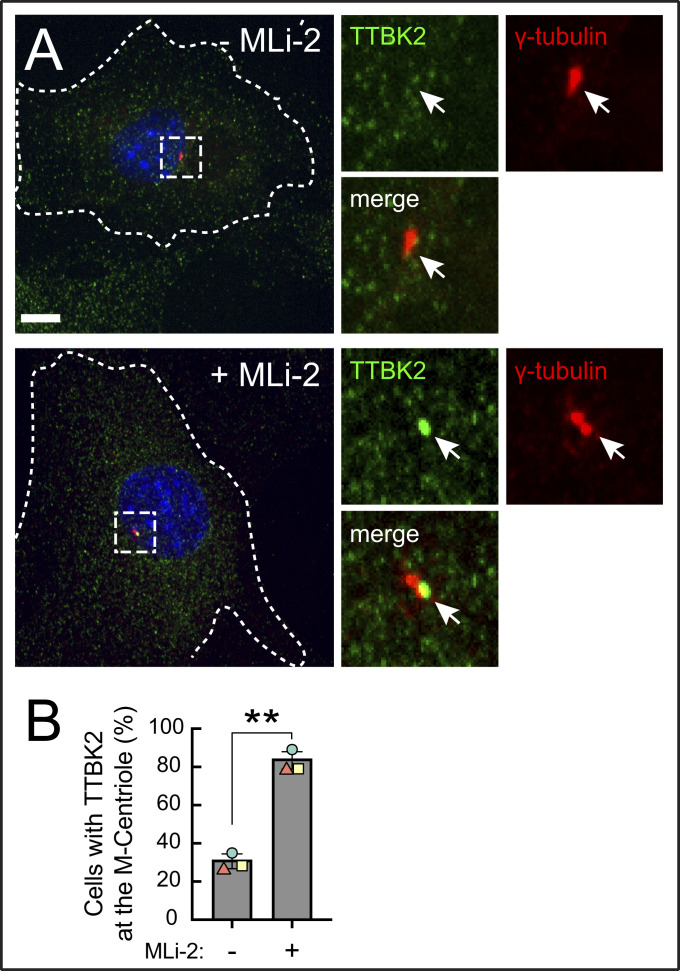
To test if pathogenic LRRK2 influenced the recruitment of TTBK2 to the mother centriole, we stably introduced GFP tagged TTBK2 at barely detectable levels into R1441C LRRK2 MEF cells and monitored its recruitment to the mother centriole upon serum starvation. (Endogenous TTBK2 was not detectable by immunofluorescence staining or by Western blot using a commercial antibody in this cell type). In addition to R1441C LRRK2 expression blocking release of CP110 from mother centrioles in MEF cells (Fig. 4), hyperactive LRRK2 also blocked the recruitment of TTBK2 to the centriolar region marked by gamma-tubulin (Fig. 5) and, more specifically, to the mother centriole, marked by distal appendage protein CEP164 (Fig. 6). In contrast, in cells treated overnight with the LRRK2-specific MLi-2 inhibitor, 90% of cells showed a bright TTBK2 spot at the mother centriole, as expected for cells in which CP110 is released (Fig. 6B).
Fig. 5.
Pathogenic LRRK2 activity blocks TTBK2 recruitment to the mother centriole. (A) R1441C LRRK2 MEF cells stably expressing GFP-TTBK2 were starved for 24 h ± MLi-2. Cells were then fixed with cold methanol and stained with rabbit anti-TTBK2 (green) and mouse anti-γ-tubulin (red) antibodies. At right are enlarged regions boxed in the larger images at left. (B) Percent of cells with TTBK2 at the centrosome, marked by γ-tubulin staining. Values represent the mean and SEM of three independent experiments (n > 60 in each experiment). Significance was determined by the paired t test; *P = 0.004. (Scale bars, 10 µm.)
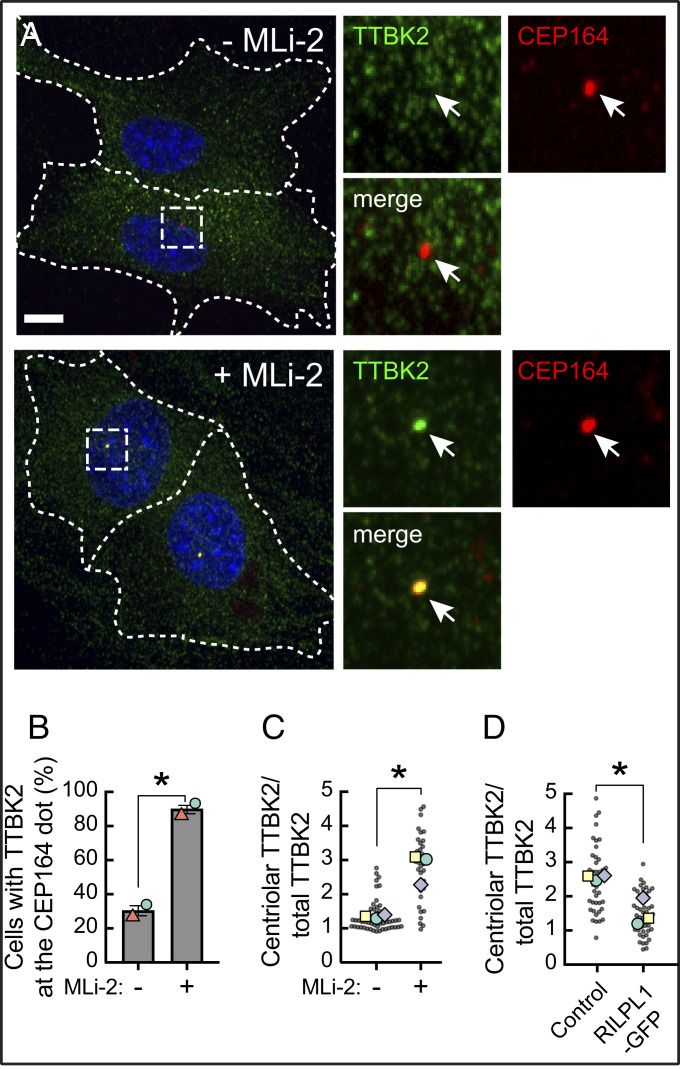
Fig. 6.
Pathogenic LRRK2 activity blocks TTBK2 recruitment without affecting CEP164. (A) R1441C LRRK2 MEF cells stably expressing GFP-TTBK2 were starved for 24 h with 200 nM MLi-2 or DMSO. Cells were then fixed with cold methanol and stained with rabbit anti-TTBK2 (green) and mouse anti-CEP164 (red) antibodies. At right are enlarged regions boxed in the larger images at left. (B) Percent of cells with TTBK2 at the mother centriole, marked by CEP164 staining, by visual inspection. Values represent the mean and SEM of two independent experiments (n > 30 in each experiment). Significance was determined by the paired t test; *P = 0.034. Colored shapes represent the mean of each separate experiment. (C) Intensity of TTBK2 staining over the centriole as a function of its overall expression in cells. Each dot represents the ratio of mean TTBK2 signal intensity at the centriolar area divided by the mean signal intensity throughout the cell (n > 30). Colored shapes represent the mean of each separate experiment. P = 0.036. (D) Effect of RILPL1 expression on TTBK2 levels at the mother centriole. RPE cells were transfected with RILPL1-GFP; its expression was induced with 1 µg/mL doxycycline, and cells were serum starved for 24 h, methanol fixed, and stained for endogenous TTBK2 and CEP164. Cells were then imaged and analyzed as in C. Each dot represents the ratio of mean TTBK2 signal intensity at the centriolar area divided by the mean signal intensity throughout the cell (n > 40). Colored shapes represent the mean of each experiment. Significance was determined by the paired t test, P = 0.034. (Scale bars, 10 µm.)
In untreated R1441C LRRK2 MEF cells, we often saw a weak signal of TTBK2 adjacent to gamma-tubulin positive centrioles, which we ascribe to the established localization of TTBK2 to the plus ends of microtubules (22, 23). Lo et al. (14) also report a scattered radial signal of TTBK2 around the mother centriole in cells without serum starvation. Thus, in addition to manual scoring centriolar staining, we quantified the actual enrichment of TTBK2 over the mother centriole (marked by CEP164) using CellProfiler (24). Using an unbiased image analysis pipeline, we detected an average twofold enrichment of TTBK2 over the mother centriole upon MLi-2 treatment (Fig. 6C; same conditions as in Figs. 5A and 6A).
We have shown previously that LRRK2-phosphorylated Rab10 blocks ciliation in concert with its binding partner, RILPL1; RILPL1 overexpression is also sufficient to dominantly block cilia formation in the absence of pathogenic LRRK2 (5). Since a portion of RILPL1 localizes adjacent to the mother centriole (5, 25), we tested whether RILPL1 overexpression can also influence TTBK2 recruitment. In wild-type retinal pigment epithelium (RPE) cells where (unlike MEFs) it was possible to quantify endogenous TTBK2 at the CEP164-marked mother centriole, expression of RILPL1-GFP reduced TTBK2 recruitment to the mother centriole twofold compared with control cells (Fig. 6D and SI Appendix, Fig. S6). Thus, exogenous expression of RILPL1-GFP in wild-type cells phenocopies the effect of pathogenic LRRK2 on centriolar TTBK2 recruitment.
Early membrane trafficking events that include recruitment of distal appendage vesicles and ciliary vesicles are critical for axoneme extension (26). EHD1 is a membrane-associated protein needed for both ciliary vesicle formation and the release of CP110 from the distal end of mother centriole (27). Despite the retention of CP110 at the mother centriole in mutant LRRK2 expressing cells, we found that most cells retained EHD1 staining near centrosomes, and centrosomal EHD1 staining levels did not change upon LRRK2 inhibition (SI Appendix, Fig. S7). These data suggest that the ability of LRRK2 to block CP110 release from the mother centriole is independent of EHD1 recruitment to the centrosomal region and is more closely linked to failure of TTBK2 recruitment rather than to a block in the ciliary vesicle fusion pathway.
Hyperactive LRRK2 decreases cilia number via Rab10 phosphorylation and RILPL1, which binds phosphoRab10 with high affinity (1, 5). Thus, we tested whether the LRRK2 blockade of CP110 release and TTBK2 recruitment require endogenous Rab10 or RILPL1 proteins. As shown in Fig. 7, knockdown of Rab10 or RILPL1 proteins in R1441C MEF cells decreased the amount of CP110 on the mother centriole (Fig. 7 A and B).
Fig. 7.
Loss of Rab10 or RILPL1 restored CP110 release and TTBK2 recruitment in R1441C LRRK2 cells. (A) R1441C LRRK2 MEF cells depleted of Rab10 or RILPL1 (SI Appendix, Fig. S1) were serum starved for 24 h ± MLi-2. Cells were stained with rabbit anti-CP110 (green) and mouse anti-CEP164 (red) antibodies. Individual cells are outlined; enlarged regions are boxed and shown below. (B) Quantitation of CP110 intensity at the mother (M) centriole compared to the daughter (D) centriole. Colored shapes represent the mean of each separate experiment. Significance was determined by the repeated measures one-way ANOVA with Dunnett post hoc test. Scr versus shRab10 (P = 0.028), Scr versus shRILPL1 (P = 0.018). (C) Percent of cells with a eGFP-TTBK2 dot at the mother centriole, marked by CEP164 staining and scored visually. Values represent the mean and SEM of two to five independent experiments (n > 15 in each experiment). Colored shapes represent the mean of each separate experiment. Significance was determined by the ordinary one-way ANOVA with Dunnett post hoc test. shScr −MLi-2 versus shScr +MLi-2 (P < 0.001), shScr −MLi-2 versus shRab10 −MLi-2 (P < 0.001), shScr −MLi-2 versus shRab10 +MLi-2 (P < 0.001), shScr −MLi-2 versus shRILPL1 −MLi-2 (P = 0.002), and shScr −MLi-2 versus shRILPL1 +MLi-2 (P = 0.003). (D) Intensity of TTBK2 staining over the centriole as a function of its overall expression in cells. Each dot represents the ratio of mean TTBK2 signal intensity at the centriolar area divided by the mean signal intensity throughout the cell (n > 15). Colored shapes represent the mean of each experiment. Significance was determined by the ordinary one-way ANOVA with Dunnett post hoc test. shScr −MLi-2 versus shScr +MLi-2 (P = 0.038), shScr −MLi-2 versus shRab10 −MLi-2 (P = 0.011), shScr −MLi-2 versus shRab10 +MLi-2 (P = 0.026), shScr −MLi-2 versus shRILPL1 −MLi-2 (P = 0.077), and shScr −MLi-2 versus shRILPL1 +MLi-2 (P = 0.064). (Scale bar, 10 µm.)
Cells stably expressing eGFP-TTBK2 were also serum starved ± MLi-2. As shown in Fig. 6, control cells without MLi-2 had significantly less TTBK2 at the mother centriole, but TTBK2 recruitment was restored upon MLi-2 treatment. In Rab10- or RILPL1-depleted cells, TTBK2 recruitment was the same as that seen in MLi-2 treated control cells. Moreover, MLi-2 treatment appeared to have little if any additional effect in Rab10/RILPL1 depleted cells (Fig. 7 C and D). Altogether, these results indicate that pathogenic LRRK2 blocks cilia formation via Rab10 phosphorylation and RILPL1 binding.
Discussion
We have shown here that pathogenic LRRK2 decreases the probability that a cilium will form under conditions of serum starvation and increases the probability of cilia loss in mouse embryo fibroblasts. This is because when cells are starved to initiate ciliogenesis, TTBK2 fails to be recruited to the mother centriole in the presence of pathogenic LRRK2 activity, and Rab10 and RILPL1 are needed for this blockade. Under these conditions, CP110 continues to cap both mother and daughter centrioles, thereby inhibiting axoneme extension. When deciliation was examined by live-cell microscopy, we found that R1441C LRRK2 MEF cells lost cilia with increased probability by a process of retraction and ectocytosis or decapitation. We saw no effect of LRRK2 activity on the timing of the first decapitation event or the number or timing of all subsequent decapitation events. Moreover, LRRK2 did not influence the rate of cell cycle progression up to the time at which cilia loss was detected. Rather, LRRK2 mutant-expressing cells seem more sensitive to serum-triggered initiation of deciliation. Our study showed further that Rab10 and RILPL1 are not needed for the initiation of cilia loss, strongly suggesting that this process occurs by an independent, LRRK2-mediated pathway.
CP110 is a negative regulator of cilia formation that functions with CEP97 to restrict the ciliogenesis program to the appropriate stage of the cell cycle (21). Multiple cellular components drive CP110 release from the mother centriole prior to microtubule extension via TTBK2 recruitment and the formation of a ciliary vesicle. Although EHD1 is essential for ciliary vesicle-associated decapping (26), we saw no apparent change in EHD1 accumulation adjacent to the mother centriole. This suggests that the inability to recruit TTBK2 to the mother centriole and to interact with CEP164 is the predominant pathway by which primary cilia formation is blocked in LRRK2 mutant MEF cells.
We had previously shown that LRRK2 generated, phospho-Rab10 GTPase and its novel binding partner RILPL1 are both required for the process by which LRRK2 blocks cilia (5). RILPL1 is centriolar (25), as are complexes of pRab10 and RILPL1 protein (5). In addition, overexpressed RILPL1 is localized in part to the centriole, even in cells lacking Rab10 protein (5). Indeed, we found here that simple overexpression of RILPL1 that phenocopies mutant LRRK2 expression (5) also decreases TTBK2 recruitment to the mother centriole. Our data support a model in which the pRab10-RILPL1 complex acts as a dominant-negative inhibitor of TTBK2 recruitment, possibly by occluding interaction of this kinase with CEP164. Other recent work from our laboratory has shown that pRab10 sequesters myosin Va at the mother centriole together with RILPL2, perhaps interfering with myosin Va’s essential role in ciliogenesis (28).
The precise signals that lead to TTBK2 recruitment to the mother centriole and regulate its subsequent interaction with CEP164 are still unclear. Xu et al. (29) presented a model in which phosphatidylinositol 4-phosphate (PtdIns(4)P) levels adjacent to the mother centriole regulate the interaction of TTBK2 with CEP164. INPP5E phosphatase generates PtdIns(4)P from PtdIns(4,5)P2 adjacent to the mother centriole; PIPKI gamma reconverts the PtdIns(4)P to PtdIns(4,5)P2. It is possible that LRRK2 phosphorylation changes the levels of phosphoinositide kinases and phosphatases present at the mother centriole to impact this interaction. In this regard, OCRL and INPP5B are both Rab8 effectors that may be lost upon Rab8 phosphorylation, as phosphorylation decreases the ability of Rab8 to bind either of these proteins (2). Future experiments will enable us to explore these various scenarios in deeper mechanistic detail.
Materials and Methods
Plasmids.
DNA constructs were amplified in Escherichia coli DH5α and purified using mini prep columns (Enzymax). DNA sequence verification of all plasmids was performed by Sequetech (www.sequetech.com). PACT-mKO2 was generated by amplifying the PACT domain of pericentrin from complementary DNA of RPE cells using the primers cggatatccatcacactggcggccgca-tggaccccggccggct and ttgatccctcgatgttaactctagattaatcatcgggtggcaggatttct and cloned into Not1/Xba1 linearized lentiviral vector CSII-EF-mKO2 (gift of Aaron Straight, Stanford) using Gibson assembly cloning. The PACT domain was determined as described by Gillingham and Munro (16) . mApple-SSTR3-N-17 was from Addgene (#54949). SSTR3 was shuttled into CSII-EF lentiviral vector into the XhoI/XbaI site by Gibson cloning using the primers aacacgcta-ccggtctcgagaattcatggccactgttacctatcctt and tgctcaccatcagatggctcagtgtgctgg for SSTR3 and gagccatctgatggtgagcaagggcgag and ttgatccctcgatgttaactctag-attacttgtacagctcgtccatgcc for the GFP sequence from the eGFP-N1 vector. eGFP-TTBK2 (DU17481) was obtained from the Medical Research Council Protein Phosphorylation and Ubiquitylation Unit and cloned into lentiviral vector pMCB306 into the added restriction site AscI/EcoRI by Gibson assembly using primers cgagctgtacaaggggtccggactcagatccac and attaggtccctcgacgctatctgctgagtttactggctgg. RILPL1-GFP was cloned into pcDNA5D-TO and expression induced with doxycycline (1, 5).
Antibodies and Reagents.
For immunostaining, rabbit anti-TTBK2 (Sigma, HPA018113, 1:2,000), mouse anti-Arl13b (NeuroMab, 75–287, 1:2,000), mouse anti–gamma-tubulin (Invitrogen, MA1-19421, 1:2,000), rabbit anti-CP110 (Proteintech, 12780–1-AP, 1:2,000), mouse anti-CEP164 (Santa Cruz, sc-515403, 1:1,000), rabbit anti-EHD1 (Abcam, ab109747, 1:1,000), and chicken anti-GFP (Aves, GFP1020, 1:5,000) were used. For Western blotting, mouse anti-Rab10 (Abcam clone 4E2, ab104859, 1:1,000), rabbit anti-phospho(T72) Rab10 (Abcam, MJFR-22–5, ab230261, 1:1,000), rabbit anti-RILPL1 (Sigma, HPA-014314 1:500), rabbit anti-LRRK2 (Abcam, UDD3, ab133518, 1:1,000), mouse anti-alpha tubulin (Sigma, DM1a, 1:4,000), and MLi-2 was obtained from Tocris Bioscience (catalog number 5756) and dissolved in dimethyl sulfoxide (DMSO) to 1 mM stock and kept in −20 °C.
Cell Culture, Transfection, and Viral Transduction.
MEFs derived from WT or LRRK2-R1441C knock-in mice, control (scramble shRNA), Rab10 knockdown, and RILPL1 knockdown cells were created as described (1, 5). Cells were cultured in Dulbecco’s Modified Eagle Medium (Thermo Fisher Scientific, catalog number 12100046) containing 10% fetal bovine serum and penicillin (100 U/mL)/streptomycin (100 µg/mL). MEF and RPE cells were transfected with Lipofectamine 3000 according to the manufacturer. Lentivirus particles were produced in 293T cells and employed as described previously (5). After lentiviral transduction, cells were sorted on a Sony SH800Z cell sorter to collect the lowest expressing cells to avoid overexpression artifacts. In the case of GFP-TTBK2, viral transduced cells were selected using puromycin selection. RILPL1-GFP was induced by 1 µg/mL doxycycline 24 h after transfection.
Cell Cycle Analysis.
MEF R1441C cells were starved for 24 h in cell culture media without serum. Media containing 10% fetal bovine serum (FBS) with 200 nM MLi-2 or DMSO was added and cells were trypsinized and collected at indicated times. Cells were fixed with 70% ethanol at −20 °C for 15 min. After rehydrating cells with phosphate-buffered saline (PBS) for 15 min at room temperature (RT), cells were stained with 5 µg/mL propidium iodide (MP Biomedicals) with 100 µg/mL of RNase A (Sigma). Cells were sorted using a Flow Cytometer (BD Accuri C6) and analyzed with FlowJo software v10. Graphs were prepared using Illustrator software.
Live Imaging.
MEF R1441C cells were plated on 8-well dishes at 3 × 104 cells per each well (Nunc Lab-Tek II Chambered Coverglass from Thermo Fisher Scientific). Cells were cultured overnight with complete media including 10% FBS. To observe cilia formation, cells were washed three times and transferred to Leibovitz’s L-15 Medium, no phenol red (Thermo Fisher Scientific) with 200 nM MLi-2 or DMSO for 2 h before imaging. For monitoring cilia loss, cells were washed with PBS three times and cultured with complete media lacking serum for 20 h. Then cells were transferred to L-15 Medium with 10% FBS ± MLi-2; after a 2 h culture, cells were imaged for 8 h. For MLi-2 washout, ciliated cells were washed and transferred to L-15 Medium ± MLi-2. Images were captured using MetaMorph software at every 15 min in 37 °C (PeCon Tempcontrol 37-2) using a confocal spinning disk system (Leica DMI6000 B equipped with Yokogawa CSU-W1 Confocal Scanner Unit) with the Andor iXon Life 897 EMCCD camera and 63× glycerol immersion objective (HC PL APO CS2 63×/1.30 GLYC, Leica).
Light Microscopy.
Cells were plated on glass coverslips and starved for 24 h. Cells were then fixed with cold methanol for 5 min or 3.5% paraformaldehyde/PBS followed by permeabilization with 0.1% Triton X-100 in PBS, blocked with 1% bovine serum albumin in PBS, and stained with indicated antibodies. Primary and secondary antibody incubations were for 1 h at RT. Highly cross-absorbed H+L secondary antibodies (Life Technologies) conjugated to Alexa Fluor 488, 568, or 647 were used at 1:5,000. Nuclei were stained with 0.1 µg/mL DAPI (Sigma). Glass coverslips were mounted onto slides using Mowiol. All images were obtained using MetaMorph software with a spinning disk confocal microscope with a 100 × 1.4 numerical aperture (NA) oil-immersion objective or a 63 × 1.3 NA glycerol immersion at RT.
Image Analysis.
For live-cell imaging analysis, Z-stacks of captured pictures were subject to maximum intensity projections, and areas of interest were cropped by Fiji (NIH). A decapitation event was designated if a vesicular SSTR3-GFP+ structure appeared near the tip of the cilium. Numbers and timing of cilia generation, loss, or decapitation were counted manually. Cilium length was measured using CellProfiler (24). After highlighting cilia structures by enhancing neurite features, cilia were identified as objects. Cilia were skeletonized, and then pixel lengths were calculated. For quantifying TTBK2 at the centrosome, CellProfiler software was used as follows. Maximum intensity projection images of CEP164 staining were used to identify mother centrioles. The mean intensities of TTBK2 staining were calculated within the circular areas around the M centriole of 1.5 µm diameter and 10 µm diameter. The inner ring was used to determine TTBK2 local concentration, whereas the outer ring provided information related to overall TTBK2 levels. The ratios of these two values were used to determine TTBK2 concentration at the M centriole. EHD1 was analyzed by the same method except that gamma-tubulin staining was used to determine centrosomal localization, and therefore each cell had two gamma-tubulin circles that were analyzed.
Statistics.
Graphs were created using GraphPad Prism 6 and RStudio software. Error bars indicate SEM. To test significance, a Student’s paired or unpaired t test was used where two experimental conditions were compared, and ordinary or repeated measures one-way ANOVA with Dunnett and Tukey’s post hoc tests were used when three or more experimental conditions were compared. Two-tailed P values < 0.05 were considered statistically significant (30).
Supplementary Material
Acknowledgments
This research was funded by grants to S.R.P. from the US NIH (DK37332) and from the Michael J. Fox Foundation for Parkinson’s research. We are grateful to Dario Alessi for encouragement and support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005894118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Steger M., et al., Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. eLife 6, e31012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steger M., et al., Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeffer S. R., Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25, 414–419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeffer S. R., Rab GTPases: Master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell 28, 712–715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhekne H. S., et al., A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain. eLife 7, e40202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berndsen K., et al., PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins. eLife 8, e50416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lara Ordónez A. J., et al., RAB8, RAB10 and RILPL1 contribute to both LRRK2 kinase-mediated centrosomal cohesion and ciliogenesis deficits. Hum. Mol. Genet. 28, 3552–3568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goetz S. C., Liem K. F. Jr, Anderson K. V., The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151, 847–858 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouskila M., et al., TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, activity, localization and development. Biochem. J. 437, 157–167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowie E., Goetz S. C., TTBK2 and primary cilia are essential for the connectivity and survival of cerebellar Purkinje neurons. eLife 9, e51166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Čajánek L., Nigg E. A., Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. U.S.A. 111, E2841–E2850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oda T., Chiba S., Nagai T., Mizuno K., Binding to Cep164, but not EB1, is essential for centriolar localization of TTBK2 and its function in ciliogenesis. Genes Cells 19, 927–940 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Huang N., et al., M-Phase Phosphoprotein 9 regulates ciliogenesis by modulating CP110-CEP97 complex localization at the mother centriole. Nat. Commun. 9, 4511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo C. H., et al., Phosphorylation of CEP83 by TTBK2 is necessary for cilia initiation. J. Cell Biol. 218, 3489–3505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berbari N. F., Johnson A. D., Lewis J. S., Askwith C. C., Mykytyn K., Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell 19, 1540–1547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillingham A. K., Munro S., The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524–529 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirvis M., Siemers K. A., Nelson W. J., Stearns T. P., Primary cilium loss in mammalian cells occurs predominantly by whole-cilium shedding. PLoS Biol. 17, e3000381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nager A. R., et al., An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phua S. C., et al., Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168, 264–279.e15 (2017).Corrected in: Cell178, 261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford M. J., et al., A cell/cilia cycle biosensor for single-cell kinetics reveals persistence of cilia after G1/S transition is a general property in cells and mice. Dev. Cell 47, 509–523.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spektor A., Tsang W. Y., Khoo D., Dynlacht B. D., Cep97 and CP110 suppress a cilia assembly program. Cell 130, 678–690 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Jiang K., et al., A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800–1807 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Watanabe T., et al., TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J. Cell Biol. 210, 737–751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter A. E., et al., CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaub J. R., Stearns T., The Rilp-like proteins Rilpl1 and Rilpl2 regulate ciliary membrane content. Mol. Biol. Cell 24, 453–464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorokin S., Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15, 363–377 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Q., et al., Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17, 228–240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanatori I., et al., LRRK2-phosphorylated Rab10 sequesters Myosin Va with RILPL2 during ciliogenesis blockade. bioRxiv [Preprint] (2020). 10.1101/2020.04.28.065664 (Accessed 18 February 2021). [DOI] [PMC free article] [PubMed]
- 29.Xu Q., et al., Phosphatidylinositol phosphate kinase PIPKIγ and phosphatase INPP5E coordinate initiation of ciliogenesis. Nat. Commun. 7, 10777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lord S. J., Velle K. B., Mullins R. D., Fritz-Laylin L. K., SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.