Exome-based identification of TCR genes in a tumor microenvironment
Keywords: CTLA4, PD-1, PD-L2, TCR recombinations, tumor specimen exomes
Abstract
Renal cell carcinoma exome-derived, V(D)J recombination reads had an elevated presence and variability, for both TcR-α and -β, when compared to marginal tissue, reflecting an opportunity to assess tumor immunogenicity by comparison with marginal tissue T cells. PD-1, PD-L2, CTLA4 and FOXP3, all of which are implicated in the evasion of an anti-tumor immune response, had a significantly higher expression for samples representing co-detection of productive TcR-α and -β recombination reads. Samples representing tumors with productive TcR-α recombination reads but no detectable, productive TcR-β recombination reads, reflected a 20% survival advantage, and RNASeq data indicated an intermediate level of immune checkpoint gene expression for those samples. These results raise the question of whether relatively high levels of detection of productive TcR-α recombination reads, in comparison with detection of reads representing the TcR-β gene, identify a microenvironment that has not yet entered a T-cell exhaustion phase and may thereby represent conditions for immune enhancements that do not require anti-immune checkpoint therapies.
Introduction
Immune cells infiltrate the tumor microenvironment and modulate various stages of tumor development such as malignant conversion and metastasis (1). T cells and macrophages are a significant part of the tumor microenvironment and exhibit distinct phenotype classes in renal cell carcinoma (RCC) that correspond to progression-free survival (2). Although T-cell type 1 immune responses are critical in preventing tumor growth by targeting antigenic tumor cells, tumors can show reduced immunogenicity and a corresponding T-cell response suppression (3). This exhausted T-cell state is characterized by the loss of effector functions, among which include reduced IL-2 production, defects in production of IFN-γ and chemokines and failure to acquire antigen-independent memory responsiveness (4). T-cell exhaustion is associated with increased regulatory CD4-positive T-cell activity (FOXP3-positive) and increased activation of co-inhibitory receptors like PD-1 and CTLA4, a state in which exhausted T cells are ineffective at eliminating tumors (4). Accordingly, further characterization of the tumor immune microenvironment with respect to T-cell exhaustion will contribute to the ongoing development of immune-based interventions for RCC.
It has recently become apparent that genomics approaches to tumor characterization have the potential of revealing aspects of the tumor microenvironment. For example, RNASeq data can reveal the expression levels of HLA class II genes (5), presumably representing antigen-presenting cells, and of immune checkpoint protein genes, presumably actively transcribed in tumor-resident T cells. Furthermore, lymphocyte immune receptor recombinations can be detected in RNASeq and in (whole genome or whole exome) DNA sequence files (6, 7). The RNASeq files permit detection of a relatively large number of immune receptor recombination reads, allowing some efficiencies of approach, such as building contigs across the V(D)J recombination sites with overlapping sequence reads (8). The DNA sequence files have fewer immune receptor recombination reads but provide opportunities for detecting unproductive recombinations (9), which could indicate lymphocyte influence on tumor growth independent of tumor immunogenicity. In addition, the opportunity to detect immune receptor recombination reads in whole exome sequencing (WXS) files in particular offers the hope of exploiting this now common evaluation of patient tumors for multiple aspects of tumor characterization (9).
Methods
Processing the human cancer exome (WXS) files
The original computer programs used to recover the immune receptor recombination reads from the tumor specimen WXS files are updated versions of previously published, scripted algorithms (Supplementary Table S1, available at International Immunology Online). Briefly, the program searches WXS files for 10 nucleotide sequences representing V and J regions of immune receptors. The 10 nucleotide sequences for the V region are located close to the 3′ end and close to the 5′ end for the J regions. The algorithm steps away from the 3′ and 5′ ends, respectively, employing 10 nucleotide search strings, 3 bases at a time, to move away from the V-J junction region and thereby obtain recombination reads that do not precisely match the 3′ and 5′ ends of the V and J regions, due to N-region diversity. The nucleotide sequences for human TcR V and J regions were obtained from NCBI and IMGT/GENE-DB (10). WXS for RCCs and for barcode (TCGA sample)-matched, surrounding tissue were collected from the National Cancer Institute’s Genomic Data Commons (TCGA-KIRC), under approved project number 6300 (Supplementary Table S2, available at International Immunology Online). See also ref. (11) and the following link (https://github.com/martinda/gnu-parallel/blob/master/CITATION).
Obtaining RNASeq data
The RNASeq raw values for the genes indicated in ‘Results’ were obtained from cBioPortal.org, and the values for each barcode used in this study are provided in Supplementary Table S3 (available at International Immunology Online) (12, 13).
Generation of Kaplan–Meier curves via IBM SPSS
IBM SPSS (statistics program for the social sciences) was used to generate Kaplan–Meier (KM) curves for barcodes representing the recovery of different sets of recombination reads from the WXS files, as detailed in ‘Results’. Clinical data were obtained from cBioPortal.org.
Results
Tumor tissue yields a greater range of recoveries of TcR recombination reads in comparison to surrounding, marginal tissue
To determine how recovery of immune receptor recombination reads from marginal tissue could reflect tumor T-cell infiltration, we searched TCGA-KIRC for TcR-α, -β and -δ recombination reads in the WXS files representing 341 tumor samples and 265 marginal tissue samples (Table 1; Supplementary Table S2, available at International Immunology Online). Reads representing both productive and unproductive recombinations were detected for TcR-α and -β, in both tumors and surrounding tissue. The α/β TcR recombination reads are more dominant than TcR-δ recombinations, as expected. Also, barcodes (samples) representing productive TcR-α recombination reads occurred in a greater proportion, overall, in tumors (64%) compared to the marginal tissue (39%). In contrast, the rate of identification of barcodes representing unproductive TcR-α recombinations was similar for tumor and marginal tissue. Almost every barcode represented detection of unproductive TcR-α recombination reads, but the rate for unproductive TcR-β recombinations is much lower, at 11% for tumor and 5% for marginal tissue. This very low detection of unproductive TcR-β recombination reads is consistent with previous findings for melanoma (14). In particular, the ratio of productive to unproductive TcR-α reads for tumor approximates a one to one ratio in RCC, melanoma and other tumors, whereas the ratio of productive to unproductive TcR-β recombination reads for tumor approximates four to one in the same set of tumors (14). Lastly, the proportion of samples with detection of productive TcR-β recombination rates was similar between tumor and marginal tissue (Table 1) but as discussed in the next paragraph, tumors had higher recombination read counts per sample.
Table 1.
Barcode count for recombinations identified for TcR genes in primary tumor and marginal tissue, in TCGA-KIRC WXS files
| TcR gene | 341 tumor barcodes | 265 marginal tissue barcodes | ||||||
|---|---|---|---|---|---|---|---|---|
| Productive | Unproductive | Productive | Unproductive | |||||
| α | 217 | 64% | 311 | 91% | 103 | 39% | 263 | 99% |
| β | 126 | 37% | 36 | 11% | 80 | 30% | 12 | 5% |
| δ | 2 | 0.6% | 9 | 3% | 1 | 0.4% | 15 | 6% |
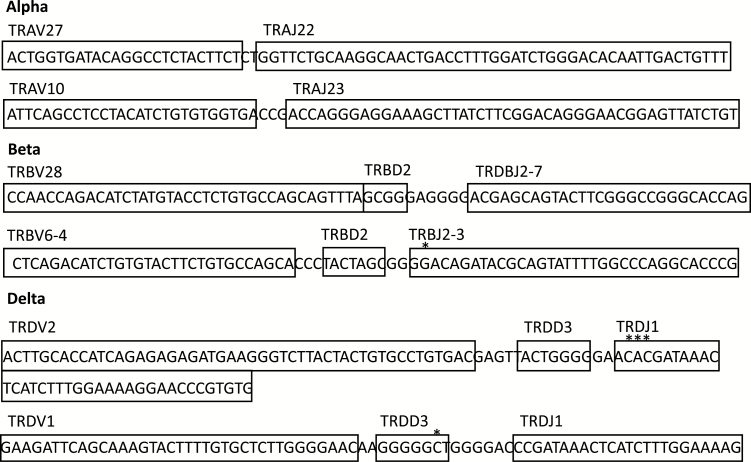
The VJ usage for each productive read recovered is given in Supplementary Table S4 (available at International Immunology Online). Examples of the TcR-α, -β and -δ recombination reads, and their VDJ usage, are shown in Fig. 1.
Fig. 1.
Examples of TcR-α, TcR-β, TcR-δ productive reads from WXS KIRC files.
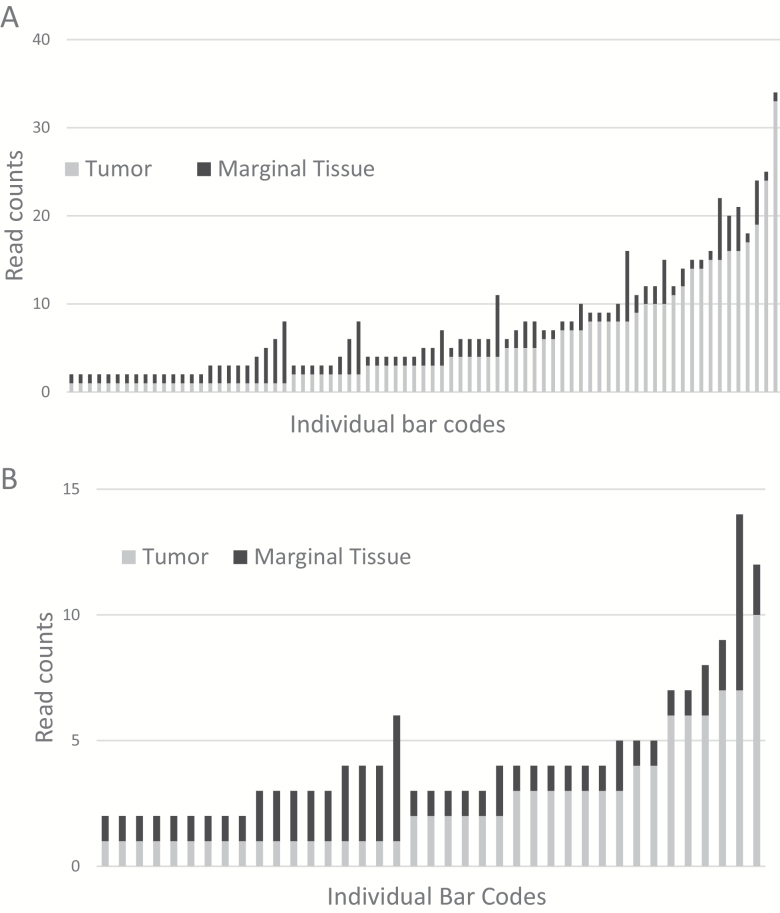
To further assess differences in the recovery of immune receptor recombination reads from WXS files representing tumor and marginal tissue, we analyzed the recombination read counts for TcR-α and -β (Table 2; Supplementary Table S5, available at International Immunology Online). Results indicated that the mean read count is higher in tumor samples than in the marginal tissue samples, for both TcR-α and -β (TcR-α difference between tumor and marginal tissue, P value <0.0001; TcR-β difference between tumor and marginal tissue, P value <0.002). To ensure that the mean read counts were independent of a potential difference between total number of mapped reads for each WXS tumor or tissue file, we normalized the read counts to the mapped read totals (Supplementary Table S6, available at International Immunology Online). The results remained significant for the difference in mean number of reads between tumor and marginal tissue for both TcR-α and -β recombination read counts. The distribution of the recoveries of recombination reads for TcR-α and -β shows a greater range of variability of T-cell infiltrate for the tumor than in the marginal tissue. For example, TcR-α VJ recombination read counts ranged between 1 and 33 for tumors, whereas the range of the recombination reads for marginal tissue was from 1 to 8 (Fig. 2A). This result was also true for recovery of the TcR-β recombination reads, but to a lesser extent (Fig. 2B).
Table 2.
TcR-α and TcR-β V(D)J recombination read counts: comparison of RCC tumor and marginal tissue
| TcR gene | Tumor samples | Marginal tissue samples | P value | ||
|---|---|---|---|---|---|
| Mean | # of samples | Mean | # of samples | ||
| α | 4.29 | 217 | 2.01 | 103 | 7.97 E-06 |
| β | 2.33 | 126 | 1.59 | 80 | 0.0017 |
Fig. 2.
TcR read counts for WXS files representing tumor and surrounding, marginal tissue. (A) TcR-α read counts in tumor and marginal tissue WXS files, with the x-axis representing individual TCGA barcodes (samples). (B) TcR-β read counts in tumor versus marginal tissue, with the x-axis representing individual barcodes.
Co-detection of TcR-α/β recombination reads define a state of T-cell exhaustion
To obtain a more comprehensive picture of the immune activity in the tumor microenvironment, the RNASeq raw values for the following genes were compared across different barcode sets, following previously published approaches (5, 14): PDCD1 (PD-1), CD274 (PD-L1), PDCD1LG2 (PD-L2), CTLA4, CD28, FOXP3, IL-2, IL2RA, CD3D (CD3), CD4, CD8, HLA-DRA, CTSS, CTSL and CIITA (Table 3; Supplementary Table S3, available at International Immunology Online). [The preceding genes are identified by Human Genome Organization (HUGO) symbols, with common symbols in parentheses.] Set 1 represented barcodes with recovery of both productive TcR-α and -β recombination reads in the same WXS file, i.e. for the same barcode; and Set 2 represented barcodes with no detection of either productive α or β. Comparison of Set 1 and Set 2 demonstrated a statistically significant difference in expression for genes that play a role in tumor evasion of immune surveillance (PD-1, PD-L2, CTLA4, CD28 and FOXP3) with Set 1 representing co-detection of TcR-α/β recombination reads having the higher level of expression. (No difference in the expression of PD-L1 was obtained, in the comparison of the two sets of barcodes.) In addition, IL-2 and IL-2 receptor (IL2RA) RNASeq values were analyzed because high-dose IL-2 is a common treatment option for metastatic RCC (15). IL-2 and IL2RA were also found to be expressed at a significantly higher level in Set 1 barcodes representing co-detection of productive α and β recombination reads (Table 3; Supplementary Table S3, available at International Immunology Online).
Table 3.
RNASeq values for immune function genes for TCGA-KIRC barcodes representing co-detection of TcR-α and TcR-β recombination reads in WXS files
| Set number for matching P value comparisons | KIRC barcode sets and set comparisons | PD-1 | PD-L1 | PD-L2 | CTLA4 | CD28 | FOXP3 | IL-2 |
|---|---|---|---|---|---|---|---|---|
| 1 | Productive α and productive β | 153.53 | 121.25 | 146.52 | 58.40 | 154.26 | 84.23 | 1.38 |
| 2 | No detection of productive α or β | 26.50 | 49.25 | 66.28 | 15.12 | 46.63 | 15.27 | 0.27 |
| 3 | Productive α and no productive β | 97.74 | 56.66 | 111.01 | 34.40 | 100.02 | 50.22 | 0.67 |
| 1 versus 2 | P values for comparisons of sets 1 and 2 | 2.62 E-06 | 0.40 | 1.18 E-05 | 1.24 E-05 | 3.86 E-13 | 1.80 E-07 | 2.14 E-04 |
| 1 versus 3 | P values for comparisons of sets 1 and 3 | 0.013 | 0.24 | 0.011 | 0.0011 | 1.28 E-05 | 0.0016 | 0.0017 |
As a verification that the recovery of productive TcR-α and -β recombination reads represented T-cell infiltrates, the T-cell markers, CD3, CD4 and CD8 were also compared between Sets 1 and 2, with results indicating a clearly higher level of expression for each of these T-cell markers in Set 1 (Supplementary Table S3, available at International Immunology Online). We also obtained RNASeq values for genes representing antigen-presenting functions, in particular, HLA-DRA, CTSS (cathepsin S), CTSL (cathepsin L) and CIITA (class II transactivator). There was a positive correlation between co-detection of productive TcR-α and -β recombination reads and increased levels of expression of these antigen-presenting genes, with the exception of CTSL (Supplementary Table S3, available at International Immunology Online).
Survival rates for barcode sets representing the recovery of different sets of TcR recombination reads
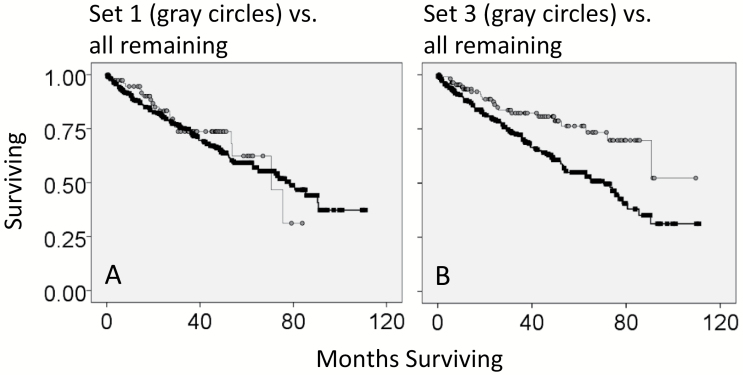
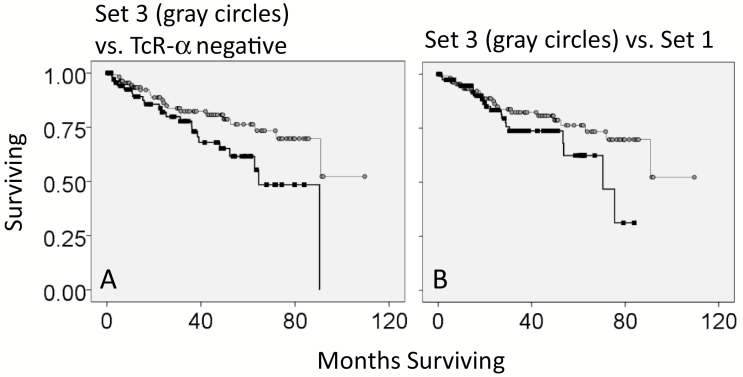
To determine whether there was a survival advantage for patients that had tumors with co-detection of productive TcR-α and -β recombination reads in the KIRC WXS files, we obtained the clinical data representing overall months of survival and overall survival status (living or deceased), to create KM curves (Supplementary Table S7, available at International Immunology Online). In the comparison of Set 1 (co-detection of productive TcR-α and -β recombination reads) with all remaining samples, there was no survival advantage (Fig. 3A). However, patients with tumors that had productive α and absence of productive β (defined here as Set 3) had a survival advantage over the remaining patients (P value = 0.001, Fig. 3B). At the 5-year mark, there was a 20% survival advantage for Set 3. In addition, Set 3 patients (with tumors that had productive α and absence of productive β) also had greater survivability when compared to patients without productive α (P value = 0.028, Fig. 4A).
Fig. 3.
Survival rates for different sets of barcodes representing different recoveries of TcR recombination reads, in comparison to all remaining barcodes. (A) Kaplan–Meier (KM) survival curve for barcodes with both productive TcR-α and TCR-β recombination reads (Set 1; gray circles) against the remaining samples (black squares). Log rank P value = 0.637. (B) KM survival curve for barcodes with only productive TcR-α (absence of productive TcR-β) (gray circles) against all remaining samples (black squares). Log rank P value = 0.001.
Fig. 4.
Comparison of specific subsets of barcodes, representing different recoveries of TcR recombination reads. (A) Kaplan–Meier (KM) survival curve for barcodes with only productive TcR-α (absence of productive TcR-β) (Set 3) (gray circles) against barcodes with the absence of productive TcR-α (black squares). Log rank P value = 0.028. (B) KM survival curve for barcodes with productive TcR-α and absence of productive TcR-β (Set 3) (gray circles) against barcodes with the co-detection of productive TcR-α and TCR-β (Set 1) (black squares). Log rank P value = 0.107.
In consideration of the above indicated survival results, we examined the RNAseq data for productive α and absence of productive β (Set 3) in comparison to co-detection of productive α and β (Set 1) and in comparison to no detection of either productive α or β (Set 2) (Fig. 4B). We found that the Set 3 RNASeq values representing the immune genes for PD-1, PD-L2, CTLA4, CD28, FOXP3 and IL-2 were intermediate between Set 1 (highest expression levels) and Set 2 (lowest expression levels) (Table 3).
Discussion
The most important conclusions from the above work are: (i) for the first time, knowledge of the difference in the recovery of immune receptor recombination reads, between marginal and tumor tissue, can be used to distinguish the non-self nature of the tumor; (ii) high level recovery of immune receptor recombination reads, in particular co-detection of reads representing productive recombination of TcR-α and -β, correlates with a relatively high level of immune checkpoint protein expression, similar to findings with melanoma (14); and (iii) finally, recovery of productive TcR-α recombination reads in the absence of recovery of productive TcR-β recombination reads indicates an intermediate state of expression of T-cell exhaustion markers.
With regard to point (i) above, the distinction between tumor tissue and marginal tissue, with regard to recovery of TcR recombination reads, and the indications that tumor tissue immunogenicity can be thereby distinguished from surrounding normal tissue, represent a reminder regarding the value of tissue-resident lymphocytes for research and potential near-term therapies, such as CAR T-cell therapies. In particular, the human ‘control’ tissue behaves as would be expected, and the human experimental tissue behaves as would be expected, keeping in mind tumor immunogenicity. This observation is important because it supports the use of animal models, where tissue-resident lymphocytes can be obtained more readily than in a human setting, for research advances, in particular for recovery and characterization of antigen-specific immune receptors. Equally importantly, it emphasizes the potential of the occasional human setting where tissue-resident lymphocytes, representing both diseased (experimental) and normal (control) tissue, can be had as a result of standard of care procedures, such as tissue removal for flesh-eating bacteria infections.
Considering the second and third observations above, which have not been reported for any other cancer dataset, immune checkpoint genes such as PD-1, PD-L2 and CTLA4 were expressed at an intermediate level for a newly defined classification of tumor immunogenicity based on T-cell recombinations. This intermediate level of expression was established for barcodes with TcR-α recombination reads in the absence of recovery of productive TcR-β recombinations. This is in comparison to the barcodes representing co-detection of productive α and β recombination reads (highest immune checkpoint expression levels) and to the barcodes representing the absence of productive α or β (lowest expression levels). Notably, the survival data suggest that a high level presence of TcR-β (relatively frequent detection via exome data mining) is disadvantageous to survivability. Thus, keeping in mind that these data were obtained by mining tumor specimen exomes, co-detection of TcR-α recombination reads and TcR-β recombination reads may represent a high level of T-cell activity and indicate that proportionately more T cells have progressed to the exhausted state of high level PD-1 and CTLA4 expression. Presumably, such a state would represent a lower survival outcome. This raises the question of whether detection of productive α recombination reads, in the absence of productive β, is a pre-T-cell exhaustion phase that may be sub-optimal for treatments like anti-immune checkpoint therapies.
The reasons for lack of TcR-β recombination reads, when TcR-α reads are recovered, are unknown, but there are several interesting possibilities. (i) First, the chances of detecting a TRB recombination is technically less than for a TRA recombination due to the fact that the computer code (‘Methods’) searches for a V and J region on the same sequencing read, i.e. a read of between 75 and 100 nucleotides. In the case of TRB, there is a D region segment between the V and the J, making it less probable that a V and J will be detected on the same read, particularly keeping in mind the IMGT confirmation step of the processing, which required that a minimum number of (20) nucleotides be used in the V and J identifications. (ii) Second, on the basis of recombination frequencies, from a biological standpoint, TRA is more common, i.e. both alleles can recombine in the case of TRA but only one allele in the case of TRB (16). In particular, both of these TRA chains can be functional (16). Thus, there may be cases where the exome prep includes a very small number of T cells and in those cases, just by random chance of inclusion of any one DNA segment in the exome sequencing process, detection of TRA recombinations would be more likely. (iii) Finally, there have been reports of T cells with the recombination of only one TCR chain in the periphery (17–19). Thus, it remains possible that the detection of only the TRA recombination, which ordinarily would precede the TRB recombination, represents a fundamental aspect of T-cell function in the periphery that has simply been under-investigated.
Further investigation into T-cell recombination characterization of RCC tumors and treatment may help to refine the treatment protocol for RCC that currently includes first-line therapies such as high-dose IL-2 and PD-1/PD-L1 pathway blockers (20). For example, an immunogenomics assay related to the above approaches could be used to identify patients with a pre-exhaustion lymphocyte infiltrate that would be preferential to high-dose IL-2 treatment. In contrast, genomics-based indications of a state of T-cell exhaustion, for example, a relatively low ratio of productive TcR-α to productive TcR-β recombination reads, may justify treatment with an immune checkpoint inhibitor instead. While it is possible for current diagnostic tools to establish relative immune checkpoint inhibitor protein levels, the immunogenomics-based opportunities suggested by this report could augment current approaches by adding a level of sensitivity to the treatment model for RCC. In addition, the genomics-based approach alone may facilitate treatment decisions in settings where multiple diagnostic approaches are cost prohibitive.
An additional noteworthy finding is that PD-L2, not PD-L1, is significantly up-regulated in RCC tumor samples with productive TcR-α and -β recombination reads. This adds complexity to the conventional understanding that PD-L1 is the primary ligand for PD-1 implicated in RCC anti-tumor immunity (21). This finding corroborates Erlmeier et al.’s conclusion that PD-L2’s expression in clear cell RCC correlates inversely with overall survival and may be an important prognostic factor for RCC (22). These findings suggest further investigation into PD-L2 as a potential target for RCC treatment.
Supplementary data
Supplementary data are available at International Immunology online.
Conflicts of interest statement: The authors declared no conflicts of interest.
Acknowledgements
The authors would like to gratefully acknowledge the support of USF research computing and the taxpayers of the State of Florida.
References
- 1. Grivennikov, S. I., Greten, F. R. and Karin, M. 2010. Immunity, inflammation, and cancer. Cell 140:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chevrier, S., Levine, J. H., Zanotelli, V. R. T.et al. 2017. An immune atlas of clear cell renal cell carcinoma. Cell 169:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Speiser, D. E., Ho, P. C. and Verdeil, G. 2016. Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 16:599. [DOI] [PubMed] [Google Scholar]
- 4. Wherry, E. J. and Kurachi, M. 2015. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butler, S. N. and Blanck, G. 2016. Immunoscoring by correlating MHC class II and TCR expression: high level immune functions represented by the KIRP dataset of TCGA. Cell Tissue Res. 363:491. [DOI] [PubMed] [Google Scholar]
- 6. Brown, S. D., Raeburn, L. A. and Holt, R. A. 2015. Profiling tissue-resident T cell repertoires by RNA sequencing. Genome Med. 7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill, T. R., Samy, M. D., Butler, S. N., Mauro, J. A., Sexton, W. J. and Blanck, G. 2016. Detection of productively rearranged TcR-α V-J sequences in TCGA exome files: implications for tumor immunoscoring and recovery of antitumor T-cells. Cancer Inform. 15:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li, B., Li, T., Wang, B.et al. 2017. Ultrasensitive detection of TCR hypervariable-region sequences in solid-tissue RNA-seq data. Nat. Genet. 49:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samy, M. D., Tong, W. L., Yavorski, J. M., Sexton, W. J. and Blanck, G. 2017. T cell receptor gene recombinations in human tumor specimen exome files: detection of T cell receptor-β VDJ recombinations associates with a favorable oncologic outcome for bladder cancer. Cancer Immunol. Immunother. 66:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alamyar, E., Duroux, P., Lefranc, M. P. and Giudicelli, V. 2012. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882:569. [DOI] [PubMed] [Google Scholar]
- 11. Malaspinas, A. S., Tange, O., Moreno-Mayar, J. V.et al. 2014. bammds: a tool for assessing the ancestry of low-depth whole-genome data using multidimensional scaling (MDS). Bioinformatics 30:2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cerami, E., Gao, J., Dogrusoz, U.et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao, J., Aksoy, B. A., Dogrusoz, U.et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tu, Y. N., Tong, W. L., Samy, M. D., Yavorski, J. M., Kim, M. and Blanck, G. 2017. Assessing microenvironment immunogenicity using tumor specimen exomes: co-detection of TcR-α/β V(D)J recombinations correlates with PD-1 expression. Int. J. Cancer 140:2568. [DOI] [PubMed] [Google Scholar]
- 15. Itsumi, M. and Tatsugami, K. 2010. Immunotherapy for renal cell carcinoma. Clin. Dev. Immunol. 2010:284581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Padovan, E., Casorati, G., Dellabona, P., Meyer, S., Brockhaus, M. and Lanzavecchia, A. 1993. Expression of two T cell receptor alpha chains: dual receptor T cells. Science 262:422. [DOI] [PubMed] [Google Scholar]
- 17. Eichelberger, M., McMickle, A., Blackman, M., Mombaerts, P., Tonegawa, S. and Doherty, P. C. 1995. Functional analysis of the TCR alpha- beta+ cells that accumulate in the pneumonic lung of influenza virus-infected TCR-alpha-/- mice. J. Immunol. 154:1569. [PubMed] [Google Scholar]
- 18. Hillion, S., Rochas, C., Youinou, P. and Jamin, C. 2005. Expression and reexpression of recombination activating genes: relevance to the development of autoimmune states. Ann. NY Acad. Sci. 1050:10. [DOI] [PubMed] [Google Scholar]
- 19. Hillion, S., Rochas, C., Devauchelle, V., Youinou, P. and Jamin, C. 2006. Central and peripheral RAG protein re-expression: underestimate mechanisms of tolerance?Scand. J. Immunol. 64:185. [DOI] [PubMed] [Google Scholar]
- 20. Rini, B. I., McDermott, D. F., Hammers, H.et al. 2016. Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of renal cell carcinoma. J. Immunother. Cancer 4:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bardhan, K., Anagnostou, T. and Boussiotis, V. A. 2016. The PD1:PD-L1/2 pathway from discovery to clinical implementation. Front. Immunol. 7:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erlmeier, F., Weichert, W., Autenrieth, M.et al. 2017. PD-L2: a prognostic marker in chromophobe renal cell carcinoma?Med. Oncol. 34:71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.