Abstract

Covalently acting inhibitors constitute a large and growing fraction of approved small-molecule therapeutics as well as useful tools for a variety of in vitro and in vivo applications. Here, we aimed to develop a covalent antagonist of CC chemokine receptor 2 (CCR2), a class A GPCR that has been pursued as a therapeutic target in inflammation and immuno-oncology. Based on a known intracellularly binding CCR2 antagonist, several covalent derivatives were synthesized and characterized by radioligand binding and functional assays. These studies revealed compound 14 as an intracellular covalent ligand for CCR2. In silico modeling followed by site-directed mutagenesis confirmed that 14 forms a covalent bond with one of three proximal cysteine residues, which can be engaged interchangeably. To our knowledge, compound 14 represents the first covalent ligand reported for CCR2. Due to its unique properties, it may represent a promising tool for ongoing and future studies of CCR2 pharmacology.
Introduction
CC chemokine receptor 2 (CCR2) is a G protein-coupled receptor (GPCR) expressed on the surface of various immune cells, including monocytes, basophiles, and natural killer cells.1 Activation of CCR2 by its endogenous chemokine ligands, such as CCL2, results in leukocyte trafficking toward sites of inflammation as part of the immune response.2 However, dysregulation of CCR2 signaling can lead to leukocyte accumulation—a hallmark of the inflammatory response—and ultimately to a variety of inflammatory and immune diseases.3 Preclinical studies have suggested a critical role of CCR2/CCL2 signaling in atherosclerosis,4 diabetes,5 neuropathic pain,6 and cancer,7 among others. However, despite extensive efforts to develop CCR2 antagonist therapeutics, most clinical candidates have failed due to lack of efficacy.8 Thus, a better understanding of the structure and biological function of CCR2, both in vitro and in vivo, is necessary for the development of clinically successful CCR2 antagonists.
Covalent ligands have recently re-emerged as valuable tool compounds and therapeutic agents for various targets and diseases.9−11 Covalent ligands contain a reactive chemical group, or “warhead”, which allows them to bind to their target in an irreversible manner.12 In the field of GPCRs, covalent ligands have been primarily used as tool compounds due to major safety concerns; however, recent studies have highlighted several potential advantages for the development of covalent drugs, including insurmountability and prolonged duration of action.13−15 The later advantage may be particularly useful in disease contexts in which high receptor occupancy and prolonged inhibition are required for an effective response.16 As tool compounds, covalent ligands have been increasingly used for structure elucidation as they can stabilize inherently flexible receptor–ligand complexes.9 Examples include the recent crystal structures of the cannabinoid CB1 receptor17 and the adenosine A1 receptor.18 Furthermore, covalent probes represent valuable starting points for a wide variety of chemical biology and proteomic profiling applications.9,12
Recently, the X-ray structures of CCR2 isoform A (CCR2a)19 and isoform B (CCR2b)20 have been solved in complex with small-molecule antagonists. Besides providing structural insight into the binding poses of the antagonists, the crystal structure of CCR2b revealed an intracellular binding site, which accommodates small molecules that inhibit CCR2 without directly competing with the binding of chemokines.20 Due to their noncompetitive manner of inhibition, these intracellular allosteric modulators might be more efficacious in the treatment of inflammatory diseases characterized by high levels of endogenous chemokines.21 An intracellular covalent ligand could be valuable for further investigation of CCR2 pharmacology and role in diseases. Thus, based on previously described high-affinity intracellular ligands for CCR2,22 we aimed to design and synthesize a novel covalent intracellular ligand for this receptor. Biochemical characterization, in silico modeling, and a mutational study resulted in the identification of compound 14 as a covalent, negative allosteric modulator (NAM) that binds intracellularly and forms a covalent bond with one of three proximal cysteine residues of CCR2. Compound 14 represents a promising starting point for a variety of applications to further study CCR2.
Results
Covalent Derivatization of a Sulfonamide Antagonist Yields Novel CCR2 Inhibitors
The design of covalent ligands for CCR2 was based on a previously described sulfonamide series, whose representative SD-2422 was identified as a high-affinity, intracellular ligand for CCR2 (Figure 1a).23 Previous structure–activity studies also showed that it is possible to extend these derivatives on the “left hand” phenyl moiety, particularly at the para position, as CCR2 activity is not affected after introduction of cyclic amines linked by an ethylacetamide moiety.24 Thus, we decided to introduce a reactive warhead in the para position, using the described ethylacetamide linker. A close analogue of SD-24, with the carboxylic group in the para position (sulfonamide 34 by Peace et al.,22 corresponding to compound 7 in our study (Figure 1a)) was used as a non-covalent control. Because the predicted binding mode of compound 7 featured a hydrogen-bond between its carboxyl group and Cys75 in the first intracellular loop (ICL1) of CCR2 (Cys75ICL1, Figure 1b), we envisioned four warheads with varying selectivity toward cysteine: isothiocyanate, which targets both cysteine and lysine residues;25,26 acryloyl, a Michael acceptor that preferentially targets cysteine over lysine;27 a non-selective fluorosulfonyl, with reactivity toward serine, threonine, tyrosine, lysine, cysteine, and histidine;28 and thiocyanate, which selectively reacts with cysteine.12,29 In addition, a trifluoromethyl group was incorporated in the design as this moiety has been shown to improve the activity of sulfonamide derivatives toward CCR224 (Figure 1a).
Figure 1.
Chemical structures and binding mode of sulfonamide derivatives. (a) Chemical structures of the parent compound SD-24, non-covalent control 7, and the sulfonamide scaffold used for the design of putative covalent ligands. (b) Binding mode of compound 7 in the intracellular binding pocket of CCR2. (c) Docking of compound 7 into the crystal structure of CCR2 (PDB 5T1A); compound 7 makes a hydrogen bond with Cys75ICL1.
The synthesis of final compounds 7 and 11–14 is shown in Scheme 1. Briefly, commercially available starting materials (methyl 4-hydroxybenzoate and 5-chloro-2-fluoronitrobenzene) were combined under basic conditions to generate compound 1 in high yield. Reduction of the aromatic nitro group using tin(II) chloride as a reducing agent30 followed by sulfonylation of the release amine using indium in acetonitrile31 or DMAP in pyridine as a catalyst yielded 5 or 6, respectively. Compounds 5 and 6 were subjected to saponification with sodium hydroxide in dioxane to yield the final compound 7 and the intermediate compound 8. The ethylacetamide linkers were attached to 8 by a peptide coupling reaction32 followed by the introduction of the different electrophilic warheads. Transformation of the amine in 9 to an isothiocyanate in 11 was performed using carbon disulfide and tosyl chloride (TsCl) as previously described.33 Reaction of the terminal amine in 9 with acryloyl chloride in the presence of triethylamine yielded the final compound 12, while the fluorosulfonyl warhead on 13 was attached by peptide coupling of 9 with 4-(fluorosulfonyl)benzoic acid.32 Finally, the thiocyanate warhead was incorporated by SN2 substitution on the bromide intermediate 10, yielding the final compound 14 (Scheme 1).
Scheme 1. Synthesis of Compounds 7 and 11–14.
Final compounds were synthesized using the following reagents and conditions: (a) K2CO3, DMF, 70 °C, 3.5 h or overnight; (b) SnCl2·2H2O, EtOAc, rt, overnight; (c) 3, indium, MeCN, rt, overnight, or 4, DMAP, pyridine, microwave 95 °C, 3 h; (d) NaOH, dioxane, rt, 2 h, or NaOH, dioxane, 60 °C, 2.5 h; (e) i. tert-butyl-N-(2-aminoethyl)carbamate, EDC, HOBt, dioxane, rt; ii. TFA, DCM, rt, for 9; or 2-bromoethan-1-amine, EDC, HOBt, dioxane, rt for 10; (f) 9, TEA, CS2, TsCl, THF for 11; (g) 9, acryloyl chloride, TEA, acetonitrile, −78 °C to rt for 12; (h) 9, 4-(fluorosulfonyl) benzoic acid, EDC, HOBt, TEA, dioxane, rt for 13; (i) 10, KSCN, EtOH, reflux, for 14.
The binding affinities of compounds 7 and 11–14 were determined in membranes from U2OS cells stably expressing hCCR2b (U2OS-CCR2). [3H]-CCR2-RA, a racemic version of the CCR2-RA-[R] antagonist previously shown to target the intracellular allosteric site in CCR2,20,23,34,35 was used as a radiolabeled probe. In competition binding assays, all compounds fully displaced [3H]-CCR2-RA in a concentration-dependent manner, displaying high to moderate binding affinities for hCCR2b (Ki < 100 nM, Table 1). Of note, for compounds 11–14, binding affinities are reported as “apparent (p)Ki” values due to the putative covalent interaction between these compounds with CCR2, which precludes the determination of equilibrium binding parameters. The non-covalent control, compound 7, displaced [3H]-CCR2-RA with a Ki value of 6 nM (Table 1). Compound 13 with a non-selective fluorosulfonyl warhead exhibited the highest (15-fold) loss of affinity compared to compound 7 (apparent Ki, 89 nM). Compound 13 was followed by compound 11 with the isothiocyanate warhead, which showed a 5-fold reduction in affinity compared to compound 7 (apparent Ki, 31 nM). Compounds containing an acryloyl (12) or thiocyanate (14) warhead displayed the highest apparent affinities toward CCR2. Compound 12 displaced [3H]-CCR2-RA with an apparent Ki of 22 nM, while 14 displaced it with an apparent Ki of 4 nM, the highest affinity in this series of compounds (Table 1). Based on the apparent affinities, we decided to continue with compound 14 for further characterization.
Table 1. Binding Affinities (pKi) of Synthesized Sulfonamide Derivatives Determined in [3H]-CCR2-RA Displacement Assaysb.
| compound | pKi ± SEM (Ki, nM) |
|---|---|
| 7 | 8.2 ± 0.03 (6) |
| 11 | 7.5 ± 0.04 (31)a |
| 12 | 7.7 ± 0.14 (22)a |
| 13 | 7.2 ± 0.16 (89)a |
| 14 | 8.4 ± 0.06 (4)a |
As these compounds might bind covalently, we only refer to the affinities as apparent affinities.
Data are presented as mean ± SEM of at least three individual experiments performed in duplicate.
Because putative covalent drugs can be intrinsically unstable, the stability of 7 and 14 was assessed under the conditions used in radioligand binding assays (data not shown). Both compounds (25 μM) remained intact for over 6 h in assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.1% CHAPS) at room temperature. Furthermore, to demonstrate that compound 14’s reactivity with CCR2 is significantly higher than the intrinsic reactivity of the warhead, compound 14 (25 μM) was coincubated with 80 μM reduced glutathione (GSH) at room temperature for over 6 h. This treatment did not result in the formation of a GSH-adduct, suggesting a low intrinsic reactivity of the thiocyanate warhead under the assay conditions. Finally, increasing the concentration of compound to 1 mM and the incubation temperature to 37 °C, resulted in gradual degradation of compound 14 over 4 h, indicating lower stability at higher temperatures. Compound 7 remained intact under these conditions.
Radioligand Binding Assays with Preincubation and Washing Suggest Covalent Binding of Compound 14 to CCR2
To determine whether compound 14 binds irreversibly to CCR2, the time dependency of its affinity, in comparison with the non-covalent control, was determined using the (R)-pure isomer [3H]-CCR2-RA-[R]. Specifically, the affinity of compounds 7 and 14 to U2OS-CCR2 membranes was measured in a [3H]-CCR2-RA-[R] displacement assay with a short incubation time of 20 min, which was preceded or not by a longer preincubation of the membranes with compounds 7 or 14 (Figure 2a,b). Compared to the coincubation experiment, the affinity of compound 7 was similar following a 4 h preincubation, in agreement with its reversible mode of interaction (Figure 2a and Table 2). In contrast, the apparent affinity of compound 14 increased almost 4-fold after preincubation, from a Ki of 2.4 nM to 0.7 nM (Table 2), which is apparent as a shift to the left in the concentration-displacement curve (Figure 2b). This indicates that, over time, more of the compound is covalently bound to the receptor, and thus, less of the compound is needed to achieve similar levels of displacement.
Figure 2.

Radioligand binding assays with preincubation and washing suggest a covalent interaction. (a, b) Time-dependent characterization of affinity of compounds 7 (a) and 14 (b). Concentration-dependent inhibition of [3H]-CCR2-RA-[R] binding to U2OS-CCR2 cell membrane preparations following a 20 min coincubation which was preceded (4 h) or not (0 h) by preincubation with unlabeled ligands. Preincubation shifts the apparent affinity of compound 14. pKi values obtained from non-linear regression fits of these data are described in Table 2. (c) [3H]-CCR2-RA-[R] binding to U2OS-CCR2 membranes pretreated or not with iodoacetamide (IA) followed by preincubation with 10×IC50 concentration of compounds 7 and 14 and repeated washouts. Following the washouts, the inhibition of radioligand binding by compound 14 persists in non-IA-treated membranes but is recovered in membranes pretreated with IA, indicating the involvement of cysteine residues. Data represent the mean ± SEM of at least three independent experiments performed in duplicate. Significant differences between indicated groups were determined using an unpaired t-test with Welch’s correction: **p < 0.01, ***p < 0.001 or ns, not significant.
Table 2. Time-Dependent Characterization of the Affinity (pKi) of Compounds 7 and 14 Obtained from [3H]-CCR2-RA-[R] Displacement Assays with U2OS-CCR2 Membranesd.
| compound | pKi,0h ± SEM (Ki,0h, nM)a | pKi,4h ± SEM (Ki,4h, nM)b | Ki shiftc |
|---|---|---|---|
| 7 | 8.4 ± 0.08 (4.4) | 8.2 ± 0.02 (6.1) | 0.7 ± 0.1 |
| 14 | 8.7 ± 0.10 (2.4) | 9.2 ± 0.15 (0.7)** | 3.8 ± 0.5 |
Affinity after 20 min coincubation of unlabeled ligands with [3H]-CCR2-RA-[R] and no preincubation.
Affinity after 4 h preincubation with unlabeled ligands followed by 20 min coincubation with radioligand.
Ki shift represents the ratio of Ki,4h/Ki,0h calculated per experiment. Data shown are the mean ± SEM of the individual Ki ratios.
Data are presented as mean ± SEM of at least three individual experiments performed in duplicate. Differences in pKi,0h versus pKi,4h values were analyzed using a paired, two-tailed, Student’s t-test, with significance indicated as **p < 0.01.
To assess the irreversibility of the interaction with the receptor, a [3H]-CCR2-RA-[R] washout experiment was employed. In this assay, U2OS-CCR2 cell membranes were preincubated with 10×IC50 concentration of compounds 7 or 14 for 2 h followed by four extensive washing and centrifugation cycles in order to remove the non-covalently bound ligands. After the washing steps, [3H]-CCR2-RA-[R] was added and the sample further incubated for 2 h before measuring radioligand binding. Radioligand binding was compared to vehicle control in the absence of compounds 7 or 14 (set as 100% binding). [3H]-CCR2-RA-[R] binding was fully recovered in membranes preincubated with 7 (117 ± 6% binding), indicating that this compound was completely washed away during the washing and centrifugation cycles (Figure 2c). In contrast, less than 30% binding of [3H]-CCR2-RA-[R] was recovered in membranes preincubated with compound 14 (27 ± 0.4%), indicating that, even after extensive washing, a significant amount of this compound remained bound to the receptor (Figure 2c). As the thiocyanate warhead is expected to interact with cysteine residues, we performed the same washout assay following membrane pretreatment with iodoacetamide (IA), a highly reactive alkylating agent that modifies cysteine sulfhydryl groups and makes them unavailable for covalent interactions. When U2OS-CCR2 membranes were pretreated with IA for 30 min in the dark before preincubation with compounds 7 or 14, [3H]-CCR2-RA-[R] binding was fully recovered for both compounds: 93 ± 5% for compound 14 and 104 ± 5% for compound 7 (Figure 2c). A comparable effect was observed by pretreating the membranes with 1 mM N-ethylmaleimide, another common cysteine modifier36 (Figure S1). This loss of wash-resistant capacity of compound 14 in membranes pretreated with IA confirmed that cysteine residues were indeed responsible for the irreversible binding of 14 with CCR2.
Compound 14 Is More Potent than Its Non-covalent Analogue and Wash-Resistant in Functional Assays
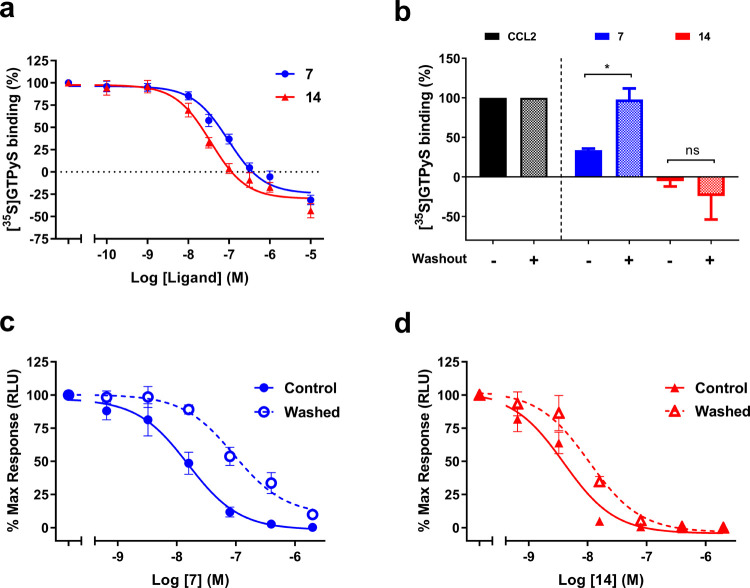
After demonstrating that the interaction of compound 14 with CCR2 was wash-resistant, we investigated whether this compound was able to inhibit the receptor in a functional assay. First compounds 7 and 14 were characterized in a [35S]GTPγS binding assay on U2OS-CCR2 cell membranes. In this assay, both compounds behaved as negative allosteric modulators (NAMs) as they were able to inhibit, in a concentration-dependent manner, [35S]GTPγS binding to the membranes induced by a submaximal concentration of CCL2 (20 nM). The non-covalent control 7 inhibited [35S]GTPγS binding with a pIC50 of 7.1 ± 0.05, while compound 14 showed a higher pIC50 of 7.5 ± 0.07 (Figure 3a and Table 3). Of note, both compounds decreased the basal activity of CCR2 at the highest concentrations (Figure 3a).
Figure 3.
Functional assays suggest a covalent interaction. (a) Inhibition of CCL2-stimulated [35S]GTPyS binding by increasing concentrations of compounds 7 and 14. (b) After washout, compound 7 is not able to inhibit CCL2-induced [35S]GTPyS binding, while compound 14 remains capable of completely inhibiting [35S]GTPyS binding. *p < 0.05 or ns, not significant, according to unpaired t- test with Welch’s correction. (c, d) Inhibition of CCL2-stimulated β-arrestin recruitment by increasing concentrations of compounds 7 (c) and 14 (d) with or without two washing steps before addition of CCL2. Data represent the mean ± SEM of at least three independent experiments performed in duplicate. pIC50 values obtained from these graphs are described in Table 3.
Table 3. Functional Characterization of Compounds 7 and 14 from [35S]GTPγS and β-Arrestin Recruitment Assaysd.
| pIC50 ± SEM (IC50, nM) |
|||
|---|---|---|---|
| compound | [35S]GTPγS bindinga | β-arrestinb | β-arrestin_washedc |
| 7 | 7.1 ± 0.05 (87) | 7.8 ± 0.15 (16) | 7.0 ± 0.09 (103)** |
| 14 | 7.5 ± 0.07 (33) | 8.4 ± 0.12 (4) | 8.0 ± 0.08 (10)* |
Inhibition of [35S]GTPyS binding to U2OS-CCR2 cell membranes at 25 °C, after stimulation with 20 nM CCL2.
Inhibition of β-arrestin recruitment determined with a NanoBiT CCR2 assay, after stimulation with 200 nM CCL2.
Inhibition of β-arrestin recruitment determined with a NanoBiT CCR2 assay, after two washing steps followed by stimulation with 200 nM CCL2. Differences in pIC50 values between unwashed and washed samples were analyzed using a paired, two-tailed, Student’s t-test, with significance shown as *p < 0.05 and **p < 0.01.
Data are presented as mean ± SEM of at least three individual experiments performed in duplicate.
At a single saturating compound concentration (600 and 250 nM for compounds 7 and 14, respectively), [35S]GTPγS binding to CCR2 was reduced to 34 ± 2% by compound 7 and fully inhibited to −5 ± 7% by compound 14 (Figure 3b). However, when the same measurement was performed after four cycles of membrane washing and centrifugation, [35S]GTPγS binding to membranes preincubated with 7 was recovered to 98 ± 14%, which is comparable to levels in the presence of CCL2 alone and significantly different from the “unwashed” situation, (p < 0.05, Figure 3b). In contrast, the inhibition of [35S]GTPγS binding to membranes preincubated with compound 14 persisted and remained at the level of −24 ± 30%, comparable to the “unwashed” situation (Figure 3b). This indicates that, as for the competition binding experiments, compound 7 was washed away and lost its effectiveness in this functional assay, whereas the binding of compound 14 was wash-resistant and led to persistent inhibition of the receptor activity.
Next, we explored the functional effects of irreversible compound binding in live HEK293t cells transiently transfected with hCCR2b. We used a variation of a previously described NanoBiT luminescence complementation assay measuring chemokine-induced β-arrestin recruitment to CCR2,37 to which we added a washout step to distinguish rapidly dissociating compounds from long residence time binders. Increasing concentrations of compounds 7 and 14 were preincubated with the cells prior to two washing steps and a 90 min incubation with compound-free assay buffer, which was followed by cell stimulation with a fixed concentration of CCL2 (200 nM) for another 10 min. In parallel, a control experiment was performed where cells were stimulated with CCL2 in the presence of compounds 7 or 14. In the unwashed control situation, compounds 7 and 14 inhibited CCL2-induced β-arrestin recruitment with 5-fold and 8-fold higher potencies, respectively, than those measured in the [35S]GTPγS binding assay (Table 3). After two washing steps, compound 7 displayed a pIC50 of 7.0 ± 0.09, corresponding to a 6-fold reduction in potency compared to the unwashed control (pIC50 of 7.8 ± 0.15) (Table 3 and Figure 3c). For compound 14, we observed a smaller shift in potency after the washing steps: from a pIC50 of 8.4 ± 0.12 in unwashed cells to a pIC50 of 8.0 ± 0.08 after washing, corresponding to only a 2.5-fold reduction in potency (Table 3 and Figure 3d). In other words, even in the context of a live-cell assay where covalent compound binding to CCR2 is affected by the reducing intracellular environment, compound 14 was more resistant to washing than compound 7, consistent with its covalent mechanism of action.
Structural Basis of the Covalent Binding of Compound 14 to CCR2 Elucidated by Molecular Docking
As suggested by the results of the washout experiment in the presence of IA (Figure 2c), compound 14 is likely to form an irreversible interaction with a cysteine residue in the intracellular binding pocket of CCR2. In the crystal structure of CCR2-RA-[R] bound to hCCR2b-T4L (PDB 5T1A(20)), there are two cysteine residues in proximity of the ligand: Cys75ICL1 within 4 Å and Cys70ICL1 at 6.4 Å. Additionally, following the removal of T4L and re-building of the native ICL3 of CCR2, we discovered that it contains Cys232ICL3 at 14 Å from the intracellular binding pocket. Although this distance (14 Å) is too large to be spanned by the ethylacetamide linker, the flexible nature of ICL3 may enable closer approach of Cys232ICL3 to the pocket (Figure 4a). The binding pose of compound 14 was predicted via covalent docking, assuming that the closest of the three cysteines, Cys75ICL1, is the covalent attachment point. The predicted pose was indeed (i) consistent with that of non-covalent analogs from the same series and (ii) compatible with the linker attachment to Cys75ICL1 (Figure 4b).
Figure 4.
Cysteine 75 seems to be involved in a covalent bond with compound 14. (a) Docking of compound 14 into the crystal structure of CCR2 (PDB 5T1A), showing the cysteine residues with potential to interact with this ligand: Cys75ICL1 within 4 Å; Cys70ICL1 at 6.4 Å; and Cys232ICL3 at 14 Å. (b) Proposed binding mode of compound 14, displaying the covalent interaction between the thiocyanate warhead and Cys75ICL1. (c) Washout-radioligand experiments performed after preincubation of compound 14 (60 or 160 nM for triple mutant) or compound 7 (200 nM) in membranes from CHO cells transiently transfected with CCR2 mutants. Data represent the mean ± SEM of at least three independent experiments performed in duplicate. ****p < 0.0001, one-way ANOVA with Dunnett’s post hoc test.
Covalent Binding of 14 to CCR2 Is Mediated by a Unique Switching Mechanism between Cysteine Residues
To assess which of the three cysteines suggested by docking was responsible for the covalent binding of compound 14 (Figure 4a,b), we designed binding experiments using CCR2 variants in which single-point mutations of cysteine to serine were made: C70ICL1S, C75ICL1S, and C232ICL3S. These CCR2 mutants, together with CCR2 wild-type (WT) as a control, were transiently transfected into CHO cells, and cell membranes were prepared. We first determined if the mutations affected the binding of the radiolabeled probe, [3H]-CCR2-RA-[R], to CCR2 using homologous displacement assays. As shown in Table 4, the affinity of CCR2-RA-[R] for the single CCR2 mutants was similar to WT CCR2 (pIC50 of 7.9 ± 0.06), confirming the integrity of the intracellular pocket in all mutants. Next, we used [3H]-CCR2-RA-[R] displacement to characterize mutant receptor binding to compounds 7 and 14. The affinity of compound 14 for both the C70S and C75S CCR2 mutants was significantly decreased compared to WT CCR2 (pIC50 of 8.8 ± 0.10, Table 4). In the case of compound 7, its affinity was significantly decreased for the C70S mutant and increased for the C75S mutant in comparison to the WT CCR2 (pIC50 of 7.9 ± 0.02). The affinities of both compounds 7 and 14 were not affected by the C232S mutation, indicating that this cysteine might not be involved in the binding of these compounds to CCR2 (Table 4).
Table 4. Binding Affinities (pIC50) of 7 and 14 to Membranes of CHO Cells Expressing the Different CCR2 Constructs, Obtained from [3H]CCR2-RA-[R] Displacement Assaysa.
| pIC50 ± SEM (IC50, nM) |
|||
|---|---|---|---|
| construct | 7 | 14 | CCR2-RA-[R] |
| WT | 7.9 ± 0.02 (12) | 8.8 ± 0.10 (2) | 7.9 ± 0.06 (12) |
| C70S | 7.7 ± 0.09 (22)* | 8.4 ± 0.04 (4)* | 7.9 ± 0.02 (13) |
| C75S | 8.2 ± 0.05 (6)** | 8.4 ± 0.10 (5)* | 8.0 ± 0.05 (11) |
| C232S | 7.9 ± 0.005 (12) | 8.8 ± 0.06 (1) | 8.0 ± 0.09 (9) |
| C70S/C75S/C232S | 8.3 ± 0.02 (5)** | 7.8 ± 0.02 (16)**** | 8.1 ± 0.02 (8) |
Data are presented as mean ± SEM of at least three individual experiments performed in duplicate. Differences in pIC50 values compared to WT were analyzed using a one-way ANOVA with Dunnett’s post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Next, a [3H]-CCR2-RA-[R] washout assay was performed using membrane preparations from WT and mutant CCR2 expressing CHO cells (Figure 4c). Similar to the washout assays in U2OS-CCR2 membranes, compound 7 was completely washed away from the WT receptor, leading to full recovery of [3H]-CCR2-RA-[R] binding (∼100%), while compound 14 only led to 9% binding of [3H]-CCR2-RA-[R], in agreement with its irreversible nature (Figure 4c). For compound 7, full recovery of radioligand binding was observed in the three mutants. In the case of compound 14, mutants C70S and C232S showed similar [3H]-CCR2-RA-[R] binding levels as for the WT receptor, indicating that compound 14 still binds covalently despite these mutations. However, [3H]-CCR2-RA-[R] binding was significantly increased to 30% in the C75S mutant (p < 0.0001), indicating a partial loss of covalent binding in this mutant. These data suggest that Cys75ICL1 is partially responsible for the irreversible nature of compound 14. However, the recovery of [3H]-CCR2-RA-[R] binding was not complete, i.e., to similar levels as compound 7. To rule out the possibility that the serine group introduced at position 75 covalently interacts with the thiocyanate warhead through its nucleophilic hydroxyl group, we mutated Cys75ICL1 to alanine; however, alanine mutation of this residue (C75A) did not improve the recovery of [3H]-CCR2-RA-[R] binding in comparison with the C75S mutant (Figure S2). These results suggest that other residues might also be involved in the formation of a covalent bond with compound 14 or become involved when Cys75ICL1 is not available. Hence, we mutated all three cysteine residues to serine, yielding the triple C70S/C75S/C232S mutant. The [3H]-CCR2-RA-[R] binding assay showed that the affinity of compound 14 was significantly decreased in this triple mutant (Table 4) from a pIC50 of 8.8 for the WT receptor to a pIC50 of 7.8 for the mutant. In contrast, the affinity of compound 7 was increased to a similar level as for the C75S mutant (Table 4). Lastly, washout experiments with compound 14 revealed that binding of [3H]-CCR2-RA-[R] was recovered to levels comparable to compound 7 for this mutant (Figure 4c). This recovery indicates that the irreversible interaction between compound 14 and CCR2 was lost when the three cysteine residues were mutated to serine. Overall, these data suggest that compound 14 interacts primarily with Cys75ICL1 but can switch to Cys70ICL1 or Cys232ICL3 as secondary residues when Cys75ICL1 is not available.
Compound 14 Is Selective toward CCR2 vs Closely Homologous Chemokine Receptors CCR1 and CCR5
Some CCR2 intracellular antagonists also bind to the highly homologous CCR148 and CCR5.23,52 Therefore, we investigated whether compounds 7 and 14 are also able to inhibit CCL3-induced [35S]GTPγS binding in U2OS-CCR1 and U2OS-CCR5 cell membranes. Compared to CCR2, both compounds displayed a much lower inhibitory potency on CCR1 and CCR5, with pIC50 values of 5.3 and 5.4 (Figure S3 and Table S1). This is remarkable since CCR5 has two out of three candidate cysteines in the intracellular binding site (Figure S3A): only Cys75ICL1 of CCR2 is substituted by a serine in CCR5. This again underlines the role of CCR2 Cys75ICL1 as a covalent attachment point for compound 14. On the contrary, in CCR1, none of the three intracellular pocket cysteines are conserved, which may also partially explain the lack of activity of compound 14 toward this receptor.
Discussion and Conclusions
Many studies have suggested a critical role of CCR2 in the pathology of inflammatory and immune diseases;3 yet, no CCR2 antagonists have been approved for clinical use so far. Thus, novel approaches are needed to better study and target this receptor. Covalent ligands represent useful tools to study the structure and function of GPCRs;9,12 furthermore, due to their “infinite residence time”, covalent inhibitors may lead to enhanced in vivo efficacy as a result of their extended duration of action and insurmountability.13−15 Several orthosteric and allosteric antagonists have been previously described for CCR2,38 but no covalent binders have been described so far. Some of the existing CCR2 antagonists bind to an allosteric binding site located in the intracellular region of the receptor, from where they inhibit the receptor in a noncompetitive and insurmountable manner.20,23,35 In an attempt to combine the advantages provided by covalent inhibition and those of intracellular ligands, we designed, synthesized, and characterized a novel covalent intracellular ligand for CCR2 based on a previously described sulfonamide scaffold.
Previous research in our group identified sulfonamide derivatives, similar to compound 7 in the present study, as intracellular antagonists for CCR2.22,23 Based on the predicted proximity of these compounds to cysteine residues in the intracellular region of CCR2, we synthesized sulfonamide derivatives containing four electrophilic warheads with different reactivity profiles: isothiocyanate, acryloyl, fluorosulfonyl, and thiocyanate. Compound 14 with a thiocyanate warhead12,29 was prioritized for further characterization based on the results of the equilibrium binding assays. However, equilibrium parameters (Ki or IC50) are poor indicators of the “true” binding affinity or potency of covalent ligands due to their two-step mechanism of inhibition, which results in a time-dependent shift in affinity.39 Therefore, we further characterized compound 14 in binding assays with a preincubation step. Preincubation led to an increase in the apparent affinity of compound 14 to CCR2, consistent with an increase in receptor occupancy, and thus covalent binding, over time. However, a shift in affinity over time can also be obtained with slowly dissociating, long residence time ligands, as observed with long residence time antagonists for CCR5 and cannabinoid CB1 receptors.40,41 Thus, we next confirmed the irreversible nature of the interaction of compound 14 with CCR2 using “washout” radioligand binding assays, which rely on extensive washing to ensure the removal of unbound, non-covalent ligands. Such assays have been previously used to validate the irreversible nature of covalent ligands for other GPCRs.42−46 The binding of compound 14 was resistant to washing and in fact was not completely abolished even with repeated extensive washes, consistent with a covalent mechanism.
Two functional assays, one involving GTPγS binding to receptor-expressing membranes and another involving β-arrestin recruitment to the receptor in live cells, provided further evidence of the covalent nature of compound 14. In the former assay, repeated membrane washes completely abrogated the ability of the non-covalent analogue compound 7 to inhibit CCL2-induced GTPγS binding to CCR2-expressing membranes; however, the inhibition by compound 14 was unaffected. In the latter assay, the inhibitory potency of compound 14 toward CCL2-induced β-arrestin recruitment to CCR2 was reduced by washing to a significantly lesser extent than that of compound 7. However, and in contrast to the GTPγS binding assay, compound 14 was not fully resistant to washing in the β-arrestin assay. This may be because the shorter compound preincubation in the β-arrestin recruitment assay (20 min versus 2 h in the [35S]GTPγS assays) was not sufficient to allow compound 14 to fully bind CCR2 in an irreversible manner. Another reason could be the highly reducing nature of the intracellular environment in live cells, which might limit the formation of the disulfide bond between the cysteine residue and the compound warhead.47 Interestingly, in the [35S]GTPγS binding assay, both compounds 7 and 14 displayed inverse agonism: a behavior also observed with other CCR2 intracellular ligands.48
While searching for the target amino acid of the thiocyanate warhead, we unexpectedly discovered that compound 14 is able to interact with more than one cysteine residue in the intracellular pocket of CCR2. First, the radioligand washout assays in the presence of IA narrowed the potential list of candidates to cysteine residues (as opposed to, e.g., serines or lysines). Docking of compound 14 into an in silico model of CCR2 suggested three proximal candidate cysteines: Cys70ICL1, Cys75ICL1 (the closest and most likely to interact), and Cys232ICL3. The irreversibility of compound 14 was indeed affected when Cys75ICL1 was mutated to serine; however, a complete loss of covalent binding required elimination of all three cysteines. This suggests that the flexibility of the compound linker and the receptor loops allows compound 14 to adopt multiple binding poses and interact with alternative nucleophilic residues when Cys75ICL1 is removed. A similar phenomenon has been described for other GPCRs, such as adenosine A2A receptor,44 cannabinoid CB2 receptor,49 and muscarinic receptors,50 in which more than one residue was found to mediate the covalent binding of the chemical probes. Furthermore, a recent study suggested that CCR2 antagonists can induce several inactive conformations of the receptor, further supporting receptor flexibility.51
With intracellularly acting antagonists for chemokine receptors, receptor selectivity can be a challenge due to the high degree of residue conservation at their binding sites.21 Many existing intracellularly acting antagonists of CCR2 also inhibit related chemokine receptors such as CCR148 and CCR5.23,52 Compound 14 performed very favorably in this respect: we demonstrated that it is highly selective toward CCR2 compared to CCR1 and CCR5 in a [35S]GTPγS binding assay. This is likely because the action of compound 14 is mediated by Cys75ICL1 that is unique to CCR2 among all chemokine receptors: most of them possess a serine residue in this position. Despite the ability of compound 14 to interact with secondary sites when Cys75ICL1 is not present, the compound was specific for CCR2, further supporting Cys75ICL1 as the main site for covalent interaction. Thus, targeting of Cys75ICL1 may lead to improved selectivity of intracellularly acting CCR2 antagonists.
To conclude, we report the design, synthesis, and pharmacological characterization of compound 14, the first intracellular covalent NAM for CCR2. The results of binding and functional assays support the idea of an irreversible interaction between compound 14 and CCR2, which leads to a long-lasting inhibitory effect. In silico structure-based docking and receptor mutagenesis studies suggest Cys75ICL1 in the intracellular binding site of CCR2 as the primary site of covalent binding for compound 14, although secondary interaction sites were also identified. Overall, compound 14 may represent a useful tool for further studies of CCR2 structure and function in a variety of in vitro and in vivo systems, which might ultimately assist the translation of preclinical findings into the clinic.
Experimental Section
Chemistry
General Methods
All solvents and reagents were purchased from commercial sources and were used without further purification. 1H spectra were recorded on a Bruker AV 400 MHz liquid spectrometer at room temperature (rt) using CDCl3, MeOD, or DMSO as a solvent. Chemical shifts are reported in ppm relative to internal standard tetramethylsilane (TMS) or solvent resonance. Purity of the compounds was determined by HPLC with a C18 column (50 × 4.6 mm, 3 μm), flow rate = 1.3 mL/min, using a gradient of 10–90% MeCN/H2O (0.1% TFA) and measuring UV absorbance at 254 nm. Purity of all final compounds used in biological assays was at least 95%. HPLC traces of all final compounds 7 and 11–14 are shown in Figures S4 and S5. Reactions were monitored by TLC using Merck TLC Silica gel 60 F254 aluminum sheets. Compounds were visualized by UV irradiation or by staining with a KMnO4 solution in H2O. A Biotage Initiator microwave synthesizer was used for the reactions performed in a microwave reactor. For the flash chromatography, Davisil silica gel (40–63 μm) was used. The automatic flash chromatography was performed on an Isolera One Automatic Flash Chromatography System by Biotage with pre-packed flash cartridges (ISCO RediSep or Biotage ZIP Sphere). Mass spectra were measured using a Shimadzu Prominence LCMS-2020 system and a Gemini C18 Phenomenex column (50 × 3 mm, 3 μm).
Experimental Procedures
Methyl 4-(4-Chloro-2-nitrophenoxy)benzoate (1)
Commercially available methyl 4-hydroxybenzoate (1 eq) and 5-chloro-2-fluoronitrobenzene (1 eq) were dissolved in dimethylformamide (DMF, 1 mL/mmol) together with potassium carbonate (2 eq) in a round-bottom flask. The mixture was stirred at 70 °C under nitrogen atmosphere for 3.5 h. After the solvent was evaporated, the residue was dissolved in EtOAc and washed with water. The organic layer was dried over MgSO4 and concentrated in vacuum to afford 1. Yield: 13.9 g, 45.1 mmol, 90%. 1H NMR (400 MHz, DMSO) δ 8.29 (d, J = 2.6 Hz, 1H), 8.03–7.96 (m, 2H), 7.86 (dd, J = 8.9, 2.6 Hz, 1H), 7.42 (d, J = 8.9 Hz, 1H), 7.20–7.13 (m, 2H), 3.84 (s, 3H).
Methyl 4-(2-Amino-4-chlorophenoxy)benzoate (2)
Intermediate 1 was dissolved in EtOAc (0.05 M) before addition of SnCl2·2H2O (5 eq). The reaction was stirred overnight at rt under a nitrogen atmosphere. Upon completion, the reaction was quenched with 1 M NaOH and extracted with EtOAc. The organic layer was then washed with H2O and dried over MgSO4 to yield 2. Yield: 4.13 g, 14.9 mmol, 75%. 1H NMR (400 MHz, DMSO) δ 7.98–7.88 (m, 2H), 7.01–6.93 (m, 2H), 6.89 (d, J = 8.5 Hz, 1H), 6.85 (d, J = 2.5 Hz, 1H), 6.57 (dd, J = 8.5, 2.6 Hz, 1H), 5.36 (s, 2H).
Methyl 4-(4-Chloro-2-((3,4-dichlorophenyl)sulfonamido)phenoxy)benzoate (5)
Intermediate 2 was dissolved in CH3CN before addition of indium (0.1 eq) and commercially available 3,4-dichloro-benzenesulfonylchloride 3 (1 eq). The mixture was stirred overnight at rt after purging the system with nitrogen. The solvents were then evaporated, and the residue extracted with EtOAc and washed successively with H2O and brine. The organic layer was dried over MgSO4, filtered, and evaporated, after which the product was purified by flash column chromatography (CHCl3/petroleum ether 3:1) in order to obtain pure 5. Yield: 3.61 g, 7.41 mmol, 50%. 1H NMR (400 MHz, DMSO) δ 10.60 (s, 1H), 7.79 (d, J = 8.8 Hz, 2H), 7.76 (d, J = 1.9 Hz, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.54 (dd, J = 11.1, 2.2 Hz, 2H), 7.33 (dd, J = 8.8, 2.5 Hz, 1H), 7.07 (d, J = 8.8 Hz, 1H), 6.64 (d, J = 8.8 Hz, 2H), 3.83 (s, 3H).
4-(4-Chloro-2-((3,4-dichlorophenyl)sulfonamido)phenoxy)benzoic Acid (7)
A mixture of 2 M NaOH (10 mL) and dioxane (10 mL) was added to intermediate 5, and the reaction mixture was stirred at rt for 2 h. Solvents were evaporated, and the residue was extracted with DCM. The organic layer was extracted with H2O. The aqueous layers were pooled and acidified with 6 M HCl. Filtration of the precipitate gave final compound 7. Yield: 37 mg, 8%. 1H NMR (400 MHz, DMSO) δ 12.81 (s, 1H), 10.60 (s, 1H), 7.81 (s, 1H), 7.81–7.75 (m, 2H), 7.63 (d, J = 8.4 Hz, 1H), 7.57 (dd, J = 8.4, 2.0 Hz, 1H), 7.51 (d, J = 2.4 Hz, 1H), 7.33 (dd, J = 8.4, 2.4 Hz, 1H), 7.04 (d, J = 9.6 Hz, 1H), 6.63 (d, J = 8.8 Hz, 2H); MS: ESI [M – H]−: 469.8; HPLC: 10.2 min.
Methyl 4-(4-Chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzoate (6)
Intermediate 2 (1 eq) and commercially available 4-chloro-3-(trifluoromethyl)benzenesulfonyl chloride 4 (1.1 eq) were dissolved in pyridine (0.4 M) and N,N-dimethylaminopyridine (0.1 eq) was added. The mixture was stirred at 95 °C for 3 h under microwave irradiation. The mixture was extracted with DCM, and the organic layer was washed with aqueous HCl, water, and brine. The organic phase was dried over MgSO4 and evaporated, after which precipitation with DCM and PE gave 6. Yield: 1.15 g, 63%. 1H NMR (400 MHz, CDCl3) δ 8.03 ( d, J = 2.1 Hz, 1H), 7.93 (d, J = 8.8 Hz, 2H), 7.79 (dd. J = 8.4, 2.1 Hz, 1H), 7.73 (d, J = 2.4 Hz, 1H), 7.51 (d, J = 8.4 Hz, 1H), 7.12 (dd, J = 8.8, 2.5 Hz, 1H), 7.00 (s, 1H), 6.77 (d, J = 8.8 Hz, 1H), 6.65–6.60 (m, 2H), 3.92 (s, 3H).
4-(4-Chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzoic Acid (8)
A mixture of 2 M NaOH (6 mL) and dioxane (30 mL) was added to intermediate 6, and the reaction mixture was stirred for 2.5 h at 60 °C. Upon completion, the reaction was diluted with H2O and acidified with aqueous HCl. The product was then extracted with EtOAc (2 × 150 mL), dried over MgSO4, and evaporated to yield 8. Yield: 1.64 g, 85%. 1H NMR (400 MHz, CDCl3) δ 8.03 (d, J = 1.8 Hz, 1H), 7.99 (d, J = 8.6 Hz, 2H), 7.80 (dd, J = 8.3, 2.0 Hz, 1H), 7.76 (d, J = 2.5 Hz, 1H), 7.52 (d, J = 8.4 Hz, 1H), 7.13 (dd, J = 8.8, 2.5 Hz, 1H), 6.98 (s, 1H), 6.80 (d, J = 8.7 Hz, 1H), 6.68–6.63 (m, 2H).
N-(2-Aminoethyl)-4-(4-chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzamide Hydrochloride (9)
A mixture of intermediate carboxylic acid 8 (1 eq), N-Boc-ethylenediamine (1 eq), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC, 2 eq), and hydroxybenzotriazole (HOBt, 0.1 eq) was dissolved in dioxane (0.015 M) and stirred at rt for at least 20 h until completion. The reaction mixture was diluted with EtOAc, and the organic phase was washed with water and brine, dried over MgSO4, and concentrated. The crude product was purified on a silica gel column chromatography with DCM/MeOH to give the Boc-protected intermediate compound tert-butyl(2-(4-(4-chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzamido)ethyl)carbamate (581 mg, 61%). 1H NMR (400 MHz, CDCl3) δ 8.10 (d, J = 1.5 Hz, 1H), 7.83 (dd, J = 8.4, 1.8 Hz, 1H), 7.75–7.68 (m, 3H), 7.54 (d, J = 8.4 Hz, 1H), 7.46 (s, 1H), 7.33 (s, 1H), 7.08 (dd, J = 8.8, 2.5 Hz, 1H), 6.70 (d, J = 8.8 Hz, 1H), 6.64 (d, J = 8.7 Hz, 2H), 5.07 (s, 1H), 3.56–3.53 (m, 2H), 3.40 (d, J = 4.6 Hz, 2H), 1.43 (s, 9H).
The intermediate was dissolved in DCM (0.1 M), and trifluoroacetic acid (TFA, 4 mL/mmol) was added dropwise. The mixture was stirred at rt for 3 h. After completion, the solvents were evaporated and the residue was dissolved in H2O and alkalized with 2 M NaOH. The aqueous phase was then extracted with EtOAc and washed successively with water and brine. After drying over MgSO4 and concentrating, the residue was taken up in EtOAc and 1 M HCl (1 mL/mmol) was added. Evaporation gave 9. Yield: 358 mg, 92%. 1H NMR (400 MHz, MeOD) δ 8.66 (t, 1H), 8.03 (d, J = 2.1 Hz, 1H), 7.84 (dd, J = 8.3, 2.1 Hz, 1H), 7.75 (d, J = 8.8 Hz, 2H), 7.63 (d, J = 2.5 Hz, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.21 (dd, J = 8.8, 2.6 Hz, 1H), 6.82 (d, J = 8.8 Hz, 1H), 6.63 (d, J = 8.8 Hz, 2H), 3.64 (q, J = 5.9 Hz, 2H), 3.14 (t, J = 5.9 Hz, 2H).
4-(4-Chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)-N-(2-isothiocyanatoethyl)benzamide (11)
Intermediate 9 (1 eq) was dissolved in THF (0.02 M), and triethylamine (6 eq) was added. The mixture was cooled to 0 °C before adding carbon disulfide (4 eq), and the reaction mixture was stirred for approximately 2 h at 0 °C until complete conversion to an intermediate. Tosyl chloride (3.5 eq) was added, and the mixture was stirred for an additional 19 h. The reaction was quenched with a phosphate buffer, and the products were extracted into DCM. The organic phase was washed with brine dried over MgSO4 and concentrated. The crude compound was purified with flash column chromatography using DCM/MeOH as eluents to yield final compound 11. Yield: 6 mg, 6%. 1H NMR (400 MHz, CDCl3) δ 8.07 (d, J = 2.1 Hz, 1H), 7.83 (dd, J = 8.4, 2.2 Hz, 1H), 7.72 (d, J = 2.5 Hz, 1H), 7.71–7.67 (m, 2H), 7.54 (d, J = 8.4 Hz, 1H), 7.32 (s, 1H), 7.11 (dd, J = 8.8, 2.5 Hz, 1H), 6.75 (d, J = 8.8 Hz, 1H), 6.69–6.62 (m, 2H), 6.54 (t, J = 5.9 Hz, 1H), 3.83–3.76 (m, 2H), 3.76–3.69 (m, 2H); MS: ESI [M – H]−: 587.9; HPLC: 13.0 min.
N-(2-Acrylamidoethyl)-4-(4-chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzamide (12)
Intermediate 9 (1 eq) was dissolved in acetonitrile (5 mL) and cooled to −78 °C in an acetone/dry ice bath. Triethylamine (3.5 eq) and acryloyl chloride (1.1 eq) were added. The reaction mixture was then allowed to warm to room temperature and stirred for 2 h. The mixture was poured into water and acidified with aqueous HCl, and the aqueous layer was extracted with ethyl acetate twice. The organic layers were washed with water, dried over MgSO4, and concentrated. Purification by preparative TLC with EtOAc/acetone 100:1 as an eluent gave the final compound 12. Yield: 10 mg, 20%. 1H NMR (400 MHz, CDCl3) δ 8.09 (d, J = 2.1 Hz, 1H), 7.82 (dd, J = 8.4, 2.2 Hz, 1H), 7.71 (d, J = 2.1 Hz, 1H), 7.70–7.68 (m, 2H), 7.53 (d, J = 8.4 Hz, 1H), 7.44 (s, 1H), 7.43 (s, 1H), 7.09 (dd, J = 8.8, 2.5 Hz, 1H), 6.73 (d, J = 8.8 Hz, 1H), 6.64 (d, J = 8.8 Hz, 2H), 6.51 (s, 1H), 6.30 (dd, J = 17.0, 1.3 Hz, 1H), 6.14 (dd, J = 17.0, 10.2 Hz, 1H), 5.69 (dd, J = 10.2, 1.3 Hz, 1H), 3.60 (s, 4H); MS: ESI [M – H]−: 600.0; HPLC: 11.7 min.
4-((2-(4-(4-Chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzamido)ethyl) carbamoyl)benzenesulfonyl Fluoride (13)
A mixture of intermediate 9 (1 eq), commercially available 4-fluorosulfonylbenzoic acid (1 eq), EDC (2 eq), and HOBt (0.1 eq) was dissolved in dioxane (0.015 M) and stirred at rt for at least 20 h until completion. The reaction mixture was diluted with EtOAc, and the organic phase was washed with water and brine, dried over MgSO4, and concentrated. The crude product was purified on a silica gel column chromatography with DCM/MeOH to give 13. Yield: 8 mg, 31%. 1H NMR (400 MHz, CDCl3) δ 8.07 (s, 4H), 8.06 (d, J = 2.1 Hz, 1H), 7.89 (t, J = 4.4 Hz, 1H), 7.82 (dd, J = 8.4, 2.1 Hz, 1H), 7.71 (d, J = 2.5 Hz, 1H), 7.67 (d, J = 8.8 Hz, 2H), 7.52 (d, J = 8.4 Hz, 1H), 7.30 (s, 1H), 7.10 (dd, J = 8.8, 2.5 Hz, 1H), 6.93 (t, J = 5.2 Hz, 1H), 6.74 (d, J = 8.8 Hz, 1H), 6.65 (d, J = 8.8 Hz, 2H), 3.81–3.69 (m, 4H); MS: ESI [M – H]−: 732.0; HPLC: 12.8 min.
N-(2-Bromoethyl)-4-(4-chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)benzamide (10)
A mixture of carboxylic acid 8 (1 eq), 2-bromoethylamine (1 eq), EDC (2 eq), and HOBt (0.1 eq) was dissolved in dioxane (0.015 M) and stirred at rt for at least 20 h until completion. The reaction mixture was diluted with EtOAc, and the organic phase was washed with water and brine, dried over MgSO4, and concentrated. The crude product was purified on a silica gel column chromatography with EtOAc/petroleum ether to yield 10. Yield: 62 mg, 51%. 1H NMR (400 MHz, CDCl3) δ 8.06 (s, 1H), 7.85–7.77 (m, 3H), 7.73–7.69 (m, 1H), 7.51 (d, J = 8.4 Hz, 1H), 7.09 (dd, J = 8.7, 2.5 Hz, 1H), 6.73 (d, J = 8.8 Hz, 1H), 6.60 (d, J = 8.8 Hz, 2H), 4.44 (t, J = 9.5 Hz, 2H), 4.05 (t, J = 9.5 Hz, 2H).
4-(4-Chloro-2-((4-chloro-3-(trifluoromethyl)phenyl)sulfonamido)phenoxy)-N-(2-thiocyanatoethyl)benzamide (14)
A mixture of KSCN (6 eq) and the intermediate (1 eq) was dissolved in EtOH (0.025 M), and the mixture was refluxed for 3 days. After completion of the reaction, the solvent was evaporated and the residue was dissolved in EtOAc, washed with water and brine, dried over MgSO4, and concentrated. Preparative TLC with DCM/MeOH 25:1 as an eluent gave the final compound 14. Yield: 4 mg, 21%. 1H NMR (400 MHz, CDCl3) δ 8.05 (d, J = 2.1 Hz, 1H), 7.82 (dd, J = 8.4, 2.1 Hz, 1H), 7.73 (d, J = 2.5 Hz, 1H), 7.73–7.68 (m, 2H), 7.54 (d, J = 8.4 Hz, 1H), 7.19 (s, 1H), 7.11 (dd, J = 8.7, 2.5 Hz, 2H), 6.75 (d, J = 8.8 Hz, 1H), 6.68–6.63 (m, 2H), 6.62 (t, J = 5.8 Hz, 1H), 3.88 (q, J = 6.0 Hz, 2H), 3.26 (t, J = 6.0 Hz, 2H); MS: ESI [M – H]−: 587.9; HPLC: 12.5 min.
Biology
Chemicals and Reagents
[3H]-CCR2-RA-[R] (specific activity 59.6 Ci mmol–1) was custom-labeled by Vitrax (Placentia, CA), and [35S]GTPγS (specific activity 1250 Ci mmol–1) was purchased from PerkinElmer (Groningen, The Netherlands). The CCR2 ligands CCR2-RA-[R] and JNJ-27141491 were synthesized as previously described.53,54 Human CCL2 was purchased from PeproTech (Rocky Hill, NJ). Bovine serum albumin (BSA, fraction V), guanosine 5′-diphosphate (GDP), and iodoacetamide (IA) were from Sigma Aldrich (St. Louis, MO, USA). Pierce bicinchoninic acid (BCA) protein assay kit and coelenterazine (CTZ-n) were obtained from Thermo Fisher Scientific (Rockford, IL). Polyethyleneimine (PEI) was purchased from Polysciences Inc. (Warrington, Pennsylvania). Tango CCR2-bla osteosarcoma cells stably expressing human CCR2b (U2OS-CCR2) were obtained from Invitrogen (Carlsbad, CA). Chinese hamster ovary (CHO) cells were kindly provided by Hans den Dulk (Leiden University, the Netherlands) and originally obtained from ATCC. All other chemicals were from standard commercial sources.
Plasmids Used in the Study
pCDNA3.1+ plasmids contacting the FLAG-tagged wild-type human CCR2, the human CCR2 mutants C70SICL1, C75SICL1, and C232SICL3, and the triple mutant C70S/C75S/C232S were cloned in-house.
The NanoBiT luminescence complementation system has been previously described.55,56 Here, we used the pCAGGS LgBiT-β-arrestin1-EE construct containing the NanoBiT LgBiT fused to the N-terminus of clathrin-binding-deficient variant57 of human β-arrestin1, first described by Shihoya et al..58 We also used the pCAGGS hCCR2b-SmBiT construct in which the SmBiT was fused to the C-terminus of human CCR2b37 with a flexible 15 AA linker (GGSGGGGSGGSSSGG). The plasmids were propagated in E. coli using ampicillin (100 μg/mL) as a bacterial selection marker.
Cell Culture
U2OS-CCR2 and CHO cells were grown as a monolayer in 10 cm ø or 15 cm ø culture plates at 37 °C and 5% CO2. U2OS-CCR2 cells were cultured in McCoy’s 5A medium containing 10% fetal calf serum (FCS), 2 mM glutamine, 0.1 mM non-essential amino acids, 25 mM HEPES, 1 mM sodium pyruvate, 200 IU/mL penicillin, 200 μg/mL streptomycin, 100 μg/mL G418, 40–50 μg/mL hygromycin, and 125 μg/mL zeocin. Cells were subcultured twice a week after trypsinization at a ratio of 1:3 to 1:8. Dialyzed fetal calf serum was used before membrane preparation of U2OS-CCR2 cells. Empty CHO cells were cultured in DMEM/F12 medium supplemented with 10% (v/v) newborn calf serum, 200 IU/mL penicillin, 200 μg/mL streptomycin, and 2 mM glutamine and subcultured twice a week after trypsinization at a ratio of 1:30 to 1:50. HEK293t cells for NanoBiT β-arrestin recruitment experiments were cultured in 75 mL tissue culture treated flasks in DMEM supplemented with 10% FCS and subcultured three times a week at a ratio of 1:10 after trypsinization.
Transfections
Transfections of CHO cells with FLAG-tagged WT or mutant CCR2 were performed using a previously described PEI method.23 Briefly, empty CHO cells were grown to ∼50% confluence in 15 cm ø culture plates and transfected with a DNA/PEI mixture in 150 mM NaCl solution containing 10 μg of plasmid mixed with PEI (1 mg/mL) at a mass ratio of 1:6. Before transfection, the DNA/PEI mixture was incubated for 20 min and the culture medium of the cells was refreshed. After 24 h, sodium butyrate (final concentration of 3 mM) was added to the plates to increase receptor expression.59 Finally, cells were incubated for another 24 h at 37 °C and 5% CO2. For NanoBiT β-arrestin recruitment experiments, HEK293t cells were transiently transfected with CCR2b-SmBiT and LgBiT-β-arrestin1-EE plasmids using TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI, USA). Empty cells were grown in 6 cm ø culture plates to 60–80% confluence. Prior to transfection, the culture medium on the cells was replaced with fresh DMEM +10% FBS. DNA/reagent mixture was prepared with 3 μg of CCR2b-SmBiT DNA and 3 μg of LgBiT-β-arrestin1-EE DNA mixed with 18 μL of TransIT-LT1 reagent in 600 μL of serum-free OptiMEM medium, according to the manufacturer’s instructions. The DNA/reagent mixture was incubated for 30 min at room temperature and added dropwise to the cells. The transfected cells were incubated for another 24 h at 37 °C and 5% CO2 before performing the NanoBiT assays.
Membrane Preparation
Membranes from U2OS-CCR2 or CHO cells transiently transfected with wild-type (WT) human CCR2 or human CCR2 mutants were prepared as previously described, using several centrifugation and homogenization steps.35 Final membrane pellets were resuspended in ice-cold Tris buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2), homogenized with an Ultra Turrax homogenizer (IKA-Werke GmbH & Co. KG, Staufen, Germany), and stored in aliquots of 100 or 250 μL at −80 °C. A standard BCA protein determination assay was used to measure the membrane protein concentrations (Pierce BCA protein assay kit).60
Radioligand Binding Assays
[3H]-CCR2-RA and [3H]-CCR2-RA-[R] Binding Assays
For all radioligand binding assays, membranes from U2OS-CCR2 cells or CHO cells transiently transfected with WT or mutant CCR2 were first thawed and homogenized using an Ultra Turrax homogenizer (IKA-Werke GmbH & Co.KG, Staufen, Germany). Membranes were then diluted in assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.1% CHAPS) to a final concentration of 5–30 μg membrane protein in a total volume of 100 μL. For displacement assays, membranes were coincubated for 2 h at 25 °C with multiple concentrations of competing ligand, ranging from 0.01 nM to 10 μM, and a fixed concentration of radioligand (∼6 nM [3H]-CCR2-RA or [3H]-CCR2-RA-[R]). At this concentration, we ensured that the total radioligand binding did not exceed 10% of the total radioactivity added in order to prevent radioligand depletion. In all cases, nonspecific binding was determined using 10 μM JNJ-27141491. In the case of preincubation experiments, membranes were preincubated for 4 h with increasing concentrations of competing ligand before addition of ∼6 nM [3H]-CCR2-RA-[R] and further coincubation of 20 min. In all assays, incubations were terminated by rapid vacuum filtration through pre-wetted GF/B filter plates using a PerkinElmer FilterMate harvester (PerkinElmer, Groningen, The Netherlands). Filters were subsequently washed 10 times with ice-cold wash buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.01% CHAPS) and dried at 55 °C for at least 30 min. Finally, filter-bound radioactivity was measured in a P-E 2450 Microbeta2 counter (PerkinElmer) after addition of 25 μL of Microscint scintillation cocktail (PerkinElmer).
Washout Assays with [3H]-CCR2-RA-[R]
For washout assays, membrane homogenates (80–100 μg) were preincubated in 1.5 mL Eppendorf tubes with a single concentration of compounds 7 and 14 in a final volume of 300 μL of assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.1% CHAPS). In the case of washout assays using U2OS-CCR2 membranes, a 10×IC50 concentration was chosen for 7 (90 nM) and 14 (12 nM). In the case of washout assays using CHO-CCR2 mutant receptor membranes, concentrations of 200 and 60 nM were used for 7 and 14, respectively, to ensure a saturating concentration in all mutants despite changes in affinity. For the triple mutant, the concentration of 14 was increased to 160 nM to account for the loss in affinity. In the case of washout assays with IA, U2OS-CCR2 membranes were pretreated with 2 mg/mL IA for 30 min at room temperature and protected from light prior to incubation with ligands. After incubation for 2 h at 25 °C with shaking at approximately 800 rpm, the mixture was centrifuged at 13,000 rpm for 5 min at 4 °C and the supernatant containing unbound ligand was removed. The remaining pellet was resuspended in 1 mL assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.1% CHAPS) and incubated for an extra 20 min before another cycle of centrifugation and washing. After four cycles, the final membrane pellet was resuspended in 300 μL of assay buffer, transferred to test tubes, and incubated with 100 μL of ∼6 nM [3H]-CCR2-RA-[R] for 2 h at 25 °C while shaking. Incubations were terminated by rapid filtration through a pre-wetted Whatman GF/B filter using a Brandel harvester 24 (Brandel, Gaithersburg, MD, USA). Filters were washed three times with 2 mL of ice-cold wash buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 0.01% CHAPS) and transferred to polyethylene Pony vials (PerkinElmer) before measurement of filter-bound radioactivity in a Tri-Carb 2810TR liquid scintillation analyzer (PerkinElmer).
[35S]GTPγS Binding Assays
[35S]GTPγS binding assays in CCR1, CCR2 and CCR5 were performed as previously described.48,52 Briefly, U2OS-CCR1 or U2OS-CCR2 membrane homogenates (10 μg) were diluted in assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA, and 0.05% BSA) supplemented with saponin (0.5 mg/mL) and 10 μM GDP to a total volume of 100 μL. In the case of U2OS-CCR5, 10 μg of membrane homogenates was diluted in assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA, and 0.05% BSA) supplemented with 0.25 mg/mL saponin and 5 μM GDP. To determine the (p)IC50 values of 7 and 14, membranes were preincubated for 30 min at 25 °C with increasing concentrations of ligand in the presence of an EC80 concentration of CCL2 (20 nM) or CCL3 (8 nM for CCR1 and 25 nM for CCR5), as previously determined.48,52 Basal activity was determined in the absence of any ligand or chemokine; maximal activity in the presence of CCL2 or CCL3 only. After addition of 20 μL of [35S]GTPγS (0.3 nM), the mixture was incubated for 90 more minutes at 25 °C before stopping the reaction with ice-cold wash buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2). Filtration and radioactivity measurement were performed as described under the Binding Assays section.
Washout Assays with [35S]GTPγS
Washout assays with [35S]GTPγS were performed as described under the Washout Assays section. Briefly, U2OS-CCR2 membrane homogenates (40 μg) were preincubated in the absence or presence of a single concentration of 7 or 14 (600 or 250 nM, respectively) in a final volume of 400 μL of buffer containing 50 mM Tris-HCl (pH 7.4) and 5 mM MgCl2. After four cycles of centrifugation and washing, the remaining pellets were transferred to test tubes in a final volume of 320 μL containing [35S]GTPγS assay buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1 mM EDTA, and 0.05% BSA), saponin (0.5 mg/mL), GDP (10 μM), and an EC80 concentration of CCL2 (20 nM). For unwashed samples, only the last centrifugation step was performed in order to resuspend the sample in the same volume of 320 μL. All samples were then preincubated for 30 min at 25 °C before addition of 80 μL of [35S]GTPγS (0.3 nM). Reactions were stopped after 90 min at 25 °C by rapid filtration as described under the Washout Assays section, but using [35S]GTPγS wash buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2).
NanoBiT Assays for Quantifying β-arrestin1 Recruitment to CCR2 in Live Cells
Twenty-four hours post-transfection of HEK293t cells with CCR2b-SmBiT and LgBiT-β-arrestin1-EE plasmids, the cells were detached with PBS/EDTA (0.2 mM), centrifuged, and resuspended in assay buffer (1× HBSS, 5 mM HEPES pH 7.2, 0.05% BSA) to a concentration of 1.3 × 106 cells/mL. The cells were aliquoted into a conical-bottom 96-well plate (Greiner Bio-One, #651201) at 120,000 cells per well in 90 μL of assay buffer. Serial dilutions of test compounds in assay buffer, at 10 × final concentrations, were prepared from 10 mM DMSO stocks. Then, 10 μL of assay buffer (unwashed wells) or diluted test compounds (washout wells) was added to the plate. After incubating the plate at room temperature for at least 20 min, cells were washed twice by spinning the plate at 400g for 5 min and discarding the supernatant. Afterward, cells were resuspended in 80 μL of assay buffer and transferred to a Falcon 96-well black/clear flat-bottom tissue-culture treated plate (Corning, NY, USA). Ten microliters of assay buffer or 10 μL of test compounds was added to washout and unwashed wells, respectively. Ten microliters of coelenterazine (CTZ-n, Fisher Scientific, Waltham, MA, USA), diluted in assay buffer from a 5 mM stock in ethanol, was added to the plate to achieve a final concentration of 10 μM in a 100 μL final volume in each well. The plate was then incubated at room temperature for approximately 90 min, protected from light. Backing tape (PerkinElmer, #6005199) was applied to the bottom of the plate, after which basal luminescence from each well was read using a PerkinElmer Victor X Light 2030 apparatus (1 s, no filter). Next, 10 μL of 2 μM CCL2 in assay buffer was added to each well and the cells were further incubated at room temperature for 10 min, protected from light, after which the plate was read again for endpoint luminescence generated by structural complementation.
Molecular Modeling and Ligand Docking
Generation of a Model of WT CCR2
The crystal structure of engineered human CCR2 in complex with an orthosteric and an allosteric antagonist (PDB 5T1A(20)) was used as a template for the generation of the CCR2 model. All modeling and docking was performed in ICM v3.8-7a (Molsoft LLC, San Diego, CA61). The T4 lysozyme (T4L) fusion protein present in the structure was removed, the mutated residues in the intracellular part of TM6 was reverted to WT, and the intracellular loop 3 (ICL3) was rebuilt. For rebuilding ICL3, a peptide containing residues 223:243 of CCR2 was built ab initio as follows: the backbones of residues 223:231 and 236:243 and the side chains of residues 223:226 and 241:243 were tethered to their respective positions in the crystal structure, the receptor converted into a set of three-dimensional (3D) grid maps representing van der Waals, electrostatic, hydrogen bonding, and surface energy potentials, and the peptide conformation was optimized using the biased probability Monte Carlo (BPMC) sampling in internal coordinates as implemented in ICM.61 The simulation simultaneously optimized the intramolecular energy of the peptide and its interaction with the context in the form of potential grid maps. The best scoring conformation of the peptide was then merged with the rest of the receptor coordinates, and the system was minimized in its full-atom representation, with harmonic restraints of gradually decreasing strength imposed between the model and either the X-ray coordinates or the best prediction conformations of the ab initio modeled ICL3. Toward the end of the optimization, the restraints were released entirely. The simulations were performed in the presence of crystallographic ligands.
Generation of a Model of CCR2:14 Complex
Alternative conformers of the allosteric pocket in CCR2 were generated by systematic sampling of aliphatic side-chain rotamers and translations of the intracellular ends of TM helices 6 and 7. The conformer that best predicted the binding poses of the sulfonamide series of allosteric CCR2 antagonists (analogous to 14) was chosen. The binding pose of 14 was predicted by covalent docking in ICM under the assumption that the covalent attachment residue is C75 due to its proximity. For docking, the receptor atoms in the 4 Å vicinity of the allosteric pocket were represented as 3D grid potential maps as above, with the exception of C75 for which the explicit representation was used. The covalent bond between 14 and C75 was imposed, and the system was sampled as described above to generate a ranked list of alternative conformations for the ligand. The top 10 conformations were merged with the full-atom model of the receptor and re-scored using the ICM ligand scoring function previously optimized for ligand geometry prediction on a diverse benchmark of crystallographic protein–ligand complexes.62 The top-scoring pose was selected.
Data Analysis
Data analyses were performed using Prism 7.00 (GraphPad software, San Diego, CA, USA). (p)IC50 values from radioligand displacement assays, [35S]GTPγS binding assays, and NanoBiT assays were obtained by non-linear regression curve fitting into a sigmoidal concentration–response curve using the equation: Y = Bottom + (Top – Bottom)/(1 + 10^(X – LogIC50)). pKi values were obtained from pIC50 values using the Cheng–Prusoff equation.63 Data are shown as mean ± standard error of the mean (SEM) of at least three individual experiments performed in duplicate. Statistical analyses were performed as indicated. If p-values were below 0.05, observed differences were considered statistically significant.
Acknowledgments
This work is supported by NIH grants R01 AI118985, R01 GM117424, R01 GM136202, and R21 AI149369 to I.K. and T.M.H. K.K.C. is supported by a Training Fellowship from the Cancer Cell Mapping Initiative Consortium U54 CA209891. L.H.H. and N.V.O.Z. were also supported by funding from Oncode Institute, The Netherlands. A.I. was funded by the LEAP JP19gm0010004 from the Japan Agency for Medical Research and Development (AMED).
Glossary
ABBREVIATIONS
- BCA
bicinchoninic acid
- BPMC
biased probability Monte Carlo
- BSA
bovine serum albumin (BSA)
- CCR2
CC chemokine receptor 2
- CCR2a
CCR2 isoform A
- CCR2b
CCR2 isoform B
- CHO
Chinese hamster ovary
- CTZ-n
coelenterazine
- FCS
fetal calf serum
- GDP
guanosine 5′-diphosphate
- GPCRs
G protein-coupled receptors
- [35S]GTPyS
guanosine 5′-O-[γ-thio]-triphosphate
- IA
iodoacetamide
- ICL3
intracellular loop 3
- NanoBiT
NanoLuc binary technology
- NAM
negative allosteric modulator
- PEI
polyethyleneimine
- SEM
standard error of the mean
- T4L
T4 lysozyme
- U2OS
human osteosarcoma cells
- WT
wild-type
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c01137.
Washout-radioligand experiments in presence of N-ethylmaleimide (Figure S1); washout-radioligand experiments in CCR2-C75A mutant (Figure S2); selectivity assessment of compounds (Figure S3); HPLC traces of final compounds 7 and 14 (Figure S4); HPLC traces of final compounds 11, 12, and 13 (Figure S5); and potency of compounds 7 and 14 in U2OS-CCR1 and U2OS-CCR5 membranes using [35S]GTPγS binding assays (Table S1) (PDF)
Molecular formula strings (CSV)
Homology model of CCR2b with docking of compounds 7 and 14 (PDB, PDB)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Notes
The authors will release the atomic coordinates of the docking models upon article publication.
Supplementary Material
References
- Bachelerie F.; Ben-Baruch A.; Burkhardt A. M.; Combadiere C.; Farber J. M.; Graham G. J.; Horuk R.; Sparre-Ulrich A. H.; Locati M.; Luster A. D.; Mantovani A.; Matsushima K.; Murphy P. M.; Nibbs R.; Nomiyama H.; Power C. A.; Proudfoot A. E.; Rosenkilde M. M.; Rot A.; Sozzani S.; Thelen M.; Yoshie O.; Zlotnik A. International union of basic and clinical pharmacology. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 2014, 66, 1–79. 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola A.; Luster A. D. Chemokines and their receptors: drug targets in immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 171–197. 10.1146/annurev.pharmtox.48.121806.154841. [DOI] [PubMed] [Google Scholar]
- O’Connor T.; Borsig L.; Heikenwalder M. CCL2-CCR2 signaling in disease pathogenesis. Endocr. Metab. Immune Disord. Drug Targets 2015, 15, 105–118. 10.2174/1871530315666150316120920. [DOI] [PubMed] [Google Scholar]
- Bot I.; Ortiz Zacarias N. V.; de Witte W. E. A.; de Vries H.; van Santbrink P. J.; van der Velden D.; Kroner M. J.; van der Berg D. J.; Stamos D.; de Lange E. C. M.; Kuiper J.; IJzerman A. P.; Heitman L. H. A novel CCR2 antagonist inhibits atherogenesis in apoE deficient mice by achieving high receptor occupancy. Sci. Rep. 2017, 7, 52. 10.1038/s41598-017-00104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. S.; Lee M. H.; Song H. K.; Ko G. J.; Kwon O. S.; Lim T. K.; Kim S. H.; Han S. Y.; Han K. H.; Lee J. E.; Han J. Y.; Kim H. K.; Cha D. R. CCR2 antagonism improves insulin resistance, lipid metabolism, and diabetic nephropathy in type 2 diabetic mice. Kidney Int. 2010, 78, 883–894. 10.1038/ki.2010.263. [DOI] [PubMed] [Google Scholar]
- Piotrowska A.; Kwiatkowski K.; Rojewska E.; Slusarczyk J.; Makuch W.; Basta-Kaim A.; Przewlocka B.; Mika J. Direct and indirect pharmacological modulation of CCL2/CCR2 pathway results in attenuation of neuropathic pain - In vivo and in vitro evidence. J. Neuroimmunol. 2016, 297, 9–19. 10.1016/j.jneuroim.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Qian B. Z.; Li J.; Zhang H.; Kitamura T.; Zhang J.; Campion L. R.; Kaiser E. A.; Snyder L. A.; Pollard J. W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horuk R. Chemokine receptor antagonists: overcoming developmental hurdles. Nat. Rev. Drug Discovery 2009, 8, 23–33. 10.1038/nrd2734. [DOI] [PubMed] [Google Scholar]
- Weichert D.; Gmeiner P. Covalent molecular probes for class A G protein-coupled receptors: advances and applications. ACS Chem. Biol. 2015, 10, 1376–1386. 10.1021/acschembio.5b00070. [DOI] [PubMed] [Google Scholar]
- Ghosh A. K.; Samanta I.; Mondal A.; Liu W. R. Covalent inhibition in drug discovery. ChemMedChem 2019, 14, 889–906. 10.1002/cmdc.201900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L. H. Cell permeable affinity- and activity-based probes. Future Med. Chem. 2015, 7, 2131–2141. 10.4155/fmc.15.100. [DOI] [PubMed] [Google Scholar]
- Jörg M.; Scammells P. J. Guidelines for the synthesis of small-molecule irreversible probes targeting G protein-coupled receptors. ChemMedChem 2016, 11, 1488–1498. 10.1002/cmdc.201600066. [DOI] [PubMed] [Google Scholar]
- Bauer R. A. Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discovery Today 2015, 20, 1061–1073. 10.1016/j.drudis.2015.05.005. [DOI] [PubMed] [Google Scholar]
- González-Bello C. Designing irreversible inhibitors--worth the effort?. ChemMedChem 2016, 11, 22–30. 10.1002/cmdc.201500469. [DOI] [PubMed] [Google Scholar]
- Baillie T. A. Targeted covalent inhibitors for drug design. Angew. Chem., Int. Ed. 2016, 55, 13408–13421. 10.1002/anie.201601091. [DOI] [PubMed] [Google Scholar]
- Schall T. J.; Proudfoot A. E. I. Overcoming hurdles in developing successful drugs targeting chemokine receptors. Nat. Rev. Immunol. 2011, 11, 355–363. 10.1038/nri2972. [DOI] [PubMed] [Google Scholar]
- Hua T.; Vemuri K.; Nikas S. P.; Laprairie R. B.; Wu Y.; Qu L.; Pu M.; Korde A.; Jiang S.; Ho J. H.; Han G. W.; Ding K.; Li X.; Liu H.; Hanson M. A.; Zhao S.; Bohn L. M.; Makriyannis A.; Stevens R. C.; Liu Z. J. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 2017, 547, 468–471. 10.1038/nature23272. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Glukhova A.; Thal D. M.; Nguyen A. T.; Vecchio E. A.; Jörg M.; Scammells P. J.; May L. T.; Sexton P. M.; Christopoulos A. Structure of the adenosine A1 receptor reveals the basis for subtype selectivity. Cell 2017, 168, 867–877.e13. 10.1016/j.cell.2017.01.042. [DOI] [PubMed] [Google Scholar]
- Apel A. K.; Cheng R. K. Y.; Tautermann C. S.; Brauchle M.; Huang C. Y.; Pautsch A.; Hennig M.; Nar H.; Schnapp G. Crystal structure of CC chemokine receptor 2A in complex with an orthosteric antagonist provides insights for the design of selective antagonists. Structure 2019, 27, 427–438.e5. 10.1016/j.str.2018.10.027. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Qin L.; Ortiz Zacarías N. V.; de Vries H.; Han G. W.; Gustavsson M.; Dabros M.; Zhao C.; Cherney R. J.; Carter P.; Stamos D.; Abagyan R.; Cherezov V.; Stevens R. C.; Ijzerman A. P.; Heitman L. H.; Tebben A.; Kufareva I.; Handel T. M. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature 2016, 540, 458–461. 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz Zacarias N. V.; Lenselink E. B.; IJzerman A. P.; Handel T. M.; Heitman L. H. Intracellular receptor modulation: novel approach to target GPCRs. Trends Pharmacol. Sci. 2018, 39, 547–559. 10.1016/j.tips.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace S.; Philp J.; Brooks C.; Piercy V.; Moores K.; Smethurst C.; Watson S.; Gaines S.; Zippoli M.; Mookherjee C.; Ife R. Identification of a sulfonamide series of CCR2 antagonists. Bioorg. Med. Chem. Lett. 2010, 20, 3961–3964. 10.1016/j.bmcl.2010.04.142. [DOI] [PubMed] [Google Scholar]
- Zweemer A. J. M.; Bunnik J.; Veenhuizen M.; Miraglia F.; Lenselink E. B.; Vilums M.; de Vries H.; Gibert A.; Thiele S.; Rosenkilde M. M.; IJzerman A. P.; Heitman L. H. Discovery and mapping of an intracellular antagonist binding site at the chemokine receptor CCR2. Mol. Pharmacol. 2014, 86, 358–368. 10.1124/mol.114.093328. [DOI] [PubMed] [Google Scholar]
- Wang G. Z.; Haile P. A.; Daniel T.; Belot B.; Viet A. Q.; Goodman K. B.; Sha D.; Dowdell S. E.; Varga N.; Hong X.; Chakravorty S.; Webb C.; Cornejo C.; Olzinski A.; Bernard R.; Evans C.; Emmons A.; Briand J.; Chung C. W.; Quek R.; Lee D.; Gough P. J.; Sehon C. A. CCR2 receptor antagonists: optimization of biaryl sulfonamides to increase activity in whole blood. Bioorg. Med. Chem. Lett. 2011, 21, 7291–7294. 10.1016/j.bmcl.2011.10.038. [DOI] [PubMed] [Google Scholar]
- Burke T. R.; Bajwa B. S.; Jacobson A. E.; Rice K. C.; Streaty R. A.; Klee W. A. Probes for narcotic receptor mediated phenomena. 7. Synthesis and pharmacological properties of irreversible ligands specific for μ or δ opiate receptors. J. Med. Chem. 1984, 27, 1570–1574. 10.1021/jm00378a008. [DOI] [PubMed] [Google Scholar]
- Nakamura T.; Kawai Y.; Kitamoto N.; Osawa T.; Kato Y. Covalent modification of lysine residues by allyl isothiocyanate in physiological conditions: plausible transformation of isothiocyanate from thiol to amine. Chem. Res. Toxicol. 2009, 22, 536–542. 10.1021/tx8003906. [DOI] [PubMed] [Google Scholar]
- Doorn J. A.; Petersen D. R. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem. Res. Toxicol. 2002, 15, 1445–1450. 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- Narayanan A.; Jones L. H. Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 2015, 6, 2650–2659. 10.1039/C5SC00408J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F.; Bikbulatov R. V.; Mocanu V.; Dicheva N.; Parker C. E.; Wetsel W. C.; Mosier P. D.; Westkaemper R. B.; Allen J. A.; Zjawiony J. K.; Roth B. L. Structure-based design, synthesis, and biochemical and pharmacological characterization of novel salvinorin A analogues as active state probes of the kappa-opioid receptor. Biochemistry 2009, 48, 6898–6908. 10.1021/bi900605n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy F. D.; Ou K. Selective reduction of aromatic nitro compounds with stannous chloride in non acidic and non aqueous medium. Tetrahedron Lett. 1984, 25, 839–842. 10.1016/S0040-4039(01)80041-1. [DOI] [Google Scholar]
- Kim J.-G.; Jang D. Mild and efficient indium metal catalyzed synthesis of sulfonamides and sulfonic esters. Synlett 2007, 2007, 2501–2504. 10.1055/s-2007-986632. [DOI] [Google Scholar]
- El-Faham A.; Albericio F. Peptide coupling reagents, more than a letter soup. Chem. Rev. 2011, 111, 6557–6602. 10.1021/cr100048w. [DOI] [PubMed] [Google Scholar]
- Wong R.; Dolman S. J. Isothiocyanates from tosyl chloride mediated decomposition of in situ generated dithiocarbamic acid salts. J. Org. Chem. 2007, 72, 3969–3971. 10.1021/jo070246n. [DOI] [PubMed] [Google Scholar]
- Dasse O.; Evans J.; Zhai H.-X.; Zou D.; Kintigh J.; Chan F.; Hamilton K.; Hill E.; Eckman J.; Higgins P.; Volosov A.; Collart P.; Nicolas J. M.; Kondru R. K.; Schwartz C. E. Novel, acidic CCR2 receptor antagonists: lead optimization. Lett. Drug Des. Discovery 2007, 4, 263–271. 10.2174/157018007784619989. [DOI] [Google Scholar]
- Zweemer A. J. M.; Nederpelt I.; Vrieling H.; Hafith S.; Doornbos M. L. J.; de Vries H.; Abt J.; Gross R.; Stamos D.; Saunders J.; Smit M. J.; IJzerman A. P.; Heitman L. H. Multiple binding sites for small-molecule antagonists at the CC chemokine receptor 2. Mol. Pharmacol. 2013, 84, 551–561. 10.1124/mol.113.086850. [DOI] [PubMed] [Google Scholar]
- Garritsen A.; IJzerman A. P.; Beukers M. W.; Soudijn W. Chemical modification of adenosine A1 receptors: implications for the interaction with R-PIA. Biochem. Pharmacol. 1990, 40, 835–842. 10.1016/0006-2952(90)90324-E. [DOI] [PubMed] [Google Scholar]
- Regan D. P.; Coy J. W.; Chahal K. K.; Chow L.; Kurihara J. N.; Guth A. M.; Kufareva I.; Dow S. W. The angiotensin receptor blocker losartan suppresses growth of pulmonary metastases via AT1R-independent inhibition of CCR2 signaling and monocyte recruitment. J. Immunol. 2019, 202, 3087–3102. 10.4049/jimmunol.1800619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. H. Progress in the discovery of CC chemokine receptor 2 antagonists, 2009 – 2012. Expert Opin. Ther. Pat. 2013, 23, 549–568. 10.1517/13543776.2013.771168. [DOI] [PubMed] [Google Scholar]
- Strelow J. M. A perspective on the kinetics of covalent and irreversible inhibition. SLAS Discov 2017, 22, 3–20. 10.1177/1087057116671509. [DOI] [PubMed] [Google Scholar]
- Xia L.; de Vries H.; Yang X.; Lenselink E. B.; Kyrizaki A.; Barth F.; Louvel J.; Dreyer M. K.; van der Es D.; IJzerman A. P.; Heitman L. H. Kinetics of human cannabinoid 1 (CB1) receptor antagonists: structure-kinetics relationships (SKR) and implications for insurmountable antagonism. Biochem. Pharmacol. 2018, 151, 166–179. 10.1016/j.bcp.2017.10.014. [DOI] [PubMed] [Google Scholar]
- Swinney D. C.; Beavis P.; Chuang K. T.; Zheng Y.; Lee I.; Gee P.; Deval J.; Rotstein D. M.; Dioszegi M.; Ravendran P.; Zhang J.; Sankuratri S.; Kondru R.; Vauquelin G. A study of the molecular mechanism of binding kinetics and long residence times of human CCR5 receptor small molecule allosteric ligands. Br. J. Pharmacol. 2014, 171, 3364–3375. 10.1111/bph.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss S. M.; Jayasekara P. S.; Paoletta S.; Gao Z. G.; Jacobson K. A. Structure-based design of reactive nucleosides for site-specific modification of the A2A adenosine receptor. ACS Med. Chem. Lett. 2014, 5, 1043–1048. 10.1021/ml5002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone R. P.; Khanolkar A. D.; Xu W.; Ayotte L. A.; Thakur G. A.; Hurst D. P.; Abood M. E.; Reggio P. H.; Fournier D. J.; Makriyannis A. (−)-7’-Isothiocyanato-11-hydroxy-1′,1′-dimethylheptylhexahydrocannabinol (AM841), a high-affinity electrophilic ligand, interacts covalently with a cysteine in helix six and activates the CB1 cannabinoid receptor. Mol. Pharmacol. 2005, 68, 1623–1635. 10.1124/mol.105.014407. [DOI] [PubMed] [Google Scholar]
- Yang X.; Dong G.; Michiels T. J. M.; Lenselink E. B.; Heitman L.; Louvel J.; IJzerman A. P. A covalent antagonist for the human adenosine A2A receptor. Purinergic Signalling 2017, 13, 191–201. 10.1007/s11302-016-9549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; van Veldhoven J. P. D.; Offringa J.; Kuiper B. J.; Lenselink E. B.; Heitman L. H.; van der Es D.; IJzerman A. P. Development of covalent ligands for G protein-coupled receptors: a case for the human adenosine A(3) receptor. J. Med. Chem. 2019, 62, 3539–3552. 10.1021/acs.jmedchem.8b02026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos M. L. J.; Wang X.; Vermond S. C.; Peeters L.; Perez-Benito L.; Trabanco A. A.; Lavreysen H.; Cid J. M.; Heitman L. H.; Tresadern G.; IJzerman A. P. Covalent allosteric probe for the metabotropic glutamate receptor 2: design, synthesis, and pharmacological characterization. J. Med. Chem. 2019, 62, 223–233. 10.1021/acs.jmedchem.8b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietsch A.; Beckwith J. The genetics of disulfide bond metabolism. Annu. Rev. Genet. 1998, 32, 163–184. 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- Ortiz Zacarías N. V.; van Veldhoven J. P. D.; Portner L.; van Spronsen E.; Ullo S.; Veenhuizen M.; van der Velden W. J. C.; Zweemer A. J. M.; Kreekel R. M.; Oenema K.; Lenselink E. B.; Heitman L. H.; IJzerman A. P. Pyrrolone derivatives as intracellular allosteric modulators for chemokine receptors: selective and dual-targeting inhibitors of CC chemokine receptors 1 and 2. J. Med. Chem. 2018, 61, 9146–9161. 10.1021/acs.jmedchem.8b00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R. W.; Pei Y.; Pandarinathan L.; Janero D. R.; Zhang J.; Makriyannis A. hCB2 ligand-interaction landscape: cysteine residues critical to biarylpyrazole antagonist binding motif and receptor modulation. Chem. Biol. 2010, 17, 1132–1142. 10.1016/j.chembiol.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H.; Sawyer G. W.; Ehlert F. J. Mutagenesis of nucleophilic residues near the orthosteric binding pocket of M1 and M2 muscarinic receptors: effect on the binding of nitrogen mustard analogs of acetylcholine and McN-A-343. Mol. Pharmacol. 2010, 78, 745–755. 10.1124/mol.110.065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. C.; Lee C. T.; Amaro R. E. Structural basis for ligand modulation of the CCR2 conformational landscape. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 8131. 10.1073/pnas.1814131116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz Zacarias N. V.; van Veldhoven J. P. D.; den Hollander L. S.; Dogan B.; Openy J.; Hsiao Y. Y.; Lenselink E. B.; Heitman L. H.; IJzerman A. P. Synthesis and pharmacological evaluation of triazolopyrimidinone derivatives as noncompetitive, intracellular antagonists for CC chemokine receptors 2 and 5. J. Med. Chem. 2019, 62, 11035–11053. 10.1021/acs.jmedchem.9b00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon J.; Coesemans E.; Boeckx S.; Buntinx M.; Hermans B.; Van Wauwe J. P.; Gilissen R. A. H. J.; De Groot A. H. J.; Corens D.; Van Lommen G. Discovery of potent, orally bioavailable small-molecule inhibitors of the human CCR2 receptor. ChemMedChem 2008, 3, 660–669. 10.1002/cmdc.200700276. [DOI] [PubMed] [Google Scholar]
- Zou D.; Zhai H.-X.; Eckman J.; Higgins P.; Gillard M.; Knerr L.; Carre S.; Pasau P.; Collart P.; Grassi J. Novel, acidic CCR2 receptor antagonists: from hit to lead. Lett. Drug. Des. Discov. 2007, 4, 185–191. 10.2174/157018007780077381. [DOI] [Google Scholar]
- Dixon A. S.; Schwinn M. K.; Hall M. P.; Zimmerman K.; Otto P.; Lubben T. H.; Butler B. L.; Binkowski B. F.; Machleidt T.; Kirkland T. A.; Wood M. G.; Eggers C. T.; Encell L. P.; Wood K. V. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016, 11, 400–408. 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- England C. G.; Ehlerding E. B.; Cai W. NanoLuc: a small luciferase is brightening up the field of bioluminescence. Bioconjugate Chem. 2016, 27, 1175–1187. 10.1021/acs.bioconjchem.6b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-M.; Benovic J. L. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J. Biol. Chem. 2002, 277, 30760–30768. 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- Shihoya W.; Izume T.; Inoue A.; Yamashita K.; Kadji F. M. N.; Hirata K.; Aoki J.; Nishizawa T.; Nureki O. Crystal structures of human ETB receptor provide mechanistic insight into receptor activation and partial activation. Nat. Commun. 2018, 9, 4711. 10.1038/s41467-018-07094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuisset L.; Tichonicky L.; Jaffray P.; Delpech M. The effects of sodium butyrate on transcription are mediated through activation of a protein phosphatase. J Biol Chem 1997, 272, 24148–24153. 10.1074/jbc.272.39.24148. [DOI] [PubMed] [Google Scholar]
- Smith P. K.; Krohn R. I.; Hermanson G. T.; Mallia A. K.; Gartner F. H.; Provenzano M. D.; Fujimoto E. K.; Goeke N. M.; Olson B. J.; Klenk D. C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Abagyan R.; Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994, 235, 983–1002. 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Schapira M.; Totrov M.; Abagyan R. Prediction of the binding energy for small molecules, peptides and proteins. J. Mol. Recognit. 1999, 12, 177–190. . [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Prusoff W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.