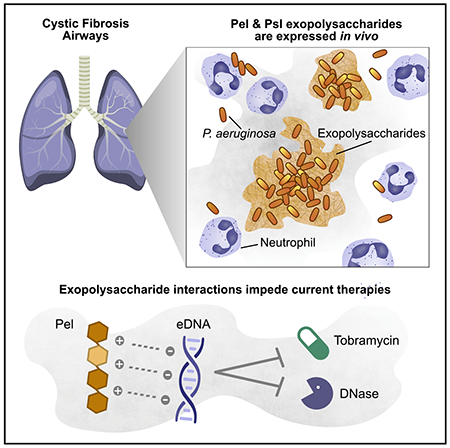
SUMMARY
In cystic fibrosis (CF) airways, Pseudomonas aeruginosa forms cellular aggregates called biofilms that are thought to contribute to chronic infection. To form aggregates, P. aeruginosa can use different mechanisms, each with its own pathogenic implications. However, how they form in vivo is controversial and unclear. One mechanism involves a bacterially produced extracellular matrix that holds the aggregates together. Pel and Psl exopolysaccharides are structural and protective components of this matrix. We develop an immunohistochemical method to visualize Pel and Psl in CF sputum. We demonstrate that both exopolysaccharides are expressed in the CF airways and that the morphology of aggregates is consistent with an exopolysaccharide-dependent aggregation mechanism. We reason that the cationic exopolysaccharide Pel may interact with some of the abundant anionic host polymers in sputum. We show that Pel binds extracellular DNA (eDNA) and that this interaction likely impacts current therapies by increasing antimicrobial tolerance and protecting eDNA from digestion.
In brief
Pseudomonas aeruginosa forms biofilm aggregates in cystic fibrosis airways. Jennings et al. demonstrate that Pel and Psl exopolysaccharides are produced by P. aeruginosa in cystic fibrosis sputum and that aggregates are consistent with a polysaccharide-dependent aggregation mechanism. Exopolysaccharide interactions with extracellular DNA increase antimicrobial tolerance and protect DNA from digestion.
Graphical Abstract
INTRODUCTION
Cystic fibrosis (CF) is caused by mutations in the gene encoding the CF transmembrane conductance regulator (CFTR), a membrane anion channel (Stoltz et al., 2015). The vast majority of individuals with CF will eventually become chronically infected with the Gram-negative pathogen Pseudomonas aeruginosa (Lyczak et al., 2002; Moreau-Marquis et al., 2008). Persistence of CF airway infections is associated with non-resolving inflammation, accelerated lung disease, and earlier mortality (Gibson et al., 2003; Lyczak et al., 2002). In vitro antibiotic susceptibility of P. aeruginosa clinical isolates by itself does not predict treatment outcomes (Somayaji et al., 2019). Thus, it is not clear which host and microbiological factors influence antibiotic efficacy.
In the CF airways, P. aeruginosa is thought to reside as a biofilm or aggregate of cells embedded in a polymer-rich matrix (Bjarnsholt et al., 2009; Costerton et al., 1995; Singh et al., 2000). Exopolysaccharides (EPSs) are key components of this matrix (Flemming and Wingender, 2010; Pamp et al., 2007; Sutherland, 2001). P. aeruginosa is capable of producing at least three distinct matrix EPSs: alginate, Psl, and Pel (Franklin et al., 2011). Alginate is the primary EPS used by mucoid strains. Nonmucoid clinical and environmental isolates vary the amount and type of EPSs that they produce (Colvin et al., 2012; Ma et al., 2006). Psl is a neutral polymer containing mannose, glucose, and rhamnose in a repeating pentameric unit (Byrd et al., 2009). In vitro, Psl facilitates surface adherence and proinflammatory signaling in lung epithelial cells (Byrd et al., 2010) and inhibits opsonization and bacterial killing by phagocytes (Mishra et al., 2012). Pel, a polycationic polymer composed of partially acetylated glucosamine and galactosamine (Jennings et al., 2015), can bind extracellular DNA (eDNA) via an ionic binding mechanism (Jennings et al., 2015) and play an additional protective role by enhancing resistance to aminoglycoside antibiotics (Colvin et al., 2011). Whether in vivo P. aeruginosa aggregates produce Pel and Psl, or any other features of the matrix described in vitro, remains unknown.
However, recent reports suggest that in vitro findings regarding P. aeruginosa aggregate formation may not apply to aggregates produced during chronic infection (Bjarnsholt et al., 2013). For example, some studies found that functions that contribute to aggregate formation in vitro are not required to produce aggregates under conditions that mimic the CF environment. Staudinger et al. (2014) showed that P. aeruginosa growing in a viscous medium in vitro could produce aggregates independent of EPS production. These aggregates exhibited many of the phenotypic attributes associated with matrix-mediated aggregation. Subsequently, Secor et al. (2018) demonstrated that a polymer-rich environment (like that found in the CF airways) could drive bacterial aggregation through a passive mechanism termed depletion aggregation. Depletion aggregation occurs in environments where bacterial aggregation reduces the entropy of the system by providing more space for the polymers. Collectively, these data call into question the relevance of some in vitro biofilm findings to chronic CF airway infections and highlight the importance of studying aggregates generated in the human disease environment.
In this study, we apply immunohistochemistry (IHC) using antibodies specific to Pel and Psl EPSs to directly visualize the P. aeruginosa biofilm matrix in CF sputum. We provide evidence that Pel and Psl are produced in the CF airways. We demonstrate that Pel is partially acetylated and has a positive charge at CF-relevant airway pH. This charge allows Pel to bind an abundant host component of the airways, eDNA. The presence of Pel-bound eDNA increased tolerance to aminoglycoside antibiotics. Moreover, the ability of Pel to bind eDNA protects the polymer from DNase digestion. Collectively, our data indicate that, in CF, a subpopulation of P. aeruginosa produces aggregates expressing Pel and/or Psl EPSs. This finding is significant. It shows that aggregate growth in vivo is consistent with the biofilm mode of growth as defined by in vitro laboratory research. Our work also suggests that Pel produced by aggregates likely diminishes the efficacy of the most commonly used current CF therapeutics: inhaled tobramycin and DNase. Thus, patient-to-patient differences in EPS production by bacterial aggregates could help explain the heterogeneous responses to these therapies in the clinic.
RESULTS
CF isolates of P. aeruginosa have the genetic capacity for Pel and Psl production
Sputum samples were collected with consent from eight CF patients at the University of Washington (UW) Adult CF Clinic. Five sputum samples (hereafter indicated as P1—P5) were identified to contain P. aeruginosa using culture-based techniques. Three control sputum samples (C1–C3) did not contain detectable levels of P. aeruginosa. Clinical descriptions of patient samples are listed in Table S1.
We verified the presence or absence of P. aeruginosa in sputum samples using endpoint PCR with P. aeruginosa-specific primers. We probed sputum samples for the presence of genomic DNA harboring pel or psl genes (Figure S1). We found that all sputum samples that contained P. aeruginosa also tested positive for pel genes and that psl genes were detected in four of five samples, suggesting that the genetic capability of sputum pathogens to produce these EPSs varies between patients. As expected, pel and psl genes were not detected in the control samples.
Pel and Psl are associated with P. aeruginosa aggregates in CF sputum
We next sought to determine whether EPSs were being produced in sputum samples. Toward this end, we used IHC with antibodies directed specifically to the Pel and Psl EPSs. These antibodies were validated and optimized by spiking EPS-producing cells into sputum samples lacking P. aeruginosa (Figures S2A and S2B). We then applied our IHC assay to sputum samples obtained from people with CF that we had documented to contain P. aeruginosa (Figure 1). All samples contained abundant neutrophils in which the multi-lobed nuclei stained dark blue due to the hematoxylin counterstain, which is consistent with an intense inflammatory response in the CF airways that fails to clear the infection (Cantin et al., 2015). Visualization of P. aeruginosa in these samples showed abundant single cells and small aggregates as well as a few larger aggregates. Pel, Psl, or both were associated with P. aeruginosa clusters and detected in all of the clinical specimens containing P. aeruginosa (see P. aeruginosa aggregates in sputum indicate heterogeneity in aggregate size and matrix composition). Dual staining of Pel and P. aeruginosa illustrated some aggregates in which the matrix extends beyond cell clusters, similar to in vitro biofilms (Figure 1A). In many cases, some of the larger aggregates appeared to consist of several smaller clusters separated by matrix material (Figure 1B, second panel from left).
Figure 1. Characteristics and observations of the in vivo biofilm matrix.
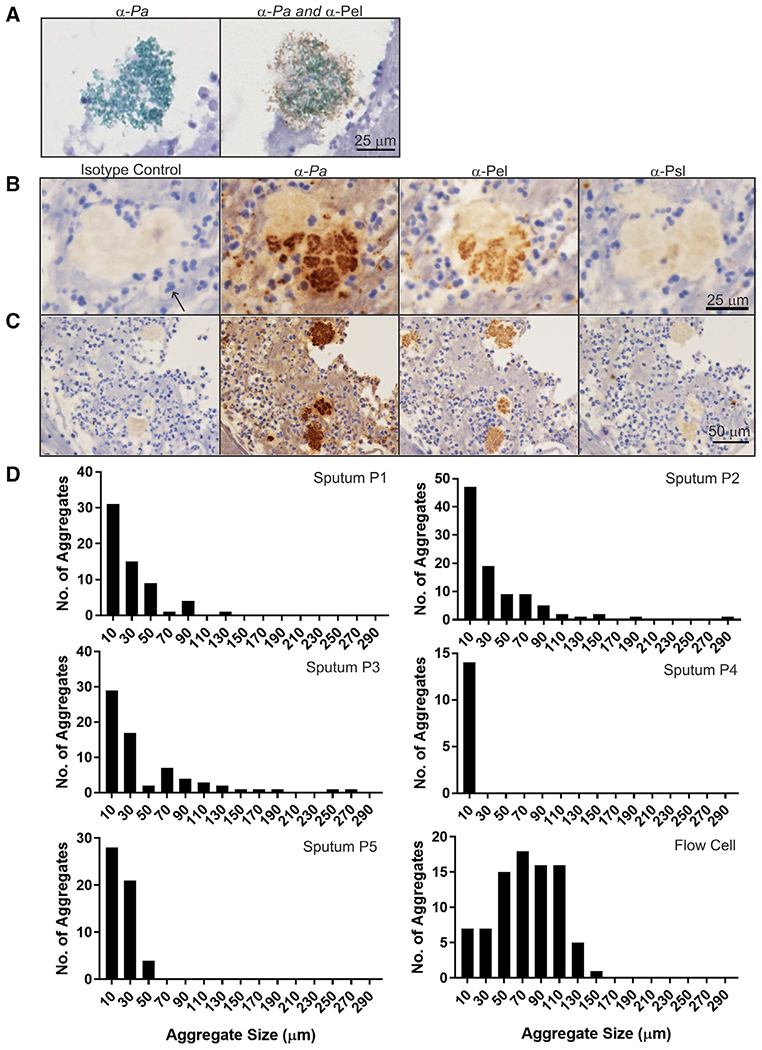
(A) Dual IHC staining of a sputum aggregate showing Pel (brown) and P. aeruginosa (green). (B and C) Representative IHC images of Pel, Psl, and P. aeruginosa (Pa) antibodies (brown) applied to sequential slices of Pa-positive sputum. Hematoxylin counterstain is blue. A neutrophil is highlighted (arrow).
(B) Images showing larger Pa cell cluster that appears to consist of several smaller clusters.
(C) Images showing heterogeneity in Pa cell cluster size.
(D) Size distribution of Pa aggregates in sputum samples (P1–P5) and in vitro flow-cell biofilms from reference Colvin et al. (2012).
P. aeruginosa aggregates in sputum indicate heterogeneity in aggregate size and matrix composition
Abundant single cells and small aggregates, as well as a few larger aggregates, were visible (Figure 1C). Cluster size analysis of P. aeruginosa aggregates was conducted on five sputum samples and compared with biofilm aggregates of clinical and environmental isolates of P. aeruginosa (PAO1, PA14, MSH3, and 19660) cultivated in vitro under continuous flow as described by Colvin et al. (2012). Overall, in vivo aggregates in sputum are smaller than aggregates commonly observed in in vitro studies (Figure 1D). The upper limit of sputum aggregates was ~300 μm (compared with 150 μm for flow cells), with the most abundant number of aggregates in the smallest size range (<20 μm). By contrast, flow cell aggregates were more uniform in size.
Analysis of IHC images by visual inspection showed the presence or absence of Pel and Psl in sputum samples (Figures 2A and 2B). Pel was detected in all five P. aeruginosa-positive sputum samples tested (P1–P5). Psl was detected in three of five P. aeruginosa-positive sputum samples (P1, P3, and P5). Pel and Psl were not detected in controls without P. aeruginosa (C1–C3). Detection of Pel and Psl in sputum by IHC was consistent with PCR results. Gram-positive cocci, which were presumably Staphylococcus, did not stain with P. aeruginosa, Pel, or Psl antibodies (Figure S2C). In addition to detecting Pel and Psl by visual inspection, we sought to objectively quantify the antibody signal using VisioPharm image analysis software. A global quantification of antibody signal across the entire slide could be used to determine the presence/absence of the target antigen for controls (Figure S2B) and most sputum samples (Figure 2C). Quantification with Visio-Pharm has its limitations. The main limitation is that it is challenging to detect polysaccharides in samples with only a few aggregates staining. For these reasons, the global quantification method likely results in an underestimation of EPS detection. Within sputum samples, most P. aeruginosa aggregates stained positive for Pel, Psl, or both (Figure 2D). In the samples that we tested, Pel staining is more prevalent than Psl staining. Overall, our analysis indicates all sputum samples contained at least one EPS and that the expression of Pel and Psl can vary between patients.
Figure 2. IHC indicates presence and localization of Pel and Psl in CF sputum.
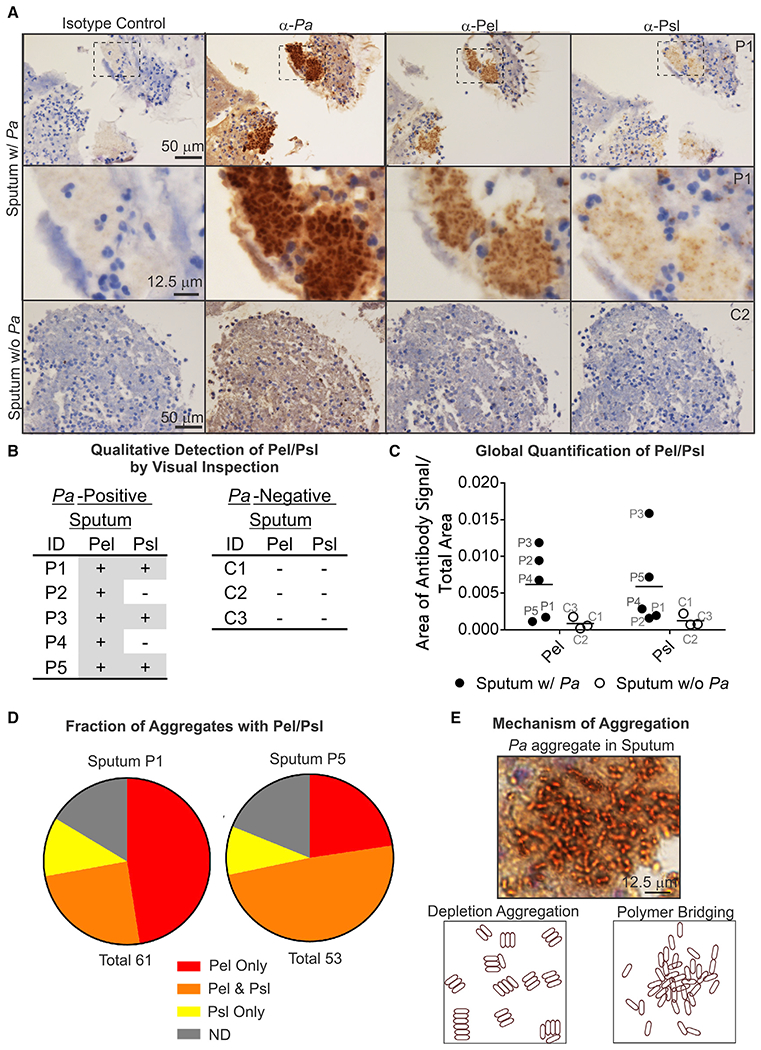
(A) Representative IHC images of Pel, Psl, and Pa antibodies (brown) applied to sequential slices of sputum with Pa (P1) and sputum without Pa (C2).
(B) Qualitative detection of Pel or Psl in sputum by visual inspection. +, detected; −, not detected.
(C) Global quantification of the area of antibody signal/total area of sample for sputum samples.
(D) Fraction of aggregates within a sputum sample with Pel or Psl. ND, aggregate not detected; Total, number of aggregates.
(E) Representative image showing morphology of Pa aggregates in sputum is consistent with a polymer-bridging mechanism.
Cellular orientation within clusters suggests aggregation occurs by an EPS-mediated polymer-bridging mechanism
Bacterial aggregation can occur by different mechanisms: restricted motility (Staudinger et al., 2014), depletion aggregation (Dorken et al., 2012; Schwarz-Linek et al., 2010; Secor et al., 2018), and polymer bridging (Strand et al., 2003). The morphology of aggregates in the sputum samples can be used to distinguish between different mechanisms of aggregation (Secor et al., 2018). Non-motile cells produce aggregates that are densely packed and uniformly distributed throughout the aggregate, compared with polymer bridging in which aggregates have a higher cell density in the center with fewer cells toward the edges (Staudinger et al., 2014). In depletion aggregation, rod-shaped bacteria laterally align to minimize aggregate surface area (Figure 2E, bottom left) (Secor et al., 2018), compared with the less ordered orientation of cells aggregated by polymer bridging (Figure 2E, bottom right). IHC reveals that P. aeruginosa cells in sputum are not laterally aligned and that the cell density is greater toward the center of most clusters (Figure 2E, top). The cellular packing and orientation of P. aeruginosa within aggregates in sputum are consistent with a polymer-bridging mechanism.
Pel is highly deacetylated and aggregates CF-relevant polymers and sputum in a pH-dependent manner in vitro
The evidence that Pel and Psl are produced by P. aeruginosa in the CF airways raises the possibility that these EPSs might mediate functionally important interactions with host-derived airway components. Of particular interest is Pel, which is polycationic and could potentially contribute to interactions in airways that are rich in host-derived anionic constituents. Our previous work showed that Pel was enriched in 1,4-glycosidic linkages of N-acetylgalactosamine (GalNAc) and N-acetylglucosamine (GlcNAc) sugars (Jennings et al., 2015). Deacetylation of Pel sugars by the enzyme PelA results in its overall positive charge (Colvin et al., 2013).
Previous attempts to apply solution NMR to Pel were hindered by the polymer’s limited solubility. To assess the degree of deacetylation, we applied solid-state NMR to Pel from a Pel-overproducing strain (PAO1 ΔwspF Δpsl PBADpel), herein designated as PBADpel (Jennings et al., 2015). The 13C cross-polarization magic-angle spinning (13C-CPMAS) NMR spectrum of Pel was acquired (Figures 3A and S3C) and contained peaks consistent with GlcNAc/GalNAc (Vårum et al., 1991). To determine the extent of Pel deacetylation, the CP contact time was arrayed, which permitted quantitative peak integration (Figures S3A and S3B). This experiment determined that there is ~0.5 acetyl group per sugar. The extent of deacetylation of Pel is greater than that observed for poly-N-acetylglucosamine (PNAG) produced by Staphylococcus spp. (10%–20% deacetylated) (Little et al., 2014).
Figure 3. Pel is highly deacetylated and aggregates CF-relevant polymers in a pH-dependent manner.
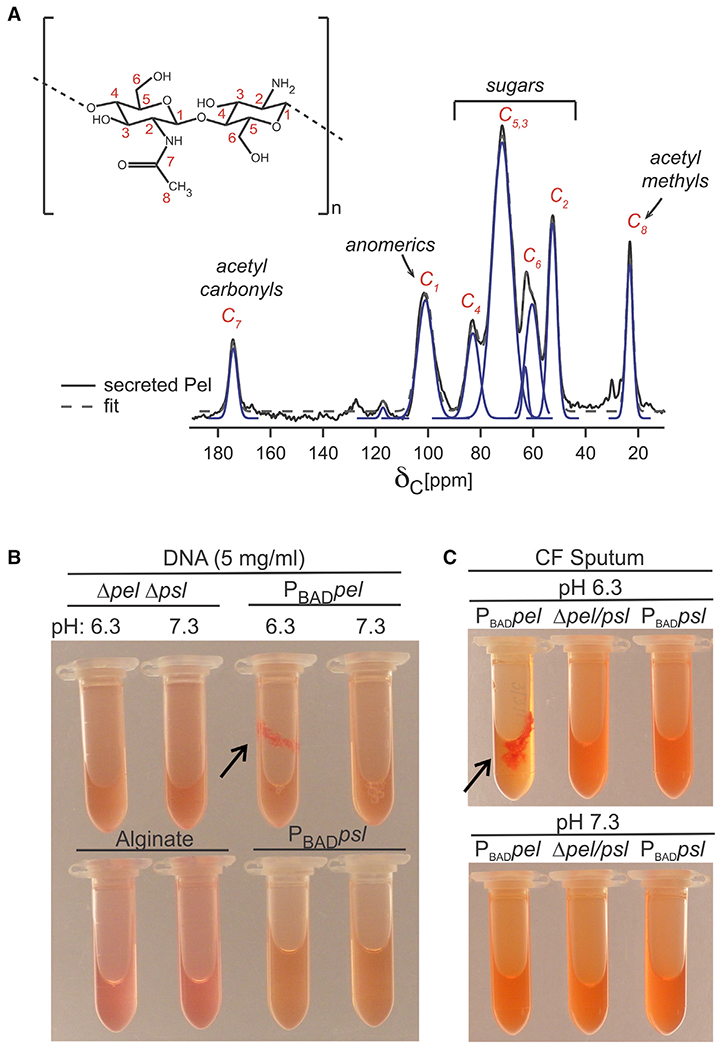
(A) NMR structural analysis of Pel is consistent with a 50% acetylated polymer of GIcNAc and GalNAc. The 13C-CPMAS spectrum of Pel was acquired and compared with poly-β-1,4-GlcNAc (inset). Biological replicate spectra shown in Figure S3C.
(B) Pel, but not Psl or alginate, binds eDNA. Supernatant from PBADpel culture combined with salmon sperm DNA forms a large aggregate (arrow) at pH 6.3, where Pel is positively charged. Aggregation did not occur for supernatant from PBADpsl or alginate solution or no polysaccharide control (Δpel Δpsl).
(C) Pel aggregates CF sputum at pH 6.3 (arrow). (B and C) Representative images shown for clarity. Triplicate biological replicates behaved similarly.
Based on the high degree of deacetylation determined from NMR, we reasoned that Pel would bind anionic polymers in the CF airways (Fischer and Widdicombe, 2006). The pH of the CF airways is mildly acidic (pH 6.0) (Coakley et al., 2003) and would impart a positive charge on Pel, facilitating the formation of ionic interactions. In addition, we had evidence that Pel can bind to both DNA and mucin, which are some of the most abundant anionic polymers in CF sputum (Button et al., 2012; Mrsny et al., 1996; Shak et al., 1990). To follow up on these observations, we wanted to verify that it was Pel that bound eDNA and not any other exopolymers that P. aeruginosa produces. Therefore, we analyzed the ability of the additional P. aeruginosa EPS Psl and alginate to interact with eDNA. As previously reported, pH-dependent aggregation (due to binding) was observed when positively charged Pel was mixed with salmon sperm DNA (Figure 3B). No aggregation was observed with alginate, Psl, or a polysaccharide-free control (Δpel Δpsl) at either pH tested. Pel and Psl immunoblots suggest Pel and Psl supernatants had similar amounts of polysaccharide (Figure S3D). Next, we sought to determine whether Pel could directly aggregate macromolecules present in sputum recovered from a person with CF. We found that Pel aggregated sputum in a pH-dependent manner, while no aggregation was observed for Psl or a polysaccharide-free control (Δpel Δpsl) (Figure 3C).
Pel interactions with host-airway components reduce P. aeruginosa’s susceptibility to antibiotic and mucolytic treatments
CF airways are rich in eDNA, which can be incorporated into the P. aeruginosa biofilm matrix (Moreau-Marquis et al., 2008; Walker et al., 2005). To determine whether the biofilm matrix composition impacts the ability of aggregates to incorporate eDNA, we tested in-vitro-grown biofilms using either Pel- or Psl-rich matrices in growth medium supplemented with salmon sperm DNA. Propidium iodide (PI) staining of eDNA showed that biofilms overexpressing Pel incorporated large amounts of DNA throughout the biofilm matrix, while DNA observed in Psl overexpression biofilms was near background levels (Figure 4A).
Figure 4. Pel-eDNA interactions enhance bacterial tolerance to aminoglycoside antibiotics and resist nuclease digestion.
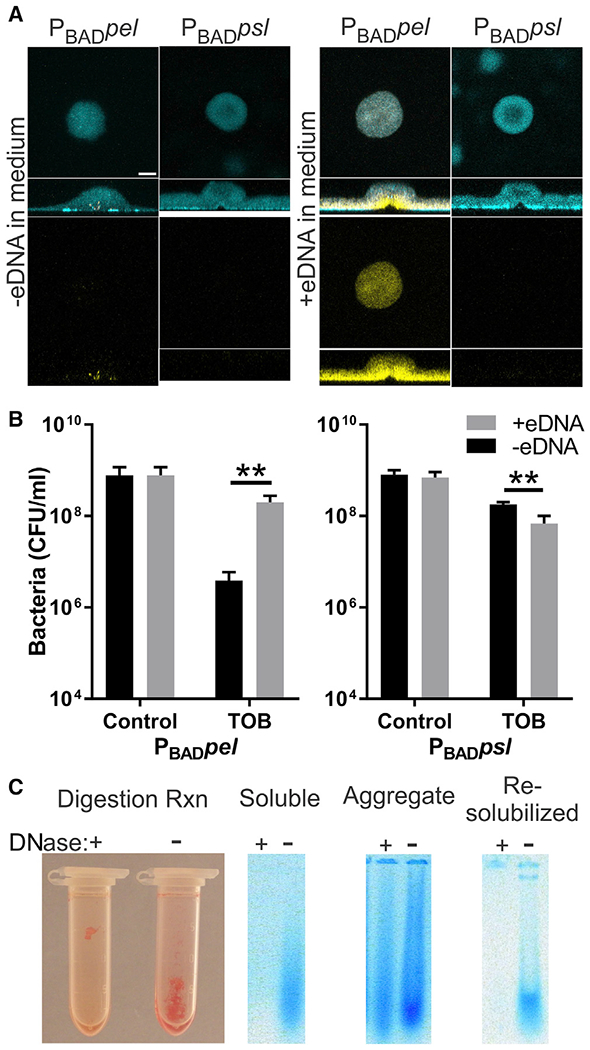
(A) Pel-dependent biofilms retain more eDNA than Psl-dependent biofilms. Representative confocal microscopy images of extracellular DNA stained with propidium iodide (yellow) and biomass stained with SYTO62 (cyan). Scale bar, 30 μm.
(B) eDNA enhances tolerance to tobramycin (TOB) in a Pel-dependent manner. Plots show viable bacterial counts from static biofilms treated with TOB compared with no antibiotic controls. Results are the mean ± SD (n = 3), p value = 0.0048 and 0.0049 for Pel and Psl, respectively.
(C) Pel protects DNA from nuclease digestion. Image shows Pel-DNA aggregates after 48-h DNase digestion. Ethidium-bromide-stained agarose gel shows DNA (blue) from sonicated Pel-DNA aggregate and soluble portion. Re-solubilizing the aggregates by increasing the pH to disrupt ionic interactions enables digestion with DNase. Representative images shown for clarity. Biological replicate behaved similarly.
We then assessed whether Pel-bound eDNA might influence antibiotic susceptibility. Colvin et al. (2011) demonstrated that Pel enhances tolerance to the aminoglycoside antibiotic tobramycin, while Psl did not affect susceptibility. Protection only occurred with the charged antibiotic tobramycin and not the neutral antibiotic ciprofloxacin. We cultivated static Pel- or Psl-rich biofilms in medium supplemented with or without eDNA (Figure 4B). We found that Pel-rich biofilms supplemented with eDNA showed decreased susceptibility to tobramycin. This is consistent with the observation that eDNA increases tolerance to aminoglycosides (Chiang et al., 2013).
A feature of EPS interactions is that they protect polymers from enzymatic degradation (Reichhardt et al., 2018). One common treatment for CF patients is the application of aerosolized recombinant human deoxyribonuclease I (rhDNase I) to digest eDNA that, in turn, reduces sputum viscosity and improves airway clearance and lung function (Frederiksen et al., 2006; Shak et al., 1990). Thus, we explored whether Pel-eDNA interactions might protect the DNA from nuclease digestion. To test this, Pel-eDNA aggregates were formed by mixing supernatant from a Pel-overproducing culture with salmon sperm DNA and adjusting the pH to 6.3 to induce aggregation. We found that Pel-DNA aggregates resisted digestion with DNase I even after extended digestion (Figure 4C). Re-solubilizing the aggregates by sonication or increasing the pH to disrupt ionic interactions between Pel and eDNA rendered the eDNA DNase sensitive. Taken together, the data suggest that Pel-eDNA interactions will not only promote tobramycin tolerance but also shield aggregates from disruption by nucleases.
DISCUSSION
A key unanswered question in the field of CF microbiology relates to why aggressive antibiotic treatments are ineffective at eradicating established P. aeruginosa infections. These chronic infections persist even after initiation of highly effective CFTR modulator therapy (Hisert et al., 2017); thus, a better understanding of how P. aeruginosa eludes killing by both host immune factors and antibiotics remains a critical area of research. Physical disruption of P. aeruginosa aggregates in laboratory experiments can restore susceptibility to killing with antibiotics (Alhede et al., 2011) and cells of the immune system (Jensen et al., 1992; Kharazmi, 1991). Thus, understanding mechanisms that promote cellular aggregation is an important focus of research efforts.
To investigate whether aggregates observed in CF have features of a matrix-dependent mechanism of formation, we developed a method to directly visualize the biofilm matrix in CF sputum using IHC with antibodies targeting Pel and Psl polysaccharides. Our results illustrate that in vivo aggregates found in CF appear to share some of the properties of commonly studied in vitro biofilms. We found evidence that both Psl and Pel are produced and can be found associated with these aggregates. The directional orientation and packing of P. aeruginosa cells within aggregates is consistent with aggregation by the bridging of matrix polysaccharides. Additional findings indicate that aggregate matrix composition can vary from patient to patient with some containing Pel and others containing both Pel and Psl. Our work is consistent with aggregation in the CF airways being mediated by a bacterially produced matrix of Pel and/or Psl.
There are several limitations of this study. First, IHC was the primary line of evidence used for the detection of Pel and Psl. We undertook significant efforts to ensure that the antibodies were specific to their target antigens by applying appropriate controls and showing that the presence/absence of IHC targets was consistent with DNA evidence from PCR. In addition to detection of Pel and Psl by IHC, the presence of matrix polysaccharides was also supported by the observation of cells aggregating by a bridging mechanism of matrix polysaccharides as opposed to a matrix-independent mechanism. Second, sputum sample sizes were small and limit the extension of our results. The vast majority of P. aeruginosa aggregates found in sputum appear to produce Pel or Psl. It is possible that some in vivo aggregates do not contain Pel or Psl, but they were not detected in our small patient cohort. Finally, there is evidence that mucoid strains can also produce Pel and Psl. Whether alginate produced by aggregates from patients harboring mucoid strains influenced Pel and Psl staining and localization is unclear.
Evidence that P. aeruginosa produces Pel during infection is potentially significant. At a CF-airway-relevant pH, Pel retains a positive charge and can interact with many of the abundant, negatively charged, host-derived compounds found in the airways, such as eDNA. We show two mechanisms by which Pel-eDNA complexes could impact disease. First, Pel-dependent biofilms sequester eDNA in the matrix, which increases tolerance to the aminoglycoside antibiotic tobramycin. This is consistent with a study that showed eDNA could mediate antibiotic tolerance to biofilm aggregates (Wilton et al., 2015). Second, Pel binding to eDNA protects it from DNase digestion. Currently, a major therapeutic used to treat CF is called Pulmozyme, which consists primarily of aerosolized, inhaled DNase and is meant to reduce airway secretion viscosity by degrading the eDNA-rich airway secretions.
This study sheds light on the nature of aggregation in vivo. Our findings are supportive of a role for Pel and/or Psl in aggregation of P. aeruginosa in disease. in vitro, Pel is only expressed when P. aeruginosa is grown as a biofilm, where it promotes cellular aggregation (Colvin et al., 2011). However, our results do not rule out the possibility that multiple mechanisms might contribute to the aggregation of P. aeruginosa in different areas/environments within the CF airways. In addition to bacterial cells, Pel is also capable of aggregating mucin, eDNA, and CF sputum. We also show that eDNA-Pel interactions have the potential to reduce the efficacy of two therapeutics commonly used in clinic: Pulmozyme (DNase) and tobramycin. Collectively, our work supports the matrix-driven aggregation hypothesis and suggests that targeting matrix-host compound interactions may be an important step toward improving currently used treatments in the clinic.
STAR ★METHODS
Lead contact
Further information and requests for reagents may be directed to, and will be fulfilled by, the corresponding author Dr. Matthew Parsek (parsem@u.washington.edu).
Materials availability
This study did not generate unique reagents.
Data and code availability
This study did not generate unique datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains and Growth Conditions
Bacterial strains and primers used in this study are listed in the Key Resources Table (Borlee et al., 2010; Colvin et al., 2011,2013; Irie et al., 2012; Secor et al., 2015). Planktonic cultures were routinely cultivated on Jensen’s chemically defined medium at 37°C with constant shaking (225 rpm) unless otherwise indicated. Jensen’s medium (pH 7.3) contained NaCl (85.6 mM), K2HPO4 (14.4 mM), sodium glutamate (92 mM), valine (24 mM), phenylalanine (8 mM), glucose (70 mM), MgSO4 (1.33 mM), CaCl2 (0.14 mM), FeSO4 (0.0039 mM), and ZnSO4 (0.0085 mM). The first five ingredients were mixed, the pH was adjusted to 7.3, and the resulting solution was subsequently autoclaved. Jensen’s medium was supplemented with glucose and metals after autoclaving. Biofilms were cultivated on glucose minimal media (pH 7), which was made by modifying the Jensen’s glucose recipe as follows: the glucose concentration was reduced to 0.3 mM, ammonium sulfate was added (15.1 mM final concentration), and the amino acids valine, phenylalanine, and glutamic acid were removed. Media for induced strains was supplemented with arabinose at 0.5% (vol/vol).
Sputum Sample Collection and Processing
Spontaneously expectorated sputum specimens were collected at the University of Washington (UW) Adult Cystic Fibrosis Clinic and kept on ice (0.5 - 4 hours) until processing. Informed consent was obtained from all subjects, and the study was approved by the UW Human Subjects Division. Patient identification number, age, forced expiratory volume (FEV1), clinical lab microbiology, and cystic fibrosis transmembrane conductance regulator (CFTR) genotype are indicated in Table S1. Sex and gender identity for patients is not known. For each sputum sample, part was fixed in formalin for IHC analysis, part was used to extract bacterial DNA, and the remaining sample was frozen at –20°C.
METHOD DETAILS
Detection of Pel and Psl Genes in Sputum
Bacterial DNA was extracted directly from fresh expectorated sputum using a QIAamp DNA Microbiome Kit (QIAGEN) to reduce contamination with human DNA. Endpoint PCR was used to confirm clinical findings that sputum samples were P. aeruginosa positive or negative and to determine if pel or psl genes were present. PCR was performed using GoTaq® Flexi DNA polymerase (Promega) according to the manufacturer’s instructions with primers specific for Pel (pelA-F, pelA-R), Psl (pslA-F, pslA-R), and P. aeruginosa (PA-rplU-F, PA-rplU-R) (see Key resources table). An annealing temperature of 61°C was used for each of the PCR reactions.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pseudomonas aeruginosa antibody, guinea pig polyclonal, isotype IgG | GeneTex | GTX41031; RRID:AB_424383 |
| Bacterial and virus strains | ||
| PAO1 ΔwspF Δpel PBADpsl - arabinose-inducible psl operon | Irie et al., 2012 | N/A |
| PAO1 ΔwspF Δpsl PBADpel - wspF, nonpolar mutation; pslBCD, polar mutant of the psl operon; arabinose-inducible pel operon | Colvin et al., 2013 | N/A |
| PAO1 ΔwspF Δpsl Δpel - wspF, nonpolar mutation; pelA, polar mutant of the pel operon; pslBCD, polar mutant of the psl operon | Borlee et al., 2010 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium alginate | Sigma-Aldrich | W201502; CAS: 9005-38-3 |
| DNA, salmon sperm | USB | 14377 |
| Congo red | Sigma-Aldrich | C6767 |
| SYTO 62 red fluorescent nucleic-acid stain | Thermo Fisher | S11344 |
| Propidium iodide (PI) | Thermo Fisher | BMS500PI |
| Triton X-100 detergent | Roche | 11332481001; CAS: 9002-93-1 |
| Harris hematoxylin | Leica Biosystems | 3801562 |
| GoTaq Flexi DNA polymerase | Promega | M8291 |
| DNase I, recombinant RNase-free | Sigma-Aldrich | 4716728001 |
| Proteinase K | New England Biolabs | P8107S |
| Critical commercial assays | ||
| QIAamp DNA Microbiome Kit | QIAGEN | 19091 |
| Oligonucleotides | ||
| Pa-rplU-F: CGT CAC GAC AAG GTC CGC | Secor et al., 2015 | N/A |
| Pa-rplU-R: GGC CTG AAT GCC GGT GAT C | Secor et al., 2015 | N/A |
| pelA-F: CCT TCA GCC ATC CGT TCT | Colvin et al., 2011 | N/A |
| pelA-R: TCG CGT ACG AAG TCG ACC TT | Colvin et al., 2011 | N/A |
| pslA-F: CTG CCC TCA CCT TTC GCC | This Study | N/A |
| pslA-R: GGA AGG ATC AGC TGC G | This Study | N/A |
| Software and algorithms | ||
| Igor Pro | WaveMetrics | https://www.wavemetrics.com/products/igorpro |
| Velocity | Quorum Technologies | https://www.quorumtechnologies.com |
| Prism | GraphPad | https://www.graphpad.com:443/ |
| VisioPharm Image Analysis Software | VisioPharm | https://www.visiopharm.com/ |
IHC
IHC targeting Pel, Psl, and P. aeruginosa was performed by the Histology and Imaging Core Facility at UW. Following expectoration, sputum samples were placed in 50 mL conical vials containing 10% formalin. Fixed specimens were embedded in paraffin and sectioned. 5 μm sections were collected and placed onto charged slides, deparaffinized and rehydrated through graded ethanol. The signal-to-noise ratio for each antibody was optimized by varying the antigen retrieval and primary antibody concentration. Antigen retrieval was performed with citrate buffer at pH 6 for 10 minutes followed by blocking with endogenous peroxidase activity with 3.0% hydrogen peroxide for 5 minutes. The sections were incubated with rabbit anti-Pel antisera, rabbit anti-Psl antisera, guinea pig anti-Pseudomonas aeruginosa antisera (GeneTex) or rabbit non-specific control IgGs. Antisera for P. aeruginosa, Pel, and Psl was applied at 1:5000, 1:60, and 1:2000 dilutions, respectively. Sections were washed, incubated with secondary antibodies conjugated to horseradish peroxidase. Counter-staining was performed using Harris hematoxylin (Leica Biosystems).
Antibodies for IHC were validated using positive and negative controls. Negative controls consisted of sputum without P. aeruginosa. PCR confirmed that negative controls did not contain P. aeruginosa, pel or psl genes. To create positive controls, 100 ul of a 16-hour culture of a Pel or Psl overproducing strain cultivated in Jensen’s minimal media was spiked into sputum without P. aeruginosa (Figures S2A and S2B). The sputum with culture was allowed to stand for an hour at room temperature prior to fixing in formalin for IHC analysis. Sputum without P. aeruginosa did not demonstrate robust staining with Pel or Psl antibodies. Some non-specific binding to abiotic material and host cells was observed in controls and could be distinguished from matrix staining which co-localized with anti-P. aeruginosa staining of cell aggregates. Sputum exogenously spiked with cultures that overproduce Pel or Psl showed the presence of rod-shaped bacteria associated with Pel and Psl staining. Variability was noted between sputum samples both in composition and staining. Some samples contained large amounts of saliva, while others relatively little. The P. aeruginosa antibody (GeneTex) targeted P. aeruginosa outer membrane protein fraction and could not be applied to some P. aeruginosa-containing samples due to high background staining.
Polysaccharide Isolation for NMR
Crude preparations of Pel and Psl polysaccharides were prepared by harvesting the supernatant from strains engineered to overproduce Pel or Psl in the presence of arabinose. Jensen’s medium was inoculated with 1 mL of overnight culture of the Pel overexpression strain PAO1 ΔwspF Δpsl PBADpel or Psl overexpression strain PAO1 ΔwspF Δpel PBADpsl (Jennings et al., 2015). Cultures were grown for 20 h at 37°C with constant shaking. To obtain secreted Pel or Psl, the supernatant was harvested by centrifugation (8,300 x g for 15 min at 20°C).
Secreted Pel polysaccharides from culture supernatant were precipitated for 1 h at 4°C with ethanol (final concentration 75% v/v). The precipitate was washed three times with 95%–100% (v/v) ethanol, resuspended in 20 mL of buffer (1 mM CaCl2and 2 mM MgCl2 in 50 mM Tris, pH 7.5), and treated with 100 mg DNase I (Sigma Aldrich) overnight at 37°C, followed by 100 mg of proteinase K(NEB) overnight at 37°C. Sodium dodecyl sulfate (SDS) was added to the enzyme-treated samples at a final concentration of 4% (w/v) and heated to 95°C for 10 min. The polysaccharide sample was centrifuged at 8,300 x g for 15 min at 20°C, and the resulting pellet was resuspended in water. The water washing step was repeated until the SDS was removed. The polysaccharides were precipitated again for 1 h at 4°C with ethanol (final concentration 75% v/v). The samples were extensively dialyzed against water (100 kDa molecular-weight cut off), and then flash-frozen and lyophilized for NMR analysis.
Solid-state NMR
All solid-state NMR experiments were performed using an 89-mm wide-bore Varian magnet at 11.7 T (499.12 MHz for 1H, 125.52 MHz for 13C, and 50.58 MHz for 15N) and an 89-mm wide-bore Agilent magnet at 11.7 T (500.92 MHz for 1H, 125.97 MHz for 13C, and 50.76 MHz for 15N), Varian/Agilent consoles, and home-built four-frequency transmission-line probes with a 13.66 mm long, 6-mm-inner-diameter sample coil and a Revolution NMR MAS Vespel stator. Samples were spun in thin-wall 5-mm-outer-diameter zirconia rotors (Revolution NMR, LLC) at 7,143 ± 2 Hz, using a Varian MAS control unit. The temperature was maintained at –10°C. For all NMR experiments, p-pulse lengths were 7 μs for 1H and 10 μs for 13C. Proton-carbon cross-polarization occurred at 50 kHz for 1.5 ms unless otherwise noted. Proton dipolar coupling was performed at 90 kHz with TPPM modulation. The chemical shift of 13C spectra was referenced to external adamantane. Multi-peak fitting and integration was performed in software written for Igor Pro (WaveMetrics, Lake Oswego, OR, USA).
To confirm the polysaccharide composition and to assess the degree of deacetylation, we applied solid-state NMR to Pel from an overproducing strain PBADpel (Jennings et al., 2015). The 13C cross-polarization magic-angle spinning (13C-CPMAS) NMR spectrum of secreted Pel was acquired. To determine the extent of Pel deacetylation, the cross-polarization (CP) contact time was arrayed, which permitted quantitative peak integration.
Polymer Binding Assay
To determine if P. aeruginosa EPS other than Pel could ionically bind exogenous DNA, a method was used similar to that outlined in Jennings et al. (2015). Sodium alginate (5 mg/ml, Sigma W201502) and culture supernatant (1.5 ml) from Pel or Psl overproducing strains and a Δpel Δpsl deletion mutant were combined with salmon sperm DNA (5 mg/ml, USB 14377). Samples were vortexed, placed on a rotator for at least 20 minutes and drawn through a 22-gauge needle to ensure that polymers were fully in solution. To induce aggregation, the pH of the EPS/DNA solution was adjusted to 6.3 or 7.3 by the addition of 1 N HCl. Congo Red (Sigma-Aldrich, C6767, 40 μg/ml final concentration) was added to enhance visualization of aggregates, although unstained samples behaved similarly.
To test for interactions between Pel and sputum, supernatant from Pel and Psl overexpression strains and a Δpel Δpsl deletion mutant (1.4 ml) was mixed with homogenized sputum (0.1 ml). The pH of the corresponding solutions was adjusted to 6.3 and 7.3.
DNase Digestion of Pel/DNA Aggregates
To determine if Pel/DNA aggregates resisted nuclease digestion, Pel/DNA aggregates were formed by mixing supernatant from a Pel overexpression culture with 5 mg/ml DNA and lowering the pH to 6.3. Aggregates were physically removed and placed in a buffer (1 mM CaCl2 and 2 mM MgCl2 in 50 mM Tris, pH 7.5) with or without DNase I (Sigma-Aldrich, 20 μg/ml). Digestion was carried out at 37°C for 1 hour or up to 48 h. Congo Red was added prior to imaging. Half of the aggregates were broken-up by sonication in 50 μl of water. The sonicated aggregate and the soluble portion of the enzyme digest reaction was run on a 1% agarose gel to detect DNA in the samples.
Flow-Cell Biofilm and Confocal Microscopy
Biofilms were cultivated in flow cell chambers essentially as described by Jennings et al. (2015). Flow cells were inoculated from a mid-log LB culture that was diluted with glucose minimal media to an optical density at 600 nm of 0.01. Strains that aggregated in mid-log phase were homogenized before dilution for inoculation. Cells were allowed to attach under static conditions to an inverted flow cell for 1 h before induction of flow. Biofilms were grown on glucose minimal media supplemented with or without exogenous salmon sperm DNA (USB) at 40 μg/ml for 2 d at room temperature under a constant flow rate (10 mL/h). Biofilms were stained for 15 min with Syto62 red fluorescent nucleic-acid stain (5 μM; ThermoFisher) for biomass and PI (30 μM; ThermoFisher)for eDNA. After staining, flow cells were washed with media at 10 mL/h for 5 min and then visualized on a Zeiss LSM 510 scanning confocal laser microscope. Image analysis was conducted using Velocity software (Quorum Technologies). Microscopy images were artificially colored as follows: Syto62 biomass, cyan; and PI eDNA, yellow.
Antimicrobial Tolerance in Biofilm Assay
Static biofilms were used to measure the effect of eDNA on the sensitivity of PBAD pel and PBAD psl to tobramycin (Sigma). 48-well plates containing LB with and without exogenous DNA at 100 μg/ml were inoculated with exponentially growing PBADpel and PBADpsl cells. Biofilms were cultivated statically for 8 hours at 37°C. At which point, the spent medium was decanted, and biofilms were washed once with phosphate buffered saline solution (PBS) followed by replacement with medium supplemented with or without tobramycin (50 μg/ml). Biofilms were incubated with antibiotic for 30 min at 37°C followed by removal of medium and washing of cells with PBS. After the second washing, biofilms were treated with 0.5% Triton X-100 detergent (Roche) and shaken at room temperature for 30 minutes to dislodge cells from the plate surface. Viable bacterial counts were determined by making serial dilutions into PBS and dropping 10 μl dilutions onto LB agar plates. Colony forming units were counted after overnight incubation at 37°C.
QUANTIFICATION AND STATISTICAL ANALYSIS
Antibiotic Susceptibility
The statistical significance of the effect of DNA on antibiotic susceptibility was determined in Prism (GraphPad) using an unpaired, nonparametric Mann-Whitney test to calculate approximate p-values comparing conditions with and without exogenous DNA. The mean ± SD of three experiments (n = 3 for each experiment) were calculated. A p-value less than or equal to 0.05 was considered significant.
IHC Image Analysis
IHC slides were scanned and visualized on a Hamamatsu NanoZoomer Digital Pathology (NDP) System. Images of IHC slides were captured by microscopy. Biofilm aggregate size was estimated in sputum samples containing P. aeruginosa by measuring the longest diameter or length directly on scanned IHC slides on aggregates greater than 4 mm. The presence of Pel and Psl in sputum was determined by visual inspection and quantification of antibody signal using VisioPharm image analysis software. A global quantification of antibody signal across the entire slide was normalized to total specimen area.
Supplementary Material
Highlights.
Pel and Psl are produced by P. aeruginosa aggregates in cystic fibrosis sputum
Attributes of in vitro biofilms are seen in clinical aggregates
Aggregate morphology is consistent with polysaccharide-dependent aggregation
Pel-DNA interactions reduce susceptibility to antibiotic and mucolytic treatments
ACKNOWLEDGMENTS
This work was supported by NIH grants 5R01AI077628 (M.R.P. and D.J.W.), R01AI143916 (M.R.P. and D.J.W.), R01AI134895 (M.R.P. and D.J.W.), and in part by P20GM103546 and R01AI138981 (P.R.S.), and CFF grant PARSEK17IO (M.R.P.). L.K.J. is the recipient of an AHA fellowship (14POST20130017). C.R. is the recipient of an NIH K99 (5K99GM134121-02) and CFF award (REICHH19F5). We thank Lynette Cegelski for NMR spectrometer access.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108782.
DECLARATION OF INTERESTS
The authors declare no conflicts of interests.
REFERENCES
- Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen PØ, Nielsen AK, Parsek M, Wozniak D, et al. (2011). Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS ONE 6, e27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, and Høiby N (2009). Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, and Høiby N (2013). The in vivo biofilm.Trends Microbiol. 21,460–474. [DOI] [PubMed] [Google Scholar]
- Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, and Parsek MR (2010). Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75, 827–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button B, Cai L-H, Ehre C, Kesimer M, Hill DB, Sheehan JK, Boucher RC, and Rubinstein M (2012). A periciliary brush promotesthe lung health by separating the mucus layer from airway epithelia. Science 337, 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Pang B, Mishra M, Swords WE, and Wozniak DJ (2010).The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappaB activation in A549 cells. MBio 1, e00140–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, et al. (2009). Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin AM, Hartl D, Konstan MW, and Chmiel JF (2015). Inflammation in cystic fibrosis lung disease: Pathogenesis and therapy. J. Cyst. Fibros. 14, 419–430. [DOI] [PubMed] [Google Scholar]
- Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, and Tolker-Nielsen T (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley RD, Grubb BR, Paradiso AM, Gatzy JT, Johnson LG, Kreda SM, O’Neal WK, and Boucher RC (2003). Abnormal surface liquid pH regulation by cultured cystic fibrosis bronchial epithelium. Proc. Natl. Acad. Sci. USA 100, 16083–16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, and Parsek MR (2011). The pel polysaccharidecan serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7, e1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, and Parsek MR (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Alnabelseya N, Baker P, Whitney JC, Howell PL, and Parsek MR (2013). PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 195, 2329–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, and Lappin-Scott HM (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. [DOI] [PubMed] [Google Scholar]
- Dorken G, Ferguson GP, French CE, and Poon WC (2012).Aggregation by depletion attraction in cultures of bacteria producing exopolysaccharide. J. R. Soc. Interface 9, 3490–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, and Widdicombe JH (2006). Mechanisms of acid and base secretion by the airway epithelium. J. Membr. Biol. 211, 139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, and Wingender J (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. [DOI] [PubMed] [Google Scholar]
- Franklin MJ, Nivens DE, Weadge JT, and Howell PL (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen B, Pressler T, Hansen A, Koch C, and Høiby N (2006). Effect of aerosolized rhDNase (Pulmozyme) on pulmonary colonization in patients with cystic fibrosis. Acta Paediatr. 95, 1070–1074. [DOI] [PubMed] [Google Scholar]
- Gibson RL, Burns JL, and Ramsey BW (2003). Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. CareMed. 168, 918–951. [DOI] [PubMed] [Google Scholar]
- Hisert KB, Heltshe SL, Pope C, Jorth P, Wu X, Edwards RM, Radey M, Accurso FJ, Wolter DJ, Cooke G, et al. (2017). Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am. J. Respir. Crit. Care Med. 195, 1617–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, and Parsek MR (2012). Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 109, 20632–20636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, et al. (2015). Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 112, 1135311358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ET, Kharazmi A, Høiby N, and Costerton JW (1992). Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. APMIS 100, 727–733. [PubMed] [Google Scholar]
- Kharazmi A (1991). Mechanisms involved in the evasion of the host defence by Pseudomonas aeruginosa. Immunol. Lett. 30, 201–205. [DOI] [PubMed] [Google Scholar]
- Little DJ, Li G, Ing C, DiFrancesco BR, Bamford NC, Robinson H, Nitz M, Pomès R, and Howell PL (2014). Modification and periplasmic translocation of the biofilm exopolysaccharide poly-β-1,6-N-acetyl-D-glucosamine. Proc. Natl. Acad. Sci. USA 111, 11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, and Pier GB (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jackson KD, Landry RM, Parsek MR, and Wozniak DJ (2006). Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, and Wozniak DJ (2012). Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell. Microbiol. 14, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Marquis S, Stanton BA, and O’Toole GA (2008). Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrsny RJ, Daugherty AL, Short SM, Widmer R, Siegel MW, and Keller G-A (1996). Distribution of DNA and alginate in purulent cystic fibrosis sputum: implications to pulmonary targeting strategies. J. Drug Target. 4, 233–243. [DOI] [PubMed] [Google Scholar]
- Pamp SJ, Gjermansen M, and Tolker-Nielsen T (2007). The biofilm matrix: a sticky framework. In The biofilm mode of life: mechanisms and adaptations, Kjelleberg S and Givskov M, eds. (Horizon Bioscience), pp. 37–69 [Google Scholar]
- Reichhardt C, Wong C, Passos da Silva D, Wozniak DJ, and Parsek MR (2018). CdrA interactions within the Pseudomonas aeruginosa biofilm matrix safeguard it from proteolysis and promote cellular packing. MBio 9, e01376–e01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Linek J, Dorken G, Winkler A, Wilson L, Pham N, French C, Schilling T, and Poon W (2010). Polymer-induced phase separation in suspensions of bacteria. EPL 89, 68003. [Google Scholar]
- Secor PR, Michaels LA, Ratjen A, Jennings LK, and Singh PK (2018). Entropically driven aggregation of bacteria by host polymers promotes antibiotic tolerance in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 115, 10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR, et al. (2015). Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shak S, Capon DJ, Hellmiss R, Marsters SA, and Baker CL (1990). Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 87, 9188–9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, and Greenberg EP (2000). Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407, 762–764. [DOI] [PubMed] [Google Scholar]
- Somayaji R, Parkins MD, Shah A, Martiniano SL, Tunney MM, Kahle JS, Waters VJ, Elborn JS, Bell SC, Flume PA, and VanDevanter DR; Antimicrobial Resistance in Cystic Fibrosis InternationalWorking Group (2019). Antimicrobial susceptibility testing (AST) and associated clinical outcomes in individuals with cystic fibrosis: A systematic review. J. Cyst. Fibros. 18, 236–243. [DOI] [PubMed] [Google Scholar]
- Staudinger BJ, Muller JF, Halldórsson S, Boles B, Angermeyer A, Nguyen D, Rosen H, Baldursson O, Gottfreðsson M, Guðmundsson GH, and Singh PK (2014). Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 189, 812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz DA, Meyerholz DK, and Welsh MJ (2015). Origins of cystic fibrosis lung disease. N. Engl. J. Med. 372, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand SP, Vårum KM, and Østgaard K (2003). Interactions between chitosans and bacterial suspensions: adsorption and flocculation. Colloids Surf. B Biointerfaces 27, 71–81. [Google Scholar]
- Sutherland I (2001). Biofilm exopolysaccharides: a strong and sticky framework. Microbiology (Reading) 147, 3–9. [DOI] [PubMed] [Google Scholar]
- Vårum KM, Anthonsen MW, Grasdalen H, and Smidsrød O (1991). Determination of the degree of N-acetylation and the distribution of N-acetyl groups in partially N-deacetylated chitins (chitosans) by high-field NMR spectroscopy. Carbohydr. Res. 211, 17–23. [DOI] [PubMed] [Google Scholar]
- Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, and Nick JA (2005). Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 73, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M, Charron-Mazenod L, Moore R, and Lewenza S (2015). Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate unique datasets or code.


