Next-generation sequencing retrospectively identified rilpivirine-resistant variants with a 1% threshold in 8.8% and 5.7% of 489 treatment-naive patients according to the ANRS and Stanford algorithms, respectively. Minority resistant variants had no impact on the virological response to a rilpivirine-based regimen.
Keywords: minority resistant variants, rilpivirine, first-line antiretroviral therapy, ultra-deep sequencing
Abstract
Background
Minority resistant variants of human immunodeficiency virus type 1 (HIV-1) could influence the virological response to treatment based on nonnucleoside reverse transcriptase inhibitors (NNRTIs). Data on minority rilpivirine-resistant variants are scarce. This study used next-generation sequencing (NGS) to identify patients harboring minority resistant variants to nucleos(t)ide reverse transcriptase inhibitors and NNRTIs and to assess their influence on the virological response (VR).
Methods
All the subjects, 541 HIV-1–infected patients started a first-line regimen containing rilpivirine. VR was defined as a HIV-1 RNA load <50 copies/mL at month 6 with continued suppression at month 12. NGS was performed at baseline (retrospectively) on the 454 GS-FLX platform (Roche).
Results
NGS revealed resistance-associated mutations accounting for 1% to <5% of variants in 17.2% of samples, for 5%–20% in 5.7% of samples, and for >20% in 29% of samples. We identified 43 (8.8%) and 36 (7.4%) patients who harbored rilpivirine-resistant variants with a 1% sensitivity threshold according to the French National Agency for Research on AIDS and Viral Hepatitis and Stanford algorithms, respectively. The VR was 96.9% at month 12. Detection of minority rilpivirine resistant variants was not associated with virological failure (VF). Multivariate analysis indicated that VF at month 12 was associated with a CD4 count <250 cells/µL at baseline, a slower decrease in viral load at month 3, and rilpivirine resistance at baseline using the Stanford algorithm with a 20% threshold.
Conclusions
Minority resistant variants had no impact on the VR of treatment-naive patients to a rilpivirine-based regimen.
Human immunodeficiency virus type 1 (HIV-1) is genetically highly variable, which enables it to adapt and thus escape destruction by the host immune system and antiretroviral treatment. Infected individuals harbor a virus population comprising many genetically distinct but related variants, which define the quasi-species. HIV-1–infected patients are screened for resistance mutations that may decrease sensitivity to certain drugs before they are given antiretroviral drugs. Genotypic resistance testing is conventionally carried out by direct sequencing that identifies variants accounting for at least 20% of the total virus population. However, minority variants accounting for <20% of the population may harbor resistance mutations and may emerge under drug selection pressure. Newly available methods more sensitive than direct sequencing have shown that resistant variants are more prevalent than previously thought [1, 2]. The 454 GS FLX system (Roche Diagnostic, Meylan, France) can analyze long fragments of virus quasi-species genes (400–800 base pairs [bp]) [3]. Minority variants that are resistant to nonnucleoside reverse transcriptase inhibitors (NNRTIs) may influence the virological response (VR) to NNRTI-based combination therapy [4–6]. A meta-analysis has shown that patients harboring resistant minority variants are at greater risk of virological failure when treated with a first-line antiretroviral regimen containing an NNRTI [7].
Rilpivirine is a second-generation NNRTI indicated as first-line regimen [8, 9]. Population sequencing of infected antiretroviral-naive patients indicated that genotypic resistance to rilpivirine (RPV) accounted for <5% of the viral variants globally, 3.7% in subtype B–infected patients, and 6% in patients infected with non-B subtypes [10]. There are only limited data from clinical cohorts relating baseline RPV resistance patterns to therapy failure [11]. As a result, there is a lack of knowledge on the predictive value of RPV-associated mutations at baseline [12, 13].
This study was designed to identify minority resistant variants to nucleos(t)ide reverse transcriptase inhibitors (NRTIs) and NNRTIs in a cohort of antiretroviral-naive patients and to assess their influence on the VR to a first-line regimen containing RPV.
METHODS
Study Population and Samples
We studied HIV-1–infected patients from 24 centers in France who were being treated with an RPV-NRTI combination, in accordance with French recommendations. They were all antiretroviral-naive before this treatment. Inclusion criteria were HIV-1 infection, age ≥18 years, first-line treatment with RPV, a pretreatment HIV plasma RNA load >1000 copies/mL, and a frozen pretreatment plasma sample. Rilpivirine is authorized for antiretroviral treatment–naive patients with a viral load ≤100000 HIV-1 RNA copies/mL according to the European Medicines Agency.
The 24 participating laboratories belong to the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) AC11 network and participate in the annual ANRS quality control assessment of HIV-1 drug resistance sequencing [14]. The pretreatment plasma samples were collected and analyzed in the Toulouse Virology Laboratory for ultra-deep sequencing of the reverse transcriptase (RT) gene. We collected data from each participating laboratory: plasma HIV-1 RNA, CD4+ T-cell counts, and RT gene sequences (bulk sequencing) at baseline, and plasma HIV-1 RNA 1, 3, and 6 months after treatment initiation and during follow-up (6–42 months). The VR at months 6 and 12 was defined as a plasma HIV-1 RNA load <50 copies/mL after 6 months and 12 months of treatment, respectively. Virological failure (VF) was defined by a viral load >50 copies/mL on 2 consecutive measurements after >6 months of treatment.
Ultra-Deep Sequencing of the Reverse Transcriptase Gene
HIV-1 RNA was extracted from 500 µL plasma with viral loads >1000 copies/mL using MagNA Pure 96 DNA and Viral NA Large Volume kits (Roche Diagnostic, Meylan, France). Ultra-deep sequencing of the HIV-1 RT gene was performed as previously described [13]. We used the GS FLX Titanium Sequencing Kit XL+ to obtain long reads and analyzed one 723-bp fragment of the RT gene. First-strand complementary DNA was generated with 2 gene-specific oligonucleotides. Nested polymerase chain reaction produced 1 amplicon covering the HIV-1 RT gene (codons 19–259), which was purified and quantified. Equimolar amounts of amplicon from each sample were pooled and subjected to clonal amplification on beads using reagents that enabled sequencing in both the forward and reverse directions. The beads were isolated and those bearing enriched DNA were counted. Ultra-deep sequencing was carried out on 230000 beads loaded onto one region of a PicoTiter plate fitted with an 8-lane gasket and sequenced on a Genome Sequencer FLX (Roche-454 Life Sciences).
Bioinformatic Analysis
A median of 1629 (interquartile range [IQR], 1169–2252) reads per nucleotide position was obtained for this set of samples. GS Amplicon Variant Analyzer software was used to analyze the ultra-deep sequencing results. The amplicon nucleotide sequence reads were aligned with the consensus sequence HXB2. The generated files were then analyzed using DeepChek-HIV version 3.27 software (Advanced Biological Laboratories, TherapyEdge). As the errors generated during deep sequencing are sequence-dependent, we determined specific error rates at each amino acid position in the RT gene and calculated the sensitivity per position. The amino acid variants above the sensitivity threshold and with a frequency >1% were kept for analyses.
We used the 2017 International AIDS Society (IAS) list of mutations [15] to identify resistance-associated mutations (RAMs), the ANRS resistance algorithm (2016, version 26, available at http://www.hivfrenchresistance.org), and the Stanford HIV Drug Resistance Database interpretation system (version 8.3) to interpret HIV resistance to NRTIs and NNRTIs. Reduced susceptibility to an antiretroviral was defined as intermediate or high-level resistance. The genotypic sensitivity score (GSS) was calculated by DeepChek-HIV using either the ANRS resistance algorithm or the Stanford resistance algorithm: Each antiretroviral was assigned a score of 1 if there is no resistance, 0.5 for intermediate resistance, and 0 for high resistance.
Statistical Analysis
Previous studies on NNRTI-based antiretroviral therapy have indicated that the frequency of virological failure is 15% in the absence of minority variants resistant to NNRTI and 35% in the presence of minor variants [7]. The frequency of minor variants resistant to RPV was 15% in treatment-naive patients at the primary infection stage [13]. We needed a sample containing 487 patients to show a 20% difference in the VR of patients with and without minority variants with α = .05 and 1 − β = .8. We used Stata version 9.0 software (StataCorp, College Station, Texas) for statistical analyses. Fisher exact test and the χ2 test were used for between-group comparisons. Continuous variables were tested with nonparametric tests. A statistically significant difference was defined as a P value <.05. We analyzed the factors associated with the VR at months 6 and 12 using univariate and multivariate logistic regressions.
RESULTS
Patient Characteristics
A total of 541 patients who began an RPV-based regimen between 2012 and 2015 were included, but 52 of the samples taken before treatment initiation could not be amplified. The patients with failing samples had similar virological and clinical characteristics to the patients with amplified samples. Thus, we studied samples from 489 antiretroviral-naive patients who were given RPV in combination with other antiretrovirals. The background regimen for 484 patients (99%) was tenofovir plus emtricitabine (TDF/FTC). Three patients were given abacavir plus lamivudine instead of TDF/FTC, 1 patient was given maraviroc in addition to TDF/FTC, and the third was given raltegravir in addition to TDF/FTC. The median patient age was 36 years (IQR, 29–44 years), and 79% were men. Most patients were infected with subtype B (61%) or CRF02-AG (19%). They had a median viral load of 4.3 log copies/mL (IQR, 3.9–4.6 copies/mL) and a median CD4 cell count of 470 cells/µL (IQR, 353–601 cells/µL) at treatment initiation.
HIV-1 Genotypic Resistance at Baseline
Next-generation sequencing (NGS) identified 288 RAMs accounting for at least 1% of the virus population in 229 of 489 (46.8%) pretreatment samples. These RAMs were based on the 2017 IAS list, the ANRS resistance algorithm (2016 version 26), and the Stanford resistance algorithm.
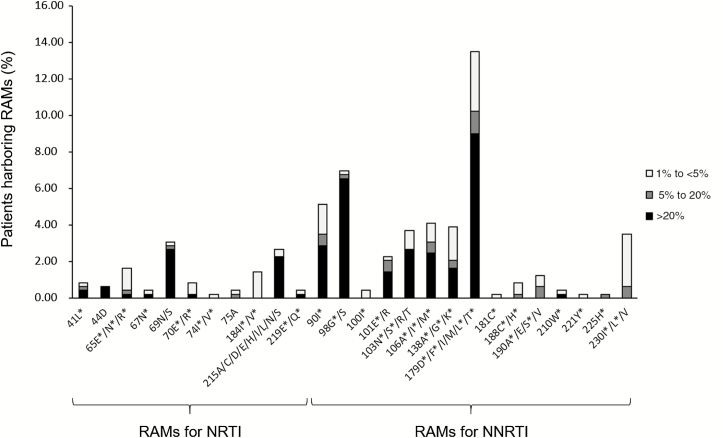
NGS identified 164 RAMs accounting for >20% of the virus population in 142 samples (29%), 30 RAMs accounting for 5%–20% in 28 samples (5.7%), and 94 RAMs accounting for 1% to <5% in 84 samples (17.2%). Therefore, 142 samples harbored mutant variants with a 20% threshold, 165 with a 5% threshold, and 229 with a 1% threshold (Table 1). The percentage of patients with RAMs is detailed in Figure 1.
Table 1.
Impact of Resistance-Associated Mutations on the Genotypic Sensitivity Score
| Threshold of Virus Variants | Samples With RAMs for NRTIs and NNRTIs | Samples With Rilpivirine Resistance According to ANRS Algorithm | Samples With Rilpivirine Resistance According to Stanford Algorithm | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypic Sensitivity Score According to ANRS | Genotypic Sensitivity Score According to Stanford | ||||||||||||
| GSS = 0 | GSS = 1 | GSS = 2 | GSS = 2.5 | GSS = 3 | GSS = 0 | GSS = 1.5 | GSS = 2 | GSS = 2.5 | GSS = 3 | ||||
| 20% | 142 | 13 | 9 | 0 | 0 | 8 | 5 | 128 | 0 | 0 | 0 | 10 | 132 |
| 5% | 165 | 18 | 16 | 0 | 1 | 13 | 5 | 146 | 0 | 1 | 3 | 14 | 147 |
| 1% | 229 | 43 | 36 | 1 | 4 | 48 | 5 | 171 | 0 | 2 | 13 | 33 | 181 |
Abbreviations: ANRS, French National Agency for Research on AIDS and Viral Hepatitis; GSS, genotypic sensitivity score; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleos(t)ide reverse transcriptase inhibitor; RAM, resistance-associated mutation.
Figure 1.
Patients harboring resistance-associated mutations (RAMs) in the reverse transcriptase gene identified by next-generation sequencing. The bars represent the percentage of the patients harboring mutations indicated on the horizontal axis: black bars for RAMs accounting for >20% of the virus population, dark gray bars for RAMs at 5%–20%, and light gray bars for RAMs accounting for 1% to <5%. *Mutations in the International AIDS Society list. Abbreviations: NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleos(t)ide reverse transcriptase inhibitor.
Analysis using the ANRS resistance algorithm showed that RAMs were responsible for RPV resistance or intermediate resistance in 43 samples (8.8%) with a 1% threshold (22 subtypes B, 11 CRF02, and 10 other subtypes). The GSS was calculated taking into account the resistant variants at different thresholds (Table 1). Majority resistant variants (accounting for >20%) were found in 13 samples: 8 harbored the mutation E138A and 5 harbored the mutation V179D, which is associated with intermediate resistance to RPV. Minority resistant variants harbored mutations E138A/G/K, V179L, Y181C, H221Y, and M230I/V. The absolute copy numbers of minority resistant variants to RPV were 1.3–3.9 log copies/mL (median, 2.8 log copies/mL). One sample harbored a mutation conferring resistance to tenofovir (K65E, 22.9%). Sixteen samples had variants resistant to TDF and/or FTC (K65E/N/R; K70E; M184V/I) with frequencies of 1%–11.1% and absolute copy numbers of 1.1–3.7 log copies/mL (median, 2.6 log copies/mL).
Analysis using the Stanford algorithm showed that 36 samples (7.4%) harbored resistance to RPV with a 1% threshold (21 subtype B, 6 CRF02, and 9 other subtypes) (Table 1). Majority RPV RAMs were 138A (8 samples) and 98G (1 sample). Minority resistant variants harbored mutations L100V, E138A/G/K, V179L, Y181C, Y188F, G190E, H221Y, and M230I.
Agreement Between NGS and Sanger Sequencing
We analyzed the 425 samples for which Sanger sequences were available at the time of treatment initiation to determine the agreement between NGS and Sanger sequencing. Sanger sequencing identified 96 of the 110 RAMs (87%) identified by NGS (20% threshold), 3 of 24 RAMs (12%) accounting for 5%–20% using NGS, and 0 of 83 RAMs accounting for 1% to <5%. The 14 RAMs at >20% identified using NGS and not detected by Sanger had a median frequency of 90.9% (IQR, 41.9%–98.1%). The 3 RAMs at 5%–20% identified using NGS and detected by Sanger had frequencies of 15.4%, 12.4%, and 17.4%.
NGS identified 100 of the 106 RAMs (94%) identified by Sanger sequencing. Six mixed wild/mutant populations were not identified by NGS while double peaks were visible on the Sanger electropherograms.
Virological Outcome to Rilpivirine-Based Treatment
Almost all patients obtained a VR at 6 months (97.9%). Univariate analysis of the factors associated with VR at month 6 indicated that a CD4 cell count >250 cells/µL at baseline and a viral load slope >2 log copies/mL at month 3 were associated with a greater VR (Table 2). A subtype B of HIV-1 also tended to be associated with VR. Multivariate analysis indicated that only the CD4 cell count and the decrease in viral load at month 3 were associated with VR at month 6.
Table 2.
Factors Influencing the Virological Response at Months 6 and 12 on a Rilpivirine-Based First-line Therapy
| Variable | Virological Response at Month 6 | Virological Response at Month 12 | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysisa | Multivariate Analysisa | Univariate Analysisa | Multivariate Analysisa | |||||
| P Value | OR | (95% CI) | P Value | P Value | OR | (95% CI) | P Value | |
| Age | .1653 | … | … | … | … | … | … | … |
| Sex | .148 | … | … | … | … | … | … | … |
| HIV-1 subtype (B vs non-B) | .053 | … | … | … | .078 | … | … | … |
| Baseline viral load | .3495 | … | … | … | … | … | … | … |
| Viral load slope at month 3b (>2 log copies/mL) | <.0001 | 18.14 | (3.51–93.79) | <.001 | .0319 | 17.26 | (3.36–88.52) | <.001 |
| Baseline CD4 cell count (>250 cells/µL) | <.001 | 17.61 | (3.38–91.72) | <.001 | .040 | 10.89 | (1.89–62.69) | <.01 |
| Baseline resistance to rilpivirine (ANRS, threshold 20%) | .161 | … | … | … | .246 | … | … | … |
| Baseline resistance to rilpivirine (ANRS, threshold 5%) | .307 | … | … | … | .459 | … | … | … |
| Baseline resistance to rilpivirine (ANRS, threshold 1%) | .910 | .839 | ||||||
| GSS <3 (ANRS, threshold 20%) | .155 | … | … | … | .238 | … | … | … |
| GSS <3 (ANRS, threshold 5%) | .331 | .492 | ||||||
| GSS <3 (ANRS, threshold 1%) | .366 | .649 | ||||||
| Baseline resistance to rilpivirine (Stanford, threshold 20%) | .061 | … | … | … | <.001 | 0.028 | (.003–(.270) | <.01 |
| Baseline resistance to rilpivirine (Stanford, threshold 5%) | .248 | … | … | … | .014 | … | … | … |
| Baseline resistance to rilpivirine (Stanford, threshold 1%) | .784 | .307 | ||||||
| GSS <3 (Stanford, threshold 20%) | .083 | … | … | … | <.001 | … | … | … |
| GSS <3 (Stanford, threshold 5%) | .315 | .029 | ||||||
| GSS <3 (Stanford, threshold 1%) | .291 | .134 | ||||||
Abbreviations: ANRS, French National Agency for Research on AIDS and Viral Hepatitis; CI, confidence interval; GSS, genotypic sensitivity score; HIV, human immunodeficiency virus; OR, odds ratio.
Logistic regression.
Viral load difference between month 3 and baseline.
At 12 months, VR was 96.9%. Factors associated with VR at month 12 in the univariate analysis included the CD4 cell count, the viral load slope at month 3, the absence of resistance to RPV at baseline and a GSS of 3 using the Stanford interpretation system with a 20% threshold and with a 5% threshold (Table 2). Multivariate analysis indicated that the CD4 cell count, the decrease in viral load at month 3, and the absence of resistance to RPV at baseline using Stanford with a 20% threshold were associated with VR at month 12.
Fifteen patients experienced virological failure. We detected no RAMs in 7 of them before treatment using either NGS or the Sanger sequencing, 5 of them had majority mutations (patients 8–12), and the remaining 3 had minority mutations (patients 13–15) (Table 3). The E138A mutation was found in 8 patients using NGS and in 4 by Sanger sequencing.
Table 3.
Baseline Characteristics of Patients With Virological Failure
| Patients With VF | RAM (Frequency) | Resistance Using ANRS Algorithm | Resistance Using Stanford Algorithm |
|---|---|---|---|
| Patients 1–7 | None | None | None |
| Patient 8 | 138A (99.9%) | Rilpivirine | Rilpivirine |
| Patient 9 | 106I (99.2%) | None | None |
| Patient 10 | 106I (98.6%) | None | None |
| Patient 11 | 98S (97.9%) | Nevirapine | None |
| Patient 12 | 98G (90.7%) | None | Rilpivirine |
| Patient 13 | 179I (17.4%) | None | None |
| Patient 14 | 65N (1.3%) | Tenofovir | Tenofovir |
| Patient 15 | 90I (1.1%) | None | None |
Abbreviations: ANRS, French National Agency for Research on AIDS and Viral Hepatitis; RAM, resistance-associated mutation; VF, virological failure.
DISCUSSION
Next-generation sequencing technologies provide new data on the frequency of minority HIV-1 resistant variants. Their clinical impact on the response of HIV-1–infected patients to treatment must be assessed before routine testing for minority resistant variants. We therefore used the 454 GS-FLX NGS platform to determine the prevalence of NRTI and NNRTI resistant minority variants in a cohort of antiretroviral-naive patients. All the mutations identified in the RT gene were used to evaluate their impact on the VR to a first-line regimen containing RPV. Although the 454 technology is no longer supported, genotypic resistance results provided by the Illumina platform have been shown to be highly concordant with those obtained with the 454 platform in previous studies [16, 17]. The demographic characteristics of our patients were similar with those previously described [1, 18]. The subtypes of HIV-1 were representative of the subtype distribution in France [19, 20], with a predominance of subtype B and CRF02-AG. However, the baseline median CD4 cell count was higher (470 cells/µL) than that of other cohorts of patients given first-line NNRTIs (259 and 227 cells/µL) [18, 21]. Almost all (99%) of our patients were given the same antiretroviral therapy, which consisted of RPV, TDF, and FTC.
We analyzed the amino acid positions involved in resistance to NRTIs and NNRTIs of the virus RT gene. The prevalence of minority resistant variants at 1%–20% (18%) was similar to that found in a similar cohort (21%) [18]. The 20% threshold RAMs were more prevalent (29%) than those reported in other studies analyzing the same RAMs on patients given first-line NNRTIs (6%–18%) [11, 18]. The majority RAMs identified by NGS were also identified in most cases by bulk sequencing. Their prevalence was high, but most of them must be combined with other mutations to affect drug susceptibility. The ANRS algorithm used at the time of screening by bulk sequencing did not take into account the mutations 138A and 179D/L. Therefore, patients harboring those mutations were given an antiretroviral regimen containing RPV. However, the new version of the ANRS algorithm used for our retrospective study did include these 2 mutations.
The agreement between NGS and Sanger sequencing indicates the robustness of the NGS mutations analysis for the >20% RAMs, in line with previous observations [13, 22]. In addition, NGS identified 104 <20% RAMs that were not seen by Sanger sequencing. NGS identified RPV-resistant variants at a 1% threshold in 8.8% according to the ANRS algorithm and 7.4% of patients according to the Stanford algorithm. This prevalence was lower than that previously published for a smaller cohort of patients with primary infection (15%) [13], but higher than that found in a case-control study including antiretroviral-naive patients (4%) [11].
We found an association between baseline RPV-majority RAMs according to the Stanford algorithm and virological failure in our patients despite a high virological success rate. RAMs for RPV detected by bulk sequencing and interpreted with the Stanford algorithm were also associated with VF. By contrast, we found no association between VF and minority RAMs. In those cohort studies that found a relationship between primary mutations and VF [18, 23], the primary mutations were not observed as natural polymorphisms with bulk sequencing. The RPV-associated mutations are polymorphic in nature and can easily be found as dominant variants without selection pressure. Other studies found no association between the detection of minority resistant variants and VF of first-generation NNRTI-based treatment [24–27]. VF seems to be more frequent in studies that found that minority resistant variants influenced the response to treatment [28, 29]. One case-control study found that the baseline frequencies of minority RPV-resistant variants in patients on RPV-based treatment with virological success and failure were similar [11].
We identified 2 other factors associated with the virological response despite the low VF. A baseline CD4 cell count <250 cells/µL and a shallower slope of viral load at month 3 were associated with VR at months 6 and 12. While the baseline viral load was not predictive of VR, all of the viral loads were <100000 copies/mL, as recommended by the European Medicines Agency for RPV administration. Non-B subtype viruses tended to be associated with a higher VF in univariate analysis, while the baseline viral loads and the prevalence of RAMs in B and non-B subtypes were similar (data not shown). However, the patients infected with non-B subtypes had a lower CD4 cell count (467 cells/µL) than those with a subtype B infection (512 cells/µL) (P = .03). We therefore believe that minority resistant variants alone do not influence the virological response to RPV-based treatment, but that cofactors like adherence and CD4 cell count play a major role in the virological outcome.
One limit of our study is that we have no adherence reports. Though adherence may partly explain the VF [4, 28], other studies have found that minority resistant variants influence VF without reporting adherence data [21, 30]. Drug concentrations were not available for analyzing possible reasons for VF. Another limitation is that the difference in the virological responses of patients harboring minority resistant variants and those harboring none might be below the expected 20%. However, many more patients would be required to show a smaller difference. Last, the long-term influence of minority variants was not assessed owing to insufficient follow-up.
In conclusion, NGS identified RPV-resistant variants with a 1% threshold in 8.8% of patients according to the ANRS algorithm and 7.4% of patients according to the Stanford algorithm. The minority resistant variants to NRTIs and NNRTIs (<20% of the virus population) had no impact on the virological response to an RPV-based regimen. Rilpivirine seems to be safe for use in antiretroviral-naive patients, even if minority resistant variants are not searched before treatment initiation. However, resistance testing by bulk sequencing or NGS with a 20% threshold is recommended before treatment initiation.
Notes
Acknowledgments. We thank C. Lefebvre and R. Carcenac for technical assistance with next-generation sequencing.
Financial support. This work was supported by the Agence Nationale de Recherches sur le SIDA et les hépatites virales (French National Agency for Research on AIDS and Viral Hepatitis; ANRS). Roche Laboratories provided nucleic acid extraction reagents and performed NGS on the 454 GS-FLX. Janssen-Cilag SAS financed the laboratories reagents used in this work and sample transport.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
ANRS AC11 Resistance Study Group. Aix en Provence (Dr E. Lagier); Amiens (Dr C. Roussel); Angers (Dr H. Le Guillou-Guillemette); Avicenne (Dr C. Alloui); Besançon (Dr D. Bettinger); Bicêtre (Dr C. Pallier); Bordeaux (Pr H. Fleury, Dr S. Reigadas, Dr P. Bellecave, Dr P. Recordon-Pinson); Brest (Pr C. Payan, Dr S. Vallet); Caen (Pr A. Vabret, Dr J. Dina); Clermont-Ferrand (Dr C. Henquell, Dr A. Mirand); Créteil (Dr M. Bouvier-Alias); Dijon (Dr A. de Rougemont); Fort de France (Dr G. Dos Santos); Grenoble (Pr P. Morand, Dr A. Signori-Schmuck); Lille (Dr L. Bocket); Limoges (Pr S. Rogez); Lyon (Dr P. Andre, Dr J. C. Tardy, Dr M. A. Trabaud); Marseille (Dr C. Tamalet); Metz-Thionville (Dr C. Delamare); Montpellier (Dr B. Montes); Nancy (Pr E. Schvoerer); Nantes (Pr V. Ferré, Dr E. André-Garnier); Nice (Dr J. Cottalorda); Orléans (Dr J. Guinard, Dr A. Guiguon); Paris-Bichat Claude Bernard (Pr D. Descamps, Pr F. Brun-Vézinet, Dr C. Charpentier, Dr B. Visseaux, Dr G. Peytavin); Paris-Cochin (Dr A. Krivine); Paris-HEGP (Dr A. Si-Mohamed); Paris-Necker (Dr V. Avettand-Fenoel); Paris-Pitié- Salpêtrière (Pr A. G. Marcelin, Pr V. Calvez, Dr S. Lambert-Niclot, Dr C. Soulié, Dr M. Wirden); Paris Saint-Antoine (Dr L. Morand-Joubert); Paris-Saint Louis (Pr C. Delaugerre, Dr M. L. Chaix); Paris-Tenon (Dr C. Amiel, Dr V. Schneider); Poitiers (Dr G. Giraudeau, Dr A. Beby- Defaux); Reims (Dr V. Brodard); Rennes (Dr A. Maillard); Rouen (Pr J. C. Plantier); Saint Denis (Dr C. Chaplain); Saint Etienne (Dr T. Bourlet); Strasbourg (Pr S. Fafi-Kremer, Pr F. Stoll-Keller, Dr M. P. Schmitt, Dr H. Barth); Suisse (Dr S. Yerly); Toulon (Dr C. Poggi); Toulouse (Pr J. Izopet, Dr S. Raymond); Tours (Pr F. Barin, Dr A. Chaillon); Versailles (Dr S. Marque-Juillet); Villejuif (Pr A. M. Roque-Afonso, Dr S. Haïm-Boukobza); INSERM UMR-S1136 (Dr P. Flandre, M. Grudé, Dr L. Assoumou, Dr D. Costagliola).
ANRS Clinical Centers. Aix en Provence (Dr T. Allegre); Amiens (Pr J. L. Schmit); Angers (Dr J. M. Chennebault); Avicenne (Pr O. Bouchaud); Besançon (Pr N. Magy-Bertrand); Bicêtre (Pr J. F. Delfraissy); Bordeaux (Pr M. Dupon, Pr P. Morlat, Pr D. Neau); Brest (Pr S. Ansart, Dr S. Jaffuel); Caen (Pr R. Verdon); Clermont-Ferrand (Dr C. Jacomet); Créteil (Pr Y. Lévy, Dr S. Dominguez); Dijon (Pr P. Chavanet; Pr L. Piroth); Fort de France (Dr A. Cabié); Grenoble (Dr P. Leclercq); Lille-Tourcoing (Dr F. Ajana, Dr A. Cheret); Limoges (Pr P. Weinbreck); Lyon (Dr L. Cotte); Marseille (Dr I. Poizot-Martin, Dr I. Ravaud); Metz-Thionville (Dr B. Christian, Dr F. Truchetet, Dr M. Grandidier); Montpellier (Pr J. Reynes); Nancy (Pr T. May, Dr F. Goehringer); Nantes (Pr F. Raffi); Nice (Pr P. Dellamonica); Orléans (Dr T. Prazuck, Dr L. Hocqueloux); Paris-Bichat Claude Bernard (Dr R. Landman, Pr Yazdanpanah); Paris-Cochin (Dr O. Launay); Paris- HEGP (Pr L. Weiss); Paris-Necker (Dr J. P. Viard); Paris-Pitié-Salpêtrière (Pr C. Katlama, Dr A. Simon); Paris-Saint Antoine (Pr P. M. Girard, Dr J. L. Meynard); Paris-Saint Louis (Pr J. M. Molina); Paris Tenon (Pr G. Pialoux); Pointe-á-Pitre (Pr B. Hoen, Dr M.T. Goeger-Sow, Dr I. Lamaury, Dr G. Beaucaire); Poitiers (Dr G. Le Moal); Reims (Pr R. Jaussaud, Dr C. Rouger); Rennes (Pr C. Michelet); Rouen (Dr F. Borsa-Lebas, Pr F. Caron); Saint Denis (Dr M. A. Khuong); Saint Etienne (Pr F. Lucht); Strasbourg (Dr D. Rey); Suisse (Dr A. Calmy); Toulon (Dr A. Lafeuillade); Toulouse (Pr B. Marchou, Pr P. Delobel); Tours (Dr G. Gras), Versailles (Dr A. Greder-Belan); Villejuif (Pr D. Vittecoq, Dr E. Teicher).
Contributor Information
French National Agency for Research on AIDS and Viral Hepatitis (ANRS) AC11 Resistance Study Group:
E Lagier, C Roussel, H Le Guillou-Guillemette, C Alloui, D Bettinger, C Pallier, H Fleury, S Reigadas, P Bellecave, P Recordon-Pinson, C Payan, S Vallet, A Vabret, J Dina, C Henquell, A Mirand, M Bouvier-Alias, A de Rougemont, G Dos Santos, P Morand, A Signori-Schmuck, L Bocket, S Rogez, P Andre, J C Tardy, M A Trabaud, C Tamalet, C Delamare, B Montes, E Schvoerer, V Ferré, E André-Garnier, J Cottalorda, J Guinard, A Guiguon, D Descamps, F Brun-Vézinet, C Charpentier, B Visseaux, G Peytavin, A Krivine, A Si-Mohamed, V Avettand-Fenoel, A G Marcelin, V Calvez, S Lambert-Niclot, C Soulié, M Wirden, L Morand-Joubert, C Delaugerre, M L Chaix, C Amiel, V Schneider, G Giraudeau, A Beby- Defaux, V Brodard, A Maillard, J C Plantier, C Chaplain, T Bourlet, S Fafi-Kremer, F Stoll-Keller, M P Schmitt, H Barth, S Yerly, C Poggi, J Izopet, S Raymond, F Barin, A Chaillon, S Marque-Juillet, A M Roque-Afonso, S Haïm-Boukobza, P Flandre, M Grudé, L Assoumou, D Costagliola, T Allegre, J L Schmit, J M Chennebault, O Bouchaud, N Magy-Bertrand, J F Delfraissy, M Dupon, P Morlat, D Neau, S Ansart, S Jaffuel, R Verdon, C Jacomet, Y Lévy, S Dominguez, P Chavanet, L Piroth, A Cabié, P Leclercq, F Ajana, A Cheret, P Weinbreck, L Cotte, I Poizot-Martin, I Ravaud, B Christian, F Truchetet, M Grandidier, J Reynes, T May, F Goehringer, F Raffi, P Dellamonica, T Prazuck, L Hocqueloux, R Landman, Yazdanpanah, O Launay, L Weiss, J P Viard, C Katlama, A Simon, P M Girard, J L Meynard, J M Molina, G Pialoux, B Hoen, M T Goeger-Sow, I Lamaury, G Beaucaire, G Le Moal, R Jaussaud, C Rouger, C Michelet, F Borsa-Lebas, F Caron, M A Khuong, F Lucht, D Rey, A Calmy, A Lafeuillade, B Marchou, P Delobel, G Gras, A Greder-Belan, D Vittecoq, and E Teiche
References
- 1. Messiaen P, Verhofstede C, Vandenbroucke Iet al. Ultra-deep sequencing of HIV-1 reverse transcriptase before start of an NNRTI-based regimen in treatment-naive patients. Virology 2012; 426:7–11. [DOI] [PubMed] [Google Scholar]
- 2. Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res 2007; 17:1195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loman NJ, Misra RV, Dallman TJet al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 2012; 30:434–9. [DOI] [PubMed] [Google Scholar]
- 4. Li JZ, Paredes R, Ribaudo HJet al. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS 2012; 26:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Halvas EK, Wiegand A, Boltz VFet al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment- experienced patients. J Infect Dis 2010; 201:672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lecossier D, Shulman NS, Morand-Joubert Let al. Detection of minority populations of HIV-1 expressing the K103N resistance mutation in patients failing nevirapine. J Acquir Immune Defic Syndr 2005; 38:37–42. [DOI] [PubMed] [Google Scholar]
- 7. Li JZ, Paredes R, Ribaudo HJet al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011; 305:1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Molina JM, Clumeck N, Redant K, Rimsky L, Vanveggel S, Stevens M; ECHO Study Group; THRIVE Study Group. Rilpivirine vs. efavirenz in HIV-1 patients with baseline viral load 100000 copies/ml or less: week 48 phase III analysis. AIDS 2013; 27:889–97. [DOI] [PubMed] [Google Scholar]
- 9. Cohen CJ, Molina JM, Cassetti Iet al. ; ECHO and THRIVE Study Groups. Week 96 efficacy and safety of rilpivirine in treatment-naive, HIV-1 patients in two phase III randomized trials. AIDS 2013; 27:939–50. [DOI] [PubMed] [Google Scholar]
- 10. Lambert-Niclot S, Charpentier C, Storto Aet al. Prevalence of pre-existing resistance-associated mutations to rilpivirine, emtricitabine and tenofovir in antiretroviral-naive patients infected with B and non-B subtype HIV-1 viruses. J Antimicrob Chemother 2013; 68:1237–42. [DOI] [PubMed] [Google Scholar]
- 11. Van Eygen V, Thys K, Van Hove Cet al. Deep sequencing analysis of HIV-1 reverse transcriptase at baseline and time of failure in patients receiving rilpivirine in the phase III studies ECHO and THRIVE. J Med Virol 2016; 88:798–806. [DOI] [PubMed] [Google Scholar]
- 12. Asahchop EL, Wainberg MA, Oliveira Met al. Distinct resistance patterns to etravirine and rilpivirine in viruses containing nonnucleoside reverse transcriptase inhibitor mutations at baseline. AIDS 2013; 27:879–87. [DOI] [PubMed] [Google Scholar]
- 13. Nicot F, Saliou A, Raymond Set al. Minority variants associated with resistance to HIV-1 nonnucleoside reverse transcriptase inhibitors during primary infection. J Clin Virol 2012; 55:107–13. [DOI] [PubMed] [Google Scholar]
- 14. Descamps D, Delaugerre C, Masquelier Bet al. Repeated HIV-1 resistance genotyping external quality assessments improve virology laboratory performance. J Med Virol 2006; 78:153–60. [DOI] [PubMed] [Google Scholar]
- 15. Wensing AM, Calvez V, Günthard HFet al. 2017 update of the drug resistance mutations in HIV-1. Top Antivir Med 2017; 24:132–3. [PMC free article] [PubMed] [Google Scholar]
- 16. Li JZ, Chapman B, Charlebois Pet al. ; ACTG A5262 Study Team. Comparison of Illumina and 454 deep sequencing in participants failing raltegravir-based antiretroviral therapy. PLoS One 2014; 9:e90485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thys K, Verhasselt P, Reumers J, Verbist BM, Maes B, Aerssens J. Performance assessment of the Illumina massively parallel sequencing platform for deep sequencing analysis of viral minority variants. J Virol Methods 2015; 221:29–38. [DOI] [PubMed] [Google Scholar]
- 18. Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo Fet al. ; CHAIN Minority HIV-1 Variants Working Group. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother 2015; 70:930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Descamps D, Assoumou L, Chaix MLet al. National sentinel surveillance of transmitted drug resistance in antiretroviral-naive chronically HIV-infected patients in France over a decade: 2001–2011. J Antimicrob Chemother 2013; 68:2626–31. [DOI] [PubMed] [Google Scholar]
- 20. Frange P, Assoumou L, Descamps Det al. ; French ANRS CO 6 PRIMO Cohort, the ANRS 147 OPTIPRIM Clinical Trial and the AC11 Resistance Study Groups; French ANRS CO 6 PRIMO Cohort the ANRS 147 OPTIPRIM Clinical Trial and the AC11 Resistance Study Groups. HIV-1 subtype B-infected MSM may have driven the spread of transmitted resistant strains in France in 2007-12: impact on susceptibility to first-line strategies. J Antimicrob Chemother 2015; 70:2084–9. [DOI] [PubMed] [Google Scholar]
- 21. Goodman DD, Zhou Y, Margot NAet al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS 2011; 25:325–33. [DOI] [PubMed] [Google Scholar]
- 22. Mohamed S, Penaranda G, Gonzalez Det al. Comparison of ultra-deep versus Sanger sequencing detection of minority mutations on the HIV-1 drug resistance interpretations after virological failure. AIDS 2014; 28:1315–24. [DOI] [PubMed] [Google Scholar]
- 23. Li JZ, Paredes R, Ribaudo HJet al. Impact of minority nonnucleoside reverse transcriptase inhibitor resistance mutations on resistance genotype after virologic failure. J Infect Dis 2013; 207:893–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peuchant O, Thiébaut R, Capdepont Set al. ; ANRS CO3 Aquitaine Cohort. Transmission of HIV-1 minority-resistant variants and response to first-line antiretroviral therapy. AIDS 2008; 22:1417–23. [DOI] [PubMed] [Google Scholar]
- 25. Metzner KJ, Rauch P, Braun Pet al. Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naive patients. J Clin Virol 2011; 50:156–61. [DOI] [PubMed] [Google Scholar]
- 26. Jakobsen MR, Tolstrup M, Søgaard OSet al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis 2010; 50:566–73. [DOI] [PubMed] [Google Scholar]
- 27. Nicot F, Sauné K, Raymond Set al. Minority resistant HIV-1 variants and the response to first-line NNRTI therapy. J Clin Virol 2015; 62:20–4. [DOI] [PubMed] [Google Scholar]
- 28. Paredes R, Lalama CM, Ribaudo HJet al. ; AIDS Clinical Trials Group (ACTG) A5095 Study Team. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 2010; 201:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balduin M, Oette M, Däumer MP, Hoffmann D, Pfister HJ, Kaiser R. Prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naive patients and their impact on the virological failure. J Clin Virol 2009; 45:34–8. [DOI] [PubMed] [Google Scholar]
- 30. Simen BB, Simons JF, Hullsiek KHet al. ; Terry Beirn Community Programs for Clinical Research on AIDS. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199:693–701. [DOI] [PubMed] [Google Scholar]