Abstract
Chloroplasts cannot develop normally without the coordinated action of various proteins and signaling connections between the nucleus and the chloroplast genome. Many questions regarding these processes remain unanswered. Here, we report a novel P-type pentatricopeptide repeat (PPR) factor, named Albino Cotyledon Mutant1 (ACM1), which is encoded by a nuclear gene and involved in chloroplast development. Knock-down of ACM1 transgenic plants displayed albino cotyledons but normal true leaves, while knock-out of the ACM1 gene in seedlings was lethal. Fluorescent protein analysis showed that ACM1 was specifically localized within chloroplasts. PEP-dependent plastid transcript levels and splicing efficiency of several group II introns were seriously affected in cotyledons in the RNAi line. Furthermore, denaturing gel electrophoresis and Western blot experiments showed that the accumulation of chloroplast ribosomes was probably damaged. Collectively, our results indicate ACM1 is indispensable in early chloroplast development in Arabidopsis cotyledons.
Keywords: ACM1, biogenesis, chloroplast, cotyledon, development, PPR protein, ribosome accumulate
1. Introduction
Chloroplasts are well known as the most important organelle in higher plants. Beyond photosynthesis, chloroplasts are also the main sites of the biosynthesis of fatty acids, hormones, amino acids, and metabolites, and the assimilation of nitrate [1,2]. Because of its ancestry, the chloroplast is generally considered to have originated through endosymbiosis with species of cyanobacteria. The process of chloroplast biogenesis and development is highly complex and ordered; while the molecular mechanisms have not been fully elucidated yet [3]. Development of the chloroplast involves an intricate interplay between both the chloroplast and the nuclear-cytoplasmic synthetic systems. A lot of signaling between the nucleus and chloroplast occurs to guide the proper formation and assembly of functional and photosynthetically active chloroplasts [4]. In addition, chloroplast gene expression is also vital for the chloroplast development process, which is mediated by two distinct types of RNA polymerases; plastid-encoded RNA polymerase (PEP) and nuclear-encoded RNA polymerases (NEP) [5,6].
There is mounting evidence that chloroplast biogenesis and development differ between monocotyledons and dicotyledons [3]. Furthermore, in dicotyledonous plants, chloroplast differentiation follows distinct paths in both the cotyledons and the true leaves [7]. The development of cotyledons formed during embryogenesis is different from the development of true leaves, which arise as the result of apical meristem activity [8,9]. Usually, in cotyledons, plastids partially develop during embryogenesis but stop at seed maturation and dormancy. When germinated in light, the plastids can further develop into functional chloroplasts. Contrastingly, within true leaves, proplastids differentiate into mature chloroplasts. In other words, fully differentiated chloroplasts in cotyledons are similar to young true leaf chloroplasts [8,10]. Studies on mutants with albino or pale-green cotyledons but normal green true leaves suggest, however, that there are indeed differences in the regulation of plastid development between these two organs. For examples, soc1 and soc2 [11,12]; sig2 and sig6 [13,14]; ys1 [15]; ecb2 [16] and some lines with gene down-regulation like PRDA1-RNAi [17] and ECD1-RNAi [18]. Several different mechanisms result in similar phenotypes. Among these, we are very interested in ECB2 and ECD1, which belong to the PPR family proteins.
Chloroplasts are semiautonomous organelles, retaining their own genomes and gene expression apparatuses but controlled by nucleus genome encoded protein factors. The PPR protein family is one of the largest protein families in land plants, with more than 400 members in most species; Arabidopsis has 450 and maize has 600 PPR proteins [19,20]. PPR proteins play crucial roles in plant function and development, especially in RNA regulation in chloroplasts and mitochondria. Through the examination of RNA editing, maturation, stabilization, or intron splicing, and in transcription and translation processes of organellar genes, more evidence of the diverse roles of PPR proteins in plants is gained every year [21,22]. Defective or damaged PPR proteins, therefore, usually lead to very severe phenotypes, such as embryo development delay, pigmentation defect, and abnormal chloroplast biogenesis [23,24,25]. Focusing on only chloroplasts, defective and damaged PPR proteins lead to several phenotypes: pale-yellowish, grana thylakoid formation altered, chlorophyll synthesis hindered, and even severe defects in photosynthesis [26,27]. In conclusion, PPR proteins can influence chloroplast biogenesis or development by diverse action modes and functional types.
The PPR family can be classified into two subfamilies, PLS and P, according to the motif structure. The P-class PPR proteins always contain from 2 to over 30 conserved 35 amino acid PPR (P) motifs [28]. While the PLS-class subfamily proteins contain arrays of P, L, and S triplets. Beyond the P motif, the L and S motifs are related to the 35/36 amino acid and 31 amino acid PPR motifs, respectively [20,29]. Generally speaking, P-type PPR proteins are responsible for RNA stabilization, translational activation and also promote the splicing of group II introns [20]. AtBFA2 is proven to be needed for the stabilization of atpH/F transcripts in chloroplasts [30] and two newly discovered P-type PPR proteins, PDM3 and PDM4, are necessary for several group II intron splicing [31,32]. PLS-type PPR proteins are considered to be involved in RNA editing at specific sites [33]. Both QED1 in Arabidopsis and PPR756 in rice, are members of the PLS subfamily, which affect RNA editing at different sites [34,35]. Of course, some PPR proteins do not follow this rule strictly [36,37,38,39] and require further examination.
In this study, a novel PPR protein was screened and identified to affect chloroplast development in Arabidopsis thaliana. The mutant shows an embryo lethality phenotype and RNAi lines display albino cotyledons but normal true leaves, so this new protein was named ALBINO COTYLEDON MUTANT1 (ACM1). Within the mutant, abnormal chloroplast development occurred, and further analyses proved that ACM1 is essential for several group II introns splicing and plastid gene expression in cotyledons.
2. Results
2.1. Characterization of the acm1 Mutant
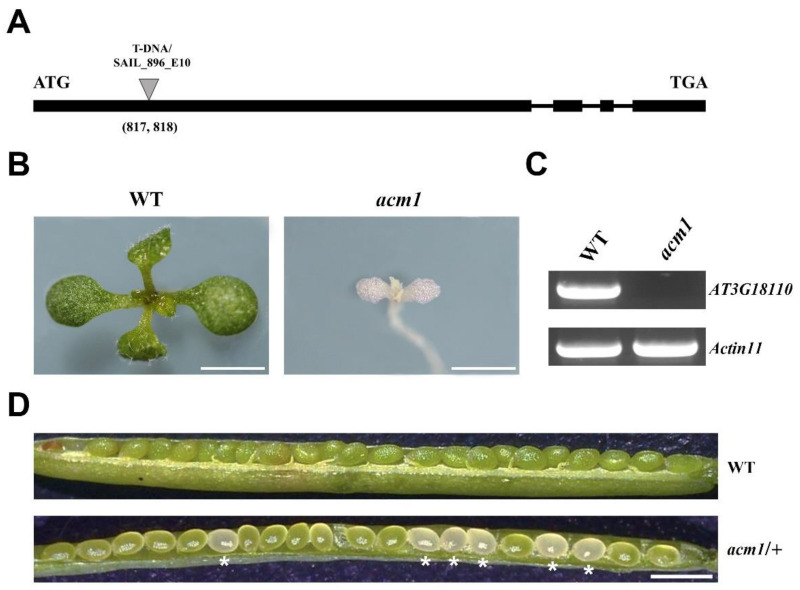
In order to further study the molecular biosynthesis mechanism and development regulation of chloroplasts, we screened a series of T-DNA insertion lines related to chloroplast development defects from the Arabidopsis Biological Resource Center (ABRC). Among these, a novel PPR (AT3G18110) mutant with albino phenotype, designated as acm1, was chosen for the following experiments. The sequencing of T-DNA flanking sequences showed that the T-DNA was inserted at 817 bp site from the start codon (Figure 1A). The mutant was seedling lethal under autotrophic growth conditions (survived about one week in soil, data not shown) and even in medium with sucrose, the acm1 mutant showed an albino phenotype with a little purple coloring (Figure 1B). Reverse transcription PCR (RT-PCR) analysis showed that the expression of the AT3G18110 was undetectable under 29 PCR cycles (Figure 1C). When we observed the siliques between the wild-type (WT) and acm1 mutant we found that the ovules had developed normally in WT, but in the heterozygous acm1/+ siliques, some of the ovules were white (Figure 1D). In 42 siliques from the heterozygous plants, 224 ovules were white and 658 ovules were green, resulting in a ratio of white to green of about 1:3 (data not shown).
Figure 1.
Phenotype of the acm1 mutant. (A) Schematic diagram of the ACM1 gene. Exons and introns are indicated by black boxes and lines respectively. The T-DNA insertion by the triangle; ATG represents the initiation codon and TGA represents the stop codon. (B) Albino phenotype of the acm1 mutant. Ten-day-old plants were grown on 1/2 MS medium with sucrose. Scale bar: 3 mm. (C) Reverse transcription PCR analysis. RT-PCR was performed using specific primers for AT3G18110 and Actin 11 for 29 cycles for WT and acm1. (D) The ovules of heterozygous acm1/+ mutant silique. The asterisks indicate albino ovules. Scale bar is 1 mm.
2.2. Phenotypic Characterizations of ACM1 Knock-Down Lines
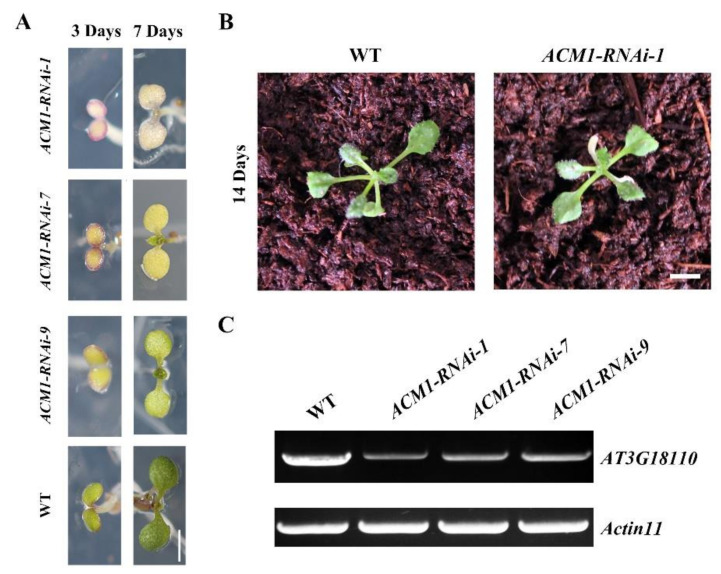
We next created RNAi lines to weaken the lethal phenotype of gene deletion to further investigate the ACM1 gene functions. We obtained 36 out of 65 transgenic lines (data not shown) with the albino cotyledon phenotype. Three lines, line 1, line 7, and line 9 had a range of stable white cotyledons and stunted plant growth and were selected for further experimentation (Figure 2A). When grown in soil for about 2 weeks, these lines could develop normal green true leaves and exhibited a normal growth state, we selected line 1 as a representative showed in Figure 2B. After this growing time, we tested again the ACM1 expression by RT-PCR in the WT and the three RNAi lines, the results were consistent with the phenotype: the expression level of ACM1 was the lowest in the ACM1-RNAi-1 line, which showed the most severe phenotype (Figure 2B,C). These results can confirm that disruption of the ACM1 resulted in abnormal cotyledons but normal true leaves. The RNAi-1 plants can mimick the phenotypes of knock-out mutant acm1, in which the ACM1 transcripts were decreased significantly. Subsequently, a subset of experiments could be suitably performed on RNAi-1 plants due to the most severely affected phenotype, although the heterotrophic ability remained.
Figure 2.
Characterization of the ACM1-RNAi transgenic plants. (A) Identification and isolation of RNAi lines with different degrees of inhibition of ACM1 expression. Plants were grown on 1/2 MS medium with 2% (w/v) sucrose for 3 d and 7 d. Scale bar is 1 mm. (B) Albino cotyledon but normal true leaves phenotype of ACM1-RNAi-1. Plants were grown on soil (14 d). Scale bar: 2 mm. (C) Reverse transcription PCR analysis. Using specific primers for AT3G18110 and Actin 11 for 29 cycles for WT and RNAi lines with different degrees of inhibition of ACM1.
2.3. ACM1 Encodes a P-Type PPR Protein Localized in the Chloroplast
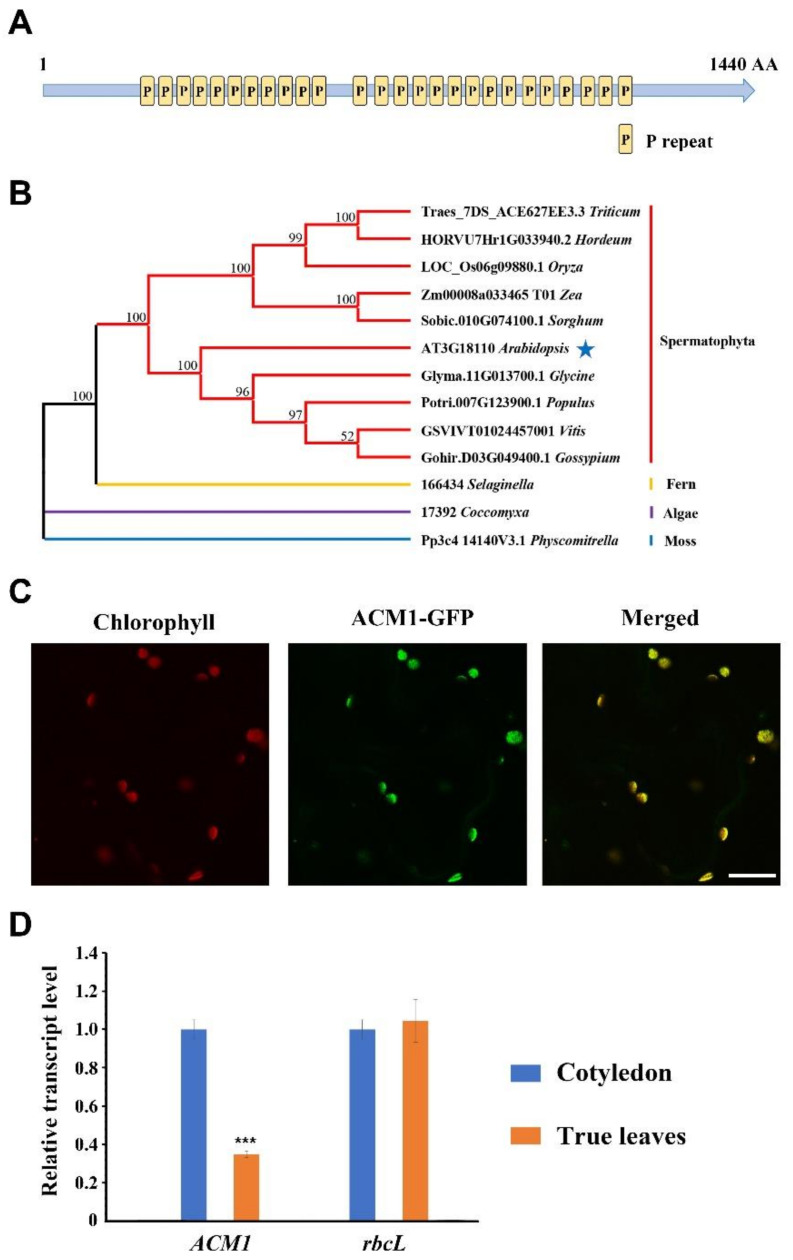
According to the classification of PPR motifs, we find ACM1 belongs to the P-type PPR proteins and the ACM1 gene encodes a polypeptide of 1440 amino acids. Protein sequence analysis in the Pfam database showed that ACM1 contains a tandem repeat of 26 PPR motifs (Figure 3A). A BLAST search in the NCBI database was performed to identify ACM1 homologs in various plant species. Several species belonging to Spermatophyta, fern, algae, and moss were chosen to do multiple sequence alignment with ACM1. The results showed ACM1 exhibits a high level of similarity in Populus trichocarpa (Potri.007G123900.1, 66.62%) and Gossypium hirsutum (Gohir.D03G049400.1, 66.60%), while it showed a relatively low level of similarity in Physcomitrella patens (Pp3c4_14140V3.1; 16.85%) and in Coccomyxa subellipsoidea (17392; 10.26% similarity) (Figure S1). According to the alignment, we created a phylogenetic tree (www.phylogeny.fr (accessed on 26 February 2021)) of these amino acid sequences from Figure S1 to reveal the relationship among these species (Figure 3B). The above results confirm that ACM1-like proteins are present in most plants but are more conservative in Spermatophyta.
Figure 3.
Sequence analysis and subcellular localization of ACM1. (A) Schematic diagram of ACM1 protein with 26 PPR domains (P). (B) Phylogenetic tree of ACM1 and other 12 PPR family members from Figure S1. Maximum likelihood (ML) tree was inferred with RAxML using the PROTGAMMALGF model. The numbers on the branches refer to the bootstrap values (%) for 1000 replications. Complete deletion was adopted for the treatment of gaps and missing data. (C) Localization of the ACM1 protein within the chloroplast using the GFP assay by transient transformation into tobacco leaf epidermal cells. Scale bar: 10 μm. (D) Quantitative real-time PCR to detect the expression profile of ACM1 transcript between cotyledons and true leaves in WT. RNA was extracted from 7-day-old cotyledons and 14-day-old true leaves from WT, and then reverse-transcribed. *** P < 0.001, by Student’s t-test.
To examine the subcellular localization of ACM1, a construct containing 35S:ACM1-GFP was transiently transformed into tobacco leaf epidermal cells. Transient expression observed by confocal laser scanning microscopy showed that the fusion proteins co-localized with Chlorophyll (Figure 3C), indicating that ACM1 proteins indeed are chloroplast-localized proteins. Moreover, we performed the quantitative real-time PCR to detect the expression profile of ACM1 transcript between cotyledons and true leaves in WT. The result showed that the transcript levels of ACM1 in cotyledon are higher than in true leaves (Figure 3D), the transcript levels of rbcL were tested as a control.
2.4. Chloroplast Development Was Affected in Cotyledons in the ACM1 Knock-Down Line
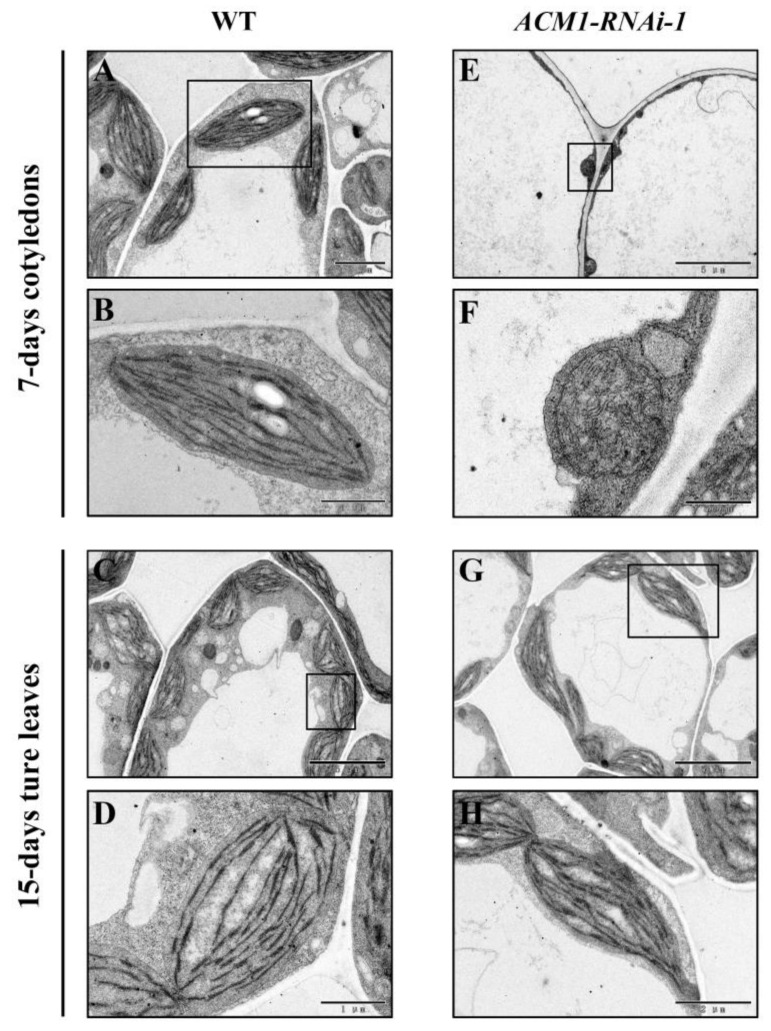
The obvious phenotype of ACM1 T-DNA insertion mutant plants and knock-down lines (ACM1-RNAi) suggest that ACM1 might play an essential role in chloroplast development based on previous research [31,32,36]. Thus, we examined the ultrastructure of chloroplasts from 7-day-old cotyledons and 14-day-old true leaves of RNAi-1 and WT by a transmission electron microscope. In WT plants, the chloroplasts from 7-day-old cotyledons and 14-day-old true leaves were oval-shaped and with well-developed thylakoids (Figure 4A–D). However, in the ACM1-RNAi-1 line, the chloroplasts of 7-day-old cotyledons were much smaller in size and were abnormally shaped, being nearly circular. Furthermore, the chloroplasts from the 7-day-old cotyledons of ACM1-RNAi-1 lacked internal membrane structures, such as stromal thylakoids and stacked grana thylakoids (Figure 4E,F). In contrast, the chloroplasts of 14-day-old true leaves from the ACM1-RNAi-1 line were completely normal (Figure 4G,H). These results prove that ACM1 is indeed required for chloroplast development in cotyledons.
Figure 4.
Ultrastructure of chloroplasts in RNAi-1 and WT. (A,B) The chloroplast ultrastructure in cotyledons from 7-day-old WT plants. Scale bar: 2 μm for A and 1 μm for B. (C,D) The chloroplast ultrastructure in true leaves from 14-day-old WT plants. Scale bar: 5 μm for C and 1 μm for D. (E,F) The chloroplast ultrastructure in cotyledons from 7-day-old ACM1-RNAi-1 line. Scale bar: 5 μm for E and 500 nm for F. (G,H) The chloroplast ultrastructure in true leaves from 14-day-old ACM1-RNAi-1 line. Scale bar: 5 μm for G and 2 μm for H.
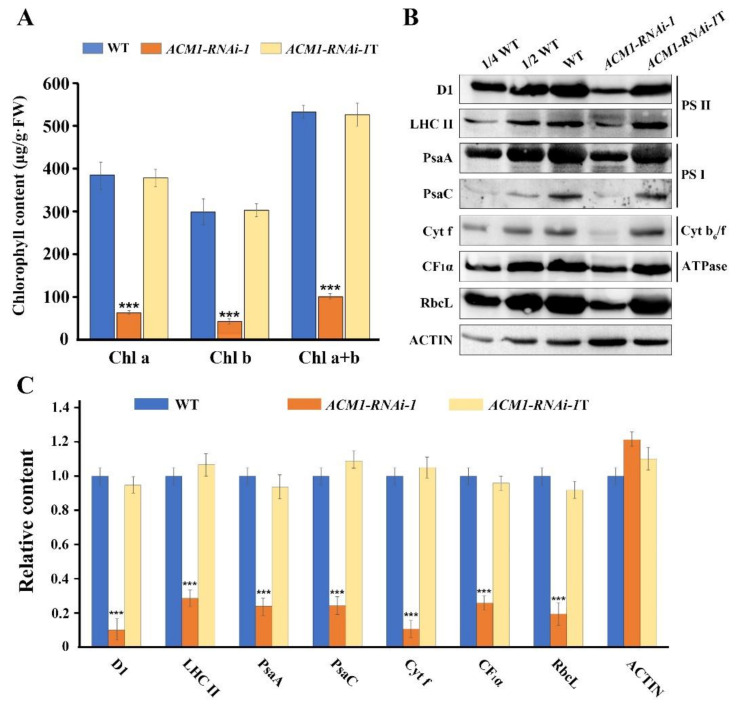
2.5. Knock-Down of ACM1 Affects the Accumulation of Chlorophyll and Photosynthetic Proteins
Because the level of chlorophyll is one of the most important indicators of the change of leaf color [31]. we tested the chlorophyll content in the RNAi line and WT. The chlorophyll content in the albino ACM1-RNAi-1 cotyledons was significantly reduced compared with WT, but chlorophyll content in the 14-day-old true leaves was normal compared to the control (Figure 5A). Combined with the chlorophyll content reduction and defective chloroplast development in RNAi lines, we inferred that the accumulation of photosynthetic complexes was also heavily affected. We then analyzed the protein profile between the RNAi line and WT. D1 and LHC II, the two main subunits of PSII; and PsaA and PsaC, representatives of PS I, were markedly reduced in RNAi-1 plants. Furthermore, Cyt f, the subunits of Cyt b6/f, the CF1α of ATP synthase, and RbcL (large subunit of the stromal protein ribulose biphosphate carboxylase) were also decreased to a very low level and were present at less than 20% of that of the WT (Figure 5B,C). All these data indicate that the ACM1 protein is indispensable for the accumulation of chlorophyll and photosynthetic proteins in cotyledons.
Figure 5.
The content of chlorophyll and photosynthetic proteins. (A) Chlorophyll content of WT and ACM1-RNAi-1 seedlings. Chlorophyll was quantified from 7-day-old seedlings cotyledons and 14-day-old seedlings’ true leaves. Values given are μg/g fresh weight ± SD. The asterisks indicate significant differences (Student’s t-test; *** p < 0.001). The average of three replicates is shown. (B) Immunoblot analysis of photosynthetic proteins. Total proteins from 7-day-old cotyledons and 14-day-old true leaves of WT and ACM1-RNAi-1 were separated by 10% SDS-PAGE (Sodium dodecyl sulfate polyacrylamide gel electrophoresis). Probed using specific anti-D1, anti-LHCII, anti-PsaA, anti-PsaC, anti-Cyt f, anti-CF1α, and anti-RbcL. ACTIN is used as a control. The experiments were repeated three times at least with similar results. (C) Proteins immunodetected from (A) were analyzed by ImageJ software. Values (means ± SE; n = 3 independent biological replicates) are given as ratios to protein amounts of the WT and ACM1-RANi-1. *** P < 0.001, by Student’s t-test. T refers to the proteins in true leaves of ACM1-RNAi-1 seedlings.
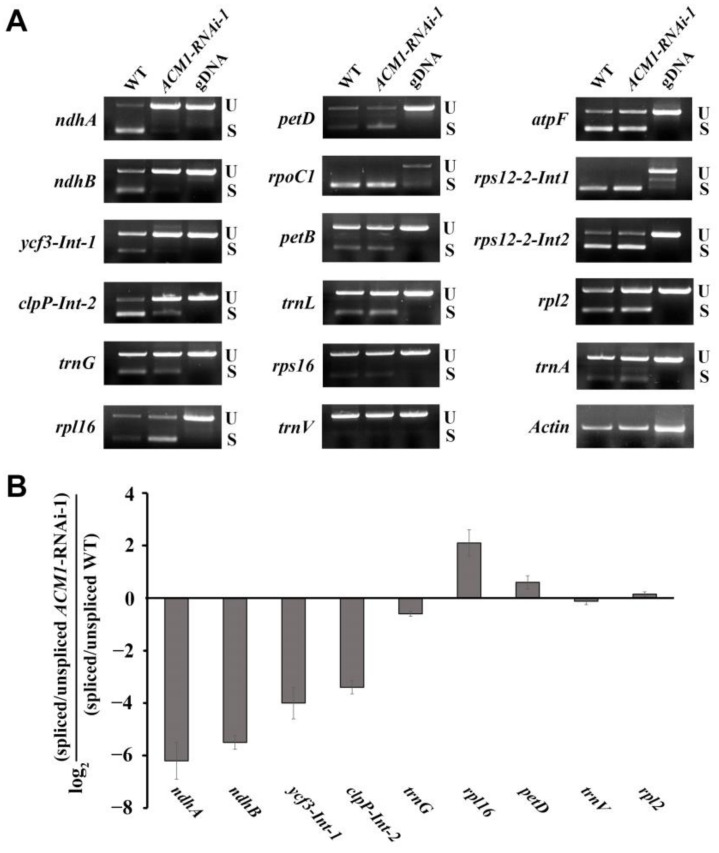
2.6. ACM1 Is Responsible for Several Group II Intron Splicing in the Cotyledon Chloroplast
P-type PPR proteins are usually responsible for RNA splicing of group II introns and the few studies examining this indeed confirm this view [31,32]. To further confirm this function in our new P-type PPR protein ACM1, we assayed some splicing events in the RNAi line by performing RT-PCR analyses. From the results (Figure 6) we found the unspliced precursors of ndhA, ndhB, ycf3-int-1, and clpp-int-2 accumulated visibly to an increased level in the ACM1 down-regulation line. The unspliced precursors of trnG were slightly accumulated compared with the WT (Figure 6). In contrast, the splicing efficiency of rpl16 and petD was increased to some degree according to our primary result. Other splicing events were also assayed but no obvious differences were found, these were rpoC1, petB, trnL, rps16, trnV, atpF, rps12-2-int1, rps12-2-int2, rpl2, and trnA (Figure 6). Furthermore, we performed the quantitative real-time PCR to confirm the splicing efficiency of several transcripts from the splicing events which were detected. The results of quantitative real-time PCR were basically consistent with the results of RT-PCR. In summary, these results provide evidence that ACM1 is involved in RNA splicing in cotyledon chloroplasts.
Figure 6.
Splicing of several group II introns in the ACM1-RNAi-1 chloroplast. (A) The splicing pattern of chloroplast intron-containing genes was analyzed by RT -PCR in the WT and ACM1-RNAi-1 plants. Actin was considered a control. The experiments were repeated three times with similar results. The cDNA of WT and ACM1-RNAi-1 was reverse-transcribed by total RNA from 7-day-old seedlings cotyledon. U, unsplicing; S, splicing. (B) Quantitative RT-PCR analysis to confirm the splicing efficiency of several transcripts as representative from (A). Data obtained from at least three independent experiments. RNA was extracted from 7-day-old seedlings cotyledons and reverse-transcribed.
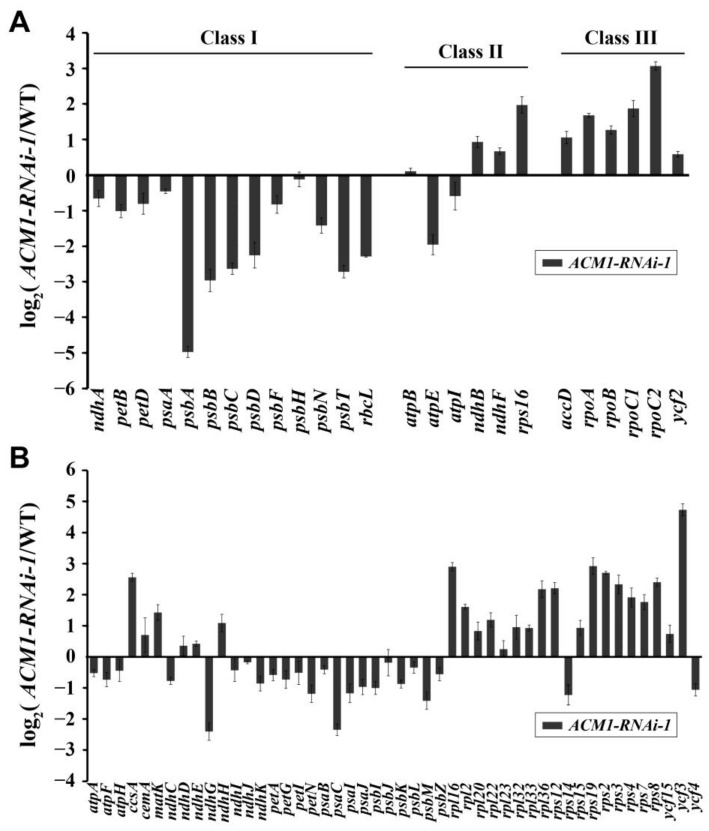
2.7. Knock-Down of ACM1 Affects Transcript Level of Plastid Gene
The development of chloroplasts was significantly impacted by chloroplast gene expression. According to the requirement of different RNA polymerases, plastid genes can divide into three classes: genes mainly synthesized by PEP are Class I; Class II genes are transcribed by both NEP and PEP and Class III genes are exclusively transcribed by NEP. To investigate the ACM1 functions in plastid gene expression, we examined the transcript abundance of plastid genes in the cotyledon between RNAi line and WT by quantitative RT-PCR analysis, the results are shown with log2 indexes in Figure 7. The transcript levels of Class I genes were significantly reduced in RNAi-1 compared with the WT. In contrast, transcript levels of Class III genes were increased and Class II genes were differentially regulated in ACM1-RNAi-1 (Figure 7A). The transcript levels of other not clearly classified chloroplast genes were also detected and are shown in Figure 7B.
Figure 7.
Chloroplast gene expression in ACM1-RNAi-1 relative to the WT. (A) Transcript levels of chloroplast genes were measured by quantitative RT-PCR. Data are given as log2 of ACM1-RNAi-1/WT ratios from at least three independent experiments. (B) Transcript levels of not clearly classified chloroplast genes. Data are given as log2 of ACM1-RNAi-1/WT ratios from at least three independent experiments. RNA was extracted from 7-day-old seedlings cotyledons and reverse-transcribed.
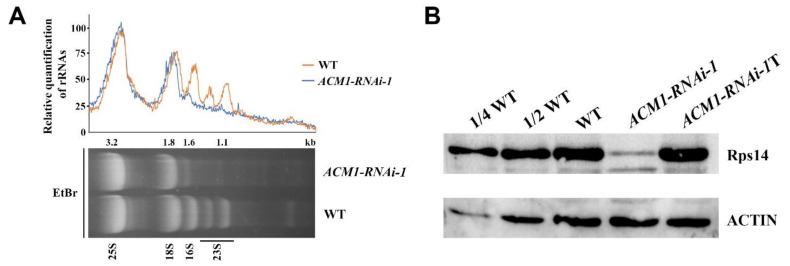
2.8. Accumulation of Chloroplast rRNAs and Ribosome Subunit RPS14 Was Disrupted in RNAi Line
The expression of chloroplast proteins in higher plants is largely regulated by the translation level. Proteins encoded by the plastid genome are synthesized by plastidic prokaryotic-type 70S ribosomes composed of 30S and 50S subunits. The 30S subunit consists of 16S rRNA, and about 20 ribosomal proteins; the 50S subunit contains 23S rRNA, 5S rRNA, 4.5S rRNA and about 30 ribosomal proteins [40]. After total RNA extraction from cotyledons of WT and the ACM1-RNAi-line, the RNA was subjected to denaturing agarose gel electrophoresis (Figure 8A). From the electrophoresis results, lower transcript levels of the 1.6 kb RNA corresponding to chloroplast 16S rRNA and 1.1 kb RNA corresponding to a breakdown product of the chloroplast 23S rRNA were detected in the cotyledons of the RNAi line (Figure 8A). At the protein level, we detected the content of PRS14, which is required for the accumulation of ribosomal 30S subunits, in cotyledons of the RNAi line and WT by Western blot analysis. An obvious difference is shown in Figure 8B, with the protein being substantially reduced in the RNAi line.
Figure 8.
The abundance of rRNA and accumulation of ribosomal subunit RPS14. (A) Five μg of total RNA from 7-day-old WT and ACM1-RNAi-1 seedlings was separated on a denaturing gel. rRNA was quantified using ImageJ software. The value of cytoplasmic 25S rRNA in ACM1-RNAi-1 was set to 100, and the relative values of other RNAs were obtained by comparing with 25S rRNA. (B) Immunoblot of chloroplast ribosomal subunits Rps14 in the 7-day-old cotyledon of WT and ACM1-RNAi-1 seedlings and 14-day-old true leaves in ACM1-RNAi-1 line. ACTIN was considered a control. All experiments were repeated three times. T refers to the proteins in true leaves of ACM1-RNAi-1 plants.
3. Discussion
In this study, we screened an embryo lethality mutant and isolated the gene ACM1, which encodes a P-type PPR protein. Further analyses were conducted on a selected RNAi line that exhibited a cotyledon-specific albino phenotype.
It is known that most seedling-lethal mutants are defective in chloroplast function or development, which reveals that the development of functional chloroplasts is tightly associated with plant growth and development [41,42]. In this study, the pigment-defective and seedling-lethal phenotypes of the acm1 mutant (Figure 1B,D) suggest that the ACM1 plays an important role in chloroplast development. The phenotype of RNAi lines of ACM1 (Figure 2A,B) also supports this point. Subcellular location analysis found that the ACM1 is a chloroplast-localized protein (Figure 3C), which provides a premise for its functions in chloroplasts [20,31,32]. Further observation of the plastid ultrastructure indicated that the plastids of the ACM1-RNAi line did not fully develop, stacked thylakoid membranes were disrupted and grana and stroma thylakoids were fuzzy (Figure 4). There is a very interesting possibility that ACM1 may affect ubiquitin-mediated chloroplast degradation because the vacuoles of the RNAi line are full of electron-dense material (Figure 4E) that well fits with the degradation hypothesis. Maybe similar to a proven example, the absence of the PPR protein GUN1 triggers the ubiquitination pathway and the cytosolic proteasome activity [43]. Of course, this point needed a series of experiments to verify in our research. To sum up, combined with the huge accumulation defect of chlorophyll and photosynthetic proteins in RNAi plants (Figure 5), we can conclude that ACM1 is indeed indispensable for early plastid development.
It is well known that the chloroplast developmental state is strongly linked with the expression level of plastid-encoded genes of photosynthesis [44]. In addition, functional chloroplast development needs the coordinated expression of genes encoded by both the nuclear and plastid genomes [45]. Therefore, we performed the quantitative RT-PCR and the results were very convincing. We found that PEP-dependent gene transcripts (Class I) were dramatically reduced, whereas those of NEP-dependent genes (Class III) were increased (Figure 7A), suggesting that ACM1, a nuclear gene, influences the regulation of plastid gene expression. This result indicates that it is likely that the early arrest of chloroplast development in the acm1 mutant and the ACM1-RNAi lines is due to the reduced PEP activity [46]. Similar results have been demonstrated in previous studies, like on other PPR proteins PDM2, PDM3, and SEL1 [31,36,46]; and the type protein MRL7 [47], which all lead to a loss in PEP activity. Interestingly, MRL7 might somehow affect the function of PPR proteins, and thereby, indirectly affect RNA metabolism, resulting in damage to PEP activity [47].
An intriguing feature of the PPR protein is that it is comprised of a relatively small range of protein architectures, yet exhibits diverse molecular functions [20]. Integrating previous study results of PPR proteins, we can summarize that P-type PPR proteins can stabilize specific RNAs and position processed RNA termini [48,49], activate and repress the translation of specific mRNAs [50,51], stimulate RNA cleavage in plant mitochondria [52] and promote the splicing of group II introns [31,32,53]. Analyses of ACM1 down-regulated lines showed the splicing efficiency of several transcripts’ introns was decreased (Figure 6), however, the splicing efficiency of rpl16 and petD was improved in this study (Figure 6). The possibility of this phenomenon may be due to the lack of barriers (ACM1) thus increasing the splicing efficiency. The above situation is similar to the function of another P-type PPR protein, BFA2 [30].
Proteins that are encoded by the plastid genome are synthesized by plastidic prokaryotic type 70S ribosome composed of 30S and 50S subunits [40,54]. The composition of plant ribosomes is based on the developmental stage and the type of tissue and is altered when stimulated by environmental stimuli [55,56]. Mutants of essential ribosome biogenesis factors (RBFs) mainly show defects in female and male gametophyte development or embryo lethality. Whereas mutants of nonessential RBFs show more specific phenotypes related to cotyledon, root, or leaf development [57], and damage to the 70S ribosome leads to abnormal chloroplast development. For example, in Arabidopsis thaliana, the mutant of RNA HELICASES 22 (RH22) showed an embryo lethal phenotype, while knock-down lines of RH22 accumulated precursors of 23S and 4.5S rRNA, and displayed a pale green phenotype. This study revealed that RH22 indirectly affected ribosome assembly due to its role in rRNA metabolism [58]. However, the situation was not quite the same in pdm4, the mutant was proved defective in chloroplast rRNA accumulation, but showed a seedling lethal phenotype rather than embryo lethal [32]. It is very interesting that maybe there are significant differences between individual ppr mutants. Based on a previous study, they found the defect in the accumulation of RPS14 protein in the ECD1-RNAi lines resulted in compromised ribosome accumulation, and the albino phenotype of cotyledons was presumably duo to deficiency in translation [18]. Similar to the ECD1, our result exhibits that the rRNAs sharply decrease in RNAi-1 plants (Figure 8A), and the content of RPS14, which is needed for the accumulation of ribosomal 30S subunits is almost undetectable (Figure 8B). In conclusion, we could logically infer that the accumulation of chloroplast ribosome was may be affected in the ACM1-RNAi line.
In the ACM1-RNAi line, chloroplast development is severely damaged and the transcription level of PEP-dependent genes was seriously affected. The ACM1 also affecting the splicing of multiple chloroplast group II introns and chloroplast rRNA accumulation. These processes influence and restrict each other, which may together lead to the loss of chloroplast development in cotyledon [32]. Based on our research, the key or the direct reason for this phenomenon is still not clear. So, further study of ACM1 function could facilitate the general understanding of the mechanism of chloroplast development.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The Arabidopsis thaliana plants used in this study were all Columbia-0 background. The mutant acm1 (SAIL_896_E10) was obtained from the ABRC. Seeds were sown on vermiculite or Murashige and Skoog (MS) medium containing 2% sucrose, 0.4% phytagel, and allowed to vernalize for 2 days at 4°C after sterilized by 75% (v/v) ethanol. Plants were then grown at 22 °C under 16-h light/8-h dark cycles. The T-DNA insertion was confirmed by PCR analysis and subsequent sequencing with the primers LB2 (5′-GCTTCCTATTATATCTTCCCAAATTACCAATACA-3′) and acm1-RP. The homozygous acm1 mutant line was verified by PCR using specific primers acm1-LP 5′-TCCACTGTTCGGTTTGC-3′ and acm1-RP 5′-TCATTGGCACTCTCCTA-3′.
4.2. Creation of RNAi Lines
For the RNAi lines, a fragment of ACM1 (379 bp) was subcloned into the cloning sites of pFGC5941 to create a binary vector. The recombinant plasmid was then mobilized into the Agrobacterium tumefaciens strain GV3101, as described previously [59]. In addition, the floral-dip method was used to produce transgenic plants according to Clough and Bent. (1998) [60]. The transgenic plants were selected on MS medium containing 20 mg/L BASTA. Transgenic kanamycin-resistant seedlings were sown as described before. Three transgenic lines, named RNAi 1, RNAi 7, and RNAi 9 are presented in our results.
4.3. Determination of Chlorophyll Content
For the measurement of the chlorophyll content, cotyledons from 7-day-old Arabidopsis seedlings and true leaves from 14-day-old seedlings were collected. The chlorophyll was extracted in 80% acetone and quantified on a UV2800 spectrophotometer (Unico, Dayton, NJ, USA). Methods are further described in a previous study [61]. Three biological replicates, each with three repeats, were analyzed for each sample.
4.4. Transmission Electron Microscope Analysis
To assess the changes in chloroplast structure, we conducted a TEM analysis. For this, 7-day-old cotyledons and 14-day-old true leaves of WT and RNAi plants were prepared for electron microscopic observation. The method was according to the following study with minor modifications [62]. Arabidopsis leaves were fixed by glutaraldehyde and osmium tetroxide, dehydrated in an ethanol series before being infiltrated with Spurr’s resin, and then stained with alkaline lead citrate and uranyl acetate. Finally, the samples were examined with a transmission electron microscope (JEM 1200EX, JEOL, Japan).
4.5. Subcellular Localization
To create a GFP subcellular localization vector, a fragment encoding the N-terminal 1–279 amino acids of ACM1 was amplified and subcloned into the pBSK-35s-EGFP vector to generate a fusion protein with green fluorescent protein as a reporter in the C terminus. The ACM1-GFP fusion construct plasmids were transferred into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method. Then, the fusion construct was transiently expressed in Nicotiana benthamiana leaf epidermal cells. Then, 48 h after injection, the green fluorescence signal was detected by using a confocal laser microscope (TCS-SP5; Leica), where red fluorescence represents the autofluorescence of chlorophyll.
4.6. RNA Isolation, RT-PCR and Quantitative RT-PCR Analysis
Total RNA was extracted from 7-day-old cotyledons tissue using the RNeasy Plant Mini Kit (Qiagen). After removing DNA (treated with DNase I, NEB, https://www.neb.com/ (accessed on 26 February 2021)) and conducting a quality test, total RNA was used to synthesize cDNA with the TranScript One-Step gDNA Removal and cDNA Synthesis Super Mix Kit (Transgen).
To examine gene expression, RT-PCR and quantitative real-time PCR (using the SYBR Premix ExTaq Kit, Takara) analyses were carried out according to Jiang et al. (2018) [18]. ACTIN 11 was selected as the internal control and each experiment had three biological repeats, each with three technical replicates. The primers used in quantitative RT-PCR are described in Chateigner-Boutin et al. (2008) [63].
4.7. Total Protein Extraction and Western Blot
Total protein was separated from 7-day-old cotyledons and 14-day-old true leaves of WT and RNAi plants with NB1 buffer (1 mM MgCl2, 5 mM DTT, 0.5 M sucrose, 50 mM Tris MES, 10 mM EDTA, and protease inhibitor cocktail, pH 8.0). Total protein samples were separated by 10% SDS-PAGE and then transferred onto PVDF membranes. The antibodies used in this study were according to Xiao et al. (2012) [64], and the signals were detected using enhanced chemiluminescence method according to Du et al (2017) [36].The signal intensity of the protein band was analyzed by “ImageJ” software. These experiments were repeated at least three times independently.
4.8. Data Analysis
Data were analyzed using Graphpad Prism (v8.0.2, CA, USA) and Microsoft Excel 2016. Values are expressed as mean ± SD. Two-tailed unpaired Student’s t-tests were used to evaluate differences between WT and mutant or RNAi line. A p-value of 0.05 was considered significant (* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001).
5. Conclusions
Functional chloroplast development, together with gene expression and regulation, are fundamental cellular activities that require coordination between the nucleus and the chloroplast. In this study, we identified a novel factor that affected the chloroplast development in cotyledons of Arabidopsis. ACM1 has 26 PPR motifs and belongs to the P subfamily, which is localized within the chloroplast. Down-regulation of ACM1 led to the abnormal chloroplast structure and reduction of chlorophyll. Compared with WT, the transcription levels of PEP-dependent genes were decreased in cotyledons of the ACM1-RNAi line, and splicing of several group II introns was disturbed. Furthermore, the accumulation of chloroplast ribosomes was probably defective in the ACM1-RNAi cotyledons.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the acm1 mutant line. And we thank the EditorBar (https://www.editorbar.com/ (accessed on 2 January 2021)) for its linguistic assistance during the preparation of this manuscript.
Abbreviations
| ABRC | Arabidopsis Biological Resource Center |
| CDS | Coding Sequence |
| Cyt b6f | Cytochrome b6f complex |
| GFP | Green fluorescent protein |
| MS | Murashige and Skoog culture medium |
| NEP | Nuclear-encoded RNA polymerases |
| PEP | Plastid-encoded RNA polymerase |
| PPR | Pentatricopeptide repeat |
| PS I | Photo System I |
| PS II | Photo System II |
| qRT-PCR | Quantitative real-time PCR |
| RBFs | Ribosome biogenesis factors |
| RNAi | RNA interference |
| RT-PCR | Reverse transcription PCR |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| TEM | Transmission electron microscope |
| WT | Wild type |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/22/5/2512/s1 online. Figure S1: Amino acid sequence alignment of ACM1. The amino acid sequence of ACM1 was aligned to homologous proteins from different species using ClustalW 2.0. Lines above the sequences show the predicted PPR motifs. Table S1: A list of the primers used in this study. File S2: The position of the primers used in splicing events.
Author Contributions
J.X. designed the research. X.W., and Y.A. performed research. J.X. and X.W. wrote the manuscript. J.X. and Y.L. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31970653).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sakamoto W., Miyagishima S.Y., Jarvis P. Chloroplast biogenesis: Control of plastid development, protein import, division and inheritance. Arab. Book. 2008;6:e0110. doi: 10.1199/tab.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters M.T., Langdale J.A. The making of a chloroplast. EMBO J. 2009;28:2861–2873. doi: 10.1038/emboj.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pogson B.J., Ganguly D., Albrecht-Borth V. Insights into chloroplast biogenesis and development. Biochim Biophys Acta. 2015;1847:1017–1024. doi: 10.1016/j.bbabio.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Jarvis P., López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 5.Liere K., Börner T. Cell and Molecular Biology of Plastids. Springer; Berlin/Heidelberg, Germany: 2007. Transcription and transcriptional regulation in plastids. [Google Scholar]
- 6.Börner T., Aleynikova A.Y., Zubo Y.O., Kusnetsov V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim Biophys Acta. 2015;1847:761–769. doi: 10.1016/j.bbabio.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Mansfield S.G., Briarty L.G. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int. J. Plant Sci. 1996;157:280–295. doi: 10.1086/297347. [DOI] [Google Scholar]
- 8.Shimada H., Mochizuki M., Ogura K., Froehlich J.E., Osteryoung K.W., Shirano Y., Shibata D., Masuda S., Mori K., Takamiya K. Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell. 2007;19:3157–3169. doi: 10.1105/tpc.107.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tominaga J., Mizutani H., Horikawa D., Nakahara Y., Takami T., Sakamoto W., Sakamoto A., Shimada H. Rice CYO1, an ortholog of Arabidopsis thaliana cotyledon chloroplast biogenesis factor AtCYO1, is expressed in leaves and involved in photosynthetic performance. J. Plant Physiol. 2016;207:78–83. doi: 10.1016/j.jplph.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Deng X.W., Gruissem W. Control of plastid gene expression during development: The limited role of transcriptional regulation. Cell. 1987;49:379–387. doi: 10.1016/0092-8674(87)90290-X. [DOI] [PubMed] [Google Scholar]
- 11.Albrecht V., Ingenfeld A., Apel K. Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 2006;60:507–518. doi: 10.1007/s11103-005-4921-0. [DOI] [PubMed] [Google Scholar]
- 12.Albrecht V., Ingenfeld A., Apel K. Snowy cotyledon 2: The identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol. Biol. 2008;66:599–608. doi: 10.1007/s11103-008-9291-y. [DOI] [PubMed] [Google Scholar]
- 13.Privat I., Hakimi M.A., Buhot L., Favory J.J., Mache-Lerbs S. Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol. Biol. 2003;51:385–399. doi: 10.1023/A:1022095017355. [DOI] [PubMed] [Google Scholar]
- 14.Ishizaki Y., Tsunoyama Y., Hatano K., Ando K., Kato K., Shinmyo A., Kobori M., Takeba G., Nakahira Y., Shiina T. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou W., Cheng Y., Yap A., Chateigner-Boutin A.L., Delannoy E., Hammani K., Small I., Huang J. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 2009;58:82–96. doi: 10.1111/j.1365-313X.2008.03766.x. [DOI] [PubMed] [Google Scholar]
- 16.Cao Z.L., Yu Q.B., Sun Y., Lu Y., Cui Y.L., Yang Z.N. A point mutation in the pentatricopeptide repeat motif of the AtECB2 protein causes delayed chloroplast development. J. Integr. Plant Biol. 2011;53:258–269. doi: 10.1111/j.1744-7909.2011.01030.x. [DOI] [PubMed] [Google Scholar]
- 17.Qiao J., Li J., Chu W., Luo M. PRDA1, a novel chloroplast nucleoid protein, is required for early chloroplast development and is involved in the regulation of plastid gene expression in Arabidopsis. Plant Cell Physiol. 2013;54:2071–2084. doi: 10.1093/pcp/pct148. [DOI] [PubMed] [Google Scholar]
- 18.Jiang T., Zhang J., Rong L., Feng Y., Wang Q., Song Q., Zhang L., Ouyang M. ECD1 functions as an RNA-editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J. Exp. Bot. 2018;69:3037–3051. doi: 10.1093/jxb/ery139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barkan A., Small I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 21.Stern D.B., Goldschmidt-Clermont M., Hanson M.R. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 22.Rovira A.G., Smith A.G. PPR proteins-orchestrators of organelle RNA metabolism. Physiol. Plant. 2019;166:451–459. doi: 10.1111/ppl.12950. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y., Huang J., Zhong S., Gu H., He S., Qu L.J. Novel DYW-type pentatricopeptide repeat (PPR) protein BLX controls mitochondrial RNA editing and splicing essential for early seed development of Arabidopsis. J. Genet. Genom. 2018;45:155–168. doi: 10.1016/j.jgg.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Yuan N., Wang J., Zhou Y., An D., Xiao Q., Wang W., Wu Y. EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes. Plant Sci. 2019;287:110203. doi: 10.1016/j.plantsci.2019.110203. [DOI] [PubMed] [Google Scholar]
- 25.Williams-Carrier R., Brewster C., Belcher S.E., Rojas M., Chotewutmontri P., Ljungdahl S., Barkan A. The Arabidopsis pentatricopeptide repeat protein LPE1 and its maize ortholog are required for translation of the chloroplast psbJ RNA. Plant J. 2019;99:56–66. doi: 10.1111/tpj.14308. [DOI] [PubMed] [Google Scholar]
- 26.Chen L., Huang L., Dai L., Gao Y., Zou W., Lu X., Wang C., Zhang G., Ren D., Hu J., et al. PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant Cell Physiol. 2019;60:587–598. doi: 10.1093/pcp/pcy229. [DOI] [PubMed] [Google Scholar]
- 27.Lv J., Shang L., Chen Y., Han Y., Yang X., Xie S., Bai W., Hu M., Wu H., Lei K., et al. OsSLC1 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling survival. Rice. 2020;13:25. doi: 10.1186/s12284-020-00385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M.F., Bai F., Castleden I., Song Y., Song B., Huang J., et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016;85:532–547. doi: 10.1111/tpj.13121. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Lu C. The enigmatic roles of PPR-SMR proteins in plants. Adv. Sci. 2019;6:1900361. doi: 10.1002/advs.201900361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., Zhou W., Che L., Rochaix J.D., Lu C., Li W., Peng L. PPR Protein BFA2 is essential for the accumulation of the atpH/F transcript in chloroplasts. Front. Plant Sci. 2019;10:446. doi: 10.3389/fpls.2019.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Xiao J., Li Y., Su B., Xu H., Shan X., Song C., Xie J., Li R. PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 2017;68:5615–5627. doi: 10.1093/jxb/erx360. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Zhao L., Man Y., Li X., Wang L., Xiao J. PDM4, a pentatricopeptide repeat protein, affects chloroplast gene expression and chloroplast development in Arabidopsis thaliana. Front. Plant Sci. 2020;11:1198. doi: 10.3389/fpls.2020.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shikanai T. RNA editing in plants: Machinery and flexibility of site recognition. Biochim Biophys Acta. 2015;1847:779–785. doi: 10.1016/j.bbabio.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Wagoner J.A., Sun T., Lin L., Hanson M.R. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 2015;290:2957–2968. doi: 10.1074/jbc.M114.622084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q., Xu Y., Huang J., Zhang K., Xiao H., Qin X., Zhu L., Zhu Y., Hu J. The rice pentatricopeptide repeat protein PPR756 is involved in pollen development by affecting multiple RNA editing in mitochondria. Front. Plant Sci. 2020;11:749. doi: 10.3389/fpls.2020.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du L., Zhang J., Qu S., Zhao Y., Su B., Lv X., Li R., Wan Y., Xiao J. The pentratricopeptide repeat protein Pigment-Defective Mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 2017;58:747–759. doi: 10.1093/pcp/pcx004. [DOI] [PubMed] [Google Scholar]
- 37.Liu X., Lan J., Huang Y., Cao P., Zhou C., Ren Y., He N., Liu S., Tian Y., Nguyen T., et al. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 2018;69:3949–3961. doi: 10.1093/jxb/ery214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z.W., Lv J., Xie S.Z., Zhang Y., Ren D.Y. OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul. 2018;84:249–260. doi: 10.1007/s10725-017-0336-6. [DOI] [Google Scholar]
- 39.Tan J., Tan Z., Wu F., Sheng P., Heng Y., Wang X., Ren Y., Wang J., Guo X., Zhang X., et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant. 2014;7:1329–1349. doi: 10.1093/mp/ssu054. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Wang C., Zheng M., Lyu J., Xu Y., Li X., Niu M., Long W., Wang D., Wang H., et al. WHITE PANICLE1, a val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol. 2016;170:2110–2123. doi: 10.1104/pp.15.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meinke D., Muralla R., Sweeney C., Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 2008;13:483–491. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Meinke D.W., Meinke L.K., Showalter T.C., Schissel A.M., Mueller L.A., Tzafrir I. A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol. 2003;131:409–418. doi: 10.1104/pp.014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tadini L., Peracchio C., Trotta A., Colombo M., Mancini I., Jeran N., Costa A., Faoro F., Marsoni M., Vannini C., et al. GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. 2020;101:1198–1220. doi: 10.1111/tpj.14585. [DOI] [PubMed] [Google Scholar]
- 44.Chi W., Mao J., Li Q., Ji D., Zou M., Lu C., Zhang L. Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 2010;64:14–25. doi: 10.1111/j.1365-313X.2010.04304.x. [DOI] [PubMed] [Google Scholar]
- 45.Ma F., Hu Y., Ju Y., Jiang Q., Cheng Z., Zhang Q., Sodmergen A novel tetratricopeptide repeat protein, WHITE TO GREEN1, is required for early chloroplast development and affects RNA editing in chloroplasts. J. Exp. Bot. 2017;68:5829–5843. doi: 10.1093/jxb/erx383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyo Y.J., Kwon K.C., Kim A., Cho M.H. Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 2013;163:1844–1858. doi: 10.1104/pp.113.227199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao J., Ma C., Wimmelbacher M., Börnke F., Luo M. Two novel proteins, MRL7 and its paralog MRL7-L, have essential but functionally distinct roles in chloroplast development and are involved in plastid gene expression regulation in Arabidopsis. Plant Cell Physiol. 2011;52:1017–1030. doi: 10.1093/pcp/pcr054. [DOI] [PubMed] [Google Scholar]
- 48.Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5’ and 3’ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prikryl J., Rojas M., Schuster G., Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoschke R., Watkins K.P., Barkan A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013;25:2265–2275. doi: 10.1105/tpc.113.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suárez M., Börner T., Barkan A. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012;40:3092–3105. doi: 10.1093/nar/gkr1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hölzle A., Jonietz C., Törjek O., Altmann T., Binder S., Forner J. A RESTORER OF FERTILITY-like PPR gene is required for 5’-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 2011;65:737–744. doi: 10.1111/j.1365-313X.2010.04460.x. [DOI] [PubMed] [Google Scholar]
- 53.Khrouchtchova A., Monde R.A., Barkan A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA. 2012;18:1197–1209. doi: 10.1261/rna.032623.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manuell A.L., Quispe J., Mayfield S.P. Structure of the chloroplast ribosome: Novel domains for translation regulation. PLoS Biol. 2007;5:e209. doi: 10.1371/journal.pbio.0050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koyama K., Wada A., Maki Y., Tanaka A. Changes in the protein composition of cytoplasmic ribsomes during the greening of etiolated barley leaves. Physiol. Plant. 1996;96:85–90. doi: 10.1111/j.1399-3054.1996.tb00187.x. [DOI] [Google Scholar]
- 56.Hummel M., Cordewener J.H., de Groot J.C., Smeekens S., America A.H., Hanson J. Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MSE proteomics. Proteomics. 2012;12:1024–1038. doi: 10.1002/pmic.201100413. [DOI] [PubMed] [Google Scholar]
- 57.Weis B.L., Kovacevic J., Missbach S., Schleiff E. Plant-specific features of ribosome biogenesis. Trends Plant Sci. 2015;20:729–740. doi: 10.1016/j.tplants.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Chi W., He B., Mao J., Li Q., Ma J., Ji D., Zou M., Zhang L. The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol. 2012;158:693–707. doi: 10.1104/pp.111.186775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J., Yang H., Lu Q., Wen X., Chen F., Peng L., Zhang L., Lu C. PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell. 2012;24:4992–5006. doi: 10.1105/tpc.112.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 61.Huang X., Zhang X., Yang S. A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res. 2009;19:1205–1216. doi: 10.1038/cr.2009.84. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y., Kollmann R. Vascular differentiation in the graft union of in-vitro grafts with different compatibility-structural and functional aspects. J. Plant Physiol. 1996;147:521–533. doi: 10.1016/S0176-1617(96)80041-1. [DOI] [Google Scholar]
- 63.Chateigner-Boutin A.L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L.F., Lurin C., Small I., et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008;56:590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- 64.Xiao J., Li J., Ouyang M., Yun T., He B., Ji D., Ma J., Chi W., Lu C., Zhang L. DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis. Plant Physiol. 2012;160:1911–1922. doi: 10.1104/pp.112.204891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.