Abstract
Transcription factors control, coordinate, and separate the functions of distinct network modules spatially and temporally. In this review, we focus on the transcription factor 21 (TCF21) network, a highly conserved basic-helix-loop-helix (bHLH) protein that functions to integrate signals and modulate gene expression. We summarize the molecular and biological properties of TCF21 control with an emphasis on molecular and functional TCF21 interactions. We suggest that these interactions serve to modulate the development of different organs at the transcriptional level to maintain growth homeostasis and to influence cell fate. Importantly, TCF21 expression is epigenetically inactivated in different types of human cancers. The epigenetic modification or activation and/or loss of TCF21 expression results in an imbalance in TCF21 signaling, which may lead to tumor initiation and, most likely, to progression and tumor metastasis. This review focuses on research on the roles of TCF21 in development and tumorigenesis systematically considering the physiological and pathological function of TCF21. In addition, we focus on the main molecular bases of its different roles whose importance should be clarified in future research. For this review, PubMed databases and keywords such as TCF21, POD-1, capsulin, tumors, carcinomas, tumorigenesis, development, and mechanism of action were utilized. Articles were selected within a historical context as were a number of citations from journals with relevant impact.
Keywords: TCF21, Cancer suppressor genes, Growth and development, Cancer, Tumorigenesis, Neoplasm metastases
Introduction
Definition and classification of basic helix-loop-helix (bHLH) proteins
The bHLH genes are conserved transcription factors that have been reported in eukaryotic organisms and unicellular organisms. When bHLH proteins are expressed in determined cell types, they induce a number of genes and specific phenotypes. A bHLH domain is composed of two regions, a basic region for binding to a target DNA sequence and a helix‐loop‐helix (HLH) motif, a structure comprising two α‐helices separated by a loop of variable length. bHLH motifs mediate dimerization, producing homo- or heterodimers (1,2). bHLHs have been categorized into different classes based on molecular phylogenetic relationships, expression patterns, dimerization selectivity, DNA-binding specificities, and protein structures (3,4). The classification that divides bHLH proteins into seven classes based on the presence of additional domains, the expression pattern, and the observed transcriptional function has been commonly employed (1,5). Class I HLH proteins are abundantly expressed and bind DNA as either homodimers or heterodimers. These HLH proteins were named E proteins (E12, E47, E2-2, HEB, TCF4), since they bind to Ephrussi-box (E-box) sequences (CANNTG) (6). Class II HLH proteins (MyoD, myogenin, NeuroD1-D2, MYF5-6) bind DNA as either homodimers or heterodimers with E proteins. In contrast to Class I HLH protein, which is expressed in many tissues, Class II protein expression is tissue-specific. Both Class I and Class II of the bHLH transcription factors (TFs) do not possess additional domains. Class III proteins (c-Myc) also contain a leucine zipper dimerization domain and function as either transcriptional activators or repressors, which regulate oncogenic transformation, apoptosis, and cellular differentiation. Class IV HLH proteins (Mad, Max, and Mxi) form heterodimers with c-Myc and regulate its activity but are also able to create homo- and heterodimers with each other. These proteins bind to CACGTG or CATGTTG E-box sites, but Mad/Max dimers are transcription regulators lacking a transactivation domain (TAD) that act on transcription otherwise. In Class V, HLH proteins (Id proteins) lack a basic region and antagonize the DNA activity of their bHLH partners. Class VI HLH proteins (Hes protein) present proline residues in their basic regions. Class VI HLH proteins recognize a unique sequence, CACGCG or CACGAG, and predominantly achieve transcriptional repression by binding to Groucho. Finally, the class VII HLH proteins (including members of circadian clock proteins and hypoxia) show PAS domains and bind predominantly to ACGTG or GCGTG sequences and function as transcriptional repressors (5,7).
TCF21 identification
In 1998, a novel member of the bHLH transcription factor family was identified as essential during murine embryonic development of mesodermal tissues. Firstly, it was known as epicardin (8), capsulin (9,10), or POD-1 (11), and currently as transcription factor 21 (TCF21). TCF21 is a bHLH class II transcription factor expressed in brachial muscle precursors and mesenchymal cells at sites of epithelial-mesenchymal interactions in the kidney, lung, intestine, pancreas, and spleen and in the developing respiratory, gastrointestinal, urogenital, and cardiovascular systems during embryogenic development. The expression levels of Tcf21 rapidly decrease in postnatal tissues, except in interstitial cells of kidney, lung, and heart. Knockout Pod1 mice die at birth due to lung, kidney, and cardiac defects and display gonadal, gastric, and splenic dysgenesis, suggesting a crucial role in embryogenesis (12–16). TCF21 is encoded by a gene located on human chromosome 6q23-q24 and can bind to the consensus E-box sequence to form a heterodimer with E12, also known as HEB or TCF12 (17,18). However, there are other putative interactants that have not been fully validated to date as shown in Table 1 (19–23).
Table 1. Possible interactions of transcription factor 21 (TCF21).
| Interactant | Full name | Experiment type | Experimental model | References |
|---|---|---|---|---|
| TCF12; *HEB; CRS3 | Transcription factor 12 | Affinity Capture Western; Yeast 2 Hybrid | HEK293 cells | 18 |
| APLP1 | Amyloid-like protein 1 | Yeast 2 Hybrid | COS-1 and HEK293 cells | 19 |
| LMO4 | LIM domain only 4 | Yeast 2 Hybrid | HepG2 cells | 20 |
| TCF3, E2A, E47, ITF1, VDIR, bHLHb21 | Transcription factor 3 | In vivo, In vitro, Yeast 2 Hybrid | Mouse, HEK293 cells | 18–21 |
| APEX1 | Apurinic/apyrimidinic endodeoxyribonuclease1 APEX nuclease | Yeast 2 Hybrid | CHO-K1 cells | 22 |
| GTF3C5 | General transcription factor IIIC subunit 5 | Yeast 2 Hybrid | CHO-K1 cells | 22 |
| HOXB6 | Homeobox B6 | Yeast 2 Hybrid | CHO-K1 cells | 22 |
| KLF15 | Kruppel-like factor 15p | Yeast 2 Hybrid | HEK293 cells | 23 |
| TCF4 | E2-2; transcription factor 4 | Affinity Capture Western; Yeast 2 Hybrid | HEK293 cells | 18 |
*HEB: transcription factor 12; CRS3: craniosynostosis-3; E2A/E47: immunoglobulin enhancer-binding factors E12/E47; ITF1: immunoglobulin transcription factor 1; VDIR: VDR interacting repressor; bHLHb21: class B basic helix-loop-helix protein 21.
TCF21 function
The functions and/or activities attributed to TCF21, inferred from sequence or structural similarity, sequence alignment, sequence model, direct assay, or physical interaction, are DNA-binding transcription activator, RNA polymerase II-specific, E-box binding, androgen receptor binding, bHLH transcription factor binding, histone deacetylase binding, and protein dimerization activity (24,25). Moreover, TCF21 is related to distinct and specific biological processes, such as development of the reproductive system (Sertoli cell differentiation and sex determination), urinary system (glomerulus development, metanephric mesenchymal cell differentiation, and glomerular capillary formation), respiratory system (lung morphogenesis and bronchiole and diaphragm development), spleen, adrenal gland, and digestive tract. In addition, TCF21 is involved in the negative and positive regulation of transcription by RNA polymerase and the negative regulation of the androgen receptor signaling pathway. In PubMed in early 2020, TCF21 was cited in 53 high-impact articles, and in 33 of them, a physiological or molecular function of TCF21 was cited.
Regulation of TCF21 expression
The expression of TCF21 seems to be regulated by epigenetic mechanisms, such as methylation, histone acetylation, and SUMOylation, as well as noncoding RNAs (ncRNAs), mostly described in tumor cells. The three exons encoding the human TCF21 gene are associated with methylation of CpG islands. There is also evidence that the long non-coding RNA (lncRNA) TCF21 antisense RNA-inducing demethylation, called TARID transcribed by exon 3, is associated with the CGI3 methylation island and is responsible for the demethylation of the CGI1 island in TCF21 and activates TCF21 expression by inducing promoter demethylation in melanomas (26) as described in Figure 1. TARID associates with the TCF21 promoter, forming an R-loop, a structure that acts as an anchoring support for adapter protein GADD45A, which recruits essential components of the base excision repair (BER) pathway, TDG and TET, responsible for promoting DNA demethylation by oxidation and replacement of 5-methylcytosine with 5-cytosine (27,28). TARID also recruits histone acetyltransferase enzymes that add three methyl groups to lysine 4 of the histone 3 protein (H3K4me3), enabling the remodeling and opening of chromatin to access transcription factors (29). MicroRNAs (miRNAs), such as miRNA-21, miRNA-224, miRNA-205, miRNA-30-3p, and miR-3648, are also related to TCF21 regulation (Figure 1) and are involved in the silencing of genes in human tumors (30–32). Inhibition of TCF21 expression was also associated with an increase of risk of coronary artery disease in a Chinese population (33). More recently, a study suggested that CXC chemokine ligand 12 (CXCL12) activates the GSK3β/β-catenin/TCF21 signaling pathway through CXCR4, a specific receptor for CXCL12, inhibiting cholesterol efflux from macrophages and promoting atherosclerosis (34).
Figure 1. Transcription factor 21 (TCF21) participates in development and tumorigenesis through distinct mechanisms. A, In development, TARID recruits components of the BER demethylation pathway and activates TCF21 expression by inducing promoter demethylation. Expression of TCF21 contributes to mesenchymal-epithelial transition (MET) and embryogenic development/differentiation of organs through different factors. B, In tumorigenesis, TCF21 is hypermethylated, and silencing by microRNAs (miRNAs) may contribute to epithelial-mesenchymal transition (EMT) and tumor progression. TARID: TCF21 antisense RNA inducing promoter demethylation; BER: TDG dependent base excision repair; GADD45A: growth arrest and DNA damage inducible alpha; TET: ten eleven-translocation family of dioxygenases; TDG: thymine DNA glycosylase and trimethylation of histone H3 at lysine 4 (H3k4me3).
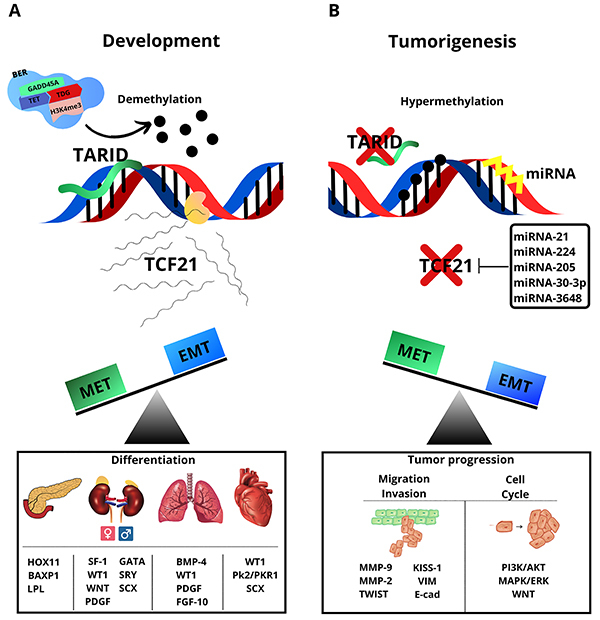
TCF21 in development
First detected in visceral glomerular epithelial cells (podocytes) in the mouse kidney and with its expression related to the onset of podocyte differentiation, the observed bHLH transcript factor was named POD-1 (11). In the meantime, a bHLH transcription factor was called capsulin after its expression had been identified specifically in mesodermic-derived cells that encapsulate the heart and tubular structures in the lungs, kidneys, and intestine during mouse embryogenesis (9,10). In murine heart development, a bHLH transcription factor was found to be expressed in the epicardium and endocardium and was given the name epicardin, since it is involved in cardiac organogenesis (8). This section will discuss the important findings about the TCF21 transcription factor in the development of different tissues and organs illustrated in Figure 1.
TCF21 in kidney development
TCF21 is persistently expressed from the early to the later stages of mouse kidney development in mesenchymal cells adjacent to the ureteric epithelium and in podocytes (11,35). The presence of TCF21 induces mesenchymal cells to condense followed by the mesenchymal-epithelial transition process. To determine the function of Tcf21 in vivo, a lacZ-expressing null Pod1 allele was generated. Null mutant mice are born but die with severely hypoplastic lungs and kidneys (12). The inhibition of Tcf21 expression resulted in decreased condensation of mesenchymal cells, which are necessary for the formation of nephron tubules. Additionally, TCF21 seems to be important for podocyte migration and differentiation around glomerular capillaries (36). During glomerular maturation, the Wilms tumor suppressor gene (WT1) is expressed in the metanephrogenic mesenchyme, which positively regulates Tcf21, inducing podocyte differentiation (36,37). TCF21 may regulate differentiation through the inhibition of cell cycle arrest-promoting factors in the HEK293 cell line, such as p21, which promotes the expansion of undifferentiated precursor cells and Gdnf-Ret-Wnt1 pathway activation required for mouse branching morphogenesis (25,38).
TCF21 in heart development
Capsulin/Tcf21 was identified in different cell populations during murine heart development. Initially detected in pericardial mesoderm at approximately embryonic day (E) E8.5 dpc (days post-coitum), the cells expressing capsulin migrate to the heart surface and originate the visceral pericardium, smooth muscle and endothelial cells of coronary vessels (9,10). During mouse epicardial development, WT1, TCF21, TBX18, SEMA3D, and SCX mediate differentiation in a group of pro-epicardium cells (39,40), which provide multipotent progenitor cells for cardiac lineages, including pericytes, fibroblasts, and coronary smooth muscle cells (41,42). TCF21 seems to inhibit prockineticin-2/PKR1 signaling (Figure 1), promoting the differentiation of cardiac progenitor cells (39). Depletion of Tcf21 results in disruption of pro-epicardial cell specification and in failure of mature epithelial epicardium formation during early stages of epicardial development in Xenopus (43).
TCF21 in lung development
Mice with the lacZ-expressing null Pod1 allele present severely hypoplastic lungs and die at birth (12). During pulmonary embryogenesis, Tcf21 is expressed in progenitor cells at the embryonic stage of E11.5 mesenchymal cells and regulates the expression of bone morphogenetic protein-4 (BMP-4), which is responsible for epithelial respiratory tract differentiation (12) (Figure 1). Tcf21 is expressed in a subpopulation of fibroblasts and lipofibroblasts, and is co-expressed with fibroblast growth factor 10 (FGF10), a fibroblast differentiation marker, and platelet-derived growth factor α receptor (PDGFs) (44,45). TCF21 regulates human lipofibroblast activation and fetal lung maturation (46). TCF21 overexpression in primary neonatal pulmonary fibroblasts increases intracellular lipids responsible for activating the synthesis of surfactants and phospholipids in adult rat alveolar type II cells in culture (47).
TCF21 in gonads and adrenal development
The murine adrenal and gonads have a common developmental origin, known as adrenogonadal primordium (AGP), detected at approximately E9.5 dpc (48). By E10.5 dpc, adrenal (AP) and gonadal primordium differentiate separately. During gonadal development, TCF21 is indispensable for ovarian and testicular differentiation and sexual differentiation. Tcf21 knockout (KO) mice show gonadal failures, vascular abnormalities, and sex indistinction during embryogenesis, while mutant Tcf21 mice show gonadal dysgenesis (15,16). During mouse male sex determination, the testis-determining factor SRY promotes Sertoli cell differentiation and consequent gonadal development. Tcf21 is a direct target of SRY (49), while the bHLH gene scleraxis (Scx) is a target of TCF21 (Figure 1). Therefore, the SRY-TCF21-SCX cascade participates in the initial molecular events of Sertoli cell differentiation and mouse testis development (50).
After differentiation of gonads, the expression of Tcf21 increases in mouse males after E11.5 dpc and in females after birth. TCF21 regulates the expansion of progenitor gonadal cells by inhibiting the steroidogenic factor Sf-1, which is involved in the sexual differentiation of the gonads and the regulation of steroid hormones (15,16).
During mouse adrenal gland development, an AGP cell group that expresses higher levels of SF-1 migrates to the dorsomedial region to form AP, which settles ventrolateral to the aorta (51). Approximately 48 dpc and E13 in mice, neural crest cells migrate from the neural tube and invade the AP during development, differentiating into the catecholamine-producing chromaffin cells of the adrenal medulla (52). At approximately 52 dpc in humans and E14.5 in mice, the adrenal cortex becomes encapsulated with a fibrous layer of cells (53). While SF-1 is continuously expressed during adult life, Sf-1 expression is driven by fetal adrenal-specific enhancer (FAdE) during AP development. FAdE becomes inactive when the adrenal gland differentiates, suggesting that distinct mechanisms are involved in SF-1 expression in mouse fetal and adult adrenal glands (54).
Different studies have shown the presence of adrenocortical stem and/or progenitor/proliferative cells located in the outermost layers of mouse adrenal cortex in the subcapsular region. However, cell lineage tracing studies have shown definitive evidence of a centripetal change in adrenocortical cells between zones (55). Cellular and molecular studies provided evidence that the specific paracrine/autocrine signaling pathways activated are involved in adrenocortical homeostasis. The Wnt and hedgehog (HH) pathways are well-studied and characterized pathways activated to play a role in mouse adrenocortical homeostasis. Pathway activity is regulated by cortical and stromal cell crosstalk that maintains organ homeostasis. In addition to Wnt and HH pathways, other mesenchymal-like cell lineages have also been identified in the capsule and the stroma. In the adrenal capsule, cells expressing the Wilms tumor protein homolog (WT1), a transcriptional regulator of adrenal and gonad organogenesis, with AGP features were observed (37). Ectopic expression of WT1 prevents the differentiation locking of adrenocortical steroidogenic cells in progenitor status. In addition, Tcf21 and Gli1 are direct targets of WT1. Moreover, cell lineage tracing analyses identified a long-living progenitor population in the adrenal cortex expressing WT1, GATA4, GLI1, and TCF21 that can generate steroidogenic cells in vivo (Figure 1). These cells may also have important roles in adrenal gland maintenance. Wood et al. (56) identified cells expressing TCF21 in the mouse adrenal cortex at E9.5 until E14.5 when TCF21 expression is restricted to the adrenal capsule. Lineage-tracing experiments showed that before encapsulation, TCF21-expressing cells give rise to both non-steroidogenic capsular cells and steroidogenic cortical cells. The TCF21-expressing capsular cells give rise to a population of stromal cells in adult mice.
TCF21 in the development of other tissues
Lu et al. (13 ) reported that mice homozygous for the Tcf21 null mutation fail to form a spleen. TCF21 seems to control morphogenetic expansion of splenic differentiation, and when it is absent, spleen precursor cells undergo apoptosis. Moreover, TCF21 controls an essential early step in spleen organogenesis among the Hoxl1 and Bapx1 genes, with both playing a critical role in murine spleen embryogenesis (57,58).
TCF21 may play an important role in regulating white adipose tissue development. Microarray analyses showed that Tcf21 is expressed in murine white pre-adipocytes, but it is not expressed in brown pre-adipocytes (59). Subsequent reports have shown that TCF21 mRNA decreases during the transition from white to brown human adipocytes (60). Moreover, TCF21 expression is significantly higher in the visceral adipose tissue of obese mice than of normal mice (61). Recently, a study using chicken lines showed a significant decrease in TCF21 levels in the abdominal fat of lean broilers compared with fat broilers. In vitro, Tcf21 increased pre-adipocyte differentiation, whereas Tcf21 knockdown and overexpression attenuated and promoted pre-adipocyte differentiation, respectively, which changed lipid droplet accumulation. Moreover, ChIP and luciferase analyses showed that TCF21 controls the transcription of lipoprotein lipase (LPL) by directly binding to the E-box motif in the Lpl promoter (62).
TCF21 in the tumorigenic process
This part of the review focuses on advances in the understanding of the role of TCF21 in various tumors. TCF21 is involved in tumor initiation, invasion, metastasis, and apoptosis with the molecular mechanisms largely undefined (Figure 1). The pathological function and molecular basis of TCF21 have been considered, and studies have shown distinct biological roles and some directions for research. In 2006, Smith et al. (63) described for the first time that the promoter region on human chromosome 6q23-q24 of TCF21 was hypermethylated in non-small-cell lung cancers (NSCLC) and head and neck squamous cell carcinomas. Since then, TCF21 has been studied in other types of human cancers and is considered a tumor suppressor gene in melanomas, renal cancer, adrenocortical carcinomas, colorectal tumors, gastric cancer, and hepatocellular carcinomas (18,). In general, the proposition is that TCF21 is downregulated primarily by DNA hypermethylation, rather than genetic mutations, in malignant tumors. DNA methylation can be reversed, and transcription can be restored, enabling TCF21 to be a therapeutic target. This section will discuss the important findings about the TCF21 transcription factor in tumorigenesis, what is unknown about its function, and what needs to be elucidated. These issues are illustrated in Figure 1.
TCF21 in lung cancer
The application of genomic scanning to obtain DNA methylation profiling in the region of recurrent loss of heterozygosity at human chromosome 6q23-q24 identified the TCF21 gene in lung cancer (63). In NSCLC, TCF21 is aberrantly methylated. The prevalence of TCF21 methylation in primary lung adenocarcinoma samples was 81%, higher than other methylated genes found in the 6q12-q27 locus (70). In addition, the ability of TCF21 to regulate mesenchymal-epithelial transition is lost in lung carcinomas. Moreover, exogenous expression of TCF21 in cells that have silenced endogenous TCF21 showed impaired tumorigenic properties in vitro and in vivo (63). In 2011, Richards et al. (71) concluded that TCF21 hypermethylation and impaired TCF21 expression are specific and frequent in NSCLC cells, even in early-stage disease, thereby making TCF21 a potential biomarker for early-stage NSCLC. Demethylation of the TCF21 promoter in H1299 cells (NSCLC cell line) by 5-aza-2'-deoxycytine (5-Aza) resulted in a higher level of TCF21 expression, leading to a loss of cell viability and invasion ability and an increase in cell apoptosis. In addition, inhibition of autophagy by 3-methyladenine (3-MA) increased TCF21 expression, suggesting that autophagy may also regulate TCF21 expression (72).
Analysis of LINC00163 levels revealed that a novel lncRNA was significantly downregulated in metastatic lung cancer tissues compared with non-metastatic tissues. Functional studies showed that LINC00163 induces TCF21 expression by recruiting ARID1A to the TCF21 promoter, suggesting a mechanism of lung cancer progression (73).
TCF21 in genitourinary tumors
After gene expression assessment from 12 human cancer cell lines and 318 clinical samples of three urological cancers, prostate, bladder, and kidney, two genes were identified as novel DNA methylation biomarkers, PCDH17 and TCF21 (74). PCDH17 and TCF21 promoter methylation levels provided a sensitivity rate of 96% for prostate cancer, 92% for bladder cancer, and 67% for renal cell tumors. However, in urine samples, the sensitivity was only 38% overall with absolute specificity.
For clear cell renal carcinoma (ccRCC), by far the most common histological subtype of renal cancer, TCF21 was an independent prognostic factor for poor survival in 186 samples of ccRCC patients, which were related to aberrant methylation of TCF21 (75). In a ccRCC cell line, Caki-2 cells, miR-21 is upregulated compared with its expression in normal renal cells. miR-21 upregulation by pre-miR-21 decreased TCF21 expression, whereas anti-miR-21 showed the opposite effects. Moreover, siRNA-TCF21 downregulates the expression of KISS1 and the invasion ability in Cak-1 cells, suggesting that aberrantly expressed miR-21 regulates the invasion pathway through TCF21-KISS1 association in renal cell carcinoma (30). In addition, TCF21 overexpression in ccRCC 786-O cells augmented E-cadherin expression (64).
In children, clear cell sarcoma of the kidney (CCSK) is a rare tumor with an unknown molecular pathogenesis. Analysis of 13 CCSKs showed no chromosomal copy number changes or somatic variants but identified promoter hypermethylation and low expression of TCF21 in 12 CCSKs. TARID, the lncRNA responsible for TCF21 demethylation, was undetectable in most CCSK samples, suggesting that TCF21 hypermethylation and reduced TARID expression are involved in the pathogenic pathway of CCSK (76).
Similar to miR-21 activity in ccRCC, in human invasive bladder cancer cells, UMUC3 and T24T cell lines, miR-3648 inhibits TCF21 protein expression by reducing its mRNA stability. Additionally, the increase in miR-3648 regulates the TCF21/KISS1 association, resulting in the promotion of invasion and metastasis of human bladder cancer (32). In human ovarian cancer, miR-205 is increased, resulting in inhibition of TCF21, which is a direct target of miR-205, and promotion of cell invasion of the ovarian cell lines OVCAR-5, OVCAR-8, and SKOV-3 (31). Li et al. (77) described that TCF21 can be a transcriptional target of p53 in response to hypoxia in uterine corpus endometrial carcinoma (UCEC) carrying wild type p53. Moreover, TCF21 interferes with the MAPK pathway by reducing ERK levels in UCEC cells expressing higher levels of TCF21.
TCF21 in breast cancer
Resembling the tumors described to date, TCF21 is less expressed in human breast cancer cell lines and tissues and is associated with larger tumor mass size and lymphoid metastases (78). Overexpression of TCF21 in MDA-MB-231 breast cancer cells decreased cell proliferation, invasion, and migration capacity and inhibited angiogenesis, epithelial-mesenchymal transition, and apoptosis (79). The TCF21 mechanism of action in human breast tumor cell lines showed that TCF21 is able to regulate aberrant estrogen receptor-α signaling (Erα), an important factor for the progression of breast cancer, thereby reducing its functionality. The SUMOylation of TCF21 at lysine residue 24 (K24) by the small ubiquitin-like modifier SUMO1 stabilizes the TCF21 protein and enhances its interaction with histone deacetylases (HDAC1/2). The SUMOylation of TCF21 represses the activity of Erα and decreases the percentage of S-phase cells (80).
Analysis of the genetic polymorphism of TCF21 and the risk of breast cancer in Chinese women suggests that the TCF21 rs12190287 polymorphism can regulate TCF21 expression and may serve as a potential marker for genetic susceptibility to breast cancer (81). Interestingly, the same polymorphism is considered a prognostic factor in osteosarcoma (82).
TCF21 in adrenocortical tumors
Adrenocortical carcinoma (ACC) is a rare disease with an incidence of 1 to 2 cases per million (83). Despite few defined markers for adrenocortical tumors (ACTs), TCF21 is less expressed in ACC than in adenomas (ACA) and normal tissue (65,66). TCF21 binds directly in the E-box promoter sequence of steroidogenic factor 1 (SF1/NR5A1), decreasing SF-1 transcription in both the human ACC cell line (H295R) and in ACC cell culture obtained from patient tumor fragment (ACC-T36). Increased expression of TCF21 in H295R and ACC-T36 cells and in primary culture of rat ACC decreased SF-1/NR5A1 and steroidogenic enzyme StAR expression (65,84). In cultures of ACA cells obtained from pediatric tumor fragments, ACA-T7 cells, TCF21 silencing by small interfering RNA led to an increase in SF1 mRNA expression (85). Although SF-1 regulates steroid production, it is also related to adrenocortical cell proliferation (86,87). In fact, an increase in SF-1 dosage activates adrenocortical cell proliferation and induces adrenocortical neoplasia in mice (88). However, TCF21 overexpression showed no difference in proliferative capacity in ACC cells (65). KEGG pathway analysis has shown that BUB1B, among other genes, is negatively correlated with TCF21 expression in ACC (66). More recently, analysis of the combined expression of TCF21 and BUB1B in adult and pediatric adrenocortical tumors showed a negative correlation between the expression levels of TCF21 and BUB1B in adult ACCs. Additionally, the combined expression of TCF21 and BUB1B was a predictor of overall survival and was able to separate ACCs into two subgroups: one with poor prognosis and the other with good prognosis. In agreement with the observation that pediatric ACT appear to have a biphasic age distribution with a poor clinical outcome in the group aged >5 years, it was found that the combination of TCF21 and SF1 was a good predictor of malignancy for children less than 5 years old (65).
TCF21 in other tumors
By using quantitative DNA methylation analysis in melanoma biopsies of patients and in their derived cell lines, it was demonstrated that TCF21 expression is downregulated in metastatic melanoma by promoter hypermethylation. Moreover, TCF21 promoter DNA methylation is correlated with decreased survival in metastatic melanoma. Functionally, TCF21 binds to the KISS1 promoter, a melanoma metastasis-suppressor gene, increasing its expression. Finally, overexpression of TCF21 inhibits the motility of the C8161 human melanoma cell line (18).
In colorectal and gastric cancer cells, downregulation of TCF21 by hypermethylation induces cell proliferation, migration, and invasion, probably through inactivation of PI3K/AKT signaling (Figure 1). The overexpression of TCF21 in two human colorectal tumor cells, HCT116 and HT29, increased KISS1 protein expression and reduced the expression of both metalloproteinases MMP2 and MMP9 (Figure 1), which are involved in cell invasion (89). As in the vast majority of tumors analyzed, TCF21 is downregulated in esophageal squamous carcinoma (ESCC) and correlated with poor prognosis. Enhancement of TCF21 expression levels in ESCC cells inhibits migration, invasion, and proliferation, as well as epithelial-mesenchymal transition, which may be partly through the increase in Kiss-1 and downregulation of TWIST expression (90). As the epigenetic regulation of TCF21 through different levels of methylation is a common observation in different malignancies, these data are summarized in Table 2.
Table 2. Transcription factor 21 (TCF21) in tumorigenesis.
| TCF21 status/function | Tumor | References |
|---|---|---|
| Epigenetic modification | ||
| Promoter hypermethylation | lung, head and neck squamous cell carcinomas, renal cell, bladder, prostate, melanomas, colorectal, clear cell sarcoma of the kidney | 26,63,68 ,70,74,76 |
| rs12190287 polymorphism | breast, osteosarcoma | 81,82 |
| Clinical biomarker | ||
| Prognostic factor | lung adenocarcinoma, clear cell renal cell carcinoma, adult ACTs, metastatic melanoma, colorectal, esophageal squamous cell carcinoma | 18,66,70,75,89,90 |
| Diagnostic factor | bladder, prostate, renal, pediatric ACTs | 66,74 |
| Tumor mass size | breast | 78 |
| Lymphoid metastases | breast, colorectal | 78 |
| Biological effect | ||
| Reduction of tumor growth | gastric | 67 |
| Cell death by apoptosis | NSCLC | 72 |
| Decreased cell viability | NSCLC | 72 |
| Apoptosis reduction | breast | 79 |
| Inhibition of cell invasion | NSCLC, breast, colorectal, gastric, esophageal squamous cell carcinoma | 67,68,72,79,89,90 |
| Inhibition of cell migration | renal, breast, melanomas, colorectal, gastric, esophageal squamous cell carcinoma | 18,64,67,68,79,89,90 |
| Inhibition of cell proliferation | breast, colorectal, gastric, esophageal squamous cell carcinoma | 67,68,79,89,90 |
| Angiogenesis inhibition | breast | 79 |
TCF21: transcription factor 21; ACTs: adrenocortical tumors; NSCLC: non-small-cell lung carcinoma cells.
Conclusion
Taken together, these data demonstrate that TCF21 is a tumor suppressor gene epigenetically regulated by promoter methylation. TCF21 is capable of substantially interfering with important biological processes in migration, invasion, and metastasis. However, functional studies on the main pathways of TCF21 action and their partner molecules in development and tumorigenic processes are lacking. Further studies are warranted to assign to the transcription factor TCF21 the condition of tumor biomarkers and therapeutic targets.
Acknowledgments
B.S. Passaia and J.L. Kremer are recipients of scholarships from Fundação de Amparo è Pesquisa do Estado de São Paulo (FAPESP, 2016/12381-7 and 2016/17285-6). C.F.P. Lotfi received funding from FAPESP (2018/19035-9) and from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
References
- 1.Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, et al. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 2.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murre C. Helix-loop-helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev. 2019;33:6–25. doi: 10.1101/gad.320663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 7.Carroll PA, Freie BW, Mathsyaraja H, Eisenman RN. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front Med. 2018;12:412–425. doi: 10.1007/s11684-018-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robb L, Mifsud L, Hartley L, Biben C, Copeland NG, Gilbert DJ, et al. Epicardin - a novel basic helix-loop-helix transcription factor gene expressed in epicardium, branchial arch myoblasts, and mesenchyme of developing lung, gut, kidney, and gonads. Dev Dyn. 1998;213:105–113. doi: 10.1002/(SICI)1097-0177(199809)213:1<105::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Richardson JA, Olson EN. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Dev. 1998;73:23–32. doi: 10.1016/S0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 10.Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous EE. Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech Dev. 1998;73:33–43. doi: 10.1016/S0925-4773(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 11.Quaggin SE, Vanden Heuvel GB, Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech Dev. 1998;71:37–48. doi: 10.1016/S0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- 12.Quaggin SE, Schwartz L, Cui S, Igarashi P, Deimling J, Post M, et al. The basic-helix-loop-helix protein Pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–5783. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, Chang P, Richardson JA, Gan L, Weiler H, Olson EN. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc Natl Acad Sci USA. 2000;97:9525–9530. doi: 10.1073/pnas.97.17.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson M, Giraud AS, Familari M. The role of capsulin in the morphogenesis and differentiation of fetal rat gastric mucosa. Int J Dev Biol. 2001;45:887–893. doi: 10.1387/ijdb.11804032. [DOI] [PubMed] [Google Scholar]
- 15.Tamura M, KannoY, Chuma S, Saito T, Nakatsuji N. Pod-1/Capsulin shows a sex-and stage-dependent expression pattern in the mouse gonad development and represses expression of Ad4BP/SF-1. Mech Dev. 2001;102:135–144. doi: 10.1016/S0925-4773(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 16.Cui S, Ross A, Stallings N, Parker KL, Capel B, Quaggin SE. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development. 2004;131:4095–4105. doi: 10.1242/dev.01266. [DOI] [PubMed] [Google Scholar]
- 17.Miyagishi M, Hatta M, OhshimaT, Ishida J, Fujii R, Nakajima T, et al. Cell type-dependent transactivation or repression of mesoderm-restricted basic helix-loop-helix protein, POD-1/Capsulin. Mol Cell Biochem. 2000;5:141–147. doi: 10.1023/A:1007057611868. [DOI] [PubMed] [Google Scholar]
- 18.Arab K, Smith LT, Gast A, Weichenhan D, Huang JPH, Claus R, et al. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32:1467–1473. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, et al. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki H, Fukunishi Y, Kagawa I, Saito R, Oda H, Endo T, et al. Protein-protein interaction panel using mouse full-length cDNAs. Genome Res. 2001;11:1758–1765. doi: 10.1101/gr.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Huo K, Ma L, Tang L, Li D, Huang X, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2017;13:965. doi: 10.15252/msb.20178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 25.Funato N, Ohyama K, Kuroda T, Nakamura M. Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription. J Biol Chem. 2003;278:7486–7493. doi: 10.1074/jbc.M212248200. [DOI] [PubMed] [Google Scholar]
- 26.Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 28.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine. Mol Cell Biol. 2012;32:2323–2334. doi: 10.1128/MCB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Guo Y, Shang C, Song Y, Wu B. MiR-21 downregulated TCF21 to inhibit KISS1 in renal cancer. Urology. 2012;80:1298–1302.e1. doi: 10.1016/j.urology.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Wei J, Zhang L, Li J, Zhu S, Tai M, Mason CW, et al. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J Ovarian Res. 2017;10:33. doi: 10.1186/s13048-017-0328-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun W, Li S, Yu Y, Jin H, Xie Q, Hua X, et al. 2019. MicroRNA-3648 is upregulated to suppress TCF21, resulting in promotion of invasion and metastasis of human bladder cancer. Mol Ther Nucleic Acids. 2019;16:519–530. doi: 10.1016/j.omtn.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao JH, He LH, Yu XH, Zhao ZW, Wang G, Zou J, et al. CXCL12 promotes atherosclerosis by downregulating ABCA1 expression via the CXCR4/GSK3β/β-catenin/TCF21 pathway. J Lipid Res. 2019;60:2020–2033. doi: 10.1194/jlr.RA119000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui S, Schwartz L, Quaggin SE. Pod1 is required in stromal cells for glomerulogenesis. Dev Dyn. 2003;226:512–522. doi: 10.1002/dvdy.10244. [DOI] [PubMed] [Google Scholar]
- 36.Quaggin SE. Transcriptional regulation of podocyte specification and differentiation. Microsc Res Tech. 2002;57:208–211. doi: 10.1002/jemt.10076. [DOI] [PubMed] [Google Scholar]
- 37.Bandiera R, Vidal VPI, Motamedi FJ, Clarkson M, Sahut-Barnola I, vonGise A, et al. WT1 maintains adrenal-gonadal primordium identity and marks a population of AGP-like progenitors within the adrenal gland. Dev Cell. 2013;27:5–18. doi: 10.1016/j.devcel.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ide S, Finer G, Maezawa Y, Onay T, Souma T, Scott R, et al. Transcription factor 21 is required for branching morphogenesis and regulates the Gdnf-Axis in kidney development. J Am Soc Nephrol. 2018;29:2795–2808. doi: 10.1681/ASN.2017121278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qureshi R, Kindo M, Arora H, Boulberdaa M, Steenman M, Nebigil CG. Prokineticin receptor-1-dependent paracrine and autocrine pathways control cardiac TCF21+ fibroblast progenitor cell transformation into adipocytes and vascular cells. Sci Rep. 2017;7:12804. doi: 10.1038/s41598-017-13198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupu IE, Redpath AN, Smart N. Spatiotemporal analysis reveals significant overlap of key proepicardial markers in the developing murine heart. bioRxiv. 2019 doi: 10.1101/666610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, et al. The bHLH transcription factor Tcf21 is required for lineage specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, Conlon FL. Tcf21 regulates the specification and maturation of proepicardial cells. Development. 2012;140:2409–2421. doi: 10.1242/dev.093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Agha E, Herold S, Alam D Al, Quantius J, MacKenzie BA, Carraro G, et al. Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development. 2014;141:296–306. doi: 10.1242/dev.099747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast Growth Factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 46.McGowan SE, Torday JS. The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development. Annu Rev Physiol. 1997;59:43–62. doi: 10.1146/annurev.physiol.59.1.43. [DOI] [PubMed] [Google Scholar]
- 47.Sugahara K, Rubin JS, Mason RJ, Aronsen EL, Shannon M. Keratinocyte growth factor increases mRNAs for SP-A and SP-B in adult rat alveolar type II cells in culture. Am J Physiol. 1995;269:L344–L350.. doi: 10.1152/ajplung.1995.269.3.l344. [DOI] [PubMed] [Google Scholar]
- 48.Hatano O, Takakusu A, Nomura M, Morohashi K. Identical origin of adrenal cortex and gonad revealed by expression profiles of Ad4BP/SF‐1. Genes Cells. 1996;1:663–671. doi: 10.1046/j.1365-2443.1996.00254.x. [DOI] [PubMed] [Google Scholar]
- 49.Bhandari RK, Sadler-Riggleman I, Clement TM, Skinner MK. Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLoS One. 2011;6:e19935. doi: 10.1371/journal.pone.0019935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhandari RK, Haque MM, Skinner MK. Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS One. 2012;7:e43380. doi: 10.1371/journal.pone.0043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 52.Doupe AJ, Patterson PH, Landis SC. Small intensely fluorescent cells in culture: Role of glucocorticoids and growth factors in their development and interconversions with other neural crest derivatives. J Neurosci. 1985;5:2143–2160. doi: 10.1523/JNEUROSCI.05-08-02143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keegan CE, Hammer GD. Recent insights into organogenesis of the adrenal cortex. Trends Endocrinol Metab. 2002;13:200–208. doi: 10.1016/S1043-2760(02)00602-1. [DOI] [PubMed] [Google Scholar]
- 54.Zubair M, Parker KL, Morohashi K. Developmental links between the fetal and adult zones of the adrenal cortex revealed by lineage tracing. Mol Cell Biol. 2008;28:7030–7040. doi: 10.1128/MCB.00900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.King P, Paul A, Laufer E. Shh signaling regulates adrenocortical development and identifies progenitors of steroidogenic lineages. Proc Natl Acad Sci USA. 2009;106:21185–21190. doi: 10.1073/pnas.0909471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood MA, Acharya A, Finco I, Swonger JM, Elston MJ, Tallquist MD, Hammer GD. Fetal adrenal capsular cells serve as progenitor cells for steroidogenic and stromal adrenocortical cell lineages in M. musculus. Development. 2013;140:4522–4532. doi: 10.1242/dev.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- 58.Lettice LA, Purdie LA, Carlson GJ, Kilanowski F, Dorin J, Hill RE. The mouse bagpipe gene controls development of axial skeleton, skull, and spleen. Proc Natl Acad Sci USA. 1999;96:9695–9700. doi: 10.1073/pnas.96.17.9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci USA. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guennoun A, Kazantzis M, Thomas R, Wabitsch M, Tews D, Seetharama Sastry K, et al. Comprehensive molecular characterization of human adipocytes reveals a transient brown phenotype. J Transl Med. 2015;13:135. doi: 10.1186/s12967-015-0480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stadion M, Schwerbel K, Graja A, Baumeier C, Rödiger M, Jonas W, et al. Increased Ifi202b/IFI16 expression stimulates adipogenesis in mice and humans. Diabetologia. 2018;61:1167–1179. doi: 10.1007/s00125-018-4571-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, Cheng B, Liu C, Du Z, Zhang H, Wang N, et al. A novel regulator of preadipocyte differentiation, transcription factor TCF21, functions partially through promoting LPL expression. Front Physiol. 2019;10:458. doi: 10.3389/fphys.2019.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith LT, Lin M, Brena RM, Lang JC, Schuller DE, Otterson GA, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci USA. 2006;103:982–987. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gooskens SL, Klasson TD, Gremmels H, Logister I, Pieters R, Perlman EJ, et al. TCF21 hypermethylation regulates renal tumor cell clonogenic proliferation and migration. Mol Oncol. 2018;12:166–179. doi: 10.1002/1878-0261.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.França MM, Ferraz-de-Souza B, Santos MG, Lerario AM, Fragoso MCBV, Latronico AC, et al. POD-1 binding to the E-box sequence inhibits SF-1 and StAR expression in human adrenocortical tumor cells. Mol Cell Endocrinol. 2013;371:140–147. doi: 10.1016/j.mce.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Passaia BS, Dias MH, Kremer JL, Antonini SRR, de Almeida MQ, Fragoso MCBV, et al. TCF21/POD-1, a transcriptional regulator of SF-1/NR5A1, as a potential prognosis marker in adult and pediatric adrenocortical tumors. Front Endocrinol (Lausanne) 2018;9:38. doi: 10.3389/fendo.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, Li DM, Xie Q, Dai DQ. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol. 2014;141:211–220. doi: 10.1007/s00432-014-1809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai Y, Duan H, Duan C, Zhou R, He Y, Tu Q, et al. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration and invasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469:430–436. doi: 10.1016/j.bbrc.2015.09.109. [DOI] [PubMed] [Google Scholar]
- 69.Lu W, Yang C, Du P, Zhang JL, Zhang JC. Expression tendency and prognostic value of TCF21 in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol. 2019;47:1466–1470. doi: 10.1080/21691401.2019.1601102. [DOI] [PubMed] [Google Scholar]
- 70.Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 71.Richards KL, Zhang B, Sun M, Dong W, Churchill J, Bachinski LL, et al. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer. 2011;117:606–617. doi: 10.1002/cncr.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen B, Zeng C, Ye Y, Wu D, Mu Z, Liu J, et al. Promoter methylation of TCF21 may repress autophagy in the progression of lung cancer. J Cell Commu Signal. 2018;12:423–432. doi: 10.1007/s12079-017-0418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo X, Wei Y, Wang Z, Liu W, Yang Y, Yu X, et al. LncRNA LINC00163 upregulation suppresses lung cancer development though transcriptionally increasing TCF21 expression. Am J Cancer Res. 2018;8:2494–2506. [PMC free article] [PubMed] [Google Scholar]
- 74.Costa VL, Henrique R, Danielsen SA, Eknaes M, Patrício P, Morais A, et al. TCF21 and PCDH17 methylation: an innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6:1120–1130. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]
- 75.Ye YW, Jiang ZM, Li WH, Li ZS, Han YH, Sun L, et al. Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma. 2012;59:599–605. doi: 10.4149/neo_2012_076. [DOI] [PubMed] [Google Scholar]
- 76.Gooskens SL, Gadd S, Auvil JMG, Gerhard DS, Khan J, Patidar R, et al. TCF21 hypermethylation in genetically quiescent clear cell sarcoma of the kidney. Oncotarget. 2015;6:15828–15841. doi: 10.18632/oncotarget.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Q, Lei Y, Du W. A novel target of p53, TCF21, can respond to hypoxia by mapk pathway inactivation in uterine corpus endometrial carcinoma. DNA Cell Biol. 2018;37:473–480. doi: 10.1089/dna.2017.4062. [DOI] [PubMed] [Google Scholar]
- 78.Wang J, Gao X, Wang M, Zhang J. Clinicopathological significance and biological role of TCF21 mRNA in breast cancer. Tumor Biol. 2015;36:8679–8683. doi: 10.1007/s13277-015-3476-1. [DOI] [PubMed] [Google Scholar]
- 79.Li PW, Wang HY, Xu LL. Effect of TCF21 on proliferation, migration and vascular production of breast cancer cells [in Chinese] Zhonghua Bing Li Xue Za Zhi. 2019;48:307–311. doi: 10.3760/cma.j.issn.0529-5807.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 80.Ao X, Li S, Xu Z, Yang Y, Chen M, Jiang X, et al. Sumoylation of TCF21 downregulates the transcriptional activity of estrogen receptor-alpha. Oncotarget. 2016;7:26220–26234. doi: 10.18632/oncotarget.8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao X, Yang J, Wang M, Zhang J. TCF21 genetic polymorphisms and breast cancer risk in Chinese women. Oncotarget. 2016;7:55757–55764. doi: 10.18632/oncotarget.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang Z, Zhang W, Chen Z, Shao J, Chen L, Wang Z. Transcription factor 21 (TCF21) rs12190287 polymorphism is associated with osteosarcoma risk and outcomes in East Chinese population. Med Sci Monit. 2017;23:3185–3191. doi: 10.12659/MSM.905595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Libé R. Adrenocortical carcinoma (ACC): diagnosis, prognosis, and treatment. Front Cell Dev Biol. 2015;3:45. doi: 10.3389/fcell.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.França MM, Abreu NP, Vrechi TA, Lotfi CF. POD-1/Tcf21 overexpression reduces endogenous SF-1 and StAR expression in rat adrenal cells. Braz J Med Biol Res. 2015;48:1087–1094. doi: 10.1590/1414-431x20154748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.França MM, Lerario AM, Fragoso MCBV, Lotfi CFP. New evidences on the regulation of SF-1 expression by POD1/TCF21 in adrenocortical tumor cells. Clinics. 2017;72:391–394. doi: 10.6061/clinics/2017(06)10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morohashi KI, Zanger UM, Honda SI, Hara M, Waterman MR, Omura T. Activation of CYP11A and CYP11B gene promoters by the steroidogenic cell-specific transcription factor, Ad4BP. Mol Endocrinol. 1993;7:1196–1204. doi: 10.1210/mend.7.9.8247022. [DOI] [PubMed] [Google Scholar]
- 87.Lalli E, Doghman M, Latre de Late P, El Wakil A, Mus-Veteau I. Beyond steroidogenesis: novel target genes for SF-1 discovered by genomics. Mol Cell Endocrinol. 2013;371:154–159. doi: 10.1016/j.mce.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 88.Doghman M, Karpova T, Rodrigues GA, Arhatte M, De Moura J, Cavalli LR, et al. Increased steroidogenic factor-1 dosage triggers adrenocortical cell proliferation and cancer. Mol Endocrinol. 2007;21:2968–2987. doi: 10.1210/me.2007-0120. [DOI] [PubMed] [Google Scholar]
- 89.Dai Y, Duan H, Duan C, Zhu H, Zhou R, Pei H, et al. TCF21 functions as a tumor suppressor in colorectal cancer through inactivation of PI3K/AKT signaling. Onco Targets Ther. 2017;10:1603–1611. doi: 10.2147/OTT.S118151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Y, Zhang C, Chen J, Zhang B, Zhang H, Yang X, et al. Expression of transcription factor 21 (TCF21) and upregulation its level inhibits invasion and metastasis in esophageal squamous cell carcinoma. Med Sci Monit. 2018; 24:4128–4136. doi: 10.12659/MSM.909138. [DOI] [PMC free article] [PubMed] [Google Scholar]


