Abstract
Background
High-mobility group box 1 (HMGB1) is a multifunctional redox-sensitive protein involved in various intracellular (eg, chromatin remodeling, transcription, autophagy) and extracellular (inflammation, autoimmunity) processes. Regarding its role in cancer development/progression, paradoxical results exist in the literature and it is still unclear whether HMGB1 mainly acts as an oncogene or a tumor suppressor.
Methods
HMGB1 expression was first assessed in tissue specimens (n=359) of invasive breast, lung and cervical cancer and the two distinct staining patterns detected (nuclear vs cytoplasmic) were correlated to the secretion profile of malignant cells, patient outcomes and the presence of infiltrating immune cells within tumor microenvironment. Using several orthotopic, syngeneic mouse models of basal-like breast (4T1, 67NR and EpRas) or non-small cell lung (TC-1) cancer, the efficacy of several HMGB1 inhibitors alone and in combination with immune checkpoint blockade antibodies (anti-PD-1/PD-L1) was then investigated. Isolated from retrieved tumors, 14 immune cell (sub)populations as well as the activation status of antigen-presenting cells were extensively analyzed in each condition. Finally, the redox state of HMGB1 in tumor-extruded fluids and the influence of different forms (oxidized, reduced or disulfide) on both dendritic cell (DC) and plasmacytoid DC (pDC) activation were determined.
Results
Associated with an unfavorable prognosis in human patients, we clearly demonstrated that targeting extracellular HMGB1 elicits a profound remodeling of tumor immune microenvironment for efficient cancer therapy. Indeed, without affecting the global number of (CD45+) immune cells, drastic reductions of monocytic/granulocytic myeloid-derived suppressor cells (MDSC) and regulatory T lymphocytes, a higher M1/M2 ratio of macrophages as well as an increased activation of both DC and pDC were continually observed following HMGB1 inhibition. Moreover, blocking HMGB1 improved the efficacy of anti-PD-1 cancer monoimmunotherapy. We also reported that a significant fraction of HMGB1 encountered within cancer microenvironment (interstitial fluids) is oxidized and, in opposite to its reduced isoform, oxidized HMGB1 acts as a tolerogenic signal in a receptor for advanced glycation endproducts-dependent manner.
Conclusion
Collectively, we present evidence that extracellular HMGB1 blockade may complement first-generation cancer immunotherapies by remobilizing antitumor immune response.
Keywords: breast neoplasms, translational medical research, tumor microenvironment
Introduction
High-mobility group box 1 (HMGB1) was originally identified in the early 70s as an ubiquitous and abundant non-histone chromatin-binding protein.1 Highly conserved (>95% identity) among all mammals, nuclear HMGB1 is involved in replication, gene expression, DNA repair and contributes actively to genome stability. Thirty years later, compelling data revealed the implication of HMGB1 in the pathogenesis of a variety of non-infectious, inflammatory/autoimmune disorders (eg, sepsis, rheumatoid arthritis, systemic lupus erythematosus, myositis),2–5 supporting its additional role in immune responses. Actively secreted under inflammatory/stress conditions or passively released from any type of injured cells, HMGB1 triggers and amplifies these diverse inflammation-associated diseases through both stimulating proinflammatory cytokine synthesis (eg, tumor necrosis factor-α (TNFα), interleukin (IL)-1β, IL-6) and inducing immature immune cell activation and chemotaxis.6–8 In addition to operate as an ‘alarmin’ via the receptor for advanced glycation endproducts (RAGE) and/or Toll-like receptor (TLR) 2, 4 and 9 signaling(s), extracellular HMGB1 may also regulate vascular growth and axonal sprouting,9 10 extending its activity well beyond its pro-inflammatory function. More recently, the crucial importance of redox status of three cysteine residues at positions 23, 45 and 106 in the regulation of HMGB1 cytokine activity was emphasized.11–14 As a result, three redox forms of HMGB1 have been described: reduced, disulfide (Cys-23 and Cys-45 forming an intramolecular bond) and oxidized HMGB1. Of note, this latter form can be partially (Cys-106) or completely oxidized at the critical amino acids. Reduced and disulfide HMGB1 have been shown to have chemotactic and pro-inflammatory activity, respectively,13 whereas reactive oxygen species (ROS)-dependent oxidation of HMGB1 at Cys-106 abrogates its immunostimulatory activity and contributes to apoptotic cell-mediated immunotolerance through still unknown mechanisms.11 These opposite roles displayed by mutually exclusive forms of HMGB1 are likely to explain the paradoxical results collected over the years, especially in the context of cancer development. Although increasing evidence reports that HMGB1 could enhance drug resistance and contribute to tumor progression and metastasis,15–20 a defective immunogenic cell death associated with a reduced efficacy of both anthracyclines and oxaliplatin following HMGB1 knockdown was also described.21 22 Therefore, it remains unclear whether extracellular HMGB1 acts as a pro-tumor or a tumor-suppressor protein during carcinogenesis.23
In the past decade, immunotherapy using immune checkpoint inhibitors (especially drugs blocking the programmed cell death protein 1 (PD-1)and its ligand (PD-L1)) has raised considerable interest for treating patients with advanced (metastatic) cancers or unresponsive/resistant to targeted therapies. Despite tremendous clinical successes with some cancer (sub)types (eg, Merkel cell carcinoma, melanoma, microsatellite instability-high colorectal cancer) and the progressive increase of eligible patients for anti-PD-1/PD-L1 monoclonal antibodies (from ~1% in 2011 to >40% in 2018), the overall response rate to checkpoint immunotherapy remains modest for most cancers (12.46% in 2018 among US patients).24 As an example, basal-like (also commonly called ‘triple negative’ due to the lack of expression of HER2 and nuclear hormone receptors) breast cancer has been shown to exhibit a high immunogenicity25 but the proportion of responders did not exceed 15% in several phase I–II clinical trials,26 27 reflecting that individual patient immunity is complex and, undoubtedly, leads to variability in response to treatment. Therefore, the combination of immune checkpoint inhibitors with standard cancer treatments (eg, chemotherapy) or drugs targeting another tumor-induced immune defect is frequently considered as a promising strategy for improving the efficacy of future therapies (without, hopefully, excessively increasing the incidence of immune-related adverse events).28 In this regard, recent clinical data reported the beneficial effect of chemoimmunotherapy in the context of various solid tumors (eg, non-small-cell lung cancer and basal-like breast cancer).29 30 The combination of two immunomodulatory drugs (the therapy concurrently targeting PD-1 and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) is, by far, the most vigorously examined) has also showed to improve both durable response rate and disease-free survival of responding patients compared with monotherapies.31 However, of note is that the combinational approach of several immunoregulatory blockers can also cause antagonistic instead of synergistic effects (eg, concurrent immunotherapy using anti-PD-1 antibody and CD134/OX40 agonist).32
The present study aims at clearly determining whether targeting extracellular HMGB1 with both direct and indirect inhibitors may be efficient in cancer therapy. Particularly relevant for studying novel anticancer drugs with potential immunotherapeutic properties, several syngeneic mouse models were used and the tumor immune microenvironment was precisely characterized in each condition. The efficacy of HMGB1 inhibitors in combination with immune checkpoint blockade antibodies (anti-PD-1/PD-L1) was also assessed. Finally, the redox state of HMGB1 in tumor-extruded fluids as well as the influence of different forms on antigen presenting cell activation were determined.
Materials and methods
Tissue samples and clinical data retrieval
A total of 359 paraffin-embedded cancer specimens and 180 paired normal tissues from the same patients were retrieved from the Tissue Biobank of the University Hospital Center of Liege (Belgium). These tissue samples included 120 normal breast tissues, 40 HPV-negative ectocervical mucosae, 20 normal bronchial epithelia, 275 invasive breast tumors (35 luminal A, 31 luminal B, 37 HER2+ and 172 basal-like), 57 cervical squamous cell carcinomas (SCC) and 27 non-small-cell lung cancers (adenocarcinomas). All cases were re-examined by experienced histopathologists to confirm the diagnosis. Clinicopathological data (age at diagnosis, TNM classification, proliferation index, treatment details (surgery and/or (neo)adjuvant therapies) and follow-up data) for all women diagnosed with basal-like breast cancer were extracted from patient’s medical records (online supplemental table 1).
jitc-2020-001966supp001.pdf (104.2KB, pdf)
Mouse and human cell cultures
The 4T1, 67NR (mouse basal-like breast cancer cells) and TC-1 (mouse lung cancer cells expressing HPV16 E6 and E7 oncoproteins) cells were kindly provided by Dr Nathalie Bendriss-Vermare (INSERM U1052, Claude Bernard Lyon 1 University, France) and Dr Sophie Hallez (Laboratory of Chemical Biology, Université Libre de Bruxelles, Belgium), respectively. These latter cell lines were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Thermo Fisher Scientific, Waltham, Massachusetts, USA) containing 10% fetal calf serum (Lonza, Basel, Switzerland), 1% non-essential amino acid (Gibco, 100X), 1 mM sodium pyruvate (Gibco), 2 mM L-glutamine (Gibco), 50 µM β-mercaptoethanol (Gibco) and 1% penicillin-streptomycin (Gibco, 100X). Neu15 cells (kindly provided by Dr Nathalie Bendriss-Vermare) were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 1% non-essential amino acids, 1 mM sodium pyruvate, 2 mM L-glutamine, 1% insulin-transferrin-selenium (Gibco, 100X) and 1% penicillin-streptomycin. Both EpRas and EpH4 (kindly provided by Professor Hartmut Beug, University of Vienna, Austria) as well as RAW264.7 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal calf serum and supplied with 1% non-essential amino acids, 1 mM sodium pyruvate and 1% penicillin-streptomycin. Normal human mammary epithelial cells (HMEC and MCF10A) and human breast cancer cell lines (Luminal A: T-47D and MCF7; HER2+: SK-BR-3; basal-like: MDA-MB-468, BT-549, MDA-MB-231, MDA-MB-157, HCC70, HCC1143 and Hs578T) were obtained from the American Type Culture Collection (Manassas, Virginia, USA) or were kindly provided by Dr Akeila Bellahcene (Metastasis Research Laboratory, University of Liege, Belgium), Dr Christine Gilles (Laboratory of Tumor and Development Biology, University of Liege, Belgium) or Dr Alain Chariot (Laboratory of Medical Chemistry, University of Liege, Belgium). MDA-MB-231, HCC70 and HCC1143 were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 1% non-essential amino acids, 1 mM sodium pyruvate, 50 µM β-mercaptoethanol and 1% penicillin-streptomycin. MCF7 and Hs578T were maintained in DMEM containing 10% fetal calf serum, 1% non-essential amino acids, 1 mM sodium pyruvate, 2 mM L-glutamine, 10 µg/mL insulin (Sigma Aldrich, Saint Louis, Michigan, USA) and 1% penicillin-streptomycin. MDA-MB-157 and MDA-MB-468 were grown in DMEM containing 10% fetal calf serum and supplied with 1% non-essential amino acids, 2 mM L-glutamine and 1% penicillin-streptomycin. HMEC cells were cultured in Mammary Epithelial Cell Growth Basal Medium (Lonza) containing all requested supplements and growth factors (0.25% bovine pituitary extract, 20 ng/mL epidermal growth factor (EGF), 5 µg/mL insulin, 0.2 µg/mL hydrocortisone, 30 µg/mL gentamicin and 15 ng/mL amphotericin B (Lonza)). MCF10A was maintained in 1:1 mixture of DMEM and Ham’s F12 medium (Gibco) containing 5% horse serum (Sigma Aldrich), 1% non-essential amino acids, 0.5 µg/mL hydrocortisone, 2.5 mM L-glutamine, 20 ng/mL EGF, 0.1 µg/mL cholera toxin, 10 µg/mL insulin and 1% penicillin-streptomycin. T-47D cells were cultured in RPMI 1640 containing 10% fetal calf serum, 7.692 µg/mL insulin, 1% non-essential amino acids, 1 mM sodium pyruvate and 1% penicillin–streptomycin. SK-BR-3 cells were grown in McCoy 5A (Gibco) supplemented with 10% fetal calf serum, 1% non-essential amino acid, 1 mM sodium pyruvate and 1% penicillin-streptomycin. BT-549 cells were cultured in RPMI 1640 containing 10% fetal calf serum and supplied with 0.88 µg/mL insulin, 1% non-essential amino acids, 1 mM sodium pyruvate and 1% penicillin–streptomycin. All cell lines were authenticated by Short Tandem Repeat (STR) analysis (Eurofins Genomics, Ebersberg, Germany) and tested negative for mycoplasma contamination (MycoAlert Mycoplasma Detection Kit, Lonza).
Assessing HMGB1 inhibitor efficacy in vitro
The neutralizing activity of glycyrrhizin (direct HMGB1 inhibitor) (Sigma Aldrich) as well as RAGE antagonist peptide (RAP) (Merck Millipore, Burlington, Massachusetts, USA) and truncated N-terminal domain of HMGB1 (A box) (HMGBiotech, Milan, Italy) was indirectly determined through analyzing their ability to attenuate HMGB1-induced TNFα release by macrophages/monocytes. To do this, 4×105 murine RAW 264.7 macrophages per well of a twenty four-well plate were seeded in appropriate growth medium. Twenty-four hours later, the cells were stimulated with 1 µg/mL recombinant mouse HMGB1 (R&D systems, Minneapolis, Minnesota, USA) alone or in combination with inhibitors (several concentrations were tested). After 16 hours, TNFα level in cell culture supernatant was quantified using the mouse TNFα DuoSet ELISA purchased from R&D systems. In order to determine whether the dose range for each inhibitor also inhibited HMGB1 secreted by both lung (TC-1) and basal-like breast cancer cells (4T1, 67NR, EpRas) used in our in vivo models, similar experiments were performed in which the stimulation by recombinant HMGB1 was replaced by conditioned media from these latter cell lines (1×106 cells per well of a six-well plate for 48 hours in 2 mL of growth medium). As described in previous articles,14 33 a similar procedure was used for analyzing the neutralizing effect of a polyclonal anti-HMGB1 antibody raised in rabbit (Eurogentec, Seraing, Belgium). Titer for this latter antibody was determined by immunoblotting.
For ethyl pyruvate (EP) (Sigma Aldrich), its inhibitory effect on HMGB1 secretion was determined by ELISA (HMGB1 Detection kit, Chondrex, Redmond, WA, USA). Forty-8 hours after adding EP (concentration range: 0.1–10 mM), conditioned medium from basal-like breast cancer cell lines was harvested and HMGB1 concentration was measured.
Syngeneic mouse models
C57BL/6J and BALB/c female mice aged 6–8 weeks (n=10 per condition) were used in the present study. Five×105 4T1, 1×106 67NR and 2×106 EpRas (in 300 µL appropriate growth medium) were orthotopically injected into the mammary fat pad of mice. Regarding TC-1 cells (8×105), they were subcutaneously injected (mouse flank). In some peculiar cases, a similar procedure was used with Nude RJ:ATHYM-Foxn1nu mice (n=6 per condition). When tumor volume reached 50–100 mm3, treatments with HMGB1 inhibitors alone or in combination with checkpoint blockade antibodies were initiated. One nM/kg glycyrrhizin, 10 µM/kg RAP, 500 µg/kg A box and 1 mM/kg EP were administrated by intraperitoneal (i.p.) injection at 3-day intervals for 3 weeks. Similar administration frequencies were used for both rabbit (8.6 mg/kg, Eurogentec) and mouse (5 mg/kg, clone 3E8, Biolegend, San Diego, California, USA) anti-HMGB1 antibodies. When indicated (combination therapy), mice received 200 µg of in vivo plus monoclonal anti-mouse PD-1 (clone 29F.1A12, Bio X cell, West Lebanon, New Hampshire, USA) or 100 µg of antimouse PD-L1 (clone 10F.9G2, Bio X cell) antibody by i.p. injection at days 4, 7 and 11. Tumor size was monitored with a digital caliper (Thermo Fisher Scientific) every 2–3 days and mice were euthanized when the average tumor volume [(length × width2)× π /6] of the control group exceeded 1500 mm3. Tumor growth inhibition index was calculated using the following formula: [1-(mean volume of treated tumors)/(mean volume of control tumors)] × 100%. Half of retrieved tumors were fixed in 10% formalin for 48 hours, embedded in paraffin and sections were subjected to immunohistochemical analysis. The other unfixed tumors were crudely dissected into small pieces, incubated with an enzymatic cocktail (1 mg/mL collagenase A (Sigma Aldrich) and 20 µg/mL DNase (Sigma Aldrich)) for 30 min at 37°C and then passed through a 70 μm pore size membrane filter (BD Biosciences, San Jose, California, USA). The cells were finally resuspended in phosphate-buffered saline (PBS) 3% BSA (Merck Millipore) before CD45+ cell isolation and flow cytometry analysis (procedure and fluorochrome-conjugated antimouse antibodies detailed below). Due to mortality in some tumor-bearing mice treated with anti-PD-1 or anti-PD-L1 inhibitors, in order to have the same number of analyses per condition, some resected tumors were cut into halves (the first half was embedded in paraffin, while the immune cells were retrieved from the second part). The animals were bred in-house (GIGA-Mouse facility and Transgenics platform) or purchased from Janvier Labs (Le Genest-Saint-Isle, France) and the experiments were performed in strict compliance with the ethical rules/recommendations established by the Federation of European Laboratory Animal Sciences Associations.
Antibody-mediated granulocytic MDSC depletion in mice
Granulocytic MDSC were depleted by i.p. injection of 100 µg of ultra-LEAF purified anti-Ly6G antibody (clone 1A8, Biolegend) at day −1, 3 and 7. Promoting target cell phagocytosis through Fc-dependent opsonization and allowing a durable and controlled depletion,34 mice were treated using a recently published double antibody-based strategy (100 µg anti-Ly6G+50 µg in vivo antirat kappa immunoglobulin light chain (clone MAR 18.5, Bio X cell)) during the second week of depletion (at days 11–12 and 15–16).
Flow cytometry (for in vivo experiments)
Flow cytometry analyses were performed using a FACSCanto II flow cytometer and collected data were analyzed with FACSDiva software, V.6.1.2 (BD Biosciences). Immune cells from digested tumors were first isolated using CD45 MicroBeads according to manufacturer’s protocol (Miltenyi Biotec, Bergisch Gladbach, Germany). For precisely determining the number of CD45+ cells per milligram of tumor, this latter positive selection was not performed in this particular case. In order to minimize non-specific binding, CD45+ cells were then incubated with anti-CD16/CD32 antibody (clone 2.4G2, BD Biosciences). All cell surface reactions were performed at 4°C for 30 min. The fluorochrome-conjugated antimouse antibodies used in the present study are listed in online supplemental table 2. The permeabilization step allowing intracellular Foxp3 staining was performed according to the manufacturer’s recommendations (eBioscience, Thermo Fisher Scientific). The following immune cell populations were analyzed: DC (CD45+, CD11b+, I-A/I-Ehigh, F4/80-), pDC (CD45+, CD11bint, CD11c+, BST2+, Siglec H+), CD4+ T cells (CD45+, CD3+, CD4+), CD8+ T cells (CD45+, CD3+, CD8+), monocytic MDSC (CD45+, CD11b+, I-A/I-Eint, Ly6G-, Ly6Chigh), granulocytic MDSC (CD45+, CD11b+, I-A/I-Eint, Ly6G+, Ly6C-), neutrophils (CD45+, CD11b+, I-A/I-Eint, Ly6G+, Ly6Cint), M1 macrophages (CD45+, CD11b+, CD11c+, F4/80+, CD206-), M2 macrophages (CD45+, CD11b+, CD11c-, F4/80+, CD206+), CD4+ T regulatory cells (CD45+, CD3+, CD4+, CD25+, FoxP3+), CD8+ T regulatory cells (CD45+, CD3+, CD8+, CD25+, FoxP3+), NK cells (CD45+, NKp46+), B cells (CD45+, CD19+) and eosinophils (CD45+, Ly6C-, Siglec F+). Of note, the proportion of DC is likely slightly overestimated (<2%) in some experiments given that the expression of F4/80 was not systematically evaluated. The activation status of DC and pDC was also determined by evaluating CD80, CD86, I-A/I-E, ILT3 and CD80, CD86, I-A/I-E, ICOSL, respectively. In order to properly segregate negative from positive cell populations, PMT voltages were set optimally. Cell fragments and debris were eliminated based on both forward-scatter and side-scatter values. Gating strategy is shown in online supplemental figure 1.
jitc-2020-001966supp002.pdf (89.4KB, pdf)
jitc-2020-001966supp003.pdf (958KB, pdf)
Assessing HMGB1 inhibitor toxicity in vivo
Eight-week-old female cancer-free BALB/c mice (n=5 per condition) were treated at 3-day intervals by i.p. injections of glycyrrhizin (1 nM/kg), RAP (10 µM/kg), A box (500 µg/kg) and EP (1 mM/kg) (similarly to what is done with the syngeneic mouse models). Mice were weighed every 2–3 days. After 3 weeks, mice were euthanized and lungs, heart, spleen, liver and kidneys were retrieved, weighed and then embedded in paraffin for histological evaluation. The levels of serum urea and alanine aminotransferase (ALT) were measured using a cobas 6000 analyzer (Roche, Basel, Switzerland).
Immunohistochemistry
Immunohistochemical analyses were performed using a standard protocol detailed previously.35 36 Antigens were retrieved in 10 mM citrate (pH 6) (Sigma Aldrich) or EDTA (pH 9) (Zytomed Systems, Berlin, Germany) buffer. The primary antibodies used in the present study were as followed: anti-HMGB1 (clone 1D5, Abnova, Taipei City, Taiwan), anti-cleaved caspase 3 (clone C92-605, BD Biosciences), anti-CD31 (ab28364, Abcam, Cambridge, UK), anti-CD3 (clone 2GV6, Ventana Medical Systems, Tucson, Arizona, USA), anti-Foxp3 (clone 236 A/E7; eBioscience, San Diego, California, USA), anti-CD68 (clone KP-1, Ventana Medical Systems), anti-CD206 (clone E2L9N, Cell Signaling Technology, Danvers, MA, USA), anti-PD-1 (clone NAT105, Abcam), anti-human PD-L1 (clone 28–8, Abcam) and anti-mouse PD-L1 (orb10162, Biorbyt, Cambridge, UK). The secondary reaction was performed using the mouse or rabbit Envision+system (Dako, Glostrup, Denmark) according to the manufacturer’s recommendations. Positive cells were visualized using a 3,3’-diaminobenzidine (DAB) substrate (Cell Signaling Technology). Mouse or rabbit control IgGs (Santa Cruz Biotechnology, Santa Cruz, California, USA) were used as negative controls.
Assessment of immunohistochemical stainings
Experienced histopathologists evaluated the percentage of epithelial (normal or cancer) cells displaying a cytoplasmic HMGB1 immunoreactivity. Collected results were stratified as followed: negative, <10% and >10%. PD-L1 expression was considered as positive when membranous immunostaining was detected in ≥1% of cancer cells or inflammatory cells within tumor microenvironment. As previously described,37–39 staining quantification for apoptotic (cleaved caspase 3+) cancer cells as well as CD3+, Foxp3+, CD68+, CD206+ and PD-1+ immune cells infiltrating the stroma surrounding tumor cells was performed by computerized counts (QuPath 0.2.0 open source software for digital pathology image analysis). Slides were scanned using 3 dimensional Histech Pannoramic scanner (Sysmex, Villepinte, France). The number of positive cells was reported to cancer area (mm2). For CD31 immunoreactivity, QuPath software was also used (positive pixel count algorithm) and, as previously described,40 the ratio of DAB-stained pixels to total cancer area was determined.
Generation of DC and pDC from human CD34+ hematopoietic stem/progenitor cells and cocultures
Both DC and pDC were generated from CD34+ cells isolated from cord blood samples using the MACS CD34 MicroBead kit (Miltenyi Biotec, Bergisch Gladbach, Germany). As previously described,41 for the generation of DC, CD34+ precursor cells were cultured for 7 days in the presence of 20 ng/mL human SCF (Peprotech, Cranbury, New Jersey, USA), 10 ng/mL TPO (Peprotech), 25 ng/mL Flt3L (Peprotech), 200 U/mL GM-CSF (Amoytop Biotech, Xiamen, China) and 100 U/mL IL-4 (ImmunoTools, Friesoythe, Germany). Regarding the production of pDC, CD34+ cells were cultured for 21 days in the presence of 10 ng/mL TPO (Peprotech), 100 ng/mL Flt3L (Peprotech) and 20 ng/mL IL-3 (Peprotech). The detailed protocol was also previously described.42 Where indicated, DC were treated with 3 µg/mL fully reduced, disulfide or terminally oxidized HMGB1 (HMGBiotech, Milan, Italy) for 1 day, followed by a lipopolysaccharide (LPS)-induced maturation (1 µg/mL, Sigma-Aldrich) for 24 hours. The procedure was identical with pDC except that maturation was induced by adding 12 µg/mL CpG ODN (Eurogentec) in the cell cultures for 24 hours. In selected experiments, an inhibitor of RAGE (10 µM RAP) or TLR4 (2 µM LPS-RS, Invitrogen, Carlsbad, California, USA) was added in the culture system. The expression of several maturation markers (CD40, CD80, CD83, CD86, HLA-DR, HLA-ABC and CCR7) was finally determined by flow cytometry (FACSCanto II flow cytometer, GIGA-in vitro imaging platform). The fluorochrome-conjugated anti-human antibodies used in this study are listed in online supplemental table 2.
Characterization of the redox state of released HMGB1 in tumor microenvironment
Mouse breast cancer cells (4T1, 67NR and EpRas) were orthotopically injected into the mammary fat pad of mice. When the volume reached 1000 mm3, tumors were retrieved and soluble proteins/interstitial fluids were collected using a pressure-assisted innovative methodology (EXPEL) recently described.43 In order to irreversibly freeze the redox state of HMGB1 molecules contained in tumor-extruded fluids, an alkylation step (50 mM iodoacetamide for 30 min at 25°C) was then directly conducted. Proteins were quantified using the Pierce BCA protein assay (ThermoFisher Scientific) and 15 µg were separated by electrophoresis on 10% polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. After saturation with TBS-Tween 20 0.1% supplemented with 5% skim milk for 30 min, the membrane was incubated overnight at 4°C with anti-HMGB1 antibody (1/1000, A00066-1, Boster Biological Technology, Pleasanton, California, USA). After washing and incubation with the horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody, the protein bands were detected by chemoluminescence and quantified using ImageJ software (National Institute of Health (NIH), Bethesda, Maryland, USA). As oxidized and reduced controls, recombinant HMGB1 was exposed to either 10 mM H2O2 or DTT for 1 hour and then alkylated.
Gene expression analysis
The expression of HMGB1, PD-1 (PDCD1 gene) and PD-L1 (CD274 gene) according to cancer subtypes, grades, lymph node and metastatic statuses was evaluated using the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) public dataset (Illumina HT-12 v3 platform for transcriptional profiling).44 45 Breast cancers were categorized into the four current major molecular subtypes based on proliferative index (Ki67), hormone receptor expression (estrogen receptor (ER), progesterone receptor (PR)) and HER2 positivity: Luminal A (ER+/PR+, HER2−, Ki67low), Luminal B (ER+/PR+, HER2−, Ki67high and ER+/PR+, HER2+), HER2+ (ER−/PR−, HER2+) and basal-like (ER−/PR−, HER2−).
Metabolic extracellular flux analysis
Mouse basal-like breast cancer cells (10 000 cells per well) were seeded in Seahorse XFp mini-plates (Agilent, Santa Clara, California, USA) and analyzed using the mitochondrial stress test as previously described.46 HMGB1 inhibitors were added in growth culture medium for 12 hours and removed before the assay. For the optimal measurement of both oxygen consumption (OCR) and extracellular acidification (ECAR), cells were maintained in unbuffered serum-free DMEM (pH 7.4) containing 1 mM pyruvate, 2 mM glutamine and 10 mM glucose during the assay. Cells were successively stressed with 1 µM oligomycin, 1 µM FCCP and 0.5 µM rotenone/antimycin A and collected results were normalized to cell number (evaluated by Hoechst).
HMGB1 measurement by ELISA
One×106 cells per well of a six-well plate were cultured in appropriate growth medium during 48 hours. Cell culture supernatant was then harvested and HMGB1 release by both human and mouse breast cancer cells was quantified by ELISA using the following commercially available kit: HMGB1 Detection kit (Chondrex). After 48 hours, the number of attached cells in each condition was also determined in order to normalize HMGB1 measurements (ng/mL per 106 cells).
ROS measurement
Mitochondrial ROS production by cancer cells was measured using CellROX Flow Cytometry kit (Life Technologies, Carlsbad, California, USA) according to the manufacturer’s protocol. N-acetylcysteine (5 mM) and Tert-butyl hydroperoxide (100 µM) were used as negative and positive controls, respectively.
Cell proliferation and apoptosis/necrosis
Cell proliferation under indicated culture conditions was monitored for 6 days using live-cell imaging analysis (IncuCyte ZOOM system, Essen BioScience, Welwyn Garden City, UK). The percentage of apoptotic/necrotic cells was determined by annexin V-FITC and propidium iodide staining according to the manufacturer’s recommendations (BD Biosciences). Results were acquired by flow cytometry (FACSCalibur flow cytometer, BD Biosciences).
Statistical analysis
Collected experimental data were analyzed using the GraphPad Prism V.8 software (San Diego, California, USA). The comparison of both patient characteristics and immunohistochemical variables (negative, <10% and >10% of cells exhibiting a cytoplasmic HMGB1 immunostaining) between independent groups was performed using a Fisher’s exact test or a χ2 test according to the number of variables. Two group comparisons were performed using (Welch-corrected) unpaired t-tests. Experiments containing more than two groups were compared using one-way analysis of variance (ANOVA), followed by Dunnett’s multiple comparison test or Bonferroni post-test. Regarding METABRIC gene expression data, both graphs and statistical analyses (one-way ANOVA followed by Bonferroni post-test or unpaired t-test) were done using the open source statistical language R (V.3.5). Disease-free survival was defined as the time from the date of original biopsy/diagnosis to the date of local or distant recurrence (metastasis). The Kaplan-Meier method (with log-rank (Martel-Cox) test) was used. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Results
HMGB1 is highly secreted by basal-like breast cancer cells and its cytoplasmic immunoreactivity is associated with both regulatory T (Treg) cell infiltration within tumor stroma and poor outcome
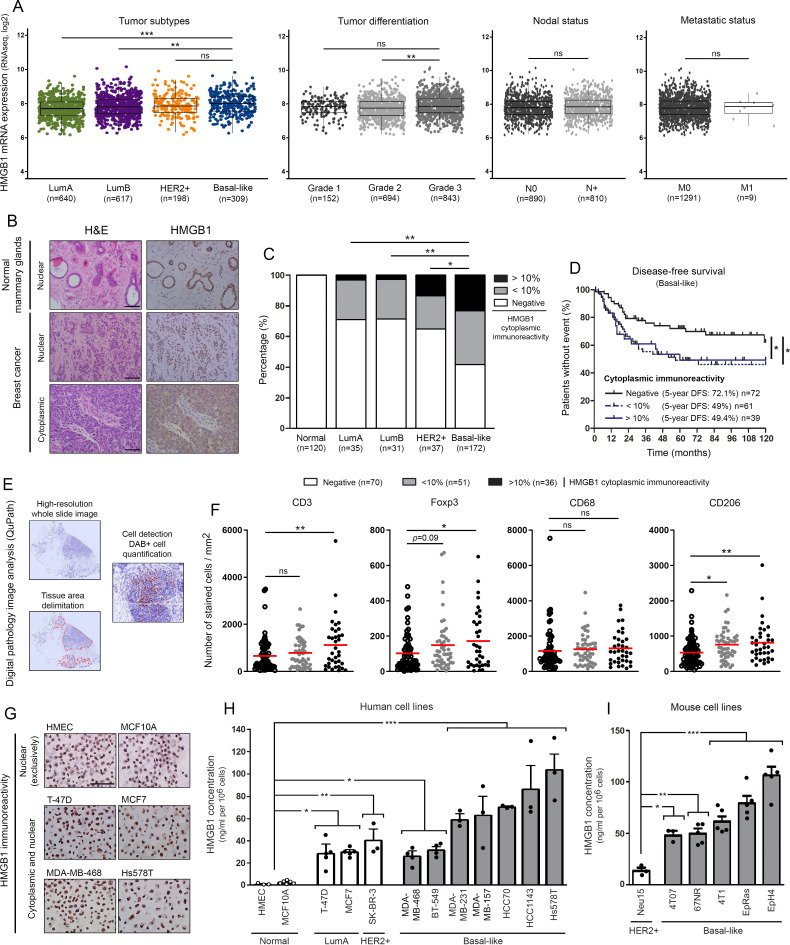
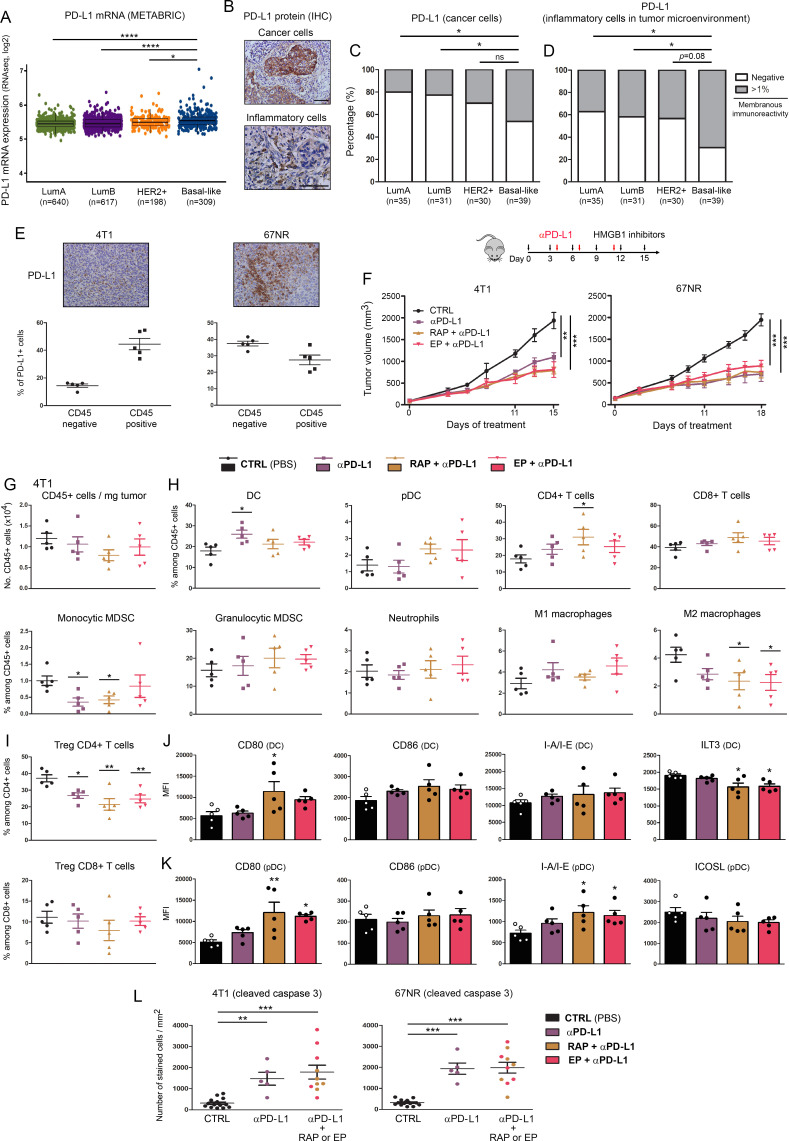
By using the METABRIC public dataset, the mRNA expression level of HMGB1 according to breast cancer molecular subtypes, histologic grades, nodal and metastatic statuses was first assessed. Although the statistical significance was reached between some cancer subtypes (basal-like vs LumA (p<0.001) or LumB (p<0.01)) and grades (grade 2 vs grade 3 (p<0.01)) (very likely due to the high number of analyzed samples), no obvious difference was observed (figure 1A). Kaplan-Meier analyses demonstrated no difference in 5-year patient survival according to HMGB1 gene expression either (online supplemental figure 2). A large cohort of both normal and neoplastic breast tissues was then examined by immunohistochemistry and, interestingly, two distinct HMGB1 staining patterns were identified: nuclear versus cytoplasmic (figure 1B). Whereas normal mammary glands always displayed a nuclear immunoreactivity, a cytoplasmic expression of HMGB1 (in up to 40% tumor cells) was detected in approximately half of analyzed breast cancer specimens (132/275, 48%). Similar results were collected in the context of other solid tumors (cervical SCC and non-small-cell lung cancer) (online supplemental figure 3A, B). As shown in figure 1C, diffuse cytoplasmic HMGB1 staining was more frequently observed in triple negative/basal-like cancers compared with their counterparts expressing HER2 (p<0.05) and/or hormone receptors (p<0.01). Remarkably, the prognostic value of this latter expression pattern was also highlighted. Indeed, patients with basal-like breast cancer displaying a cytoplasmic HMGB1 expression in either 0%–10% or >10% cells were significantly associated with an unfavorable outcome (figure 1D). As detailed in online supplemental table 1, these shorter disease-free survivals cannot be explained by differences in age at diagnosis, tumor size, nodal/metastatic status, tumor stage, proliferative index or treatment modality. Of note, a significant increased density of both Foxp3+ Treg lymphocytes (102.6 vs 148.8 and 171.6 cells/mm2) and CD206+ M2 macrophages (533.4 vs 757.1 and 811.1 cells/mm2) was noticed within tumor microenvironment of cytoplasmic HMGB1-positive cancers compared with basal-like neoplasms exhibiting exclusively a nuclear immunoreactivity (figure 1F). Quantified by computerized counts (figure 1E), the global number of lymphocytes (CD3+) also augmented (657 vs 1120 cells/mm2) in case of diffuse (in >10% cancer cells) cytoplasmic HMGB1 expression. In contrast, the global number of macrophages (CD68+) was quite similar for all the analyzed groups (1161, 1256 and 1309 cells/mm2) (figure 1F). Immune cell densities were undetermined in 15 out of 172 (8.7%) basal-like breast cancer patients (mainly due to the lack of remaining slides). Confirming our ex vivo data, cytoplasmic staining pattern for HMGB1 was only detected in malignant cell lines (especially those derived from basal-like breast cancers) and associated to protein concentration in both human and mouse cell culture media (figure 1G–I). Given that the percentages of apoptotic/necrotic cells detected in vitro (online supplemental figure 4) did not correlate with the HMGB1 concentrations in culture supernatants, the differences measured by ELISA are likely related to the active secretion of HMGB1 rather than its passive release from injured cells.
Figure 1.
HMGB1 is highly secreted by basal-like breast cancer cells and its tumor-specific cytoplasmic expression is associated with immune tolerance and poor outcome. (A) The METABRIC dataset was used for analyzing HMGB1 expression level in breast cancers according to molecular subtypes, histologic grades, nodal and metastatic statuses. (B) Representative pictures of normal mammary glands and breast cancers stained for HMGB1. Note the two distinct HMGB1 staining patterns detected in tumor specimens: nuclear versus cytoplasmic. (C) Semiquantitative evaluation of cytoplasmic HMGB1 immunoreactivity (negative, 0%–10% or >10% positive cells) in both normal mammary glands (n=120) and neoplasms (LumA, n=35; LumB, n=31; HER2+, n=37; basal-like, n=172). (D) Disease-free survival of patients treated for basal-like breast cancer according to cytoplasmic HMGB1 expression (negative, n=72; 0%–10%, n=61; >10%, n=39). This latter parameter was clearly found to be an independent prognostic factor. (E) Illustration of the different steps for DAB-positive cell quantification using computerized image analysis (QuPath). (F) CD3+, Foxp3+, CD68+ and CD206+ cell infiltrations in microenvironment of basal-like breast tumors. Whereas the global number (CD68+) did not significantly change, an increased density of CD206+ M2 macrophages was detected in cytoplasmic HMGB1-positive cancers. A similar increase was also reported with Foxp3+ Treg lymphocytes. the number of positive cells was reported to tumor area (mm2). (G) Representative examples of normal (HMEC and MCF10A) and malignant cells (LumA: T-47D and MCF7; basal-like: MDA-MB-468 and Hs578T) stained for HMGB1. Note the exclusive nuclear immunoreactivity displayed by normal mammary cells. Secretion/release of HMGB1 analyzed by ELISA in (H) human and (I) mouse cell culture supernatants. High concentrations were especially detected in cell cultures derived from triple negative/basal-like tumors. The means±SEM (plus each individual data point) for at least three independent experiments are represented. The scale bar represents 100 µm. Asterisks indicate statistically significant differences (*p<0.05; **p<0.01; ***p<0.001). P values were determined using one-way ANOVA followed by Bonferroni post-test (A), unpaired t-test (A), χ2 test (C), log-rank (Mantel-Cox) test (D) and one-way ANOVA followed by Dunnett’s multiple comparison post-test (F, H, I). ANOVA, analysis of variance; HMGB1, high-mobility group box 1; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium.
jitc-2020-001966supp004.pdf (663.5KB, pdf)
jitc-2020-001966supp005.pdf (1.1MB, pdf)
jitc-2020-001966supp006.pdf (129.1KB, pdf)
Both direct and indirect HMGB1 inhibitors efficiently neutralize extracellular HMGB1 in vitro without affecting tumor cell proliferation/viability
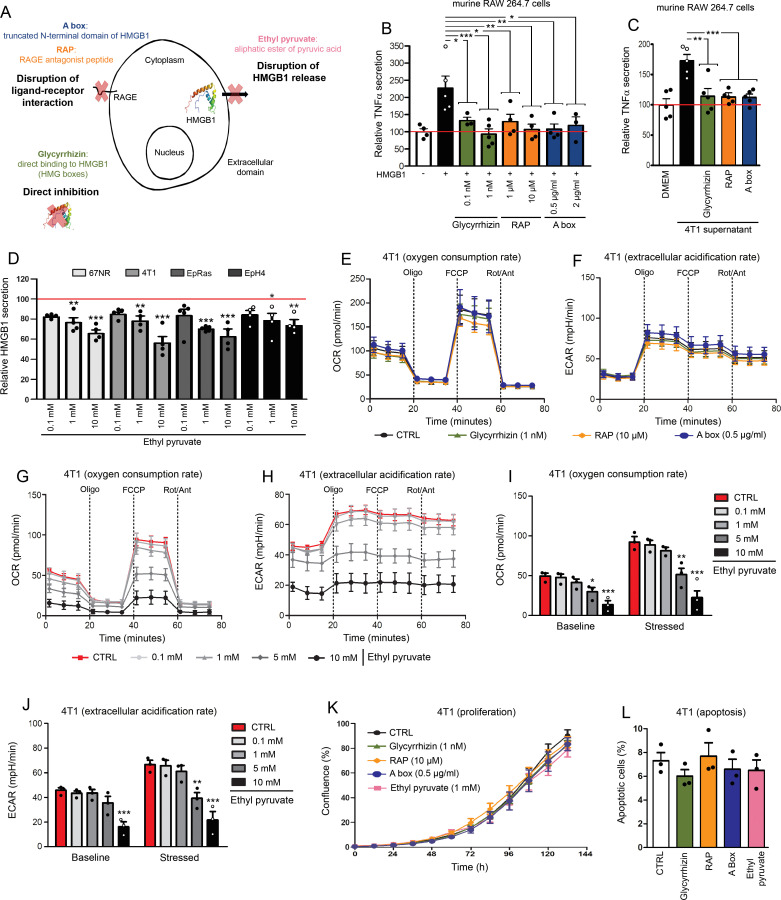
In the last decade, different modes of inhibition for extracellular HMGB1 as well as several direct or indirect inhibitors have been highlighted (figure 2A). Consisting of the truncated N-terminal domain of HMGB1, A box has previously been shown to efficiently reverse established sepsis in vivo.33 A box acts as a competitive antagonist of HMGB1 and, given the sequence similarity between both human and mouse HMGB1, it is presumed to elicit very low neutralizing immunological responses. Composed of 10 amino acids of the RAGE-binding domain of HMGB1, RAP competes with natural HMGB1 for a site on the extracellular domain of RAGE that is required for ligand-receptor interaction,47 resulting in RAGE signaling pathway inhibition. Derived from the licorice plant (Glycyrrhiza glabra) and administered at high doses to patients with chronic hepatitis B or C infection, glycyrrhizin has been shown to have anti-inflammatory properties through binding directly to extracellular HMGB1 and, consequently, blocking its activity.48 Regarding EP, a stable aliphatic esther of pyruvic acid, its ability both to inhibit HMGB1 release in a dose-dependent manner and subsequently ameliorate mouse models of inflammatory diseases (eg, colitis) was recently demonstrated by several research groups.49 50 The importance of SIRT1/STAT signaling pathway inhibition in this mechanism was proposed.50 In order to determine the effective concentration of glycyrrhizin, RAP and A box that inhibited HMGB1 activity by >80%, murine RAW 264.7 cells were stimulated with recombinant HMGB1 (synthesized in mouse tumor cells) alone or in the presence of each inhibitor. HMGB1-dependent TNFα secretion was then determined by ELISA. As shown in figure 2B, all three inhibitors neutralize very efficiently extracellular HMGB1. Similar results were obtained when the stimulation by recombinant HMGB1 was replaced by conditioned media from mouse tumor cells used in our in vivo models (figure 2C and online supplemental figure 5). To analyze the ability of EP to reduce HMGB1 release, this latter compound was directly added in mouse basal-like breast cancer cell cultures for 48 hours and HMGB1 concentrations were measured. As shown in figure 2D, a significant decrease was observed at a concentration as low as 1 mM. The effect of each HMGB1 inhibitor on cell metabolism, proliferation and apoptosis was then precisely assessed using Seahorse flux analyzer, IncuCyte live cell analyzing system and annexin V-propidium iodide staining assay, respectively (figure 2E–L and online supplemental figure 6). Whereas glycyrrhizin (1 nM), RAP (10 µM) and A box (0.5 µg/mL) did not affect these three cellular parameters, significant changes in both OCR and extracellular pH were observed with 5 and 10 mM EP (figure 2G–J), suggesting that high doses of this derivative of pyruvic acid alters cell energetic metabolism (glycolysis) (very likely by competing with endogenous pyruvate). Of note is that the metabolism, proliferation and apoptosis of tumor cells were not affected by the addition of 1 mM EP that has been shown above to be enough for exerting a significant inhibitory effect on HMGB1 release (figure 2D–L and online supplemental figure 6).
Figure 2.
Glycyrrhizin, RAP, A box and EP exert efficient neutralizing effects on extracellular HMGB1 without altering tumor cell proliferation and apoptosis/necrosis. (A) Schematic representation of different modes of inhibition for extracellular HMGB1. Mouse RAW 264.7 cells were stimulated with recombinant HMGB1 (B) or conditioned media from 4T1 basal-like breast cancer cells (C) in the absence or presence of glycyrrhizin, RAP or a box (several concentrations were tested). Note the significant decrease of HMGB1-induced TNFα secretion when HMGB1 inhibitors were added in the cell cultures, indicating their efficient neutralizing effect. (D) EP was directly added in the culture medium of 4 different mouse basal-like breast cancer cell lines (4T1, 67NR, EpRas and EpH4). Forty-eight hours later, HMGB1 concentrations were determined by ELISA and the ability of EP to inhibit HMGB1 release in a dose-dependent manner was highlighted. (E) Oxygen consumption rate (OCR) and (F) extracellular acidification rate (ECAR) in 4T1 cells in the absence or presence of glycyrrhizin (1 nM), RAP (10 µM) and a box (0.5 µg/mL) were determined using Seahorse extracellular flux analyzer. No modification of OCR/ECAR was detected with these three HMGB1 inhibitors. (G) OCR and (H) ECAR in 4T1 cells following EP addition (concentration range: 0.1–10 mM). Histograms representing OCR (I) and ECAR (J) before (baseline) and after (stressed) oligomycin and FCCP addition in the absence or presence of EP. Both OCR and ECAR were strongly decreased with 5 and 10 mM EP. No significant change was detected with lower concentrations (0.1–1 mM). (K) Cell proliferation and (L) apoptosis of mouse 4T1 cells cultured without or with HMGB1 inhibitors (glycyrrhizin (1 nM), RAP (10 µM), a box (0.5 µg/mL) and EP (1 mM)) were determined using IncuCyte live cell analyzing system and annexin V-propidium iodide staining assay, respectively. No significant change was reported. The means±SEM (plus each individual data point) for at least three independent experiments are represented. Asterisks indicate statistically significant differences (*p<0.05; **p<0.01; ***p<0.001). P values were determined using one-way ANOVA, followed by Dunnett’s multiple comparison post-test (B, C, D, I, J, K, L). ANOVA, analysis of variance; ECAR, extracellular acidification rate; EP, ethyl pyruvate; HMGB1, high-mobility group box 1; RAGE, receptor for advanced glycation endproducts; RAP, RAGE antagonist peptide; TNFα, tumor necrosis factor-α.
jitc-2020-001966supp007.pdf (466.4KB, pdf)
jitc-2020-001966supp008.pdf (1.6MB, pdf)
Extracellular HMGB1 blockade inhibits tumor growth in syngeneic mouse tumor models through activating adaptive immune responses
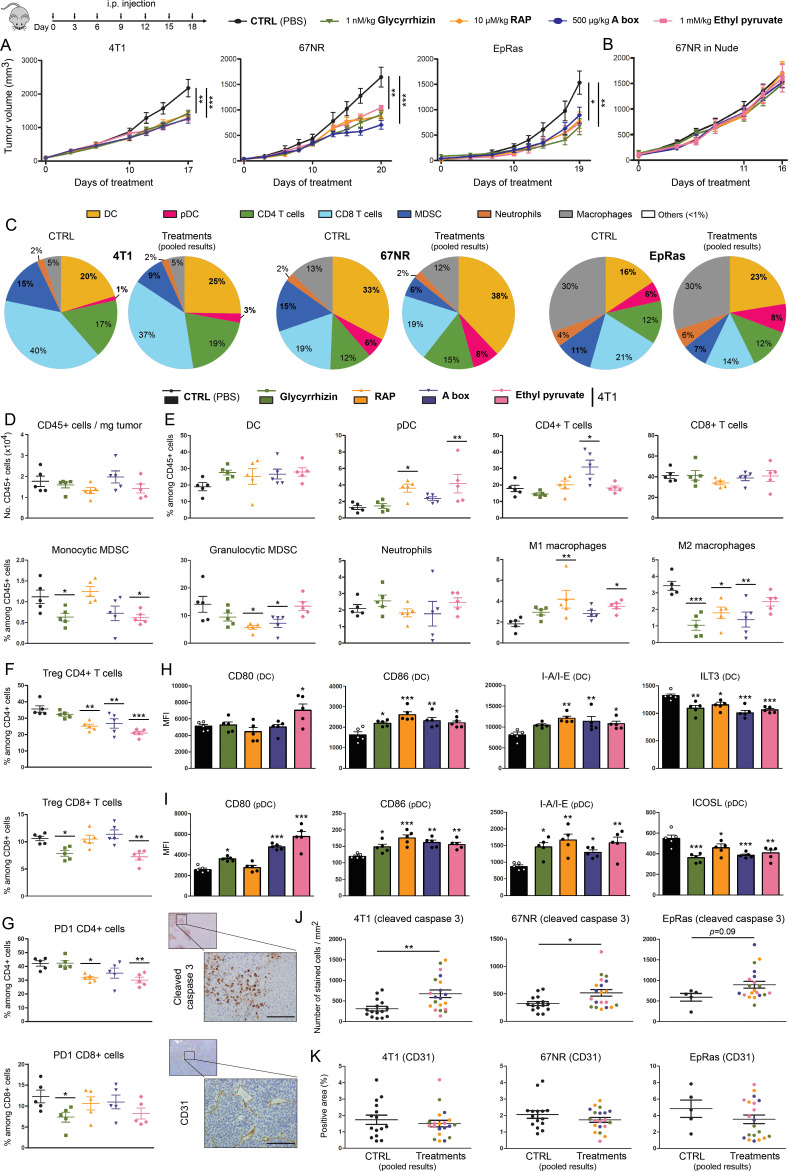
Next, we sought to determine whether extracellular HMGB1 blockade had antitumor effects in vivo. Three syngeneic mouse models of basal-like breast cancer (4T1, 67NR and EpRas) as well as one additional of non-small cell lung cancer (TC-1) were used (figure 3 and online supplemental figure 3). Due to the stable transfection of HPV16 E6 and E7, these latter cells are also frequently used as a model of cervical cancer. To mimic human disease closely, all breast cancer cell lines were orthotopically injected (into the mammary fat pad) and treatments with HMGB1 inhibitors (injections at 3-day intervals for 15–20 days) began when solid tumors (50–100 mm3) were established. As commonly practiced, the concentration used in vivo for each HMGB1 inhibitor (glycyrrhizin (1 nM/kg), RAP (10 µM/kg), A box (500 µg/kg) and EP (1 mM/kg)) was extrapolated from our in vitro data (figure 2) by assuming that mol/L~mol/kg for compounds diluted in liquid solutions (PBS). Strikingly, in all four models, a significant decrease in tumor growth was observed with extracellular HMGB1 blockade (figure 3A and online supplemental figure 3D). Although having different modes of action, these four HMGB1 inhibitors displayed quite similar therapeutic antitumor activities. When collected data were pooled, mean tumor growth inhibitions of 38%, 46%, 49% and 31% were obtained for treated mice transplanted with 4T1, 67NR, EpRas and TC-1 cells, respectively. Importantly, this latter beneficial effect was not observed in immunocompromised Nude mice, indicating its dependence on the adaptive immune responses (figure 3B). When the average tumor volume reached 1500 mm3 in the control group, CD45+ cells were isolated from harvested tumors and both the number of immune cells per milligram of tumor and the percentage of each individual immune cell population (DC, pDC, CD4+ and CD8+ T cells, monocytic and granulocytic MDSC, neutrophils, M1 and M2 macrophages) among total CD45+ cells were determined by flow cytometry in the different treatment groups. As shown in figure 3C and online supplemental figure 3E, it is interesting to notice that each syngeneic mouse model displays a specific immune environment dominated by CD8+ T cells (4T1), DC (67NR) or macrophages (EpRas and TC-1). Despite minor variations (related to models and/or drugs used), four main observations were made in treated groups compared with the controls: (1) a diminution of both monocytic and granulocytic MDSC was observed following extracellular HMGB1 inhibition, (2) without affecting the global number of macrophages, a higher M1/M2 ratio was detected in treated tumors, (3) reduced proportions of Treg cells among total CD4+/CD8+ cells and 4) an increased activation of antigen-presenting cells (DC and pDC), as demonstrated by the higher expression of CD80, CD86, I-A/I-E associated to the lower presence of ILT3 (DC) or ICOSL (pDC) (figure 3D–I, online supplemental figures 3 and 7). To evaluate whether the strong reduction of intratumoral granulocytic MDSC actively participate to the beneficial (anti-tumor) effect of HMGB1 inhibitors, depletion experiments were performed. Importantly, anti-Ly6G antibody treatment significantly reduced tumor growth, mimicking the clinical effect of extracellular HMGB1 blockade (online supplemental figure 8). Beside the immune cell infiltration, the apoptosis of cancer cells as well as angiogenesis were also assessed in retrieved tumors by quantifying cleaved caspase 3+ cells and CD31 expression (by computerized counts), respectively. Whereas no difference was observed for the density of blood vessels within tumor microenvironment (figure 3K), the number of apoptotic tumor cells was significantly higher (up to 2.2-fold increase) in treated cancers compared with control groups (figure 3J), indicating even more an enhancement of immune responses in vivo following extracellular HMGB1 blockade. Finally, no weight loss or sign of toxicity (determined by both the histological evaluation of several organs and the levels of serum urea and ALT) was observed with any of the HMGB1 inhibitors (online supplemental figure 9).
Figure 3.
Extracellular HMGB1 blockade inhibits the growth of pre-established solid tumors in immunocompetent mice through activating anticancer immune responses. (A) Mouse basal-like breast cancer cells (4T1, 67NR and EpRas) were orthotopically injected into the mammary fat pad of immunocompetent BALB/c mice. Tumor-bearing mice were then treated at 3-day intervals with PBS (control) or HMGB1 inhibitors (glycyrrhizin (1 nM/kg), RAP (10 µM/kg), a box (500 µg/kg) and EP (1 mM/kg)). The mean tumor volumes±SEM are represented. (B) HMGB1 inhibitors were tested in nude mice implanted with 67NR cells. Note the absence of beneficial effect in these latter immunocompromised mice, indicating the dependence on the adaptive immune responses. (C) At day 17, 19 or 20 (depending on the analyzed cell line), tumors were harvested, CD45+ immune cells were isolated and analyzed by flow cytometry. The proportions of each analyzed immune cell population in both control and treated groups (pooled results) are shown. Note the drastic reduction of MDSC following extracellular HMGB1 blockade. (D) Total number of (CD45+) immune cells per milligram of tumor in both control and treated groups. (E) Scatter dot plots showing the percentage of each individual immune cell population (DC, PDC, CD4+ and CD8+ T cells, monocytic and granulocytic MDSC, neutrophils, M1 and M2 macrophages) among CD45+ cells in the different treatment groups. an increased M1/M2 ratio of macrophages was observed in most HMGB1 inhibitor-treated tumors. The intratumoral immune cells were analyzed in five mice per condition. (F) Scatter dot plots illustrating the percentage of tumor-infiltrating Treg (Foxp3+) CD4+ and CD8+ cells among total CD4+ and CD8+ populations in the different treatment groups. (G) Scatter dot plots illustrating the percentage of tumor-infiltrating PD-1+ CD4+ and PD-1+ CD8+ cells among total CD4+ and CD8+ populations in the treatment groups. The activation status of both DC (H) and PDC (I) in the different treatment groups was also determined by analyzing the expression of several cell surface markers (CD80, CD86, I-A/I-E, ILT3 and ICOSL). Data represent the mean fluorescent intensity (MFI)±SEM of 5 independent experiments in each group (each individual data point is shown). The number of apoptotic cancer cells (cleaved caspase 3+) (J) as well as the density of blood vessels within tumor microenvironment (CD31+) (K) were determined by computerized counting (using QuPath software). The number of cleaved caspase 3+ cells and the percentage of CD31+ pixels were reported to tumor area. The scale bar represents 100 µm. Asterisks indicate statistically significant differences (*p<0.05; **p<0.01; ***p<0.001). P values were determined using one-way ANOVA followed by Dunnett’s multiple comparison post-test (A, B, D, E, F, G, H, I) and (Welch-corrected) unpaired t-test (J, K). ANOVA, analysis of variance; DC, dendritic cell; EP, ethyl pyruvate; HMGB1, high-mobility group box 1; i.p, intraperitoneal; MDSC, myeloid-derived suppressor cells; pDC, plasmacytoid DC; RAP, RAGE antagonist peptide.
jitc-2020-001966supp009.pdf (2.1MB, pdf)
jitc-2020-001966supp010.pdf (221.8KB, pdf)
jitc-2020-001966supp011.pdf (855.6KB, pdf)
Extracellular HMGB1 blockade enhances therapeutic effectiveness of anti-PD-1 antibody in immunocompetent mice
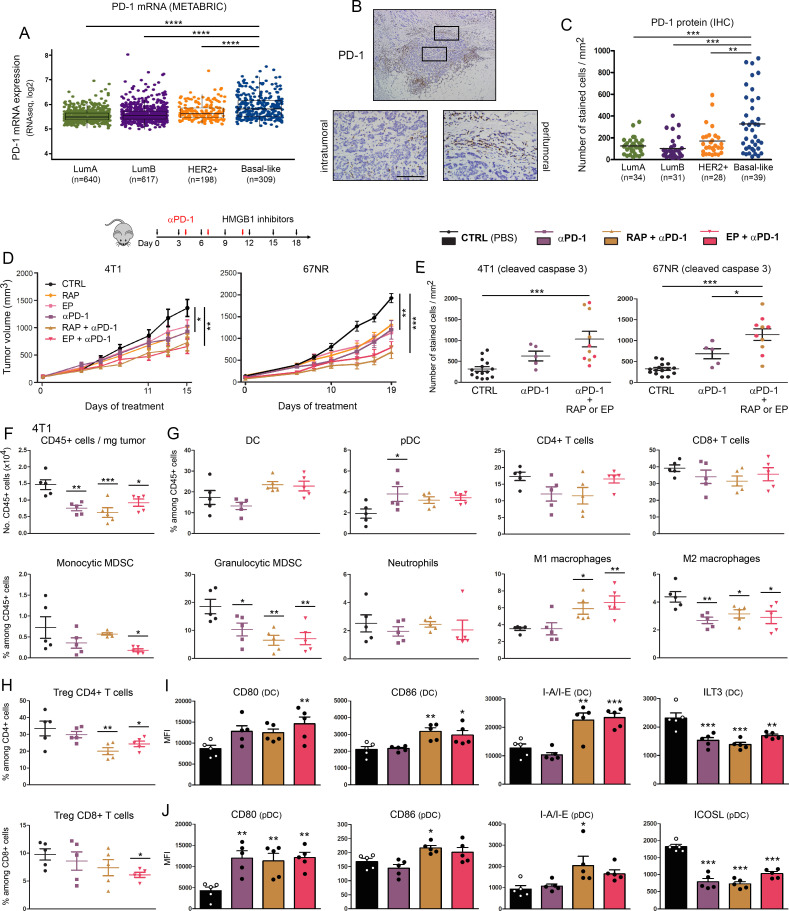
The expression of the immune checkpoint receptor PD-1 and its ligand PD-L1 was first assessed in human breast specimens as well as in our syngeneic mouse models (4T1 and 67NR). As shown in figures 4A–C and 5A–D, both the mRNA and protein levels of PD-1/PD-L1 were significantly increased in triple negative/basal-like cancers compared with the other molecular breast cancer subtypes. Due to the absence of PD-L1 immunoreactivity in a substantial proportion of human samples (figure 5C, D), the reported differences were less evident with this latter protein compared with PD-1. Importantly, the cell surface expression of both proteins was revealed in harvested (untreated) 4T1/67NR tumors by flow cytometry (figures 3G and 5E), supporting the adequacy of our in vivo models for analyzing the anti-cancer efficacy of HMGB1 blockade and anti-PD-1/PD-L1 combination therapy. Detected on tumor and/or immune cells, it is interesting to notice that 4T1 and 67NR tumors displayed distinct PD-L1 expression profiles (mainly on inflammatory cells: 4T1 vs on cancer cells: 67NR) (figure 5E). As shown in figure 4D, E and online supplemental figure 10, the combination of anti-PD-1 antibody and HMGB1 inhibition (RAP and EP) was associated with increased numbers of apoptotic tumor cells as well as significantly higher tumor growth inhibitions compared with anti-PD-1 alone (49.5% vs 33% (4T1, p=0.12) and 61.5% vs 34% (67NR, p<0.05)) or each HMGB1 inhibitor used in monotherapy [49.5% vs 30% (4T1, p<0.05) and 61.5% vs 35% (67NR, p<0.01)]. As illustrated by these two latter parameters (figures 3A–D and 4D, E), anti-PD-1 and extracellular HMGB1 blockade displayed close efficacy in our immune-competent mouse models. Although the significance was not reached in all conditions, the addition of one HMGB1 inhibitor to anti-PD-1 antibody also resulted in a higher percentage of M1 macrophages, a reduction of intratumoral Foxp3+ Treg cells and an increased activation of DC/pDC (figure 4G–J and online supplemental figure 11), suggesting even more that HMGB1 blockade boosts anti-PD1-mediated enhancement of (T cell) immune responses. Due to the strong antitumor effect of anti-PD-L1 monotherapy in our models, the benefit of the combination therapy was less marked (tumor growth inhibition observed with 4T1 tumor-bearing mice: 45% (anti-PD-L1) vs 58% (anti-PD-L1 plus RAP or EP)) or absent (67NR: 64% (anti-PD-L1) vs 59% (combination of anti-PD-L1 and HMGB1 inhibition)) (figure 5F–L, (online supplemental figures 10 and 12). Although anti-PD-L1 displayed impressive therapeutic effectiveness (with an elimination below palpable detection observed in 3 mice-bearing 67NR tumors], anti-PD-L1 treatment, however, was associated with rapid and fatal hypersensitivity reactions in ~30% of both 4T1 and 67NR tumor-bearing mice. Observed with anti-PD-1 as well, these latter reactions were mainly observed within 20–30 min after the third administration. As previously published,51 one extra dose caused the mortality of virtually all animals (data not shown).
Figure 4.
Extracellular HMGB1 blockade enhances anti-PD-1-induced inhibition of tumor growth in vivo. (A) PD-1 mRNA expression (PDCD1 gene) in the four major molecular subtypes of breast cancer was determined using the METABRIC public dataset. (B) Representative example of breast cancer stained for PD-1. Positive cells were observed in the epithelial component of the tumor as well as in the stroma surrounding cancer cells. (C) PD-1+ cell infiltration within tumor microenvironment was determined by computerized counting. Each point represents the number of positive cells/mm2 for one independent tumor specimen. (D) Mouse breast cancer cells (4T1 and 67NR) were orthotopically injected into the mammary fat pad of immunocompetent BALB/c mice. Anti-PD-1 antibody was tested alone (i.p. injection of 200 µg at days 4, 7 and 11) and in combination with HMGB1 inhibitors (RAP (10 µM/kg) and EP (1 mM/kg), treatment at 3 day intervals). In parallel, the anticancer efficacy of these combination regimens was also compared with that displayed by each individual HMGB1 inhibitor used in monotherapy. The mean tumor volumes±SEM are represented. (E) The apoptotic cancer cells (cleaved caspase 3+) were detected by immunohistochemistry and quantified using QuPath software. The number of positive cells was reported to tumor area (mm2). (F) The total number of (CD45+) immune cells per milligram of tumor was determined in the different treatment groups. (G) Scatter dot plots illustrating the percentage of each individual immune cell population (DC, PDC, CD4+ and CD8+ T cells, monocytic and granulocytic MDSC, neutrophils, M1 and M2 macrophages) among CD45+ cells in both control and treated groups. Reduced densities of granulocytic MDSC as well as an increase of M1 macrophages were especially observed in case of combination therapy. The intratumoral immune cells were analyzed in five mice per condition. (H) Scatter dot plots showing the percentage of tumor-infiltrating Treg (Foxp3+) CD4+ and CD8+ cells among total CD4+ and CD8+ populations in the different treatment groups. the activation status of DC (I) and pDC (J) was determined by flow cytometry. the expression of several surface markers (CD80, CD86, I-A/I-E, ILT3 and ICOSL) was analyzed. Data represent the mean fluorescent intensity (MFI)±SEM of 5 independent experiments in each group (each individual data point is shown). The scale bar represents 100 µm. Asterisks indicate statistically significant differences (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001). P values were determined using one-way ANOVA followed by Bonferroni post-test (A, C, E) or Dunnett’s multiple comparison post-test (D, F, G, H, I, J). ANOVA, analysis of variance; DC, dendritic cell; HMGB1, high-mobility group box 1; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; MDSC, myeloid-derived suppressor cells; pDC, plasmacytoid DC; RAP, RAGE antagonist peptide.
Figure 5.
Combination of anti-PD-L1 with HMGB1 inhibitors strongly inhibits tumor growth in syngeneic mouse models of basal-like breast cancer. (A) mRNA level of PD-L1 (CD274 gene) in the four major molecular subtypes of breast cancer was determined using the METABRIC public dataset. (B) Representative example of breast cancer stained for PD-L1. Positive signals were detected on cancer cells and/or on inflammatory cells within tumor microenvironment. Semiquantitative evaluation of PD-L1 immunoreactivity (negative or >1% membrane staining) displayed by cancer cells (C) or inflammatory cells infiltrating the tumor microenvironment (D). The analyzed cancer specimens were categorized into the four molecular subtypes of breast cancer (LumA, LumB, HER2+ and basal-like). (E) The percentage of PD-L1+ cells in both epithelial/cancer (CD45-) and inflammatory (CD45+) components of untreated harvested 4T1/67NR tumors was determined by flow cytometry. Note the distinct profile displayed by these two cell lines. (F) Mouse breast cancer cells (4T1 and 67NR) were orthotopically injected into the mammary fat pad of immunocompetent BALB/c mice. Anti-PD-L1 antibody was tested alone (i.p. injection of 100 µg at days 4, 7 and 11) and in combination with HMGB1 inhibitors (RAP (10 µM/kg) and EP (1 mM/kg), treatment at 3-day intervals). The mean tumor volumes±SEM are represented. (G) The total number of (CD45+) immune cells per milligram of tumor was determined in the different treatment groups by flow cytometry. (H) Scatter dot plots illustrating the percentage of each individual immune cell population (DC, PDC, CD4+ and CD8+ T cells, monocytic and granulocytic MDSC, neutrophils, M1 and M2 macrophages) among CD45+ cells in both control and treated groups. The intratumoral immune cell infiltration was analyzed in five mice per condition. (I) scatter dot plots showing the percentage of tumor-infiltrating Treg (Foxp3+) CD4+ and CD8+ cells among total CD4+ and CD8+ populations in the different treatment groups. The activation status of DC (J) and pDC (K) was determined by flow cytometry. the expression of several surface markers (CD80, CD86, I-A/I-E, ILT3 and ICOSL) was assessed. Data represent the mean fluorescent intensity (MFI)±SEM of five independent experiments in each group (each individual data point is shown). (L) The apoptotic cancer cells (cleaved caspase 3+) were detected by immunohistochemistry and quantified using QuPath software. The number of positive cells was reported to tumor area (mm2). The scale bar represents 100 µm. Asterisks indicate statistically significant differences (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001). P values were determined using one-way ANOVA, followed by Bonferroni post-test (A, L), Fisher’s exact test (C, D) and one-way ANOVA followed by Dunnett’s multiple comparison post-test (F, G, H, I, J, K). ANOVA, analysis of variance; DC, dendritic cell; EP, ethyl pyruvate; HMGB1, high-mobility group box 1; i.p, intraperitoneal; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; pDC, plasmacytoid DC; RAP, RAGE antagonist peptide.
jitc-2020-001966supp012.pdf (217.5KB, pdf)
jitc-2020-001966supp013.pdf (621.3KB, pdf)
jitc-2020-001966supp014.pdf (602.3KB, pdf)
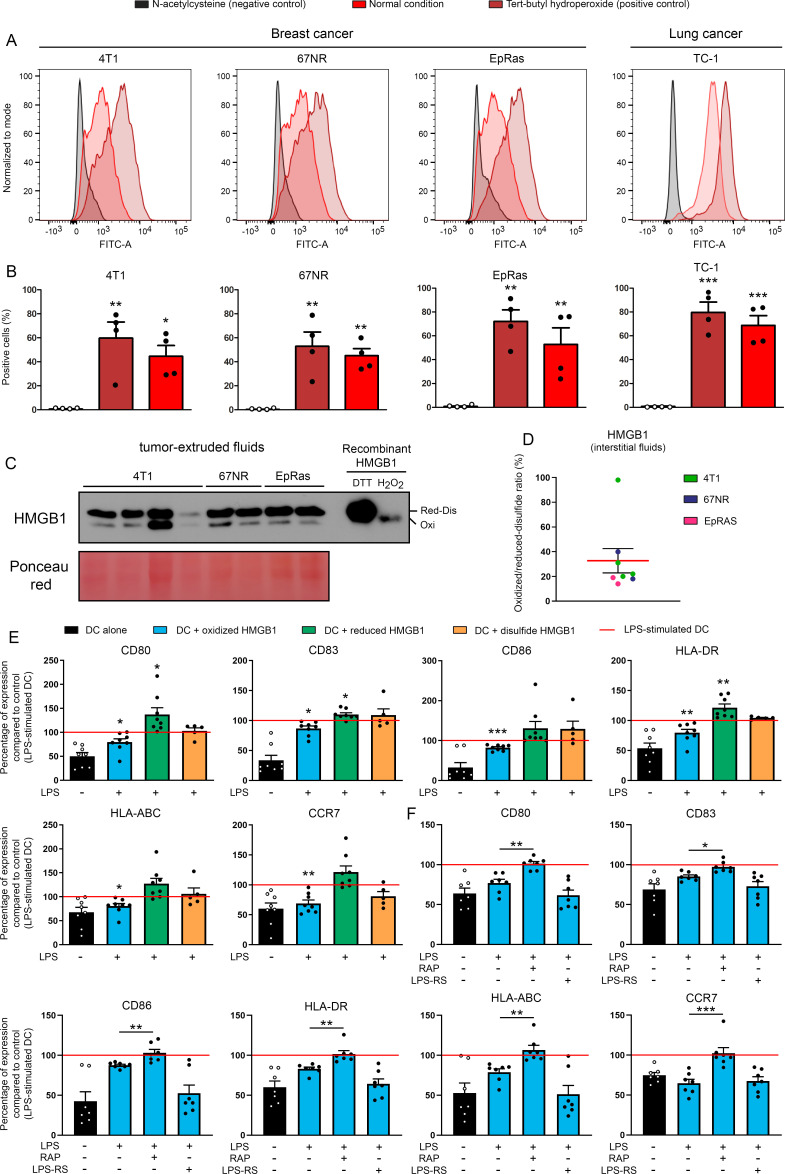
Oxidized HMGB1 is encountered in cancer microenvironment (interstitial fluids) and induces DC tolerogenicity in a RAGE-dependent manner
To further characterize the immunosuppressive activity of HMGB1 during cancer progression, the redox state as well as the impact on DC/pDC maturation of each HMGB1 isoform detected within the tumor extracellular compartment were determined. Causing the oxidation of different types of macromolecules (eg, proteins, DNA and lipids), we first measured the production of mitochondrial ROS by both breast (4T1, 67NR, EpRas) and lung (TC-1) cancer cells. Consistent with the long-time observations that most tumors cells display an altered redox status,52 the malignant cells used in this study have been shown to represent a major source of ROS (figure 6A, B). Using a novel innovative methodology (called EXPEL),43 soluble proteins contained in tumor-extruded fluids were retrieved and directly alkylated in order to maintain their original redox state. As shown in figure 6C, D, all HMGB1 forms are encountered within tumor microenvironment with an approximate 2:1 reduced-disulfide/oxidized ratio. However, it is important to notice that the proportion of oxidized HMGB1 is very likely underestimated by the lower sensitivity displayed by the anti-HMGB1 antibody for this latter redox form (as indicated by control bands). To precisely determine the effect of each individual form of HMGB1 on antigen-presenting cell phenotype/functional activity, human DC were incubated with oxidized, reduced or disulfide HMGB1 for 24 hours and then stimulated for maturation (LPS during 24 hours). While disulfide HMGB1 displayed no activity on DC phenotype, the reduced and oxidized forms significantly induced the activation and tolerogenicity of DC, respectively (figure 6E). Similar results were obtained with pDC generated from human CD34+ hematopoietic progenitor cells (online supplemental figure 13). Importantly, the tolerogenic properties of oxidized HMGB1 were significantly blocked by the inhibition of RAGE, leading to the complete restoration of DC activation (figure 6F). In contrast, TLR4 did not contribute to DC functional impairment in the presence of oxidized HMGB1 (figure 6F).
Figure 6.
A significant fraction of HMGB1 contained in tumor-extruded fluids is in its oxidized form and displays RAGE-dependent tolerogenic properties. (A, B) The ROS accumulation in both breast (4T1, 67NR, EpRas) and lung (TC-1) cancer cells used in the present study was assessed by flow cytometry. N-acetylcysteine (5 mM) and tert-butyl hydroperoxide (100 µM) were used as negative and positive controls, respectively. Results represent the means±SEM of four independent experiments (each individual data point is shown). (C) The redox state of extracellular HMGB1 contained in tumor-extruded fluids was analyzed by Western blot. All samples were directly alkylated in order to ‘freeze’ the redox state of HMGB1 molecules. Recombinant HMGB1 (0.5 µg) incubated with either H2O2 or DTT (and then alkylated) were used as controls. (D) Oxidized/reduced-disulfide HMGB1 ratio (%) was calculated from the Western blot bands using ImageJ software. (E) DCs were incubated with terminally oxidized, fully reduced or disulfide HMGB1 for 24 hours before being stimulated with LPS for 24 hours. The expression of cell-surface molecules (CD80, CD83, CD86, HLA-DR, HLA-ABC and CCR7) was then measured by flow cytometry. All data were normalized to LPS-stimulated DC. Data represent the relative mean fluorescent intensity (MFI)±SEM of at least five independent experiments (each individual data point is shown). (F) DCs were incubated with terminally oxidized HMGB1 for 24 hours before being stimulated with LPS for 24 hours. When indicated, an inhibitor of RAGE (10 µM RAP) or TLR4 (2 µM LPS-RS) was added in the cell culture. The expression of DC activation markers was determined by flow cytometry. All data were normalized to LPS-stimulated DC. The relative MFI±SEM of 7 independent experiments are shown. Asterisks indicate statistically significant differences (*p<0.05, **p<0.01, ***p<0.001). P values were determined using one-way ANOVA, followed by Dunnett’s multiple comparison post-test (A, B, E, F). ANOVA, analysis of variance; DC, dendritic cell; HMGB1, high-mobility group box 1.
jitc-2020-001966supp015.pdf (278.9KB, pdf)
Discussion
The role of HMGB1 in cancer progression has been a matter of debate for over ten years.23 In this context, quite a few theoretical and translational questions remained unanswered. Does HMGB1 expression affect patient outcome? Is HMGB1 a promising target for cancer therapy or, in contrast, should we promote its cellular release? Does extracellular HMGB1 positively or negatively impact antitumor immune responses? Which redox form is detected in cancer microenvironment? In the present study, we first reported that active HMGB1 secretion/cytoplasmic immunoreactivity is both a specific trait of malignant cells (not detected in normal conditions) and a predictor for unfavorable prognosis in patients with locally advanced cancer. In an era of large-scale transcriptome analyses, it is interesting to notice that next-generation sequencing results did not highlight the predictive value of HMGB1 due to their unability to discriminate dual effects/locations of a same target. Lacking a leader sequence, HMGB1 has been shown to be actively secreted through lysosome-mediated exocytosis following its JAK/STAT1-regulated nuclear-cytoplasmic translocation.53 54 Nevertheless, the upstream components of this non-classical secretory pathway are still largely unknown as well as the reasons why some cancer (sub)types highly secrete HMGB1 (and display large areas of HMGB1 cytoplasmic immunoreactivity) while others do not. These latter points certainly merit further investigations given the clear protumor activity of HMGB1 demonstrated here in vivo.
Using several syngeneic mouse models of basal-like or lung cancer, both direct and indirect extracellular HMGB1 blockade significantly resulted in tumor growth inhibition. In each case, the antitumor effect of HMGB1 inhibitors was only detected 7–10 days following the first injection. This latency period very likely corresponds to the necessary time for the adaptive immune responses to be remobilized. In agreement, the dependence on adaptive immunity was distinctly demonstrated by the absence of anticancer effect when immunedeficient mice were used. Besides glycyrrhizin, RAP, A box and EP, a purified rabbit anti-HMGB1 antibody was also tested (online supplemental figure 14). Despite an in vitro neutralizing efficiency comparable to other HMGB1 inhibitors, and while a protective effect against sepsis lethality has previously been demonstrated,33 the administration of an anti-HMGB1 antibody produced in rabbit was inefficient for reducing tumor growth. The different posologies/study designs used in our orthotopic models of cancer (injection of anti-HMGB1 antibody at 3-day intervals for 18 days) and those of murine sepsis (administration twice daily for 3 days and then monitoring for 10 days) are susceptible to explain the divergent results. Indeed, we cannot exclude that repeated doses for 3 weeks led to the generation of mouse antibodies against foreign epitopes, blocking ultimately the function of injected rabbit anti-HMGB1 antibody and/or shortening its half-life. Supporting this hypothesis, tumor-bearing mice were treated with a mouse anti-HMGB1 antibody and an important tumor growth inhibition (similar to that reported with the other tested HMGB1 inhibitors) was observed (online supplemental figure 14).
jitc-2020-001966supp016.pdf (375.5KB, pdf)
Without affecting the global number of CD45+ cells (arguing against an ‘alarmin’ role of HMGB1 in cancer), we found that extracellular HMGB1 blockade alone elicited a drastic remodeling of tumor immune microenvironment. Overall, reduced proportions of immunosuppressive cells (especially MDSC but also M2 macrophages and Treg cells) associated with an increased activation of antigen-presenting cells (DC and pDC) were observed, indicating that targeting HMGB1 therapeutically enhances significantly local antitumor immune responses. Although the effector mechanism of HMGB1 inhibitors involves very likely several immune cell subtypes, the key role played by the reduction of intratumoral granulocytic MDSC was clearly demonstrated in anti-Ly6G-mediated depletion experiments. Confirming our observations on human samples (lower densities of both Foxp3+ and CD206+ cells within tumor microenvironment of nuclear HMGB1-positive cancers compared with their counterparts displaying a cytoplasmic immunoreactivity), the improvement/activation of T cell immune response following HMGB1 inhibition was further supported by the increased number of apoptotic (cleaved caspase 3+) tumor cells in treated cancers compared with control groups. Although the vast majority of studies focusing on HMGB1 did not pay attention to its redox state, it is intriguing to notice that, when an immune suppression (through promoting tolerogenic functionalities by DC/pDC, enhancing suppressive activities of MDSC or stimulating M2 polarization of macrophages) was reported in vitro,55–57 the authors consistently used recombinant HMGB1 produced in cancer cell lines (g, myeloma (NS0)) and not in bacteria. Therefore, as extensively discussed below, it is reasonable to think that cysteine residues within the protein were mostly oxidized due to the high endogenous oxidative stress displayed by almost all tumors.
Given the numerous immunosuppressive molecules which can be encountered within tumor microenvironment, it is unrealistic to believe that treatment regimens using only one drug (eg, humanized anti-PD-1 or PD-L1 antibody) will display therapeutic efficacy in all patients with cancer. Therefore, both treatment personalization and the combinatorial targeting of several tumor-induced immune defects make sense intuitively for improving the efficacy of future therapies. Here, we showed that concurrent HMGB1 inhibition plus anti-PD-1 antibody resulted in significantly higher tumor growth inhibitions than did anti-PD-1 alone. Similarly to most studies analyzing the efficacy of immune checkpoint inhibitors in preclinical mouse models, it is important to emphasize that a quite high interindividual variability in anti-PD-1 therapeutic response was observed (online supplemental figure 10). Although still not completely understood, this latter phenomenon is very likely multifactorial (eg, differences in gut microbiota, upregulation/downregulation of the targeted proteins during treatment, expression of other immunosuppressive factors constituting compensatory mechanisms). Regarding the combination therapy using anti-PD-L1 antibody, the benefit was relatively modest (with 4T1 tumor cells) or absent (67NR). Displaying over 20% PD-L1+ inflammatory and/or cancer cells, the ‘super responder’ profile of both models to anti-PD-L1 monotherapy is likely to explain these latter results. Altogether, the oversecretion of HMGB1 by cancer cells, its immunosuppressive/protumor activity as well as both the absence of toxicity and the long-standing use of some HMGB1 inhibitors in the context of other diseases (eg, glycyrrhizin in chronic hepatitis C patients)58 59 make it a very promising target for cancer treatment. Furthermore, blocking extracellular HMGB1 with humanized antibodies or antagonists is unlikely to alter the beneficial nuclear functions (replication, DNA repair, genome stability) of the protein.
It is well established that tumor cells produce higher levels of ROS compared with normal/non-transformed cells. Still commonly called ‘free radicals’, these oxygen-containing chemically reactive molecules cause the oxidation of proteins, DNA and lipids, promoting ultimately many aspects of cancer progression.60 61 To the best of our knowledge, this report provides the first evidence that HMGB1 is encountered on its oxidized form in cancer microenvironment/interstitial fluids (at least one third of total extracellular HMGB1) and acts as a tolerogenic signal on antigen-presenting cells in a RAGE-dependent manner. Based on our in vivo results, there is little doubt that the overall effect of HMGB1 in cancer is largely dictated by this latter form. Interestingly, without having identified the involved receptor, the induction of immunological tolerance by oxidized HMGB1 was previously proposed in the context of programmed cell death (apoptosis).11 However, by lack of use of in vitro/in vivo models allowing the assessment of acquired immunotolerance development, most authors are still only focusing on the chemotactic/proinflammatory activities of HMGB1, and therefore, are considering its oxidized form as inactive.
In case of injury or pathogen infection, as a damage-associated molecular pattern molecule, extracellular reduced/disulfide HMGB1 stimulates innate immune cell chemotaxis and activation, mediating immunogenic cell death and triggering acute inflammatory responses. However, within the highly oxidative cancer microenvironment, the dark side of HMGB1 is revealed. Similarly to other secreted immunosuppressive molecules, the oxidation of HMGB1 is likely to be essential during the inflammation resolution but the persistence/chronicity of its tolerogenic activity is noxious and could be therapeutically targeted.
Acknowledgments
The authors thank the Biobank of the University of Liege as well as the GIGA-Immunohistochemistry and in vitro imaging facilities (University of Liege) for their assistance. We are also grateful to Dr Stephanie Gofflot, Raphael Thonon, Kamilia El Kandoussi and Laurence Poma for their technical assistance.
Footnotes
Contributors: PH and MH designed the study; PH, PR, SD, CP, MA, CR, TL, DB, AL, CR, MM, EH, OP and MH performed experiments; PH, SD, M-JN, OP and MH interpreted the data; PD provided resources; PH and MH generated the figures; MH wrote the manuscript. All authors discussed the results and commented on the manuscript.
Funding: This work was supported in part by the University of Liege (Fonds spéciaux de la recherche and credits sectoriels), the Belgian Fund for Scientific Research (FNRS, MIS F.4520.20), the Televie (PDR 7.8507.19 and 7.4577.19), the Leon Fredericq Foundation and the Belgian Foundation against Cancer.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: All procedures and animal experiments were initially reviewed and approved by the institutional ethics committee of the University of Liege (#13-1572). All cancer specimens used in the present study were retrieved from the Tissue Biobank of the University Hospital Center of Liege (Belgium) with the approval of the local ethics committee (#2015-68).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as online supplemental information. Any further information about resources and reagents should be directly requested to the corresponding author and will be fulfilled on reasonable request.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem 1973;38:14–19. 10.1111/j.1432-1033.1973.tb03026.x [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Bloom O, Zhang M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999;285:248–51. 10.1126/science.285.5425.248 [DOI] [PubMed] [Google Scholar]
- 3.Pullerits R, Jonsson I-M, Verdrengh M, et al. High mobility group box chromosomal protein 1, a DNA binding cytokine, induces arthritis. Arthritis Rheum 2003;48:1693–700. 10.1002/art.11028 [DOI] [PubMed] [Google Scholar]
- 4.Popovic K, Ek M, Espinosa A, et al. Increased expression of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in skin lesions of patients with lupus erythematosus. Arthritis Rheum 2005;52:3639–45. 10.1002/art.21398 [DOI] [PubMed] [Google Scholar]
- 5.Zong M, Bruton JD, Grundtman C, et al. TLR4 as receptor for HMGB1 induced muscle dysfunction in myositis. Ann Rheum Dis 2013;72:1390–9. 10.1136/annrheumdis-2012-202207 [DOI] [PubMed] [Google Scholar]
- 6.Park JS, Svetkauskaite D, He Q, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem 2004;279:7370–7. 10.1074/jbc.M306793200 [DOI] [PubMed] [Google Scholar]
- 7.Andersson U, Wang H, Palmblad K, et al. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J Exp Med 2000;192:565–70. 10.1084/jem.192.4.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, Chen Q, Yang H, et al. High mobility group box-1 protein induces the migration and activation of human dendritic cells and acts as an alarmin. J Leukoc Biol 2007;81:59–66. 10.1189/jlb.0306180 [DOI] [PubMed] [Google Scholar]
- 9.Yang S, Xu L, Yang T, et al. High-mobility group box-1 and its role in angiogenesis. J Leukoc Biol 2014;95:563–74. 10.1189/jlb.0713412 [DOI] [PubMed] [Google Scholar]
- 10.Fang P, Schachner M, Shen Y-Q. HMGB1 in development and diseases of the central nervous system. Mol Neurobiol 2012;45:499–506. 10.1007/s12035-012-8264-y [DOI] [PubMed] [Google Scholar]
- 11.Kazama H, Ricci J-E, Herndon JM, et al. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity 2008;29:21–32. 10.1016/j.immuni.2008.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Lundbäck P, Ottosson L, et al. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol Med 2012;18:250–9. 10.2119/molmed.2011.00389 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Venereau E, Casalgrandi M, Schiraldi M, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med 2012;209:1519–28. 10.1084/jem.20120189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Hreggvidsdottir HS, Palmblad K, et al. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc Natl Acad Sci U S A 2010;107:11942–7. 10.1073/pnas.1003893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Ni J, Liu K, et al. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res 2012;72:230–8. 10.1158/0008-5472.CAN-11-2001 [DOI] [PubMed] [Google Scholar]
- 16.Yan W, Chang Y, Liang X, et al. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology 2012;55:1863–75. 10.1002/hep.25572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv D-J, Song X-L, Huang B, et al. Hmgb1 promotes prostate cancer development and metastasis by interacting with Brahma-related gene 1 and activating the Akt signaling pathway. Theranostics 2019;9:5166–82. 10.7150/thno.33972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S, Cheng J, Sun L, et al. HMGB1 released by irradiated tumor cells promotes living tumor cell proliferation via paracrine effect. Cell Death Dis 2018;9:648. 10.1038/s41419-018-0626-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng H, Chen J-N, Yu X, et al. Hmgb1 enhances drug resistance and promotes in vivo tumor growth of lung cancer cells. DNA Cell Biol 2016;35:622–7. 10.1089/dna.2016.3360 [DOI] [PubMed] [Google Scholar]
- 20.Li B, Song T-N, Wang F-R, et al. Tumor-derived exosomal HMGB1 promotes esophageal squamous cell carcinoma progression through inducing PD1+ TAM expansion. Oncogenesis 2019;8:17. 10.1038/s41389-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki T, Hannani D, Poirier-Colame V, et al. Defective immunogenic cell death of HMGB1-deficient tumors: compensatory therapy with TLR4 agonists. Cell Death Differ 2014;21:69–78. 10.1038/cdd.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 2010;29:482–91. 10.1038/onc.2009.356 [DOI] [PubMed] [Google Scholar]
- 23.Kang R, Zhang Q, Zeh HJ, et al. Hmgb1 in cancer: good, bad, or both? Clin Cancer Res 2013;19:4046–57. 10.1158/1078-0432.CCR-13-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open 2019;2:e192535. 10.1001/jamanetworkopen.2019.2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Li M, Jiang Z, et al. A comprehensive immunologic portrait of triple-negative breast cancer. Transl Oncol 2018;11:311–29. 10.1016/j.tranon.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanda R, Chow LQM, Dees EC, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol 2016;34:2460–7. 10.1200/JCO.2015.64.8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dirix LY, Takacs I, Jerusalem G, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1B javelin solid tumor study. Breast Cancer Res Treat 2018;167:671–86. 10.1007/s10549-017-4537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zitvogel L, Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 2012;1:1223–5. 10.4161/onci.21335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 30.Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med 2018;379:2108–21. 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 31.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res 2019;38:255. 10.1186/s13046-019-1259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrimali RK, Ahmad S, Verma V, et al. Concurrent PD-1 blockade negates the effects of OX40 agonist antibody in combination immunotherapy through inducing T-cell apoptosis. Cancer Immunol Res 2017;5:755–66. 10.1158/2326-6066.CIR-17-0292 [DOI] [PubMed] [Google Scholar]
- 33.Yang H, Ochani M, Li J, et al. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc Natl Acad Sci U S A 2004;101:296–301. 10.1073/pnas.2434651100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boivin G, Faget J, Ancey P-B, et al. Durable and controlled depletion of neutrophils in mice. Nat Commun 2020;11:2762. 10.1038/s41467-020-16596-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herfs M, Herman L, Hubert P, et al. High expression of PGE2 enzymatic pathways in cervical (pre)neoplastic lesions and functional consequences for antigen-presenting cells. Cancer Immunol Immunother 2009;58:603–14. 10.1007/s00262-008-0584-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herfs M, Vargas SO, Yamamoto Y, et al. A novel blueprint for 'top down' differentiation defines the cervical squamocolumnar junction during development, reproductive life, and neoplasia. J Pathol 2013;229:460–8. 10.1002/path.4110 [DOI] [PubMed] [Google Scholar]
- 37.Herfs M, Roncarati P, Koopmansch B, et al. A dualistic model of primary anal canal adenocarcinoma with distinct cellular origins, etiologies, inflammatory microenvironments and mutational signatures: implications for personalised medicine. Br J Cancer 2018;118:1302–12. 10.1038/s41416-018-0049-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruyere D, Monnien F, Colpart P, et al. Treatment algorithm and prognostic factors for patients with stage I-III carcinoma of the anal canal: a 20-year multicenter study. Mod Pathol 2021;34:116–30. 10.1038/s41379-020-0637-6 [DOI] [PubMed] [Google Scholar]
- 40.Kim K, Watson PA, Lebdai S, et al. Androgen deprivation therapy potentiates the efficacy of vascular targeted photodynamic therapy of prostate cancer xenografts. Clin Cancer Res 2018;24:2408–16. 10.1158/1078-0432.CCR-17-3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demoulin SA, Somja J, Duray A, et al. Cervical (pre)neoplastic microenvironment promotes the emergence of tolerogenic dendritic cells via RANKL secretion. Oncoimmunology 2015;4:e1008334. 10.1080/2162402X.2015.1008334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demoulin S, Roncarati P, Delvenne P, et al. Production of large numbers of plasmacytoid dendritic cells with functional activities from CD34(+) hematopoietic progenitor cells: use of interleukin-3. Exp Hematol 2012;40:268–78. 10.1016/j.exphem.2012.01.002 [DOI] [PubMed] [Google Scholar]
- 43.Costanza B, Turtoi A, Bellahcène A, et al. Innovative methodology for the identification of soluble biomarkers in fresh tissues. Oncotarget 2018;9:10665–80. 10.18632/oncotarget.24366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis C, Shah SP, Chin S-F, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira B, Chin S-F, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rademaker G, Hennequière V, Brohée L, et al. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene 2018;37:4398–412. 10.1038/s41388-018-0287-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arumugam T, Ramachandran V, Gomez SB, et al. S100P-derived RAGE antagonistic peptide reduces tumor growth and metastasis. Clin Cancer Res 2012;18:4356–64. 10.1158/1078-0432.CCR-12-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mollica L, De Marchis F, Spitaleri A, et al. Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem Biol 2007;14:431–41. 10.1016/j.chembiol.2007.03.007 [DOI] [PubMed] [Google Scholar]
- 49.Davé SH, Tilstra JS, Matsuoka K, et al. Ethyl pyruvate decreases HMGB1 release and ameliorates murine colitis. J Leukoc Biol 2009;86:633–43. 10.1189/jlb.1008662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim YM, Park EJ, Kim JH, et al. Ethyl pyruvate inhibits the acetylation and release of HMGB1 via effects on SIRT1/STAT signaling in LPS-activated RAW264.7 cells and peritoneal macrophages. Int Immunopharmacol 2016;41:98–105. 10.1016/j.intimp.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 51.Mall C, Sckisel GD, Proia DA, et al. Repeated PD-1/PD-L1 monoclonal antibody administration induces fatal xenogeneic hypersensitivity reactions in a murine model of breast cancer. Oncoimmunology 2016;5:e1075114. 10.1080/2162402X.2015.1075114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res 1991;51:794–8. [PubMed] [Google Scholar]
- 53.Lu B, Antoine DJ, Kwan K, et al. Jak/Stat1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc Natl Acad Sci U S A 2014;111:3068–73. 10.1073/pnas.1316925111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardella S, Andrei C, Ferrera D, et al. The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep 2002;3:995–1001. 10.1093/embo-reports/kvf198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demoulin S, Herfs M, Somja J, et al. Hmgb1 secretion during cervical carcinogenesis promotes the acquisition of a tolerogenic functionality by plasmacytoid dendritic cells. Int J Cancer 2015;137:345–58. 10.1002/ijc.29389 [DOI] [PubMed] [Google Scholar]
- 56.Parker KH, Sinha P, Horn LA, et al. Hmgb1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res 2014;74:5723–33. 10.1158/0008-5472.CAN-13-2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rojas A, Delgado-López F, Perez-Castro R, et al. Hmgb1 enhances the protumoral activities of M2 macrophages by a RAGE-dependent mechanism. Tumour Biol 2016;37:3321–9. 10.1007/s13277-015-3940-y [DOI] [PubMed] [Google Scholar]
- 58.Arase Y, Ikeda K, Murashima N, et al. The long term efficacy of glycyrrhizin in chronic hepatitis C patients. Cancer 1997;79:1494–500. [DOI] [PubMed] [Google Scholar]
- 59.Ikeda K. Glycyrrhizin injection therapy prevents hepatocellular carcinogenesis in patients with interferon-resistant active chronic hepatitis C. Hepatol Res 2007;37 Suppl 2:S287–93. 10.1111/j.1872-034X.2007.00199.x [DOI] [PubMed] [Google Scholar]
- 60.Liou G-Y, Storz P. Reactive oxygen species in cancer. Free Radic Res 2010;44:479–96. 10.3109/10715761003667554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aggarwal V, Tuli HS, Varol A, et al. Role of reactive oxygen species in cancer progression: molecular mechanisms and recent advancements. Biomolecules 2019;9:735. 10.3390/biom9110735 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001966supp001.pdf (104.2KB, pdf)
jitc-2020-001966supp002.pdf (89.4KB, pdf)
jitc-2020-001966supp003.pdf (958KB, pdf)
jitc-2020-001966supp004.pdf (663.5KB, pdf)
jitc-2020-001966supp005.pdf (1.1MB, pdf)
jitc-2020-001966supp006.pdf (129.1KB, pdf)
jitc-2020-001966supp007.pdf (466.4KB, pdf)
jitc-2020-001966supp008.pdf (1.6MB, pdf)
jitc-2020-001966supp009.pdf (2.1MB, pdf)
jitc-2020-001966supp010.pdf (221.8KB, pdf)
jitc-2020-001966supp011.pdf (855.6KB, pdf)
jitc-2020-001966supp012.pdf (217.5KB, pdf)
jitc-2020-001966supp013.pdf (621.3KB, pdf)
jitc-2020-001966supp014.pdf (602.3KB, pdf)
jitc-2020-001966supp015.pdf (278.9KB, pdf)
jitc-2020-001966supp016.pdf (375.5KB, pdf)