Abstract
Like RNA viruses in general, coronaviruses (CoV) exhibit high mutation rates which, in combination with their strong tendency to recombine, enable them to overcome the host species barrier and adapt to new hosts. It is currently known that six CoV are able to infect pigs. Four of them belong to the genus Alphacoronavirus [transmissible gastroenteritis coronavirus (TEGV), porcine respiratory coronavirus (PRCV), porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV)], one of them to the genus Betacoronavirus [porcine hemagglutinating encephalomyelitis virus (PHEV)] and the last one to the genus Deltacoronavirus (PDCoV). PHEV was one of the first identified swine CoV and is still widespread, causing subclinical infections in pigs in several countries. PRCV, a spike deletion mutant of TGEV associated with respiratory tract infection, appeared in the 1980s. PRCV is considered non-pathogenic since its infection course is mild or subclinical. Since its appearance, pig populations have become immune to both PRCV and TGEV, leading to a significant reduction in the clinical and economic importance of TGEV. TGEV, PEDV and PDCoV are enteropathogenic CoV and cause clinically indistinguishable acute gastroenteritis in all age groups of pigs. PDCoV and SADS-CoV have emerged in 2014 (US) and in 2017 (China), respectively. Rapid diagnosis is crucial for controlling CoV infections and preventing them from spreading. Since vaccines are available only for some porcine CoV, prevention should focus mainly on a high level of biosecurity. In view of the diversity of CoV and the potential risk factors associated with zoonotic emergence, updating the knowledge concerning this area is essential.
Keywords: Coronavirus, Pig, Emerging, Re-emerging
Introduction
Coronaviruses (CoV) are the largest positive-sense RNA viruses. They belong to the Coronaviridae family, order Nidovirales, and owe their name to their specific structure resembling a crown in an electron microscope image. CoV are responsible for a number of respiratory, digestive and nervous infections in mammals and birds. Their tendency to recombine and the inherently high mutation rates of RNA viruses allow them to occasionally transmit and adapt to new hosts and ecological niches (Herrewegh et al. 1998; Woo et al. 2006). Their prevalence in nature is very high. In pigs, the CoV causing gastrointestinal infections are the most important ones from a clinical and epidemiological standpoint. Importantly, porcine CoV infection in humans has not been confirmed yet.
To date, based on genomic criteria, four genera of CoV can be distinguished: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus (Chen et al. 2020) So far, six different CoV infecting pigs have been identified, including four belonging to the genus Alphacoronavirus [transmissible gastroenteritis coronavirus (TGEV), porcine respiratory coronavirus (PRCV), porcine epidemic diarrhea virus (PEDV) and swine acute diarrhea syndrome coronavirus (SADS-CoV)], one to the genus Betacoronavirus (porcine hemagglutinating encephalomyelitis virus (PHEV) and one to the genus Deltacoronavirus (PDCoV) (Wang et al. 2019). Among them, TGEV, PRCV and PHEV have been circulating in pigs for decades, while PEDV, PDCoV and SADS-CoV are considered newly emerging CoV. In addition, chimeric TGEV and PEDV strains have been found in Italy, Germany, Slovakia and Spain (Akimkin et al. 2016; Boniotti et al. 2016; Mandelik et al. 2018, de Nova et al. 2020). Chimeric swine enteric coronavirus (SeCoV), which is a novel recombinant between TGEV and PEDV, isolated in Italy and Germany has a similar recombination pattern and 99.5% nucleotide identity. The presence of SeCoV has not been reported in any other country. No detailed data on the importance, virulence and spread of SeCoV are available (Akimkin et al. 2016; Boniotti et al. 2016; Mandelik et al. 2018). The appearance of a new CoV in humans, SARS-CoV-2, which is attributed to zoonotic origin, holds interest of many scientists in the possibility of its occurrence and pathogenicity for domestic animals, including pigs as a host of different coronaviruses and one of the most important food-producing animals that has the potential to impact public health significantly. It has previously been found that a similar pathogen, SARS-CoV, responsible for SARS in humans, did not cause clinical symptoms or pathological lesions in pigs (Weingartl et al. 2004). Available data on the newly emerged SARS-CoV-2 also indicate that pigs are probably not susceptible to infection and do not play any role in the epidemiology of COVID-19 (Shi et al. 2020).
Porcine Respiratory Coronavirus
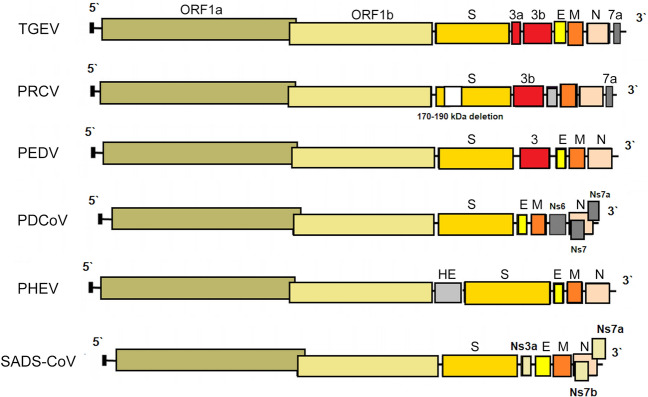
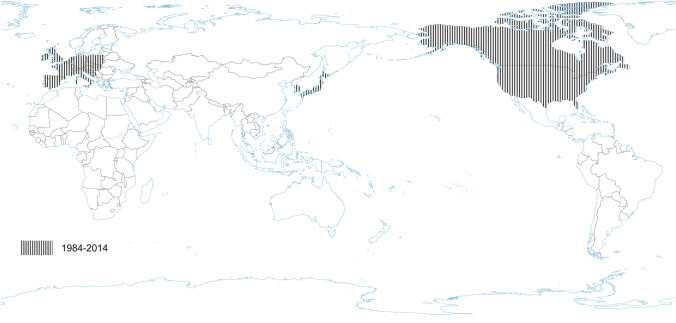
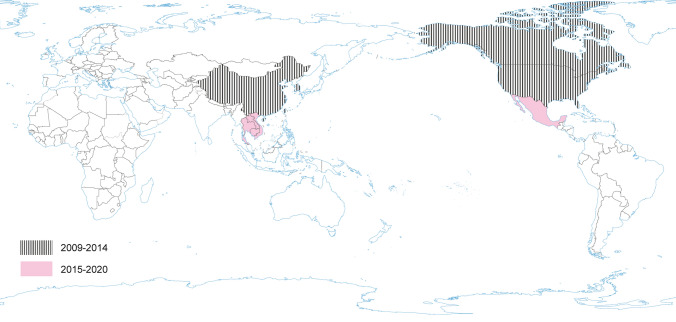
PRCV is a member of the genus Alphacoronavirus, and species Alphacoronavirus 1 (Saif et al. 2012). PRCV is common in the swine population and does not cause noticeable problems. The 5′ two thirds of the PRCV genome contain two large ORFs, 1a and 1b, that encode two nonstructural polyproteins, pp1a and pp1ab, that direct genome replication and transcription. The remaining genome contains ORFs specifying structural and non-structural proteins in the following order: spike (S), ORF 3, envelope (E), membrane (M) and nucleoprotein (N) (Bridgen et al. 1993; Duarte and Laude 1994; Vlasova et al. 2014) (Fig. 1). The genome structure described above is typical for all CoVs. It is a naturally occurring spike gene deletion (170 to 190 kDa) mutant of TGEV (Magtoto et al. 2019), described in the 1980s (Fig. 1). PRCV, unlike TGEV, has an affinity to the respiratory system. The discovery of this virus was based on a survey performed in 1984 in Belgium. It showed an increase of animals with antibodies to TGEV (up to 68%), with no increase in the incidence of TGE the following winter, in the absence of vaccination. Three years later, the virus spread to 100% of the swine farms in Belgium (Enjuanes and Van der Zeijst 1995). The PRCV has been observed in many European countries, i.e. the Netherlands, Denmark, Great Britain, Spain, and France (Enjuanes and Van der Zeijst 1995). In the United States, serological tests detected PRCV for the first time in Indiana in 1989 (Enjuanes and Van der Zeijst 1995). Since then, other PRCV isolates have been reported in the United States and Canada (Enjuanes and Van der Zeijst 1995) (Fig. 2). This virus has spread very quickly in the European pig population, without causing significant health problems. What is extremely important from the epidemiological point of view is that cross-resistance exists between PRCV and TGEV (the antibodies formed as a result of infection with PRCV protect pigs against TGEV infection), and this is probably how TGE has been eliminated and is no longer a significant problem for pig farmers in Europe (Magtoto et al. 2019; Pejsak 2007).
Fig. 1.
The genome structures of porcine coronaviruses. S—Spike S structural gene, E—envelope, M—membrane, N—nucleoprotein, HE—hemagglutinin-esterase, Ns3a, Ns6, Ns7, Ns7a, Ns7b—accessory genes.
Fig. 2.
The occurrence of PRCV. No published data in recent years.
While the parental TGEV is strongly enteric, PRCV infects the upper respiratory tract, tonsils, or lungs, with limited intestinal replication. The tropism change is presumably caused by the deletion in the spike removing the domain which in TGEV mediates binding to sialic acids (Krempl et al. 1997). It was shown in recent studies, that PRCV preferentially targets non-ciliated and among them the non-mucus-producing cells. Aminopeptidase N (APN), the cellular receptor for PRCV was also more abundantly expressed on this type of cell suggesting that APN is a determinant of the cell tropism (Peng et al. 2020). Common clinical findings of PRCV are dyspnea, tachypnea, sneezing, coughing, fever, anorexia and delayed growth (Halbur et al. 2003; Jung et al. 2009; Vannier 1990) (Table 1). Type 2 pneumocytes are the major target cells for PRCV replication in the lungs, but PRCV antigens have also been found in the cytoplasm of bronchiolar epithelial cells and macrophages. Yet, in the case of mild or subclinical infection, PRCV is a relatively good inducer of several proinflammatory cytokines, i.e. interferon (IFN)-α and interleukin (IL)-6. High levels of IFN-γ and IL-12 during the first 5–7 days post infection (DPI) have also been described (Van Gucht et al. 2006). Massive production of proinflammatory cytokines, or perhaps other factors in addition to cytokines may be necessary for the development of clinical symptoms (Atanasova et al. 2008).
Table 1.
Selected parameters characteristics for porcine coronaviruses.
| Species | Year of emergence (new-emergence) | Mortality in newborn piglets | Pathogenicity for other species | Seroconversion | Humoral immunity duration | Clinical symptoms | Tissue tropism | Vaccine |
|---|---|---|---|---|---|---|---|---|
| TGEV | 1946 | Almost 100% | Not reported | 6–7 dpi | Several months | Anorexia, diarrhea, vomiting, dehydration, weight loss, lethargy | Digestive system (intestinal epithelial cells) | Available (prosystem® tge/rota; prosystem® trec) |
| PEDV | 1971 reemerged in 2010 | Up to 100%, depending on the strain | Not reported | 10 dpi | Up to 16 weeks or more | Anorexia, diarrhea, vomiting, dehydration, weight loss, lethargy | Digestive system (intestinal epithelial cells) | Live, inactivated and subunit developed in China, Japan, Korea, US (not sufficient to control PEDV outbreaks) |
| PDCoV | 2014 initially detected in 2009 | Up to 40% | Cattle, poultry | 5–7 dpi | Six months | Anorexia, diarrhea, vomiting, dehydration, weight loss, lethargy | Digestive system (intestinal epithelial cells) | No |
| SADS-CoV | 2017 | Up to 5th day of life 90–100%; above the 8th day of life—5% | Not reported | Unknown | Unknown | Anorexia, diarrhea, vomiting, dehydration, weight loss, lethargy | Digestive system (intestinal epithelial cells) | No |
| PRCV | 1984 | Negligible | Not reported | Around 6 dpi | Around 1 month | Dyspnea, tachypnea, sneezing, coughing, fever, anorexia and delayed growth | Respiratory tract (epithelial cells), tonsil, lung, limited intestinal replication | No |
| PHEV | 1957 | Up to 100% | Not reported | 6–7 dpi | 4–18 weeks | Anorexia, constipation, vomiting, wasting, incoordination, ataxia, stiffness, hyperesthesia, posterior paralysis, respiratory distress | Central nervous system | No |
Despite the differences in pathogenesis and tissue tropism, TGEV and PRCV are closely related antigenically. PRCV induces an antibody response in pigs that is difficult to distinguish from TGEV-specific antibody by common serological assays (i.e. indirect immunofluorescence (IF), virus neutralization test) (Magtoto et al. 2019; Valkó et al. 2019), therefore various ELISAs have been developed using monoclonal antibodies targeting the different epitopes of the S protein (Carman et al. 2002). In the study of Valkó et al. (2019) the differentiating ELISA showed that almost all anti-TGEV antibodies found in the IF test, were produced against PRCV. Only one sample contained antibodies produced against TGEV and three IF positive samples remained negative for both viruses with ELISA. Differentiating ELISA can be used to monitor the TGEV/PRCV epidemiological situation, herd status or newly transported animals, as TGEV/PRCV-seronegative farms are at risk of the disease. However, TGE outbreaks have occurred only sporadically since the appearance of PRCV acting antibodies, causing difficulties in serological diagnostics (Valkó et al. 2019).
Serum neutralizing antibodies can be detected from around 6 DPI upon primary infection with the peak approximately at 14 DPI. Then the antibody response decreases subsequently (van Nieuwstadt et al. 1989). The duration of an effective PRCV-induced immunity appears to be relatively short: PRCV-induced neutralizing antibody titers are low by 36 DPI and minimal to absent one year post-infection, which indicates that a new infection with PRCV can take place during that time (Wesley 2002). Additionally, passive immunity decreases within 1–2 weeks post-weaning making piglets susceptible to PRCV infection (Callebaut et al. 1990; Saif 1993; Sestak et al. 1996).
A single experimental PRCV vaccine has been described. A recombinant adenovirus expressing the PRCV spike glycoprotein has been found to be antigenic and partially protected vaccinated piglets upon PRCV challenge, nevertheless, no subsequent studies have been published using this vaccine (Callebaut and Pensaert 1995). No vaccines are available commercially. As PRCV generally causes a subclinical infection, identification of PRCV positive herds requires regular monitoring by serological tests. Maintenance of a PRCV-negative facility may be achieved using strict biosecurity protocols and it may be important economically as some export markets require swine to be PRCV-negative (Killoran and Leedom-Larson 2016). Early weaning of piglets from seropositive sows and moving them to a clean facility may help establish a PRCV-negative herd (Burlatschenko and Arsenault 2015). Due to the close relation between PRCV and TGEV, disinfection procedures may be extrapolated. Hence, PRCV is susceptible to iodides, quaternary ammonium compounds, phenols, phenol plus aldehyde, betapropiolactone, ethylenamine, formalin, sodium hydroxide and sodium hypochlorite (Saif et al. 2012).
Porcine Hemagglutinating Encephalomyelitis Virus
PHEV, an etiological agent of vomiting and wasting disease (VWD) and/or encephalomyelitis was the first swine CoV identified (Mora-Díaz et al. 2019). It is also the only known neurotropic CoV that affects pigs (Mora-Díaz et al. 2019). The first clinical outbreak was reported in 1957 in Ontario, Canada (Roe and Alexander, 1958). However, the virus was first isolated in 1962, in primary pig kidney (PK) cells from the brains of 7–8 days old piglets showing histopathological evidence of viral poliencephalomyelitis (Greig et al. 1962). As most CoVs, PHEV contain four structural proteins: a large surface spike glycoprotein (S), a small membrane protein (E), a transmembrane glycoprotein (M), and a nucleocapsid protein (N). However, hemagglutinating coronaviruses like PHEV also possess an envelope-associated glycoprotein, the hemagglutinin-esterase (HE; ~140 kDa), which is made of two subunits (~65 kDa each) linked together by disulfide bonds (Mora-Díaz et al. 2019) (Fig. 1).
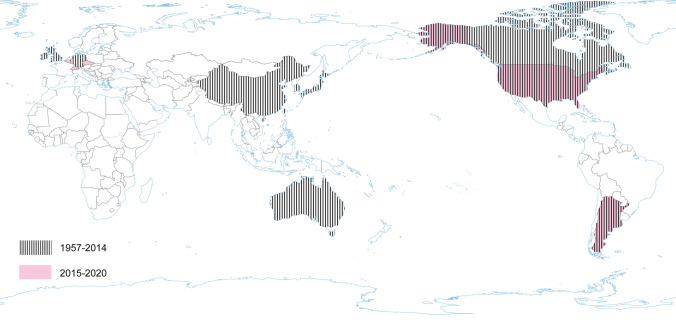
Serologic surveys (1960–1990) demonstrated that PHEV is highly prevalent and circulates without clinical manifestation in most swine herds worldwide and can be found in most swine-producing regions of the world, including Europe, the Americas, Asia, and Australia (Hirano and Ono 1998; Saif et al. 2012; Mora-Díaz et al. 2019). Clinical cases have also been reported in various countries: Canada, Belgium, China, Argentina, South Korea, USA (Mora-Díaz et al. 2019). However, the current worldwide seroprevalence of PHEV is unknown. A recent study determined the seroprevalence of PHEV in sow herds in the USA, using 2756 serum samples from breeding females on 104 commercial farms with no history of PHEV-associated disease. The overall seroprevalence at individual and herd levels was 53.35% (51.5%–55.2%) and 96.15% (92.4%–99.8%), respectively. Among positive farms, within-herd prevalence ranged from 1% to 50%, 51% to 70%, and 71% to 100% in 41.3%, 26.9%, and 28.8% of herds, respectively. The study confirms that PHEV is endemic in USA swine herds (Mora-Díaz et al. 2020). Another serological survey was performed in Argentina. A total of 961 serum samples collected from 14 breeding herds and three farrow-to-finish farms were assessed: the overall seroprevalence was 41.62% (38.5%–44.74%). The within herd prevalence varied from 12.5% to 86.6% for sows, 25% to 85.7% for gilts, and 3.7% to 90% for grower/fattener pigs in positive farms, indicating that PHEV is widespread and is circulating subclinically in Argentina (Mora-Díaz et al. 2019) (Fig. 3).
Fig. 3.
The occurrence of PHEV during the last five years and earlier.
Pigs are the only species susceptible to PHEV natural infection (Mora-Díaz et al. 2019). In experimental infections, the incubation period can vary from 4 to 7 days in pigs less than 4-weeks-of-age (Andries and Pensaert 1980). PHEV replicates primarily in the respiratory tract: nasal mucosa, tonsils, and lungs. The virus spreads from the primary sites of replication through the peripheral nervous system to the central nervous system. It may involve the trigeminal, inferior vagal, and superior cervical ganglia; intestinal nervous plexus, and the celiac and dorsal root ganglia in the lower thorax. The virus may be found in the submucosal and myenteric nervous plexuses of the small intestine following the infection of villus epithelium and the spread to the local sensory ganglia of the spinal cord (Andries and Pensaert 1980). Vomiting is induced by viral replication in the vagal sensory ganglion or by stimulation of the vagal ganglia infected neurons and impulses from the vomiting center (Andries et al. 1978). The spread of the virus may be strain specific and associated with clinical signs: anorexia, constipation, vomiting, wasting, incoordination, ataxia, stiffness, hyperesthesia, posterior paralysis, respiratory distress, and impaired weight gain (Mengeling and Cutlip 1972; Mora-Díaz et al. 2019). Access to nerve pathways seems to play the key role in the development of clinical signs, while viremia does not (Andries et al. 1978; Hirano et al. 1993). Usually, the infection is acute and subsequently cleared in pigs, but it may also result in mild or subclinical disease.
Hemagglutination inhibition (HI) antibodies against PHEV first appear 6–7 DPI and in vivo studies reported that animals with high HI antibody titers (HI titer ≥ 256) were not susceptible to PHEV infection (Andries et al. 1978; Pensaert and Callebaut 1974). Serum-neutralizing antibodies are detectable from 7–9 DPI, soon after the clinical signs are present and coincident with histopathological changes in central nervous system and tonsils crypts (Andries et al. 1978; Narita et al. 1989a, 1989b). Antibody levels peak around 12 DPI (Cartwright and Lucas 1970). Immune sows protect piglets by passive transfer of PHEV neutralizing antibodies in colostrum and milk, and maternally-derived antibodies are detectable in their offspring for 4–18 weeks (Paul and Mengeling, 1984). Gilts that received passive immunity as suckling piglets require subsequent infections and seroconversion to protect their offspring from PHEV disease (Appel et al. 1965). PHEV-antibodies do not cross-neutralize other porcine CoV, i.e. TGEV or PEDV (Pensaert et al. 1981). There is no data concerning protection against PHEV infection due to an infection with another strain of PHEV (Killoran and Leedom-Larson 2018). Increasing levels of systemic INF-α (DPI 3), tumor necrosis alfa (TNF-α) (DPI 10–17), IL-8 (DPI 14), an increase in concentration of both monocytes (DPI 10) and cytotoxic T cells (DPI 21) were detected in the study conducted on grower-finisher pigs infected with PHEV under experimental conditions (Mora-Díaz et al. 2019).
In animals older than 4 weeks of age, PHEV generally causes subclinical infections, while mortality in younger piglets infected with PHEV can be as high as 100% (Saif et al. 2012). PHEV may persist endemically in breeding farms. In large, closed herds that maintain endemic PHEV, protection of piglets may be provided by lactogenic immunity transferred from PHEV seropositive dams to their offspring (Saif et al. 2012). Piglets born to non-immune sows can be protected against PHEV shortly after birth with circulating anti-PHEV antibodies (hyper-immune serum), administered parentally or intraperitoneally (Mora-Díaz et al. 2019; Saif et al. 2012). It has been shown that neutralizing monoclonal antibodies against PHEV could be useful for antibody-based treatment of the disease (Raihana et al. 2009). Strict biosecurity measures must also be implemented to prevent virus entry to the nursery in farms with low or no passive immunity. There is no PHEV vaccine, although PHEV-related disease is not clinically relevant in most of the swine-producing countries. Promoting virus circulation on farms with early exposure to gilts and young sows could induce maternal immunity and effectively prevent disease in piglets (Mora-Díaz et al. 2019). Dithiothreitol and ether are effective in PHEV disinfection (Killoran and Leedom-Larson 2018).
Porcine Enteric Coronaviruses
Porcine enteric CoV belong to the group of Alpha- and Deltacoronaviruses. They are predisposed to interspecies transmission (Chan et al. 2013; Vlasova and Saif 2013). Four of the swine CoV can cause gastrointestinal infections: TGEV, PEDV, PDCoV and SADS-CoV (Wang et al. 2019) (Table 1). PEDV, PDCoV and SADS-CoV are newly emerging or re-emerging CoV that can be a real problem in pig production. They cause acute gastroenteritis in newborn piglets. Among reasons of the highest susceptibility of neonatal piglets to CoV infection are: less acidic pH in stomach compared to older pigs; renewal of enterocytes lining the intestinal villi from progenitor cells in the intestinal crypts is less rapid than in older pigs; the neonatal immune system is not fully mature; higher vulnerability to the electrolyte and fluid imbalances that result from maldigestion and severe malabsorption diarrhea, characteristic of CoV gastroenteritis in piglets (Fenner 2017). Research suggests that PEDV and SADS-CoV probably originate from bat CoV, while PDCoV probably originates from sparrow CoV, consistent with CoV interspecies transmission.
Transmissible Gastroenteritis Virus
TGEV is an etiological agent of transmissible gastroenteritis (TGE), an acute swine enteric disease. In the past, this virus caused acute gastrointestinal symptoms in pigs worldwide. The TGEV genome is approximately 28.6 kb in size and consists of a 5′ untranslated region (UTR), open reading frame 1a/1b (ORF1a/1b), genes for the spike (S), envelope (E), membrane (M) and nucleocapsid (N) protein, and a 3′ UTR, arranged in this order, with three accessory genes—named 3a, 3b and 7—interspersed within the structural genes in the 3′ part of the genome (Putics et al. 2006; Yount et al. 2000) (Fig. 1). The ORF1a/b encodes the non-structural proteins 1–10 (NSP1–10) and NSP11–16. These non-structural proteins have various functions in the viral life cycle. Their main function involves the replication and transcription of the viral genome. In addition, many of these proteins have various other functions in interaction with cellular processes such as contributing to the regulation of host translation (Chen et al. 2019). NSP1 is the major factor associated with pathogenicity in CoV. The NSP2, 3 and 8 can incorporate into the viral articles, and NSP3 is also associated with protease and ADP-ribose 1″-monophosphatase activities (Chen et al. 2019). The functions of other NSPs of TGEV remain unknown. The S glycoprotein attaches to the host cellular receptor porcine aminopeptidase N (pAPN) or sialic acid. Moreover, the S glycoprotein has hemagglutination activity, induces cellular fusion and stimulates neutralizing antibodies (Chen et al. 2019).
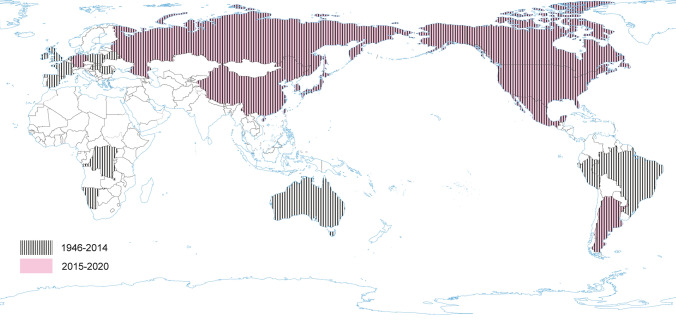
The disease with the characteristic features of TGE was first reported in 1935. However, TGEV, as an etiological agent of TGE in swine, was first identified over 10 years later (Doyle and Hutchings 1946; Enjuanes and Van der Zeijst 1995). TGEV probably circulated earlier in the pig population without significant influence on productivity or health, and became important to pig husbandry concurrently with its intensification (Enjuanes and Van der Zeijst 1995). During the next 20 years after its first identification TGE had been reported worldwide. Recent TGEV epidemiological studies are lacking in many countries, but it seems that TGE is currently nearly absent in Europe (Valkó et al. 2019). Sporadic outbreaks have been reported from China (Chen et al. 2019). TGEV is closely related to feline and canine CoV. Hence, it may be able to replicate subclinically in dogs, cats and foxes (Bohl 1989), while birds may act as mechanical vectors (Pilchard 1965) (Fig. 4).
Fig. 4.
The occurrence of TGEV during the last five years and earlier.
The decrease in the morbidity and mortality of suckling piglets due to TGE in Europe is explained by the long-term, natural immunization of the pig population with PRCV. However, the cross-reactivity of TGEV and PRCV antibody significantly hinders an objective assessment of the epidemiological situation regarding TGEV (Pejsak 2007; Valkó et al. 2019).
Porcine Deltacoronavirus
Porcine deltacoronavirus (PDCoV) is an emerging swine enteric CoV that causes diarrhea in piglets (Zhang Y et al. 2020). The enveloped PDCoV is pleomorphic with a diameter of 60–180 nm, excluding the projections (Ma et al. 2015), classified to the genus Deltacoronavirus (Wang et al. 2014a, b). PDCoV has a genome of approximately 25.4 kb in size that encodes four structural proteins [spike (S), envelope (E), membrane (M), and nucleocapsid (N)], and three accessory proteins (NS6, NS7, NS7a) (Li et al. 2014; Ma et al. 2015; Wang et al. 2015, 2019; Fang et al. 2017) (Fig. 1). Although the general characteristics of the proteins of CoV and their roles in viral replication have been identified, their functions and roles in host cells are not clear (Jung and Saif 2015; Fang et al. 2017). In relation to PDCoV, the results of molecular analyses indicate that interspecies transmission must have occurred quite recently, as a result of interaction between birds and mammals (Ma et al. 2016), and virus ancestors should be sought among birds (quail, sparrows) (Li et al. 2018). In addition, PDCoV has been shown to effectively infect a wide spectrum of host cells, including porcine, humans and chicken cells (Li et al. 2018). Recent studies have demonstrated that PDCoV can infect and kill cells of other species as well as chicks and poults (Boley et al. 2020). In vivo confirmation of avian susceptibility to PDCoV suggests that in vitro data implicating human susceptibility should be further evaluated (Boley et al. 2020).
The hemagglutination (HA) ability of PDCoV has been reported (Zhang et al. 2020b). PDCoV showed no HA activity against erythrocytes from pig, chicken, mouse, guinea pig, or human; however, it could agglutinate rabbit erythrocytes when virus was pre-treated with trypsin or neuraminidase. Additionally, the HA assay results showed a significant positive correlation with the infectious viral titer. The mentioned results suggest that assessing the HA activity of PDCoV may be a useful diagnostic method for investigating and surveilling PDCoV infections (Zhang Y et al. 2020).
Porcine deltacoronavirus (as HKU15) was identified in 2009 (reported in 2012), during molecular surveillance of CoV in avian and mammalian species in China, however, its role in swine pathology had not been known until 2014 (Wang et al. 2014a; Boley et al. 2020). In January 2014, PDCoV emerged as another new swine enteric disease in the USA (Marthaler et al. 2014; Wang et al. 2014a), when it was considered to be the causative agent of diarrhea in pigs (Woo et al. 2012; Wang et al. 2014b) (Fig. 5). A sequence analysis of the USA strains showed approximately 99% nucleotide identity to the PDCoV strains previously detected in China (Hong Kong) in 2009 (Wang et al. 2014b). Recent investigations have identified four distinct phylogenetic lineages of PDCoV (Thailand, China, USA and Early China Lineage), which differ in their geographic circulation patterns (He et al. 2020). More frequent intra- and interlineage recombination and higher virus genetic diversity has been identified in the Chinese lineages compared with the USA lineage where pigs are raised in different farming systems and ecological environments (He et al. 2020).
Fig. 5.
The occurrence of PDCoV during the last five years and earlier.
Since its introduction to the US in 2014, PDCoV has spread throughout most states, although at a lower prevalence rate than PEDV. Among diarrheic pigs in the US and China, the prevalence of PDCoV was found to be as high as 30% (US) and 7% (China) (Marthaler et al. 2014). Until the end of 2017 PDCoV had been detected in 21 states and > 540 herds (USDA 2017). Subsequent to US introduction, PDCoV has since been detected in Thailand and Korea (Janetanakit et al. 2016; Lee S and Lee C 2014).
A passive surveillance study was performed recently in Henan Province in China. The study revealed that PDCoV infection in pigs with diarrhea was high up to 23.49%, and co-infections with PEDV were common (60.40%; 61/101) (Zhang H et al. 2019). The highest prevalence of PDCoV was noted in suckling piglets (36.43%, 94/258). Sampling only from pigs with clinical signs enables to conclude the percentage of PDCoV infected pigs only among animals with diarrhea, however, it does not reflect the actual prevalence of PDCoV in pigs in this region. A randomized study focused on virus prevalence and/or seroprevalence in a given animal population is required to assess the actual spread of this pathogen. The results obtained so far have come from studies aimed at screening pigs with signs associated with gastrointestinal tract (diarrhea). Phylogenetic analysis based on the complete genome, spike and nucleocapsid gene sequences from this study revealed that the PDCoV Henan strains were closely related to other PDCoV reference strains that located in the Chinese clade (Zhang H et al. 2019). Furthermore, the phylogenetic analysis showed that the ancestor of sequenced strains may be different. The results support the understanding of the prevalence and evolution characteristics of PDCoV in China (Zhang M et al. 2019). Recent results revealed that the PDCoV EP-4E88 sequence shared very low similarity (< 22.2%) with other porcine CoV (PEDV, TGEV, PRCV, SADS-CoV, PHEV), demonstrating that it is an epitope that can be used for distinguishing PDCoV and other porcine CoV (Fu et al. 2020).
Swine Acute Diarrhea Syndrome Coronavirus
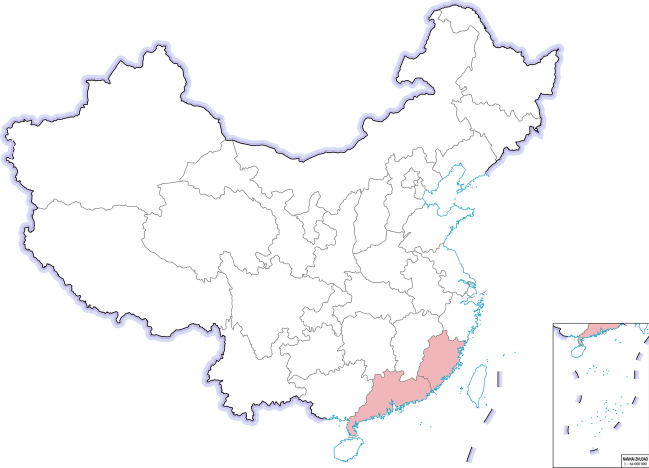
Another highly pathogenic CoV, SADS-CoV appeared in China, causing high mortality infections, especially among young pigs (Gong et al. 2017; Pan et al. 2017; Zhou et al. 2018). However, retrospective investigation showed that SADS‐CoV had been present in China at least since August 2016 (Zhou et al. 2019b). SADS-CoV is a novel member of the genus Alphacoronavirus and was first detected and identified as the etiological agent of a devastating swine disease outbreak in southern China in 2017, causing death of approximately 24.500 piglets and huge economic losses (Pan et al. 2017). It emerged again in January 2019 in pig herds in Guangdong (Zhou et al. 2019a). SADS-CoV has not been reported outside of China yet (Fig. 6).
Fig. 6.
The occurrence of SADS-CoV in Chinese provinces in 2017–2019.
The SADS-CoV genome is approximately 27 kb and contains nine open reading frame (ORFs) that are ORF1a, ORF1b, spike (S), envelope (E), membrane (M), nucleocapsid (N) and three accessory genes, NS3a, NS7a and NS7b (Zhou et al. 2018) and shows great similarity (95% identity) to CoV found in bats (HKU2-CoV), identified in 2007. However, the S gene sequence identity of only 86% suggests that HKU2-CoV may not be a direct progenitor of SADS-CoV, but they may have a common ancestor (Fig. 1). The results obtained have prompted researchers to search for new CoVs in bats. CoVs associated with SADS (with sequence identity of 96%–98% with SADS-CoV) were identified in 9.8% of bats, mainly Rhinolophus spp., which are known as CoV reservoirs associated with SARS (Zhou et al. 2018). Researchers indicated the possibility that rodents are among the susceptible hosts of SADS-CoV, highlighting the potential cross-species transmissibility of SADS-CoV (Yang et al. 2019).
Currently, there are no licensed vaccines against SADS-CoV. However, coronaviral proteases, including main proteases and papain-like proteases (PLP), are attractive antiviral targets because of their essential roles in polyprotein processing and thus virus maturation (Wang et al. 2020).
Porcine Epidemic Diarrhea Virus
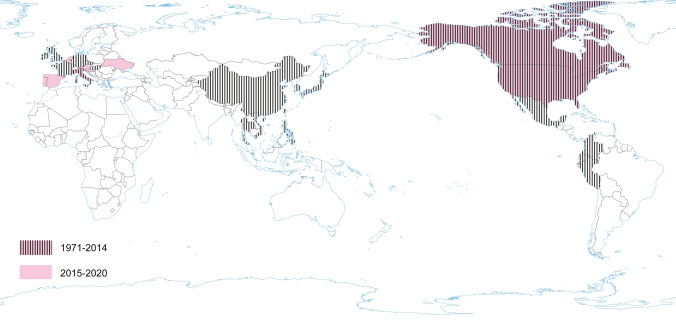
PEDV has an enveloped, single-stranded, positive-sense RNA genome of ~ 28 kb (Pensaert and de Bouck 1978; Vlasova et al. 2014) that encodes four structural proteins, namely, S, envelope (E), membrane (M) and nucleocapsid (N) proteins, sixteen nonstructural proteins (nsp1-nsp16) and an accessory protein ORF3 (Kocherhans et al. 2001) (Fig. 1). Porcine epidemic diarrhea (PED) was first recognized in the United Kingdom in 1971 as a severe enteric disease that manifests as sporadic outbreaks during the winter, leading to large economic losses on breeding farms due to the high mortality of piglets (Song and Park 2012; Wood 1977). In 1978, the etiologic agent of PED was identified as a new coronavirus and was assigned as PEDV (Pensaert and de Bouck 1978). In Asia, PEDV was first identified in 1982 and is now considered endemic. During the following 20-year period, PEDV was reported in many European countries (i.e. Belgium, England, France, the Netherlands, Germany, Hungary, Italy, the Czech Republic) (Song and Park 2012). Nowadays, PEDV does not cause significant outbreaks in Europe, but is a source of great concern in Asia, where outbreaks are more acute and severe than those observed in Europe. However, from 2014 to 2016, several emerging cases were reported in Germany, Belgium, Ukraine, France, Italy, and Austria (Boniotti et al. 2016; Hanke et al. 2015; Dastjerdi et al. 2015; Grasland et al. 2015; Theuns et al. 2015; Steinrigl et al. 2015) (Fig. 7). Since 2010, PEDV has been responsible for substantial economic losses to pork producers in many Asian countries (i.e. China, South Korea, Thailand, and Vietnam) (Song and Park 2012). It causes massive outbreaks characterized by 80%–100% illness among infected swine herds and a 50%–90% mortality rate among suckling piglets (Fan et al. 2012; Luo et al. 2012; Vlasova et al. 2014). The results of latest prevalence study in China are quite interesting: an average PEDV-positive rate in pigs from 39 farms across eight provinces, mainly around Hunan, was 38.04% (Tan et al. 2020), however the study was conducted on the 184 specimens from pigs, of which only 63 were clinically healthy. Predictably, the positive rate in pigs with diarrhea (48.76%, 59/121) was almost threefold higher than in pigs without clinical symptoms (17.46%, 11/63). Epidemiological information on PEDV infections obtained in the above mentioned study indicates that it is advisable to study larger populations using randomized procedures by which animals are selected for experimental study, thus allowing a more precise estimation of the PEDV prevalence across the tested population.
Fig. 7.
The occurrence of PEDV during the last five years and earlier.
Current data indicate that in order to confirm / exclude the presence of the virus, passive monitoring seems to be more appropriate, hence significantly more positive results are obtained in animals with diarrhea than in clinically healthy individuals. Phylogenetic and sequence analysis demonstrated that 14 representative PEDV strains from 14 swine farms belonged to the G2 group (G2-a and G2-b subgroups) and displayed a high degree of genetic variation. This data show that PED remains a severe threat to the pig industry and that variant PEDV stains are circulating in China (Tan et al. 2020). In North America PEDV emerged in April 2013 (in the US) (Stevenson et al. 2013). Since its introduction, PEDV has spread rapidly across the US, causing high rates of death among piglets and huge economic losses (Cima 2013; Mole 2013). The PEDV detected in the USA had 96.6%–99.5% identity with all known PEDV strains and the highest identity (> 99.0%) to some of the recent Chinese strains (2011–2012). The almost simultaneous outbreaks of the disease in the USA, and significant homology between the PEDV strains from unrelated farms, suggest a common source of the virus (Stevenson et al. 2013). In Canada, PEDV was first detected in January 2014; and the outbreaks have been reported in various provinces (Kochhar 2014). In Mexico, the first outbreaks suggestive of the circulation of PEDV were identified at the beginning of July 2013 (Lara-Romero et al. 2018). PEDV has become a very important swine pathogen in China, ranking third after African swine fever virus and swine reproductive and respiratory syndrome virus (PRRSV) (Wang et al. 2019). Ojkic et al. (2015) reported the first PEDV case on an Ontario farm in Canada—the samples of colon content of pigs with acute diarrhea were PCR-positive for PEDV and sequencing of these PCR products showed that this Ontario PEDV was 99% identical to recent PEDV isolates from the USA and China (Ojkic et al. 2015). Moreover, the Mexican strain of PEDV named PEDV/MEX/VER/01/2014 had a 99.8% identity with the American strain OH851 (Wang et al. 2014c). The similarity of strains found in Canada and Mexico with American strains, and the timing of PEDV introduction into these countries suggest that the outbreaks in Canada and Mexico may be associated with a PEDV emergence in the US.
The origin of PEDV is not explained clearly. There is no incontrovertible evidence confirming that it was introduced to the pig population through bats in the 1970s. However, the full-length genomic sequence of the prototype Belgian CV777 strain determined by Kocherhans et al. (2001) revealed that it was more closely related to a Scotophilus bat coronavirus (BtCoV) 512/2005 than to other known alphacoronaviruses, such as TGEV and human CoV, in phylogeny as well as in PEDV genome organization. It suggests that PEDV and BtCoV/512/2005 had a common evolutionary precursor and that cross-species transmission of CoV might have occurred between bats and pigs (Huang et al. 2013; Tang et al. 2006). In addition, PEDV infects cell lines derived from pigs, humans, monkeys, ducks and bats, which further supports the theory that it has once crossed the interspecies barrier between bats and pigs. (Liu et al. 2015; Teeravechyan et al. 2016). PEDV has also been detected in feral pigs in Korea and USA (Bevins et al. 2018; Lee et al. 2016). Research indicates that originally the virus could have been transmitted from domestic to wild pigs rather than vice versa. Since there are currently no control measures to prevent PEDV circulation in wild pigs (oral vaccines), they may remain a PEDV reservoir leading to future PED outbreaks in domestic pigs (Bevins et al. 2018; Lee et al. 2016).
Pathogenesis of Enteric Coronavirus Infections in Pigs
TGEV, PEDV, PDCoV and SADS-CoV are enteric CoV and cause clinically indistinguishable acute gastroenteritis in all age groups of pigs (Table 1). Laboratory confirmation is required to confirm the etiology. However, all viruses are antigenically distinct and cross‐protection does not occur between them (Jung et al. 2016; Ma et al. 2015; Pensaert and de Bouck, 1978).
Clinical manifestations of TGEV, PEDV, PDCoV and SADS-CoV infections are very similar and mainly include loss of appetite, diarrhea and vomiting, that lead to dehydration, weight loss, lethargy and death (Wang et al. 2019; Qian et al. 2020). Clinical symptoms appear 1–3 days after infection and usually do not last longer than 10 days. The disease is generally fatal in newborn piglets, especially those born from seronegative mothers. In older pigs, the morbidity is high but the mortality is low (Wang et al. 2019).
TGEV spreads rapidly and infects all age groups of pigs, with almost 100% mortality in piglets up to the second week of life. TGEV replicates in the cells of small intestinal epithelium, leading to the destruction of intestinal villi, which is the cause of malabsorption and maldigestion (Saif et al. 2012). The virus selectively infects and destroys the mature enterocytes lining the small intestinal villi, resulting in a significant shortening of villi. The destruction of the enterocytes lining the villi leads to the loss of critical digestive enzymes such as lactase and other disaccharidases, responsible for digestion of milk. The undigested milk increases osmolarity and results in further loss of water and electrolytes into the bowel lumen leading to diarrhea, acidosis and severe dehydration. Intestinal crypt epithelial cells remain uninfected, which is why the restoration of the villi function is rapid after recovery (Fenner 2017).
PEDV is considered more pathogenic than PDCoV (Jung et al. 2016). In the first year of the epidemic in the USA, the morbidity rate due to PEDV and PDCoV in pigs was approximately 100% and 30%, respectively (Marthaler et al. 2014; Wang et al. 2019). The mortality in newborn piglets was up to 100% and 40% for PEDV and PDCoV infections, respectively (Jung et al. 2016; Niederwerder and Hesse 2018). SADS-CoV mortality observed in China in piglets younger than 5 days old was very high (90%–100%), whereas in piglets older than 8 days it decreased to 5% (Xu et al. 2019; Zhou et al. 2018).
The primary target cells of SADS-CoV are intestinal epithelial cells, similar to other porcine enteric CoV (Zhou et al. 2018; Xu et al. 2019; Koonpaew et al. 2019; Wang et al. 2019). Several studies revealed remarkable villus atrophy and severe pathological lesions in the intestines of SADS-CoV inoculated piglets (Zhou et al. 2018; Xu et al. 2019; Koonpaew et al. 2019; Wang et al. 2019). The data regarding pathogenesis and immune response to SADS-CoV are scarce or inconsistent, although new articles and studies focusing on this area are still being published. Recent studies have demonstrated that SADS-CoV infection does not induce IFN-β production in porcine small intestinal epithelial cells (Zhou et al. 2020). SADS-CoV primarily interfered with the activity of mitochondria antiviral-signaling protein and retinoic acid-inducible gene I to inhibit the phosphorylation and nuclear translocation of IFN regulatory factor 3 to block the IFN-β production (Zhou et al. 2020). Another recent study showed that SADS-CoV infection causes characteristic morphological and biochemical changes of apoptosis such as DNA fragmentation, chromatin condensation, externalization of phosphatidylserine, caspase activation, and PARP cleavage in vitro and in vivo. Caspase-dependent FasL (extrinsic)—and mitochondria (intrinsic)—mediated apoptotic pathways play a central role in SADS-CoV-induced apoptosis that facilitates viral replication. In addition, authors highlighted that SADS-CoV is the first virus to induce direct apoptosis in vitro and in vivo (Zhang J et al. 2020).
All porcine gastrointestinal CoV probably have similar transmission routes and pathways. Infection with these viruses is mainly spread per os (the source of infection is feces of infected pigs). Inanimate objects contaminated with the virus, including vehicles or contaminated feed as well as the environment, etc. play a significant role in spreading swine gastrointestinal CoV infections (Niederwerder and Hesse 2018). A number of PEDV studies have shown that contaminated trailers and feed/ingredients can serve as a vehicle to transmit PEDV (Bowman et al. 2015; Pasick et al. 2014; Pillatzki et al. 2015). Aerogenic transmission was confirmed for both PEDV and PDCoV, probably by ingestion of aerosols containing viral particles (Niederwerder and Hesse 2018).
The pathogenesis of PEDV and PDCoV has been studied using pigs of different ages. It is known that the disease is more severe in younger pigs than in older ones. It is associated with the age-dependent resistance and is largely due to the immaturity of the immune system in piglets and the lower rate of enterocyte regeneration in younger pigs (Annamalai et al. 2015; Hammerberg et al. 1989; Jung et al. 2015; Moon et al. 1975). Deficiencies in the full functionality of the immune cells in newborn piglets may contribute to more severe PEDV in piglets compared to growers, which also occurred in TGEV infection (Derbyshire et al. 1969).
During the acute phase of infection with enteric CoV, intensive viral shedding with feces is observed. PEDV can be excreted in the faeces of infected pigs a few weeks after clinical symptoms disappear, which creates difficulties in managing the disease on the farm (Niederwerder and Hesse 2018). PDCoV-infected pigs shed high amounts of virus from 1 to 10 DPI. No histopathological lesions have been observed in the cecum and colon in PEDV and PDCoV infections, although PEDV and PDCoV antigens were detected in epithelial cells of these intestinal sections (Jung et al. 2016). PDCoV infection can induce acute transient and low-level viremia (Chen et al. 2015). During the acute infection phase, the virus was detected in large amount in small intestines especially the jejunum and ileum. Low amounts of PDCoV RNA were found in non-enteric tissues (Chen et al. 2015; Ma et al. 2015).
Li et al. (2020) reported that PDCoV infection reduced bacterial diversity and significantly altered the composition of microbiota from phylum to genus in the colon and faeces of piglets. Firmicutes (phylum), Lactobacillaceae (family), and Lactobacillus (genus) were significantly increased, while the abundance of Bacteroidetes (phylum) was markedly reduced in the colon and feces of the PDCoV-infected piglets when compared to those of the healthy piglets (Li et al. 2020). The above-mentioned findings provide new insight into the pathology and physiology of PDCoV.
Previous studies showed that PEDV neutralizing antibodies appear in the serum at approximately 10 dpi, the peak is reached at about 18 dpi and the level remains high up to 42 dpi (Diel et al. 2016). Long-term persistence of antibodies, compared to the presence of the virus in the body, indicates that the analysis of seroconversion or the presence of anti-PEDV antibodies may be useful in the diagnosis and monitoring of PED. Therefore, the development of PEDV antibody-based assays is crucial for detecting infected animals, confirming previous virus exposure, and monitoring sow herd immunity. Nevertheless, the potential assay is required to be sensitive and specific enough not to give false results related to cross-reactions, e.g. with other coronaviruses. The results obtained by Gimenez-Lirola et al. (2017) indicate that the PEDV S protein may play a key role as an antigen potentially used in such an assay. They showed that antibodies against the PEDV S protein have been shown not to cross-react with the antigens of other porcine intestinal CoV, including TGEV and PDCoV. For this reason, the S protein may be considered a sensitive and specific marker of PEDV infection (Gimenez-Lirola et al. 2017). Differences in the antibody responses to various PEDV proteins have practical implications in the development of serological tests for the monitoring and diagnostics of PEDV infections. A strong correlation between protection against PEDV infection and the level of cells secreting IgA and IgG antibodies in lymphatic tissue associated with intestines (GALT) and in the blood, as well as between protection and serum IgG and IgA titers has been demonstrated (de Arriba et al. 2002a, b). Under field conditions, in sows exposed to PEDV, cells secreting anti-PEDV IgA and IgG antibodies were detected in the intestine one month after PEDV exposure. Antibodies remained at a detectable level for the next six months (Ouyang et al. 2015). PEDV infection of sows appears to lead to the development and maintenance of effector/memory B cell response and strong virus neutralization titers in plasma up to 6–7 months post-infection. Current results regarding the correlation between antibody levels and anti-infective protection suggest that sows should be protected against reinfection during this period and that they may also provide maternal immunity to newborn piglets. Challenge studies where both sows and their offspring were exposed to the virus were performed by Goede et al. (2015). The authors evaluated the response of 3-day-old piglets born to sows exposed to a mild strain of PEDV seven months before challenge with a virulent PEDV isolate. Litters from sows with previous exposure had significantly less diarrhea (43% of litter) compared to litters from sows with no previous exposure (100% of litter) and a significantly lower mortality (0%) compared to litters from sows with no previous exposure (33%; P = 0.048). The findings indicated that sows previously exposed to a mild PEDV strain developed durable lactogenic immunity, which induced cross-protection with representative virulent PEDV, providing the piglets with significant passive immune protection for seven months against challenge with virulent PEDV. The immune response against PEDV has also been investigated by Krishna et al. (2020). Three-week-old pigs were infected with PEDV at the beginning of the experiment and subsequently challenged with a field isolate of PEDV at 30 DPI. The authors showed that PEDV infection induced both humoral and cell-mediated immune responses with an increase in PEDV-specific IgA and IgG antibodies in intestine and serum after primary exposure. This resulted in protection against challenge with a virulent field isolate. Characterization of lactogenic cross-resistance and understanding of immune response against PEDV are particularly important, especially as PEDV is endemic around the world and has recently reemerged in Europe.
Differences in the magnitude and duration of antibody responses were found between PEDV-and PDCoV-infected animals, when measured by indirect fluorescent antibody (IFA) test and serum-virus neutralization (SVN) assay (Mayo 2017). PEDV-and PDCoV-inoculated pigs seroconverted to their corresponding viruses by 5 dpi. More than 50% of pigs seroconverted at 7 DPI by IFA test and 14 DPI by SVN assay. All of the PEDV-inoculated pigs developed neutralizing antibodies by 28 DPI and remained seropositive up to 42 DPI as measured by SVN assay, whereas PDCoV antibodies had developed in 50% of the PDCoV-inoculated pigs at 14 DPI. In contrast to PEDV antibody responses, PDCoV serum antibodies were detected at much lower titers by both IFA and SVN with antibodies waning and being no longer detectable at 21 DPI (Mayo 2017).
The immune response in conventionally weaned pigs infected with PDCoV was also assessed after challenge and rechallenge (Zhao et al. 2020). After the first challenge, diarrhea and viral shedding developed successively in all pigs from 3 to 14 DPI, and all pigs recovered by 21 dpi. All pigs exhibited significantly increased PDCoV-specific IgG, IgA and virus-neutralizing antibody titers after the first challenge. All pigs were completely protected against rechallenge at 21 dpi. The serum levels of PDCoV-specific IgG, IgA, and virus-neutralizing antibody increased further after rechallenge (Zhao et al. 2020).
The obtained results indicate that pigs develop anti-infective immunity in the case of CoV infections in the gastrointestinal tract, which protects the animals against reinfection for a certain period of time. However, determining the maximum length of time that animals will be protected from reinfection requires further research, considering the different infection intervals. The role of not only humoral but also cellular immunity in protecting against reinfection with porcine coronaviruses requires further analysis. So far, it has been shown that the cell-mediated response may also play an important role in inducing protective immunity against PEDV. Pigs infected with the attenuated and highly pathogenic PEDV PC22A strain have been shown to produce similar levels of antibodies, but only pigs infected with a virulent PEDV were protected against homologous virulent virus infection, indicating a protective role of elements other than antibodies (Lin et al. 2019). However, the data on cell-mediated immune responses are limited and focus mainly on the inflammatory response (cytokines) (Annamalai et al. 2015). Following infection with TGEV or PRCV, specific antibodies can be detected in serum from 6 or 7 DPI, and persist at least for months (Callebaut et al. 1989; Enjuanes et al 2001; Garwes et al 1988; Saif and Sestak 2006; Wesley and Woods 1996). The PRCV can induce protective immunity to TGEV in neonatal pigs. Moreover, protective immunity coincides with the appearance of virus-neutralizing antibody (Wesley and Woods 1996). The antibody post-exposure response to PRCV infection is described in details in the PRCV section above. Although the PRCV and TGEV antibodies show complete neutralization of either virus, there are differences in the specificities of some of the non-neutralizing antibodies as PRCV lacks certain epitopes present on the TGEV (Callebaut et al. 1989; Enjuanes et al. 2001; Garwes et al. 1988; Saif and Sestak 2006). It was reported earlier that the sows immune after TGEV infection secrete TGEV-antibodies of IgA class in their milk. This production of TGEV-IgA antibodies in the mammary gland is the result of antigenic stimulation in the gut-associated lymphoid tissue and results in the protection of suckling piglets against TGEV (Van Deun et al. 1990). In case of a PRCV infection, neutralizing antibodies can be found in milk at minimal levels from 7–14 days post-farrowing with an increase in titers as lactation continues. Milk IgA levels vary among individuals. A single infection and reinfection with PRCV results in an increase in IgA detected in milk (Callebaut et al. 1990).
It has also been shown that toll-like receptors (TLR) play an important role in the immune response and course of PEDV infections (Cao et al. 2015). Significantly lower amounts of NK cells, with minimal killing activity, were observed in sucking piglets infected with PEDV. In addition, a lower percentage of IFN-γ producing CD3-CD4-CD8+ NK cells were found in the blood and intestines compared to weaned piglets (Annamalai et al. 2015). Incomplete cross-protection exists between different genetic clusters of PEDV variants (Lin et al. 2015). In addition, there is no cross-resistance between PEDV and PDCoV antibodies (Ma et al. 2016). The occurrence of SADS-CoV outbreaks on a pig farm where PEDV infection was previously observed also suggests a lack of cross-protection between PEDV and SADS-CoV (Zhou et al. 2018).
Pathological changes in young pigs are similar in the course of infection with all listed enteric CoV. Virus replication occurs very quickly, mainly in small intestinal epithelial cells (villi), causing destruction and villi atrophy. Therefore, diarrhea observed during infection is probably the consequence of malabsorption due to massive enterocytes destruction. In addition, PEDV-infected pigs in which vomiting was observed, showed significantly lower levels of serotonin secreting cells in the duodenum, middle jejunum, ileum and colon than in negative control pigs, suggesting that serotonin secretion may have an impact on vomiting during PEDV infection (Jung et al. 2018). Pathogenesis of recently discovered SADS-CoV infections was also described (Pan et al. 2017; Zhou et al. 2018; Xu et al. 2019). However, the results obtained are inconsistent. Pan et al. (2017) showed that SADS-CoV causes only mild and moderate bowel changes in three-day-old piglets. Research conducted by Zhou et al. (2018) indicates that SADS-CoV proteins were detected only in a small percentage of jejunum epithelial cells, which does not fully explain the 50% (3/6) mortality observed in three-day piglets. In the studies conducted by Xu et al. (2019) it was shown that SADS-CoV mainly attacks the intestine, causing 100% (12/12) and 50% (2/4) mortality in conventional piglets at 5 days of age and contact pigs infected at the age of 7 days, respectively. In the course of infection, viral RNA was also detected in the heart, liver, spleen, kidneys, stomach and lungs, but it was not found in the blood of piglets killed on the seventh day after inoculation, while the pigs still had severe diarrhea (Xu et al. 2019). It seems that these extremely different research results and a potentially serious course of the disease caused by SADS-CoV in pigs justify the need for more detailed research in this area.
Control and Prevention
Rapid diagnosis is crucial in controlling CoV infections to prevent them from spreading. A number of techniques which may be used in laboratory diagnostics for a number of CoV are currently being developed. A high level of biosecurity (strictly following the biosecurity rules) and in some cases vaccines are the first choice to prevent infection with these pathogens (Crawford et al. 2016). Due to the lack of cross-resistance between most porcine CoV (except TGE and PRCV), the use of specific immunoprophylaxis will require the development of separate vaccines specific to each virus.
Due to the fact that the course of CoV infections is the most severe in newborn piglets (up to one week), protection of this group should focus on ensuring adequate passive immunity, particularly in relation to neutralizing antibodies, which can be delivered to piglets through the colostrum and milk of immunized mothers (vaccines, contact with contagious material). It has been demonstrated that IgA or neutralizing antibody titers in milk and colostrum are better markers of lactogenic immunity than serum antibody titers (Bjustrom-Kraft et al. 2018).
There are two commercial vaccines based on a live-modified TGEV strain for combined oral-intramuscular administration (PROSYSTEM® TGE/Rota; PROSYSTEM® TREC). PROSYSTEM® TGE/Rota is dedicated for use in healthy pregnant swine as an aid in the prevention and control of rotaviral diarrhea and TGE in their nursing piglets. It contains attenuated live TGEV and two major rotavirus serotypes in desiccated form. All viruses have been modified so that they do not cause disease in baby pigs and pregnant swine. PROSYSTEM® TREC is also indicated for the vaccination of healthy pregnant swine, to provide passive protection to the nursed pigs. These vaccines can effectively stimulate a response in previously exposed pigs, but do not protect the naïve population. Live, inactivated and subunit PED vaccines have been developed in China, Japan, Korea and the US for sows (Crawford et al. 2016; Song et al. 2015), but their effectiveness is not sufficient to control PEDV outbreaks, since the disease has also appeared in vaccinated herds (Table 1). For PDCoV or SADS-CoV, there is currently no information about the specific immunoprophylaxis. Some authors recommend feedback with the use of comminuted intestines from infected pigs (Chattha et al. 2015) in order to reduce mortality in suckling piglets, as part of obtaining immunity in pregnant sows (and consequently passive immunity in piglets). These methods raise a problem with long-term persistence of coronaviruses in farms, or with the transmission of other pathogens within the herd.
Conclusion
Due to their enormous diversity, their ability to shift host and the constant emergence of new CoV, the knowledge about this family of viruses requires continual updates. Research on CoV leading to fundamental understanding of their biology is extremely important since, as recent experience with SARS-CoV-2 has revealed, viruses with high and also human pandemic potential, may emerge and lead to serious global consequences.
Acknowledgments
The scientific activity of the corresponding author was supported by the National Science Centre, Poland, under research project (DEC-2014/13/B/NZ6/02566) and statutory funding 506.514.05.00 of the Department of Preclinical Sciences and Infectious Diseases, Faculty of Veterinary Medicine and Animal Science, Poznan University of Life Sciences, Poznan, Poland. We thank Wiktoria Mól for improving the use of English in the manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Akimkin V, Beer M, Blome S, Hanke D, Höper D, Jenckel M, Pohlmann A. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg Infect Dis. 2016;22:1314–1315. doi: 10.3201/eid2207.160179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K, Pensaert MB. Immunofluorescence studies on the pathogenesis of hemagglutinating encephalomyelitis virus infection in pigs after oronasal inoculation. Am J Vet Res. 1980;41:1372–1378. [PubMed] [Google Scholar]
- Andries K, Pensaert M, Callebaut P. Pathogenicity of hemagglutinating encephalomyelitis (vomiting and wasting disease) virus of pigs, using different routes of inoculation. Zentralbl Veterinarmed B. 1978;25:461–468. doi: 10.1111/j.1439-0450.1978.tb00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai T, Saif LJ, Lu Z, Jung K. Age-dependent variation in innate immune responses to porcine epidemic diarrhea virus infection in suckling versus weaned pigs. Vet Immunol Immunopathol. 2015;168:193–202. doi: 10.1016/j.vetimm.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel M, Greig AS, Corner AH. Encephalomyelitis of swine caused by a hemagglutinating virus. IV. Transmission studies. Res Vet Sci. 1965;6:482–489. [PubMed] [Google Scholar]
- Atanasova K, Van Gucht S, Barbé F, Lefebvre DJ, Chiers K, Van Reeth K. Lung cell tropism and inflammatory cytokine-profile of porcine respiratory coronavirus infection. Open Vet Sci J. 2008;2:117–126. doi: 10.2174/1874318800802010117. [DOI] [Google Scholar]
- Bevins SN, Lutman M, Pedersen K, Barrett N, Gidlewski T, Deliberto TJ, Franklin AB. Spillover of swine coronaviruses, United States. Emerg Infect Dis. 2018;24:1390–1392. doi: 10.3201/eid2407.172077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjustrom-Kraft J, Woodard K, Gimenez-Lirola L, Setness B, Ji J, Lasley P, Nelson E, Zhang JQ, Baum D, Gauger P, Main R, Zimmerman J. Serum and mammary secretion antibody responses in porcine epidemic diarrhea-immune gilts following porcine epidemic diarrhea vaccination. J Swine Health Prod. 2018;26:34–40. [Google Scholar]
- Bohl EH (1989) Transmissible gastroenteritis virus (classical enteric variant). In: Pensaert MB (ed.) Virus infections of vertebrates, vol 2, 2nd edn. Elsevier, Amsterdam, pp 139–153
- Boley PA, Alhamo MA, Lossie G, Yadav KK, Vasquez-Lee M, Saif LJ, Kenney SP. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg Infect Dis. 2020;26:255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, Faccini S, Bonilauri P, Cordioli P, Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Krogwold RA, Price T, Davis M, Moeller SJ. Investigating the introduction of porcine epidemic Diarrhea virus into an Ohio swine operation. BMC Vet Res. 2015;11:38. doi: 10.1186/s12917-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgen A, Tobler K, Ackermann M. Identification of coronaviral conserved sequences and application to viral genome amplification. Adv Exp Med Biol. 1993;342:81–82. doi: 10.1007/978-1-4615-2996-5_13. [DOI] [PubMed] [Google Scholar]
- Burlatschenko S, Arsenault C. Elimination of porcine respiratory coronavirus by early weaning and segregation. J Swine Health Prod. 2015;23:208–213. [Google Scholar]
- Callebaut TP, Pensaert MB, Hooyberghs J. A competitive inhibition ELISA for the differentiation of serum antibodies from pigs infected with transmissible gastroenteritis virus (TGEV) or with the TGEV-related porcine respiratory coronavirus. Vet Microbiol. 1989;20:9–19. doi: 10.1016/0378-1135(89)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut P, Cox E, Pensaert M, Van Deun K. Induction of milk IgA antibodies by porcine respiratory coronavirus infection. Adv Exp Med Biol. 1990;276:421–428. doi: 10.1007/978-1-4684-5823-7_58. [DOI] [PubMed] [Google Scholar]
- Callebaut P, Pensaert M. Expression and immunogenicity of the spike glycoprotein of porcine respiratory coronavirus encoded in the E3 region of adenovirus. Adv Exp Med Biol. 1995;380:65–270. doi: 10.1007/978-1-4615-1899-0_42. [DOI] [PubMed] [Google Scholar]
- Cao L, Ge X, Gao Y, Ren Y, Ren X, Li G. Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J Gen Virol. 2015;96:1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- Carman S, Josephson G, McEwen B, Maxie G, Antochi M, Eernisse K, Nayar G, Halbur P, Erickson G, Nilsson E (2002) Field validation of a commercial blocking ELISA to differentiate antibody to transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus and to identify TGEV-infected swine herds. J Vet Diagn Invest 14:97–105 [DOI] [PubMed]
- Cartwright SF, Lucas M. Vomiting and wasting disease in piglets. Virological and epidemiological studies. Vet Rec. 1970;86:278–280. doi: 10.1136/vr.86.10.278. [DOI] [PubMed] [Google Scholar]
- Chan JF, To KK, Tse H, Jin DY, Yuen KY. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattha KS, Roth JA, Saif LJ. Strategies for design and application of enteric viral vaccines. Annu Rev Anim Biosci. 2015;3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, Madson D, Brodie J, Magstadt D, Derscheid R, Welch M, Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Knutson TP, Rossow S, Saif LJ, Marthaler DG. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci Rep. 2019;9:3953. doi: 10.1038/s41598-019-40564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cima G. Viral disease affects US pigs: porcine epidemic diarrhea found in at least 11 states. J Am Vet Med Assoc. 2013;243:30–31. [PubMed] [Google Scholar]
- Crawford K, Lager KM, Kulshreshtha V, Miller LC, Faaberg KS. Status of vaccines for porcine epidemic diarrhea virus in the United States and Canada. Virus Res. 2016;226:108–116. doi: 10.1016/j.virusres.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Dastjerdi A, Carr J, Ellis RJ, Steinbach F, Williamson S. Porcine epidemic diarrhea virus among farmed pigs, Ukraine. Emerg Infect Dis. 2015;21:2235–2237. doi: 10.3201/eid2112.150272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arriba ML, Carvajal A, Pozo J, Rubio P. Mucosal and systemic isotype-specific antibody responses and protection in conventional pigs exposed to virulent or attenuated porcine epidemic diarrhoea virus. Vet Immunol Immunopathol. 2002;85:85–97. doi: 10.1016/s0165-2427(01)00417-2. [DOI] [PubMed] [Google Scholar]
- De Arriba ML, Carvajal A, Pozo J, Rubio P. Rubio Isotype-specific antibody-secreting cells in systemic and mucosal associated lymphoid tissues and antibody responses in serum of conventional pigs inoculated with PEDV. Vet Immunol Immunopathol. 2002;84:1–16. doi: 10.1016/S0165-2427(01)00386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova PJG, Cortey M, Díaz I, Puente H, Rubio P, Martín M, Carvajal A. A retrospective study of porcine epidemic diarrhoea virus (PEDV) reveals the presence of swine enteric coronavirus (SeCoV) since 1993 and the recent introduction of a recombinant PEDV-SeCoV in Spain. Transbound Emerg Dis. 2020 doi: 10.1111/tbed.13666. [DOI] [PubMed] [Google Scholar]
- Derbyshire JB, Jessett DM, Newman G. An experimental epidemiological study of porcine transmissible gastroenteritis. J Comp Pathol. 1969;79:445–452. doi: 10.1016/0021-9975(69)90064-4. [DOI] [PubMed] [Google Scholar]
- Diel DG, Lawson S, Okda F, Singrey A, Clement T, Fernandes MHV, Christopher-Hennings J, Nelson EA. Porcine epidemic diarrhea virus: an overview of current virological and serological diagnostic methods. Virus Res. 2016;226:60–70. doi: 10.1016/j.virusres.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle LP, Hutchings LM. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc. 1946;108:257. [PubMed] [Google Scholar]
- Duarte M, Laude H. Sequence of the spike protein of the porcine epidemic diarrhoea virus. J Gen Virol. 1994;75:1195–1200. doi: 10.1099/0022-1317-75-5-1195. [DOI] [PubMed] [Google Scholar]
- Enjuanes L, van der Zeijst BAM (1995) Molecular basis of transmissible gastroenteritis virus epidemiology. Coronaviridae 337–376
- Enjuanes L, Sola I, Almazán F, Ortego J, Izeta A, González JM, Alonso S, Sánchez-Morgado JM, Escors D, Calvo E, Riquelme C, Sánchez CM. Coronavirus derived expression systems. J Biotech. 2001;88:183–204. doi: 10.1016/S0168-1656(01)00281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Zhang J, Ye Y, Tong T, Xie K, Liao M. Complete genome sequence of a novel porcine epidemic diarrhea virus in south China. J Virol. 2012;86:10248–10249. doi: 10.1128/JVI.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Fang L, Hong Y, Liu X, Dong N, Ma P, Bi J, Wang D, Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J Gen Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner F. Chapter 24. Coronaviridae. In: Maclachlan NJ, Dubovi EJ, editors. Fenner’s veterinary virology. 5. New York: Academic Press; 2017. pp. 435–461. [Google Scholar]
- Fu J, Chen R, Hu J, Qu H, Zhao Y, Cao S, Wen X, Wen Y, Wu R, Zhao Q, Ma X, Huang X. Identification of a novel linear B-cell epitope on the nucleocapsid protein of porcine deltacoronavirus. Int J Mol Sci. 2020;21:648. doi: 10.3390/ijms21020648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwes DJ, Stewart F, Cartwright SF, Brown I. Differentiation of porcine coronavirus from transmissible gastroenteritis virus. Vet Rec. 1988;122:86–87. doi: 10.1136/vr.122.4.86. [DOI] [PubMed] [Google Scholar]
- Gimenez-Lirola LG, Zhang J, Carrillo-Avila JA, Chen Q, Magtoto R, Poonsuk K, Baum DH, Pineyro P, Zimmerman J. Reactivity of porcine epidemic diarrhea virus structural proteins to antibodies against porcine enteric coronaviruses: diagnostic implications. J Clin Microbiol. 2017;55:1426–1436. doi: 10.1128/JCM.02507-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede D, Murtaugh MP, Nerem J, Yeske P, Rossow K, Morrison R. Previous infection of sows with a “mild” strain of porcine epidemic diarrhea virus confers protection against infection with a “severe” strain. Vet Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. A new Bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis. 2017;23:1607–1609. doi: 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasland B, Bigault L, Bernard C, Quenault H, Toulouse O, Fablet C, Rose N, Touzain F, Blanchard Y. Complete genome sequence of a porcine epidemic diarrhea s gene indel strain isolated in France in december 2014. Genome Announc. 2015;3:e00535–e615. doi: 10.1128/genomeA.00535-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig AS, Mitchell D, Corner AH, Bannister GL, Meads EB, Julian RJ. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can J Comp Med Vet Sci. 1962;26:49–56. [PMC free article] [PubMed] [Google Scholar]
- Halbur PG, Pallarés FJ, Opriessnig T, Vaughn EM, Paul PS. Pathogenicity of three isolates of porcine respiratory coronavirus in the USA. Vet Rec. 2003;152:358–361. doi: 10.1136/vr.152.12.358. [DOI] [PubMed] [Google Scholar]
- Hammerberg C, Schurig GG, Ochs DL. Immunodeficiency in young pigs. Am J Vet Res. 1989;50:868–874. [PubMed] [Google Scholar]
- Hanke D, Jenckel M, Petrov A, Ritzmann M, Stadler J, Akimkin V, Blome S, Pohlmann A, Schirrmeier H, Beer M, Höper D. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg Infect Dis. 2015;21:493–496. doi: 10.3201/eid2103.141165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W-T, Ji X, He W, Dellicour S, Wang S, Li L, Zhang L, Gilbert M, Zhu H, Xing G, Veit M, Huang Z, Han G-Z, Huang Y, Suchard MA, Baele G, Lemey P, Su S. Genomic epidemiology, evolution, and transmission dynamics of porcine deltacoronavirus. Mol Biol Evol. 2020;37:2641–2654. doi: 10.1093/molbev/msaa117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N, Ono K. A serological survey of human coronavirus in pigs of the Tohoku District of Japan. Adv Exp Med Biol. 1998;440:491–494. doi: 10.1007/978-1-4615-5331-1_63. [DOI] [PubMed] [Google Scholar]
- Hirano N, Haga S, Fujiwara K. The route of transmission of hemagglutinating encephalomyelitis virus (HEV) 67N strain in 4-week-old rats. Adv Exp Med Biol. 1993;342:333–338. doi: 10.1007/978-1-4615-2996-5_51. [DOI] [PubMed] [Google Scholar]
- Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng XJ. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4:e00737–e1713. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S, Nonthabenjawan N, Kesdaengsakonwut S, Amonsin A. Porcine deltacoronavirus, Thailand, 2015. Emerg Infect Dis. 2016;22:757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Renukaradhya GJ, Alekseev KP, Fang Y, Tang Y, Saif LJ. Porcine reproductive and respiratory syndrome virus modifies innate immunity and alters disease outcome in pigs subsequently infected with porcine respiratory coronavirus: implications for respiratory viral co-infections. J Gen Virol. 2009;90:2713–2723. doi: 10.1099/vir.0.014001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Annamalai T, Lu Z, Saif LJ. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Hu H, Saif LJ. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Miyazaki A, Saif LJ. Immunohistochemical detection of the vomiting-inducing monoamine neurotransmitter serotonin and enterochromaffin cells in the intestines of conventional or gnotobiotic (Gn) pigs infected with porcine epidemic diarrhea virus (PEDV) and serum cytokine responses of Gn pigs to acute PEDV infection. Res Vet Sci. 2018;119:99–108. doi: 10.1016/j.rvsc.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killoran KE, Leedom-Larson KR (2016) Porcine respiratory coronavirus. Swine Health Information Center and Center for Food Security and Public Health. http://www.cfsph.iastate.edu/pdf/shic-factsheetporcine-respiratory-coronavirus
- Killoran KE, Leedom-Larson KR (2018) Porcine hemagglutinating encephalomyelitis virus. Swine Health Information Center and Center for Food Security and Public Health. http://www.cfsph.iastate.edu/pdf/shic-factsheet-porcine-hemagglutinating-encephalomyelitis-virus
- Kocherhans R, Bridgen A, Ackermann M, Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar HS. Canada: porcine epidemic diarrhea in Canada: an emerging disease case study. Can Vet J. 2014;5:1048–1049. [PMC free article] [PubMed] [Google Scholar]
- Koonpaew S, Teeravechyan S, Frantz PN, Chailangkarn T, Jongkaewwattana A. PEDV and PDCoV pathogenesis: the interplay between host innate immune responses and porcine enteric coronaviruses. Front Vet Sci. 2019;6:34. doi: 10.3389/fvets.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C, Schultze B, Laude H, Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna VD, Kim Y, Yang M, Vannucci F, Molitor T, Torremorell M, Cheeran MC-J. Immune responses to porcine epidemic diarrhea virus (PEDV) in swine and protection against subsequent infection. PLoS ONE. 2020;15(4):e0231723. doi: 10.1371/journal.pone.0231723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Romero R, Gómez-Núñez L, Cerriteño-Sánchez JL, Márquez-Valdelamar L, Mendoza-Elvira S, Ramírez-Mendoza H, Rivera-Benítez J. Molecular characterization of the spike gene of the porcine epidemic diarrhea virus in Mexico, 2013–2016. Virus Genes. 2018;54:215–224. doi: 10.1007/s11262-017-1528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee C. Complete genome characterization of korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014;2:e01191–e1214. doi: 10.1128/genomeA.01191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DU, Kwon T, Je SH, Yoo SJ, Seo SW, Sunwoo SY, Lyoo YS. Wild boars harboring porcine epidemic diarrhea virus (PEDV) may play an important role as a PEDV reservoir. Vet Microbiol. 2016;192:90–94. doi: 10.1016/j.vetmic.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014;2:e00278–e314. doi: 10.1128/genomeA.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. 2018;115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-Y, Li B-X, Liang Q-Q, Jin X-H, Tang L, Ding Q-W, Wang Z-X, Wei Z-Y. Porcine deltacoronavirus infection alters bacterial communities in the colon and feces of neonatal piglets. Microbiologyopen. 2020;9:e1036. doi: 10.1002/mbo3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Annamalai T, Liu X, Gao X, Lu Z, El-Tholoth M, Hu H, Saif LJ, Wang Q. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet Res. 2015;46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Ghimire S, Hou Y, Langel SN, Vlasova AN, Saif LJ, Wang Q. Pathogenicity and immunogenicity of attenuated porcine epidemic diarrhea virus PC22A strain in conventional weaned pigs. BMC Vet Res. 2019;15:26. doi: 10.1186/s12917-018-1756-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Tang J, Ma Y, Liang X, Yang Y, Peng G, Qi Q, Jiang S, Li J, Du L. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhang J, Deng X, Ye Y, Liao M, Fan H. Complete genome sequence of a highly prevalent isolate of porcine epidemic diarrhea virus in South China. J Virol. 2012;86:9551. doi: 10.1128/JVI.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio. 2015;6:e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Liang X, Oglesbee M, Krakowka S, Niehaus A, Wang G, Jia A, Song H, Li J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtoto R, Poonsuk K, Baum D, Zhang J, Chen Q, Ji J, Piñeyro P, Zimmerman J, Giménez-Lirola LG. Evaluation of the serologic cross-reactivity between transmissible gastroenteritis coronavirus and porcine respiratory coronavirus using commercial blocking enzyme-linked immunosorbent assay kits. mSphere. 2019;4:e00017–e00019. doi: 10.1128/mSphere.00017-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelik R, Sarvas M, Jackova A, Salamunova S, Novotny J, Vilcek S. First outbreak with chimeric swine enteric coronavirus (SeCoV) on pig farms in Slovakia—lessons to learn. Acta Vet Hung. 2018;66:488–492. doi: 10.1556/004.2018.043. [DOI] [PubMed] [Google Scholar]
- Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20:1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo KA (2017) Emerging swine enteric coronaviruses: comparison of pathogenicity and antibody response. Graduate Theses and Dissertations 16732
- Mengeling WL, Cutlip RC. Experimentally induced infection of newborn pigs with hemagglutinating encephalomyelitis virus strain 67N. Am J Vet Res. 1972;33:953–956. [PubMed] [Google Scholar]
- Mole B. Deadly pig virus slips through US borders. Nature. 2013;499:388. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- Moon HW, Kemeny LJ, Lambert G, Stark SL, Booth GD. Age-dependent resistance to transmissible gastroenteritis of swine. III. Effects of epithelial cell kinetics on coronavirus production and on atrophy of intestinal villi. Vet Pathol. 1975;12:434–445. doi: 10.1177/0300985875012005-00610. [DOI] [PubMed] [Google Scholar]
- Mora-Díaz JC, Piñeyro PE, Houston E, Zimmerman J, Giménez-Lirola LG. Porcine hemagglutinating encephalomyelitis virus: a review. Front Vet Sci. 2019;6:53. doi: 10.3389/fvets.2019.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Díaz JC, Magtoto R, Houston E, Baum D, Carrillo-Ávila JA, Temeeyasen G, Zimmerman J, Piñeyro P, Giménez-Lirola L. Detecting and monitoring porcine hemagglutinating encephalomyelitis virus, an underresearched betacoronavirus. mSphere. 2020;5:e0019920. doi: 10.1128/mSphere.00199-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Kawamura H, Haritani M, Kobayashi M. Demonstration of viral antigen and immunoglobulin (IgG and IgM) in brain tissue of pigs experimentally infected with haemagglutinating encephalomyelitis virus. J Comp Pathol. 1989;100:119–128. doi: 10.1016/0021-9975(89)90122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Kawamura H, Tsuboi T, Haritani M, Kobayashi M. Immunopathological and ultrastructural studies on the tonsil of gnotobiotic pigs infected with strain 67N of haemagglutinating encephalomyelitis virus. J Comp Pathol. 1989;100:305–312. doi: 10.1016/0021-9975(89)90108-4. [DOI] [PubMed] [Google Scholar]
- Niederwerder MC, Hesse RA. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound Emerg Dis. 2018;65:660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic D, Hazlett M, Fairles J, Marom A, Slavic D, Maxie G, Alexandersen S, Pasick J, Alsop J, Burlatschenko S. The first case of porcine epidemic diarrhea in Canada. Can Vet J. 2015;56:149–152. [PMC free article] [PubMed] [Google Scholar]
- Ouyang K, Shyu DL, Dhakal S, Hiremath J, Binjawadagi B, Lakshmanappa YS, Guo R, Ransburgh R, Bondra KM, Gauger P, Zhang J, Specht T, Gilbertie A, Minton W, Fang Y, Renukaradhya GJ. Evaluation of humoral immune status in porcine epidemic diarrhea virus (PEDV) infected sows under field conditions. Vet Res. 2015;46:140. doi: 10.1186/s13567-015-0285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. Discovery of a novel swine enteric alphacoronavirus (seacov) in southern China. Vet Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J, Berhane Y, Ojkic D, Maxie G, Embury-Hyatt C, Swekla K, Handel K, Fairles J, Alexandersen S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound Emerg Dis. 2014;61:397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul PS, Mengeling WL. Persistence of passively acquired antibodies to hemagglutinating encephalomyelitis virus in swine. Am J Vet Res. 1984;45:932–934. [PubMed] [Google Scholar]
- Pejsak Z (2007) Koronawirusowe zapalenie żołądka i jelit świń, in: Ochrona Zdrowia Świń, Państwowe Wyd. Rolnicze, pp, 223–224
- Peng JY, Punyadarsaniya D, Shin DL, Pavasutthipaisit S, Beineke A, Li G, Wu NH, Herrler G. The cell tropism of porcine respiratory coronavirus for airway epithelial cells is determined by the expression of porcine aminopeptidase N. Viruses. 2020;23:1211. doi: 10.3390/v12111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert MB, Callebaut PE. Characteristics of a coronavirus causing vomition and wasting in pigs. Arch Gesamte Virusforsch. 1974;44:35–50. doi: 10.1007/BF01242179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert MB, de Bouck P, Reynolds DJ. An immunoelectron microscopic and immunofluorescent study on the antigenic relationship between the coronavirus-like agent, CV 777, and several coronaviruses. Arch Virol. 1981;68:45–52. doi: 10.1007/BF01315166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilchard EI. Experimental transmission of transmissible gastroenteritis virus by starlings. Am J Vet Res. 1965;26:1177–1179. [PubMed] [Google Scholar]
- Pillatzki A, Gauger P, Madson D, Burrough E, Zhang J, Chen Q, Magstadt D, Arruda P, Stevenson G, Yoon KJ. Experimental inoculation of neonatal piglets with feed naturally contaminated with porcine epidemic diarrhea virus (PEDV) J Swine Health Prod. 2015;23:317–320. [Google Scholar]
- Putics A, Gorbalenya AE, Ziebuhr J. Identification of protease and ADP-ribose 1''-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J Gen Virol. 2006;87:651–656. doi: 10.1099/vir.0.81596-0. [DOI] [PubMed] [Google Scholar]
- Qian S, Jia X, Gao Z, Zhang W, Xu Q, Li Z. Isolation and identification of porcine deltacoronavirus and alteration of immunoglobulin transport receptors in the intestinal mucosa of PDCoV-infected piglets. Viruses. 2020;12:79. doi: 10.3390/v12010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raihana RR, Hayakawa M, Sugiura E, Sugiura H, Hanaki K, Taniguchi T, Honda E. Analysis of the properties of neutralizing monoclonal antibodies against the hemagglutinating encephalomyelitis virus and inhibition of HEV infection by specific MAb. J Vet Med Sci. 2009;71:447–452. doi: 10.1292/jvms.71.447. [DOI] [PubMed] [Google Scholar]
- Roe CK, Alexander TJA. Disease of nursing pigs previously unreported in Ontario. Can J Comp Med Vet Sci. 1958;22:305–307. [PMC free article] [PubMed] [Google Scholar]
- Saif LJ. Coronavirus immunogens. Vet Microbiol. 1993;37:285–297. doi: 10.1016/0378-1135(93)90030-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif LJ, Sestak K. Transmissible gastroenteritis virus and porcine respiratory coronavirus. In: Straw BE, editor. Diseases of swine. 9. Ames: Blackwell Publishing; 2006. pp. 489–516. [Google Scholar]
- Saif LJ, Pensaert MB, Sestak K, Yeo S, Jung K. Coronaviruses. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of swine. 10. Ames: Wiley; 2012. pp. 501–524. [Google Scholar]
- Sestak K, Lanza I, Park SK, Weilnau PA, Saif LJ. Contribution of passive immunity to porcine respiratory coronavirus to protection against transmissible gastroenteritis virus challenge exposure in suckling pigs. Am J Vet Res. 1996;57:664–671. [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liang L, Cui P, Wang J, Zhang X, Guan Y, Chen H, Bu Z (2020) Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. BioRxiv:2020.03.30.015347 [DOI] [PMC free article] [PubMed]
- Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Moon H, Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vac Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrigl A, Fernández SR, Stoiber F, Pikalo J, Sattler T, Schmoll F. First detection, clinical presentation and phylogenetic characterization of Porcine epidemic diarrhea virus in Austria. BMC Vet Res. 2015;11:310. doi: 10.1186/s12917-015-0624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Tan L, Li Y, He J, Hu Y, Cai X, Liu W, Liu T, Wang J, Li Z, Yuan X, Zhan Y, Yang L, Deng Z, Wang N, Yang Y, Wang A. Epidemic and genetic characterization of porcine epidemic diarrhea virus strains circulating in the regions around Hunan, China, during 2017–2018. Arch Virol. 2020;165:877–889. doi: 10.1007/s00705-020-04532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LL, Peiris JS, Smith GJ, Chen H, Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeravechyan S, Frantz PN, Wongthida P, Chailangkarn T, Jaru-Ampornpan P, Koonpaew S, Jongkaewwattana A. Deciphering the biology of porcine epidemic diarrhea virus in the era of reverse genetics. Virus Res. 2016;226:152–217. doi: 10.1016/j.virusres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns S, Conceição-Neto N, Christiaens I, Zeller M, Desmarets LM, Roukaerts ID, Acar DD, Heylen E, Matthijnssens J, Nauwynck HJ (2015) Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium, January 2015. Genome Announc 3:pii:e00506–15 [DOI] [PMC free article] [PubMed]
- USDA (2017) Swine Enteric Coronavirus Disease (SECD) Situation Report. https://www.aphis.usda.gov/animal_health/animal_dis_spec/swine/downloads/secd_sit_rep_11_30_17.pdf. Animal and Plant Health Inspection Service
- Valkó A, Bálint Á, Bozsa Á, Cságola A. Prevalence of antibodies against transmissible gastroenteritis virus (TGEV) in Hungary. Vet Anim Sci. 2019;7:100042. doi: 10.1016/j.vas.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun K, Cox E, Cellebaut P, Pensaert MB (1990) Milk of sows infected with the porcine respiratory coronavirus: induction of IgA antibodies against transmissible gastroenteritis virus and protective capacity against intestinal infection in piglets. In: 11th Congres of the international pig veterinary society at: Lausanne
- Van Gucht S, Atanasova K, Barbé F, Cox E, Pensaert M, Van Reeth K. Effect of porcine respiratory coronavirus infection on lipopolysaccharide recognition proteins and haptoglobin levels in the lungs. Microbes Infect. 2006;8:1492–1501. doi: 10.1016/j.micinf.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nieuwstadt AP, Zetstra T, Boonstra J. Infection with porcine respiratory coronavirus does not fully protect pigs against intestinal transmissible gastroenteritis virus. Vet Rec. 1989;125:58–60. doi: 10.1136/vr.125.3.58. [DOI] [PubMed] [Google Scholar]
- Vannier P. Disorders induced by the experimental infection of pigs with the porcine respiratory coronavirus (P.R.C.V.) Zentralbl Veterinarmed B. 1990;37:177–180. doi: 10.1111/j.1439-0450.1990.tb01043.x. [DOI] [PubMed] [Google Scholar]
- Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow K, Rovira A, Collins J, Saif LJ. Distinct characteristics and complex Evolution of PEDV strains, North America, May 2013–February 2014. Emerg Infect Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova AN, Saif LJ (2013) Biological aspects of the interspecies transmission of selected coronaviruses. In: Singh SK (ed) Viral infections and global change. Wiley, pp 393–418
- Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Byrum B, Zhang Y. Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg Infect Dis. 2014;20:1594–1595. doi: 10.3201/eid2009.140756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hu W, Fan C. Structural and biochemical characterization of SADS-CoV papain-like protease 2. Protein Sci. 2020;29:1228–1241. doi: 10.1002/pro.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YW, Yue H, Fang W, Huang YW. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015;3:e00945–e1015. doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl HM, Copps J, Drebot MA, Marszal P, Smith G, Gren J, Andonova M, Pasick J, Kitching P, Czub M. Susceptibility of pigs and chickens to SARS coronavirus. Emerg Infect Dis. 2004;10:179–184. doi: 10.3201/eid1002.030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R. Neutralizing antibody decay and lack of contact transmission after inoculation of 3- and 4-day-old piglets with porcine respiratory coronavirus. J Vet Diagn Invest. 2002;14:525–527. doi: 10.1177/104063870201400617. [DOI] [PubMed] [Google Scholar]
- Wesley RD, Woods RD. Induction of protective immunity against transmissible gastroenteritis virus after exposure of neonatal pigs to porcine respiratory coronavirus. Am J Vet Res. 1996;57:157–162. [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Yip CC, Tsoi H, Chan K, Yuen K. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood EN. An apparently new syndrome of porcine epidemic diarrhoea. Vet Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhang Y, Gong L, Huang L, Lin Y, Qin J, Du Y, Zhou Q, Xue C, Cao Y. Isolation and characterization of a highly pathogenic strain of Porcine enteric alphacoronavirus causing watery diarrhoea and high mortality in newborn piglets. Transbound Emerg Dis. 2019;66:119–130. doi: 10.1111/tbed.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Qin P, Wang B, Liu Y, Xu GH, Peng L, Zhou J, Zhu SJ, Huang YW. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J Virol. 2019;93:e01448–e1519. doi: 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B, Curtis KM, Baric RS. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liang Q, Li B, Cui X, Wei X, DING Q, Wang Y, Hu H, (2019) Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China. Prev Vet Med 166:8–15 [DOI] [PMC free article] [PubMed]
- Zhang M, Li W, Zhou P, Liu D, Luo R, Jongkaewwattana A, He Q. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: a novel strategy for vaccine design. Emerg Microbes Infect. 2019;9:20–31. doi: 10.1080/22221751.2019.1701391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Han Y, Shi H, Chen J, Zhang X, Wang X, Zhou L, Liu J, Zhang J, Ji Z, Jing Z, Ma J, Shi D, Feng L. Swine acute diarrhea syndrome coronavirus-induced apoptosis is caspase- and cyclophilin D-dependent. Emerg Microbes Infect. 2020;9:439–456. doi: 10.1080/22221751.2020.1722758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Han L, Xia L, Yuan Y, Hu H. Assessment of hemagglutination activity of porcine deltacoronavirus. J Vet Sci. 2020;21:e12. doi: 10.4142/jvs.2020.21.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XLL, Wang YY, Xing SZ, Chen YS Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY (2018) Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258 [DOI] [PMC free article] [PubMed]
- Zhou L, Li QN, Su JN, Chen GH, Wu ZX, Luo Y, Wu RT, Sun Y, Lan T, Ma JY. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound Emerg Dis. 2019;66:2180–2183. doi: 10.1111/tbed.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Sun Y, Lan T, Wu R, Chen J, Wu Z, Xie Q, Zhang X, Ma J. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound Emerg Dis. 2019;66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Gao X, Zhou P, Zhang L, Zhang Y, Wang Y, Liu X. Evaluation of the immune response in conventionally weaned pigs infected with porcine deltacoronavirus. Arch Virol. 2020;12:1–6. doi: 10.1007/s00705-020-04590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Sun Y, Yan X, Tang X, Li Q, Tan Y, Lan T, Ma J. Swine acute diarrhea syndrome coronavirus (SADS-CoV) antagonizes interferon- production via blocking IPS-1 and RIG-I. Virus Res. 2020;278:197843. doi: 10.1016/j.virusres.2019.197843. [DOI] [PMC free article] [PubMed] [Google Scholar]