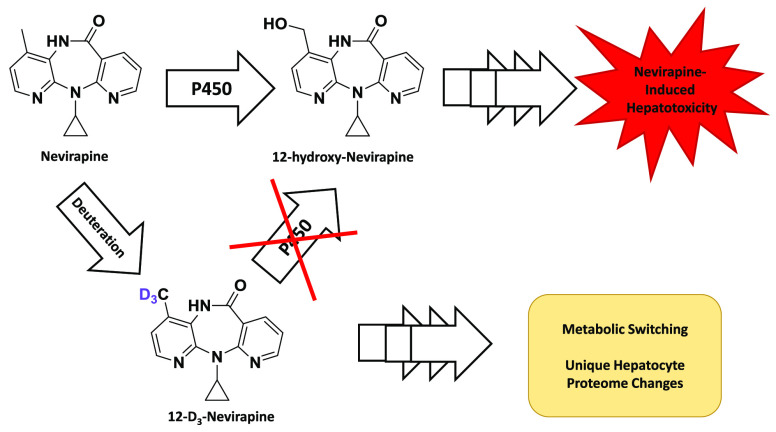
Abstract
Cytochrome P450-dependent metabolism of the anti-HIV drug nevirapine (NVP) to 12-hydroxy-NVP (12-OHNVP) has been implicated in NVP toxicities. We investigated the impact of twelfth-position trideuteration (12-D3NVP) on the hepatic metabolism of and response to NVP. Formation of 12-OHNVP decreased in human (10.6-fold) and mouse (4.6-fold) hepatocytes incubated with 10 μM 12-D3NVP vs NVP. An observed kinetic isotope effect of 10.1 was measured in human liver microsomes. During mouse hepatocyte treatment (400 μM) with NVP or 12-D3NVP, cell death was reduced 30% with 12-D3NVP vs NVP, while glucuronidated and glutathione-conjugated metabolites increased with 12-D3NVP vs NVP. Using mass spectrometry proteomics, changes in hepatocyte protein expression, including an increase in stress marker insulin-like growth factor-binding protein 1 (IGFBP-1), were observed with 12-D3NVP vs NVP. These results demonstrate that while deuteration can reduce P450 metabolite formation, impacts on phase II metabolism and hepatocyte protein expression should be considered when employing deuteration to reduce P450 metabolite-related hepatotoxicity.
Introduction
Substitution of hydrogen atoms at the site of cytochrome P450 (CYP or P450) metabolism with the heavier isotope of hydrogen, deuterium, is a useful strategy in controlling P450 oxygen insertion at the site of deuteration.1 In 2017, the first deuterated drug, deutetrabenazine, was approved by the FDA for treatment of chorea associated with Huntington’s disease as well as tardive dyskinesia.2 This compound is a deuterated form of the already approved tetrabenazine, for which deuteration was a successful strategy for reducing CYP2D6 metabolism. This increased the active drug half-life and resulted in a lower required dosage and more favorable safety profile than undeuterated tetrabenazine.3
Drug deuteration has been a technique of growing interest in drug development to tackle problems caused by cytochrome P450 metabolism, such as the extension of drug half-lives through the reduction of drug metabolism by P450s at specific sites that drive clearance or blocking sites of metabolism that result in the formation of toxic metabolites or reactive intermediates. In addition, due to the fact that deuterium atoms are still able to participate in hydrogen bonding, it is thought that this change will not have a significant effect on binding of the intended drug target.4−6 The impact of deuteration on P450 activity has been reported to vary significantly. Effects ranging from no change to a 20-fold reduction in the rate of P450 hydrogen-substrate bond breaking, a necessary step in cytochrome P450 product formation (also called the intrinsic kinetic isotope effect), have been observed.1 Deuteration at one site can also result in “metabolic switching,” in which metabolism by P450s at another position is increased following a reduction of metabolism at the deuterated site.5,7
Nevirapine (NVP) is a first-generation non-nucleoside reverse transcriptase inhibitor currently used for the treatment of human immunodeficiency virus (HIV) infections, for HIV postexposure prophylaxis, and for prevention of mother-to-child transmission of HIV during childbirth.8,9 The World Health Organization includes both NVP and NVP-containing combination therapies on its most recent List of Essential Medicines.8 Unfortunately, NVP has been shown to cause life-threatening hepatotoxicity in patients.10−13 The reported incidence of nevirapine-induced hepatotoxicity ranges greatly, with one example reporting only 6.0%10 and another demonstrating an incidence of 36.1%, with 7.7% of patients experiencing severe hepatotoxicity.12 In very rare cases, patients taking NVP have experienced fulminant hepatitis, sometimes resulting in liver transplantation or even death.11,13−16
NVP is metabolized by human P450s, resulting in the formation of four monooxygenated metabolites: 2-hydroxy- (2-OH), 3-hydroxy- (3-OH), 8-hydroxy- (8-OH), and 12-hydroxy- (12-OH) NVP.17,18 The average plasma concentration of NVP during regular dosing is 16.0 μM (95% CI [10.0, 19.2]). The major monooxygenated metabolite is 12-OHNVP at 1.3 μM (95% CI [0.9, 1.9]), with 2- and 3-OHNVP being submicromolar and 8-OHNVP at times unquantifiable.19 These metabolites are precursors to subsequent NVP metabolites, including further P450 activity to produce 4-carboxy-NVP18 as well as glucuronidation by uridine 5′-diphospho-glucuronosyltransferases,18 sulfation by sulfotransferases,20 and glutathione conjugation by glutathione S-transferases.21,22
Formation of 12-OHNVP by P450s has been implicated in the hepatotoxicity of NVP as well as in skin toxicity, another severe side effect of NVP.20,23,24 It has also been shown that P450-dependent metabolism to 12-OHNVP plays a role in the formation of drug adducts with proteins and DNA, either by way of promoting downstream metabolism to a potentially reactive 12-sulfoxy-NVP conjugate or through the formation of a reactive intermediate during the production of 12-OHNVP itself.20,23−28 Interestingly, treatment of rats with a version of NVP in which the twelfth-position has been trideuterated (12-D3NVP) was demonstrated to reduce both circulating concentrations of 12-OHNVP and the incidence of skin rash vs rats treated with NVP.23 Other work has demonstrated a reduction in 12-OHNVP-protein adducts in the livers of mice and rats treated with 12-D3NVP as compared to NVP and during coincubations with NVP and a pan-CYP inhibitor preventing metabolism.24
Here, we probed the difference in P450-dependent production of monooxygenated metabolites of NVP vs 12-D3NVP in primary human and mouse hepatocytes. Given that these monooxygenated metabolites undergo subsequent phase II conjugations, we measured the levels of secondary glucuronidated and glutathione conjugated metabolites of NVP. In addition, we characterized the contribution of various human P450s in NVP metabolism and quantified the observed kinetic isotope effect of this trideuteration on 12-OHNVP production by human liver microsomes. From here, we probed the difference in hepatocyte cell death levels in response to NVP or 12-D3NVP and performed relative-quantitation proteomics analysis of hepatocytes treated with NVP and 12-D3NVP in order to probe whether there is a differential cellular response to this trideuterated compound.
Results and Discussion
Primary Human and Mouse Hepatocyte Metabolism of NVP and 12-D3NVP
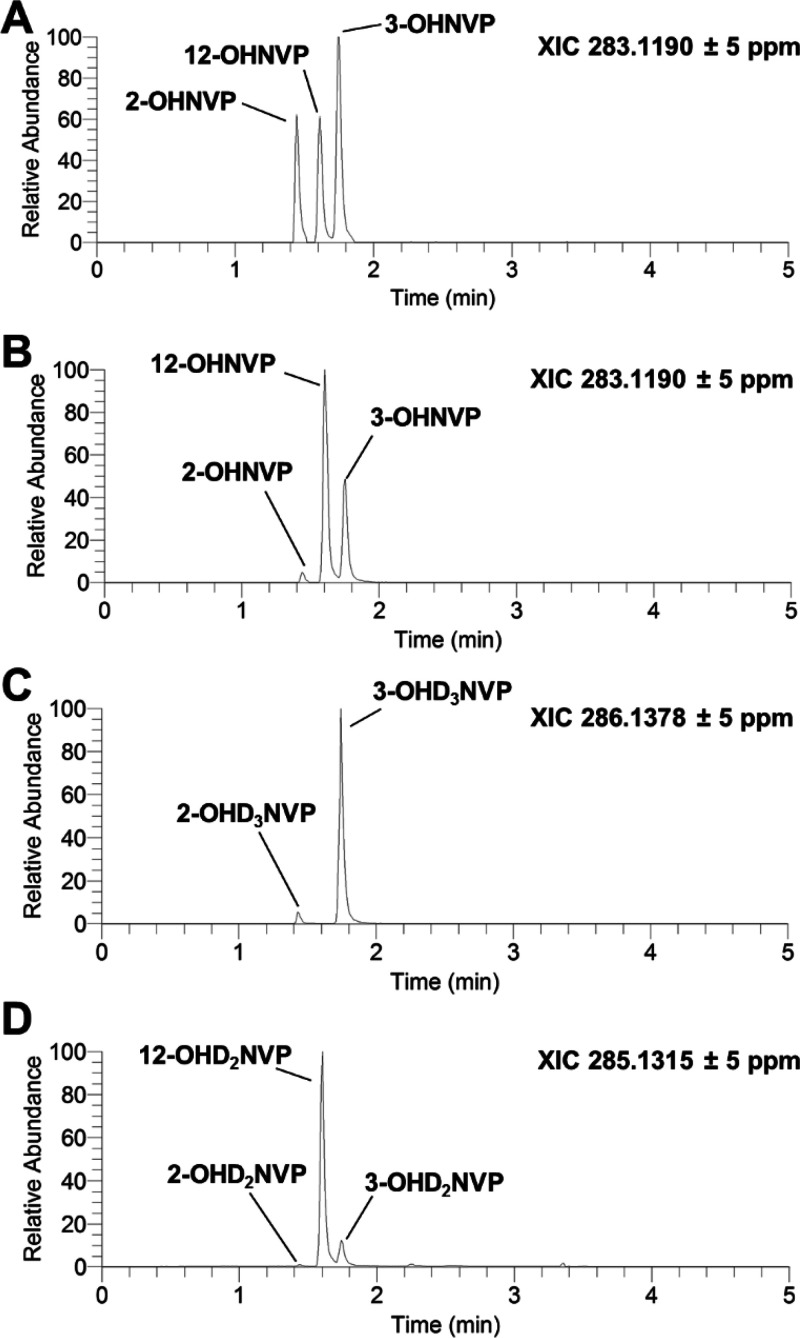
To assay the impact of twelfth-position deuteration on hepatic metabolism of NVP, we first characterized the production of P450-dependent metabolites of NVP or 12-D3NVP in the culture medium of primary hepatocytes treated with 10 μM of either of these compounds for 24 h. In our initial method development, we used C57BL/6J mouse primary hepatocytes, which have been previously employed as a model to study NVP hepatotoxicity.24 Using high resolution ultrahigh-performance liquid chromatography (uHPLC)-MS Orbitrap detection and a comparison with known standards of monooxygenated NVP metabolites (Figure 1A), we were able to identify three monooxygenated metabolites of NVP formed in primary mouse hepatocytes: 2-OHNVP, 3-OHNVP, and 12-OHNVP (Figure 1B) as well as three main monooxygenated metabolites produced from 12-D3NVP: 2-OHD3NVP, 3-OHD3NVP (Figure 1C), and 12-OHD2NVP (Figure 1D). We also identified peaks corresponding to 2- and 3-OHD2NVP, though the peak areas of these dideuterated metabolites were <10% of the trideuterated versions. Because of this, only the production of 12-OHD2NVP, 2-OHD3NVP, and 3-OHD3NVP was monitored in subsequent experiments with 12-D3NVP. No peaks corresponding to 8-OHNVP or 8-OHD3NVP were detected. No monooxygenated NVP or 12D3-NVP metabolites were detected in vehicle-treated hepatocyte medium (data not shown). With the metabolite peaks identified, we developed specific MS/MS transitions to detect these metabolites in future assays of P450 metabolism of NVP and 12-D3NVP (Supplemental Figure 1). Monitoring these fragments for quantitation, as opposed to the high-resolution parent masses, allowed for improved baseline separation of metabolite peaks in extracted ion chromatograms (XICs), especially for 12-OHNVP and 12-OHD2NVP production (Supplemental Figure 1D,H). For simplicity, in the following results and discussion, comparisons of the production of 12-OHNVP from NVP or 12-OHD2NVP from 12-D3NVP are referred to only as 12-OHNVP, with the deuteration for the product from 12-D3NVP implied. The same abbreviation is used for 2-OHNVP vs 2-OHD3NVP and 3-OHNVP vs 3-OHD3NVP production from NVP and 12-D3NVP, respectively.
Figure 1.
Characterization of P450-dependent NVP and 12-D3NVP metabolite formation in primary mouse hepatocytes. Fresh primary mouse hepatocytes from male C57BL/6J mice were cultured and treated with 10 μM NVP or 12-D3NVP for 24 h. Metabolites were extracted from the medium from these treatments and subjected to uHPLC-MS (Orbitrap) analysis. The following high resolution ions were observed to assay for the presence of P450 metabolites: 283.1190 ± 5 ppm for monooxygenated, undeuterated NVP, 286.1378 ± 5 ppm for monooxygenated, trideuterated NVP, and 285.1315 ± 5 ppm for monooxygenated, dideuterated NVP. Representative extracted ion chromatograms (XlCs) from this analysis are shown for a mixture of authentic standards for undeuterated 2-, 3-, and 12-OHNVP (A, each at 50 nM), monooxygenated, undeuterated metabolites formed during incubations with NVP (B), monooxygenated, trideuterated metabolites formed during incubations with 12-D3NVP (C), and monooxygenated, dideuterated metabolites formed during incubations with 12-D3NVP (D). Results are representations of four experimental replicates.
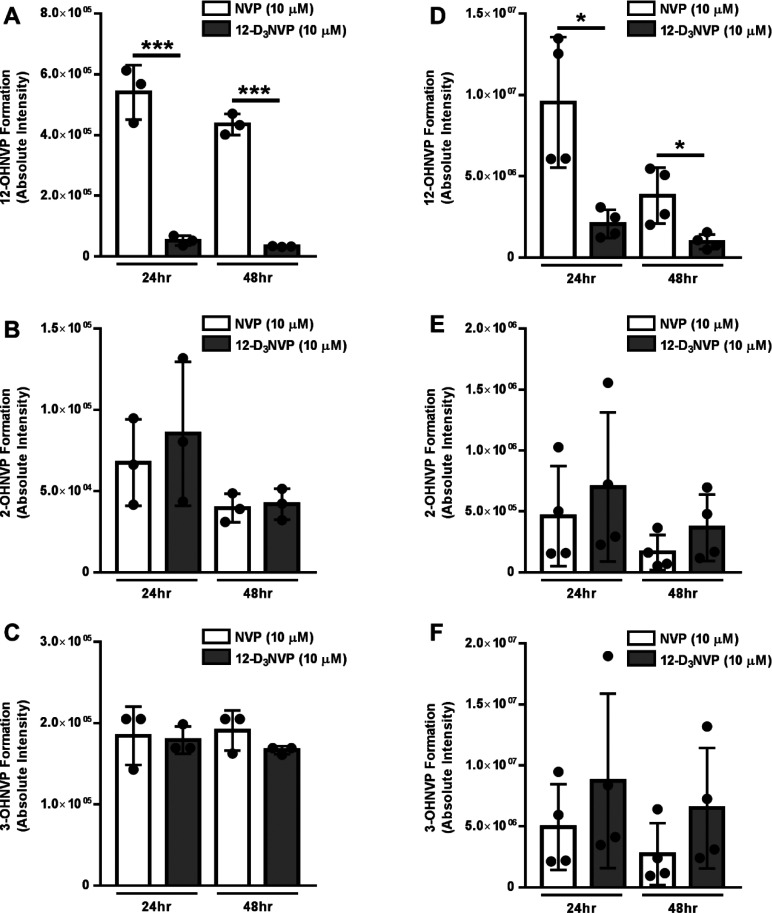
Using this method, we compared the production of these three metabolites by primary human hepatocytes treated with 10 μM of either NVP or 12-D3NVP for 24 or 48 h. Levels of 12-OHNVP production with 12-D3NVP decreased 10.6-fold (95% CI [6.7, 14.6]) at 24 h and 13.2-fold (95% CI [11.2, 15.3]) at 48 h, as compared to production with NVP (Figure 2A). Because of this reduction in 12-OHNVP production, we probed whether other P450-dependent metabolites of NVP would increase in response. No difference was observed in the production of 2- or 3-OHNVP (Figure 2B,C) and, as with mouse hepatocytes, no peak corresponding to 8-OHNVP formation was identified (data not shown). At these same treatment times and concentrations, we assayed for differences in P450-dependent metabolism in C57BL/6J mice. As with human hepatocytes, we noted decreases in 12-OHNVP production: 4.6-fold (95% CI [3.6, 5.7]) and 4.0-fold (95% CI [2.7, 5.3]) at 24 and 48 h of treatment, respectively (Figure 2D). Again, no difference was observed in either 2- or 3-OHNVP production (Figure 2E,F). These results demonstrate that trideuteration at the twelfth-position NVP is an effective method of reducing hepatic production of the 12-OHNVP metabolite in both a human hepatocyte model and in a mouse model previously employed in the study of NVP toxicities.20,24
Figure 2.
NVP and I2-D3NVP cytochrome P450-dependent metabolism in primary human and mouse hepatocytes. Cryopreserved primary human hepatocytes (A–C) and fresh primary mouse hepatocytes (D–F) were incubated with 10 μM NVP or 12-D3NVP for 24 or 48 h. Cytochrome P450 metabolites extracted from hepatocyte culture medium were measured using uHPLC-MS/MS (Orbitrap) detection. 12-OHNVP (A, D), 2-OHNVP (B, E), and 3-OHNVP (C, F) were monitored using MS/MS scans for the following transitions: 283.1190 → 223.1104 m/z (12-OHNVP), 285.1315 → 225.1230 m/z (12-OHD2NVP), 283.1190 → 161.0709 m/z (2-OHNVP), 286.1378 → 161.0709 m/z (2-OHD3NVP), 283.1190 → 242.0798 m/z (3- OHNVP), and 286.1378 → 245.0987 m/z (3-OHD3NVP). Data are representative of the mean ± standard deviation of three (human) or four (mouse) experimental replicates. Significant differences between metabolite production with NVP and 12-D3NVP were determined using an unpaired t test generating two-tailed P values (*P < 0.05 and ***P < 0.001).
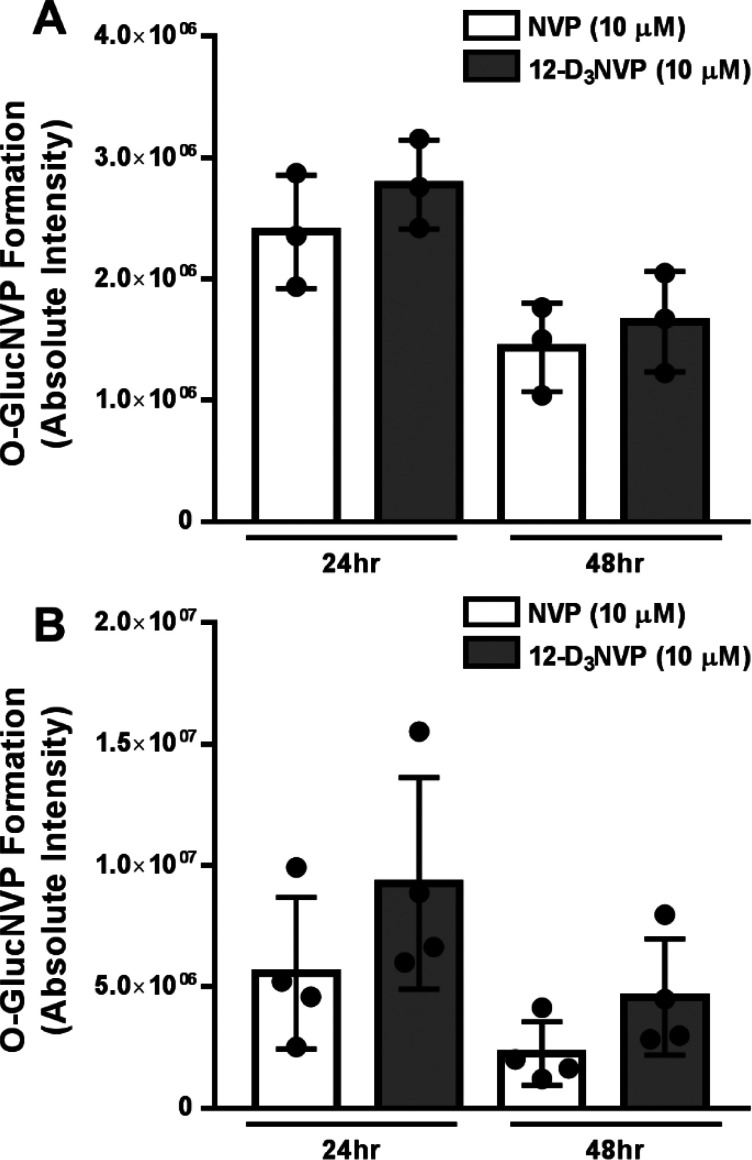
Monooxygenated metabolites of NVP can be subsequently glucuronidated to form O-glucuronide-NVP (O-GlucNVP).18 Because of this, we were interested in monitoring any impact of deuterium substitution on this subsequent metabolism of monooxygenated NVP. Again, we used C57BL/6J hepatocytes in our initial method development for the detection of these metabolites. One uHPLC-MS/MS peak corresponding to an O-GlucNVP metabolite was observed in the medium of primary mouse hepatocyte incubations with 10 μM NVP or 12-D3NVP for 24 h (Supplemental Figures 2 and 3). This peak was not detected in vehicle-treated hepatocyte medium (data not shown). In incubations with 12-D3NVP, the high-resolution accurate mass of this peak was that of a trideuterated O-glucuronidated metabolite (O-GlucD3NVP, 462.1700 m/z), which suggests that this metabolite is likely from 2- or 3-OHNVP glucuronidation, as these are the two metabolites that retain all three deuterium atoms following metabolism by cytochrome P450s. No peak corresponding to the mass of 12-O-GlucD2NVP was observed (data not shown). In our treatments of primary human and primary mouse hepatocytes with 10 μM NVP or 12-D3NVP, no difference was observed in the levels of this O-glucuronidated metabolite (Figure 3).
Figure 3.
O-glucuronidated NVP and 12-D3NVP formation in primary human and mouse hepatocytes. Cryopreserved primary human hepatocytes (A) and fresh primary mouse hepatocytes (B) were incubated with 10 μM NVP or 12-D3NVP for 24 or 48 h. O-glucuronidated metabolites extracted from hepatocyte culture medium were measured using uHPLC-MS (Orbitrap) detection. The peak area of the XICs for the following high resolution ions was observed to assay metabolite formation: 459.1510 ± 5 ppm for O-glucuronidated, undeuterated NVP (O-GlucNVP) and 462.1700 ± 5 ppm for O-glucuronidated, trideuterated NVP (O-GlucD3NVP). Data are representative of the mean ± standard deviation of three (human) or four (mouse) experimental replicates.
Identifying Human Cytochrome P450s Involved in the Metabolism of NVP
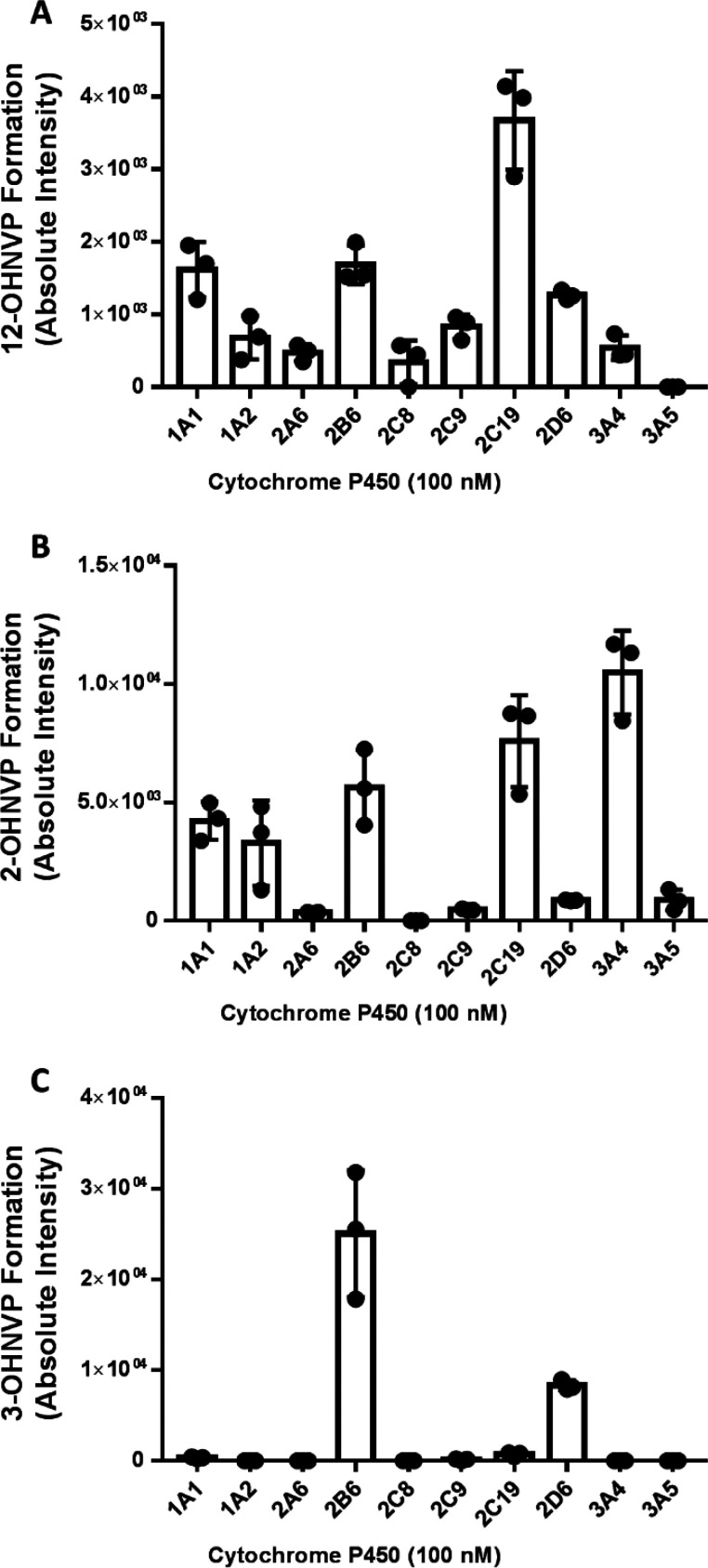
Given our observed reduction in P450-mediated formation of 12-OHNVP with 12-D3NVP as compared to NVP, we became interested in determining which human P450s are responsible for the formation of 12-OHNVP. We performed incubations of NVP (10 μM) with individual human cytochrome P450 enzymes (100 nM active enzyme) and NADPH regenerating reagents. We employed a panel of ten cytochrome P450 enzymes commonly implicated in drug metabolism.29 As in our incubations with hepatocytes, peaks corresponding to 2-, 3-, and 12-OHNVP were observed in these incubations, and no peak corresponding to 8-OHNVP was observed (data not shown). For 12-OHNVP (Figure 4A), all but one P450 tested (CYP3A5) formed this metabolite, with CYP2C19 incubations resulting in the highest 12-OHNVP formation. CYP3A4 produced the greatest levels of 2-OHNVP (Figure 4B) and CYP2B6 most readily formed 3-OHNVP (Figure 4C).
Figure 4.
NVP monooxygenated metabolites produced during incubations with individual cDNA-expressed human P450 enzymes. Insect cell microsomes containing one the following cDNA-expressed cytochrome P450s (100 nM): CYP1A1, -1A2, -2A6, -2B6, -2D6, -2C8, -2C9, -2C19, -3A4, and -3A5, were incubated with NVP (10 μM) and NADPH regenerating reagents for 30 min. 12-OHNVP (A), 2-OHNVP (B), and 3-OHNVP (C) were extracted from these incubations and measured using uHPLC-MS/MS (triple quadrupole) detection for the following transitions: 283.1 → 223.0 m/z (12-OHNVP), 283.1 →160.9 m/z (2-OHNVP), and 283.1 → 241.9 m/z (3-OHNVP). Data are representative of the mean ± standard deviation of three experimental replicates.
Cytochrome P450s do not exist in equal concentrations within the human liver. In order to determine the contribution of these P450s in a model system that is more representative of the relative abundances of P450s in the human liver, we measured monooxygenated NVP metabolite formation in human liver microsomes. Liver microsomes are endoplasmic reticulum fractions from liver homogenate, representing the diversity and relative abundances of P450s in the hepatic endoplasmic reticulum.30 We performed 1 h co-incubations with human liver microsomes (0.5 mg/mL), NADPH regenerating reagents, and NVP (10 μM) with and without the addition of small molecule P450 inhibitors (using concentrations resulting in selective inhibition of specific P450 enzymes) as indicated in Supplemental Figure 4.31,32 Interestingly, no P450 inhibitor was able to reduce the formation of 12-OHNVP, including (+)-N-3-benzyl-nirvanol, a CYP2C19 inhibitor (Supplemental Figure 4A). P450 production of 2-OHNVP was decreased 8.2-fold (95% CI [2.5, 14.0]) by ketoconazole, a CYP3A inhibitor (Supplemental Figure 4B), and 3-OHNVP production was decreased 2.6-fold (95% CI [1.0, 4.2]) by PPP, a CYP2B6 inhibitor (Supplemental Figure 4C).
These results demonstrate that multiple P450s may be responsible for the production of 12-OHNVP at a clinically relevant concentration (10 μM) and that CYP3A4 and CYP2B6 are the P450s responsible for 2-OHNVP and 3-OHNVP production, respectively. In the initial work characterizing the P450 metabolism of NVP, similar results were observed using a smaller sampling (four) of human P450s.17 Erickson et al. observed 12-OHVNP production by multiple P450s and no impact of antibody-based P450 inhibition on 12-OHNVP production during concentrations with 25 μM NVP. However, there was an impact on the production of 12-OHNVP using anti-CYP3A4 with 400 μM NVP and of ketoconazole (CYP3A inhibitor) with 100 μM NVP. From their work, Erickson et al. concluded that several P450s (including CYP3A4, -2D6, and -2C9) may contribute to 12-OHNVP formation at lower, more clinically relevant concentrations of NVP. They also attributed 2-OHNVP to the CYP3A subfamily and 3-OHNVP to CYP2B6.17 Though we could have continued with a more extensive characterization by varying substrate concentrations or trying different modes of inhibition, we feel results supported quantitation of the kinetic isotope effect of the twelfth-position deuterium substitution in a multi-P450 system such as human liver microsomes.
Quantifying the Kinetic Isotope Effect of Twelfth-Position Deuteration of NVP on 12-OHNVP Production in Human Liver Microsomes
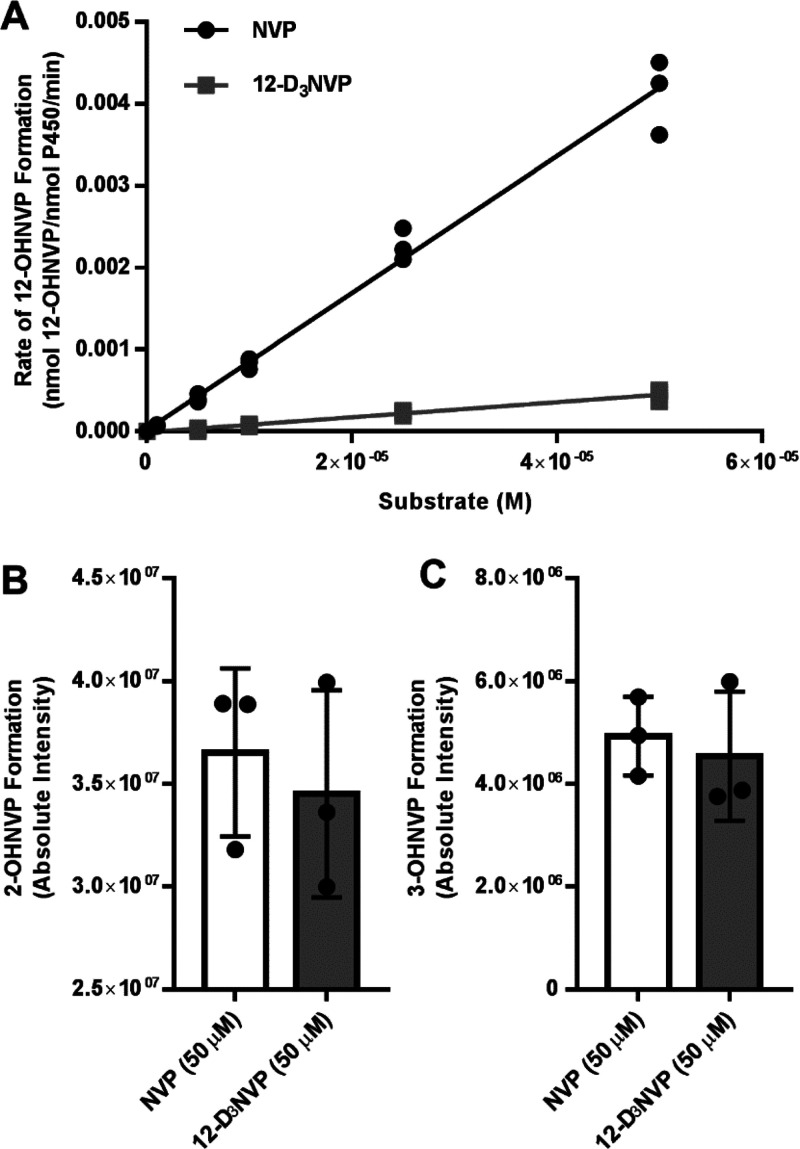
Preliminary incubations were performed in human liver microsomes to determine optimal microsomal protein concentrations and incubation time points to obtain linear rates of 12-OHNVP formation from NVP (data not shown). We performed noncompetitive intermolecular experiments during which liver microsomes were incubated with NADPH regenerating reagents and a range of concentrations (1–400 μM) of either NVP or 12-D3NVP. We were unable to achieve saturation of the rate of 12-OHNVP production at 400 μM substrate, and beyond this concentration, substrate solubility became limiting (data not shown). Because of this, we monitored the rate of 12-OHNVP production, using uHPLC-MS/MS Orbitrap detection, with lower concentrations of NVP and 12-D3NVP (0, 1, 5, 10, 25, and 50 μM), where the curve of rate of 12-OHNVP production vs substrate concentration maintained linearity for both NVP and 12-D3NVP.
The terminology established by Norththrop is used to distinguish the different parameters observed in this work.33 Levels of 12-OHNVP were quantifiable in incubations with all concentrations of NVP and in incubations with 5, 10, 25, and 50 μM 12-D3NVP (Figure 5A). Catalytic efficiency was observed (Kcat/Km, the slope of the rate vs substrate concentration line at nonsaturating conditions, Figure 5A) for 12-OHNVP formation with both NVP and 12-D3NVP with values of 83.7 (95% CI [78.2, 89.1]) M–1 min–1 and 9.2 (95% CI [8.1, 10.3]) M–1 min–1, respectively (Figure 5A). The average fold (NVP/12-D3NVP) change in Kcat/Km, known as D(V/K), was 9.1 (95% CI [7.4, 10.9]). A decrease in the rate of 12-OHNVP production (with 12-D3NVP as compared to NVP across all concentrations tested) indicates a primary, normal observed kinetic isotope effect (DK) of 10.1 (95% CI [9.4, 10.8]). We were unable to calculate an intrinsic kinetic isotope effect (Dk), which is a measure of the impact on the P450 hydrogen bond-breaking step specifically.1 However, it is our understanding that our observed D(V/K) is fairly large considering the same measurements for other P450 reactions: 2–3,34 3.1–3.18,35 4.5,36 5–11,37 and >10.34 Of note, one previous study observed DV/K to be 10.9 in microsomal incubations but only 3.7 in CYP2E1-specific incubations.38 In the future, it would be interesting to survey the diversity of kinetic isotope effects for 12-OHNVP from 12-D3NVP across different P450s, given that NVP appears to be a substrate for multiple P450s.
Figure 5.
Rate of 12-OHNVP production as well as 2- and 3-OHNVP formation from NVP and 12-D3NVP in human liver microsome incubations. Liver microsomes (2.5 mg/mL) prepared from pooled human donor livers were incubated with a range of concentrations of NVP (0, 1, 5, 10, 25, and 50 μM) or 12-D3NVP (0, 5, 10, 25, and 50 μM) and NADPH regenerating reagents for 30 min. Cytochrome P450 metabolites extracted from these incubations were measured using uHPLC-MS/MS (Orbitrap) detection. Metabolites were monitored using MS/MS scans for the following transitions: 283.1190 → 223.1104 m/z (12-OHNVP), 285.1315 → 225.1230 m/z (12-OHD2NVP), 283.1190 → 161.0709 m/z (2-OHNVP), 286.1378 → 161.0709 m/z (2-OHD3NVP), 283.1190 → 242.0798 m/z (3- OHNVP), and 286.1378 → 245.0987 m/z (3-OHD3NVP). The rate of formation of 12-OHNVP (A) was quantified by comparing peak areas of 12-OHNVP or 12-OHD2NVP to those of a standard curve of 12-OHNVP prepared in liver microsomal incubations without NADPH regenerating reagents. Formation of 2-OHNVP (B) and 3-OHNVP(C) from 50 μM NVP or 12-D3NVP is also shown. For rates of 12-OHNVP production, all data points are shown, in addition to results of linear curve fitting for the graphs of rate vs substrate concentration. For 2- and 3-OHNVP production, data are representative of the mean ± standard deviation of three experimental replicates.
As in our hepatocyte incubations, we also measured 2- and 3-OHNVP with NVP and 12-D3NVP to probe for potential metabolic switching. No statistically significant changes were observed in 2- or 3-OHNVP formation using 50 μM 12-D3NVP vs NVP (Figure 5B,C, respectively) or with any of the other NVP concentrations tested (5, 10, 25, 100, 200, and 400 μM, data not shown). We found these results as well as the lack of change observed for both 2- and 3-OHNVP production in hepatocyte medium intriguing, as metabolic switching, in which other drug metabolite levels increase in response to decreased metabolism at one position, has been known to occur with deuterium substitution.5,7,39−41 Our observation may be due to differences in P450 contributions to metabolism at these different positions of the NVP scaffold. Since we did not see a significant decrease in 12-OHNVP with inhibitors that reduced production of 2-OHNVP and 3-OHNVP, this suggests that the enzymes responsible for 2- and 3-OHNVP production (CYP3A4 and CYP2B6, respectively) do not play a measurable role in the conversion of NVP to 12-OHNVP. With that, modifying the dynamics of this specific conversion may not impact the rates of CYP3A4/CYP2B6 production of 2- or 3-OHNVP.
Hepatocyte Viability during Treatments with NVP and 12-D3NVP
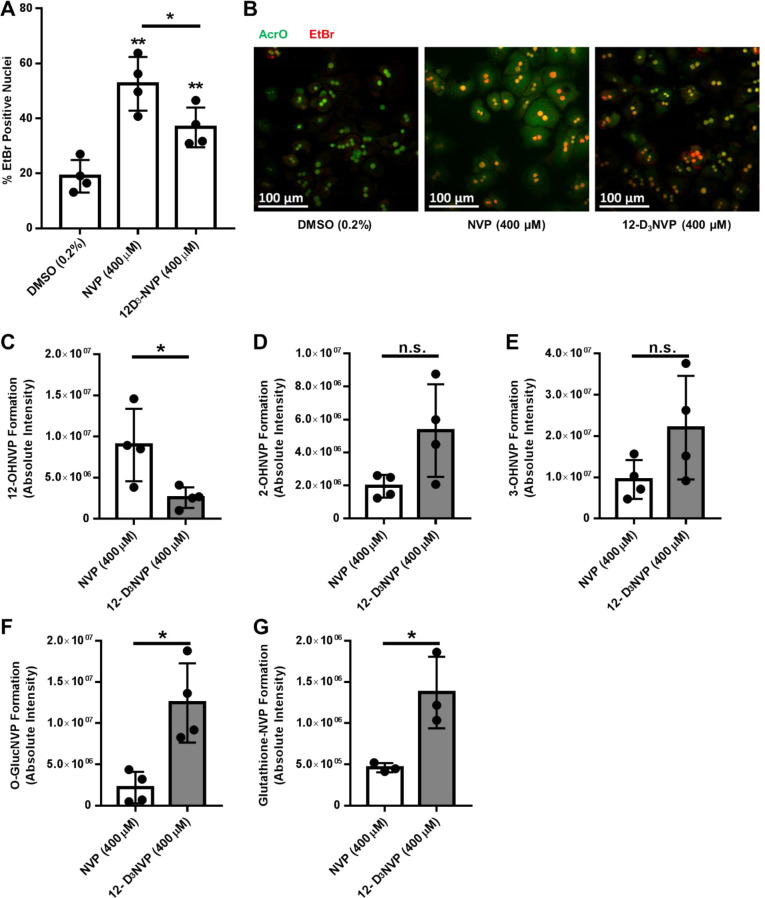
Given the observed reduction in formation of 12-OHNVP from 12-D3NVP using hepatocytes, we probed whether there is a difference in hepatocyte viability during treatments with NVP and 12-D3NVP. Primary mouse hepatocytes were treated with 400 μM NVP or 12-D3NVP for 8 h, and their viability was measured using ethidium bromide (EtBr)/acridine orange (AcrO) costaining and compared to treatment with vehicle alone (Figure 6A,B). EtBr-positive nuclei were monitored as a marker for cell death. Treatment with 10 μM staurosporine for 8 h was used as a positive control (data not shown). During treatment with NVP, the percentage of EtBr-positive cell nuclei was 52.6% (95% CI [37.0, 68.2]), which is notably greater than the 19.0% (95% CI [9.5, 28.4]) observed for vehicle treatments. The percentage of EtBr-positive cell nuclei in treatments using 12-D3NVP was 36.8% (95% CI [25.3, 48.2]), a reduction from levels measured following treatments with NVP, though still elevated above vehicle control.
Figure 6.
EtBr/AcrO viability staining and metabolite formation in primary mouse hepatocytes treated with NVP or 12-D3NVP. Fresh primary mouse hepatocytes were incubated with vehicle (0.2% DMSO), 400 μM NVP, or 400 μM 12-D3NVP for 8 h. Quantitation of nuclear EtBr incorporation from EtBr/AcrO viability costaining (A) as well as representative images of EtBr/AcrO costaining (B) are shown for these incubations. Cytochrome P450 metabolites extracted from hepatocyte culture medium were measured using uHPLC-MS/MS (Orbitrap) detection. 12-OHNVP (C), 2-OHNVP (D), and 3-OHNVP (E) were monitored using MS/MS scans for the following transitions: 283.1190 → 223.1104 m/z (12-OHNVP), 285.1315 → 225.1230 m/z (12-OHD2NVP), 283.1190 → 161.0709 m/z (2-OHNVP), 286.1378 → 161.0709 m/z (2-OHD3NVP), 283.1190 → 242.0798 m/z (3- OHNVP), and 286.1378 → 245.0987 m/z (3-OHD3NVP). One O-glucuronidated metabolite (F) was extracted from the cell culture medium and subjected to uHPLC-MS (Orbitrap) analysis. The following high resolution ions were observed to assay for the presence of O-glucuronidated metabolite: 459.1510 ± 5 ppm for O-GlucNVP and 462.1700 ± 5 ppm for O-GlucD3NVP. One glutathione conjugate (G) was extracted from cell pellets and subjected to uHPLC-MS (Orbitrap) analysis. The following high resolution ions were observed to assay for the presence of this metabolite: 572.1922 ± 5 ppm for glutathione-NVP and 575.2110 ± 5 ppm for glutathione-D3NVP. Data are representative of the mean ± standard deviation of four experimental replicates, except for glutathione detection, which has three experimental replicates. Significant differences from vehicle (A) or for the indicated comparisons (A, C, D, E, F,G) were determined using an unpaired t test generating two-tailed P values (*P < 0.05; **P < 0.01).
As with previously published in vitro studies involving NVP, we had to use high, supra-therapeutic concentrations to observe any cell death with NVP.28,42,43 We did see a reduction in C57BL/6J hepatocyte death using 12-D3NVP as compared to NVP, though the clinical implications of this degree of change are difficult to ascertain (from 52.6% of cells dead/dying with NVP to 36.8% with 12-D3NVP). Though the observed reduction suggests that 12-OHNVP formation may play a role in hepatocyte death with NVP, given the supra-therapeutic concentrations needed to stimulate cell death and the degree of decrease observed, future experimentation is needed. Employing other in vivo animal and in vitro cell culture models may prove useful in further elucidating the role of 12-OHNVP formation in NVP toxicities. C57BL/6 mice, from which we prepared our primary hepatocytes, have been previously employed in the study of NVP toxicity, with varying results. Previously, NVP-protein adduct formation has been observed in NVP incubations with C57BL/6 liver microsomal fractions, and adduct formation was then decreased when microsomes were instead incubated with 12-D3NVP. This work also demonstrated that in vivo NVP dosing of C57BL/6 mice resulted in mild hepatotoxicity, though the severity and duration of toxicity was greater in in vivo rat models.24 Probing the effects of this trideuteration on NVP-induced hepatotoxicity in hepatocytes derived from other animal models may prove useful in understanding the role of 12-OHNVP production in NVP hepatotoxicity. In addition, advances in the culture of primary human hepatocytes models, such as 3-D spheroid culture, may facilitate more pharmacologically relevant incubations with NVP. Spheroid culture would allow for long-term incubations that may demonstrate cell death during incubations with therapeutically relevant concentrations of NVP.44 This technique also allows for coculture with immune cells.45 Both of these aspects may be useful in profiling NVP-induced hepatotoxicity in a human-derived cell culture model, especially given the proposed involvement of immune hypersensitivity in NVP-related adverse events.46
Given that incubations with 400 μM NVP were needed to induce NVP-dependent death, it is reasonable to propose that the differences in P450 metabolism between NVP and 12-D3NVP observed using lower (10 μM) hepatocyte treatment incubations may not be the same with 400 μM. Because of this, we probed NVP and 12-D3NVP metabolite formation in our primary mouse hepatocyte cell death incubations. After 8 h, production of 12-OHNVP was reduced 3.5-fold (95% CI [3.2, 3.8]) in incubations with 400 μM 12-D3NVP as compared to 400 μM NVP (Figure 6C). The formation of both 2- and 3-OHNVP trended toward an increase with 12-D3NVP as compared to NVP, though the observed changes did not rise to the level of statistical significance (Figure 6D,E). Given this trend, we also measured the formation of the downstream O-GlucNVP metabolite, likely formed from 2- or 3-OHNVP, and observed a 2.8-fold (95% CI [1.9, 3.4]) increase with 12-D3NVP as compared to NVP (Figure 6F). With this observed increase, we became interested in quantifying other subsequent metabolites of monooxygenated NVP. Glutathione conjugation has been previously observed at both the 3- and 12- positions.22 We were able to profile one glutathione conjugated metabolite in our primary mouse hepatocyte incubations (Supplemental Figures 5 and 6). When 12-D3NVP was used as the substrate, this metabolite retained all three deuterium atoms (Supplemental Figures 5 and 6). Because of this, we propose that this is likely 3-glutathione-NVP, rather than 12-glutathione-NVP, which would only have two deuterium atoms remaining following the metabolism by P450s, sulfotransferases, and glutathione-S-transferases necessary to produce 12-glutathione-NVP. Interestingly, during these treatments, we observed a 3.0-fold increase (95% CI [0.486, 5.514]) in this glutathione-conjugated metabolite with 12-D3NVP as compared to NVP (Figure 6G). We were unable to detect sulfoxy-NVP conjugates in these incubations (data not shown).
These increases in glucuronide and glutathione metabolites suggest that during these 400 μM incubations NVP deuteration may result in metabolic switching. In incubations of primary mouse hepatocytes with 10 μM NVP or 12-D3NVP, we saw no difference in 2- or 3-OHNVP production or in O-GlucNVP formation. This may be explained by differences in various P450 affinities for NVP. In incubations using 10 μM substrate incubations, several P450s may bind and metabolize NVP each to distinct monooxygenated metabolites and would not be subject to switching. At 400 μM substrate concentrations, there may be a mouse P450 with a low affinity for NVP that can bind and form all three −OHNVP metabolites and, therefore, is subject to switching. The potential for metabolic switching could prove a hurdle for scientists interested in applying deuteration to an already approved therapeutic. For instance, there is some evidence that the 3-OHNVP metabolite may also be bioreactive and contribute to adverse events with NVP.19,22 If metabolic switching to this metabolite does occur with 12-D3NVP, this compound could prove to be equally if not more toxic than NVP. Previous work by Pinheiro et al. regarding NVP metabolism in rat hepatocyte culture has demonstrated an increased detection in both the number of different NVP metabolites and in the concentrations of those metabolites formed using 3-D spheroid culture vs 2-D culture. In this work, glucuronide and sulfoxy metabolites of 12-OHNVP were only detectable in the 3-D culture system.28 It would be informative to probe metabolite formation with NVP and 12-D3NVP in a 3-D spheroid system, which may allow us to profile the impact of this trideuteration on the levels of glucuronide and glutathione conjugates of 12-OHNVP as well as of sulfoxy-NVP.
Relative-Quantitation Proteomics Analysis of NVP and 12-D3NVP Treated Hepatocytes
Though deuterium substitution in this case is intended to slow metabolism at a specific position, it can also lead to other unintended consequences such as increases in other metabolites, as we have observed with 400 μM incubations with NVP and 12-D3NVP.23 This altered exposure to drug metabolites with NVP vs 12-D3NVP may result in alternative cell signaling in response to exposure to NVP vs 12-D3NVP. To probe for differential responses to these two compounds in hepatocytes, we performed relative-quantitation label-free proteomics analysis of primary mouse hepatocytes treated with vehicle (0.2% DMSO), 400 μM NVP, or 400 μM 12-D3NVP for 8 h. In addition, to identify potential changes at more therapeutically relevant concentrations in a human model, we also performed this analysis on primary human hepatocytes treated with vehicle (0.1% DMSO), 10 μM NVP, or 10 μM 12-D3NVP for a longer treatment duration, 48 h. Our discussion will focus mainly on the differences between NVP and 12-D3NVP treatment (Tables 1 and 2). However, we also have included changes with each treatment as compared to vehicle (Supplementary Tables 1–4). Comparisons of housekeeping proteins, actin and glyceraldehyde-3-phosphate dehydrogenase, are also shown in all the tables, though no change in their expression was observed across treatments.
Table 1. Relative-Quantitation Proteomics Analysis of Protein Expression Changes with 12-D3NVP Treatment, as Compared to NVP, of Primary Mouse Hepatocytesa.
| changes
with 12-D3NVP treatment (in comparison to NVP) | ||||
|---|---|---|---|---|
| UniProt accession | UniProt entry | gene name | fold change | P value |
| P47876 | insulin-like growth factor-binding protein 1 | Igfbp1 | 4.7 ± 1.38 | 0.0048 |
| E9QPl2 | putative methyltransferase NSUN7 | Nsun7 | 3.89 ± 2.73 | 0.0244 |
| Q642L7 | MCG13441 | Rps27a | 3.03 ± 2.51 | 0.0144 |
| D3Z5F7 | protein Gm20521 | Gm20521 | 2.64 ± 0.79 | 0.0079 |
| P70441 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | Slc9a3r1 | 2.5 ± 0.86 | 0.0319 |
| O08600 | endonuclease G, mitochondrial | Endog | 2.07 ± 0.47 | 0.0336 |
| Q6A0A9 | constitutive coactivator of PPAR-gamma-like protein 1 | Fam120A | 2.06 ± 0.6 | 0.0204 |
| Q91XE4 | N-acyl-aromatic-l-amino acid amidohydrolase (carboxylate-forming) | Acy3 | 1.6 ± 0.07 | 0.0067 |
| Q8K2C6 | NAD-dependent protein deacylase sirtuin-5, mitochondrial | Sirt5 | 1.56 ± 0.29 | 0.0481 |
| Q3TMX5 | arginine-rich, mutated in early stage tumors, isoform CRA_b | Manf | 1.3 ± 0.06 | 0.0066 |
| A0A0A0MQF6 | glyceraldehyde-3-phosphate dehydrogenase | Gapdh | 1.4 ± 1.01 | 0.7024 |
| P60710 | actin, cytoplasmic 1 | Actb | 1.05 ± 0.4 | 0.7289 |
| Q4VBG1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 | Ddx47 | –1.5 ± 0.1 | 0.0094 |
| Q921l1 | serotransferrin | Tf | –1.52 ± 0.17 | 0.0440 |
| B2RT89 | predicted gene, EG434674 | Slc22a28 | –1.66 ± 0.16 | 0.0287 |
| Q99N96 | 39S ribosomal protein L1, mitochondrial | Mrpl1 | –1.87 ± 0.19 | 0.0295 |
| Q4VA32 | thioesterase superfamily member 2 | Acot13 | –1.97 ± 0.16 | 0.0055 |
| Q3KNM2 | E3 ubiquitin-protein ligase MARCH5 | March5 | –2.14 ± 0.17 | 0.0061 |
| Q6ZWY3 | 40S ribosomal protein S27-like | Rps27l | –5.67 ± 0.08 | 0.0416 |
Freshly isolated primary mouse hepatocytes were treated for 8 h with 400 μM NVP or 12-D3NVP. Cell lysate was then prepared and subjected to Top10 nanoLC-MS-based proteomics with data analysis performed in Proteome Discoverer 2.1, using Sequest HT for peptide spectral matching/protein identification and precursor ion area detection for relative quantitation. Up to 10 unique or razor peptides were used in protein precursor ion-based quantitation. Statistically significant relative-quantitation changes are shown for treatment with 12-D3NVP as compared to NVP as well as the results for housekeeping proteins actin and GAPDH. UniProt proteome Mus musculus10090 was used for protein identification profiling, with UniProt accession numbers, protein entry (protein name), and gene name’s provided. P values were generated using an unpaired t test, and the effect size was calculated using Cohen’s d equation. Data are representative of four biological replicates.
Table 2. Relative-Quantitation Proteomics Analysis of Protein Expression Changes with 12-D3NVP Treatment, as Compared to NVP, of Cryopreserved Human Hepatocytesa.
| changes
with 12-D3NVP treatment (in comparison to NVP) | ||||
|---|---|---|---|---|
| UniProt accession | UniProt entry | gene name | fold change | P value |
| Q16540 | 39S ribosomal protein L23, mitochondrial | MRPL23 | 8.32 ± 2.21 | 0.0159 |
| Q8Nl60 | atypical kinase ADCK3, mitochondrial | COQ8A | 1.95 ± 0.2 | 0.0168 |
| Q9HA77 | probable cysteine-tRNA ligase, mitochondrial | CARS2 | 1.85 ± 0.2 | 0.0041 |
| Q14498 | RNA-binding protein 39 | RBM39 | 1.7 ± 0.2 | 0.0339 |
| Q9Y2H5 | pleckstrin homology domain-containing family A member 6 | PLEKHA6 | 1.59 ± 0.05 | 0.0188 |
| Q8lYS2 | isoform 2 of uncharacterized protein KlAA2013 | KlAA2013 | 1.36 ± 0.01 | 0.0051 |
| P60709 | actin, cytoplasmic 1 | ACTB | –1.09 ± 0.19 | 0.4937 |
| P04406 | glyceraldehyde-3-phosphate dehydrogenase | GAPDH | –1.35 ± 0.16 | 0.1836 |
| P34897 | serine hydroxymethyltransferase, mitochondrial | SHMT2 | –1.28 ± 0.05 | 0.0225 |
| P08574 | cytochrome c1, heme protein, mitochondrial | CYC1 | –1.4 ± 0.09 | 0.0478 |
| Q15046 | isoform mitochondrial of lysine-tRNA ligase | KARS | –1.42 ± 0.06 | 0.0490 |
| Q96l24 | far upstream element-binding protein 3 | FUBP3 | –1.47 ± 0.03 | 0.0198 |
| P00167 | cytochrome b5 | CYB5A | –1.5 ± 0.11 | 0.0361 |
| Q13438 | protein os-9 | OS9 | –1.53 ± 0.09 | 0.0187 |
| Q9Y315 | deoxyribose-phosphate aldolase | DERA | –1.63 ± 0.06 | 0.0229 |
| Q4G176 | Acyl-CoA synthetase family member 3, mitochondrial | ACSF3 | –1.63 ± 0.09 | 0.0278 |
| P82979 | SAP domain-containing ribonucleoprotein | SARNP | –1.65 ± 0.07 | 0.0043 |
| O75431 | metaxin-2 | MTX2 | –1.67 ± 0.09 | 0.0465 |
| Q9H6R4 | nucleolar protein 6 | NOL6 | –1.85 ± 0.12 | 0.0121 |
| P13716 | isoform 2 of delta-aminolevulinic acid dehydratase | ALAD | –1.86 ± 0.08 | 0.0467 |
| O15321 | transmembrane 9 superfamily member 1 | TM9SF1 | –1.86 ± 0.08 | 0.0366 |
| P10635 | cytochrome P450 2D6 | CYP2D6 | –1.92 ± 0.07 | 0.0317 |
| Q9BVK6 | transmembrane emp24 domain-containing protein 9 | TMED9 | –1.93 ± 0.14 | 0.0218 |
| Q8N163 | cell cycle and apoptosis regulator protein 2 | CCAR2 | –2.05 ± 0.09 | 0.0164 |
| Q9UJ68 | mitochondrial peptide methionine sulfoxide reductase | MSRA | –2.05 ± 0.14 | 0.0358 |
| P11712 | cytochrome P450 2C9 | CYP2C9 | –2.06 ± 0.16 | 0.0498 |
| Q9NYL9 | tropomodulin-3 | TMOD3 | –2.1 ± 0.01 | 0.0356 |
| Q93096 | protein tyrosine phosphatase type IVA 1 | PTP4A1 | –2.42 ± 0.17 | 0.0466 |
| Q9Y2D5 | isoform 2 of A-kinase anchor protein 2 | AKAP2 | –2.67 ± 0.06 | 0.0249 |
| Q9Y3B2 | exosome complex component csl4 | EXOSC1 | –2.73 ± 0.09 | 0.0086 |
Cryopreserved pooled-donor (10, mixed-sex) primary human hepatocytes were treated for 48 h with 10 μM NVP or 12-D3NVP. Cell lysate was then prepared and subjected to Top10 nanoLC-MS-based proteomics with data analysis performed in Proteome Discoverer 2.1, using Sequest HT for peptide spectral matching/protein identification and precursor ion area detection for relative quantitation. Up to 10 unique or razor peptides were used in protein precursor ion-based quantitation. Statistically significant relative-quantitation changes are shown for treatment with 12-D3NVP as compared to NVP as well as the results for housekeeping proteins actin and GAPDH. UniProt proteome Homo sapiens9606 was used for protein identification profiling, with UniProt accession numbers, protein entry (protein name), and gene name’s provided. P values were generated using an unpaired t test, and the effect size was calculated using Cohen’s d equation. Data are representative of experiments with three different 10-donor pools of hepatocytes.
Interestingly, one protein was differentially expressed in mouse hepatocytes treated with 12-D3NVP as compared to both NVP (Table 1) and vehicle (Supplemental Table 2): increased insulin-like growth factor-binding protein 1 (IGFBP-1, 4.18-fold higher than vehicle and 4.70-fold higher than with NVP treatment). Increases in this protein have been found in the serum of patients with advanced liver fibrosis, and it has been demonstrated to be elevated during general hepatic stress.47 Two cytochrome P450 enzymes, CYP2D6 and CYP2C9, were both decreased during 12-D3NVP treatment in human hepatocytes as compared to NVP, by around 1.92- and 2.06-fold, respectively (Table 2). Reduction of P450 expression with deuterated NVP as well as with reduced clearance via 12-OHNVP metabolite could contribute to increased drug half-life, something that must be considered when using deuteration to reduce production of a toxic metabolite. The two greatest magnitude changes were with 12-D3NVP treatment: a 15.96-fold increase in the expression of mitochondrial pyruvate carrier protein 1 (MPC1) during treatment of human hepatocytes with 12-D3NVP as compared to vehicle and an 8.32-fold increase in the mitochondrial 39S ribosome protein L23 (MRPL23) with 12-D3NVP treatment in human hepatocytes as compared to NVP treatment. In fact, many of the observed changes with either NVP or 12-D3NVP were unique to these individual compounds, and not shared across NVP and 12-D3NVP treatment (Supplemental Tables 1–4). In general, differences in hepatocyte response, beyond metabolic impacts between NVP and 12-D3NVP treatment, are useful to consider in using this trideuterated compound as way to probe the impacts of P450 metabolism on NVP toxicities. These data suggest that there are distinct cellular responses to NVP and 12-D3NVP, including at least one unique hepatic stress marker (IGFBP-1) and changes in P450 expression.
Interestingly, among all of the identified changes in human hepatocytes, there is an abundance of mitochondrial proteins (Table 2 and Supplemental Tables 3 and 4). This suggests that perhaps mitochondrial dysregulation may play a role in the toxicities of NVP or 12-D3NVP. Previously, mitochondrial damage has been observed during in vivo rat and in vitro Hep G2 cell treatment with NVP.48,49 The implications of the various changes observed in the protein abundances warrant further exploration. One example, FAST kinase domain-containing protein 5 (FASTKD5, which was significantly decreased in human primary hepatocytes treated with NVP and 12-D3NVP), has been previously demonstrated to be involved in noncanonical mitochondrial mRNA processing. FASTKD5 silencing results in assembly defects of complex IV of the electron transport chain.50 The formation of quinone and quinone methide reactive intermediates from 2-, 3-, and 12-OHNVP has been proposed,23,24,51,52 with the formation of quinone derivatives from 2- and 3-OHNVP being previously captured and structurally characterized.51 Quinone and quinone methide metabolites have been implicated in the mitochondrial toxicity of other drugs, such as tamoxifen,53 and are thought not only to be bioreactive24,51 but also to contribute to the toxic formation of mitochondrial superoxide anions.54 The formation of protein-NVP adducts in the mitochondria or of damaging reactive oxygen species resulting from the formation of these quinone metabolites could explain our observed changes in mitochondrial protein expression. In addition, the fact that these reactive metabolites are proposed to derive from not only 12-OHNVP but also 2- and 3-OHNVP may explain why changes in mitochondrial protein expression were observed with both NVP and 12-D3NVP treatment.
Conclusions
In this work, we have demonstrated that deuterium substitution is effective at reducing hepatic P450 production of 12-OHNVP in both human and C57BL/6J mouse hepatocytes. We have measured a high observed kinetic isotope effect of this substitution on the production of 12-OHNVP in human liver microsomes. Metabolic switching was observed during 400 μM incubations with mouse primary hepatocytes, though not during 10 μM incubations with human hepatocytes and microsomes or with mouse hepatocytes. That being said, while NVP-induced primary mouse hepatocyte cell death was reduced, it was not completely reversed with 12-D3NVP, and in proteomics analysis, unique proteomic shifts were detected with 12-D3NVP. Taken together, these results indicate that this trideuterated compound is very effective at reducing P450 metabolism to NVP, though unintended effects, such as metabolic switching and differential impacts on hepatocyte protein expression, should be taken into consideration when using this deuterated compound and others to mitigate drug metabolite-induced hepatotoxicity.
Experimental Section
Reagents
All compounds used in this work had a purity of ≥95%. NVP (with a purity of >99.9%, assessed by the manufacturer using HPLC compared to a USP standard) was provided by the National Institutes of Health AIDS Research and Reference Reagent Program. 12-D3NVP (98% compound purity, 99% isotopic purity), 2-OHNVP (98% compound purity), 3-OHNVP (98% compound purity), and 12-OHNVP (96% compound purity) standards were synthesized by Toronto Research Chemicals, and their purity was assessed by the manufacturer using 1H nuclear magnetic resonance (for compound purity) and mass spectrometry (for isotopic purity). Staurosporine was purchased from Cell Signaling Technology. Furafylline, tranylcypromine, sulfaphenozole, (+)-N-3-benzyl-nirvanol, quinidine, and ketoconazole were all purchased from Sigma. 2-Phenyl-2-(1-piperidinyl) propane (PPP) was purchased from Santa Cruz. LC-MS-grade water, acetonitrile, methanol, acetone, and formic acid were all purchased from Fisher Scientific. Bovine pancreas trypsin was purchased from MilliporeSigma. Dithiothreitol and iodoacetamide were purchased from Pierce.
Primary Mouse Hepatocyte Isolation and Culture
The Johns Hopkins Animal Care and Use Committee approved all experiments using mice, and all procedures were in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the US National Institutes of Health. Hepatocytes were isolated from male C57BL/6J mice aged 8–13 weeks (The Jackson Laboratory). Primary hepatocytes were isolated as previously described.55 Cells were ≥85% viable upon plating and were cultured as previously described.56 Following adherence overnight, fresh treatment-containing medium was added.
Cryoplateable Human Hepatocyte Culture
LIVERPOOL cryoplateable human hepatocytes (three different lots of pools from 10 human, mixed-sex donors) were obtained from BioIVT. Cells were thawed and plated according to the manufacturer’s protocol into 12-well collagen coated plates (Corning) with a viability of ≥90% upon plating. Following adherence overnight, fresh treatment-containing InVitroGRO CP medium supplemented with Torpedo Antibiotic Mix (BioIVT) was added.
Extraction of Monooxygenated and O-Glucuronidated Metabolites of NVP and 12-D3NVP from the Culture Medium of Primary Human and Mouse Hepatocytes
Fresh mouse hepatocytes and cryoplateable human hepatocytes were treated in a 12-well format with vehicle (0.1% DMSO), 10 μM NVP, or 10 μM 12-D3NVP for 24 or 48 h. Fresh mouse hepatocytes were also treated with vehicle (0.2% DMSO), 400 μM NVP, or 400 μM 12-D3NVP for 8 h. Following these treatments, 100 μL of culture medium was obtained and metabolites were extracted via protein precipitation with the addition of acetonitrile to a final concentration of 50%. Samples were vortexed and centrifuged (3 min, 10 000g, 4 °C). Supernatant was then dried under vacuum centrifugation and reconstituted in 50 μL (mouse samples) or 17 μL (human samples) of methanol, and 2 μL was injected for uHPLC-Orbitrap analysis.
Extraction of Intracellular Glutathione-Conjugated NVP and 12-D3NVP Metabolites from Primary Mouse Hepatocytes
Fresh primary mouse hepatocytes were treated with vehicle (0.2% DMSO), 400 μM NVP, or 400 μM 12-D3NVP for 8 h in a 6-well format. Three wells of each treatment (720 000 cells) were collected and pooled via cell scraping into PBS, followed by centrifugation (5 min, 500g, 4 °C). Cell pellets were flash frozen and stored at −80 °C prior to metabolite extraction. Pellets were lysed and metabolites extracted in 50 μL of 50% methanol. Samples were incubated for 10 min at room temperature. Centrifugation was used to pellet insoluble debris (10 min, 10 000g, 4 °C), and 25 μL of supernatant was injected for uHPLC-Orbitrap analysis.
Metabolism by Individual cDNA-Expressed Cytochrome P450s
NVP (10 μM) was incubated with microsomes containing the following individual cDNA-expressed cytochrome P450 enzymes (Supersomes, BD Biosciences): CYP1A1, -1A2, -2A6, -2B6, -2C8, -2C9, -2C19, -2D6, -3A4, and -3A5. NVP was warmed in potassium phosphate buffer and an NADPH-regeneration system (Corning, used as per the manufacturer’s protocol) for 5 min at 37 °C. Reactions were initiated with the addition of microsomes for final concentrations of 0.1 M potassium phosphate buffer, 100 nM P450, and 10 μM NVP, a final reaction volume of 100 μL, and a final vehicle concentration of 0.1% DMSO. Following a 30 min incubation at 37 °C, reactions were stopped, centrifuged, and dried as described above. Samples were reconstituted in 50 μL of methanol, and 2 μL was injected for uHPLC-triple quadrupole analysis.
Microsomal NVP Metabolism during Co-incubation with P450 Inhibitors
Human liver microsomes (0.5 mg/mL) were incubated in potassium phosphate buffer and NADPH-regenerating reagents, along with the following panel of P450 inhibitors: 20 μM furafylline (CYP1A2 inhibitor), 2 μM tranylcypromine (CYP2A6 inhibitor), 30 μM PPP (CYP2B6 inhibitor), 20 μM sulfaphenazole (CYP2C9 inhibitor), 10 μM (+)-N-3-benzyl-nirvanol (CYP2C19 inhibitor), 1 μM quinidine (CYP2D6 inhibitor), and 1 μM ketoconazole (CYP3A4 inhibitor). These compounds and concentrations were selected on the basis of previous work regarding selective cytochrome P450 inhibitors.31,32 Following a 5 min preincubation at 37 °C, NVP (10 μM) was added to initiate the reaction. The final reaction volume was 100 μL with a final vehicle concentration of 0.2% DMSO. Following a 30 min incubation at 37 °C, reactions were stopped, centrifuged, and dried as described above. Samples were reconstituted in 50 μL of methanol, and 2 μL was injected for uHPLC-Orbitrap analysis.
Kinetics of 12-OHNVP Production in Liver Microsomes
Pilot experiments were performed to determine the linearity of 12-OHNVP production with respect to time and microsomal protein concentration, from which the subsequent reaction conditions (2.5 mg/mL protein and 30 min reaction times) were chosen. As above, NVP or 12-D3NVP were warmed in potassium phosphate buffer and an NADPH-regeneration system for 5 min at 37 °C. Following this, reactions were initiated through the addition of liver microsomes. Substrate concentrations of 1, 5, 10, 25, 50, 100, 200, or 400 μM NVP or 12-D3NVP were tested in a final reaction volume of 250 μL, and a final vehicle concentration of 0.1% DMSO or 0.2% DMSO for 400 μM substrate reactions. Reactions were incubated at 37 °C for 30 min, after which they were stopped, centrifuged, and dried as described above. Samples were reconstituted in 112.5 μL of methanol, and 2 μL was injected for uHPLC-MS/MS Orbitrap analysis. For quantification of 12-OHNVP formation, a standard curve of 12-OHNVP was prepared using the same method and microsome concentrations as the reactions but with water in place of the NADPH-regeneration system. The following known final concentrations of 12-OHNVP standard were spiked into the samples in place of substrate: 1, 2, 5, 20, 50, 100, 200, 500, 1000, and 2000 nM. The 12-OHNVP peak area intensity of these samples were then used to calculate 12-OHNVP amounts in each reaction with NVP or 12-D3NVP. The standard curve was fit using GraphPad Prism software to a line with 1/Y2 weighting. An accuracy of ±15% was observed for all standard curve points.
Orbitrap Detection of NVP and 12-D3NVP Metabolites
Samples were injected for analysis using a Dionex 3000 uHPLC system coupled to a Thermo Fisher Q-Exactive high-resolution Orbitrap mass spectrometer. Samples were separated on a Waters X Bridge BEH C18 column (2.5 μm pore size, 50 mm long, 2.1 mm internal diameter) using water with 0.1% formic acid (solvent A) and 100% acetonitrile with 0.1% formic acid (solvent B). A gradient of solvents was used for separation flowing at 750 μL/min during which the concentration of B increased from 5% to 15% over 1 min and 15% to 18% over 2 min, increased immediately to 100% and was held at 100% for 30 s, and then decreased immediately to 5% and was held at 5% for 1.5 min. The heated electrospray ionization source conditions were as follows: 3.5 kV spray voltage, 600 °C (monooxygenated metabolites) or 350 °C (glucuronidated or glutathione conjugated metabolites) aux gas heater temperature, 60 sheath gas, 5 aux gas, 350 °C capillary temperature, and 60 S-Lens RF amplitude.
All metabolites were subjected to a targeted selected ion monitoring scan in positive mode (resolution of 35 000, automatic gain control target of 5 × 104, maximum injection time of 100 ms) to generate high-resolution MS spectra, and a parallel reaction monitoring scan in positive mode (resolution of 35 000, automatic gain control target of 2 × 105, maximum injection time of 100 ms, and a normalized collision energy of 50 for monooxygenated metabolites and a three-step collision energy of 20, 35, and 50 for glucuronidated and glutathione conjugated metabolites) was performed to generate high-resolution MS/MS spectra. For all scans across the various metabolites, the quadrupole isolation was set to the compound accurate mass of ±0.5 m/z. For NVP monooxygenated metabolites, fragment peak area was used for quantitation in order to increase the specificity for each of the three metabolites and achieve baseline separation between metabolites in the chromatograms used for quantitation. The expected m/z values for these metabolites were: 283.1190 (for 2-, 3-, or 12-OHNVP detection), 285.1315 (for 12-OHD2NVP detection), or 286.1378 (for 2- or 3-OHD3NVP detection). Extracted ion chromatograms were generated from the spectral intensities of the following specific high-resolution fragments (±5 ppm), and their peak areas were monitored for quantitation of the monooxygenated metabolites: 161.0709 m/z for 2-OHNVP and 2-OHD3NVP, 242.0798 m/z for 3-OHNVP, 245.0987 m/z for 3-OHD3NVP, 223.1104 m/z for 12-OHNVP, and 225.1230 m/z for 12-OHD2NVP. For O-glucuronidated or glutathione conjugated NVP metabolites, the expected accurate masses were: 459.1510 (O-GlucNVP), 572.1922 (glutathione-NVP), 462.1700 (O-GlucD3NVP), 575.2110 (glutathione-D3NVP), 461.1636 (12-O-glucD2NVP), and 574.2048 (12-glutathione-D2NVP). For quantitation of glucuronidated or glutathione metabolites, the XIC peak areas for the high-resolution accurate parent masses (±5 ppm) were observed. Parallel reaction monitoring scans of these metabolites were used only to look for identifying fragments that would provide structural confirmation.
Triple-Quadrupole Detection of NVP Metabolites
The sample was injected for analysis using a Dionex 3000 uHPLC system coupled to a Thermo Fisher TSQ Vantage triple quadrupole mass spectrometer. The sample was separated on a Polaris C18 column (5 μm pore size, 100 mm long, 2 mm internal diameter). Samples were separated using water with 0.1% formic acid (solvent A) and 100% acetonitrile with 0.1% formic acid (solvent B), using a gradient of solvents flowing at 400 μL/min during which the concentration of B increased from 5% to 15% over 1 min and 15% to 21% over 5 min and then dropped immediately back to 5% and was held at 5% for 1.5 min. The heated electrospray ionization source conditions were as follows: 5000 V spray voltage, 352 °C vaporizer temperature, 30 sheath gas, 25 aux gas, 390 °C capillary temperature, and 141 S-Lens RF amplitude. Positive mode selected reaction monitoring scans (scan width of 0.002 m/z, scan time of 0.1 s, collision energy of 33, and quadrupole widths of 0.7 fwhm) were used to detect monooxygenated NVP metabolites, using the following specific transitions, as confirmed using authentic standards: 283.1 → 160.9 for 2-OHNVP, 283.1 → 241.9 for 3-OHNVP, and 283.1 → 223.0 for 12-OHNVP. Peak height was monitored as a measure of metabolite abundance.
Cell Death Staining
Primary mouse hepatocytes were plated on collagen coated coverslips as previously described56 and treated in a 12-well format with vehicle (0.2% DMSO), 400 μM NVP, or 400 μM 12-D3NVP for 8 h. Staurosporin treatments at 10 μM were also included as a positive control for cell death activation. Primary hepatocytes were stained and imaged, and those images were processed as previously described.56
Proteomics Sample Preparation
Fresh primary mouse hepatocytes were treated in a 6-well format with vehicle (0.2% DMSO), 400 μM NVP, or 400 μM 12-D3NVP for 8 h. Cryoplateable LIVERPOOL hepatocyte cultures were treated in a 12-well format with vehicle (0.1% DMSO), 10 μM NVP, or 10 μM 12-D3NVP for 48 h. Treated cells were harvested through cell scraping into PBS, followed by centrifugation (5 min, 500g, 4 °C). Pelleted cells were flash frozen and stored at −80 °C prior to sample preparation. Frozen samples were then thawed and resuspended in 1× cell lysis buffer (Cell Signaling Technologies) supplemented with 1× Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific), 0.5 mM phenylmethylsulfonyl fluoride (MilliporeSigma), and 0.5% sodium dodecyl sulfate. Cells were lysed via 30 passages through a 27-guage syringe needle and clarified via centrifugation (10 min, 3000g, 4 °C). Protein concentration was quantified using a bicinchoninic acid assay (Pierce). Lysate (100 μg for mouse samples and 50 μg for human samples) was diluted to 1 mg/mL in 1× cell lysis buffer (Cell Signaling Technologies), supplemented with 0.5% sodium dodecyl sulfate. Dithiothreitol was added to a final concentration of 10 mM, and the sample was incubated for 45 min at 50 °C. Iodoacetamide was added to a final concentration of 55 mM, and the samples were incubated for 20 min and protected from light. Protein was then precipitated through the addition of ice-cold acetone to a final concentration of 80%, followed by incubation for 1 h at −20 °C. Protein was pelleted at 16 000g for 10 min at 4 °C, and the resulting pellet was washed once with ice-cold 90% acetone. The pellet was resuspended in 200 μL of digestion buffer (Pierce). Trypsin (in a ratio of 1:25 μg of trypsin to μg of sample) was added, and the sample was digested overnight at 37 °C. Samples were flash frozen to stop the digestion.
Following digestion, samples were desalted using Oasis HLB 1 cm3 Extraction Cartridges (Waters, according to the manufacturer’s protocol) and fractionated using an Agilent 3100 OFFGEL Fractionator with pH 3–10, 12-well gel strips (according to the manufacturer’s protocol). Fractions were then pooled into six final fractions (fraction 1 with 2, 3 with 4, 5 with 6, 7 with 8, 9 with 10, and 11 with 12) and desalted using Pierce C18 Spin Columns (according to the manufacturer’s protocol). Eluent was dried under vacuum centrifugation and resuspended (35 μL for mouse, 17.5 μL for human) in 5% acetonitrile and 0.1% formic acid.
Nanoflow LC-MS Proteomics Analysis
The sample (5 μL) was injected for analysis using a Thermo Fisher Easy nLC 1200 nanoflow liquid chromatography system coupled to a Thermo Fisher Q-Exactive high-resolution Orbitrap mass spectrometer. The sample was separated on a Thermo Scientific Easy-Spray C18 Column (2 μm pore size, 150 mm long, 0.050 mm internal diameter) equipped with a trap column (Acclaim PepMap 100 C18 LC column with a 3 μm particle size, 150 mm long, 0.075 mm internal diameter). Samples were separated using water with 0.1% formic acid (solvent A) and 80% acetonitrile with 0.1% formic acid (solvent B), using a gradient of solvents flowing at 300 nL/min during which the concentration of B increased from 2% to 24% over 60 min, 24% to 36% over 10 min, and 36% to 98% over 1 min and then held at 98% for 15 min. Samples were ionized at 300 °C and 200 V with an S-lens RF level of 80. A full scan (resolution of 70 000, automatic gain control target of 3 × 106, maximum injection time of 40 ms, and scan range of 400 to 1600 m/z) coupled to a data-dependent MS-2 top 10 scan (resolution of 17 500, automatic gain control target of 5 × 104, maximum injection time of 150 ms, 0.8 m/z isolation window, normalized collision energy of 27, and dynamic exclusion period of 10 s) was performed to generate high-resolution parent and fragment masses of ionized peptides.
Proteome Discoverer 2.1 Proteomics Data Analysis
Raw files for sample fractions were pooled for analysis with data processed using Proteome Discoverer version 2.1. Spectra (with a precursor range of 350–5000 Da, a minimum charge state of +2, and minimum signal-to-noise of 1.5) and were assigned to peptides using Sequest HT. The proteome FASTA files used in the analysis were Uniprot Mus musculus10090 and Homo sapiens9606 version 2016–05–11. The digestion enzyme was given as trypsin, with the allowance of up to two missed cleavage sites. Mass tolerances were 5 ppm for precursor ions and 0.05 Da for fragment ions. The assignment was performed on the basis of b- and y-ions. The maximum number of dynamic modifications allowed for an identified peptide was three. The following dynamic modifications were included in the analysis: methionine oxidation, deamidation of asparagine or glutamine, and n-terminal acetylation. Carbidomethyl cysteine was included as a static peptide modification. Assignments were validated using the Target Decoy Peptide Spectral Match (PSM) Validator node. PSMs demonstrating a ±2 ppm precision in mass/charge measurements across repeat scans qualified for relative quantitation on the basis of precursor ion area detection. Only high confidence peptide assignments (<0.01 FDR) were included in the protein identification analysis, with a minimum peptide length of 6 amino acids, counting only rank 1 peptides. Protein assignments were scored using the Protein FDR validator node and grouped using the strict parsimony principle. Protein relative quantitation in each sample is based on precursor ion area assignments generated in the data processing step and included up to 10 unique and razor peptides for each protein.
Only high-confidence protein hits (<0.01 FDR) of Uniprot master proteins are reported. Data was analyzed in Microsoft Excel, and hits were determined to be significant via a paired, two-tailed t test (P < 0.05) with an effect size (Cohen’s d) cutoff of 2. This alternative analysis to using multiple comparisons corrections has been previously suggested for low-power (small sample size), exploratory, relative-quantitation proteomics studies.57 In identified hits, any missing quantitation values were filled using the average observed values for other biological replicates. After this adjustment, any hits that failed to remain significant (P < 0.05) or have an effect size ≥2 were excluded from the analysis. Certain hits were not identified in any replicates of a given treatment and are listed as “not found with” a given treatment in the results.
Statistical Analysis
Data analysis for all data sets except proteomics data was performed using GraphPad Prism, version 7.00. Throughout the text, fold changes are given as means of the ratios observed for each of the individual experimental replicates and reported with 95% confidence intervals (95% CI). Means of experimental replicate values are also reported throughout the text with their 95% CIs. Statistical significance was assayed using an unpaired, two-tailed t test, and P values are reported as *P < 0.05, **P < 0.01, or ***P < 0.001.
Glossary
Abbreviations Used
- AcrO
acridine orange
- CYP
cytochrome P450
- EtBr
ethidium bromide
- FASTKD5
FAST kinase domain-containing protein 5
- IGFBP-1
insulin-like growth factor-binding protein 1
- MRPL23
mitochondrial 39S ribosome protein L23
- NVP
nevirapine
- P450
cytochrome P450
- PPP
2-phenyl-2-(1-piperidinyl) propane
- PSM
peptide spectral match
- XIC
extracted ion chromatogram
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.9b01990.
Molecular formula strings (CSV)
Supplemental Figure 1: Fragment selection for uHPLC-MS/MS-based quantitation of monooxygenated NVP and 12-D3NVP metabolites; Supplemental Figure 2: Extracted ion chromatograms of an O-glucuronidated NVP and 12-D3NVP metabolite formed in primary mouse hepatocytes; Supplemental Figure 3: Representative fragmentation spectra for an O-glucuronidated NVP and 12-D3NVP metabolite formed in primary mouse hepatocytes; Supplemental Figure 4: NVP cytochrome P450-dependent metabolism in human liver microsomes during co-incubations with cytochrome P450 inhibitors; Supplemental Figure 5: Extracted ion chromatograms of a glutathione conjugated NVP and 12-D3NVP metabolite formed in primary mouse hepatocytes; Supplemental Figure 6: Representative fragmentation spectra for a glutathione conjugated NVP and 12-D3NVP metabolite formed in primary mouse hepatocytes; Supplemental Table 1: Relative-quantitation proteomics analysis of protein expression changes with NVP treatment of primary mouse hepatocytes; Supplemental Table 2: Relative-quantitation proteomics analysis of protein expression changes with 12-D3NVP treatment of primary mouse hepatocytes; Supplemental Table 3: Relative-quantitation proteomics analysis of protein expression changes with NVP treatment of cryopreserved human hepatocytes; Supplemental Table 4: Relative-quantitation proteomics analysis of protein expression changes with 12-D3NVP treatment of cryopreserved human hepatocytes (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Guengerich F. P. Kinetic deuterium isotope effects in cytochrome P450 reactions. Methods Enzymol. 2017, 596, 217–238. 10.1016/bs.mie.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teva Pharmaceuticals. AUSTEDO [Prescribing Information]; Teva Pharmaceuticals USA, Inc.: North Wales, PA, 2017. [Google Scholar]
- Heo Y. A.; Scott L. J. Deutetrabenazine: A review in chorea associated with huntington’s disease. Drugs 2017, 77 (17), 1857–1864. 10.1007/s40265-017-0831-0. [DOI] [PubMed] [Google Scholar]
- Howland R. H. Deuterated drugs. J. Psychosoc Nurs Ment Health Serv 2015, 53 (9), 13–16. 10.3928/02793695-20150821-55. [DOI] [PubMed] [Google Scholar]
- Sun H.; Piotrowski D. W.; Orr S. T. M.; Warmus J. S.; Wolford A. C.; Coffey S. B.; Futatsugi K.; Zhang Y.; Vaz A. D. N. Deuterium isotope effects in drug pharmacokinetics II: substrate-dependence of the reaction mechanism influences outcome for cytochrome P450 cleared drugs. PLoS One 2018, 13 (11), e0206279 10.1371/journal.pone.0206279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins G. S. Deuterated drugs; updates and obviousness analysis. Expert Opin. Ther. Pat. 2017, 27 (12), 1353–1361. 10.1080/13543776.2017.1378350. [DOI] [PubMed] [Google Scholar]
- Miwa G. T.; Lu A. Y. Kinetic isotope effects and ’metabolic switching’ in cytochrome P450-catalyzed reactions. BioEssays 1987, 7 (5), 215–219. 10.1002/bies.950070506. [DOI] [PubMed] [Google Scholar]
- World Health Organization WHO Model List of Essential Medicines, 20th ed.; WHO: 2017. [DOI] [PubMed] [Google Scholar]
- Usach I.; Melis V.; Peris J. E. Non-nucleoside reverse transcriptase inhibitors: a review on pharmacokinetics, pharmacodynamics, safety and tolerability. J. Int. AIDS Soc. 2013, 16, 18567. 10.7448/IAS.16.1.18567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard R. B.; Robinson P.; Dransfield K. Safety profile of nevirapine, a nonnucleoside reverse transcriptase inhibitor for the treatment of human immunodeficiency virus infection. Clin. Ther. 1998, 20 (6), 1071–1092. 10.1016/S0149-2918(98)80105-7. [DOI] [PubMed] [Google Scholar]
- Serious adverse events attributed to nevirapine regimens for postexposure prophylaxis after HIV exposures--worldwide, 1997–2000. MMWR Morb Mortal Wkly Rep 2001, 49 (51–52), 1153–1156. [PubMed] [Google Scholar]
- Zhang C.; Wang W.; Zhou M.; Han Y.; Xie J.; Qiu Z.; Guo F.; Li Y.; Wang H.; Ghanem K. G.; Li T. The interaction of CD4 T-cell count and nevirapine hepatotoxicity in China: a change in national treatment guidelines may be warranted. JAIDS, J. Acquired Immune Defic. Syndr. 2013, 62 (5), 540–545. 10.1097/QAI.0b013e3182845cc7. [DOI] [PubMed] [Google Scholar]
- Sanne I.; Mommeja-Marin H.; Hinkle J.; Bartlett J. A.; Lederman M. M.; Maartens G.; Wakeford C.; Shaw A.; Quinn J.; Gish R. G.; Rousseau F. Severe hepatotoxicity associated with nevirapine use in HIV-infected subjects. J. Infect. Dis. 2005, 191 (6), 825–829. 10.1086/428093. [DOI] [PubMed] [Google Scholar]
- Buyse S.; Vibert E.; Sebagh M.; Antonini T.; Ichai P.; Castaing D.; Samuel D.; Duclos-Vallée J. C. Liver transplantation for fulminant hepatitis related to nevirapine therapy. Liver Transpl 2006, 12 (12), 1880–1882. 10.1002/lt.20985. [DOI] [PubMed] [Google Scholar]
- Jao J.; Sturdevant M.; del Rio Martin J.; Schiano T.; Fiel M. I.; Huprikar S. Nevirapine-induced stevens johnson-syndrome and fulminant hepatic failure requiring liver transplantation. Am. J. Transplant. 2010, 10 (7), 1713–1716. 10.1111/j.1600-6143.2010.03153.x. [DOI] [PubMed] [Google Scholar]
- Maniar J. K.; Shah S. R.; Verma R.; Kamath R.; Gupte P.; Maniar A. Nevirapine-induced fulminant hepatitis. J. Assoc Physicians India 2006, 54, 957–958. [PubMed] [Google Scholar]
- Erickson D. A.; Mather G.; Trager W. F.; Levy R. H.; Keirns J. J. Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug Metab. Dispos. 1999, 27 (12), 1488–1495. [PubMed] [Google Scholar]
- Riska P.; Lamson M.; MacGregor T.; Sabo J.; Hattox S.; Pav J.; Keirns J. Disposition and biotransformation of the antiretroviral drug nevirapine in humans. Drug Metab. Dispos. 1999, 27 (8), 895–901. [PubMed] [Google Scholar]
- Marinho A. T.; Rodrigues P. M.; Caixas U.; Antunes A. M.; Branco T.; Harjivan S. G.; Marques M. M.; Monteiro E. C.; Pereira S. A. Differences in nevirapine biotransformation as a factor for its sex-dependent dimorphic profile of adverse drug reactions. J. Antimicrob. Chemother. 2014, 69 (2), 476–482. 10.1093/jac/dkt359. [DOI] [PubMed] [Google Scholar]
- Sharma A. M.; Klarskov K.; Uetrecht J. Nevirapine bioactivation and covalent binding in the skin. Chem. Res. Toxicol. 2013, 26 (3), 410–421. 10.1021/tx3004938. [DOI] [PubMed] [Google Scholar]
- Wen B.; Chen Y.; Fitch W. L. Metabolic activation of nevirapine in human liver microsomes: dehydrogenation and inactivation of cytochrome P450 3A4. Drug Metab. Dispos. 2009, 37 (7), 1557–1562. 10.1124/dmd.108.024851. [DOI] [PubMed] [Google Scholar]
- Dekker S. J.; Zhang Y.; Vos J. C.; Vermeulen N. P.; Commandeur J. N. Different reactive metabolites of nevirapine require distinct glutathione S-transferase isoforms for bioinactivation. Chem. Res. Toxicol. 2016, 29 (12), 2136–2144. 10.1021/acs.chemrestox.6b00250. [DOI] [PubMed] [Google Scholar]
- Chen J.; Mannargudi B. M.; Xu L.; Uetrecht J. Demonstration of the metabolic pathway responsible for nevirapine-induced skin rash. Chem. Res. Toxicol. 2008, 21 (9), 1862–1870. 10.1021/tx800177k. [DOI] [PubMed] [Google Scholar]
- Sharma A. M.; Li Y.; Novalen M.; Hayes M. A.; Uetrecht J. Bioactivation of nevirapine to a reactive quinone methide: implications for liver injury. Chem. Res. Toxicol. 2012, 25 (8), 1708–1719. 10.1021/tx300172s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A. M.; Godinho A. L.; Martins I. L.; Justino G. C.; Beland F. A.; Marques M. M. Amino acid adduct formation by the nevirapine metabolite, 12-hydroxynevirapine--a possible factor in nevirapine toxicity. Chem. Res. Toxicol. 2010, 23 (5), 888–899. 10.1021/tx900443z. [DOI] [PubMed] [Google Scholar]
- Antunes A. M.; Wolf B.; Oliveira M. C.; Beland F. A.; Marques M. M. 2′-Deoxythymidine adducts from the anti-HIV drug nevirapine. Molecules 2013, 18 (5), 4955–4971. 10.3390/molecules18054955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Sharma A. M.; Uetrecht J. Identification of danger signals in nevirapine-induced skin rash. Chem. Res. Toxicol. 2013, 26 (9), 1378–1383. 10.1021/tx400232s. [DOI] [PubMed] [Google Scholar]
- Pinheiro P. F.; Pereira S. A.; Harjivan S. G.; Martins I. L.; Marinho A. T.; Cipriano M.; Jacob C. C.; Oliveira N. G.; Castro M. F.; Marques M. M.; Antunes A. M. M.; Miranda J. P. Hepatocyte spheroids as a competent in vitro system for drug biotransformation studies: nevirapine as a bioactivation case study. Arch. Toxicol. 2017, 91 (3), 1199–1211. 10.1007/s00204-016-1792-x. [DOI] [PubMed] [Google Scholar]
- Rendic S.; Guengerich F. P. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem. Res. Toxicol. 2015, 28 (1), 38–42. 10.1021/tx500444e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights K. M.; Stresser D. M.; Miners J. O.; Crespi C. L. In vitro drug metabolism using liver microsomes. Curr. Protoc Pharmacol 2016, 74, 7.8.1–7.8.24. 10.1002/cpph.9. [DOI] [PubMed] [Google Scholar]
- Suzuki H.; Kneller M. B.; Haining R. L.; Trager W. F.; Rettie A. E. (+)-N-3-Benzyl-nirvanol and (−)-N-3-benzyl-phenobarbital: new potent and selective in vitro inhibitors of CYP2C19. Drug Metab. Dispos. 2002, 30 (3), 235–239. 10.1124/dmd.30.3.235. [DOI] [PubMed] [Google Scholar]
- Khojasteh S. C.; Prabhu S.; Kenny J. R.; Halladay J. S.; Lu A. Y. Chemical inhibitors of cytochrome P450 isoforms in human liver microsomes: a re-evaluation of P450 isoform selectivity. Eur. J. Drug Metab. Pharmacokinet. 2011, 36 (1), 1–16. 10.1007/s13318-011-0024-2. [DOI] [PubMed] [Google Scholar]
- Northrop D. B. Deuterium and tritium kinetic isotope effects on initial rates. Methods Enzymol. 1982, 87, 607–625. 10.1016/S0076-6879(82)87032-8. [DOI] [PubMed] [Google Scholar]
- Yun C. H.; Miller G. P.; Guengerich F. P. Rate-determining steps in phenacetin oxidations by human cytochrome P450 1A2 and selected mutants. Biochemistry 2000, 39 (37), 11319–11329. 10.1021/bi000869u. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P.; Miller G. P.; Hanna I. H.; Sato H.; Martin M. V. Oxidation of methoxyphenethylamines by cytochrome P450 2D6. analysis of rate-limiting steps. J. Biol. Chem. 2002, 277 (37), 33711–33719. 10.1074/jbc.M205146200. [DOI] [PubMed] [Google Scholar]
- Bell-Parikh L. C.; Guengerich F. P. Kinetics of cytochrome P450 2E1-catalyzed oxidation of ethanol to acetic acid via acetaldehyde. J. Biol. Chem. 1999, 274 (34), 23833–23840. 10.1074/jbc.274.34.23833. [DOI] [PubMed] [Google Scholar]
- Pallan P. S.; Wang C.; Lei L.; Yoshimoto F. K.; Auchus R. J.; Waterman M. R.; Guengerich F. P.; Egli M. Human cytochrome P450 21A2, the major steroid 21-hydroxylase: strucutre of the enzyme progesterone substrate complex and rate-limiting C-H bond cleavage. J. Biol. Chem. 2015, 290 (21), 13128–13143. 10.1074/jbc.M115.646307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G.; Calcutt M. W.; Nagy L. D.; Guengerich F. P. Oxidation of methyl and ethyl nitrosamines by cytochrome P450 2E1 and 2B1. Biochemistry 2012, 51 (50), 9995–10007. 10.1021/bi301092c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucal S.; Häkkinen M. R.; Alanne A. L.; Alhonen L.; Vepsäläinen J.; Keinänen T. A.; Hyvönen M. T. Controlling of N-alkylpolyamine analogue metabolism by selective deuteration. Biochem. J. 2018, 475 (3), 663–676. 10.1042/BCJ20170887. [DOI] [PubMed] [Google Scholar]
- Stringer R. A.; Williams G.; Picard F.; Sohal B.; Kretz O.; McKenna J.; Krauser J. A. Application of a deuterium replacement strategy to modulate the pharmacokinetics of 7-(3,5-dimethyl-1H-1,2,4-triazol-1-yl)-3-(4-methoxy-2-methylphenyl)-2,6-dimethylpyrazolo[5,1-b]oxazole, a novel CRF1 antagonist. Drug Metab. Dispos. 2014, 42 (5), 954–962. 10.1124/dmd.114.057265. [DOI] [PubMed] [Google Scholar]
- Kim D.; Cha G. S.; Nagy L. D.; Yun C. H.; Guengerich F. P. Kinetic analysis of lauric acid hydroxylation by human cytochrome P450 4A11. Biochemistry 2014, 53 (39), 6161–6172. 10.1021/bi500710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J. L.; Beland F. A. Differential responses of human hepatocytes to the non-nucleoside HIV-1 reverse transcriptase inhibitor nevirapine. J. Toxicol. Sci. 2013, 38 (5), 741–752. 10.2131/jts.38.741. [DOI] [PubMed] [Google Scholar]
- Marinho A. T.; Dias C. G.; Pinheiro P. F.; Lemos A. R.; Antunes A. M.; Marques M. M.; Monteiro E. C.; Miranda J. P.; Pereira S. A. Nevirapine modulation of paraoxonase-1 in the liver: an in vitro three-model approach. Eur. J. Pharm. Sci. 2016, 82, 147–153. 10.1016/j.ejps.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Bell C. C.; Hendriks D. F.; Moro S. M.; Ellis E.; Walsh J.; Renblom A.; Fredriksson Puigvert L.; Dankers A. C.; Jacobs F.; Snoeys J.; Sison-Young R. L.; Jenkins R. E.; Nordling Å.; Mkrtchian S.; Park B. K.; Kitteringham N. R.; Goldring C. E.; Lauschke V. M.; Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016, 6, 25187. 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granitzny A.; Knebel J.; Müller M.; Braun A.; Steinberg P.; Dasenbrock C.; Hansen T. Evaluation of a human in vitro hepatocyte-NPC co-culture model for the prediction of idiosyncratic drug-induced liver injury: A pilot study. Toxicol Rep 2017, 4, 89–103. 10.1016/j.toxrep.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M.; Shenton J. M.; Chen J.; Baban A.; Tharmanathan T.; Mannargudi B.; Abdulla D.; Uetrecht J. P. Nevirapine hypersensitivity. Handb. Exp. Pharmacol. 2010, 196 (196), 437–451. 10.1007/978-3-642-00663-0_15. [DOI] [PubMed] [Google Scholar]
- Hagström H.; Stål P.; Hultcrantz R.; Brismar K.; Ansurudeen I. IGFBP-1 and IGF-I as markers for advanced fibrosis in NAFLD - a pilot study. Scand. J. Gastroenterol. 2017, 52 (12), 1427–1434. 10.1080/00365521.2017.1379556. [DOI] [PubMed] [Google Scholar]
- Sastry J.; Mohammed H.; Campos M. M.; Uetrecht J.; Abu-Asab M. Nevirapine-induced liver lipid-SER inclusions and other ultrastructural aberrations. Ultrastruct. Pathol. 2018, 42 (2), 108–115. 10.1080/01913123.2017.1422831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paemanee A.; Sornjai W.; Kittisenachai S.; Sirinonthanawech N.; Roytrakul S.; Wongtrakul J.; Smith D. R. Nevirapine induced mitochondrial dysfunction in HepG2 cells. Sci. Rep. 2017, 7 (1), 9194. 10.1038/s41598-017-09321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H.; Shoubridge E. A. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015, 10 (6), 920–932. 10.1016/j.celrep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- Harjivan S. G.; Pinheiro P. F.; Martins I. L.; Godinho A. L.; Wanke R.; Santos P. P.; Pereira S. A.; Beland F. A.; Marques M. M.; Antunes A. M. M. Quinoid derivatives of the nevirapine metabolites2-hydroxy- and 3-hydroxy-nevirapine: activationpathway to amino acid adducts. Toxicol. Res. 2015, 4, 1565–1577. 10.1039/C5TX00176E. [DOI] [Google Scholar]
- Antunes A. M.; Novais D. A.; da Silva J. L.; Santos P. P.; Oliveira M. C.; Beland F. A.; Marques M. M. Synthesis and oxidation of 2-hydroxynevirapine, a metabolite of the HIV reverse transcriptase inhibitor nevirapine. Org. Biomol. Chem. 2011, 9 (22), 7822–7835. 10.1039/c1ob06052j. [DOI] [PubMed] [Google Scholar]
- Theodossiou T. A.; Wälchli S.; Olsen C. E.; Skarpen E.; Berg K. Deciphering the nongenomic, mitochondrial toxicity of tamoxifens as determined by cell metabolism and redox activity. ACS Chem. Biol. 2016, 11 (1), 251–262. 10.1021/acschembio.5b00734. [DOI] [PubMed] [Google Scholar]
- Hochstein P. Futile redox cycling: implications for oxygen radical toxicity. Fundam. Appl. Toxicol. 1983, 3 (4), 215–217. 10.1016/S0272-0590(83)80128-6. [DOI] [PubMed] [Google Scholar]
- Lee P.; Peng H.; Gelbart T.; Beutler E. The IL-6- and lipopolysaccharide-induced transcription of hepcidin in HFE-, transferrin receptor 2-, and beta 2-microglobulin-deficient hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (25), 9263–9265. 10.1073/pnas.0403108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck C. J. S.; Hamlin A. N.; Bumpus N. N. Efavirenz and efavirenz-like compounds activate human, murine, and macaque hepatic IRE1. Mol. Pharmacol. 2019, 95 (2), 183–195. 10.1124/mol.118.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascovici D.; Handler D. C.; Wu J. X.; Haynes P. A. Multiple testing corrections in quantitative proteomics: a useful but blunt tool. Proteomics 2016, 16 (18), 2448–2453. 10.1002/pmic.201600044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.