Significance
Increasing evidence suggests a role for the gut microbiome in autoimmune diseases, including multiple sclerosis. However, the impact of natural genetic variation in the mammalian host and other underlying mechanisms has been largely overlooked. We used a mouse model of natural genetic diversity to explore interactions between the host and the microbiome. Our results demonstrate a complex interplay between host genotype and gut microbiota in autoimmune disease, and identify a single commensal species capable of modifying disease susceptibility in a genetically susceptible host. Our studies underscore the need to consider host genetics and baseline gut microbiota composition in autoimmunity.
Keywords: multiple sclerosis, gut microbiome, genetics
Abstract
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system. The etiology of MS is multifactorial, with disease risk determined by genetics and environmental factors. An emerging risk factor for immune-mediated diseases is an imbalance in the gut microbiome. However, the identity of gut microbes associated with disease risk, their mechanisms of action, and the interactions with host genetics remain obscure. To address these questions, we utilized the principal autoimmune model of MS, experimental autoimmune encephalomyelitis (EAE), together with a genetically diverse mouse model representing 29 unique host genotypes, interrogated by microbiome sequencing and targeted microbiome manipulation. We identified specific gut bacteria and their metabolic functions associated with EAE susceptibility, implicating short-chain fatty acid metabolism as a key element conserved across multiple host genotypes. In parallel, we used a reductionist approach focused on two of the most disparate phenotypes identified in our screen. Manipulation of the gut microbiome by transplantation and cohousing demonstrated that transfer of these microbiomes into genetically identical hosts was sufficient to modulate EAE susceptibility and systemic metabolite profiles. Parallel bioinformatic approaches identified Lactobacillus reuteri as a commensal species unexpectedly associated with exacerbation of EAE in a genetically susceptible host, which was functionally confirmed by bacterial isolation and commensal colonization studies. These results reveal complex interactions between host genetics and gut microbiota modulating susceptibility to CNS autoimmunity, providing insights into microbiome-directed strategies aimed at lowering the risk for autoimmune disease and underscoring the need to consider host genetics and baseline gut microbiome composition.
Multiple sclerosis (MS) is an inflammatory disease of the central nervous system (CNS) characterized by demyelination, gliosis, irreversible axonal loss, and progressive neurological dysfunction. Affecting ∼2.3 million people worldwide, MS is the leading cause of nontraumatic neurological disability in young adults, with increasing prevalence in developed and developing countries (1, 2). The etiology of MS is multifactorial and polygenic, whereby genetics account for ∼30% of disease susceptibility (3–5). The remaining 70% of disease risk is attributed to environmental risk factors and/or gene × environment (G×E) interactions (6, 7). How these complex networks of interactions ultimately drive disease susceptibility remains to be elucidated.
Documented environmental risk factors for MS include vitamin D intake, Epstein Barr virus infection, smoking, and potentially diet (6, 8). An important newly appreciated environment-sensitive factor in human health and disease is the gut microbiome. The gut microbiome represents a complex mutualistic relationship between host and resident commensal microorganisms, capable of modulating many aspects of host physiology (9, 10). Human diet-derived microbial metabolites can function locally within the gut to induce regulatory immune cell populations, or enter systemic circulation to directly impact distal sites, including the brain (11, 12). With regard to MS, a number of recent case-control studies have demonstrated that the gut microbiome of MS patients differs from that of their healthy control counterparts. Some broad features, such as decreased abundance of putative short-chain fatty acid (SCFA)-producing bacteria and expansion of Akkermansia, have been observed consistently across several studies (13–19). However, large variation between studies and cohorts remains (17, 18). Such variation could be driven by genetic and environmental differences between patient populations, which makes pinpointing causative commensal microbiota particularly challenging. An additional confounder is represented by the gastrointestinal (GI) disturbances, such as constipation, which commonly manifest as a symptom of MS progression (20), and in turn are known to cause imbalances in the gut microbiome (21, 22). Finally, disease-modifying therapies for MS have profound impacts on the immune system, and thus are likely to indirectly influence the gut microbiome, as supported by recent studies (23). However, a cause-and-effect relationship between the gut microbiome and MS susceptibility is supported by two recent studies that demonstrated augmented disease severity in a mouse model of MS after fecal microbiome transplantation from MS patients (19, 24). Although this demonstrates that MS-associated changes in the gut microbiome can influence susceptibility to CNS autoimmunity, the identity of responsible microbial species and their mechanism of action, as well as the role of host genetics, have not been defined.
Mechanistic interrogation of the role of G×E interactions in human disease is challenging for multiple reasons (25). Consequently, appropriate animal models are required to bridge the gap between association studies and mechanistic cause-and-effect relationships. The laboratory mouse provides a powerful and genetically tractable tool to understand the basic mechanisms of human disease, and to establish causality for genetic or environmental factors. The principal autoimmune model of MS, experimental autoimmune encephalomyelitis (EAE), is typically elicited by immunization with myelin peptides. This activates myelin-specific autoreactive CD4+ T cells, which infiltrate into the CNS to initiate a disease that recapitulates many key aspects of MS, including inflammation, gliosis, demyelination, and axonal loss, resulting in neurological disability (26). This model has been instrumental in improving our understanding of MS pathogenesis, as well as in developing new disease-modifying therapies (27, 28).
A major limitation of mouse models of human disease is the use of common laboratory inbred mouse strains, which fail to capture human genetic diversity and evolutionary pressure from infectious and commensal microbes. This problem can be overcome by use of so-called wild-derived inbred strains, which were established by trapping wild mice and breeding them in the laboratory (29). One such wild-derived inbred strain is the PWD/PhJ (PWD) strain, which is highly genetically divergent from classic inbred strains like C57BL/6J (B6) (30). A comparison of variation at common single nucleotide polymorphisms using publicly available genotype data reveals a 54% difference between B6 and PWD, and a 13% difference between classic B6 and BALB/c strains. Thus, comparative studies between common and wild-derived strains (e.g., B6 vs. PWD) can capture much more genetic variability than a comparison between classic strains. We have previously demonstrated that, compared with B6, immune cells from PWD mice exhibit highly divergent transcriptomes, with down-regulation of MS signature genes, resulting in reduced susceptibility to EAE (31). Additionally, we utilized a panel of consomic strains carrying PWD chromosomes on the B6 background (B6.ChrPWD) to demonstrate that PWD alleles regulate EAE susceptibility (31). Experimentally, environmental risk factors can be layered onto this tractable genetic model to interrogate G×E interactions, as we recently did for vitamin D status (32).
In this study, we leveraged the natural genetic variation in wild-derived PWD mice as compared to classic B6 mice, to identify host genome and commensal gut microbiome interactions governing susceptibility to CNS autoimmunity. Utilizing a panel of 27 B6.ChrPWD consomic mice, we identified strain-specific microbiota profiles and complex relationships between host genetics, microbial constituents, and microbial-driven metabolic functions associated with differential susceptibility to EAE. In parallel, using a reductionist approach focusing on two of the most disparate gut microbiota profiles from our consomic screen, manipulation of the gut microbiome by cohousing or transplantation demonstrated that transfer of these divergent microbiomes into genetically identical hosts was sufficient to yield differences in EAE susceptibility and systemic metabolite profiles. Finally, parallel approaches identified and confirmed Lactobacillus reuteri as a commensal species capable of exacerbating EAE. These results demonstrate the existence of complex interactions between host genetics and gut microbiota modulating susceptibility to CNS autoimmune disease, providing insights into microbiome-directed strategies aimed at lowering the risk for autoimmune disease, and highlighting the need to consider host genetics and baseline gut microbiome composition for such strategies.
Results
Genetic Control of the Gut Microbiome by the Wild-Derived Host Genome.
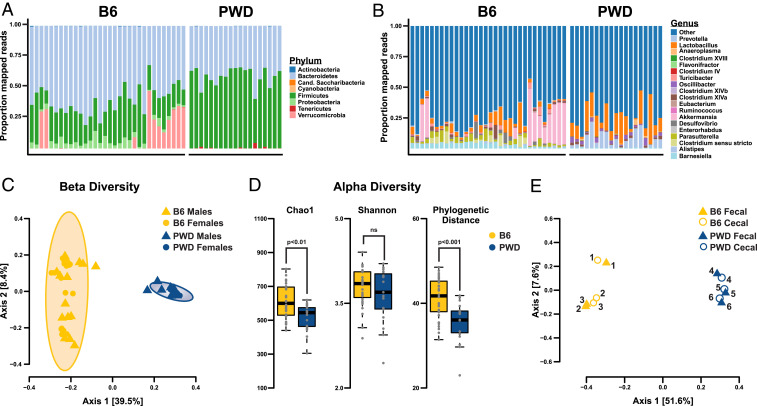
Conventional B6 and wild-derived PWD mice have widely divergent phenotypes in terms of immune cell gene expression and immune-mediated disease susceptibility (31). To assess the impact of these and other genetically driven differences on the baseline composition of the gut microbiome, we initially profiled the microbiome of these strains by 16S rRNA gene sequencing. DNA was extracted from fecal samples as a noninvasive proxy for the gut microbiome (33), followed by amplification of V3-V4 regions, high-throughput sequencing, taxonomic assignment, and bioinformatic analyses (Methods). Distribution of bacterial phyla revealed the presence of expected constituents of the mouse gut microbiome, dominated by Firmicutes and Bacteroidetes, with a significantly higher Firmicutes:Bacteroidetes ratio in PWD mice compared with B6 (Fig. 1A and SI Appendix, Fig. S1A) (Wilcoxon test P = 5.92e-12, B6 mean: 0.44, PWD mean: 1.46). Consistently, Lactobacillaceae and Lachnospiraceae (belonging to the Firmicutes phylum) were expanded in PWD mice, while the B6 gut microbiome was dominated by Porphyromonadaceae (SI Appendix, Fig. S1B). Analyses at the genus taxonomic level further revealed large differences across multiple bacterial genera (Fig. 1B and SI Appendix, Fig. S1C) with an expansion of Lactobacillus and Alistipes in PWD mice, while the B6 gut microbiome displayed high interindividual variation in Akkermansia abundance. Differences between B6 and PWD microbiomes were additionally supported by β-diversity analyses of operational taxonomic unit (OTU) level abundances, which revealed a highly significant effect of strain but no effect of sex (Adonis test strain: R2 = 0.39, P < 0.001; strain:sex R2 = 0.03, P = 0.16) (Fig. 1C and SI Appendix, Fig. S2 A and B). Species richness (α-diversity) analyses demonstrated significantly lower diversity in the PWD microbiome compared with B6 as indicated by two of three statistical metrics (Chao1 index, phylogenetic distance, but not the Shannon index) (Fig. 1D), suggesting greater selective pressure on gut microbiota from the wild-derived PWD host. To determine how well the fecal microbiome approximates the gut microbiome, we compared cecal and fecal samples from the same mice. β-Diversity analyses revealed strong correlations in microbial composition between the two sampling sites, with clear differences between B6 and PWD mice (axis 1) and strong within-individual correlation (axis 2) (Fig. 1E and SI Appendix, Fig. S2 C and D), confirming that fecal sampling provides a good approximation of at least large intestinal microbiota. These results indicate that wild-derived PWD mice have a distinct gut microbiota profile compared with conventional laboratory B6 mice.
Fig. 1.
Divergent gut microbiome phenotypes in wild-derived PWD mice and classic inbred B6 mice. Comparison of gut microbial composition in PWD and B6 mice. Distribution of phyla (A) and genera (B) comprising bacterial 16S reads. Each bar represents an individual mouse. (C) β-Diversity analysis of B6 and PWD fecal microbiomes assessed using unweighted UniFrac distance. (D) α-Diversity analysis of B6 and PWD fecal, assessed using Chao1, Shannon index, or phylogenetic distance. (E) DNA from fecal and cecal samples was isolated from the same individual mice and analyzed by 16S sequencing using unweighted UniFrac distance. Samples from individual mice of each strain are designated by numbering with a total of 32 B6 (17 female and 15 male) and 19 PWD (10 female and 9 male) mice with all 16S data rarefied to 7,000 reads.
There is ample evidence that gut microbiome composition is modulated by host genetics in both mice and humans (34–41). To assess the role of the wild-derived PWD host genome in determining gut microbial composition, we used the B6.ChrPWD consomic model. Fecal samples were collected from 27 available consomic strains, bred and housed under the same environmental conditions, followed by 16S sequencing and analysis. The composition of gut microbiota was broadly similar to parental B6 and PWD strains at the phylum, family, and genus level, with unique strain-specific phenotypes (Fig. 2 A and B and SI Appendix, Fig. S1 D–F). β-Diversity (unweighted UniFrac distance) analyses revealed a broad distribution of microbiome profiles clustering predominately by strain, with parental B6 and PWD mice representing two of the most divergent states (Fig. 2C). Statistical analyses confirmed a highly significant effect of strain on microbial community profile (Adonis test, R2 = 0.39, P < 0.001), and a more modest but significant strain-by-sex interaction effect (Adonis test, R2 = 0.05, P < 0.001). To determine relationships between microbial composition across consomic strains, multivariate homogeneity of group dispersions (PERMDISP2) analysis was applied (42). This revealed at least three distinct groupings among the strains, with B6-like, PWD-like, and B6.Chr1PWD-like profiles (Fig. 2D). These results suggest that the genetic diversity among the B6.ChrPWD consomic strains is accompanied by a diversity of genotype-specific gut microbiome states.
Fig. 2.
Genetic diversity of B6.ChrPWD consomic mice results in diversity of gut microbiome community structures. Comparisons of microbiomes across the B6.ChrPWD consomic strain panel (including B6 and PWD parental strains). DNA from fecal samples collected from 8- to 14-wk-old B6, PWD, and B6.ChrPWD consomic mice was subjected to 16S rRNA gene sequencing. All resulting reads were rarefied to 7,000. Distribution of phyla (A) and genera (B) comprising the bacterial 16S reads. Each bar represents the average distribution for each consomic strain. (C) β-Diversity analysis of B6.ChrPWD consomic microbiomes assessed using unweighted UniFrac distance. Each data point represents an individual mouse, with symbol/color combinations designating strain. (D) A heatmap of gut microbiome distribution and relationships between B6.ChrPWD consomic mice as determined using PERMDISP2 analysis. The heatmap represents Euclidean distances between strain centroids, with warmer colors indicating divergence and cooler colors similarity in gut microbial composition. Relationships between individual strains are traced across each row or column.
Gut Microbiome Composition Predicts Susceptibility to CNS Autoimmunity.
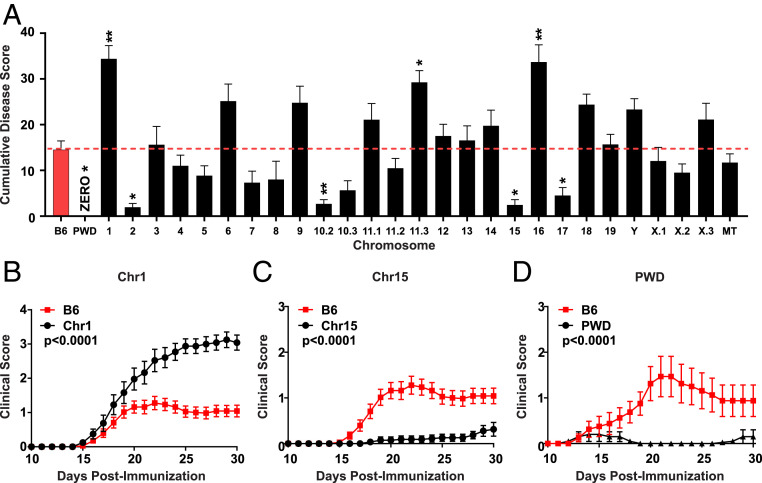
Genetics play a key role in autoimmune disease susceptibility. We previously demonstrated that the genetic diversity in the B6.ChrPWD model gives rise to divergent effects on EAE severity and susceptibility, even when small cohorts of mice were used (bred and transported from the Jackson Laboratory) (43). Here, we attempted to replicate these results with larger cohorts of mice bred and housed under stable environmental conditions within a single room in our vivarium. To this end, we employed the 2× EAE model (44), which utilizes MOG35–55 as a classic well-defined immunogen, given that each consomic strain carries the B6 H2b haplotype, including chromosome (Chr)17, which unexpectedly carries a portion of B6-derived genome (44). This model results in moderate EAE severity in B6 mice (thus optimizing for simultaneous detection of divergent effects on EAE severity) and avoids the need for pertussis toxin (PTX) as an ancillary adjuvant, the mechanism of which remains ill-defined. Our new data (Fig. 3) expanded our previous observations, and slightly reduced the total number of significant EAE loci (likely by exclusion of false positives or environment-sensitive associations in the original smaller cohort). The remaining significant loci exhibited potent and divergent effects on EAE severity and disease course, with Chr1PWD and Chr15PWD mice exhibiting the most profound increase or decrease in EAE susceptibility, respectively (Fig. 3 A–C). The parental PWD strain was completely resistant to MOG35–55-induced EAE (Fig. 3A), which is not surprising given that compared with B6 this strain carries a completely different (although not yet characterized) MHC locus. To make a more direct comparison, we used a universal induction method using mouse spinal cord homogenate as antigen to provide all possible myelin epitopes, and PTX to enhance induction. Compared with B6 mice, which developed moderate EAE with this protocol, PWD mice remained almost completely resistant (Fig. 3D). These data confirm that genetic variation distinguishing B6.ChrPWD strains robustly regulates EAE susceptibility and disease course. With these data in hand, we set out to determine how EAE pathogenesis might be modulated collectively by genetic and gut microbial variation in this model of genetic diversity.
Fig. 3.
Genetic diversity of B6.ChrPWD consomic mice results in potent and divergent regulation of EAE susceptibility. (A) CDS representing the sum of all daily EAE scores over 30 d in B6.ChrPWD consomic strains and B6 controls is shown with the dotted line representing the B6 control mean CDS. Each bar represents the strain mean, with error bars representing SEM. Asterisks indicate significant difference from B6 controls, as determined by nonparametric Kruskal–Wallis test with Dunn’s post hoc comparisons. Symbols indicate a significant difference between B6 controls and the indicated strains, as follows: *P < 0.05; **P < 0.01. Note that chromosomes 10, 11, and X, are represented by multiple subconsomic strains (e.g., 11.1, 11.2, 11.3). EAE course for representative consomic strains is shown for B6.Chr1PWD (Chr1) in B and B6.Chr15PWD (Chr15) in C, respectively. (D) Disease course of B6 and PWD parental strains using alternative EAE induction with mouse spinal cord homogenate/CFA on days 0 and 7 and PTX on days 0 and 2. Significance of difference in overall disease course in A–D were assessed using Friedman’s nonparametric two-way ANOVA.
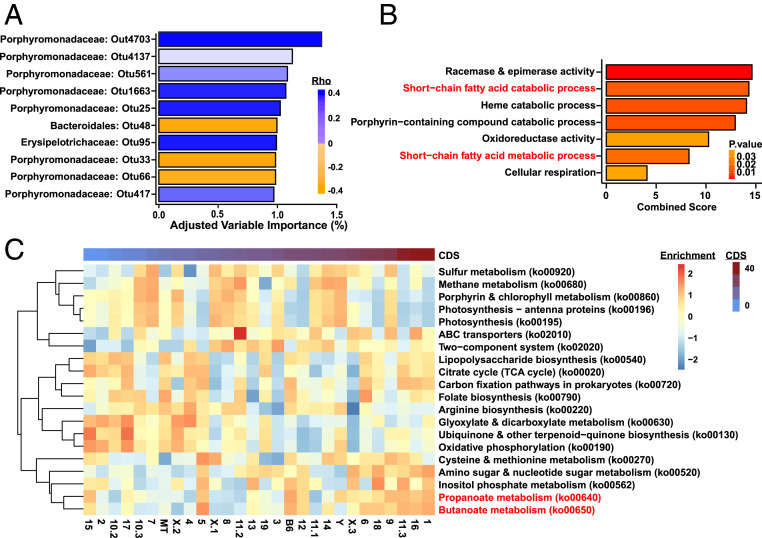
While multiple human studies have shown alterations in the gut microbiome of MS patients, it is unclear whether these represent an MS high-risk microbiome, are simply a byproduct of the GI disturbances that are highly prevalent in MS (20, 45), or are an effect of the immune modulation by MS disease-modifying therapy treatment (23). In contrast, the EAE model allows us to determine and modulate the microbiome state prior to disease onset. To detect associations between EAE outcomes and baseline gut microbiome composition, fecal samples were collected from individual consomic mice prior to EAE induction (Fig. 3) and analyzed by 16S sequencing. Using OTU-level abundance data, we applied a multivariate analysis (Microbiome Regression-based Kernel Association Test, MiRKAT) to identify associations with EAE cumulative disease score (CDS), as a single quantitative trait variable that encompasses multiple aspects of EAE severity and susceptibility (46). Association was estimated using OTUs having significant negative or positive associations with EAE CDS (Spearman rank correlation; 203 OTUs with Padj < 0.01) using both strain and sex as covariates. We found a significant association of the selected OTUs and CDS (PMiRKAT = 0.028) (SI Appendix, Fig. S3B and Dataset S1). Several OTUs were identified as having a positive association with CDS (higher abundance associated with higher EAE severity), including Acetatifactor muris, Clostridium leptum, Turicibacter sanguinis species, as well as OTUs belonging to the Clostridium cluster XlVa, Erysipelotrichaceae, and Lactobacillus families. OTUs showing a negative association with CDS included the Akkermansia muciniphila and Clostridium viride species, as well as Clostridium sensu stricto and members of the Desulfovibrio genera. Of 203 CDS-associated OTUs, 164 belonged to the Porphyromonadaceae family, which exhibited both positive and negative associations with CDS. Given the lack of species-level assignment based on 16S sequence analysis within this latter family, their significance is unclear. Interestingly, the percent variance in CDS explained by any individual OTU was modest, ranging from 0.5 to 1.3% (Fig. 4A), suggesting that in this model of genetic complexity across 28 unique host genotypes (strains), disease severity is controlled by complex interactions between numerous microbial community members. This is likely representative of the high level of variation in MS–microbiome associations seen across multiple genetically unique patient cohorts described in different reports (17–19, 24).
Fig. 4.
Identification of gut microbial species and functions associated with EAE severity in the B6.ChrPWD consomic model. (A) OTU most strongly associated with CDS where positive values in orange indicate increased abundance associated high EAE CDS and negative values in blue indicate OTU associated with low EAE CDS. Reads were rarefied to 7,000. (B) Enrichment of GO terms/pathways within the set of KEGG elements associated with EAE CDS (Dataset S2) was assessed using enrichR. All significantly enriched (uncorrected P < 0.05) GO terms for the set of KEGG elements associated with EAE CDS are shown. Combined enrichment scores and P values for enrichment are shown. (C) KEGG pathway enrichment analysis using GSVA represented as a heatmap where the relative enrichment of the significant KEGG pathways across all 27 consomic strains is ordered by CDS (low to high).
In the context of regulating mammalian host phenotypes, the identity and abundance of specific microbes in complex consortia are often less informative than their functional profiles (47, 48). Consequently, to infer microbiome functionality, we used PICRUSt2 to predict genomic content from 16S data, identifying 577 specific Kyoto Encyclopedia of Genes and Genomes (KEGG) elements (bacterial genes) whose abundance was significantly associated with EAE CDS (P ≤ 0.05) (Dataset S2). To identify biological processes connected with these microbial genes negatively or positively associated with EAE CDS, we employed two independent computational analyses: Gene ontology (GO) term enrichment using enrichR and KEGG gene set variation analysis (GSVA). GO enrichment pathway analysis identified seven biological pathways were significantly enriched in genes exhibiting associations with EAE CDS, including both SCFA catabolic and metabolic processes (Fig. 4B). Moreover, parallel analysis using GSVA identified metabolism of SCFAs propionate and butyrate as associated with CDS, with apparent higher enrichment in strains exhibiting high CDS (Fig. 4C), lending additional support for the notion that broad differences in SCFA utilization across multiple gut microbiome-genotype configurations modulate disease susceptibility. Collectively, our data demonstrate that in the B6.ChrPWD consomic model of genetic diversity, associations between microbial community structure and autoimmune disease susceptibility are complex and multifactorial, suggesting the existence of genotype-specific determinants and revealing common key microbial genetic pathways.
Genetically Determined Gut Microbiota Modulates EAE Severity and Metabolic Profiles.
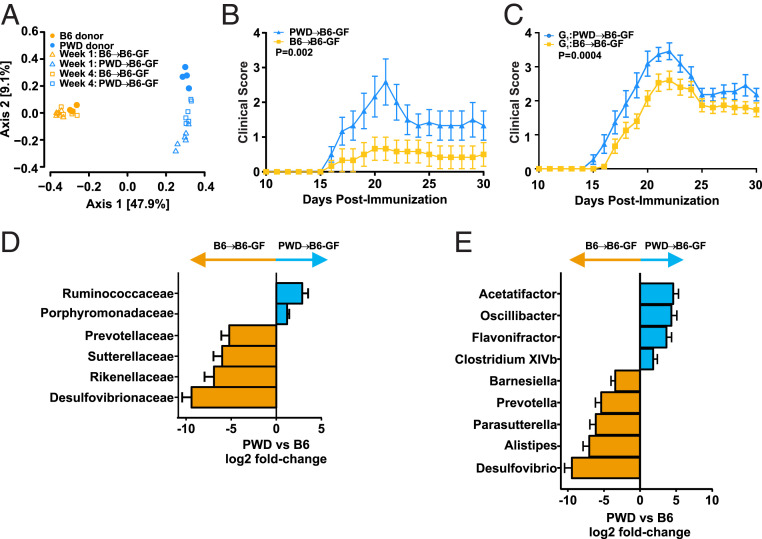
Given the complexity of genotype-microbiome community structure associations across the full B6.ChrPWD mouse panel, we focused on the two most divergent genotypes that gave rise to highly divergent gut microbiota profiles and EAE phenotypes, namely the parental strains, B6 and PWD (Figs. 1, 2C, and 3D). To determine whether the distinct gut microbiomes hosted by B6 and PWD mice have any functional effect on CNS autoimmunity, we performed gut microbiota transplant (GMT) experiments. Cecal microbiota from donor B6 and PWD mice were collected anaerobically and cryopreserved, then transplanted into 4- to 5-wk-old germ-free (GF) recipient B6 mice via oral gavage, followed by a reconstitution period of 4 wk (see schematic in SI Appendix, Fig. S4A). The 16S sequencing confirmed successful transplantation, whereby the recipients’ microbiome resembled that of donors’ (Fig. 5A). Of note, PWD→B6-GF microbiome recipients showed bigger differences between donors and recipients compared with the B6→B6-GF group (Fig. 5A), suggesting that the B6-GF host exerts selective pressure on the PWD microbiome and not the B6 microbiome, consistent with host genetic control of the microbiome. EAE was induced in GMT recipients at 4 wk posttransplantation. Unexpectedly, EAE was significantly more severe in the PWD→B6-GF group compared to the B6→B6-GF group (Fig. 5B).
Fig. 5.
GMT of divergent B6 and PWD gut microbiota into genetically identical hosts modulates EAE. B6-GF mice were inoculated with cecal contents from B6 or PWD donors. (A) β-Diversity analysis by unweighted UniFrac distance of fecal microbiomes of B6 and PWD donors (collected prior to cecal content collection) and ex-GF recipients, collected at 1 wk or 4 wk posttransplant, as indicated. (B) EAE was induced and evaluated in the ex-GF B6 GMT recipients of B6 and PWD microbiomes at 4 wk posttransplantation, as described in Fig. 3. Mean daily clinical scores are shown, with overall significance determined by Friedman’s nonparametric two-way ANOVA as in Fig. 3. (C) Vertical transmission of B6 and PWD microbiomes in B6 hosts was established by using ex-GF recipients as founder breeding pairs. EAE was induced and evaluated as above. (D and E) Comparison of differential abundance of bacterial families (D) and genera (E) in ex-GF recipients of B6 vs. PWD microbiomes at 4 wk posttransplantation, prior to EAE induction. Families and genera exhibiting differential abundance as determined using a cutoff of log2(fold change) ≥ |1| and Pcorr < 0.05 are shown with all 16S data rarefied to 10,000 reads.
To interrogate the role of these disparate microbiomes in the context of a normal developing immune system, and to circumvent the known gastrointestinal abnormalities of GF mice, a vertical transmission model was established, using B6→B6-GF and PWD→B6-GF GMT as G0 (generation zero) breeding pairs (see schematic in SI Appendix, Fig. S4B). EAE was induced in the resultant G1 offspring between 8 and 12 wk of age. Consistent with the direct GMT model, more severe EAE was observed in mice harboring the PWD gut microbiome (Fig. 5C). Of note, an overall more severe disease course in G1 offspring was observed as compared to ex-GF direct GMT recipients (Fig. 5B vs. Fig. 5C), likely due to normalized immune system development in G1 offspring compared with directly recolonized (G0) GF mice, which typically exhibit underdeveloped immune systems (49–51). To identify candidate microbes responsible for this microbiota-dependent modulation of EAE, we compared the microbiomes of ex-GF recipients of B6 and PWD microbiota. Large differences in microbiome composition were observed across several bacterial families and genera (Fig. 5 D and E), with at least 183 OTU differing significantly in abundance (Dataset S3), as expected based on the observed differences between the native microbiomes of B6 and PWD mice (Fig. 1). These data demonstrate that two divergent gut microbiomes in genetically identical hosts are sufficient to produce robust differences in EAE severity. However, these phenotypes may differ from the overall phenotype in the original host [i.e., EAE resistance of PWD mice carrying their naturally selected microbiome (Fig. 3D)], and thus are dependent upon host genotype-specific interactions with the gut-microbiome, and a susceptible genetic background.
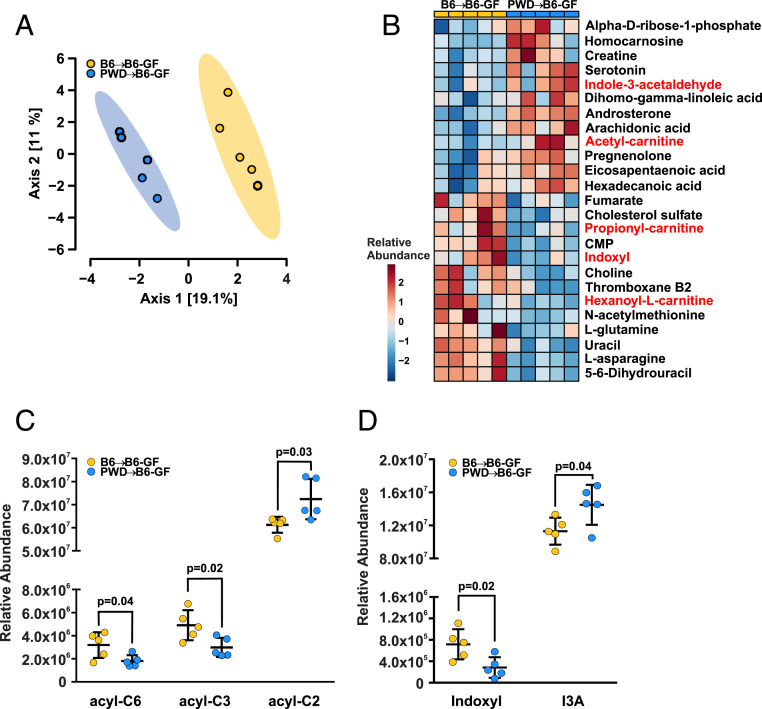
Emerging research demonstrates that gut microbiota produce an abundance of small metabolites with the capacity to enter systemic circulation and affect distal sites like CNS (52). To begin to identify the mechanisms by which distinct gut microbial profiles could impact EAE susceptibility, we analyzed a panel of ∼700 metabolites present in the serum of B6→B6 and B6→PWD ex-GF mice using mass spectrometry. Sparse partial least-squares discriminate analysis and clustering by Euclidean distance revealed that B6 and PWD gut-microbiomes result in the production of divergent circulating metabolites within genetically identical hosts (Fig. 6 A and B and Dataset S4). Among the top 23 differentially abundant metabolites, two groups were conspicuously linked to gut microbial metabolism. Two carnitine-conjugated SCFAs, propionyl- and hexanoyl-carnitines, were decreased in abundance in mice carrying the PWD microbiome, while a third, acetyl-carnitine, was elevated (Fig. 6C). Additionally, bacteria-derived tryptophan metabolites indoxyl and indole-3-acetaldehyde (I3A) displayed an inverse correlation between B6 and PWD gut microbiomes, with indoxyl at high relative abundance in mice carrying the B6 microbiome, and I3A accumulating in hosts harboring the PWD microbiome (Fig. 6D). Importantly, both represent metabolic endpoints of host dietary tryptophan with known immune-modulatory properties (52, 53). Furthermore, the two groups showed differential abundance of serotonin (Fig. 6B), another tryptophan metabolite with a well-established role in neuro- and immunomodulation (54, 55). In addition, long-chain free fatty acids (FA 16:0; 20:3; 20:4; 20:5) and arachidonic acid-derived metabolites (thromboxane B2, isobars) were significantly different between the two groups (Fig. 6B). Changes in pyrimidines (uracil, dihydrouracil, CMP) were noted as well, consistent with prior reports on alterations of pyrimidine metabolism in response to microbiome-altering interventions (56). These data suggest that divergent B6 and PWD gut microbial communities within genetically identical hosts are sufficient to modulate EAE pathogenesis and the host serum metabolome.
Fig. 6.
Metabolomic profiling identifies changes in carnitine-conjugated SCFA and tryptophan metabolites associated with GMT-mediated modulation of EAE susceptibility. Serum was collected from ex-GF recipients of B6 and PWD microbiomes, and metabolomic profiling was performed. (A) Principal component analysis of metabolic profiles of B6 and PWD microbiome recipients. (B) A heatmap of metabolites exhibiting significant differences in abundance (P < 0.05) between B6 and PWD microbiome recipients. (C) Abundance of hexanoyl-l-carnitine (acyl-C6), propionyl-carnitine (acyl-C3), and acetyl-carnitine (acyl-C2) between B6 and PWD microbiome recipients. (D) Abundance of indoxyl and I3A between B6 and PWD microbiome recipients.
Lactobacillus Species Abundance Cosegregates with EAE Severity.
Because of the large divergence between B6 and PWD gut microbiomes, identifying candidate microbial constituents responsible for driving the EAE phenotypes in Fig. 5 B and C is difficult. Therefore, we performed a complementary set of experiments wherein the gut microbiome was modulated via cohousing, which allows for a more limited degree of bidirectional transfer of microbiota (57). Littermate 3-wk-old B6 mice were randomly assigned to be cohoused with either age- and sex-matched B6 mice from a different litter (B6 controls), or with age- and sex-matched PWD mice (B6 cohoused) (see schematic in SI Appendix, Fig. S4C). As expected, 16S analysis revealed a convergence of B6 and PWD gut microbiomes after cohousing (Fig. 7A). EAE was induced after 5 wk of cohousing. In line with the GMT experiments, B6 mice with PWD exhibited higher EAE compared with B6 controls (Fig. 7B), demonstrating that partial transfer of gut microbiota from PWD to B6 hosts can modulate EAE severity.
Fig. 7.
Partial transfer of gut microbiota from PWD to B6 mice by cohousing is sufficient to modulate EAE. Three-week-old B6 mice were randomized to be cohoused with age-matched PWD mice, or a different litter of age-matched B6 mice as controls. (A) Unweighted UniFrac distance analysis of fecal microbiomes before cohousing (B6 start and PWD start), and after 5 to 6 wk of cohousing (B6 control, B6 cohoused, PWD control, PWD cohoused). (B) EAE was induced and evaluated at 5 to 6 wk postcohousing, as described in Fig. 5. (C) Indicator species analysis was used to identify OTU distinguishing microbiomes of B6 control from B6 cohoused groups. Relative abundance of the identified OTUs, as calculated using DESeq2, is shown. (D) Random forest classifier analysis was used to identify OTUs distinguishing microbiomes of B6 control from B6 cohoused groups, and the relative importance/weight of these OTUs in the model is shown with all 16S data rarefied to 12,000 reads.
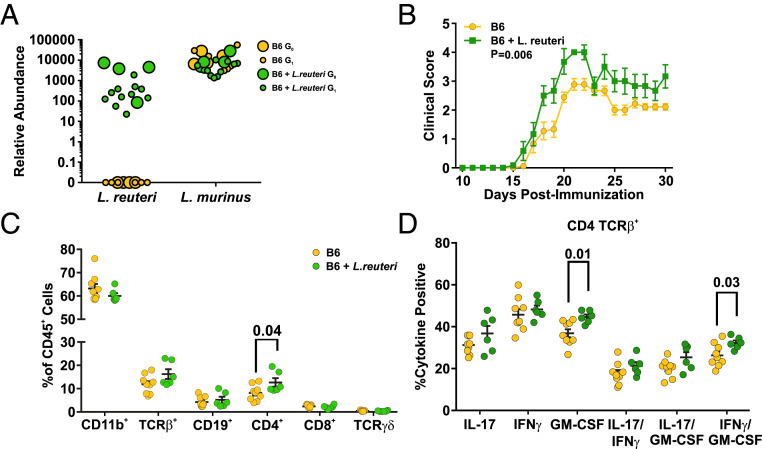
To identify putative bacterial species responsible for this EAE phenotype, we analyzed the bacterial groups that were transferred from PWD to B6 mice. Since we had employed three separate sets of experiments and analyses to characterize B6 and PWD microbiomes (Figs. 1, 5, and 7), we first performed a combined β-diversity analysis to ensure a lack of batch/experiment effect, which revealed minimal clustering by experiment, with comparable differences between B6 and PWD across all three experiments (SI Appendix, Fig. S5). Compared with GMT (Fig. 5 D and E), the transfer of bacteria from PWD by cohousing resulted in much more subtle differences between B6 control and B6 cohoused mice, which could only be detected at the OTU level. Therefore, we used indicator species analysis (58) and a machine-learning approach (random forest classifier analysis) (59) to identify OTUs that distinguished control from cohoused mice. Indicator species analysis identified 10 OTUs: 9 of them belonging to the Porphyromonadaceae family and 1 belonging to the Lactobacillus genus (Fig. 7C). Furthermore, the random forest classifier identified a single OTU as the most effective in distinguishing control from cohoused mice, which was the same Lactobacillus OTU identified during indicator species analysis (Fig. 7D). OTU sequence analysis using BLAST suggested that this OTU most likely represents L. reuteri (National Center for Biotechnology Information [NCBI] Blastn: Query coverage 100%, Evalue = 0, 99.8% identity), a common human and mouse gut commensal and a putative probiotic species, with well-documented immunomodulatory properties (60). Relative abundance analysis of 16S data showed that this species/OTU was almost completely absent in B6 mice, highly abundant in PWD mice, and transferred efficiently by cohousing (SI Appendix, Fig. S6A).
Closer examination of the major Lactobacillus OTUs identified by 16S analysis revealed the presence of another commensal Lactobacillus species, Lactobacillus murinus. Interestingly, the abundance of L. murinus was inversely correlated with that of L. reuteri, suggesting possible competition between different Lactobacillus species for the same ecological niche in the gut and selection by host genotype. In contrast to L. reuteri, L. murinus was present at high abundance in the B6 gut microbiome, almost completely absent within the PWD gut microbiome, and efficiently transferred via cohousing. To validate these 16S findings, we used qPCR with species-specific primers to confirm the identity and to quantify the relative abundance of L. reuteri and L. murinus (SI Appendix, Fig. S6B), which confirmed 16S abundance data (SI Appendix, Fig. S6A). Furthermore, both 16S and qPCR analyses of fecal samples from B6 and PWD donors and ex-GF GMT recipients (Fig. 5 A and B) demonstrated the same pattern of high L. reuteri abundance within the PWD gut microbiome and high L. murinus abundance within the B6 gut microbiome, with efficient transfer by GMT (SI Appendix, Fig. S6C). Additionally, these Lactobacillus species were efficiently transmitted vertically, with similar abundance in parental pairs and G1 offspring (SI Appendix, Fig. S6D) in conjunction with exacerbated EAE (Fig. 5C). Importantly, indole derivatives identified by metabolomics analysis as differentially associated with microbial community-specific EAE outcome (Fig. 6 B–D) are known to be produced by Lactobacillus species, including L. reuteri and L. murinus (52). These results suggest that augmentation of EAE severity in B6 mice by cohousing with PWD mice or GMT of PWD gut microbiota is associated with transfer of L. reuteri.
Commensal Colonization by L. reuteri Is Sufficient to Exacerbate EAE Pathogenesis in Conjunction with Modulating T Cell Responses.
Existing research demonstrates both pro- and antiinflammatory roles for L. reuteri, which may in part be due to variation between different strain isolates (61–64). Consequently, to determine in our model if L. reuteri is sufficient to accentuate EAE pathogenesis when introduced into the B6 gut microbiome as computationally predicted (Fig. 7 C and D), L. reuteri was isolated from PWD cecal contents (Methods). Previous efforts to understand the role of L. reuteri in EAE pathogenesis have relied on daily gavage with high doses of bacteria (63, 65), which is not representative of stable commensal colonization. Consequently, a vertical transmission model was established (see schematic in SI Appendix, Fig. S4D). Specifically, cryopreserved B6 cecal microbiota was transplanted either alone (as before) (Fig. 5), or supplemented with 109 CFU L. reuteri, into 4-wk-old GF recipient B6 mice, which subsequently served as founder (G0) breeding pairs. Breeder fecal samples were collected following a 4-wk colonization period and Lactobacillus species abundance was determined using qPCR. Experimental G1 offspring were screened by qPCR 7-d prior to EAE induction at 8 to 12 wk of age. Consistent with Lactobacillus vertical transmission in B6 and PWD GMT studies (SI Appendix, Fig. S6D), L. reuteri colonization was successfully established in breeders, and transmitted efficiently to G1 offspring (Fig. 8A). Interestingly, the high abundance of L. murinus in the B6 microbiome was not altered by colonization with L. reuteri (Fig. 8A). Strikingly, B6 mice colonized with L. reuteri demonstrated exacerbated EAE pathogenesis as compared to those devoid of this species (Fig. 8B), demonstrating that colonization with a single bacterial species from the PWD microbiome is sufficient to modulate EAE severity in the B6 host.
Fig. 8.
Colonization with L. reuteri is sufficient to modulate EAE susceptibility and encephalitogenic T cell responses. Founder G0 GF B6 mice were inoculated with cecal contents from B6 donors, supplemented or not with 109 CFU of L. reuteri, followed by fecal sample collection at 4 wk postinoculation, and establishment of breeding pairs for vertical transmission to G1 offspring. (A) Abundance of L. reuteri and L. murinus in founder breeders and G1 offspring was determined by qPCR as in Fig. 7. (B) EAE was induced and evaluated in G1 offspring at 8 to 12 wk of age, as described in Fig. 5. (C and D) At day 30 post-EAE induction, CNS-infiltrating leukocytes were isolated and analyzed by flow cytometry as described in Methods. (C) Frequencies of the major leukocyte populations within the CD45+ population. (D) Percentage of TCR-β+ CD4+ cells positive for the indicated cytokines. The percentages of cells positive for a given single cytokine indicate cells positive for the cytokine independent of coproduction of the other three cytokines (thus including single, double, and triple producers), whereas the double-positive percentages indicate cells producing specifically both of the indicated cytokines.
Both MS and EAE demyelinating disease is thought to be initiated and driven by CNS-infiltrating myelin-reactive Th1 and Th17 cells producing IFN-γ, IL-17, and granulocyte macrophage-colony stimulating factor (GM-CSF) cytokines (66), with additional contributions from IL-17 producing CD8+ T cells (Tc17) (67). To discern the cellular mechanism by which L. reuteri might be driving exacerbation of EAE pathogenesis, flow cytometric analysis of immune cell infiltrates into the spinal cord of diseased mice harboring B6 or B6+L. reuteri microbiomes was performed. Colonization with L. reuteri resulted in a higher frequency of infiltrating CD4+ T cells in the spinal cord compared with control B6 mice lacking this species (Fig. 8C). Furthermore, a higher frequency of these CD4+ T cells produced GM-CSF, as well as coproducing GM-CSF and IFN-γ together, overall indicative of a more potent GM-CSF–skewed Th1 response (Fig. 8D and SI Appendix, Fig. S7B). While the frequency of CNS-infiltrating CD8+ T cells remained unchanged upon L. reuteri colonization (Fig. 8C), a higher proportion of this subset were producing GM-CSF, IL-17, and GM-CSF and IL-17 together (SI Appendix, Fig. S7 A, C, and D). Consistently, in naïve mice, L. reuteri colonization elicited higher frequencies of CD4+ T cells producing GM-CSF and higher frequencies of both CD4+ and CD8+ T cells producing IFN-γ in the spleen (SI Appendix, Fig. S8 A and B). Furthermore, measurement of ex vivo peripheral recall response 10 d following immunization demonstrated that colonization with L. reuteri resulted in enhanced production of IL-17, IFN-γ, and GM-CSF by CD4+ T cells and a higher frequency of CD8+ T cells producing IFN-γ in the spleen (SI Appendix, Fig. S8 C and D). These data suggest that stable introduction of L. reuteri into the B6 gut microbiome is sufficient to exacerbate EAE and to amplify and prime encephalitogenic T cell responses.
Discussion
As with most complex diseases, the combined effects of genetics and the environment are thought to dictate MS risk (68). Many of the environmental risk factors for MS, including diet, vitamin D intake, smoking, stress, and previous infection can contribute to gut microbiome imbalance, suggesting that the gut microbiome may serve as an integration point for multiple risk factors (6, 8, 69, 70). Microbiome transplantation from MS patients has been shown to exacerbate EAE pathogenesis; however, differences between individual donors’ commensal microbiota (likely modified by patient genetics) drove variable EAE outcomes in these studies (19, 24), including a complete lack of effect on EAE in one study (71). Here, we have demonstrated potent effects of host genetics, on both EAE susceptibility and gut microbial composition predating disease onset. These data emphasize the profound impact of G×E interactions that ultimately synergize to influence MS pathogenesis. Additionally, defining causative commensal species within the MS dysbiotic state has remained elusive. We have demonstrated that the introduction of a single commensal species, L. reuteri, is sufficient to exacerbate EAE pathogenesis in a genetically susceptible host.
Multiple studies have now shown that MS patients exhibit distinct gut microbial composition as compared to healthy controls, suggesting that the gut microbiome may play a role in MS disease susceptibility (72–74). However, given the high prevalence of GI disturbances that occur as a symptom of disease, it is difficult to discern whether these imbalances represent a symptom of disease or rather a dysbiotic state that promotes disease onset (73, 75). Furthermore, while expansion in Akkermansia and contraction of SCFA-producing bacteria have consistently been observed across several studies (14, 76, 77), there is a lack of consensus on the exact composition of the so-called MS dysbiotic state. Broad differences at the level of genus between studies make establishing species-level MS-specific differences, let alone causative species, difficult. These differences likely reflect cohort variation in baseline commensal gut microbiome composition driven by both genetics and environmental exposures. This notion is supported by a recent report from Blaser and colleagues (78), documenting that the differences in microbiota between MS patients and controls are highly impacted by ethnic background, with the notable exception of increased Clostridium abundance. Importantly, this variation observed across clinical studies resembles the genetically driven gut microbiome variation in our consomic mouse model, underscoring the impact that host genetics play in determining baseline microbial composition prior to disease onset.
Much remains unclear regarding the impact of host genetics on the composition of the gut microbiome. Numerous studies in mouse models and human populations support the idea of a mammalian host genetic component regulating gut microbiome composition (34–41). However, newer studies in humans have challenged this concept, arguing that the environment dominates in determining the gut microbiota, while host genetics plays a negligible role (79). In this context, the precise control over environmental variables in mouse models may make delineation of genetic effects more feasible. In our B6.ChrPWD consomic model, we identified a range of microbial profiles associated with specific genotypes, suggesting that PWD alleles at specific loci modulate gut microbiome composition. This notion is supported by previous studies demonstrating a strong divergence between gut microbiomes of B6 and PWK/PhJ mice (a close relative of the PWD strain) (80), and strong genetic effects on the microbiome composition in Collaborative Cross mice, which carry B6 and PWK/PhJ alleles (81). Nonetheless, our experiments were not designed to exclude potential founder/strain isolation effects on the microbiome that can be prevalent in mouse studies (82, 83), and we cannot rule out a contribution of such effects to the observed microbiome diversity of our consomic strains. Nevertheless, and pertinent to our studies, this model provides a unique panel of genotype–microbiome combinations to interrogate phenotypic consequences.
We designed our study to profile and manipulate gut microbiota prior to EAE induction, to identify states of the microbiome that result in differential susceptibility to CNS autoimmunity, without the confounding variable of secondary effects of disease progression. In our model of genetic diversity, we identified multiple microbiome features associated with lower or higher disease susceptibility across multiple host genotypes. Interestingly, two of these taxa have been identified as differentially abundant in MS, but with what at first glance appears to be opposite directionality. Akkermansia abundance, associated with lower EAE susceptibility, was elevated in MS patients in two different United States studies (18, 19), while C. cluster XlVa, associated with higher EAE susceptibility, was depleted MS patients of Japanese descent (14). However, a subsequent study demonstrated that Akkermansia, although elevated in MS patients and during EAE progression, in fact plays a protective role in EAE (76), which is fully consistent with our studies. Interestingly, we found that the contribution of each individual taxonomic unit associated with EAE to overall susceptibility was modest, suggesting that the effect of gut bacteria on CNS autoimmunity is multifactorial and dependent on host genotype and overall microbial community structure. This suggests an explanation for the lack of consensus across human MS microbiome studies, which, with one notable exception (78), have so far largely ignored the contribution of host genetics.
In our previous studies of parental B6 and PWD strains, we demonstrated that the genetic divergence of wild-derived PWD mice from the B6 laboratory strain leads to diminished expression of MS signature genes in immune cells, and overall greatly reduced EAE susceptibility (31), as confirmed in this study. At first glance, it may seem counterintuitive that transplantation of gut microbiota from EAE-resistant PWD mice to EAE-susceptible B6 hosts exacerbates rather than suppresses EAE. However, we postulate that: 1) The resistance of PWD mice to EAE is predominantly driven by genetics, with a lesser contribution of the microbiome; and 2) the PWD gut microbiome is best adapted for its PWD host, and its “mismatched” transplantation into the B6 host leads to exacerbated autoimmunity. This accentuates the importance of host genome specific-interactions with the gut microbiome in determining susceptibility to CNS autoimmunity. In support of this notion, partial transfer of B6 microbiota via cohousing to the PWD host, which is normally resistant to EAE, resulted in moderately increased EAE susceptibility as compared with PWD noncohoused littermate controls (SI Appendix, Fig. S9). Overall, we conclude that genetic predisposition plays a primary role in setting the required “minimal baseline” of MS susceptibility, while the gut microbiota play a secondary role in modulating this susceptibility, possibly tipping the scale above or below a critical threshold for disease initiation or progression.
Many conflicting reports regarding the anti- vs. proinflammatory roles of specific members of the gut microbiota have emerged in the recent years, generating confusion and skepticism as to the impact of microbiota on human health and disease. We believe that there are four critical factors that underlie these discrepancies, namely differences in: 1) Bacterial genetics, 2) host genetics, 3) baseline existing microbiome state, and perhaps most critically, 4) timing and mode of exposure/treatment. With regard to MS/EAE pathogenesis, this is exemplified by the contradictory findings as to the impact of Lactobacillus species on modulating disease outcome when utilized as therapeutic probiotics, in studies that typically utilize daily gavage with billions of live bacteria. Orally administered L. reuteri (ML1 strain) was previously shown to exacerbate EAE in female SJL/J mice, compared with L. murinus (CNR strain) treatment, which reduced disease severity (64). A more recent study found that daily gavage with L. reuteri (DSM 17938) diminished EAE pathogenesis, reducing Th1/Th17 cells and their associated cytokines, IFN-γ/IL-17, in female C57BL/6 mice (63). While the latter study, demonstrating a protective effect of L. reuteri, appears to be in direct conflict with our findings, we believe that this is due to critical differences in the mode of delivery (probiotic gavage vs. commensal colonization), window of exposure (at EAE induction vs. life-long developmental exposure), and differences between strain isolates (probiotic strain vs. gut commensal strain). Our model addresses the role of commensal microbiota as a risk factor for MS, rather than a therapeutic probiotic intervention. Moreover, given L. reuteri’s widely accepted role as a probiotic species, our findings sound a cautionary note and highlight the need to consider both host genetics and baseline gut microbiome composition in probiotic or similar therapeutic strategies, especially given that such efforts are already underway in MS (84, 85).
Targeted probiotic treatment is not limited to MS models. In the MRL/lpr mouse lupus nephritis model, oral administration of five Lactobacillus species (including L. reuteri) decreased inflammation and improved renal function and survival (62). In contrast, a commensal L. reuteri (SP-C2-NAJ007 strain) was increased in Tlr7.1 transgenic C57BL/6 lupus-prone mice, and was sufficient to exacerbate autoimmunity when transplanted into GF or specific pathogen-free mice (61). These findings suggest a differential response to bacteria of commensal vs. probiotic origin and support our findings that commensal L. reuteri colonization may promote autoimmunity.
Similarly, conflicting reports have been presented as to the role of Lactobacilli in modulating immune function, as well as molecular mechanisms associated with this modulation. Lactobacillus members, including L. reuteri, have been shown to possess the enzymatic machinery necessary to catabolize dietary tryptophan into various indole derivatives, which can function as agonists for the aryl-hydrocarbon receptor (AhR), promoting the generation of innate-like lymphocyte populations in the gut (86, 87). In the EAE model, antibiotic treatment was shown to exacerbate clinical scores with an associated depletion of L. reuteri, an effect that could be reversed by indole supplementation (88). Additional studies demonstrated that indoles exert antiinflammatory effects via AhR activation in microglia or astrocytes (86, 88), pointing toward a potential mechanism whereby L. reuteri would attenuate (rather than exacerbate) EAE pathogenesis in our model. Furthermore, high salt diet exacerbation of EAE was associated with depletion of L. murinus, and could be reversed by daily supplementation with L. murinus or L. reuteri (89). Of note, both of these studies used therapeutic supplementation/depletion, rather than developmental commensal exposure used in our model. In contrast, we have shown that the presence of L. reuteri as a true commensal species is sufficient to exacerbate EAE disease pathogenesis. These data illustrate L. reuteri’s potential to skew host immune cell repertoires to either pro- or antiinflammatory in a context-dependent fashion, as influenced by window of exposure, host genetics, and the microbiome ecosystem within which it resides.
Despite relatively strong epidemiological associations, the role of the microbiome in MS remains complex and unclear. Our studies add several new layers to this complexity, (i.e., differential developmental exposure to commensal microbes as a risk factor) and the role of host genetics as a modulator. This notion is fully supported by two recent studies demonstrating highly individualized and sometimes deleterious responses to probiotic interventions in humans (90, 91). Additionally, we have identified and isolated a specific member of the commensal gut microbiota that is not only associated with, but functionally capable of modulating CNS autoimmunity, something that to our knowledge has only been achieved one other time, in a recent study from Weiner and colleagues (76). With clinical trials involving direct fecal microbiome transplantation, as well as antibiotic and probiotic supplementation currently underway (84, 85, 92–94), our findings highlight the need to consider timing of exposure, host genetics, and baseline gut microbiome composition in interpreting these outcomes, and designing better personalized prophylactic and therapeutic interventions.
Methods
Expanded methods for each subsection below are provided in SI Appendix, Supporting Materials and Methods.
Animals.
C57BL/6J (B6), PWD/PhJ (PWD), and B6.ChrPWD consomic mice purchased from Jackson Laboratories were bred and housed in a single room within the vivarium at the University of Vermont for two to four generations prior to experimentation. The procedures used in this study were approved by the Animal Care and Use Committee of the University of Vermont.
Induction and Evaluation of EAE.
EAE was induced in B6 and B6.ChrPWD consomic mice using the 2×MOG35–55/CFA protocol (95) or mouse spinal cord homogenate + CFA + PTX inoculation protocol (96). Starting on day 10, mice were scored visually as follows: 1, loss of tail tone; 2, loss of tail tone and weakened hind limbs; 3, hind limb paralysis; 4, hind limb paralysis and incontinence; 5, quadriplegia or death.
Microbial DNA Isolation and 16S DNA Sequencing.
DNA was extracted from fecal or cecal samples using the MoBio PowerSoil extraction kit (MoBio) with quality and quantity assessed by NanoDrop, Qubit (Life Technologies), and agarose gel electrophoresis. Amplicon libraries for the 16S rRNA hypervariable regions (V3-V4) were generated using universal forward 357-F (5′-CCTACGGGNGGCWGCAG-3′) and reverse 805-R (5′-GACTACHVGGGTATCTAATCC-3′) primers on the Fluidigm Access Array (Fluidigm) at the University of Illinois W. M. Keck Center for Comparative and Functional Genomics. Resulting pools were quantified by Qubit and qPCR at the DNA Services laboratory of the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana–Champaign following size selection on a 2% eGel (Invitrogen) and repeat qPCR. Under 400-nt products and over 400-nt products were mixed at a ratio of 1:20, denatured, spiked with 20% PhiX, and loaded onto HiSeq V2 or MiSeq flowcells at 10 pM for paired-end sequencing. Data are available at the NCBI Sequence Read Archive (BioProject ID PRJNA590851).
Microbiome Transplantation and Cohousing.
GMT protocols were based on a procedure developed by Blaser and colleagues (97). Briefly, the cecum was excised from three to four donor mice per sex/strain, tied off at proximal and distal ends, and transferred to an anaerobic chamber (Coy Labs). Contents were flushed out and resuspended in liquid dental transport medium (Anaerobe Systems), volume-adjusted with prereduced saline (Anaerobe Systems), and cryopreserved in Hungate tubes (Fisher Scientific) with sterile glycerol at 20% (vol/vol).
GMT recipient GF 4- to 5-wk-old C57BL/6J mice (National Gnotobiotic Rodent Resource Center at the University of North Carolina School of Medicine, Chapel Hill, NC) were inoculated by gastric gavage with 0.125 mL of cryopreserved cecal contents and maintained under barrier conditions with sterilized food, water, and caging. After a 4-wk stabilization period, mice were used for EAE experiments or served as founder breeding pairs for vertical transmission studies.
Colonization with L. reuteri was performed as above, with the following modifications: Four-week-old recipient B6-GF mice received 200 µL of B6 cryopreserved cecal content via oral gavage or 100 µL of B6 cecal content and 100 µL L. reuteri at 109 CFU. Fecal samples were collected 4 wk postgavage for qPCR and 16S analysis.
Lactobacillus Species Identification, Quantification, and Isolation.
Relative abundance was determined by qPCR (Dynamo ColorFlash kit, ThermoFisher) using species-specific primers for L. reuteri and L. murinus (86, 89, 98) normalized to pan-eubacterial primers (86). L. reuteri was isolated from PWD cecal contents by selection in MRS medium (ThermoFisher) containing 20 µg/mL vancomycin followed by qPCR screening of isolated colonies and cryopreservation. A pool of three independent isolates was used in final stocks for colonization studies.
Flow Cytometry.
Day 30 post-EAE mice were perfused transcardially with PBS. Spinal cords were removed, homogenized using a Dounce glass homogenizer, and filtered with a 70-µm strainer to yield a single-cell suspension. Percoll gradient (37%/70%) centrifugation was used to isolate mononuclear cells. For intracellular cytokine analysis, cells were stimulated with 5 ng/mL PMA, 250 ng/mL ionomycin, and brefeldin A (Golgi Plug reagent, BD Bioscience) for 6 h. Cells were labeled with the UV-Blue Live/Dead fixable stain (ThermoFisher), stained with surface antibodies against CD45, CD11b, CD4, CD8, TCRγδ, and TCRβ (Biolegend), fixed and permeabilized with 0.2% saponin, and labeled with anti–IL-17A, anti–IFN-γ, and anti–GM-CSF antibodies (Biolegend). Labeled cells were analyzed using an LSRII flow cytometer (BD Biosciences) using appropriate single-color controls. Data were analyzed using FlowJo software, v10 (Tree Star).
Metabolomics.
Metabolomic analysis of serum collected on day 30 post-EAE from GMT recipients was performed as described previously (99) at the University of Colorado mass spectrometry facility. Briefly, samples diluted 1:25 in methanol:acetonitrile:water (5:3:2 [vol/vol]) were analyzed following centrifugation to generate cleared supernatants using the 5-min C18 gradient method (99) on a Thermo Vanquish UHPLC coupled online to a Thermo Q Exactive mass spectrometer. Metabolite assignments and peak area measurements were performed using Maven (Princeton University) against an in-house standard library. Data were analyzed using MetaboAnalyst, v4.0 (100).
Supplementary Material
Acknowledgments
We thank Dr. Saleh Ibrahim (University of Lübeck) for his valuable input and help with coordination of personnel involved in this project and Drs. Marty Blaser (New York University) and Laurie Cox (Harvard Medical School) for the provision of detailed protocols for microbiota transplantation and helpful discussions. This work was supported by Grants R01 NS097596 from NIH/National Institute of Neurological Disorders and Stroke (to D.N.K.) and RG5170A6/1 from the National Multiple Sclerosis Society (to C.T.). Research performed at the Flow Cytometry and Cell Sorting Facility was partially supported by National Institute of General Medical Sciences P20GM103496. A.K. and H.B. acknowledge computational support from the OMICS compute cluster at the University of Lübeck. H.B. acknowledges support by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy, EXC 22167-390884018.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002817117/-/DCSupplemental.
Data Availability.
The 16S sequencing data have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject ID PRJNA590851) (101).
Note Added in Proof.
After acceptance of this manuscript, two key studies were published by two independent groups on the role of L. reuteri in EAE. The first found that commensal colonization with this microbe exacerbated EAE (102), fully consistent with our findings. The second found that continuous administration of a probiotic strain of L. reuteri suppressed EAE (103); the discordance with our study owing most likely to the mode of delivery and/or bacterial strain differences, as discussed above.
References
- 1.Frohman E. M., Racke M. K., Raine C. S., Multiple sclerosis—The plaque and its pathogenesis. N. Engl. J. Med. 354, 942–955 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Greenstein J. I., Current concepts of the cellular and molecular pathophysiology of multiple sclerosis. Dev. Neurobiol. 67, 1248–1265 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Baranzini S. E., Oksenberg J. R., The genetics of multiple sclerosis: From 0 to 200 in 50 years. Trends Genet. 33, 960–970 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodin D. S., The genetic and environmental bases of complex human-disease: Extending the utility of twin-studies. PLoS One 7, e47875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollenbach J. A., Oksenberg J. R., The immunogenetics of multiple sclerosis: A comprehensive review. J. Autoimmun. 64, 13–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebers G. C., Environmental factors and multiple sclerosis. Lancet Neurol. 7, 268–277 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Beecham A. H.et al.; International Multiple Sclerosis Genetics Consortium (IMSGC); Wellcome Trust Case Control Consortium 2 (WTCCC2); International IBD Genetics Consortium (IIBDGC) , Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 45, 1353–1360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Disanto G., Morahan J. M., Ramagopalan S. V., Multiple sclerosis: Risk factors and their interactions. CNS Neurol. Disord. Drug Targets 11, 545–555 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Grigg J. B., Sonnenberg G. F., Host-microbiota interactions shape local and systemic inflammatory diseases. J. Immunol. 198, 564–571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belkaid Y., Hand T. W., Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G., et al., Bridging intestinal immunity and gut microbiota by metabolites. Cell. Mol. Life Sci. 76, 3917–3937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin C. R., Osadchiy V., Kalani A., Mayer E. A., The brain-gut-microbiome axis. Cell. Mol. Gastroenterol. Hepatol. 6, 133–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantarel B. L., et al., Gut microbiota in multiple sclerosis: Possible influence of immunomodulators. J. Investig. Med. 63, 729–734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyake S., et al., Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One 10, e0137429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tremlett H.et al.; US Network of Pediatric MS Centers , Gut microbiota in early pediatric multiple sclerosis: A case-control study. Eur. J. Neurol. 23, 1308–1321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremlett H.et al.; US Network of Pediatric MS Centers , Gut microbiota composition and relapse risk in pediatric MS: A pilot study. J. Neurol. Sci. 363, 153–157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jangi S., et al., Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., et al., Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 6, 28484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berer K., et al., Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U.S.A. 114, 10719–10724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levinthal D. J., et al., Adding to the burden: Gastrointestinal symptoms and syndromes in multiple sclerosis. Mult. Scler. Int. 2013, 319201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parthasarathy G., et al., Relationship between microbiota of the colonic mucosa vs feces and symptoms, colonic transit, and methane production in female patients with chronic constipation. Gastroenterology 150, 367–379.e1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohkusa T., Koido S., Nishikawa Y., Sato N., Gut microbiota and chronic constipation: A review and update. Front. Med. 6, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz Sand I., et al., Disease-modifying therapies alter gut microbial composition in MS. Neurol. Neuroimmunol. Neuroinflamm. 6, e517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cekanaviciute E., et al., Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114, 10713–10718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAllister K., et al., Current challenges and new opportunities for gene-environment interaction studies of complex diseases. Am. J. Epidemiol. 186, 753–761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Constantinescu C. S., Farooqi N., O’Brien K., Gran B., Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharmacol. 164, 1079–1106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinman L., Zamvil S. S., How to successfully apply animal studies in experimental allergic encephalomyelitis to research on multiple sclerosis. Ann. Neurol. 60, 12–21 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Ben-Nun A., et al., From classic to spontaneous and humanized models of multiple sclerosis: Impact on understanding pathogenesis and drug development. J. Autoimmun. 54, 33–50 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Didion J. P., de Villena F. P., Deconstructing Mus gemischus: Advances in understanding ancestry, structure, and variation in the genome of the laboratory mouse. Mamm. Genome 24, 1–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregorová S., Forejt J., PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies—A valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. 46, 31–41 (2000). [PubMed] [Google Scholar]
- 31.Bearoff F., et al., Natural genetic variation profoundly regulates gene expression in immune cells and dictates susceptibility to CNS autoimmunity. Genes Immun. 17, 386–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krementsov D. N., Asarian L., Fang Q., McGill M. M., Teuscher C., Sex-specific gene-by-vitamin D interactions regulate susceptibility to central nervous system autoimmunity. Front. Immunol. 9, 1622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith M. I., Turpin W., Tyler A. D., Silverberg M. S., Croitoru K., Microbiome analysis—From technical advances to biological relevance. F1000Prime Rep. 6, 51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson A. K., et al., Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18933–18938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leamy L. J., et al., Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 15, 552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Org E., et al., Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25, 1558–1569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., et al., Analysis of intestinal microbiota in hybrid house mice reveals evolutionary divergence in a vertebrate hologenome. Nat. Commun. 6, 6440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodrich J. K., Davenport E. R., Waters J. L., Clark A. G., Ley R. E., Cross-species comparisons of host genetic associations with the microbiome. Science 352, 532–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., et al., Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 48, 1396–1406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turpin W.et al.; GEM Project Research Consortium , Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 48, 1413–1417 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Bonder M. J., et al., The effect of host genetics on the gut microbiome. Nat. Genet. 48, 1407–1412 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Anderson M. J., Ellingsen K. E., McArdle B. H., Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9, 683–693 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Bearoff F., et al., Identification of genetic determinants of the sexual dimorphism in CNS autoimmunity. PLoS One 10, e0117993 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krementsov D. N., et al., Sex-specific control of central nervous system autoimmunity by p38 mitogen-activated protein kinase signaling in myeloid cells. Ann. Neurol. 75, 50–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almeida M. N., Silvernale C., Kuo B., Staller K., Bowel symptoms predate the diagnosis among many patients with multiple sclerosis: A 14-year cohort study. Neurogastroenterol. Motil. 31, e13592 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Butterfield R. J., et al., Genetic analysis of disease subtypes and sexual dimorphisms in mouse experimental allergic encephalomyelitis (EAE): Relapsing/remitting and monophasic remitting/nonrelapsing EAE are immunogenetically distinct. J. Immunol. 162, 3096–3102 (1999). [PubMed] [Google Scholar]
- 47.Lozupone C. A., Stombaugh J. I., Gordon J. I., Jansson J. K., Knight R., Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heintz-Buschart A., Wilmes P., Human gut microbiome: Function matters. Trends Microbiol. 26, 563–574 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Smith K., McCoy K. D., Macpherson A. J., Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Deplancke B., Gaskins H. R., Microbial modulation of innate defense: Goblet cells and the intestinal mucus layer. Am. J. Clin. Nutr. 73, 1131S–1141S (2001). [DOI] [PubMed] [Google Scholar]
- 51.Macpherson A. J., McCoy K. D., Standardised animal models of host microbial mutualism. Mucosal Immunol. 8, 476–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L. S., Davies S. S., Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 8, 46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.La Carpia F., et al., Transfusional iron overload and intravenous iron infusions modify the mouse gut microbiota similarly to dietary iron. npj Biofilms Microbiomes 5, 26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mössner R., Lesch K.-P., Role of serotonin in the immune system and in neuroimmune interactions. Brain Behav. Immun. 12, 249–271 (1998). [DOI] [PubMed] [Google Scholar]
- 55.León-Ponte M., Ahern G. P., O’Connell P. J., Serotonin provides an accessory signal to enhance T-cell activation by signaling through the 5-HT7 receptor. Blood 109, 3139–3146 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keerthisinghe T. P., Wang M., Zhang Y., Dong W., Fang M., Low-dose tetracycline exposure alters gut bacterial metabolism and host-immune response: “Personalized” effect? Environ. Int. 131, 104989 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Goodrich J. K., et al., Conducting a microbiome study. Cell 158, 250–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rausch P., et al., Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol. 306, 343–355 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 60.Mu Q., Tavella V. J., Luo X. M., Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 9, 757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zegarra-Ruiz D. F., et al., A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe 25, 113–127.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mu Q., et al., Control of lupus nephritis by changes of gut microbiota. Microbiome 5, 73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He B., et al., Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front. Immunol. 10, 385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maassen C. B., et al., Orally administered Lactobacillus strains differentially affect the direction and efficacy of the immune response. Vet. Q. 20 (suppl. 3), S81–S83 (1998). [PubMed] [Google Scholar]
- 65.Maassen C. B. M., et al., Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 18, 2613–2623 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Becher B., Segal B. M., T(H)17 cytokines in autoimmune neuro-inflammation. Curr. Opin. Immunol. 23, 707–712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huber M., et al., IL-17A secretion by CD8+ T cells supports Th17-mediated autoimmune encephalomyelitis. J. Clin. Invest. 123, 247–260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waubant E., et al., Environmental and genetic risk factors for MS: An integrated review. Ann. Clin. Transl. Neurol. 6, 1905–1922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu F., et al., Gut microbiota in multiple sclerosis and experimental autoimmune encephalomyelitis: Current applications and future perspectives. Mediators Inflamm. 2018, 8168717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shamriz O., et al., Microbiota at the crossroads of autoimmunity. Autoimmun. Rev. 15, 859–869 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Cekanaviciute E., et al., Multiple sclerosis-associated changes in the composition and immune functions of spore-forming bacteria. mSystems 3, e00083-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freedman S. N., Shahi S. K., Mangalam A. K., The “gut feeling”: Breaking down the role of gut microbiome in multiple sclerosis. Neurotherapeutics 15, 109–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kirby T. O., Ochoa-Repáraz J., The gut microbiome in multiple sclerosis: A potential therapeutic avenue. Med. Sci. 6, 69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ochoa-Repáraz J., Kirby T. O., Kasper L. H., The gut microbiome and multiple sclerosis. Cold Spring Harb. Perspect. Med. 8, a029017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ochoa-Repáraz J., Magori K., Kasper L. H., The chicken or the egg dilemma: Intestinal dysbiosis in multiple sclerosis. Ann. Transl. Med. 5, 145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu S., et al., Oral administration of miR-30d from feces of MS patients suppresses MS-like symptoms in mice by expanding Akkermansia muciniphila. Cell Host Microbe 26, 779–794.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng Q., et al., Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem. Int. 129, 104468 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Ventura R. E., et al., Gut microbiome of treatment-naïve MS patients of different ethnicities early in disease course. Sci. Rep. 9, 16396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rothschild D., et al., Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Campbell J. H., et al., Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 6, 2033–2044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bubier J., et al., A microbe associated with sleep revealed by a novel systems genetic analysis of the microbiome in collaborative cross mice. J. Genet. 214, 719–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ubeda C., et al., Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209, 1445–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moon C., et al., Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature 521, 90–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tankou S. K., et al., Investigation of probiotics in multiple sclerosis. Mult. Scler. 24, 58–63 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Tankou S. K., et al., A probiotic modulates the microbiome and immunity in multiple sclerosis. Ann. Neurol. 83, 1147–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zelante T., et al., Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Cervantes-Barragan L., et al., Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357, 806–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rothhammer V., et al., Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilck N., et al., Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zmora N., et al., Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174, 1388–1405.e21 (2018). [DOI] [PubMed] [Google Scholar]
- 91.Suez J, et al., Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 174, 1406–1423.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 92.ClinicalTrials.gov , Fecal microbiota transplantation (FMT) in multiple sclerosis (National Library of Medicine, Bethesda, MD, 29 February 2000, Identifier NCT03975413 FMTFiMSJ) https://clinicaltrials.gov/ct2/show/NCT03975413. Accessed 14 July 2019.
- 93.ClinicalTrials.gov , Sleep disorders and gastroesophageal reflux disease (GERD) (National Library of Medicine, Bethesda, MD, 29 February 2000, Identifier NCT03797937 MBtIPiMSI) https://clinicaltrials.gov/ct2/show/NCT00287391. Accessed 14 July 2019.
- 94.ClinicalTrials.gov , Fecal microbiota transplantation (FMT) of FMP30 in relapsing-remitting multiple sclerosis (MS-BIOME) (National Library of Medicine, Bethesda, MD, 29 February 2000, Identifier NCT03594487 FMTFoFiR-R) https://clinicaltrials.gov/ct2/show/NCT03594487. Accessed 14 July 2019.
- 95.Krementsov D. N., Case L. K., Hickey W. F., Teuscher C., Exacerbation of autoimmune neuroinflammation by dietary sodium is genetically controlled and sex specific. FASEB J. 29, 3446–3457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lyons J. A., San M., Happ M. P., Cross A. H., B cells are critical to induction of experimental allergic encephalomyelitis by protein but not by a short encephalitogenic peptide. Eur. J. Immunol. 29, 3432–3439 (1999). [DOI] [PubMed] [Google Scholar]
- 97.Cox L. M., et al., Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Haarman M., Knol J., Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 72, 2359–2365 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gehrke S., et al., Red blood cell metabolic responses to torpor and arousal in the hibernator arctic ground squirrel. J. Proteome Res. 18, 1827–1841 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chong J., et al., MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Montgomery T., et al., 16S sequencing data of fecal DNA from diverse mouse strains. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra. Deposited 21 November 2019.
- 102.Miyauchi E., et al., Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 585, 102–106 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Johanson D. M. II, et al., Experimental autoimmune encephalomyelitis is associated with changes of the microbiota composition in the gastrointestinal tract. Sci. Rep. 10, 15183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S sequencing data have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject ID PRJNA590851) (101).