Significance
The tumor-associated macrophage (TAM) phenotype is continuously modulated during tumor progression to facilitate immune escape. Yet, how gene expression programs control TAM phenotypes during this process is largely unknown. Here we show that, during acquisition of a protumor phenotype, gene expression in TAMs is predominantly modulated via selective changes in mRNA translation rather than changes in mRNA abundance. Detailed studies pinpointed augmented activity of MNK2, which phosphorylates eIF4E and thereby modulates mRNA translation, as required for the antiinflammatory macrophage phenotype. Accordingly, suppression of MNK2 reeducated antiinflammatory macrophages toward a proinflammatory phenotype with the ability to activate CD8+ T cells.
Keywords: mRNA translation, tumor-associated macrophages, MNK2, T cell activation, eIF4E
Abstract
Tumor-associated macrophages (TAMs) continuously fine tune their immune modulatory properties, but how gene expression programs coordinate this immune cell plasticity is largely unknown. Selective mRNA translation, controlled by MNK1/MNK2 and mTOR pathways impinging on eIF4E, facilitates reshaping of proteomes without changes in abundance of corresponding mRNAs. Using polysome profiling developed for small samples we show that, during tumor growth, gene expression in TAMs is predominately modulated via mRNA-selective changes in translational efficiencies. These alterations in gene expression paralleled accumulation of antiinflammatory macrophages with augmented phosphorylation of eIF4E, a target of the MNK1 and MNK2 kinases, known to selectively modulate mRNA translation. Furthermore, suppression of the MNK2, but not the mTOR signaling pathway, reprogrammed antiinflammatory macrophages toward a proinflammatory phenotype with the ability to activate CD8+ T cells. Thus, selective changes of mRNA translation depending on MNK2 signaling represents a key node regulating macrophage antiinflammatory functions.
Macrophages (Mφs) are sentinel and effector cells with an inherent need to efficiently adapt to changing environmental cues to maintain homeostasis (1). TAMs accumulating during tumor progression arise from extravasating bone marrow-derived monocytes (2, 3). Mφs can acquire a spectrum of proinflammatory (also referred to as “M1”-like) to antiinflammatory (also referred to as “M2”-like) phenotypes (4, 5). In tumors, proinflammatory TAMs initially restrain tumor growth by activating cytotoxic lymphocytes (6–8). However, during tumor progression, TAMs are skewed toward an antiinflammatory phenotype promoting tumor malignancy by suppressing cytotoxic lymphocytes and stimulating blood vessel formation (9, 10). Accumulation of largely antiinflammatory TAMs therefore correlates with poor prognosis in most solid cancers (11, 12). Yet, gene expression programs governing TAM phenotypes are largely unknown.
Gene expression can be regulated at multiple levels, including transcription, mRNA stability and mRNA translation (hereafter referred to as translation). Among these, selective modulation of translation has emerged as a central node controlling gene expression in, e.g., immune cells (13) and cancer-encompassing both cancer cells (14) and the noncancerous stroma during cancer initiation (15). Translation is commonly separated into four phases: initiation, elongation, termination, and recycling (16). Although translation can be regulated by changes in elongation rate (17), most described modulations of translation are mediated via altered initiation (13). The eukaryotic initiation factor (eIF) 4F (eIF4F) complex, whose activity is primarily regulated via its 5′ mRNA cap-binding subunit eIF4E, is a central node controlling initiation of translation (18). The mTOR pathway regulates eIF4F complex formation by phosphorylating the inhibitory eIF4E binding proteins (4E-BPs) which are then released from eIF4E, thereby allowing eIF4E to associate with eIF4A and eIF4G to form the eIF4F complex. In addition, the activity of the eIF4F complex is modulated via mitogen-activated protein kinase (MAPK) interacting protein kinase (MNK, [also referred to as MKNK]) 1- and 2-dependent phosphorylation of eIF4E, which can drive tumor malignancy (19) and metastatic dissemination (20–22). Changes in eIF4F complex formation and phosphorylation of eIF4E modulate the proportion of efficiently translated mRNA transcribed from subsets of genes and thereby selectively alter protein levels without corresponding changes in mRNA levels (13). Therefore, alterations in translation initiation facilitates rapid adaptation of proteomes to environmental changes, which is believed to be required for optimal immune cell function and homeostasis in vivo (13). In proinflammatory Mφs, selective changes in mRNA translation following lipopolysaccharide (LPS) (23–25) or IFN (26) stimulation in vitro have been attributed to mTOR- and MNK1/2-dependent signaling. We examined how mRNA abundance and translation are modulated in TAMs during tumor growth. Unexpectedly, TAM gene expression was predominantly governed by selective changes in mRNA translation rather than abundance. Such alterations in gene expression associated with increased phosphorylation of eIF4E. Consistently, while MNK2 controls antiinflammatory Mφ functions, MNK1/2 suppression did not affect the proinflammatory Mφ phenotype. In contrast, suppression of mTOR signaling did not affect antiinflammatory Mφ functions. Thus, targeting MNK2 may be a novel avenue to dampen Mφ antiinflammatory functions, which in turn will augment CD8+ T cell activity.
Results
TAMs Predominantly Modulate Gene Expression via Changes in Translational Efficiencies.
To assess to what extent mRNA translation is modulated in macrophages in vivo, we selected the mouse mammary tumor virus-polyoma middle T-antigen (MMTV-PyMT) model (27) of tumor growth which is compatible with our newly developed technique to study transcriptome-wide translational efficiencies in limited numbers of cells (28). MMTV-PyMT mice (29, 30) develop mammary tumors resembling a mix of human ERlowPR−HER2+ and ER−PR−HER2− breast tumors (29). Because MMTV-PyMT mice develop multiple tumors varying in size, we used a cell line (PeRo-Bas1) derived from the MMTV-PyMT model which, following fat-pad injection in FVB mice, forms a single mammary tumor (31). Notably, even though PeRo-Bas1 is a tumor cell line, it still gives rise to heterogeneous ERlowPR−HER2+ and ER−PR−HER2− tumors (31).
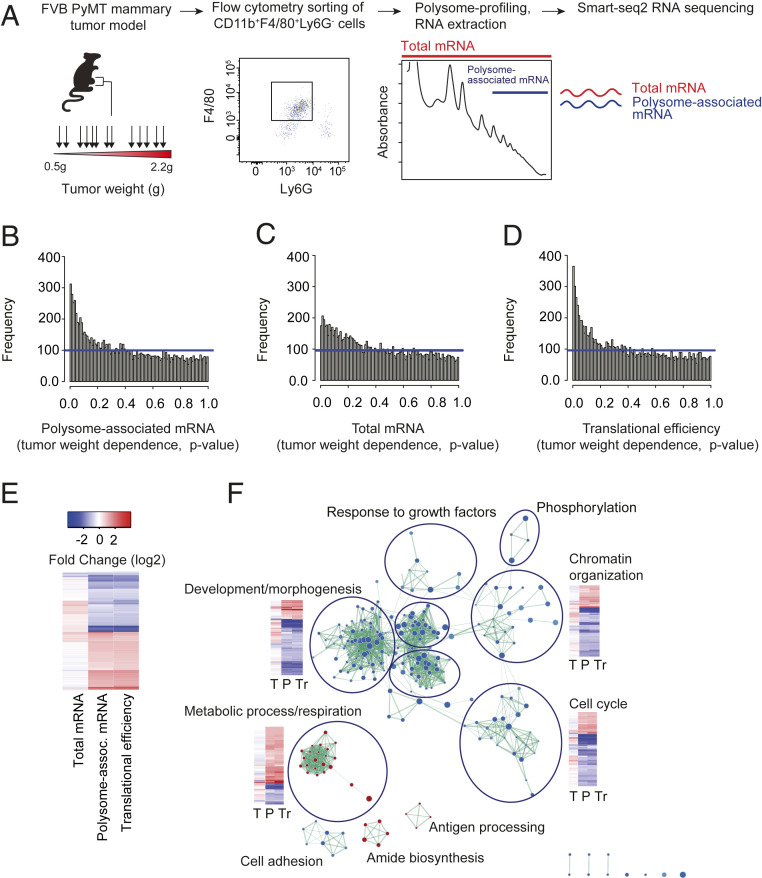
To capture gene expression changes in TAMs occurring during tumor growth, we sorted CD11b+F4/80+Ly6G− Mφs from 13 individual MMTV-PyMT tumors, weighing between 500 mg and 2,200 mg, and subjected these to polysome profiling (Fig. 1A) (28). Notably, polysome profiling was applied as this method appears to be less biased when assessing changes in translational efficiencies as compared to ribosome profiling (32, 33). During polysome profiling, efficiently translated polysome-associated mRNA (herein mRNA associated with three or more ribosomes) is quantified using RNA sequencing. As levels of polysome-associated mRNA are also affected by mechanisms modulating total mRNA levels even in the absence of changes in translational efficiencies, total mRNA is isolated and quantified in parallel. To identify bona fide alterations in translational efficiencies, changes in polysome-associated mRNA which are not paralleled by changes in total mRNA levels can then be identified using the anota2seq algorithm (34). Accordingly, polysome profiling enables quantification of changes in translational efficiencies and total mRNA levels in parallel. To analyze the resulting data, we used a variant of the anota2seq algorithm where changes in gene expression in TAMs (polysome-associated mRNA, total mRNA, or translational efficiencies) depending on tumor size were identified (Fig. 1 B–D and Dataset S1). This revealed an abundance of transcripts whose translational efficiencies associate with tumor size (Fig. 1 D and E). Such tumor-weight-associated expression was also observed for total mRNA (Fig. 1C), but appeared to be less frequent compared to modulation of mRNA translation (as judged by the number of genes with low P values and false discovery rates [FDRs]; Fig. 1 B–D and SI Appendix, Fig. S1). Therefore, during tumor growth, TAM gene expression is profoundly modulated via changes in translational efficiencies.
Fig. 1.
Abundant changes in translational efficiencies in TAMs during tumor growth. (A) Polysome profiling in macrophages sorted from tumors of different sizes. MMTV-PyMT tumor cells (PeRo-Bas1) were injected in the mammary fat pad of FVB mice. Tumors were harvested at weights ranging from 0.5 to 2.2 g (n = 13). F4/80+ macrophages were flow sorted and subjected to polysome profiling (total mRNA and polysome-associated mRNA [associated with three or more ribosomes] was isolated in each tumor and used as input for RNA sequencing). (B–D) Histograms of P values for tumor-weight-dependent changes in polysome-associated mRNA (B), total mRNA (C), or translational efficiencies (D; i.e., after adjusting changes in levels of polysome-associated mRNA for corresponding changes in total mRNA using anota2seq analysis) in TAMs. Blue lines indicate uniform frequencies of P values expected by chance. Higher frequencies of low P values as compared to chance indicate transcriptome-wide changes in gene expression depending on tumor weight. (E) Heat map of tumor-weight-dependent fold changes of polysome-associated mRNA, total mRNA, and translational efficiencies (all genes with an FDR <0.25 for any of the analyses are included [Dataset S1]). (F) Network overview of gene ontology terms enriched among proteins encoded by mRNAs showing tumor-weight-dependent translation. Red and blue nodes indicate GO terms enriched among proteins encoded by translationally activated and suppressed transcripts, respectively. The thickness of edges is proportional to the number of shared proteins between nodes. Consensus annotations for clusters of nodes are indicated together with selected heat maps for included genes (tumor-weight-dependent fold changes for total mRNA [“T”], polysome-associated mRNA [“P”], and translational efficiencies [“Tr”] [similar to E]).
Tumor-Growth-Associated Changes in Translation Target Key TAM Functions.
To assess the potential phenotypic impact of these alterations in translational efficiencies, we used gene set enrichment analysis to identify enrichment of cellular functions among all such encoded proteins (Fig. 1F and Dataset S2). This analysis revealed that proteins encoded by transcripts whose translational efficiency was positively associated with tumor growth were involved in several metabolic processes, including glutathione metabolism (e.g., Gclm, Gpx3, Gstm1, Gsto1, Gstp1, Gstp2, Gstt3, Hagh, and Sod2), ATP metabolism (e.g., Gapdh, Galt, Cyc1, Mpi, and Msh2), and mitochondrial activity (e.g., Acaa2, Timm17a, Mpc1, Aldh2, Fars2, and Atp5h, together with 16 other mitochondrial ribosome proteins). In contrast, cell cycle functions were enriched among proteins encoded by translationally suppressed mRNAs (Fig. 1F). Manual inspection, however, revealed that mRNAs encoding proteins with cell cycle functions, as well as those with apoptosis-related functions, were regulated in a manner often corresponding to enhanced proliferation or inhibited apoptosis during tumor growth. For example, during tumor growth, cell cycle drivers, including Cdk4, Cdk19, Cdc7, and Cdk12, were translationally activated; apoptosis inducers Casp3 and Bcl2l13 were translationally suppressed; and antiapoptotic factors, including Dad1 (defender against cell death 1), Triap1 (TP53 regulated inhibitor of apoptosis 1), and Aatf (apoptosis antagonizing transcription factor) were translationally activated (Dataset S1). Finally, transcripts encoding proteins regulating immune responses were enriched among mRNAs translationally suppressed during tumor growth and included Grab2-related protein (Grap), which was initially identified as a Grab mediator involved in transforming growth factor (TGFβ) release (35, 36). Thus, during tumor growth, translation of transcripts encoding proteins participating in functions related to proliferation, survival, and metabolism appears particularly modulated in TAMs.
Expression of Proteins Encoded by Transcripts Whose Translation Is Bolstered in TAMs during Tumor Growth Parallel Expansion of Antiinflammatory TAMs.
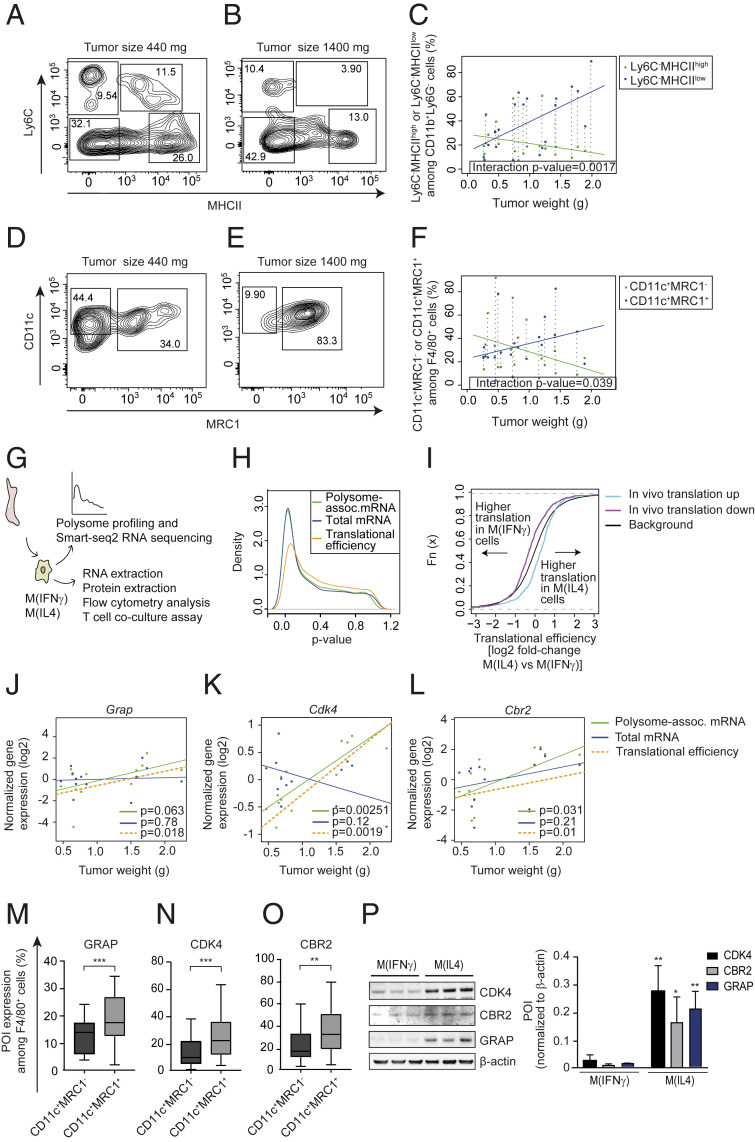
The studies above assessed changes in gene expression in TAMs during tumor growth (Fig. 1) when the TAM population is expected to shift from a predominantly proinflammatory phenotype toward the antiinflammatory phenotype. Accordingly, observed gene expression differences could be caused by this shift and/or reflect tumor-growth-associated modulation of translation in pro- and/or antiinflammatory TAMs. Flow cytometry for established pro- and antiinflammatory TAM markers (37, 38) across a range of MMTV-PyMT tumor sizes revealed a weight-associated decrease of proinflammatory TAMs (CD11b+Ly6G−Ly6C−MHCIIhigh in Fig. 2 A–C or CD11b+F4/80+CD11c+MRC1− in Fig. 2 D–F) with a concomitant increase of antiinflammatory TAMs (CD11b+Ly6G−Ly6C−MHCIIlow in Fig. 2 A–C or CD11b+F4/80+CD11c+MRC1+ in Fig. 2 D–F). Next, to assess whether observed changes in translational efficiencies (Fig. 1D) depend on the shift from proinflammatory to antiinflammatory TAMs, we performed polysome profiling using bone marrow-derived Mφs (BMDMs) polarized into a pro- and antiinflammatory phenotype using IFNγ or IL4, respectively [denoted M(IFNγ) and M(IL4)] (Fig. 2G). Analysis using anota2seq (34) to identify changes in translational efficiencies and total mRNA expression between M(IFNγ) and M(IL4) revealed an abundance of changes in total mRNA levels together with relatively fewer changes in translational efficiencies (Fig. 2H). This is in stark contrast to the in vivo studies above (Fig. 1) where changes in translational efficiencies appeared to be the predominant mechanism for modulation of gene expression. However, the discrepancy is consistent with mRNA translation as commonly modulated by factors in the tumor microenvironment (13, 39), which are absent under in vitro conditions. Nonetheless, we assessed whether transcripts whose translational efficiencies were modulated during tumor growth in vivo showed polarization-dependent differences in translational efficiencies in vitro. Indeed, mRNAs whose translational efficiencies increased in TAMs during tumor growth were more efficiently translated in M(IL4) as compared to M(IFNγ) and, conversely, transcripts whose translational efficiencies were suppressed during tumor growth in vivo were more efficiently translated in M(IFNγ) as compared to M(IL4) (Fig. 2I).
Fig. 2.
Tumor-weight-associated changes in translational efficiencies in TAMs are consistent with a shift from pro- to antiinflammatory TAMs. (A–F) Percentages of proinflammatory (CD11b+Ly6G−Ly6ClowMHCIIhigh in A and B and CD11b+F4/80+CD11c+MRC1− in D and E) and antiinflammatory (CD11b+Ly6G−Ly6ClowMHCIIlow in A and B and CD11b+F4/80+CD11c+MRC1+ in D and E) macrophages from PyMT tumors of different sizes (quantified by flow cytometry; n = 21). Representative plots of macrophages from a small (440 mg; A and D) and a large (1,440 mg; B and E) tumor together with the percentage of pro- and antiinflammatory macrophages as a function of tumor size across all analyzed tumors in C and F are shown. In C and F, the P value for an interaction between tumor weight and macrophage subtype is indicated together with the linear relationships between tumor size and percentage of pro- or antiinflammatory macrophages. Dotted vertical lines connect percentages of macrophage subtypes from the same tumor. (G) BMDMs were isolated and polarized into M(IFNγ) and M(IL4) in vitro followed by polysome profiling (n = 4). (H) Distributions (estimated using kernel densities) of P values for polarization-dependent changes in polysome-associated mRNA, total mRNA, and translational efficiencies (i.e., after anota2seq analysis). (I) Cumulative distribution functions of polarization-dependent fold changes in translational efficiencies for transcripts whose translation was activated (“translation up”) or suppressed (“translation down”) in TAMs during tumor growth in vivo (i.e., from Fig. 1). Such transcript subsets were compared to background (i.e., not in subsets) using the Wilcoxon rank-sum test (P value <0.001 for both subsets). (J–L) Tumor-weight-dependent changes in total mRNA, polysome-associated mRNA, and translational efficiency for Grap (J), Cdk4 (K), and Cbr2 (L). Points indicate normalized expression levels for polysome-associated mRNA (green) and total mRNA (blue) obtained from individual tumors (i.e., from Fig. 1) with their corresponding relationships to tumor weight from linear regressions. Shown is also the calculated linear relationship between translational efficiency (i.e., polysome-associated mRNA adjusted for total mRNA) and tumor weight (P values for weight-dependent expression are indicated). (M–O) PyMT tumor single cell suspensions were subjected to flow cytometry and the percentage of F4/80+CD11c+MRC1− and F4/80+CD11c+MRC1+ cells expressing GRAP (M), CDK4 (N), and CBR2 (O) proteins were quantified. Data are presented as mean ± SD. Student’s t test was used to compare expression between macrophage subtypes (n = 20, **P < 0.01, ***P < 0.001). (P) Expression of proteins of interest (POI) in whole cell extracts from M(IFNγ) and M(IL4) assessed by Western blotting. One out of three independent experiments is shown and the summary is presented as means + SD. Student’s t test was used to compare M(IFNγ) to M(IL4) for each POI (n = 3; *P < 0.05; **P < 0.01).
To further explore whether observed changes in translational efficiencies associated with the shift from a predominant proinflammatory to antiinflammatory TAM population, we focused on selected genes regulating metabolism (Cbr2), cell cycle (Cdk4), or cytokine production (Grap) whose translational efficiencies were modulated in a tumor-growth-dependent manner (Fig. 2 J–L; data from in vivo polysome profiling in TAMs; Fig. 1 and Dataset S1). Indeed, flow cytometry analysis of tumors ranging from 300 to 1,900 mg revealed a larger proportion of antiinflammatory (CD11b+Cd11c+MRC1+F4/80+Ly6G−) TAMs expressing CBR2, GRAP, and CDK4 as compared to proinflammatory (CD11b+CD11c+MRC1−F4/80+Ly6G−) TAMs (Fig. 2 M–O). Furthermore, these in vivo findings were supported by in vitro studies of CBR2, GRAP, and CDK4 proteins quantified by Western blotting in M(IFNγ) and M(IL4) (Fig. 2P). Altogether these data suggest changes in translational efficiencies in TAMs during tumor growth in vivo depending, at least partially, on the shift from a proinflammatory to an antiinflammatory TAM phenotype.
Protein Expression in M(IL4) Depends on the MNK/eIF4E Axis.
Previous studies in immune cells revealed selective translation governed by the eIF4F complex (13). As described above, the activity of the eIF4F complex can be modulated by the mTOR pathway via phosphorylation of 4E-BPs, leading to their inactivation, and/or via MNK1/2-dependent phosphorylation of eIF4E. We therefore assessed whether the activity of these two pathways differs between M(IFNγ) and M(IL4). Indeed, whereas 4E-BP1 (S65) was not differentially phosphorylated between M(IFNγ) and M(IL4) (Fig. 3A), eIF4E (S209) phosphorylation was augmented in M(IL4) as compared to M(IFNγ) in vitro (Fig. 3B). Consistently, flow cytometry analysis of TAMs from mammary tumors revealed increased eIF4E phosphorylation in antiinflammatory (CD11c+MRC1+F4/80+) as compared to proinflammatory TAMs (CD11c+MRC1−F4/80+) (Fig. 3C). This suggests that observed selective changes in translational efficiencies in M(IL4) as compared to M(IFNγ) may depend on the MNK/eIF4E axis. To assess this, we used the MNK1/2 inhibitor cercosporamide (MNKi). While MNKi treatment of M(IFNγ) did not affect phosphorylation of eIF4E, M(IL4) showed a strong reduction in eIF4E phosphorylation to a level comparable to M(IFNγ) (Fig. 3D). Accordingly, we focused on the role of the MNK/eIF4E axis in M(IL4). Treatment of M(IL4) with MNKi suppressed CDK4 and GRAP protein expression in a manner paralleling suppressed phosphorylation of eIF4E (Fig. 3E) while the CBR2 protein level was essentially unaffected (Fig. 3E). Consistent with modulation of translation and not mRNA abundance depending on MNK1/2 in M(IL4), Cdk4 and Grap mRNA levels were unchanged during the same treatment (Fig. 3F). In contrast, Cbr2 mRNA expression was reduced following treatment with MNKi despite unaltered protein levels (a mode of regulation recently described in mammals) (40, 41) suggesting that changes in Cbr2 mRNA translation does not depend on the MNK/eIF4E axis or that its regulation differs between in vivo and in vitro conditions. In summary, altered activation of the MNK/eIF4E axis at least partly underlies differences in gene expression between Mφs with antiinflammatory vs. proinflammatory phenotypes.
Fig. 3.
Altered activity of the MNK/eIF4E axis partly underlies differences in protein expression observed between M(IFNγ) and M(IL4). (A) Levels of 4E-BP1 protein and phosphorylated (S65) 4E-BP1 (p-4E-BP1) in whole cell extracts from M(IFNγ) or M(IL4) assessed by Western blotting. One representative membrane is shown out of three independent experiments and quantification is presented as mean + SD. Student’s t test was used to compare M(IFNγ) to M(IL4) (n = 4). (B) Levels of eIF4E protein and phosphorylated eIF4E (p-eIF4E, S209) in whole cell extracts from M(IFNγ) and M(IL4) assessed by Western blotting. β-Actin was used as a loading control. One representative membrane is shown out of three independent experiments and quantification is presented as mean + SD. Student’s t test was used to compare M(IFNγ) to M(IL4) (n = 3, ***P < 0.001). (C) Tumor single cell suspensions were subjected to flow cytometry and the percentage of F4/80+CD11c+MRC1+ (antiinflammatory) and F4/80+CD11c+MRC1− (proinflammatory) TAMs expressing p-eIF4E was quantified. Shown is a representative histogram. (D) Levels of indicated proteins in whole cell extracts from IFNγ- and IL4-treated BMDMs with or without MNKi treatment assessed by Western blotting. β-Actin was used as a loading control. One representative membrane is shown out of three independent experiments and quantifications are presented as mean + SD. One-way ANOVA was used to compare indicated groups (n = 3, ***P < 0.001). (E) Western blotting for indicated proteins in whole cell extracts from M(IL4) treated with MNKi at indicated concentrations. Fold changes refer to protein levels in treated cells compared to DMSO control, normalized to loading control (β-actin). (F) Total mRNA levels (RT-qPCR) in M(IL4) treated with MNKi at indicated concentrations. Genes of interest were normalized to β-actin using the ΔΔCt method. Data from three independent experiments are presented as mean + SD. One-way ANOVA was used to compare treatments to control (n = 4, *P < 0.05, **P < 0.01). NS, not significant.
MNK2 Regulates the Mφ Antiinflammatory Phenotype.
The above findings suggest that the MNK/eIF4E axis may govern Mφ phenotypes by selectively modulating translational efficiencies. To assess this, we determined the MNKi sensitivity of cytokines and surface markers for anti- or proinflammatory Mφs. Flow cytometry analysis revealed that MNKi-treated M(IL4) reduced cell surface expression of the antiinflammatory marker MRC1 while increasing expression of antigen-presenting, T cell costimulatory, and proinflammatory surface markers such as MHCII, CD86, CD11c, and CD25 compared to control cells (Fig. 4A and SI Appendix, Fig. S2 A–E). In contrast, MNKi treatment of M(IFNγ) did not affect expression of the aforementioned surface proteins (Fig. 4A). Moreover, MNKi treatment of M(IL4) reduced their expression of mRNAs encoding proteins associating with an antiinflammatory phenotype (Arg1, Fizz1, Ym1, Mrc1, and Il10) while expression of mRNAs encoding proinflammatory Th1 cytokines was increased (Cxcl9, -10, and -11, Il1a, and Il12a; Fig. 4B). However, not all Th1 cytokines were increased in M(IL4) upon MNKi treatment as mRNAs encoding Ifna and Ifnb were unchanged and Il6 was reduced (Fig. 4B). In line with the flow cytometry data, MNKi treatment of M(IFNγ) merely increased expression of mRNAs encoding two Th1 cytokines (Il1a and Il6) (Fig. 4B). Next, we explored whether a reduction in mTOR activity would have a similar effect on the macrophage phenotype as inhibition of eIF4E phosphorylation. To this end, we treated M(IFNγ) and M(IL4) with the active site mTOR inhibitor torin1 (42). As expected, treatment with torin1 led to complete dephosphorylation of 4E-BP1 (SI Appendix, Fig. S3A). Moreover, while MNKi did not affect global protein synthesis, torin1 treatment led to a ∼20% reduction as measured by puromycin incorporation in M(IL4) (see SI Appendix, Fig. S3 B–E; MCF7 cells were used as a positive control). Strikingly, although treatment of M(IL4) with torin1 reduced expression of mRNAs encoding antiinflammatory factors to a similar extent as MNKi, torin1 largely did not induce expression of mRNAs encoding Th1-related cytokines (except Ifna and Ifnb; Fig. 4B). In contrast, M(IFNγ) showed a torin1-dependent increase in levels of mRNAs encoding Th1 cytokines (except Il1α and Il6) (Fig. 4B). These findings show that the MNK and mTOR pathways distinctly modulate global protein synthesis and cytokine expression. To assess whether these differences affect the abilities of the inhibitors to modulate the antiinflammatory phenotype, M(IL4) were cultured with or without MNKi or torin1 and, following washout of inhibitors, these cells were cocultured with activated splenocyte-derived CD3+ T cells. Strikingly, M(IL4) subjected to MNKi but not torin1 treatment efficiently activated CD8+ T cells (measured by IFNγ expression) to a similar level as compared to M(IFNγ) (Fig. 4C). Of note, the increase in IFNγ expression in T cells following MNKi treatment was not observed in the absence of BMDMs (Fig. 4C).
Fig. 4.
Inhibition of the MNK/eIF4E axis reverses antiinflammatory macrophage functions in an Mnk2-dependent manner. (A) Expression of indicated cell surface markers in M(IL4) or M(IFNγ) cultured in presence or absence of MNKi as analyzed by flow cytometry. Shown are fold changes presented as mean + SD; treated vs. nontreated cells were compared using Student’s t test (n = 4; *P < 0.05, **P < 0.01). (B) Expression of mRNAs (RT-qPCR) encoding pro- (light blue Left side color bar) and antiinflammatory (dark red Left side color bar) cytokines in M(IL4) or M(IFNγ) treated with MNKi or torin1. Data were normalized to β-actin and presented as Student’s t test −log10 (P values) for the comparison to nontreated M(IL4) or M(IFNγ). Shades of red and blue indicate higher and lower expression, respectively, in inhibitor treated as compared to control cells (n = 3). (C) IFNγ expression evaluated by flow cytometry in CD8+ T cells with or without MNKi or torin1; CD8 T cells cocultured with control M(IFNγ) or M(IL4); and CD8+ T cells cocultured with MNKi- or torin1-pretreated M(IL4). Data are presented as mean ± SD. One-way ANOVA was used to compare indicated cell and treatment combinations (n = 3; ***P < 0.001, ****P < 0.0001). (D) Expression of Mnk2 mRNA in IL4-treated shMnk2- and shCTR-cells quantified by RT-qPCR (data are presented as mean + SD, n = 3; *P < 0.05). (E and F) Expression of indicated proteins in whole cell lysates from IL4-treated shMnk2- or shCTR cells cultured in presence or absence of MNKi as assessed by Western blotting. One representative Western blot out of three independent experiments is shown (E) together with quantification across the three independent experiments (F). One-way ANOVA was used to compare indicated cell and treatment combinations (n = 3; *P < 0.05, ***P < 0.001). (G) IFNγ expression in CD8+ T cells cocultured with IL4-treated control or MNKi-pretreated shMnk2- or shCTR as assessed by flow cytometry. A summary from three independent experiments is presented as mean ± SD. One-way ANOVA was used to compare indicated cell and treatment combinations (n = 6; *P < 0.05, ***P < 0.001). (H) CD8+ T cells expressing granzyme B or IFNγ from in vivo comingling experiments between 66cl4 cancer cells and shCTR or shMnk2 BMDMs. One-way ANOVA was used to compare indicated cell types (n = 10; ****P < 0.001). (I) CD8+ T cells expressing CD107a or IFNγ in 66cl4 tumors from bone marrow transplant experiments (KI or WT bone marrow to bone marrow-depleted WT animals). One-way ANOVA was used to compare KI to WT conditions (n = 7; *P < 0.05; **P < 0.01). NS, not significant.
As cercosporamide (MNKi) has also been described as a JAK3 inhibitor (19), we evaluated whether JAK3 inhibition could underlie these effects. Treatment of M(IL4) with MNKi only had a marginal effect on the downstream JAK3 target STAT6’s phosphorylation (SI Appendix, Fig. S4A). Moreover, suppression of JAK3 activity using the JAK inhibitor CP690550 (JAKi) affected neither Th1 cytokine expression in M(IL4) in a manner resembling that observed following MNKi treatment (SI Appendix, Fig. S4B) nor M(IL4)’s ability to suppress CD8+ T cell IFNγ expression (SI Appendix, Fig. S4C). Therefore, observed effects of MNKi on M(IL4) phenotypes seem independent of JAK3 inhibition.
Because cercosporamide (MNKi) has a lower IC50 for MNK2 as compared to MNK1 (19), we next evaluated whether effects of MNKi on M(IL4) depended on MNK2. To this end, BMDMs were transduced with lentiviral vectors encoding shRNAs targeting Mnk2 (shMnk2 BMDMs), resulting in ∼90% knockdown of the Mnk2 mRNA as compared to such cells transduced with a control vector (shCTR BMDMs; Fig. 4D). In IL4-polarized shMnk2 BMDMs, phosphorylation of eIF4E was reduced to a level comparable to IL4-polarized shCTR BMDMs treated with MNKi and was not further reduced upon MNKi treatment (Fig. 4 E and F). Moreover, IL4-polarized shMnk2 BMDMs activated CD8+ T cells to a similar extent as compared to MNKi-treated IL4-polarized shCTR BMDMs while MNKi had no additional impact on IL4-polarized shMnk2 BMDMs (Fig. 4G). To further determine the importance of MNK2 for the Mφ antiinflammatory phenotype we used the in vivo syngeneic 66cl4 mammary cancer cells and BMDMs comingling assay. In this assay, cancer cells and BMDMs were mixed in Matrigel and injected s.c. followed by harvest after 8 d. Flow cytometry analysis revealed that shMnk2 BMDMs dramatically enhanced activation of CD8+ T cells in vivo as visualized by increased levels of Granzyme B (which mediates the lytic activity of T cells) and IFNγ compared to controls (Fig. 4H).
To further establish whether the MNK/eIF4E pathway controls the Mφ phenotype in vivo, we used a transgenic mouse model wherein the MNK1/MNK2-targeted phosphorylation site in eIF4E was altered from a serine to an alanine to generate the eIF4ES209A/S209A knockin mouse (hereafter referred to as KI) (43). Bone marrow from KI or wild-type (WT) mice were transplanted into lethally irradiated WT mice (generating chimeric KI→WT or WT→WT mice, respectively). The 66cl4 mammary tumor cells were then injected into the mammary gland of these chimeric mice. Consistent with a role of the MNK/eIF4E axis in controlling the Mφs antiinflammatory phenotype, flow cytometry analysis revealed that CD8+ T cells in KI→WT tumors expressed higher levels of IFNγ and CD107a (a marker for T cell lytic activity) as compared to WT→WT tumors (Fig. 4I). Notably, the 66cl4 tumor immune landscape is dominated by TAMs (∼63% F4/80+ among CD45+ cells), while granulocytes (∼10% Ly6G+CD11b+ among CD45+ cells), monocytes (∼15% Ly6C+ CD11b+ among CD45+ cells), and lymphocytes (∼7% CD4+ T, ∼4% CD8+ T cells and ∼1% CD49b+ natural killer [NK] cells among CD45+ cells) constitute minor parts (SI Appendix, Fig. S5). Therefore, although eIF4ES209A/S209A may affect other myeloid cells (T cells were not affected by MNKi) (Fig. 4C), these data support the MNK2/eIF4E axis controlling the TAM proinflammatory function in vivo resulting in CD8+ T cell activation.
Discussion
During the last decade, antiinflammatory TAMs have emerged as supporters of tumor development, in part due to their role in suppressing cytotoxic lymphocytes while promoting expansion of antiinflammatory CD4+ T regulatory cells (44). Reciprocally, cytotoxic lymphocytes secrete IFNγ skewing TAMs toward a proinflammatory phenotype (3) while antiinflammatory CD4+ T regulatory cells secrete IL4 and thereby promote a protumor TAM phenotype (45). However, the knowledge of how gene expression programs steer and maintain various TAM phenotypes throughout tumor progression is incomplete. Transcript-selective regulation of translation has previously been linked to proinflammatory Mφ properties via IFNγ (26) and LPS (23) modulation of mTORC1 and/or MNK1/eIF4E signaling pathways. These studies identified LPS-dependent increased translation of mRNAs encoding antiinflammatory feedback inhibitors (24), IFN regulatory factor 8 (IRF8) (23), and Th1-related cytokines (25). In contrast, IFNγ caused suppressed translation of inflammatory repressors (26). To our knowledge, there are no studies of mRNA translation in antiinflammatory Mφs, although an mTORC2-dependent link between IL4 and glucose metabolism essential to maintaining the antiinflammatory phenotype has been described (46).
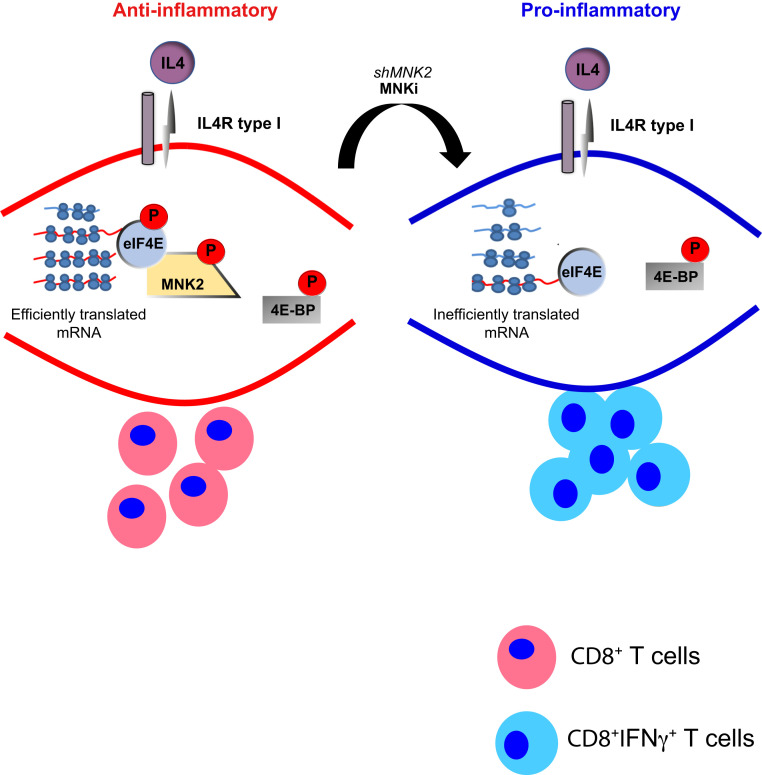
Here we provide insights into how mRNA translation is modulated in TAMs during tumor growth. For these studies we used an ex vivo approach in combination with recently developed polysome profiling for small samples (28). It is possible that isolated ex vivo cells do not completely maintain the in vivo status of gene expression programs. Nevertheless, these studies led to identification of MNK2 as key for maintaining the antiinflammatory Mφ phenotype. In vitro, suppression of the MNK2/eIF4E axis overrides M(IL4)-acquired antiinflammatory properties resulting in CD8+ T cell activation (Fig. 5). Similarly, in vivo, TAMs with suppressed MNK2 levels or expressing nonphosphorylated eIF4E boosted CD8+ T cell activity (Fig. 5). In contrast, mTOR inhibition had surprisingly limited effect on M(IL4) expression of mRNAs encoding Th1-related proteins despite leading to the previously described augmented M(IFNγ) proinflammatory properties (26). Moreover, although torin1 reduced levels of mRNAs encoding antiinflammatory genes (Il10, Arg1, Mrc1, Fizz1, and Ym1) it still potentiated M(IL4) suppression of CD8+ T cell activity. Thus, the effect on T cell activity is difficult to predict from studies of single or sets of cytokines. Taken together our data support the MNK2/eIF4E axis as central for the proinflammatory phenotype downstream of IL4 and possibly other not yet identified factors.
Fig. 5.
Schematic of MNK2-dependent control of the antiinflammatory Mφs phenotype.
Modulation of the antiinflammatory TAM phenotype via changes in translational efficiencies also included augmented proproliferative and metabolic programs which are also modulated in tumor cells during their adaptation to low oxygen and insufficient nutrients (47). Furthermore, these cellular functions are consistent with recent findings indicating that TAM metabolism (48) and selective proliferation of TAM subsets (8, 49) may determine the phenotype of the TAM population during tumor growth. Thus, mRNA translation possibly underlies multiple cellular functions which contribute to the expansion of a procancer TAM population. Accordingly, as MNK1/2 activity and eIF4E phosphorylation are dispensable for normal physiological development, our study suggests that targeting MNK2 may fine tune Mφs into a T cell activating phenotype.
Methods
Cell Culture.
Tumor cell lines.
The mouse mammary cell line (PeRo-Bas1) derived from MMTV-PyMT mice (31) and MCF7 cells (ATCC) were passaged in Dulbecco′s Modified Eagle′s Medium supplemented with l-glutamine, penicillin/streptomycin, and 10% fetal bovine serum (FBS) (Gibco/Life Technologies) at 37 °C and 5% CO2. The 66cl4 mouse mammary carcinoma cells were purchased from the Karmanos Cancer Institute at Wayne State University and passaged in RPMI-1640 (Gibco/Life Technologies) supplemented with l-glutamine, penicillin/streptomycin, and 10% FBS.
BMDMs.
Bone marrow was extracted from naïve female FVB mice, differentiated into BMDMs as previously described (8) and polarized toward a proinflammatory phenotype by treating cells with IFNγ (200 units/mL, Peprotech) for 4 h (polysome-profiling experiments) or 16 h (inhibitor experiments), or to an antiinflammatory phenotype by treating cells with IL4 (40 ng/mL, Peprotech) for 4 or 16 h. Polarized M(IFNγ) or M(IL4) were treated with 10 µM cercosporamide (MNKi; Tocris), 5 µM CP690550 citrate (JAKi; Tocris) or vehicle (i.e., cells in the presence of ∼0.1% dimethyl sulfoxide [DMSO]) for 24 h; or 250 nM torin1 (Tocris) for 4 h.
CD3+ T cells.
CD3+ T cells were purified using the EasySep mouse T cell isolation kit (Stem Cell Technologies) from spleens of naïve/tumor bearing FVB mice according to the manufacturer’s instructions. Isolated cells were cultured for 48 h in the presence of 500 ng/mL anti-CD3 (Thermo Fisher Scientific, clone 145-2C11), 20 ng/mL rmIL2 (Peprotech), and 50 μM β-mercaptoethanol (Sigma).
BMDMs and T cell coculture.
BMDMs were polarized to M(IFNγ) or M(IL4) as described above and treated with MNKi, JAKi, torin1, or vehicle (i.e., cells in the presence of ∼0.1% DMSO) in polarizing media for 24 h or torin1 for 4 h. After the indicated treatment time, media containing the inhibitor was removed and macrophages were washed three times with phosphate buffered saline (PBS). CD3+ T cells were then added at a 1:1 ratio and cocultured for 24 h. Activation of CD8+ T cells was assessed by flow cytometry analysis of intracellular IFNγ (see below).
shRNA-Mediated Mnk2 Knockdown.
We cloned the U6 promoter sequence followed by either a short hairpin RNA targeting mouse Mnk2 (clone ID: NM_021462.2-407s1c1; shMnk2) or a nontargeting control (SHC016 Sigma Aldrich; shCTR) using the XhoI restriction site of a lentiviral vector also containing GFP transcribed from the human PGK promoter. Vesicular stomatitis virus (VSV)-pseudotyped third-generation lentiviral vectors were produced by transient transfection of 293T cells, concentrated, and titered as previously described (50).
Animals and Tumor Models.
Bone marrow transplant experiment.
The eIF4ES209A/S209A phospho-eIF4E-deficient animal model was a gift from Nahum Sonenberg, McGill University, Montreal, Canada and has been previously described (43). Bone marrow donor WT and eIF4ES209A/S209A female BALB/c mice were bred in house and were 8 to 10 wk old upon bone marrow harvest. Six-week-old recipient WT animals (purchased from Charles River) were lethally irradiated with two doses of whole-body gamma radiation (4 Gy per dose) administered 6 h apart. Bone marrow from donor mice was collected by flushing the femur and tibia bones as described (8) and 107 cells were transplanted into recipient animals via retro-orbital venous sinus injection within 2 h after a second radiation treatment.
Mammary fat-pad injection.
PeRo-Bas1 (2 106) cells in 50 µL PBS were injected into the fourth inguinal mammary gland of anesthetized FVB mice (5 to 7 wk old), while 66cl4 (2 105) cells in 50 µL PBS were injected into the fourth inguinal mammary gland of anesthetized BALB/c mice 6 wk after the irradiation. Tumor growth was monitored externally, by caliper measurement, and the tumor volume was estimated using the following equation: V = 4/3π × (d/2)2 × D/2, where d is the minor tumor axis and D is the major tumor axis. Mice with tumors of different sizes were killed by cervical dislocation and the excised tumor was weighed prior to dissociation. Animals were purchased from Charles River Laboratory. These studies were approved by the Swedish Board of Agriculture (N91/15) and by the McGill University Animal Care and Use Committee (2011-6009).
Tumor-BMDM comingling assay.
shMNK2 or shCTR vector-transduced BMDMs (106 cells; of BALB/c origin) were embedded in 200 μL of growth factor-reduced Matrigel (BD Bioscience) together with 2 × 105 66cl4 tumor cells. Matrigel containing cancer cells and BMDMs were then injected s.c. in the flank of 8-wk-old female BALB/c mice and harvested 8 d postinjection.
Tumor dissociation.
Tumors or Matrigel plugs were dissociated into a single-cell solution by incubating with RPMI (Gibco, Life Technologies) containing 5% fetal bovine serum (Gibco), 2 mg/mL dispase (Sigma), 0.1 mg/mL DNaseI (Sigma), and 0.2 mg/mL Collagenase IV (Life Technologies). Cells were then passed through a 22-G syringe needle and filtering using 70-µM cell strainers (Corning) and further processed by flow cytometry.
Quantitative PCR.
Cultured BMDMs were collected in RLT buffer and homogenized using a syringe with a 20-G needle. RNA was extracted using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions and reverse transcribed with the QuantiTect Reverse Transcription kit according to the manufacturer’s instructions (Qiagen). Genomic DNA was eliminated using gDNA Wipeout Buffer (Qiagen) for 2 min at 42 °C. qPCR was performed using either TaqMan Universal MasterMix II and TaqMan gene expression assays (Applied Biosystems) or SYBR Green PCR Master Mix (Applied Biosystems) in a total volume of 10 μL. The polymerase was activated at 95 °C for 10 min and the PCR was performed for 40 cycles (95 °C for 15 s and 60 °C for 60 s). TaqMan probes are listed in SI Appendix, Table S1. Primers are listed in SI Appendix, Table S2. Gene expression was normalized to levels of β-actin using the ΔΔCt method.
Flow Cytometry.
Single cell suspensions were stained with primary antibodies (SI Appendix, Table S3A). For intracellular staining, the single cell suspensions were fixed and permeabilized with Cytoperm/Cytofix reagents (BD Biosciences) according to the manufacturer’s instructions. Following incubation with primary antibodies for intracellular proteins, cells were incubated with the appropriate secondary antibody (Invitrogen). All panels included anti-CD16/32 mAb (BioLegend) to reduce nonspecific binding. Cell viability was verified using 7AAD or the Live/Dead Fixable dead cell stain (Life Technologies). Samples were acquired with LSRII or Novocyte (BD Biosciences) and analyzed with FlowJo software (Tree Star).
Cell Sorting and Polysome Profiling.
Polysome profiling was performed as previously described (32) using macrophages isolated from 13 tumors ranging in size from 500 mg to 2,200 mg and M(IFNγ) and M(IL4) (n = 4). M(IFNγ) and M(IL4) were harvested by scraping while sorting Mφs from tumors, performed on a FACSJazz (BD Biosciences) flow sorter and gating the CD11b+, F4/80+, and Ly6G− cell population on live (7AAD−) cells. Sorted cells were kept in ice-cold PBS supplemented with 2% FBS and 100 μg/mL cycloheximide. For sorted TAMs, a fraction (∼10%) of the obtained cell population was lysed in TRIzol reagent (Thermo Fisher Scientific) and used for RNA sequencing with total RNA as input. Remaining cells were lysed in a hypotonic lysis buffer, as described (32). For BMDMs, the cytoplasmic extract was collected by centrifugation and 10% of the lysate was used to purify RNA (TRIzol) for RNA sequencing with total RNA as input. Lysates were then applied onto a 5 to 50% (wt/vol) sucrose gradient, fractionated, and fractions corresponding to mRNA associated with three or more ribosomes were collected in TRIzol reagent for subsequent RNA extraction. Such pooled samples were used for RNA sequencing of efficiently translated polysome-associated mRNA. RNA quality was assessed using the RNA 6000 Nano kit (Bioanalyzer, Agilent).
Western Blot.
Cells were lysed in Halt RIPA lysis buffer (Thermo Fisher Scientific) with freshly added Halt Protease and Phosphatase Inhibitor Mixture (Thermo Fisher Scientific). Protein concentrations were determined using the bicinchoninic acid (BCA) method (Thermo Fisher Scientific) according to the manufacturer’s instructions. A total of 2 to 3 µg of whole cell lysates were subjected to electrophoresis in 10% or 4 to 12% Bis-Tris gels and transferred onto nitrocellulose membranes using iBlot2 (Life Technologies). Membranes were probed with primary antibodies (SI Appendix, Table S3B) followed by horseradish peroxidase-conjugated secondary antibodies (Life Technologies). Membranes were then washed and visualized with an enhanced chemiluminescence (ECL) detection system (GE Healthcare) and iBright CL1000 (Thermo Fisher Scientific). Signals were quantified using ImageJ software (NIH).
Protein Synthesis Measured by Puromycin Incorporation.
M(IL4) or MCF7 cells were treated with vehicle (i.e., cells in the presence of ∼0.1% DMSO) or MNKi for 24 h or torin1 for 4 h. Cycloheximide (Sigma-Aldrich, 100 μg/mL) and/or puromycin (Sigma-Aldrich, 10 μg/mL) were added 15 min before harvest. The cells were then washed with ice-cold PBS twice and protein was collected and processed for Western blotting as indicated above.
SmartSeq2 Library Preparation and RNA Sequencing.
SmartSeq2 sequencing libraries were prepared as previously described (51) using 5 ng (BMDMs) or <1 ng (sorted cells) of total or polysome-associated mRNA as input. Pooled libraries were sequenced with Illumina HiSeq 2500 using a 50-base single-end setup. The resulting datasets were deposited at the gene expression omnibus (GSE151432 and GSE153060).
Data Analysis.
RNA sequencing reads were aligned to Mm10 using HISAT (52) and reads mapping to multiple locations in the genome were discarded (numbers of sequenced reads and unique reads mapping to protein coding mRNA are shown in Dataset S1 together with tumor characteristics such as weight, time to harvest, sequencing batch, and percentage of macrophages). Gene expression was quantified as described using default settings (53) and RefSeq gene definitions (54). Raw counts were trimmed-mean M values normalized (55) and log2 transformed using Voom (56) in R 3.5. Changes in total mRNA and polysome-associated mRNA depending on tumor size were then assessed in an ANCOVA to obtain the linear relationship between gene expression and tumor size (see SI Appendix for details).
Generally applicable gene-set enrichment for pathway analysis (57) was used to identify enrichment of genes with functions annotated by the Gene Ontology (GO) Consortium (58) using the batch-adjusted linear relationship of translational efficiency to tumor size for all genes as input. Significantly enriched GO terms among up- and down-regulated genes (FDR < 0.1) were then represented as an annotated network using the Cytoscape Enrichment Map pipeline (http://apps.cytoscape.org/apps/enrichmentmappipelinecollection). The batch-adjusted regression coefficients of gene expression to tumor size using total mRNA, polysome-associated mRNA, and translational efficiency levels for significantly regulated genes in selected clusters were used to generate heatmaps.
Supplementary Material
Acknowledgments
This study was supported by the Swedish Cancer Society (2019/281), Swedish Research Council (2018-02915), and the King Gustaf V Jubilee Fund (2018-184143) (to C.R.); and the Swedish Cancer Society (2019/314), the Swedish Research Council (2016-02891), the Stockholm Cancer Society, the Wallenberg Academy Fellow program, and the Strategic Cancer Initiative at KI (to O.L.). S.V.d.R. was funded by the Canadian Institutes for Health Research grant PJT-162260. M.B. and T.W. were supported by PhD Fellowship from Karolinska Institute, while Y.P. and H.L. were supported by the Chinese Student Council program. We would like to acknowledge support from the Science for Life Laboratory National Genomics Infrastructure core facility and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. X.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920377117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information together with Gene Expression Omnibus datasets GSE151432 (59) and GSE153060 (60).
References
- 1.Rankin L. C., Artis D., Beyond host defense: Emerging functions of the immune system in regulating complex tissue physiology. Cell 173, 554–567 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Cassetta L., Pollard J. W., Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887–904 (2018). [DOI] [PubMed] [Google Scholar]
- 3.DeNardo D. G., Ruffell B., Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chevrier S.et al., An immune Atlas of clear cell renal cell carcinoma. Cell 169, 736–749.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P., Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolny C.et al., HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Canc. Cell 19, 31–44 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Squadrito M. L.et al., miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 1, 141–154 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Wallerius M.et al., Guidance molecule SEMA3A restricts tumor growth by differentially regulating the proliferation of tumor-associated macrophages. Cancer Res. 76, 3166–3178 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Cassetta L.et al., Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35, 588–602.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil Del Alcazar C. R.et al., Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 7, 1098–1115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeNardo D. G.et al., Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1, 54–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahmoud S. M.et al., Tumour-infiltrating macrophages and clinical outcome in breast cancer. J. Clin. Pathol. 65, 159–163 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Piccirillo C. A., Bjur E., Topisirovic I., Sonenberg N., Larsson O., Translational control of immune responses: From transcripts to translatomes. Nat. Immunol. 15, 503–511 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Robichaud N., Sonenberg N., Ruggero D., Schneider R. J., Translational control in cancer. Cold Spring Harb. Perspect. Biol. 11, a032896 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandri B. J.et al., Distinct cancer-promoting stromal gene expression depending on lung function. Am. J. Respir. Crit. Care Med. 200, 348–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershey J. W., Sonenberg N., Mathews M. B., Principles of translational control: An overview. Cold Spring Harb. Perspect. Biol. 4, a011528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leprivier G., Rotblat B., Khan D., Jan E., Sorensen P. H., Stress-mediated translational control in cancer cells. Biochim. Biophys. Acta 1849, 845–860 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Sonenberg N., Hinnebusch A. G., Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konicek B. W.et al., Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Cancer Res. 71, 1849–1857 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Robichaud N.et al., Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene 34, 2032–2042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robichaud N.et al., Translational control in the tumor microenvironment promotes lung metastasis: Phosphorylation of eIF4E in neutrophils. Proc. Natl. Acad. Sci. U.S.A. 115, E2202–E2209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan Y.et al., MNK1/2 inhibition limits oncogenicity and metastasis of KIT-mutant melanoma. J. Clin. Invest. 127, 4179–4192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H.et al., Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 13, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schott J.et al., Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet. 10, e1004368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.William M.et al., Translational repression of Ccl5 and Cxcl10 by 4E-BP1 and 4E-BP2 restrains the ability of mouse macrophages to induce migration of activated T cells. Eur. J. Immunol. 49, 1200–1212 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Su X.et al., Interferon-γ regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 16, 838–849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin E. Y., Nguyen A. V., Russell R. G., Pollard J. W., Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 193, 727–740 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang S.et al., Polysome-profiling in small tissue samples. Nucleic Acids Res. 46, e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin E. Y.et al., Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guy C. T., Cardiff R. D., Muller W. J., Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roswall P.et al., Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat. Med. 24, 463–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandin V.et al., nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 26, 636–648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masvidal L., Hulea L., Furic L., Topisirovic I., Larsson O., mTOR-sensitive translation: Cleared fog reveals more trees. RNA Biol. 14, 1299–1305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oertlin C.et al., Generally applicable transcriptome-wide analysis of translation using anota2seq. Nucleic Acids Res. 47, e70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cummins T. D.et al., Quantitative mass spectrometry of diabetic kidney tubules identifies GRAP as a novel regulator of TGF-beta signaling. Biochim. Biophys. Acta 1804, 653–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palucka A. K., Coussens L. M., The basis of oncoimmunology. Cell 164, 1233–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray P. J.et al., Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laoui D.et al., Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res. 74, 24–30 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Bhat M.et al., Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 14, 261–278 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Cenik C.et al., Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res. 25, 1610–1621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorent J.et al., Translational offsetting as a mode of estrogen receptor α-dependent regulation of gene expression. EMBO J. 38, e101323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thoreen C. C.et al., An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furic L.et al., eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 107, 14134–14139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltraminelli T., De Palma M., Biology and therapeutic targeting of tumour-associated macrophages. J. Pathol. 250, 573–592 (2020). [DOI] [PubMed] [Google Scholar]
- 45.DeNardo D. G.et al., CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Canc. Cell 16, 91–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y.et al., mTOR-mediated metabolic reprogramming shapes distinct microglia functions in response to lipopolysaccharide and ATP. Glia 68, 1031–1045 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Kim J., DeBerardinis R. J., Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 30, 434–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wenes M.et al., Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 24, 701–715 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Zhu Y.et al., Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 47, 323–338.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Palma M., Naldini L., Transduction of a gene expression cassette using advanced generation lentiviral vectors. Methods Enzymol. 346, 514–529 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Picelli S.et al., Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramsköld D., Wang E. T., Burge C. B., Sandberg R., An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLOS Comput. Biol. 5, e1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coordinators N. R.; NCBI Resource Coordinators , Database resources of the national center for biotechnology information. Nucleic Acids Res. 46, D8–D13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritchie M. E.et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo W., Friedman M. S., Shedden K., Hankenson K. D., Woolf P. J., GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The Gene Ontology Consortium , Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsson O., "Selective mRNA translation governs the tumor-associated macrophage phenotype." Gene Expression Omnibus (NCBI). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151432. Deposited 30 May 2020.
- 60.Larsson O., "Polysome-profiling of IL4 or INFG polarized BMDMs." Gene Expression Omnibus (NCBI). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153060. Deposited 24 June 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information together with Gene Expression Omnibus datasets GSE151432 (59) and GSE153060 (60).