Significance
Because of the weak interactions between individual glycan-binding proteins (GBPs), such as galectin-3, and glycans, strategies that enable the direct interrogation of these interactions in living cells remain limited. Thus, the glycan and glycoprotein ligands that are physiologically relevant for galectin-3 binding are insufficiently described. Here we used a proximity labeling approach that catalytically tags interactors for galectin-3 and identified its pertinent glycan and glycoprotein counter-receptors in live cells. This study demonstrates that proximity labeling is a powerful tool for mapping GBP complexes in living cells, and when coupled with chemical inhibitors, it can discriminate between protein–protein and protein–glycan interactions.
Keywords: glycan, galectins, glycomics, proximity labeling, proteomics
Abstract
Galectin-3 is a glycan-binding protein (GBP) that binds β-galactoside glycan structures to orchestrate a variety of important biological events, including the activation of hepatic stellate cells and regulation of immune responses. While the requisite glycan epitopes needed to bind galectin-3 have long been elucidated, the cellular glycoproteins that bear these glycan signatures remain unknown. Given the importance of the three-dimensional (3D) arrangement of glycans in dictating GBP interactions, strategies that allow the identification of GBP receptors in live cells, where the native glycan presentation and glycoprotein expression are preserved, have significant advantages over static and artificial systems. Here we describe the integration of a proximity labeling method and quantitative mass spectrometry to map the glycan and glycoprotein interactors for galectin-3 in live human hepatic stellate cells and peripheral blood mononuclear cells. Understanding the identity of the glycoproteins and defining the structures of the glycans will empower efforts to design and develop selective therapeutics to mitigate galectin-3–mediated biological events.
The noncovalent interactions between glycan-binding proteins (GBPs) and glycans dictate many important biological events. Among such GBPs is galectin-3, a 26-kDa β-galactoside GBP that plays key roles in many physiological and pathological events (1). In hepatic fibrosis, a disease that manifests as the excessive buildup of scar tissue, liver-resident macrophages secrete galectin-3 (2, 3), which then binds cell surface glycans on quiescent hepatic stellate cells (HSCs), activating them to transdifferentiate into a muscle-like phenotype. Galectin-3–null mice exhibit attenuated liver fibrosis even after induced injury, highlighting its critical role (3). Galectin-3 is also known to interact with cells of the innate immune system (4, 5) to regulate apoptosis (6) or control dendritic cell differentiation (7). In these cases, as well as in other cases in which galectin-3 is involved, the full complement of interacting glycoprotein receptors remains unknown.
Despite significant advances in glycoscience, the study of GBP–glycan interactions and the identification of glycan-mediated counter-receptors remains a recurring challenge. Capturing these binding events often requires some form of artificial reconstitution to amplify individually weak interactions into high-avidity binding. Indeed, glycan microarrays with defined mixtures of homogenous glycans or recombinant GBPs have significantly propelled our understanding of glycan-mediated function (8). Conventional immunoprecipitation and lectin affinity techniques using cell lysates have similarly been used to reveal an initial catalog of 100 to 185 galectin-3–associated proteins (9–14). However, these manipulations alter the cell’s native and three-dimensional (3D) configuration and multivalent arrangement, both of which are critically important in the study of GBP–glycan interactions (15, 16).
Another key issue involves the underlying glycoprotein ligand. Although many glycoproteins carry the glycan epitope for binding a GBP, only a limited set should be recognized as physiologically relevant receptors, owing to the physical constraints imposed by the living cell (17). While often overlooked, the glycoprotein carrying the glycan can impart specific biological functions to a GBP–glycan binding event (17). Recent work has put forth the concept of “professional glycoprotein ligands,” in which a specific set of glycoproteins (instead of a broadly defined glycome) can exhibit exquisite binding and functional roles (18). Thus, determining the identity of the underlying core protein that anchors the glycan can be greatly empowering. Not only can it provide an understanding of the 3D arrangement of the glycan (if the 3D structure of the core protein is known), but it can also provide additional insight into its expression levels in different cell types and tissues, further informing strategies for selective drug development.
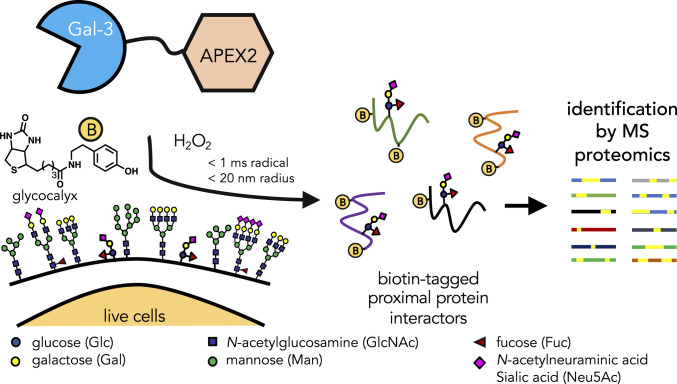
Thus, comprehensive approaches that permit the study of GBP–glycan interactions in live cells while simultaneously facilitating identification of the physiological glycoprotein receptors have great potential to impact glycoscience. We hypothesize that proximity labeling strategies (19) using an engineered ascorbate peroxidase, APEX2 (20), could be compatible for elucidating glycan-mediated GBP–glycoprotein interactions. In this approach (Fig. 1), APEX2 is fused to a protein of interest, followed by the treatment of cells with biotin-phenol and subsequently with hydrogen peroxide (H2O2). Under these conditions, APEX2 catalyzes the formation of highly reactive, short-lived (<1 ms), and proximally restricted (<20 nm) biotin-phenoxyl radicals that covalently tag nearby electron-rich residues. The biotinylated proteins can then be enriched and identified using quantitative mass spectrometry (MS)-based proteomics. Because the (glyco)proteins adjacent to the APEX2 fusion protein are preferentially biotinylated, the resulting MS data provide a readout of its immediate environment.
Fig. 1.
Schematic illustration of the identification of galectin-3 (Gal-3) interacting proteins by in situ proximity labeling. Recombinant APEX2 and galectin-3 fusion proteins are applied to living cells where galectin-3 can freely diffuse to bind its cognate ligands. On addition of biotin phenol (yellow circle with “B”; 30 min) and hydrogen peroxide (H2O2; 1 min), APEX2 catalyzes the formation of highly-reactive biotin-phenoxyl radicals that react within a short range (<20 nm) of the galectin-3 complex within a short time frame (<1 ms). The biotin-tagged protein interactors can then be identified using MS-based proteomics.
We reasoned that proximity labeling could offer significant advantages over other approaches to determining GBP–glycan interactions, including the opportunity to perform the labeling in live cells and the ability to tag weakly bound glycan-mediated interactors, as the covalent biotinylation reaction ensures that the enrichment step no longer relies on weak GBP–glycan interactions alone. When coupled with inhibitors, the proximity labeling strategy can also distinguish between glycan-mediated and non–glycan-mediated interactors. Integration of this approach with quantitative MS-based proteomics would also expedite the assignment of the interacting proteins and provide calculable measures to distinguish interactors from nonspecific binders.
Here we report that the use of an APEX2 and galectin-3 fusion protein (PX-Gal3) provides a sensitive and comprehensive approach to mapping the proteome-wide glycan-mediated galectin-3 interactome in live human HSCs and peripheral blood mononuclear cells (PBMCs). We found that the exogenous incubation of cells with PX-Gal3 in HSCs leads to glycan-dependent interactions, whereas its cellular overexpression does not. We further validated the interactions between galectin-3 and candidate proteins in vitro and discovered that some proteins are direct glycan-mediated receptors. Using MS-based glycomics, we also examined the glycomes of HSC surfaces, PX-Gal3 tagged glycoproteins, and an individual glycoprotein receptor for galectin-3. Our results highlight the utility of the in situ proximity labeling approach in discovering physiologically relevant GBP interactors in living cells.
Results
Proximity Tagging to Identify Galectin-3 Interactors in HSCs.
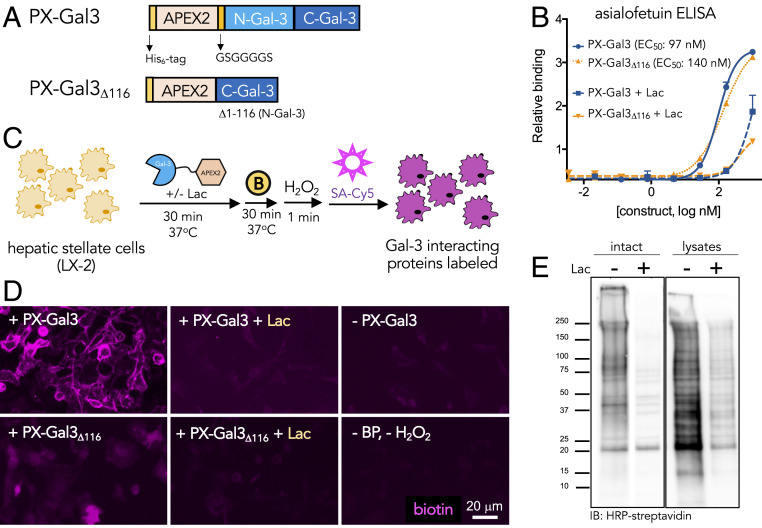
We genetically fused the peroxidase APEX2 enzyme (20, 21) to either full-length galectin-3 (PX-Gal3) or a mutant galectin-3 lacking the N-terminal domain (PX-Gal3∆116) (Fig. 2A). This mutant was produced to determine whether the proximity labeling protocol could be used to confirm the role of the N-terminal domain in facilitating homo-oligomerizing interactions to control extracellular binding activities (22, 23). We chose APEX2 over other peroxidase enzymes because of its monomeric nature, small size (28 kDa), and improved activities in various cellular compartments (20), especially for proteomic applications (24). We fused APEX2 to the N terminus of galectin-3, as previous work has shown that such fusion proteins remain active toward binding glycans (25). Both fusion proteins were generated efficiently (SI Appendix, Fig. S1 A–D) in recombinant Escherichia coli and retained their capacity to bind glycans in an enzyme-linked immunosorbent assay (ELISA) with immobilized asialofetuin (SI Appendix, Fig. S2). Coincubation with a soluble competitor, lactose, efficiently blocked binding (Fig. 2B), indicating that interactions were glycan-dependent. Apparent binding affinity constants determined for PX-Gal3 (EC50 ∼100 nM) vs. PX-Gal3Δ116 (EC50 ∼140 nM) were similar, consistent with previous reports of galectin-3 fusion proteins (25).
Fig. 2.
Design and characterization of PX-Gal3 fusion constructs for proximity labeling in LX-2 HSCs. (A) APEX2 fusion constructs of galectin-3, PX-Gal3 and PX-Gal3Δ116, were constructed. Both proteins include an N-terminal His-tag sequence, followed by the APEX2 enzyme, and either full-length galectin-3 or the C-terminal domain of galectin-3. PX-Gal3Δ116 lacks the N-terminal domain of galectin-3, which was previously implicated for its homo-oligomerization on binding of glycans at the cell surface. (B) Using an ELISA with asialofetuin as a model glycan-bearing ligand, the fusion constructs retained similar glycan-binding activities, as determined by EC50 values. (C) Application of the proximal labeling method to live adherent HSCs. The recombinant fusion proteins were first incubated with HSCs (30 min, 37 °C). After washing to remove unbound proteins, biotin phenol (yellow circle with a B; 500 μM, 30 min, 37 °C) was added, followed by H2O2 (1 mM, 1 min, RT). Biotinylated interactors (purple) were subsequently probed using Cy5-labeled streptavidin. Conditions were optimized according to reported radical-mediated biotinylation procedures (20, 21). (D) Fluorescence microscopy images of biotin-tagged (purple) HSCs showing that PX-Gal3 (100 nM; 30 min) generates significant labeling over negative controls in which a component (e.g., protein or biotin phenol and H2O2) of the labeling protocol is omitted. Coincubation with lactose (Lac; 100 mM) causes a loss of signal, suggesting that the majority of labeled interactions are glycan-dependent. PX-Gal3Δ116 fails to significantly label interactors. (E) Western blotting of (10 μg lysate per lane) biotinylated proteins produced from the proximity labeling method applied to intact live cells vs. cell lysates showing that only a subset of interactors is captured (SI Appendix, Fig. S6).
To examine whether the proximity labeling protocol could identify interactors with galectin-3, we incubated the recombinant fusion proteins with live adherent HSCs. After washing and the stepwise application of biotin-phenol and H2O2, biotin-tagged interactors were visualized using a fluorophore-conjugated streptavidin probe and fluorescence microscopy (Fig. 2C). We observed significant fluorescence in cells incubated with PX-Gal3 compared with negative controls and PX-Gal3∆116 (Fig. 2D), with signals detectable as early as after 5 min of protein incubation, which did not increase significantly even after up to 2 h (SI Appendix, Fig. S3A). We also observed a dose-dependent increase in fluorescence with increasing amounts (25 to 100 nM) of PX-Gal3 (SI Appendix, Fig. S3B). In contrast to the plate-based ELISA findings, PX-Gal3Δ116 did not elicit a significant signal compared with PX-Gal3. We attribute this observation to the differing glycan density and presentation found in live cells, establishing an important role for the N-terminal oligomerization domain not directly apparent using plate-based methods. This protocol enabled the sensitive detection of interactors with nanomolar concentrations of PX-Gal3, whereas querying for galectin-3 binding to HSCs in simple binding experiments failed to achieve a significant signal even at a concentration of 10 μM and a prolonged incubation period (SI Appendix, Fig. S3C). While fluorescence signals were mostly observed at the cell surface, some intracellular labeling was also observed with increased PX-Gal3 incubation times (SI Appendix, Fig. S3A) and concentrations (SI Appendix, Fig. S3B), presumably due to known endocytic mechanisms that occur upon galectin-3 binding (13, 26).
Proximity labeling conditions in which PX-Gal3 was coincubated with lactose (SI Appendix, Fig. S4A) or where cell surface glycosylation was inhibited, either by tunicamycin (SI Appendix, Fig. S4B) or by PNGAse F (SI Appendix, Fig. S4C), showed reduced fluorescence. Furthermore, a PX-Gal3 mutant that included three point mutations (R144S, R186A, and G182A) that abolish glycan-binding activity (27) failed to show significant fluorescence (SI Appendix, Fig. S5). These results indicate that glycan-mediated interactions are highly represented between PX-Gal3 and HSCs. We also compared labeling of live intact cells vs. cellular lysates and found that only a limited subset of potential interactors was captured and substantially blocked with excess lactose (Fig. 2E and SI Appendix, Fig. S6). Taken together, these observations indicate that the proximity labeling strategy can probe for glycan-dependent interactors in live cells with high sensitivity, and that native glycan presentation is important for the selective capture of interactions.
Identification of Biotinylated Interactors with Quantitative MS-Based Proteomics.
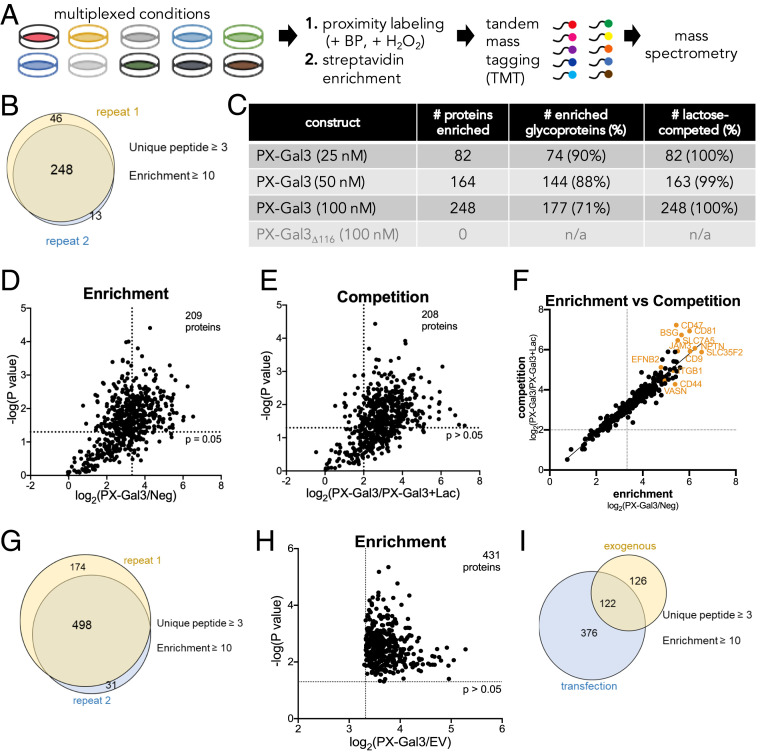
To identify the putative galectin-3 interactors, we subjected HSCs to proximity labeling, followed by cell lysis, streptavidin bead enrichment, and on-bead trypsinization (Fig. 3A). We then subjected the resulting peptides to tandem mass tag (TMT) labeling, which chemically tags the N termini and free amines of the resulting peptides from each condition with a unique isobaric tag that can be distinguished at the MS3 stage (28, 29). This method allows for a reliable comparative analysis of multiplexed samples in a single run, further ensuring the accuracy of the comparison.
Fig. 3.
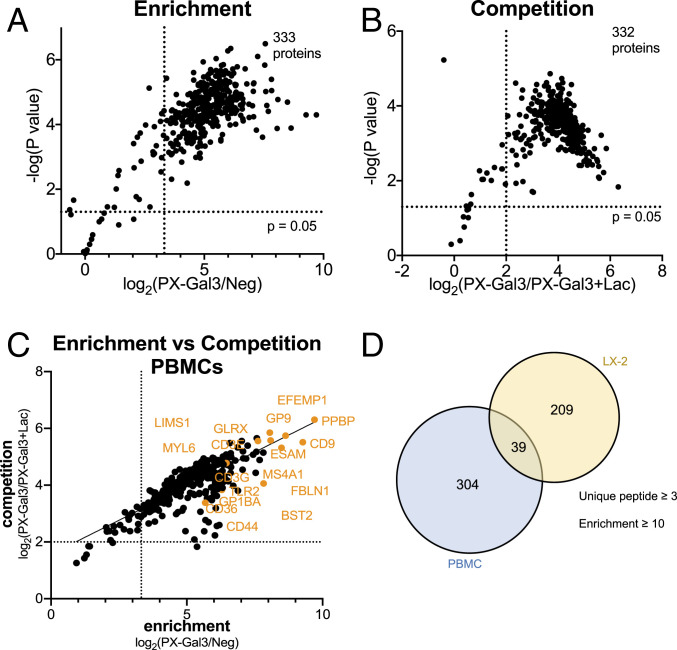
Identification and analysis of the PX-Gal3 interactome in LX-2 HSCs by quantitative MS-based proteomics. (A) Experimental workflow for the preparation of samples for analysis from LX-2 HSCs. (B) A total of 248 proteins were enriched across two replicates, defined as proteins with three or more unique peptides and with a TMT ratio (PX-Gal3/Neg) ≥10. Neg indicates conditions in which the cells were not incubated with PX-Gal3 but were still treated with biotin phenol and H2O2. (C) Analysis of proteins enriched by PX-Gal3 by dosage, glycosylation status (assigned by UniProt), and competition with lactose (100 mM). (D) Statistically significant (P < 0.05) and enriched proteins found in cells treated with PX-Gal3 (100 nM, 30 min). (E) Proteins that were statistically significant (P < 0.05) and competed (TMT ratio of PX-Gal3/PX-Gal3+Lac ≥4) on coincubation with exogenous lactose (100 mM). (F) There is high linear correlation between proteins that were enriched by and those that were competed with lactose. (G) A total of 498 proteins were found to be enriched across two replicates when PX-Gal3 was transiently overexpressed in HSCs. (H) A total of 431 proteins were found to be significantly enriched (P < 0.05). (I) There were 122 overlapping proteins between the exogenous and transfection protocols (SI Appendix, Table S1).
We identified a total of 248 proteins across two replicates (Fig. 3B, Table 1, and SI Appendix, Table S1). These proteins were consistently detected with three or more unique peptides and were highly enriched (TMT ratio ≥10) by PX-Gal3 (100 nM) over the negative control, cells that were not treated with any fusion protein but were similarly treated with biotin phenol and H2O2 (Fig. 3B). We observed a dose-dependent increase in the number of enriched proteins using PX-Gal3 (Fig. 3C), whereas no proteins were enriched by PX-Gal3∆116 (SI Appendix, Fig. S7A) or by the triple-point mutant protein (SI Appendix, Fig. S7B). A majority (71 to 90%) of enriched proteins in each dose of PX-Gal3 was previously reported to be N- or O-glycosylated (Fig. 3C) (30). Among the 248 proteins, 209 (84%) were defined as statistically significant (P < 0.05; Fig. 3D); among these, 208 (99%) were competed with lactose (TMT ratio ≥4; Fig. 3E). Proteins that were enriched generally correlated with those that were competed with lactose (Fig. 3F) and TD139 (31), a lactose analog that displays potent galectin-3 inhibition (SI Appendix, Fig. S7C). The proteins enriched by PX-Gal3 span a wide range of abundance, as determined using peptide spectral matches from unenriched proteomics in LX-2 HSCs (SI Appendix, Fig. S7D and Table S2) and reported iBAQ (intensity-based absolute quantification) values (SI Appendix, Fig. S7E) (32).
Table 1.
Abbreviated list of galectin-3 interactors determined by proximity labeling and quantitative MS
| Gene name | Protein name | UniProt ID | Glycosylation state | Subcellular location |
| BSG | BSG | P35613 | 3 N-linked sites | PM; ERS |
| NPTN | Neuroplastin | Q9Y639 | 6 N-linked sites | PM |
| JAM3 | Junctional adhesion molecule C | Q9BX67 | 2 N-linked sites | PM, ERS |
| SLC35F2 | Solute carrier family 35 member F2 | Q8IXU6 | Not known | PM |
| SLC7A5 | Large neutral amino acids transporter small subunit 1 | Q01650 | Not known | PM; Lyso; ERS |
| SLC23A2 | Solute carrier family 23 member 2 | Q9UGH3 | 2 N-linked sites | PM |
| ITGA1 | Integrin-a-1 | P56199 | 26 N-linked sites | PM; ERS |
| ITGB1 | Integrin-b-1 | P05556 | 12 N-linked sites | PM; Endo; ERS |
| CD44 | CD44 antigen | P16070 | 9 N-linked sits | PM; ERS |
| CD47 | Leukocyte surface antigen CD47 | Q08722 | 6 N-linked sites | PM; ERS |
| CD81 | CD81 antigen | P60033 | Not known | PM; ERS |
| CD9 | CD9 antigen | P21926 | 2 N-linked sites | PM; ERS |
| TSPAN6 | Tetraspanin-6 | O43657 | 1 N-linked site | PM; ERS |
| EFNB1 | Ephrin-B1 | P98172 | 1 N-linked site | PM; Nuc; ERS |
| EFNB2 | Ephrin-B2 | P52799 | 2 N-linked sites | PM |
| VASN | Vasorin | Q6EMK4 | 5 N-linked sites | ERS; Lyso; PM |
| HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | P31942 | Not known | Nuc |
| HNRNPD | Heterogeneous nuclear ribonucleoprotein D0 | Q14103 | Not known | Nuc |
| HNRNPF | Heterogeneous nuclear ribonucleoprotein F | P52597 | Not known | Nuc |
An expanded list is available in SI Appendix, Table S1. The glycosylation states are assigned according to UniProt (30). PM, plasma membrane; ERS, extracellular region or secreted; Nuc, nucleus; Endo, endosome; Lyso, lysosome.
Among the PX-Gal3 targets identified by the proximity labeling method in LX-2 HSCs (SI Appendix, Table S1) are vasorin (VASN), basigin (BSG/CD147), neuroplastin (NPTN), members of the tetraspanin family (CD9, CD81, and TSPAN6), members of the ephrin family (EPHB1 and EPHB2), members of the solute carrier family of transporters (SLC35F2, SLC23A2, and SLC7A5), and members of the integrin family (ITGA1 and ITGB1). Although many of the targets are assigned to plasma membrane or cell surface locations, several are also found intracellularly, including members of the human ribonucleoprotein (HNRNP) complex (33), which are found in the nucleus. These results indicate that despite their exogenous application, the fusion proteins traverse pathways that are expected of the dynamic nature of galectin-3, first interacting with cell surface proteins and then eventually traversing the cell membrane to associate with intracellular components (34).
We derived effective galectin-3 binding affinity constants (EC50) for selected proteins (BSG, CD47, ITGA1, JAM3, EPHB2, and VASN) using the proximity labeling protocol and TMT quantitation (SI Appendix, Fig. S7F). Such binding constants are reflective of the native cellular glycan and protein expression levels. While it is difficult to extrapolate these constants as a comparative measure of binding of galectin-3 against individual proteins, it is gratifying to see that the proximity labeling method can achieve robust nanomolar binding against these proteins in cells. We used integrated pathway analysis to derive putative binding relationships and observed that BSG lies at a nexus to other proteins enriched by PX-Gal3, such as ITGB1, SLC7A5, JAM3, and SLC23A2 (SI Appendix, Fig. S7G).
Considering that we identified intracellular protein targets with exogenously administered PX-Gal3, we set out to evaluate whether proximity labeling could also identify interactors for galectin-3 when it is expressed intracellularly. To do so, we overexpressed PX-Gal3 in HSCs by transient transfection (SI Appendix, Fig. S8 A and B). Following transfection, some cells were further treated with tunicamycin, an N-glycosylation inhibitor, to assess whether N-linked glycosylation plays a role (lactose is impermeable to cells). A total of 498 proteins (filtered by three or more unique peptides) were found to be enriched (TMT ratio over empty vector control ≥10) and to overlap across two replicates (Fig. 3G and SI Appendix, Table S3), of which 431 (87%) were statistically significant (P < 0.05; Fig. 3H). Interestingly, treatment with tunicamycin (SI Appendix, Fig. S8C) failed to compete (TMT ratio ≥4) for labeling, suggesting that intracellular galectin-3 interactions are mostly mediated by protein–protein interactions rather than by protein–glycan interactions, consistent with previous reports (34). Comparing the enriched and glycan-mediated protein interactors from the exogenous treatment (Fig. 3B) with the transient overexpression of PX-Gal3 yielded 122 proteins in common (Fig. 3I and SI Appendix, Table S1), possibly suggesting that galectin-3 may associate with some proteins in either a glycan-mediated or non–glycan-mediated manner, depending on its entry.
Validation of Interactions between Galectin-3 and Identified Proteins.
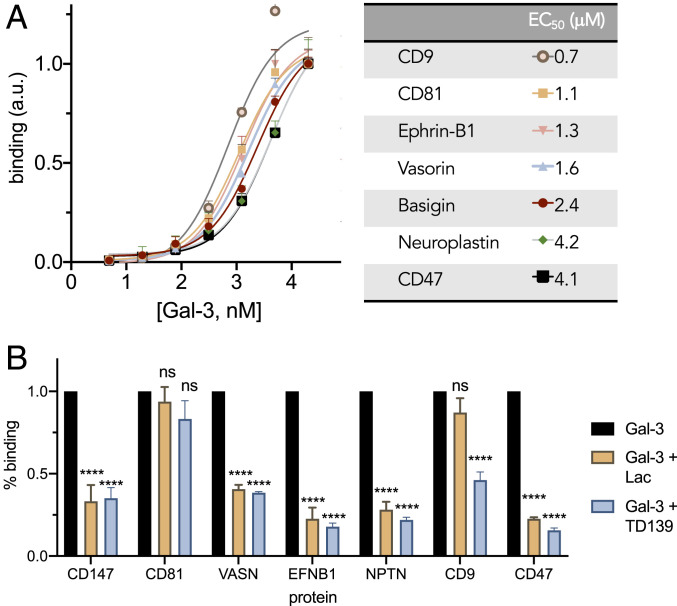
As the proximity labeling strategy only tags proteins that occur within the vicinity (<20 nm) of galectin-3, and that these might not necessarily represent direct receptors, we next examined whether selected proteins identified by the proteomic experiments are direct receptors for galectin-3. We first confirmed that BSG, CD9, EPHB1, CD47, and VASN are prominently expressed in LX-2 cells (SI Appendix, Fig. S9). Then, using immobilized human recombinant proteins (SI Appendix, Table S1) of BSG, CD9, CD47, CD81, EPHB1, NPTN, and VASN, we found that galectin-3 binds in a dose-dependent manner (Fig. 4A and SI Appendix, Table S2). Apparent binding affinities, with EC50 values ranging from 0.7 μM (CD9) to 4.2 μM (CD47), were observed (SI Appendix, Table S2). Notably, we found that the majority of the proteins were competed with the presence of either lactose or TD139, CD81 binding was not blocked by either competitor, and CD9 interactions could only be competed with TD139 (Fig. 4B). Human CD81 is not known to be glycosylated, suggesting that its binding to galectin-3 might be mediated by secondary protein–protein interactions. Human CD9 (UniProt P21926) is predicted to have two N-linked glycosylation sites, implicating potential glycan-mediated interactions with galectin-3, but these interactions may only partially contribute to galectin-3 binding, as competition with TD139 was still observed. Taken together, these results indicate that BSG, CD9, CD47, CD81, EPH-B1, VASN, and NPTN are potential receptors for galectin-3 in cells.
Fig. 4.
Validation of binding between galectin-3 and proteins identified from proteomics analysis. (A) ELISA binding curves and corresponding EC50 values determined for recombinant proteins against galectin-3. (B) Coincubation of galectin-3 (5 μM) with lactose (Lac, 100 mM) or TD139 (15.4 μM) competes for glycan-mediated binding interactions in ELISA.
MS-Based Characterization of Glycomes from LX-2 Cell Surfaces, PX-Gal3–Enriched Interactors, and Endogenous BSG.
Given the importance of cell surface glycosylation in mediating galectin-3 interactions (Figs. 2 and 3), we next sought to identify the structures of the glycans found in LX-2 HSCs. While several groups have reported that O-linked glycans from mucins (35) and glycosaminoglycans (36) can serve as ligands for galectin-3, complex galactose-terminating N-glycans (with some tolerance of sialylation) have been definitively established as the prominent high-affinity binding galectin-3 epitopes in cells (37, 38). Thus, we next focused our investigation on profiling N-glycans, with particular attention on assessing the abundance of N-glycans bearing the high-affinity terminal galactose monosaccharides (39). We further evaluated whether certain N-glycan motifs are enriched by PX-Gal3 and whether they are present on the identified glycoprotein receptors for galectin-3 (Figs. 3 and 4).
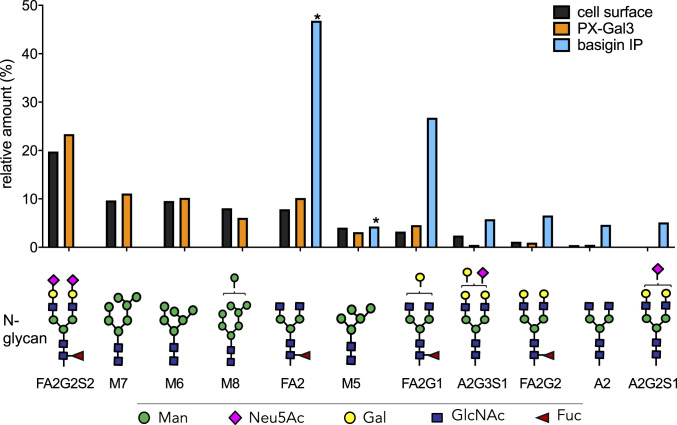
We harvested N-glycans from intact LX-2 cells by brief treatment with trypsin and PNGase F. The free N-glycans were then subjected to reduction and permethylation before MS analysis. Using the extracted ion intensities acquired during the full MS scan, relative quantitation of the most abundant N-glycans was determined (40). The most abundant N-glycans found on LX-2 cell surfaces consisted mostly of complex and oligomannose, with the most abundant being the di-sialylated FA2G2S2, composing ∼20% of the total population (Fig. 5 and SI Appendix, Figs. S10 and S11). LX-2 cells stained positively with Sambucus nigra and Maackia amurensis lectins, indicating the presence of α(2,3)- and α(2,6)-linked terminal Neu5Ac sialic acids, respectively (SI Appendix, Fig. S12). Oligomannose N-glycans (M7, M6, and M8) were the next most abundant type, representing a cumulative total of ∼27% of the cell surface N-glycome. Terminal mono- and di-galactosylated N-glycans, FA2G1 and FA2G2, composed only ∼3% and ∼1% of the population, respectively.
Fig. 5.
MS-based determination of N-glycan abundance and composition. Composition and relative amounts of the most abundant N-glycans found on LX-2 cell surfaces (black bars), glycoproteins enriched by PX-Gal3 (orange bars), and immunoprecipitated (IP) BSG (blue bars). The most abundant N-glycans found on BSG differ greatly from the cell surface samples. An asterisk indicates N-glycans that were also found in the antibody used for IP. Terminal galactose (yellow circles) presenting N-glycans were found to be enriched in BSG compared with the cell surface. Oligomannose glycans bear terminal mannose residues (green circles) and are designated with an M (M7, M6, M8, M5); all others shown here are complex-type N-glycans.
We also identified N-glycans from the resulting mixture of peptides and glycopeptides captured from proximity labeling with PX-Gal3 (Fig. 2A) by harvesting them with PNGAse F. The most abundant N-glycans from these samples were overall quite similar to those found on LX-2 cell surfaces (Fig. 5 and SI Appendix, Fig. S12). FA2G1 and FA2G2 composed ∼5% and ∼1% of the total N-glycome, respectively. We observed a modest increase in the amount of terminal galactosylated N-glycans, with the di-galactosylated FA3G2 showing an ∼57% increase compared with the cell surface (SI Appendix, Fig. S13).
Given the central role of BSG as a glycan-dependent binding receptor for galectin-3 (Fig. 4) and its ability to bind other enriched proteins (SI Appendix, Fig. S7G), we sought to determine its native N-glycosylation pattern. To this end, we immunoprecipitated BSG from LX-2 cells using an anti-BSG antibody and protein A beads (SI Appendix, Fig. S15) and observed pronounced differences compared with the cell surface N-glycome (Fig. 5). In contrast to the cell surface, which was composed of a mixture of complex and oligomannose N-glycans, the six most abundant N-glycans were found to be complex. FA2 composed ∼47% of the total population, although this N-glycan was also found in the anti-BSG antibody (SI Appendix, Fig. S16). The next most abundant glycans found exclusively in this sample were the mono- and di-galactosylated FA2G1 and FA2G2, composing ∼27% and ∼7%, respectively. The monosialylated A2G3S1 and A2G2S1 were also abundant in this sample. These observations confirm that the high-affinity galactose epitope is present on LX-2 cell surfaces, and that it is highly abundant on BSG.
Proximity Tagging to Identify Galectin-3 Interactors in PBMCs.
Encouraged by the results with LX-2 HSCs, we sought to evaluate whether the proximity tagging approach could be extended to other cell types. Given the importance of galectin-3 in the immune system, we chose to identify interactors in PBMCs, which are composed of mixtures of myeloid and lymphoid cells. We identified a total of 333 proteins that were consistently and significantly enriched across three replicates (Fig. 6A, Table 2, and SI Appendix, Table S3). We observed that 332 proteins (99.7%) interacted in a glycan-dependent manner (Fig. 6B), and that there was similarly a general correlation between enriched proteins and proteins competed with lactose (Fig. 6C). Among these 332 proteins, 39 overlapped with proteins identified in HSCs, and the remainder were expressed exclusively in PBMCs (Fig. 6D).
Fig. 6.
Identification and analysis of the PX-Gal3 interactome in PBMCs by quantitative MS-based proteomics. (A) Statistically significant (P < 0.05) and enriched proteins found in cells treated with PX-Gal3 (100 nM, 30 min). (B) Proteins that were statistically significant (P < 0.05) and competed (TMT ratio of PX-Gal3/PX-Gal3+Lac ≥4) on coincubation with exogenous lactose (100 mM). (C) There was high linear correlation between proteins that were enriched by lactose and those that were competed with lactose. (D) A total of 39 proteins were enriched in both LX-2s and PBMCs, whereas 304 proteins were exclusively identified in PBMCs.
Table 2.
Abbreviated list of galectin-3 interactors in PBMCs determined by proximity labeling and quantitative MS
| Gene name | Protein name | UniProt ID | Glycosylation state | Subcellular location |
| ESAM | Endothelial cell-selective adhesion molecule | Q96AP7 | 4 N-linked sites | PM |
| NPTN | Neuroplastin | Q9Y639 | 6 N-linked sites | PM |
| PPBP | Platelet basic protein | P02775 | Not known | ERS |
| GP9 | Platelet glycoprotein IX | P14770 | 1 N-linked site | PM |
| CD36 | Platelet glycoprotein 4 | P16671 | 10 N-linked sites | ERS; PM |
| SLC3A2 | 4F2 cell-surface antigen heavy chain | P08195 | 4 N-linked sites | PM; Lyso |
| FBLN1 | Fibulin-1 | P23142 | 3 N-linked sites | ERS |
| CCL5 | C-C motif chemokine 5 | P13501 | 2 O-linked sites | ERS |
| CD44 | CD44 antigen | P16070 | 9 N-linked sites | PM; ERS |
| CD47 | Leukocyte surface antigen CD47 | Q08722 | 6 N-linked sites | PM; ERS |
| TLR2 | Toll-like receptor 2 | O60603 | 4 N-linked sites | PM |
| CD9 | CD9 antigen | P21926 | 2 N-linked sites | PM; ERS |
| CD3G | T-cell surface glycoprotein CD3 gamma chain | P09693 | 2 N-linked sites | PM |
| FBLN1 | FBLN1 | P23142 | 3 N-linked sites | ERS |
| CD3E | T cell surface glycoprotein CD3 epsilon chain | P07766 | Not known | PM |
| SLC2A3 | Solute carrier family 2, facilitated glucose transporter member 3 | P11169 | 1 N-linked site | PM; ERS |
| MS4A1 | B-lymphocyte antigen CD20 | P11836 | Not known | PM; ERS |
| GP1BA | Platelet glycoprotein Ib alpha chain | P07359 | 2 N-linked sites and 1 O-linked site | PM; ERS |
| TLT-1 | Trem-like transcript 1 protein | Q86YW5 | Not known | PM |
An expanded list is available in SI Appendix, Table S3. The glycosylation states are assigned according to UniProt (30). PM, plasma membrane; ERS, extracellular region or secreted; Nuc, nucleus; Endo, endosome; Lyso, lysosome.
Discussion
Our overall goal was to determine whether a proximity labeling approach would be suitable for mapping the glycan-mediated interactors for galectin-3 in live cells (Fig. 1). Here we have demonstrated that a 1-min live labeling step with an exogenously applied fusion protein of APEX2 and galectin-3 covalently tagged protein interactors in HSCs (Figs. 2 and 3) and in PBMCs (Fig. 6). Integration of this method with quantitative MS-based proteomics identified 248 and 333 enriched proteins, respectively (Figs. 3 and 6). Before this study, only 100 to 185 proteins were found in complex with galectin-3 using standard affinity techniques (9–14), and our dataset extends the number of previously known interactors for galectin-3. These enriched proteins rely exclusively on glycan-dependent interactions for catalytic tagging. In contrast to in vitro experiments and tagging procedures on lysed cells, we showed that the N-terminal oligomerization domain (23) is critical for productive cell surface interactions (Fig. 2D), and despite many other possible interactors, only a subset form favorable interactions with galectin-3 (Fig. 2E).
While other labeling strategies use horseradish peroxidase fusion proteins (41) that require oxidative conditions, our method uses the APEX2 enzyme for covalent tagging. APEX2 is active in both oxidative extracellular conditions and the reducing environment of the intracellular milieu (42), permitting the identification of 498 interactors for PX-Gal3 when it is intracellularly overexpressed (Fig. 3G).
Consistent with previous reports, we also found that intracellular interactions with galectin-3 are not glycan-dependent (34). Some of the 122 proteins commonly enriched between the exogenous and the overexpressed conditions are transmembrane proteins (e.g., CD47, CD81) with the opportunity to be tagged from either side of the membrane. Others have also been found at the cell surface and intracellularly (e.g., GPC4, SLC3A2). These two points highlight the potential of galectin-3 to act via different mechanisms depending on its entry, providing a rationale for why the same proteins were tagged in a glycan-dependent manner exogenously and also in a non–glycan-dependent manner in the overexpressed model. However, we cannot exclude the possibility that tunicamycin treatment adversely affects glycoprotein expression and structural conformation.
Along with galectin-3, many of its 248 putative interactors are known to be important in HSC activation. For example, BSG and EPHB2 expression have been correlated to HSC activation (43, 44), and knockout of BSG has been shown to attenuate fibrosis (45). However, additional investigation is needed to confirm that the interactions between galectin-3 and these proteins are relevant in HSC activation. We selected seven proteins to confirm direct interactions with galectin-3 and found that recombinant BSG, CD9, CD47, CD81, EPHB1, NPTN, and VASN bound galectin-3, and that binding to CD9 and CD81 were not glycan-mediated in vitro (Fig. 4). It must be noted, however, that these recombinant proteins might not necessarily reflect the native protein sequence or glycosylation patterns found in LX-2 cells (SI Appendix, Table S1). CD47, EPH-B1, and VASN are previously unreported glycan-mediated ligands for galectin-3. While a majority of proteins enriched by the proximity labeling strategy are glycoproteins (Fig. 3C), some are not known to have sites for N-linked glycosylation. These observations highlight that while glycan-dependent interactions are responsible for the initial binding events with galectin-3, other protein–protein interactions also occur subsequent to the primary interaction. Thus, additional experiments are needed to definitively assign the direct glycoprotein ligands for galectin-3.
Both CD9 and CD81 are members of the tetraspanin family, a group of proteins that organize other proteins into discrete membrane compartments to regulate trafficking and signaling (46). They have been found in complex with each other and with other glycoproteins (47, 48), and it is intriguing to surmise how membrane organization could play a role in facilitating downstream signaling events on the binding of galectin-3 to HSCs. We speculate that galectin-3 binding either induces clustering of glycoproteins or may bind preformed clusters in HSCs, and that such organized microdomains could similarly promote galectin-3–dependent signal transduction events (35) that lead to HSC activation (49).
Changes in LX-2 cell surface glycosylation (49) and up-regulation of terminal galactosylation are associated with HSC activation (50). Using MS-based glycomics, we determined that LX-2 cell surfaces are composed mostly of complex and oligomannose N-glycans (Fig. 5). Of note, the N-glycome of endogenous BSG differs substantially from that of the cell surface, and the terminal galactose epitope is highly represented. These observations validate why LX-2 cell surfaces and BSG can serve as ligands for galectin-3. Although BSG has not previously been shown to display terminal galactosides (51, 52), such differences may reflect the natural abundance of glycan biosynthetic enzymes and sugar nucleotide precursors between different tissues. Human BSG naturally occurs in four natural isoforms (53), and our proteomics data suggest that either BSG-1 or BSG-2 is expressed in LX-2s. Endogenous BSG-2 differs from the canonical BSG-1 by missing regions 24 to 139 (SI Appendix, Fig. S17) and similarly bears three N-linked glycosylation sites. The recombinant BSG glycoprotein used for the in vitro binding experiments has only two sites for N-glycosylation (SI Appendix, Fig. S17), suggesting that tighter interactions may be achieved by the endogenous glycoprotein.
Given the importance of galectin-3 in regulating immunity, we also applied the PX-Gal3 fusion protein exogenously to PBMCs and identified glycan-mediated interactors. Of note, these proteins are highly important in both the innate and adaptive immune responses. For example, CD3G and CD3E are two of four subunits of the CD3 complex present on T cell surfaces that mediates T cell activation (54, 55). TLR-2 is one of a family of TLRs that are key for the innate immune response to various microbial agents, responsible for recognizing pathogen-associated molecular patterns (56–58). TLR-2 has also been associated with formation of a complex with CD36, another glycoprotein that we captured with high enrichment and with successful competition (59).
We have demonstrated that proximity labeling is a powerful approach to capturing glycan-mediated binding events in live cells. We show that this method is generally applicable in a cell line and freshly isolated primary immune cells. We expect that it could also be extended to studies in other biological samples, such as tissues, and could also be used to track the spatiotemporal dynamics of galectin-3 interactions (60) or to visualize interactions using electron microscopy (21). Empowered by this approach, we now have generated a priority list of proteins that will be the subject of future work to further reveal the “professional glycoprotein ligands” (18) required for the galectin-3-mediated functional activation of HSCs and regulation of immunity.
Conclusions
GBP–glycan interactions choreograph many biological events, yet they are often interrogated in nonnative environments without the identification of the glycan-bearing glycoprotein receptors. Herein, we used a proximity labeling method consisting of fusion proteins of galectin-3 and the APEX2 enzyme to map glycan-mediated galectin-3 interactions in live cells. This method is sensitive, enabling detection of galectin-3 binding at nanomolar concentrations, and we have produced an inventory of proteins that occur in complex with galectin-3 and confirmed some of these proteins as direct glycan-mediated glycoprotein receptors. Consequently, our results provide higher-resolution insight into the glycan and protein determinants that govern galectin-3 interactions in HSCs and with individual glycoproteins. Critically, although we have mapped the galectin-3 interactome with live cells, the functional consequences imparted by these discrete interactions remain unknown. Informed by our results, we are now poised to formulate new hypotheses that define the contributions of individual proteins and glycosylation sites. Importantly, we expect that the proximity labeling approach will be generally applicable to other GBPs and glycoproteins, and that it will be a powerful tool to advance our understanding of glycan-mediated cell biology.
Materials and Methods
Growth and Maintenance of HSCs.
LX-2 human HSCs (61) (Millipore) were cultured in DMEM supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. For fluorescence microscopy imaging experiments, cells were grown on plastic 24-well culture plates. To improve the adherence of LX-2 cells, dishes were precoated with poly-d-lysine and washed with Dulbecco's phosphate-buffered saline (DPBS) before cell plating.
In Situ Proximity Tagging in Live LX-2 Cells and Imaging.
The LX-2 cells were plated (precoated with poly-d-lysine) at a density of 1.5 × 105 cells per well on a plastic 24-well plate overnight at 37 °C in 5% CO2. The next day, the cells were gently washed with DPBS and then incubated with DMEM supplemented with the fusion protein of interest for defined periods (usually 30 min, unless indicated otherwise) at 37 °C in 5% CO2. After two washes with DPBS to remove unbound proteins, labeling was initiated by replacing the medium with prewarmed 10% FBS/DMEM containing 500 μM biotin-phenol. This solution was further incubated with the cells at 37 °C in 5% CO2 for 30 min. H2O2 was then added to each well to achieve a final concentration of 1 mM, and the plate was gently agitated for 1 min at room temperature (RT). After aspirating the solution, the reaction was then quenched by washing three times with quencher solution (5 mM Trolox, 10 mM sodium ascorbate, and 10 mM sodium azide in DPBS). Cells used for fluorescence imaging were then fixed with 4% p-formaldehyde in PBS at RT for 10 min and washed with PBS. To probe for biotinylated interactors, Cy5-streptavidin was used in PBS with Tween 20 for incubation for 1 h in the dark at RT. Imaging was performed on a benchtop EVOS M5000 fluorescence imager (Thermo Fisher Scientific). Experiments were repeated for at least two independent biological replicates.
Preparation and Enrichment of Sample Lysates for Proteomics.
LX-2 cells were plated onto a single 15-cm2 dish per condition. Cells were plated (precoated with poly-d-lysine) at a density of 16 to 20 × 106 cells per plate overnight at 37 °C in 5% CO2. The next day, the cells were gently washed with DPBS and incubated with DMEM supplemented with the fusion protein of interest for defined periods (usually 30 min, unless indicated otherwise) at 37 °C in 5% CO2. After two washes with DPBS to remove unbound proteins, labeling was initiated by replacing the medium with prewarmed 10% FBS/DMEM containing biotin-phenol (500 µM). This solution was further incubated with the cells at 37 °C in 5% CO2 for 30 min. H2O2 was then added to each well to achieve a final concentration of 1 mM, and the plate was gently agitated for 1 min at RT. After aspirating the solution, the reaction was then quenched by washing three times with quencher solution (5 mM Trolox, 10 mM sodium ascorbate, and 10 mM sodium azide in DPBS). Cells were then washed with DPBS, scraped and transferred to 15-mL Falcon tubes, pelleted, and stored at −80 °C until the next step.
Cell pellets were resuspended in 300 to 400 μL of DPBS and lysed by sonication (15 ms ON, 40 ms OFF, 1 s total ON, 15% amplitude ×2). Protein concentrations were normalized (1.5 to 2 mg/mL; final volume of 500 μL with PBS) using the Lowry Protein Assay (Pierce). Each lysate was transferred to a new 15-mL Falcon tube, and 2.5 mL of a cold mixture of MeOH:CHCl3 (4:1) was added to the lysate, followed by 1 mL of cold DPBS. The resulting mixture was vortexed and centrifuged (5,000 rpm, 10 min, 4 °C). The organic and aqueous layers were aspirated, and the remaining protein disk was further washed via sonication in 2 mL of cold MeOH:CHCl3 (4:1) and pelleted by centrifugation (5,000 rpm, 10 min, 4 °C). The protein pellet was aspirated and dissolved in 500 μL of freshly prepared proteomics-grade urea (6 M in DPBS) with 10 µL of 10% SDS by sonication. Disulfide bonds were reduced by adding 50 µL of freshly prepared 1:1 solution of TCEP (200 mM in DPBS) and K2CO3 (600 mM in DPBS) for 30 min at 37 °C on a shaker. Free thiols were alkylated by adding 70 µL of freshly prepared iodoacetamide (400 mM in DPBS) at RT in the dark. To each solution, 130 µL of 10% SDS in DPBS was added, and each sample was diluted with DPBS (5.5 mL) and incubated with preequilibrated streptavidin-agarose beads (100 µL of 50% slurry; Pierce) for 1.5 h at RT while rotating. The streptavidin beads were pelleted by centrifugation (2,000 rpm, 2 min, 4 °C) and sequentially washed with 0.2% SDS in DPBS (1 × 5 mL), DPBS (2 × 5 mL), and 200 mM EPPS [4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid], pH 8.4 (1 × 5 mL). The beads were transferred into LoBind microcentrifuge tubes (Eppendorf; 022431081), and bound proteins were digested for ∼14 h at 37 °C in 200 µL of trypsin premix solution containing sequence-grade trypsin (2 mg; Promega), urea (0.5 M in 200 mM EPPS, pH 8.4), and calcium chloride (1 mM in H2O). The beads were pelleted by centrifugation (2,000 rpm, 2 min, 4 °C) and the supernatant containing the digested peptides were transferred to new LoBind microcentrifuge tubes. Each digested sample was labeled with TMT (Thermo Fisher Scientific; 90406). In general, for each sample, 8 µL of the 20 µg/μL stock of TMT reagent was added along with dry MS-grade acetonitrile to achieve a final acetonitrile concentration of ∼30% (vol/vol), followed by incubation at RT for 1 h. The reaction was quenched by adding 6 µL of hydroxylamine for 15 min, followed by acidification with the addition of 5 µL of formic acid. Each TMT-labeled sample was dried via vacuum centrifugation, and the samples were combined by redissolving one sample with 400 µL of buffer A (5% MeCN in H2O, 0.1% formic acid) and transferring the solution into each sample tube until all samples were redissolved (final volume ∼600 µL). pH was adjusted to ∼3 with formic acid. The combined sample was desalted using two C-18 columns (Thermo Fisher Scientific; 89870) according to the manufacturer’s instructions. The combined sample was dried via vacuum centrifugation and stored in the −80 °C until ready for injection.
LC-MS Analysis for Proteomics.
Here, 10-plex samples were redissolved in 20 μL of buffer A (99.9% H2O, 0.1% formic acid) for MS analysis. In brief, 3 µL of each sample was loaded onto an Acclaim PepMap 100 precolumn (75 µM × 2 mm) and eluted on an Acclaim PepMap RSLC analytical column (75 µM × 15 cm) using the UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Buffer A (0.1% formic acid in H2O) and buffer B (0.1% formic acid in acetonitrile) were used to establish the 220-min gradient comprising 10 min of 2% buffer B, 192 min of 2% to 30% buffer B, 5 min of 30% to 60% buffer B, 1 min of 60% to 95% buffer B, 5 min of 95% buffer B, and 1 min of 95% to 2% buffer B, followed by reequilibration at 2% buffer B for 6 min. The flow rate was 0.3 mL/min at 35 °C.
Peptides were then analyzed with a Thermo Fisher Scientific Orbitrap Fusion Lumos proteomic mass spectrometer in a data-dependent manner with a cycle time of 3 s. Voltage applied to the nano-LC electrospray ionization source was 2.0 kV. MS1 spectra were acquired at a resolution of 120,000 with an automatic gain control (AGC) target value of 1 × 106 ions and a maximum injection time of 50 ms (dynamic exclusion enabled; repeat count 1, duration 20s). The scan range was limited from 375 to 1,500 m/z. Peptide fragmentation for MS2 was performed via collision-induced dissociation (CID), quadrupole ion trap analysis, AGC 1.8E4, CID collision energy 30%, maximum injection time 120 ms, and isolation window at 1.6. The MS3 precursor was fragmented via high-energy collision-induced dissociation (HCD) with a collision energy of 65%. Synchronous precursor selection (SPS) was enabled to include up to 10 MS2 fragment ions for the MS3 spectrum. The detector type was an Orbitrap with a resolution of 50,000, an AGC target value of 1.5 × 105, and a maximum injection time of 120 ms.
Proteomics Data Analysis.
Data processing was performed using Proteome Discoverer 2.4 software (Thermo Fisher Scientific), and peptide sequences were determined by matching protein databases with the acquired fragmentation pattern using the SEQUEST HT algorithm. The precursor mass tolerance was set to 10 ppm, and fragment ion mass tolerance was set to 0.6 Da. One missed cleavage site of trypsin was allowed. Carbamidomethyl (C, +57.02146) and TMT-tag (K and N-terminal, +229.1629) were specified as static modifications. Oxidation (M, +15.994915) was specified as a variable modification. All spectra were searched against the proteome database of Homo sapiens (42,358 sequences) using a target false discovery rate of 1% (Percolator). Identified proteins were also required to have at least three unique peptides. MS3 peptide quantitation was performed with reporter ion mass tolerence set to 20 ppm. Statistical analysis was performed with GraphPad Prism 8. TMT ratios obtained from Proteome Discoverer were transformed with log2 (x), and P values were obtained using t tests with two biological replicates. Quantitative data are listed in SI Appendix, Table S1. Normalized TMT values for each protein were calculated as the ratio of the raw TMT value to the value of PCCA (propionyl-CoA carboxylase alpha chain), an endogenously biotinylated protein present in all experiments after biotin pull-down (60). The mass spectrometry proteomics datasets have been deposited to the ProteomeXchange Consortium via the PRIDE (62) partner repository with the dataset identifier PXD021622.
Descriptions of additional experimental procedures are provided in SI Appendix.
Supplementary Material
Acknowledgments
E.J. is supported by a Skaggs Graduate Fellowship, enabled by the Henry and Jennifer Luttrell Foundation. M.L.H. is supported by an NIH K99/R00 Pathway to Independence Award (R00HD0292-03). The M.L.H. lab is supported by startup funds from Scripps Research and the Joe W. and Dorothy Dorsett Brown Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009206117/-/DCSupplemental.
Data Availability.
All MS raw files have been deposited to the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with the identifier PXD021622.
References
- 1.Di Lella S.et al., When galectins recognize glycans: From biochemistry to physiology and back again. Biochemistry 50, 7842–7857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson N. C.et al., Galectin-3 expression and secretion links macrophages to the promotion of renal fibrosis. Am. J. Pathol. 172, 288–298 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson N. C. M.et al., Galectin-3 regulates myofibroblast activation and hepatic fibrosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5060–5065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabinovich G. A., Toscano M. A., Turning “sweet” on immunity: Galectin-glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol. 9, 338–352 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Díaz-Alvarez L., Ortega E., The many roles of galectin-3, a multifaceted molecule, in innate immune responses against pathogens. Mediators Inflamm. 2017, 9247574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang R. Y., Hsu D. K., Liu F. T., Expression of galectin-3 modulates T-cell growth and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 93, 6737–6742 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung A. W.et al., Galectin-3 regulates the innate immune response of human monocytes. J. Infect. Dis. 207, 947–956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rillahan C. D., Paulson J. C., Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cederfur C.et al., Glycoproteomic identification of galectin-3 and -8 ligands in bronchoalveolar lavage of mild asthmatics and healthy subjects. Biochim. Biophys. Acta 1820, 1429–1436 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Priglinger C. S.et al., Galectin-3 induces clustering of CD147 and integrin-β1 transmembrane glycoprotein receptors on the RPE cell surface. PLoS One 8, e70011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M., Tian F., Ying W., Qian X., Quantitative proteomics reveal the anti-tumour mechanism of the carbohydrate recognition domain of galectin-3 in hepatocellular carcinoma. Sci. Rep. 7, 5189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dange M. C.et al., Mass spectrometry-based identification of galectin-3 interacting proteins potentially involved in lung melanoma metastasis. Mol. Biosyst. 13, 2303–2309 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Lakshminarayan R.et al., Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat. Cell Biol. 16, 595–606 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Obermann J.et al., Proteome-wide identification of glycosylation-dependent interactors of galectin-1 and galectin-3 on mesenchymal retinal pigment epithelial (RPE) cells. Mol. Cell. Proteomics 16, 1528–1546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L.et al., Cross-platform comparison of glycan microarray formats. Glycobiology 24, 507–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briard J. G., Jiang H., Moremen K. W., Macauley M. S., Wu P., Cell-based glycan arrays for probing glycan-glycan binding protein interactions. Nat. Commun. 9, 880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramya T. N.et al., In situ trans ligands of CD22 identified by glycan-protein photocrosslinking-enabled proteomics. Mol. Cell. Proteomics 9, 1339–1351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malaker S. A.et al., The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. U.S.A. 116, 7278–7287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trinkle-Mulcahy L., Recent advances in proximity-based labeling methods for interactome mapping. F1000 Res. 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam S. S.et al., Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung V.et al., Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepur A., Salomonsson E., Nilsson U. J., Leffler H., Ligand-induced galectin-3 protein self-association. J. Biol. Chem. 287, 21751–21756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieminen J., Kuno A., Hirabayashi J., Sato S., Visualization of galectin-3 oligomerization on the surface of neutrophils and endothelial cells using fluorescence resonance energy transfer. J. Biol. Chem. 282, 1374–1383 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Rhee H. W.et al., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Böcker S., Elling L., Binding characteristics of galectin-3 fusion proteins. Glycobiology 27, 457–468 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Gao X.et al., The two endocytic pathways mediated by the carbohydrate recognition domain and regulated by the collagen-like domain of galectin-3 in vascular endothelial cells. PLoS One 7, e52430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomonsson E.et al., Mutational tuning of galectin-3 specificity and biological function. J. Biol. Chem. 285, 35079–35091 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ting L., Rad R., Gygi S. P., Haas W., MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAlister G. C.et al., MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaine T.et al., Galectin-3-binding glycomimetics that strongly reduce bleomycin-induced lung fibrosis and modulate intracellular glycan recognition. ChemBioChem 17, 1759–1770 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Geiger T., Wehner A., Schaab C., Cox J., Mann M., Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritsch K.et al., Galectin-3 interacts with components of the nuclear ribonucleoprotein complex. BMC Cancer 16, 502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haudek K. C.et al., Dynamics of galectin-3 in the nucleus and cytoplasm. Biochim. Biophys. Acta 1800, 181–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori Y.et al., Binding of galectin-3, a β-galactoside-binding lectin, to MUC1 protein enhances phosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2) and akt, promoting tumor cell malignancy. J. Biol. Chem. 290, 26125–26140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talaga M. L.et al., Multitasking human lectin galectin-3 interacts with sulfated glycosaminoglycans and chondroitin sulfate proteoglycans. Biochemistry 55, 4541–4551 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Nielsen M. I.et al., Galectin binding to cells and glycoproteins with genetically modified glycosylation reveals galectin-glycan specificities in a natural context. J. Biol. Chem. 293, 20249–20262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patnaik S. K.et al., Complex N-glycans are the major ligands for galectin-1, -3, and -8 on Chinese hamster ovary cells. Glycobiology 16, 305–317 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Song X.et al., Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16, 36–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou S., Wooding K. M., Mechref Y., Analysis of permethylated glycan by liquid chromatography (LC) and mass spectrometry (MS). Methods Mol. Biol. 1503, 83–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang L.et al., Identification of Siglec ligands using a proximity labeling method. J. Proteome Res. 16, 3929–3941 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Hopkins C., Gibson A., Stinchcombe J., Futter C., Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 327, 35–45 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Li H.-Y.et al., Activation of TGF-β1-CD147 positive feedback loop in hepatic stellate cells promotes liver fibrosis. Sci. Rep. 5, 16552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das A.et al., Ephrin B2/EphB4 pathway in hepatic stellate cells stimulates Erk-dependent VEGF production and sinusoidal endothelial cell recruitment. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G908–G915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi W.-P.et al., CD147 promotes CXCL1 expression and modulates liver fibrogenesis. Int. J. Mol. Sci. 19, 1145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charrin S., Jouannet S., Boucheix C., Rubinstein E., Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Stipp C. S., Kolesnikova T. V., Hemler M. E., EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276, 40545–40554 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Liang Y.et al., Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J. Biol. Chem. 289, 32526–32537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu M. H.et al., Glycosylation-dependent galectin-1/neuropilin-1 interactions promote liver fibrosis through activation of TGF-β- and PDGF-like signals in hepatic stellate cells. Sci. Rep. 7, 11006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Y.et al., Alteration of protein glycosylation in human hepatic stellate cells activated with transforming growth factor-β1. J. Proteomics 75, 4114–4123 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Huang W.et al., Modulation of CD147-induced matrix metalloproteinase activity: Role of CD147 N-glycosylation. Biochem. J. 449, 437–448 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Yang L., Nyalwidhe J. O., Guo S., Drake R. R., Semmes O. J., Targeted identification of metastasis-associated cell-surface sialoglycoproteins in prostate cancer. Mol. Cell Proteomics 10, M110 007294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao C. G.et al., Characterization of basigin isoforms and the inhibitory function of basigin-3 in human hepatocellular carcinoma proliferation and invasion. Mol. Cell. Biol. 31, 2591–2604 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dietrich J.et al., Role of CD3 gamma in T cell receptor assembly. J. Cell Biol. 132, 299–310 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dietrich J., Hou X., Wegener A. M., Geisler C., CD3 gamma contains a phosphoserine-dependent di-leucine motif involved in down-regulation of the T cell receptor. EMBO J. 13, 2156–2166 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bulut Y., Faure E., Thomas L., Equils O., Arditi M., Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: Role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167, 987–994 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Aliprantis A. O.et al., Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science 285, 736–739 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Brightbill H. D.et al., Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285, 732–736 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Triantafilou M.et al., Membrane sorting of Toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J. Biol. Chem. 281, 31002–31011 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Paek J.et al., Multidimensional tracking of GPCR signaling via peroxidase-catalyzed proximity labeling. Cell 169, 338–349.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu L.et al., Human hepatic stellate cell lines, LX-1 and LX-2: New tools for analysis of hepatic fibrosis. Gut 54, 142–151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Riverol Y.et al., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MS raw files have been deposited to the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with the identifier PXD021622.