Significance
Phytoplankton are major primary producers in the marine environment that excrete a wide range of metabolites. These exudates support the growth of surrounding bacteria that in turn provide phytoplankton cells with resources and growth cofactors to proliferate. Unlike multicellular eukaryotes with dedicated structures housing microbiomes, mechanisms enabling unicellular eukaryotic phytoplankton hosts to modulate potential symbionts and opportunists within a natural microbial community are unknown. Exposure of a host phytoplankton cell to its natural microbial community triggers major transcriptional and metabolic reprogramming to release unique secondary metabolites that selectively enable growth and attachment of symbiotic taxa while simultaneously suppressing the colonization of nonsymbiont bacteria. These results suggest strong and highly selective microbiome-modulating strategies shared across the unicellular and multicellular eukaryotic lineages.
Keywords: phycosphere, microbiomes, diatoms, phytoplankton–bacteria interactions, secondary metabolism
Abstract
Unicellular eukaryotic phytoplankton, such as diatoms, rely on microbial communities for survival despite lacking specialized compartments to house microbiomes (e.g., animal gut). Microbial communities have been widely shown to benefit from diatom excretions that accumulate within the microenvironment surrounding phytoplankton cells, known as the phycosphere. However, mechanisms that enable diatoms and other unicellular eukaryotes to nurture specific microbiomes by fostering beneficial bacteria and repelling harmful ones are mostly unknown. We hypothesized that diatom exudates may tune microbial communities and employed an integrated multiomics approach using the ubiquitous diatom Asterionellopsis glacialis to reveal how it modulates its naturally associated bacteria. We show that A. glacialis reprograms its transcriptional and metabolic profiles in response to bacteria to secrete a suite of central metabolites and two unusual secondary metabolites, rosmarinic acid and azelaic acid. While central metabolites are utilized by potential bacterial symbionts and opportunists alike, rosmarinic acid promotes attachment of beneficial bacteria to the diatom and simultaneously suppresses the attachment of opportunists. Similarly, azelaic acid enhances growth of beneficial bacteria while simultaneously inhibiting growth of opportunistic ones. We further show that the bacterial response to azelaic acid is numerically rare but globally distributed in the world’s oceans and taxonomically restricted to a handful of bacterial genera. Our results demonstrate the innate ability of an important unicellular eukaryotic group to modulate select bacteria in their microbial consortia, similar to higher eukaryotes, using unique secondary metabolites that regulate bacterial growth and behavior inversely across different bacterial populations.
Large swaths of eukaryotic lineages possess associated microbiomes that play central roles in supporting host survival and ecological success (1). Several biotic and abiotic factors have been shown to drive microbiome assembly and modulation in special compartments and organelles of multicellular eukaryotes such as squid light organs (2), coral skeletons (3), mammalian guts (4), and roots and leaves of terrestrial plants (5). In contrast, unicellular eukaryotes such as diatoms lack developmental features that can harbor microbes, yet rely heavily on essential bacterial growth factors (6–8) to proliferate and thrive in their environment. Diatoms are ubiquitous primary producers in aquatic environments that account for ∼40% of marine carbon export (9). They actively release a variety of organic and inorganic compounds (10, 11) into a diffusive boundary layer that surrounds individual cells, known as the phycosphere (12). This physically sheltered microscale region is highly enriched in dissolved organic matter (DOM) and serves as the interface for diatom–bacteria associations (7, 13). Indeed, bacteria have been shown to heavily rely on phycosphere DOM to support their growth (14, 15) and must use motility, chemotaxis, and/or attachment to chase and colonize the phycosphere (16). Recent research has shown that a variety of interactions spanning mutualism, commensalism, and parasitism occur between diatoms and specific groups of bacteria (7, 17–19). As single cells floating in aquatic environments, diatoms encounter beneficial (hereafter symbiotic) and opportunistic and algicidal (hereafter opportunistic) bacteria. However, the mechanisms that enable diatoms and other phytoplankton species to actively modulate incoming microbes to evade opportunistic bacteria and nurture symbiotic ones are mostly unknown. Due to the challenges of investigating phytoplankton–bacteria interactions in the field, most studies to date have relied on laboratory-controlled coculture systems between phytoplankton and a single bacterium, an approach that has enriched our knowledge of phycosphere interactions but one that does not adequately mimic the microbial complexity in natural phycospheres. Here, we apply a holistic approach to a natural system derived from the environment by using multiomics to show that DOM secretions by the globally widespread diatom Asterionellopsis glacialis (20) modulate select bacterial behavior and growth. We hypothesize that diatom cells must adopt specific mechanisms to promote association with potentially beneficial symbionts while repelling opportunists to offset the lack of specialized compartments to house microbiomes. To this end, A. glacialis strain A3 was cultivated from its natural environment, then freed of its associated bacteria and left to acclimate until the time of reseeding, marked by the reintroduction of its natural bacterial consortium to the diatom. Transcriptional and metabolomic changes in both the diatom and the bacterial consortium at different time points were assessed, and potential representative symbiotic and opportunistic bacteria were cultivated from the consortium to further confirm hypotheses generated from multiomics experiments.
Results
To examine the interactions between the diatom and its bacterial consortium, we isolated A. glacialis A3 along with its natural microbial community (xenic A. glacialis), then cured it of bacteria using a suite of antibiotics to make it axenic, as described previously (21) (SI Appendix, Supplementary Methods). After ∼170 generations of acclimating the axenic A. glacialis A3 culture to the absence of bacteria, the true bacterial consortium composition was harvested by filtration from xenic cultures immediately before the reseeding experiment. At the time of reseeding, one portion of this natural bacterial community was added to the acclimated axenic A. glacialis A3 culture, generating a reseeded A. glacialis A3 treatment to investigate the response of the diatom to bacterial exposure and the response of bacteria to diatom exudates (SI Appendix, Fig. S1). Two additional portions of the bacterial consortium were collected and used for shotgun metagenomics and metatranscriptomics (bacterial consortium control at 0.5 h). Diatom transcriptomic samples (at 0.5 and 24 h) were collected from the control axenic A. glacialis cultures and reseeded A. glacialis treatments. In addition, samples for metabolomics at two early (0.5 and 4 h) and two late (24 and 48 h) time points were collected (SI Appendix, Supplementary Methods and Fig. S1).
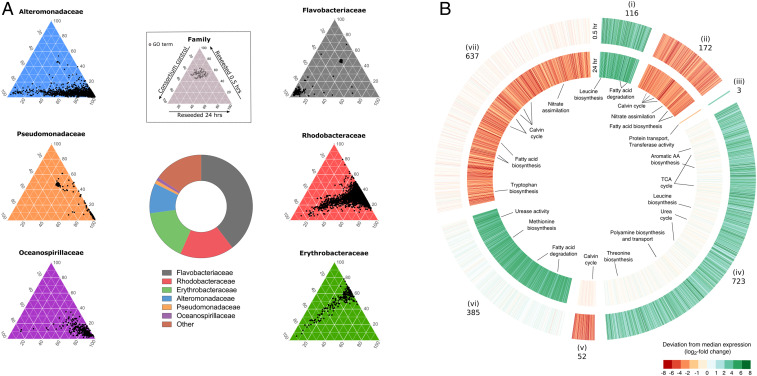
The composition of the microbial consortium collected at the time of reseeding showed the dominance of six bacterial families, with Flavobacteriaceae comprising 38.9% of all metagenomic reads, followed by Rhodobacteraceae (16.6%), Erythrobacteraceae (16%), Alteromonadaceae (9.28%), Pseudomonadaceae (1.07%), and Oceanospirillaceae (1.03%; Fig. 1A). To uncover how these families responded to diatom exudates, we assembled 10 near-complete bacterial genomes from the microbial consortium metagenome. The metagenomically assembled genomes (MAGs) belonged to most major families in the consortium, including Flavobacteriaceae (MAG9), Rhodobacteraceae (MAG3, MAG5, MAG6, and MAG11), Erythrobacteraceae (MAG10), Alteromonadaceae (MAG4 and MAG12), Oceanospirillaceae (MAG8), and Halomonadaceae (MAG13; SI Appendix, Table S1). Mapping metatranscriptome reads to all MAGs showed that the four Rhodobacteraceae MAGs recruited ∼41% of mRNA reads and were responsible for the majority of differentially expressed genes (SI Appendix, Table S2) at both early and late time points of reseeded samples relative to controls, despite representing ∼10% of the bacterial consortium metagenome (SI Appendix, Table S1).
Fig. 1.
Major reprogramming of transcriptional responses of A. glacialis A3 and Roseobacters in response to reseeding. (A) Central donut plot depicts relative abundances of the top six bacterial families in the consortium metagenomic dataset. (Inset) Key for color-coded ternary plots represents transcriptional responses of the bacterial families before (consortium 0.5 h control) and after (reseeded 0.5 and 24 h) reseeding based on biological triplicates. Each dot depicts a unique Gene Ontology (GO) annotation associated with transcripts from each of the six major families in the metatranscriptome. The position of each dot corresponds to the percent contribution of the sample (consortium control, reseeded 0.5 h, and reseeded 24 h) relative to the total normalized abundance of transcripts annotated with the same GO term in copies per million (cpm). (B) Differentially expressed (DE) genes in reseeded A. glacialis A3 after 0.5 (outer circle) and 24 (inner circle) hours relative to axenic controls. Genes are organized into seven clusters (i to vii) based on their expression pattern at the two time points. Numbers indicate the number of DE genes in each cluster. Opaque clusters indicate genes that are not DE. TCA, tricarboxylic acid; AA, amino acid.
We examined the consortium metatranscriptome and confirmed that the Rhodobacteraceae (hereafter Roseobacters) exhibited the most transcriptionally rapid and diverse response within 0.5 h of reintroduction to the diatom, as evidenced by the large number of Gene Ontology terms associated with Roseobacter genes expressed after reseeding. In stark contrast, other bacterial families either displayed no significant response to reseeding (Flavobacteriaceae), displayed a decreasing response from 0.5 to 24 h (Erythrobacteraceae), or were responsive only at 24 h after reseeding (Alteromonadaceae, Pseudomonadaceae, and Oceanospirillaceae) relative to the consortium control (Fig. 1A).
The A. glacialis A3 transcriptome showed a major reprogramming of its transcriptional profile to differentially express ∼14% of its protein-coding genes relative to axenic controls, coupled with temporal shifts in expression patterns (Fig. 1B). In response to consortium reseeding, transcripts for amino acid biosynthesis and fatty acid degradation were consistently up-regulated, while nitrate assimilation, photosynthesis, and carbon fixation were down-regulated throughout the reseeding experiment. At 0.5 h only, differentially up-regulated A. glacialis A3 transcripts included those for spermidine biosynthesis and transport and the tricarboxylic acid (TCA) and urea cycles, while transcripts for methionine biosynthesis and urease activity were differentially up-regulated at 24 h only. We also observed down-regulated transcripts involved in the Calvin cycle at both 0.5 h and 24 h and tryptophan biosynthesis-related transcripts down-regulated at 24 h only (Fig. 1B and SI Appendix, Table S3).
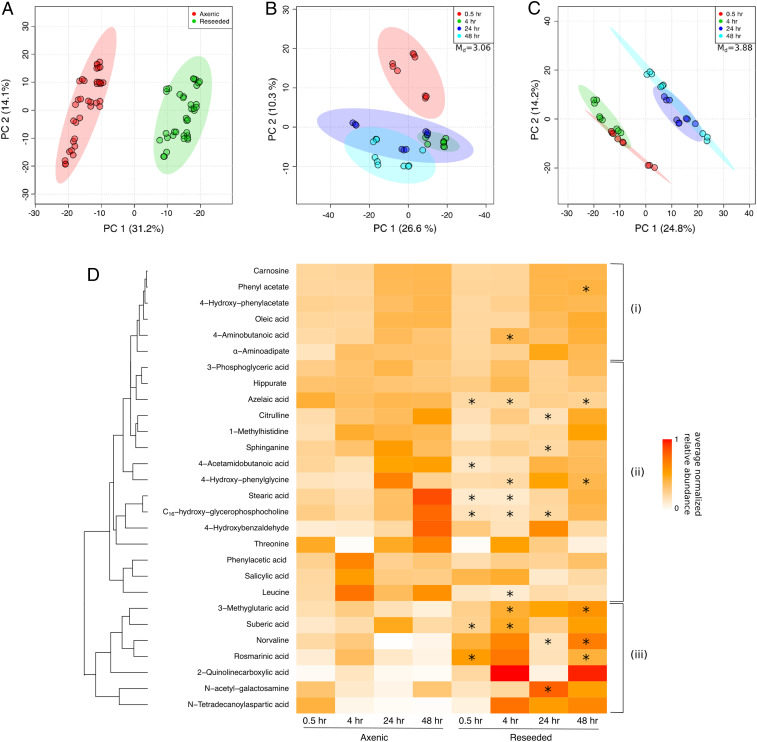
The diatom and Roseobacter transcriptional responses were coupled to major changes in the exometabolome. Exometabolomes sampled at two early and two late time points after reseeding (SI Appendix, Fig. S1B) were analyzed using a quadrupole time-of-flight mass spectrometer (Dataset S1). The DOM landscape varied between axenic and reseeded samples (Fig. 2A). Interestingly, based on Mahalanobis distances (Md), the DOM composition at early time points was significantly more distinct from late time points in the reseeded samples (Md = 3.88) than in axenic controls (Md = 3.06; Fig. 2 B and C), suggesting that DOM is temporally highly dynamic in response to consortium reseeding, similar to the diatom transcriptome. Analysis of the DOM elemental composition of extracted metabolites in axenic and reseeded samples using Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS; Datasets S2 and S3) showed ∼50% decrease in abundance of dissolved organic nitrogen (DON) in reseeded samples relative to axenic controls (SI Appendix, Fig. S2).
Fig. 2.
SPE-extracted DOM profile is highly influenced by reseeding. (A–C) Principal components analysis (PCA) plots of axenic and reseeded untargeted exometabolome samples. PCA was performed based on Mahalanobis distances (Md), comparing 1,237 SPE-extracted molecules between (A) axenic vs. reseeded samples and (B and C) early (0.5 and 4 h) and late (24 and 48 h) time points for (B) axenic and (C) reseeded conditions. Circles represent technical replicates (n = 3) of three biological replicates. (D) Euclidean hierarchical clustering of 28 exometabolites (SI Appendix, Table S4) identified in axenic and reseeded samples and confirmed using a library of in-house chemical standards. Colors represent average normalized relative abundance of each metabolite. Time points marked with an asterisk indicates a significant change in relative abundance across reseeded and axenic samples as determined with a Student’s t test (Bonferroni-adjusted P < 0.05). (i) Prospective refractory diatom metabolites, (ii) diatom metabolites possibly taken up by the consortium, and (iii) diatom metabolites with a potential signaling role.
The identity of 28 metabolites common in axenic and reseeded diatom samples was confirmed (Fig. 2D and SI Appendix, Table S4) using an in-house chemical library of >660 molecules (SI Appendix, Supplementary Methods), indicating these metabolites are secreted by the diatom. Most metabolites showed increasing relative abundance in axenic and reseeded samples as a function of time, but a markedly lower overall accumulation in reseeded samples relative to axenic controls (e.g., leucine, threonine, 3-phosphoglycerate), suggesting either diatom down-regulation of the biosynthesis of these molecules in reseeded samples and/or bacterial uptake in reseeded samples. Bacterial uptake was corroborated by the transcriptional response of the diatom to reseeding, which showed up-regulation of metabolite-specific biosynthesis genes and a concomitant up-regulation of specific Roseobacter transporters that take up these metabolites (SI Appendix, Table S5). Seven metabolites showed increases in relative abundance in reseeded samples compared to axenic controls (e.g., rosmarinic acid), suggesting either a signaling role for these diatom metabolites or coproduction by bacteria (Fig. 2D).
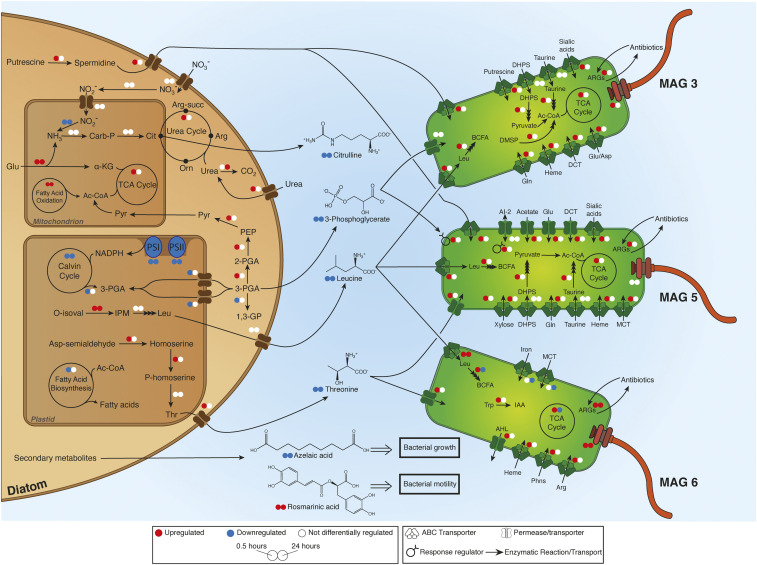
Based on the rapid response of the Roseobacters to diatom exudates, we built a conceptual model of diatom–Roseobacter interactions using the differential gene expression of A. glacialis A3, three Roseobacter MAGs (MAG3, MAG5, and MAG6), and identified exometabolites (Fig. 3 and SI Appendix, Tables S3–S5). In response to reseeding, the diatom up-regulated genes involved in the biosynthesis of spermidine (log2 fold-change = 3.8, P = 0.09) and its transport (log2 fold-change = 2.7, P = 0.04) at 0.5 h. Concomitantly, transcripts for spermidine uptake were up-regulated in both MAG3 (log2 fold-change = 5.5, P = 0.09) and MAG5 (log2 fold-change = 6.2, P = 0.08) at 0.5 h. The diatom increased transcription of glutamate dehydrogenase at 0.5 h (log2 fold-change = 2.2, P = 0.079) and 24 h (log2 fold-change = 5.4, P = 0.006) to fuel the TCA cycle and/or the urea cycle, both of which were up-regulated, by generating α-ketoglutarate and ammonia, respectively. Citrulline, a urea cycle intermediate released into the media, showed a differential decrease in abundance in reseeded samples versus axenic samples (P = 0.04 at 24 h; Fig. 2D), suggesting bacterial uptake. The diatom down-regulated homologs of phosphoglycerate kinase (22) involved in the conversion of 3-phosphoglycerate (3-PGA) to glycerate 1,3-diphosphate in the plastid (log2 fold-change = −1.6, P = 0.06) and cytoplasm (log2 fold-change = −5.3, P = 0.06). The 3-PGA transporters localized in the plastid were also down-regulated at 0.5 h after reseeding (log2 fold-change = −7.0 and −4.0, P = 0.002 and P = 0.007, respectively), suggesting reduced transport of 3-PGA across the plastid membrane and a buildup of 3-PGA in the cytoplasm. The 3-PGA was released into the media and was presumably taken up by bacteria. Transporters for 3-PGA were not differentially expressed in MAG3, while a 3-PGA response regulator was up-regulated in MAG5 at 0.5 h (log2 fold-change = 5.3, P = 0.097). Diatom transcripts involved in the biosynthesis of threonine were up-regulated at 0.5 h (log2 fold-change = 2.7, P = 0.02), and transcripts involved in the biosynthesis of leucine were up-regulated at both 0.5 h (log2 fold-change = 5.2, P = 0.001) and 24 h (log2 fold-change = 5.2, P = 0.0003). Putative neutral amino acid transporters that are either up-regulated at 0.5 h (log2 fold-change = 4.8, P = 0.03) or not differentially expressed suggest the diatom may be secreting the amino acids threonine and/or leucine. The secretion of threonine and leucine (P = 0.003 at 4 h; Fig. 2D) into the media was concomitant with an up-regulation of their transporters and subsequent assimilation of leucine into branched-chain fatty acid biosynthesis in the three Roseobacter MAGs (Fig. 3 and SI Appendix, Tables S3 and S5).
Fig. 3.
A. glacialis A3 preferentially promotes growth of Roseobacters by secreting specific metabolites that influence bacterial growth and behavior. Summary of diatom–bacteria interactions highlighting the metabolic exchanges and differentially expressed (DE) genes in A. glacialis A3 and three Roseobacter MAGs. Small colored circles (red, up-regulation; blue, down-regulation; white, no DE) represent differential expression of genes/processes at 0.5 (Left) and 24 (Right) hours after reseeding. Differential expression of metabolic cycles indicates that at least one gene was DE in one direction while no other genes were DE in the opposite direction. A complete list of genes and expression values are in SI Appendix, Tables S3 and S5. Confirmed central and secondary molecules from the exometabolome (SI Appendix, Table S4) are shown between the cells, and their relative abundance is indicated by colored circles relative to axenic controls. Multiple stacked arrows indicate several enzymatic reactions. SAMamine, S-adenosylmethionineamine; Carb-P, carbamoylphosphate; Cit, citrulline; α-KG, α-ketoglutarate; Pyr, pyruvate; Arg-succ, argininosuccinate; Arg, arginine; Orn, ornithine; PEP, phosphoenolpyruvate; 3-PGA, 3-phosphoglycerate; 1,3-GP, glycerate 1,3-diphosphate; PS, photosystem genes; O-isoval, o-isovalerate; IPM, isopropylmalate; Asp, aspartate; Glu, glutamate; Gln, glutamine; Leu, leucine; Thr, threonine; Trp, tryptophan; BCFA, branched-chain fatty acids; DHPS, 2,3-dihydroxypropanesulfonate; ARGs, antibiotic resistance genes; DCT, dicarboxylate transporter; DMSP, dimethylsulfoniopropionate; AI-2, autoinducer-2; MCT, monocarboxylate 2-oxoacid transporter; Phns, phosphonates; AHL, acyl homoserine lactones; IAA, indole-3-acetate.
To confirm the ability of Roseobacters to utilize diatom metabolites, we isolated bacteria from the bacterial consortium and sequenced their genomes (SI Appendix, Supplementary Methods). Two isolates were identified as Roseobacter species (Sulfitobacter pseudonitzschiae F5 and Phaeobacter sp. F10) and one isolate as an Alteromonadaceae species (Alteromonas macleodii F12). The average nucleotide identities (ANIs) of A. macleodii F12 were 76.8% with MAG12 and 89.9% with MAG4, while the ANI of Phaeobacter sp. F10 and MAG6 was 99.9%, suggesting MAG6 represents the same species as Phaeobacter sp. F10. The average amino acid identity between S. pseudonitzschiae F5 and all Roseobacter MAGs ranged from 61% to 66%, suggesting that they group at the genus level (cutoff 60% [23]). Phylogenomic analysis of isolate genomes and MAGs further confirmed this finding, as it clustered Phaeobacter sp. F10 close to MAG6 (SI Appendix, Fig. S3 and Table S6), while A. macleodii F12 clustered within the A. macleodii clade (SI Appendix, Fig. S4 and Table S7). Metagenomic read recruitment analysis using the three bacterial genomes indicated that, while S. pseudonitzschiae F5 represented only 0.16% of metagenomic reads, Phaeobacter sp. F10 and A. macleodii F12 represented 11.41% and 2.82% of all metagenomic reads, respectively. In addition, mapping bacterial mRNA reads to the isolates showed that S. pseudonitzschiae F5 recruited 5.3% of mRNA reads, Phaeobacter sp. F10 34.1%, and A. macleodii F12 9.1%, suggesting these bacteria are major players in the microbial community from a transcriptional perspective. Subsequently, 16 diatom metabolites from Fig. 2D were used to test the ability of S. pseudonitzschiae F5 and A. macleodii F12 (classified as a potential symbiont and an opportunist, respectively, as discussed below) to utilize these metabolites as growth substrates. Despite the more rapid transcriptional responses of Roseobacters to reseeding (Fig. 1A), both bacterial isolates were able to use most of these central metabolites as growth substrates (SI Appendix, Fig. S5).
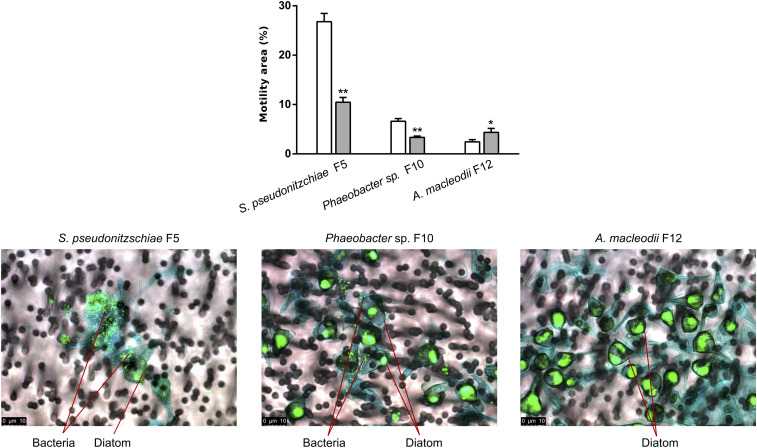
We sought to examine if diatom secondary metabolites can account for the advantage Roseobacters have over other bacterial families in the microbial consortium, like the Alteromonadaceae. Cell attachment is an important mechanism used by bacteria to remain in the phycosphere to enhance access to diatom exudates (24). The motility of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 was examined in the presence of a secondary metabolite not detected in diatoms before, rosmarinic acid, a common constituent of some terrestrial plants (25). Surprisingly, 2 μM rosmarinic acid significantly inhibited the motility of the potential symbionts S. pseudonitzschiae F5 and Phaeobacter sp. F10 and increased the motility of the potential opportunist A. macleodii F12 (Fig. 4). To confirm whether reduced motility enables the symbionts to attach to the diatom, A. glacialis A3 was cocultured with each bacterial isolate. Indeed, S. pseudonitzschiae F5 and Phaeobacter sp. F10 exhibited strong attachment in the diatom phycosphere while A. macleodii F12 showed no apparent attachment (Fig. 4).
Fig. 4.
Diatom secondary metabolite rosmarinic acid reduces motility and promotes attachment of potential Roseobacter symbionts to A. glacialis A3. (Top) Motility behavior of strains S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 grown on semisolid (0.25% wt/vol) marine agar plates with (gray bars) or without (white bars) 2 µM rosmarinic acid. Error bars represent standard deviation (SD) of three replicates. Significance was determined by Student’s t test: *P < 0.05 and **P < 0.001. (Bottom) Fluorescence microscopy images of cocultures of the diatom with the two Roseobacter strains and A. macleodii F12. SYBR Green I was used to visualize diatom and bacterial DNA; alcian blue was used to stain the diatom exopolysaccharide matrix, known as transparent exopolymeric particles (TEP; in blue). Cocultures were gently filtered prior to microscopy onto 3-μm membrane filters to remove free-living bacteria. No bacteria are visible on TEP in the vicinity of diatom cells in the A. macleodii F12 panel, indicating that most A. macleodii F12 cells were free-living and were removed by gravity filtration.
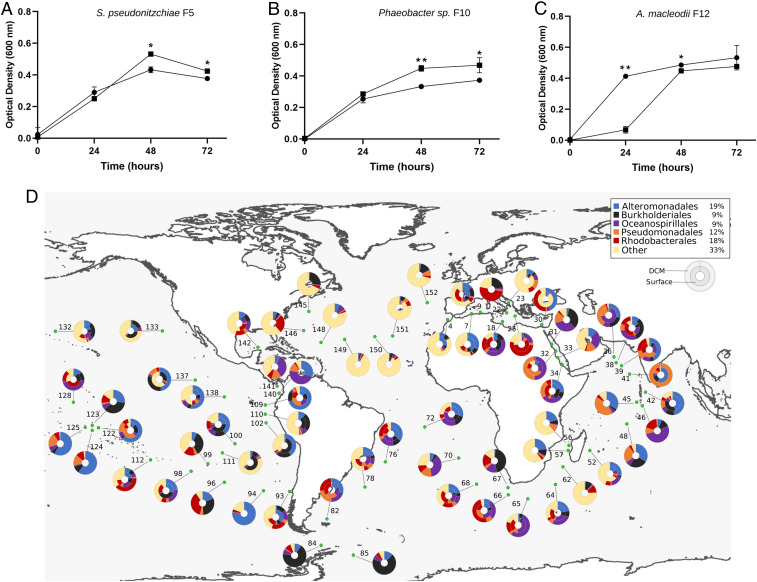
In addition to rosmarinic acid, 100 μM azelaic acid, a byproduct of oleic acid metabolism, significantly inhibited the growth of A. macleodii F12 over a 24-h period, while the same concentration promoted growth of potential symbionts over a 48-h period (Fig. 5 A–C). Bacterial response to azelaic acid was shown to be controlled by a transcriptional regulator, AzeR (26). To shed light on the prevalence of the bacterial response to azelaic acid throughout the oceans, a hidden Markov model (HMM) profile of AzeR homologs detected in all three bacterial isolates was used to search the Tara Oceans database. The average abundance of AzeR reads relative to the total reads in the database was 0.03%. AzeR homologs were consistently distributed at surface and deep chlorophyll maximum depths across the oceans, with most homologs belonging to Alteromonadales (19%) and Rhodobacterales (18%; Fig. 5D). Mining the Pfam database for AzeR homologs indicated that the response to azelaic acid in publicly available bacterial genomes is mostly limited to the Proteobacteria phylum and is mostly restricted to six orders, including Alteromonadales and Rhodobacterales, to which the Alteromonadaceae and Roseobacters belong, respectively (SI Appendix, Fig. S6).
Fig. 5.
Azelaic acid inhibits growth of A. macleodii F12 and promotes growth of Roseobacters. Growth of (A) S. pseudonitzschiae F5, (B) Phaeobacter sp. F10, and (C) A. macleodii F12 on 10% marine broth supplemented with 100 µM azelaic acid (squares) compared to controls (circles). Error bars represent SD of three replicates. Significance was determined by Student’s t test: *P < 0.05 and **P < 0.001. (D) Bacterial response to azelaic acid is geographically widespread throughout the oceans. The relative abundance of reads of the azelaic acid transcriptional regulator, AzeR, in the Tara Oceans database is 0.03%. The total percentage abundance of AzeR homologs according to their taxonomic distribution is shown in the top right box. Rhizobiales makes up the majority of hits (39%) in the “other” group. The color-coded donut plots represent the percentage taxonomic abundance of AzeR homologs from all size fractions (0 to 3 μm) at the surface (inner circle) and deep chlorophyll maximum (outer circle) from the Tara Oceans Microbiome Reference Gene Catalog. Numbers refer to the Tara Oceans stations; single donut plots depict surface samples only.
Discussion
Remineralization of phytoplankton-derived organic matter by heterotrophic bacteria plays a major role in the carbon cycle and accounts for the transformation of ∼20 gigatons of carbon per year in the ocean’s euphotic zone (27). Our current understanding of the global passive and active release of DOM by phytoplankton has been largely studied in the context of primary production, grazing events, and virus-mediated cell lysis (28, 29). Still, underlying reasons for the active excretion by phytoplankton of significant amounts of low molecular weight organic compounds into the phycosphere (6) are still being debated (27, 30). Because of their microscopic size, transport of molecules around phytoplankton cells is mostly governed by diffusion, which leads to the accumulation of phytoplankton-derived DOM within the phycosphere (13). Bacteria in the ocean expend significant energy to track and colonize these DOM-rich hotspots to fuel their growth (31), employing a variety of mechanisms to succeed in the phycosphere, including establishing symbiotic exchanges with phytoplankton cells or producing algicidal agents that harm or kill phytoplankton (7, 32, 33). Therefore, it is imperative for phytoplankton cells to control the types of bacteria that come in contact with the phycosphere, as the outcome ultimately leads to survival or death. However, the mechanisms that enable ocean-drifting phytoplankton cells to attract beneficial bacteria and repel harmful ones in the phycosphere, if any, are mostly unknown.
The microbial community composition surrounding A. glacialis A3 is typical of bacteria associated with diatom cultures and blooms (34–38). Flavobacteria, the dominant lineage in the natural bacterial community associated with the diatom, often assimilate complex organic matter (e.g., polysaccharides) that require exoenzyme activity (39), especially during phytoplankton blooms (40), partially explaining their inactivity over shorter times with A. glacialis (<24 h; Fig. 1A). Within 0.5 h of reintroducing the natural consortium to the axenic diatom culture, Roseobacters rapidly dominated the bacterial transcriptional activity (Fig. 1A). The Roseobacter group spans >70 genera (41) with a highly versatile genetic repertoire (42, 43) that often dominates microbial assemblages surrounding particulate organic matter (44–46). They have been consistently shown to establish specific symbiotic relationships with diatoms (17, 18, 47) and are especially adept at acquiring phytoplankton-derived DOM (38, 48, 49). Despite their rapid response to A. glacialis A3 exudates relative to all other families, members of the Roseobacter group only represented 16.6% of the microbial consortium of the diatom, which is in line with the average Roseobacter group abundance in phytoplankton blooms (38). This discrepancy is potentially due to competition and chemical warfare between different bacterial taxa in the consortium, manifested by the overexpression of antibiotic resistance genes in all Roseobacter MAGs (Fig. 3 and SI Appendix, Table S5), which mitigates proliferation of any one bacterial group in the phycosphere. Indeed, production of diverse antimicrobial agents in a complex microbial community has been shown to maintain bacterial diversity (50), which explains why, despite being the most active, Roseobacters cannot solely dominate the phycosphere of A. glacialis A3. Interestingly, the microbial community composition of A. glacialis A3 based on metagenomics is different from the original microbial composition recovered with A. glacialis shortly after cultivation from the field based on 16S rRNA gene amplicon sequencing (21). While dominant families like Rhodobacteraceae were common, they displayed large variation among the two datasets, either due to changes in the microbial community during laboratory cultivation or biases in DNA isolation and PCR amplification associated with amplicon sequencing.
Isolation and genome sequencing of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 from the natural diatom microbial consortium provides an ample prospect to better understand phytoplankton modulation of different bacterial taxa in the phycosphere. Sulfitobacter and Phaeobacter are considered beneficial bacteria to diatoms, dinoflagellates, and macroalgae as they provide a variety of essential nutrients and cofactors that confer metabolic advantage to their hosts (17, 18, 51–56). Remarkably, several S. pseudonitzschiae strains (16S rRNA gene sequence identity >97%) have been isolated from several diatom species originating from different oceanic regions (18, 57, 58). One such strain, S. pseudonitzschiae SA11 (clustered near S. pseudonitzschiae F5; SI Appendix, Fig. S3), is a known diatom symbiont that enhances cell division of another diatom, Pseudo-nitzschia multiseries, via the hormone indole-3-acetic acid (18). Preliminary growth experiments between S. pseudonitzschiae F5 and A. glacialis A3 indicate that it also enhances A. glacialis cell division, similar to S. pseudonitzschiae SA11 and P. multiseries (59). Thus, the current body of literature on the beneficial effects Sulfitobacter and Phaeobacter have on diatoms suggest they may be symbionts. Alteromonas has been labeled as a genus with an opportunistic lifestyle (60) due to its proliferation in resource-rich waters and during phytoplankton blooms (61, 62). They are able to outgrow native bacterial communities in coastal areas (63) and degrade a variety of algal-derived molecules such as silica (64), nutrients released during decaying phytoplankton blooms (65), and a variety of diatom-derived polysaccharides (66, 67). More specifically, A. macleodii competes with diatoms for nitrate (68), while closely related Alteromonas species contribute to the lysis of dinoflagellates (69) and produce an array of algicidal compounds effective against diatoms and other phytoplankton lineages (70–72). The placement of A. macleodii F12 within the A. macleodii clade (SI Appendix, Fig. S4) suggests it is a common copiotrophic opportunist (60).
The conceptual model presented here (Fig. 3) clearly identifies the transcriptional and metabolomic responses of the host diatom and the surrounding Roseobacters. The combination of multiomics, bacterial isolation, and examination of the effects of different metabolites on these bacterial isolates provides several lines of evidence to support our conclusions. For example, up-regulation of the biosynthesis of metabolites (Fig. 3) by the diatom in response to reseeding is corroborated by the detection of these metabolites in the exometabolome, bacterial transcriptional responses toward these metabolites supported by metatranscriptomics, and growth experiments of bacterial isolates representing the Roseobacter group in the presence of these metabolites. Although we were not able to detect polyamines (e.g., spermidine) presumably produced by the diatom in our metabolome, the up-regulation of genes involved in spermidine uptake by MAG3 and MAG5 suggests that these diatom N-rich molecules may be rapidly utilized by the Roseobacters. Consistent with this observation, genes related to polyamine transformation were shown to be expressed mostly by Roseobacters in coastal waters, where diatoms usually dominate phytoplankton composition (73). In addition to spermidine, the rapid depletion of DON relative to DOC in the reseeded exometabolome (SI Appendix, Fig. S2) is supported by previous findings showing that labile N-containing compounds are preferentially utilized by Roseobacters in estuarine waters (74). These observations suggest DON is more labile than dissolved organic carbon in the phycosphere. The significant decrease in abundance of another DON molecule, citrulline, after the reseeding of bacteria implies its potential uptake (Fig. 2D). Although citrulline has been shown to support bacterial growth as a sole carbon source, uptake mechanisms have not been yet identified (75), complicating our ability to confirm bacterial uptake. However, growth of S. pseudonitzschiae F5 on citrulline confirms the ability of some members of the Roseobacter group to use it as a carbon source (SI Appendix, Fig. S5).
Of 1,237 detected metabolites, we were able to confirm the presence of 28 using a custom-curated chemical library of >660 biomolecules (Fig. 2D). Many of these confirmed metabolites have never been shown to be produced by diatoms before, suggesting that diatoms may be a rich source of metabolites in the ocean. In addition to several central metabolites, we observe the release of obscure secondary metabolites such as quinolinecarboxylic acid, 3-methylglutaric acid, suberic acid, and carnosine (Fig. 2D), which may play a role in symbiotic interactions or defense with different marine bacteria. Interestingly, other confirmed metabolites that have not been shown to be produced by diatoms before, such as rosmarinic acid, azelaic acid, salicylic acid, hippurate, and N-acetyl-galactosamine (Fig. 2D and SI Appendix, Table S4), are involved in plant defense and interkingdom signaling mechanisms (76, 77). Production and secretion of these metabolites by the diatom hints at a defense system response (78) akin to land plants. Rosmarinic acid is one of the most frequently occurring defense compounds used by plants (25) and seagrasses (79), with a well characterized biosynthetic pathway in plants (80). Similarly, azelaic acid is known as a natural signaling compound that induces systemic changes in plant defense mechanisms (81) and is produced by a marine fungus (82) and marine angiosperms (83). Other identified metabolites that display a shift in concentration after reseeding (Fig. 2D) may have important biological functions, including supporting bacterial growth (SI Appendix, Fig. S5). The significant shift in metabolic activity over time as the diatom host came in contact with the microbial consortia (Fig. 2 A–C and SI Appendix, Fig. S2) raises the question of the presence of more specialized compounds that potentially aid in shaping the phytoplankton microbiome. We sought to validate our hypothesis by examining the bacterial response to two of these secondary metabolites, rosmarinic acid and azelaic acid, using the isolated strains.
Rosmarinic acid was one of seven molecules that showed an increase in relative abundance within 0.5 h of reseeding relative to axenic controls (P = 0.025 and P = 0.006 at 0.5 and 48 h, respectively; Fig. 2D). This increase in abundance is either due to up-regulation of its biosynthesis by the diatom in response to reseeding, suggesting an interkingdom signaling function, or due to bacterial coproduction. Bacterial coproduction can be ruled out given that rosmarinic acid is only known to be produced by some land plants and seagrasses and has never been shown to be produced by prokaryotes (84). We mined the diatom genome for rosmarinic acid biosynthesis genes using plant homologs but were unable to find any matches, suggesting that diatoms may use a unique biosynthesis pathway different from legumes. Interestingly, rosmarinic acid significantly suppressed motility and promoted attachment of potential symbionts but had the opposite effect on A. macleodii F12 (Fig. 4). Rosmarinic acid was recently reported to be produced by Arabidopsis thaliana as a mimic of pathogenic bacterial quorum sensing autoinducers (85). It is likely that rosmarinic acid is also interfering with bacterial quorum sensing to control bacterial motility and attachment in the phycosphere, a hypothesis that appears to be supported by recent findings (59).
Azelaic acid, a C9-dicarboxylic acid and a byproduct of oleic acid metabolism, is also produced by the diatom (Fig. 2D). Azelaic acid primes plant defenses (86) and leads to the production of another defense signal, salicylic acid (87), which is also released by the diatom (Fig. 2D). The decrease in abundance of azelaic acid in reseeded exometabolomes (P = 0.0002, P = 0.001, and P = 0.018 at 0.5, 4, and 48 h, respectively; Fig. 2D) and its influence on growth of bacterial isolates suggest that the compound was assimilated by Roseobacters and Alteromonadaceae. Suberic acid (C8-dicarboxylic acid), a closely related metabolite also produced by the diatom, promotes the growth of S. pseudonitzschiae F5 and A. macleodii F12 alike (SI Appendix, Fig. S5). The similar structure and activity of both congeners suggest that, while azelaic acid inhibits growth of Alteromonadaceae, suberic acid may inhibit the growth of other bacteria. While such a strategy may enable diatoms to modulate different bacterial groups, Roseobacters gain an apparent advantage by utilizing a wide range of substrates from diatoms. Analysis of transporters in the genomes of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 indicate that the Roseobacters possess a significantly higher number of transporters normalized to genome size relative to A. macleodii F12 (59). The mechanism of growth inhibition and promotion by azelaic acid remains unknown, and further work is needed to reveal its mechanism of action.
Bacteria in the phycosphere observe orders-of-magnitude higher concentrations of metabolites than cells outside (31). In contrast, these metabolites in bulk seawater potentially have much lower concentrations that fall within the nanomolar to picomolar range. This discrepancy between effective metabolite concentrations in the phycosphere and their measured concentrations in the environment and laboratory cultures is a byproduct of our inability to measure concentrations directly in the phycosphere. Recent findings show that bacterial community assembly in synthetic phycospheres can be predicted from the linear combination of taxa supported by growth on single phytoplankton central metabolites (88). Our findings further expand on our understanding of the role of metabolites in the phycosphere by incorporating host response to presence of different bacterial groups, manifested in the secretion of two unique secondary metabolites. Secretion of secondary metabolites by multicellular eukaryotes to modulate their microbiomes has been broadly reported (89–91). The ability of diatoms (and presumably other unicellular eukaryotes) to exert control over select microbial associates, suggesting a capacity to nurture microbiomes, may have evolved earlier than the rise of multicellularity in eukaryotes. More interestingly, the ability of diatom-derived metabolites to have opposite phenotypic and/or behavioral effects on two different bacterial populations has not been widely shown, to our knowledge. This ability hints at complex evolutionary trajectories of how diatoms evolved the use of these metabolites and the role of secondary metabolism in interkingdom signaling. Further work is needed to characterize the mechanisms of action of these unique molecules in bacteria and to further identify other diatom metabolites and their role in modulating bacterial populations. Shedding light on these mechanisms has the potential to expand our understanding of food web dynamics and the role of phycosphere bacteria in carbon cycling.
Summary.
Multicellular eukaryotes use diverse strategies to recruit and modulate microbiomes in specialized developmental organelles, such as the mammalian gut (92). In contrast, unicellular eukaryotes such as diatoms lack specialized organelles to house microbiomes, and, despite numerous observations that they possess unique microbial communities (93–95), it is not clear how they can modulate transient microbes. We show that, in addition to phytoplankton-derived central metabolites accessible to bacteria, the diatom A. glacialis A3 employs two unique secondary metabolites to promote the proliferation of select bacteria and demote others. The functional roles of signaling of secondary metabolites in marine environments are an important piece of the puzzle linking symbiotic exchanges between phytoplankton and bacteria with carbon cycling in the euphotic zone. Although signaling molecules are believed to constitute a minor fraction of DOM in the euphotic zone, their regulation of microbial metabolism and growth means they can exert a major influence on carbon cycling. This study provides a glimpse into the potential evolution of molecules from the same algal source that have opposite effects on two different groups of bacteria but a favorable outcome for the host. Such an efficient strategy to achieve two outcomes on symbionts and nonsymbionts in the euphotic zone (6, 96) suggests that microalgae and other unicellular eukaryotes modulate microbiomes.
Methods
Full details of the reseeding experimental design, bacterial isolation, DNA/RNA sequencing, exometabolite extraction, motility/growth assays, phylogenetic analysis, microscopy, and computational analysis are described in SI Appendix, Supplementary Methods. Briefly, the diatom A. glacialis strain A3 was isolated along with its natural bacterial community as previously described (21). A. glacialis A3 cultures were made axenic using antibiotics and left to acclimate in the absence of bacteria. To reseed the acclimated axenic A. glacialis A3 cultures, we used a consortium stock harvested from xenic A. glacialis A3 cultures after removing diatom cells. Ten near-complete bacterial genomes were assembled from the consortium metagenome. Subsequently, metatranscriptomes from the consortium were used to uncover how different bacterial families responded to diatom exudates and further examine the interactions between A. glacialis A3 and its natural bacterial associates. The A. glacialis A3 transcriptome was analyzed to reveal its gene expression profile after exposure to its natural microbial community. Exometabolites were analyzed at four different time points with a Bruker Impact II HD quadrupole time-of-flight mass spectrometer (QToF-MS), and 28 molecules were confirmed against a library of standards. In addition, Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) was used to determine the molecular composition of dissolved organic matter components to predict A. glacialis A3’s metabolic reprogramming. To confirm the ability of different taxa to utilize diatom metabolites as growth substrates, we isolated bacteria from the bacterial consortium and monitored their growth in supplemented media. Further, rosmarinic acid (2 μM) and azelaic acid (100 μM) were used to examine if diatom secondary metabolites can give Roseobacters an advantage over Alteromonadaceae by cell attachment with a bacterial motility assay or by modulation of bacterial growth, respectively. The global distribution and homology analysis of an azelaic acid transcriptional regulator was examined by querying a hidden Markov model profile against the Tara Oceans bacterial metagenomics datasets.
Supplementary Material
Acknowledgments
We thank the NYU Abu Dhabi Core Technology Platforms for support in genomics sequencing and mass spectrometry. We also thank Dain McParland for help collecting water samples and Bryndan P. Durham and Elodie Ghedin for helpful comments on the manuscript. This project was supported by a grant from the NYU Abu Dhabi Research institute to K.C.G. (ADHPG-CGSB1) and grants from NYU Abu Dhabi (AD179) and NOAA (#NA19NOS4780183) to S.A.A.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012088117/-/DCSupplemental.
Data Deposition and Materials Availability.
The A. glacialis strain A3 is available from the National Center for Marine Algae and Microbiota (NCMA) collection under the accession CCMP3542. The A. glacialis A3 genome is deposited at DDBJ/ENA/GenBank under the accession WKLE00000000 in NCBI-BioProject PRJNA588343. RNA-seq reads of A. glacialis A3 are deposited in NCBI under the BioProject PRJNA588343. Metagenomic reads and RNA-seq reads of the bacterial consortium are deposited in NCBI under the BioProject PRJNA578578. Metagenomically assembled genomes are deposited at DDBJ/ENA/GenBank under the accessions WKFI00000000–WKFN00000000 in NCBI-BioProject PRJNA588964. Whole-genome assemblies of consortium-isolated strains S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 are deposited at DDBJ/ENA/GenBank under the accessions WKFG00000000, WKFH00000000, and CP046140–CP046144, respectively, in NCBI-BioProject PRJNA588972. Mass spectral datasets are available in the MassIVE database under accession MSV000084592. All software packages used in this study are free and open-source.
References
- 1.Pennisi E., No microbiome is an island, survey reveals. Science 365, 851 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Nyholm S. V., McFall-Ngai M. J., The winnowing: Establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642 (2004). [DOI] [PubMed] [Google Scholar]
- 3.van Oppen M. J. H., Blackall L. L., Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Knight R.et al., The microbiome and human biology. Annu. Rev. Genomics Hum. Genet. 18, 65–86 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Stone B. W. G., Weingarten E. A., Jackson C. R., The role of the phyllosphere microbiome in plant health and function. Annu. Plant Rev. Online 5, 533–556 (2018). [Google Scholar]
- 6.Azam F., Malfatti F., Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Amin S. A., Parker M. S., Armbrust E. V., Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirri E., Pohnert G., Algae-bacteria interactions that balance the planktonic microbiome. New Phytol. 223, 100–106 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Tréguer P.et al., Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11, 27–37 (2017). [Google Scholar]
- 10.Wetz M. S., Wheeler P. A., Release of dissolved organic matter by coastal diatoms. Limnol. Oceanogr. 52, 798–807 (2007). [Google Scholar]
- 11.Longnecker K., Kido Soule M. C., Kujawinski E. B., Dissolved organic matter produced by Thalassiosira pseudonana. Mar. Chem. 168, 114–123 (2015). [Google Scholar]
- 12.Bell W., Mitchell R., Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277 (1972). [Google Scholar]
- 13.Seymour J. R., Amin S. A., Raina J. B., Stocker R., Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Falkowski P. G., Fenchel T., Delong E. F., The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Smriga S., Fernandez V. I., Mitchell J. G., Stocker R., Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 113, 1576–1581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raina J. B., Fernandez V., Lambert B., Stocker R., Seymour J. R., The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 17, 284–294 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Durham B. P.et al., Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amin S. A.et al., Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015). [DOI] [PubMed] [Google Scholar]
- 19.van Tol H. M., Amin S. A., Armbrust E. V., Ubiquitous marine bacterium inhibits diatom cell division. ISME J. 11, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malviya S.et al., Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U.S.A. 113, E1516–E1525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behringer G.et al., Bacterial communities of diatoms display strong conservation across strains and time. Front. Microbiol. 9, 659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S. R., Abbriano R. M., Hildebrand M., Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Res. 1, 2–16 (2012). [Google Scholar]
- 23.Luo C., Rodriguez-R L. M., Konstantinidis K. T., MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 42, e73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sule P., Belas R., A novel inducer of Roseobacter motility is also a disruptor of algal symbiosis. J. Bacteriol. 195, 637–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen M., Rosmarinic acid: New aspects. Phytochem. Rev. 12, 207–227 (2013). [Google Scholar]
- 26.Bez C.et al., AzeR, a transcriptional regulator that responds to azelaic acid in Pseudomonas nitroreducens. Microbiology (Reading) 166, 73–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran M. A.et al., Deciphering ocean carbon in a changing world. Proc. Natl. Acad. Sci. U.S.A. 113, 3143–3151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson C. A., Hansell D. A., . “DOM sources, sinks, reactivity, and budgets” in Biogeochemistry of Marine Dissolved Organic Matter, Carlson C. A., Hansell D. A., Eds. (Elsevier, Amsterdam, The Netherlands, 2015), pp. 65–126. [Google Scholar]
- 29.Benner R., Amon R. M., The size-reactivity continuum of major bioelements in the ocean. Annu. Rev. Mar. Sci. 7, 185–205 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Thornton D. C., Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46 (2014). [Google Scholar]
- 31.Stocker R., Seymour J. R., Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J., The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor J. D., Cottingham S. D., Billinge J., Cunliffe M., Seasonal microbial community dynamics correlate with phytoplankton-derived polysaccharides in surface coastal waters. ISME J. 8, 245–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teeling H.et al., Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5, e11888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H.et al., Microbial community dynamics and assembly follow trajectories of an early-spring diatom bloom in a semienclosed bay. Appl. Environ. Microbiol. 84, e01000-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossart H. P., Levold F., Allgaier M., Simon M., Brinkhoff T., Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7, 860–873 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Teeling H.et al., Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Kappelmann L.et al., Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 13, 76–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinhassi J.et al., Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon M.et al., Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newton R. J.et al., Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Moran M. A.et al., Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 73, 4559–4569 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fontanez K. M., Eppley J. M., Samo T. J., Karl D. M., DeLong E. F., Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front. Microbiol. 6, 469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mestre M., Borrull E., Sala M., Gasol J. M., Patterns of bacterial diversity in the marine planktonic particulate matter continuum. ISME J. 11, 999–1010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinta-Kanto J. M., Sun S., Sharma S., Kiene R. P., Moran M. A., Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 14, 228–239 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G., Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Landa M., Burns A. S., Roth S. J., Moran M. A., Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677–2690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landa M.et al., Sulfur metabolites that facilitate oceanic phytoplankton-bacteria carbon flux. ISME J. 13, 2536–2550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Czárán T. L., Hoekstra R. F., Pagie L., Chemical warfare between microbes promotes biodiversity. Proc. Natl. Acad. Sci. U.S.A. 99, 786–790 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao D.et al., Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73, 7844–7852 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geng H., Belas R., Molecular mechanisms underlying roseobacter-phytoplankton symbioses. Curr. Opin. Biotechnol. 21, 332–338 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Wienhausen G., Noriega-Ortega B. E., Niggemann J., Dittmar T., Simon M., The exometabolome of two model strains of the roseobacter group: A marketplace of microbial metabolites. Front. Microbiol. 8, 1985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lépinay A.et al., First insight on interactions between bacteria and the marine diatom Haslea ostrearia: Algal growth and metabolomic fingerprinting. Algal Res. 31, 395–405 (2018). [Google Scholar]
- 55.Majzoub M. E.et al., Phaeobacter inhibens controls bacterial community assembly on a marine diatom. FEMS Microbiol. Ecol. 95, fiz060 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Johansson O. N.et al., Friends with benefits: Exploring the phycosphere of the marine diatom Skeletonema marinoi. Front. Microbiol. 10, 1828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong Z.et al., Sulfitobacter pseudonitzschiae sp. nov., isolated from the toxic marine diatom Pseudo-nitzschia multiseries. Int. J. Syst. Evol. Microbiol. 65, 95–100 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Töpel M.et al., Complete genome sequence of novel Sulfitobacter pseudonitzschiae Strain SMR1, isolated from a culture of the marine diatom Skeletonema marinoi. J Genomics 7, 7–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fei C., et al., Quorum sensing regulates ‘swim-or-stick’ lifestyle in the phycosphere. Environ. Microbiol., 10.1111/1462-2920.15228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.López-Pérez M.et al., Genomes of surface isolates of Alteromonas macleodii: The life of a widespread marine opportunistic copiotroph. Sci. Rep. 2, 696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinhassi J., Berman T., Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69, 199–211 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tada Y.et al., Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl. Environ. Microbiol. 77, 4055–4065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedler B. E., Aluwihare L. I., Azam F., Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 7202–7207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bidle K. D., Azam F., Bacterial control of silicon regeneration from diatom detritus: Significance of bacterial ectohydrolases and species identity. Limnol. Oceanogr. 46, 1606–1623 (2001). [Google Scholar]
- 65.Hou S.et al., Benefit from decline: The primary transcriptome of Alteromonas macleodii str. Te101 during trichodesmium demise. ISME J. 12, 981–996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor J. D., Cunliffe M., Coastal bacterioplankton community response to diatom-derived polysaccharide microgels. Environ. Microbiol. Rep. 9, 151–157 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Koch H.et al., Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 13, 92–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diner R. E., Schwenck S. M., McCrow J. P., Zheng H., Allen A. E., Genetic manipulation of competition for nitrate between heterotrophic bacteria and diatoms. Front. Microbiol. 7, 880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X.et al., Lysis of a red-tide causing alga, Alexandrium tamarense, caused by bacteria from its phycosphere. Biol. Control 52, 123–130 (2010). [Google Scholar]
- 70.Mayali X., Azam F., Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 51, 139–144 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Wang H.et al., Characterisation of algicidal bacterial exometabolites against the lipid-accumulating diatom Skeletonema sp. Algal Res. 13, 1–6 (2016). [Google Scholar]
- 72.Umetsu S., Kanda M., Imai I., Sakai R., Fujita M. J., Questiomycins, algicidal compounds produced by the marine bacterium Alteromonas sp. D and their production cue. Molecules 24, 4522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mou X.et al., Metatranscriptomic signature of exogenous polyamine utilization by coastal bacterioplankton. Environ. Microbiol. Rep. 3, 798–806 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Mayali X., Weber P. K., Pett-Ridge J., Taxon-specific C/N relative use efficiency for amino acids in an estuarine community. FEMS Microbiol. Ecol. 83, 402–412 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Rodrigo-Torres L., Pujalte M. J., Arahal D. R., Draft genome sequence of Thalassobius gelatinovorus CECT 4357T, a roseobacter with the potential ability to degrade polycyclic aromatic hydrocarbons. Gene Rep. 9, 32–36 (2017). [Google Scholar]
- 76.van Dam N. M., Bouwmeester H. J., Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci. 21, 256–265 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Jha D., Kulshreshtha S., Chauhan S., . “Plant microbial ecology as a potential option for stress management in plants” in Plant Microbe Symbiosis, Varma A., Tripathi S., Prasad R., Eds. (Springer, Dordrecht, The Netherlands, 2020), pp. 331–360. [Google Scholar]
- 78.Durham B. P.et al., Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ. Microbiol. 19, 3500–3513 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Guan C., Saha M., Weinberger F., Chemical defence of a seagrass against microfoulers and its seasonal dynamics. Appl. Sci. (Basel) 9, 1258 (2019). [Google Scholar]
- 80.Trócsányi E., György Z., Zámboriné-Németh É., New insights into rosmarinic acid biosynthesis based on molecular studies. Curr. Plant Biol. 23, 100162 (2020). [Google Scholar]
- 81.Korenblum E.et al., Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 3874–3883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corinaldesi C., Barone G., Marcellini F., Dell’Anno A., Danovaro R., Marine microbial-derived molecules and their potential use in cosmeceutical and cosmetic products. Mar. Drugs 15, ••• (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Papazian S., Parrot D., Burýšková B., Weinberger F., Tasdemir D., Surface chemical defence of the eelgrass Zostera marina against microbial foulers. Sci. Rep. 9, 3323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bulgakov V. P., Inyushkina Y. V., Fedoreyev S. A., Rosmarinic acid and its derivatives: Biotechnology and applications. Crit. Rev. Biotechnol. 32, 203–217 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Corral-Lugo A., Daddaoua A., Ortega A., Espinosa-Urgel M., Krell T., Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 9, ra1 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Jung H. W., Tschaplinski T. J., Wang L., Glazebrook J., Greenberg J. T., Priming in systemic plant immunity. Science 324, 89–91 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Malamy J., Carr J. P., Klessig D. F., Raskin I., Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004 (1990). [DOI] [PubMed] [Google Scholar]
- 88.Fu H., Uchimiya M., Gore J., Moran M. A., Ecological drivers of bacterial community assembly in synthetic phycospheres. Proc. Natl. Acad. Sci. U.S.A. 117, 3656–3662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ochsenkühn M. A., Schmitt-Kopplin P., Harir M., Amin S. A., Coral metabolite gradients affect microbial community structures and act as a disease cue. Commun. Biol. 1, 184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pascale A., Proietti S., Pantelides I. S., Stringlis I. A., Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front Plant Sci 10, 1741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peters L.et al., Secondary metabolites of Flustra foliacea and their influence on bacteria. Appl. Environ. Microbiol. 69, 3469–3475 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McFall-Ngai M.et al., Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sapp M.et al., Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53, 683–699 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Sison-Mangus M. P., Jiang S., Tran K. N., Kudela R. M., Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J. 8, 63–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ajani P. A.et al., The microbiome of the cosmopolitan diatom Leptocylindrus reveals significant spatial and temporal variability. Front. Microbiol. 9, 2758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Azam F.et al., The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263 (1983). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The A. glacialis strain A3 is available from the National Center for Marine Algae and Microbiota (NCMA) collection under the accession CCMP3542. The A. glacialis A3 genome is deposited at DDBJ/ENA/GenBank under the accession WKLE00000000 in NCBI-BioProject PRJNA588343. RNA-seq reads of A. glacialis A3 are deposited in NCBI under the BioProject PRJNA588343. Metagenomic reads and RNA-seq reads of the bacterial consortium are deposited in NCBI under the BioProject PRJNA578578. Metagenomically assembled genomes are deposited at DDBJ/ENA/GenBank under the accessions WKFI00000000–WKFN00000000 in NCBI-BioProject PRJNA588964. Whole-genome assemblies of consortium-isolated strains S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 are deposited at DDBJ/ENA/GenBank under the accessions WKFG00000000, WKFH00000000, and CP046140–CP046144, respectively, in NCBI-BioProject PRJNA588972. Mass spectral datasets are available in the MassIVE database under accession MSV000084592. All software packages used in this study are free and open-source.