Significance
Many millions of people take probiotics over the counter, but very little is known about what they do and whether they really work. Here we show that in mice, introducing Bifidobacterium, one of the most commonly used probiotics, not only colonizes the gut, but also alters the entire microbiotic landscape. We previously found that this treatment rescues mice from an otherwise fatal inflammatory syndrome brought on by anti–CTLA-4 antibody, a checkpoint inhibitor that often causes autoimmunity in humans undergoing cancer treatment. Here we show that this is effect is due, at least in part, to the effect of this probiotic treatment on regulatory CD4+ cells, whose metabolic and immune suppressive functions are altered. These CD4+ regulatory T cells are known to be a key mechanism in the control of autoreactivity in the immune system in both mice and humans. Thus, we found a direct connection between probiotic treatment and one of the known principal mechanisms for controlling excess immune responses.
Keywords: immune checkpoint blockade, Bifidobacterium, regulatory T cell, microbiota, metabolism
Abstract
Immune checkpoint-blocking antibodies that attenuate immune tolerance have been used to effectively treat cancer, but they can also trigger severe immune-related adverse events. Previously, we found that Bifidobacterium could mitigate intestinal immunopathology in the context of CTLA-4 blockade in mice. Here we examined the mechanism underlying this process. We found that Bifidobacterium altered the composition of the gut microbiota systematically in a regulatory T cell (Treg)-dependent manner. Moreover, this altered commensal community enhanced both the mitochondrial fitness and the IL-10–mediated suppressive functions of intestinal Tregs, contributing to the amelioration of colitis during immune checkpoint blockade.
Immune checkpoint blockade therapy has become a very successful cancer treatment. The first monoclonal antibody (mAb) approved for clinical use is specific for the cytotoxic T lymphocyte-associated protein 4 (CTLA-4) for melanoma treatment (1). However, the application of immune checkpoint inhibitors (ICIs) can cause various and even fatal autoimmune responses, of which diarrhea and colitis are among the most frequent and severe (2, 3).
Components in the gut microbiota have been shown to regulate the host antitumor immune response (4–7), and several studies have implicated the function of the intestinal microbiota in modulating the efficacy of immune checkpoint blockade therapy (5, 8, 9). For example, the presence of Bifidobacterium can stimulate the host immune system to respond to anti–PD-L1 therapy in a CD8+ T cell-dependent manner (10).
Although these studies have demonstrated a role for the microbiota in antitumor immunity, the underlying events related to checkpoint antibody-induced autoimmunity remain elusive. In the clinic, patients who experienced colitis after ICI treatment harbored gut bacteria compositions that are distinct from those of colitis-free patients (11). A recent study reported the first clinical case in which reconstituting the gut microbiota with fecal microbiota transplantation successfully rescued ICI-associated colitis (9). The baseline of gut microbiota was also shown to be related to that clinical response to ipilimumab, with enrichment of Faecalibacterium consistent with long-term clinical benefit and colitis (12). We previously reported that administration of Bifidobacterium attenuated intestinal inflammation without impairing the antitumor function of CTLA-4 in mice (13). Here we dissect the fundamental principles governing the relationship between the probiotic-induced microbiome optimization and the outcome of CTLA-4 blockade. We demonstrate that Bifidobacterium systematically alters the composition of the gut microbiota, profoundly increasing the other probiotic species, Lactobacillus. This microbiome optimization is dependent on the existence of regulatory T cells (Tregs). Furthermore, we found that both the metabolic and suppressive functions of intestinal Tregs are enhanced by this altered commensal community, contributing to maintaining regional immune homeostasis under the CTLA-4 blockade condition. Taken together, our observations reveal an immunologic principle governing the complex functions of microbiota dynamics, as well as a mechanism for the relay from Bifidobacterium to Lactobacillus in ameliorating immune checkpoint blockade-related colitis.
Results
Bifidobacterium Alters Gut Microbiota Systematically in a Treg-Dependent Manner.
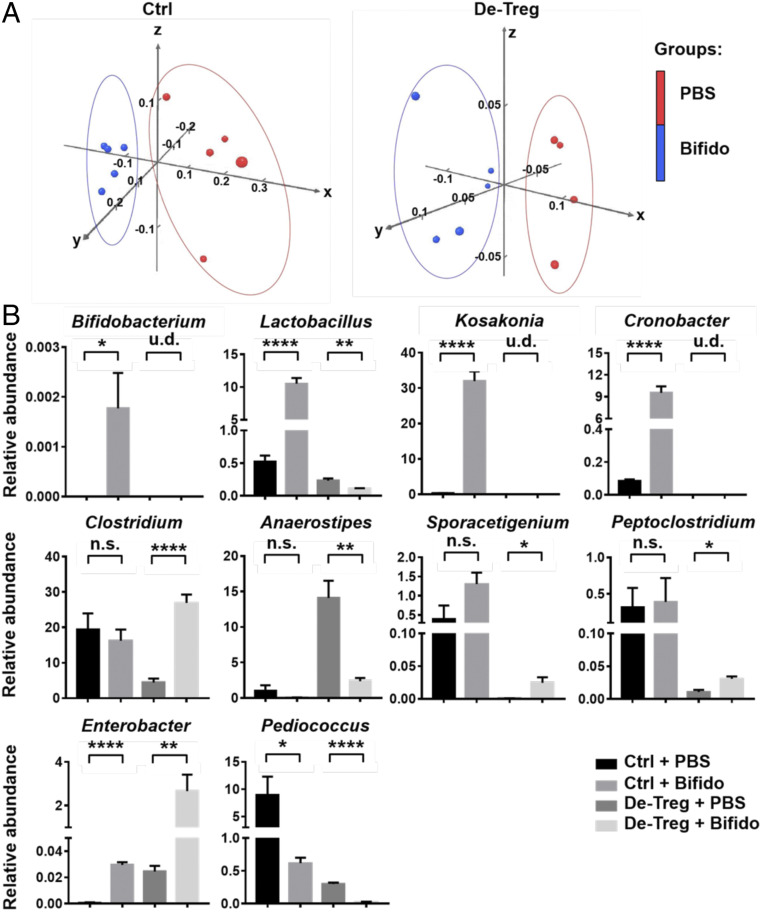
Our finding (13) that live Bifidobacterium-mediated modulation of CTLA-4–induced immunopathology requires Tregs led us to ask whether Tregs directly affect the gut microbial composition (SI Appendix, Fig. S1). To answer this question, we used Foxp3-DTR mice, which carry a diphtheria toxin receptor on Tregs, enabling the transient depletion of Tregs. We isolated DNA from stool samples and conducted 16S rRNA sequencing to compare the gut bacterial community in both control and Treg-depleted mice. Principal component analysis showed that treatment with a Bifidobacterium mixture resulted in genotype clusters distinct from those of the PBS treatment groups in both WT and Treg-depleted mice (Fig. 1A). Specifically, we found that treatment with a Bifidobacterium mixture significantly increased the abundance not only of Bifidobacterium, but also of Lactobacillus, Kosakonia, and Cronobacter, in control mice, while the abundance of these bacteria decreased dramatically, even to undetectable levels, in Treg-depleted mice (Fig. 1 B, Upper and SI Appendix, Fig. S2 A and B).
Fig. 1.
Bifidobacterium alters gut microbiota community. (A) Principal coordinate plot of microbiota composition with PBS (red) and Bifidobacterium (blue). (B) Percentage (mean ± SEM) of the total bacterial abundance of significantly changed bacteria in control and de-Treg mice after PBS or Bifidobacterium administration. n.s., not significant. u.d., undetectable. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Both Bifidobacterium and Lactobacillus are well-known probiotics that have been reported to participate in gut homeostasis (14, 15). Our previous data showed that Bifidobacterium lost its function in the Treg-depleted mice, which also lacked Lactobacillus, suggesting an immune status-related interaction between these two types of bacteria. We also found that the Bifidobacterium treatment significantly changed the percentages of Clostridium, Anaerostipes, Aporacetigenium, and Peptoclostridium in the Treg-depleted mice, while there was no significant change in the control mice (Fig. 1 B, Middle). In addition, Bifidobacterium increased the abundance of Enterobacter and Pediococcus in both control and the Treg-depleted mice (Fig. 1 B, Lower), indicating that the changes in the abundances of these bacteria induced by Bifidobacterium were independent of the gut immune environment.
Colitis-Ameliorating Strains Identified from both Bifidobacterium and Lactobacillus Genera.
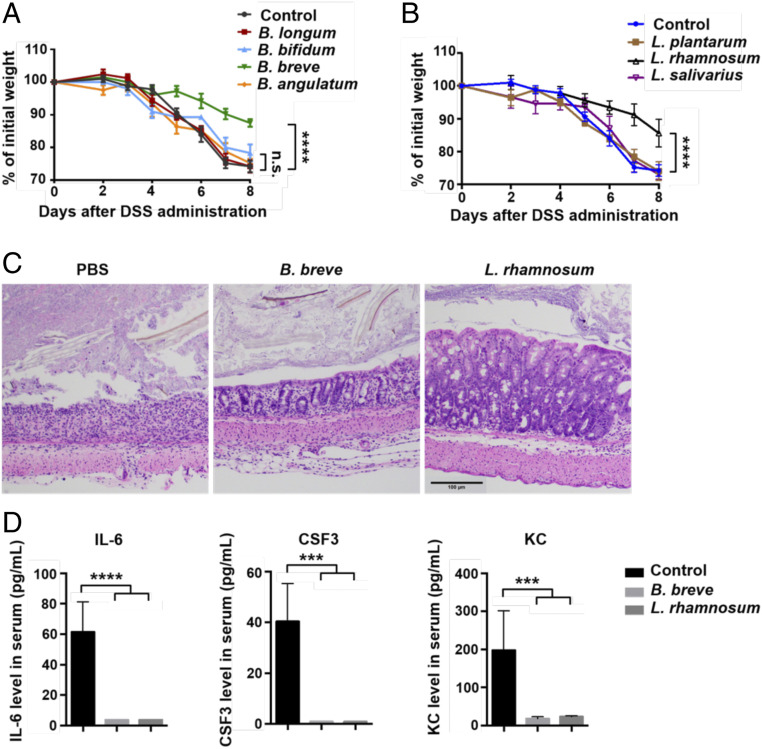
We further tested each individual Bifidobacterium strain from the mixture of four Bifidobacterium species used in previous experiments. We found that the administration of Bifidobacterium breve, but not of other Bifidobacterium strains or the PBS control, prevented weight loss in αCTLA-4–treated mice with colitis (Fig. 2A), demonstrating that B. breve is likely the key functional strain responsible for ameliorating colitis. Since our data showed a correlation between the abundance of Bifidobacterium and Lactobacillus at the genus level (Fig. 1B), we then gavaged mice with different Lactobacillus strains, including Lactobacillus plantarum, Lactobacillus rhamnosum, and Lactobacillus salivarius, to test whether they influenced the susceptibility to colitis. We found that L. rhamnosum treatment resulted in significantly less weight loss in mice with colitis (Fig. 2B). Consistent with this finding, hematoxylin and eosin (H&E) staining of colon sections revealed partial restoration of the colon structure and fewer leukocytes infiltrating into the gut tissue in both B. breve-treated and L. rhamnosum-treated mice. These two strains also resulted in decreased serum levels of the inflammatory cytokines IL-6, CSF3, and KC. Thus, we can identify B. breve and L. rhamnosum as the two functional strains that ameliorate gut immunopathology during CTLA-4 blockade.
Fig. 2.
B. breve and L. rhamnosum are potential functional strains in gut inflammation amelioration. (A) Percent initial weight of mice with 2.5% DSS-induced colitis injected with an αCTLA-4 mAb and treated with PBS control, B. longum, B. bifidum, B. breve, or Bifidobacterium angulatum. (B) Percent initial weight of mice with 2.5% DSS-induced colitis injected with an αCTLA-4 mAb and treated with PBS control, L. plantarum, L. rhamnosum, or L. salivarius. Data are presented as mean ± SEM. n = 5. n.s., not significant. ****P < 0.0001. (C) Representative colon sections of mice treated with PBS control, B. breve, or L. rhamnosum (H&E-staining; scale bar: 100 μm). (D) Concentrations of IL-6, CSF3, and KC in the serum of mice treated with PBS control, B. breve, or L. rhamnosum. ***P < 0.001; ****P < 0.0001.
Bifidobacterium Enhances Treg Function by Promoting an IL-10/IL-10Rα Self-Stimulatory Loop.
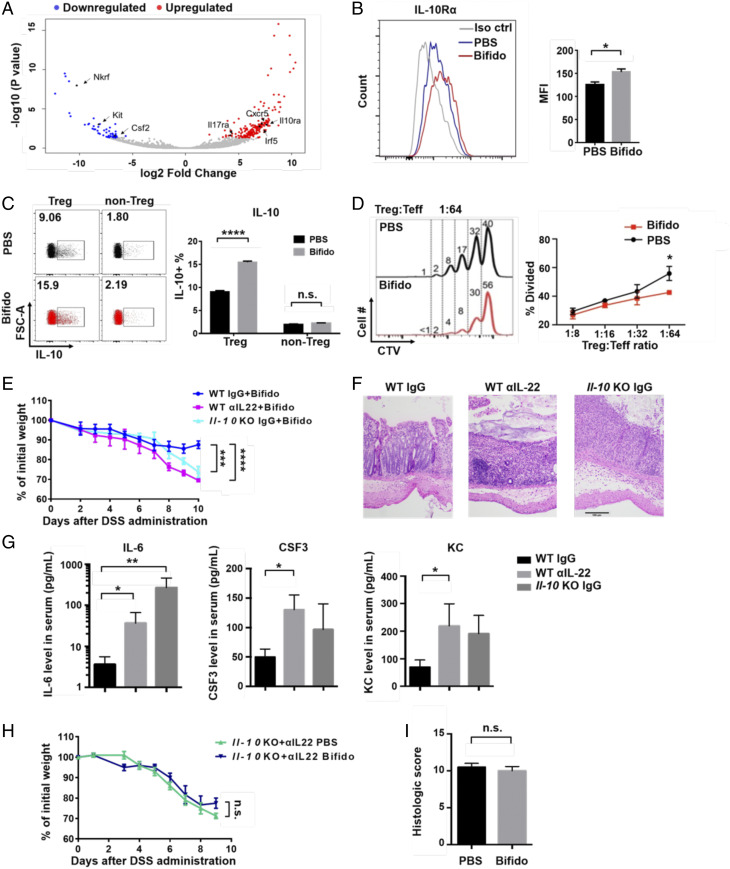
We next investigated the influence of Bifidobacterium on gut Tregs, which are required for the protective function of Bifidobacterium (13). We first analyzed the gene expression pattern of colon lamina propria (LP) Tregs from Bifidobacterium-treated and PBS-treated mice. A volcano plot shows that Bifidobacterium treatment increased several key inflammation-related genes, such as Il10rα, Cxcr5, and Il17rα (Fig. 3A). We also confirmed by flow cytometry that the expression of IL-10Rα in colon LP Tregs is increased after Bifidobacterium treatment (Fig. 3B and SI Appendix, Fig. S3), and the intracellular IL-10 level was also increased in these Tregs, but not in other cells (Fig. 3C and SI Appendix, Fig. S4). These data show that Bifidobacterium promotes an IL-10/IL-10Rα self-stimulatory loop in intestinal Tregs.
Fig. 3.
IL-10 and IL-22 are indispensable for Bifidobacterium function. (A) Volcano plot of significantly changed genes in colonic LP Tregs, comparing Bifidobacterium-treated mice with PBS-treated mice in a CTLA-4 blockade condition. (B and C) Flow cytometry analysis of IL-10Rα (B) and IL-10 (C) in colon LP Tregs isolated from CTLA-4–injected mice. Data are from two independent experiments. n = 2 mice per group in each experiment. n.s., not significant, *P < 0.05, **P < 0.01, ****P < 0.0001. (D) Treg suppression assay performed by coculturing spleen Teff cells, antigen-presenting cells, and colon Tregs from PBS- or Bifidobacterium-treated mice at 4 d after receiving anti–CTLA-4 antibody. Data are from three independent experiments. *P < 0.05. (E) Percent initial weight of WT and Il-10 KO mice with 2.5% DSS-induced colitis subjected to IgG or αIL-22 injection. The mice were treated with αCTLA-4 mAb and Bifidobacterium. n = 5. ***P < 0.001, ****P < 0.0001. (F and G) Representative colon tissue sections from Bifidobacterium-treated WT mice, αIL-22–injected WT mice, and IgG-treated Il-10 KO mice at day 10 after anti–CTLA-4 antibody injection and DSS administration (H&E-staining; scale bar: 100 μm) (F) and concentrations of cytokines in serum (G). *P < 0.05, **P < 0.01. (H) Percent initial weight of αIL-22–injected Il-10 KO mice with 2.5% DSS-induced colitis subjected to anti–CTLA-4 mAb treatment and PBS or Bifidobacterium gavage. Data are mean ± SEM. n = 5. n.s., not significant. (I) Histological scores of mice receiving PBS or Bifidobacterium.
Since IL-10 signaling is important for enhancing Treg function and maintaining gut homeostasis (16–18), we next examined the suppressive function of colon Tregs in vitro. We found that effector T cells that were cocultured with Bifidobacterium-treated Tregs showed less proliferation than those cocultured with PBS-treated Tregs, indicating that this type of intestinal microbe enhances their suppressive function (Fig. 3D and SI Appendix, Fig. S5).
Both IL-10 and IL-22 Are Involved in Bifidobacterium’s Colitis-Ameliorating Function.
To further analyze the role of IL-10 in the function of Bifidobacterium, we used Il-10 knockout (KO) mice to analyze colitis symptoms under conditions of CTLA-4 blockade with Bifidobacterium treatment. We observed more severe weight loss in Il-10 KO mice compared with wild-type (WT) mice subjected to the same treatment (Fig. 3E, blue line). IL-22 shares an immune regulatory function with IL-10R and has an important function in maintaining gut homeostasis (16, 18). Thus, we used an IL-22–neutralizing antibody to test the effect of ablating this cytokine on Bifidobacterium-mediated colitis remission during CTLA-4 blockade. We found that the antibody-treated mice lost more weight than the mice treated with an irrelevant antibody. Specifically, the average weight of Bifidobacterium-treated mice was reduced from 90% of the initial weight in the control group to ∼70% of the initial weight in the anti–IL-22 injected group on day 10 after DSS (dextran sulfate sodium) administration (Fig. 3E, red line). Consistent with this finding, H&E staining of colon sections revealed more leukocyte infiltration and more severe damage to the intestinal structure in anti-IL-22 antibody-treated or Il-10 KO mice (Fig. 3F). Anti-IL-22 treatment and Il-10 knockout also increased serum levels of the inflammatory cytokines IL-6, CSF3, and KC (Fig. 3G).
Importantly, we found that when IL-22 blockade was applied in Il-10 KO mice, Bifidobacterium treated mice showed severe weight loss, comparable to that seen in the PBS-treated control mice (Fig. 3H). Comparable histological scores were also observed in the two groups (Fig. 3I). Together, these results indicate that both IL-22 and IL-10 are required for the function of Bifidobacterium in ameliorating gut immunopathology.
Bifidobacterium Enhances the Mitochondrial Metabolism of Treg Cells.
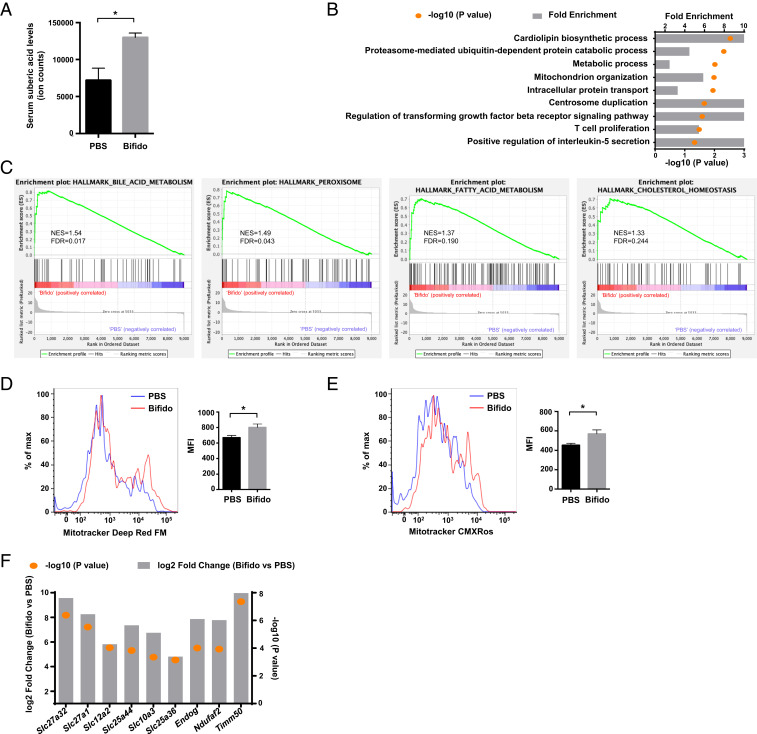
In addition to its effects on immune cell function, the gut microbiota may also impact host cell metabolism (19, 20). We performed mass spectrometry-based metabolite profiling with serum from Bifidobacterium-treated and control mice (21). Interestingly, we found that Bifidobacterium treatment increased the serum level of suberic acid (Fig. 4A), an acid that represents mitochondrial activity and is frequently detected in patients with lipid metabolism disorders (22). We then evaluated gene expression data from sorted colon LP Tregs with Gene Ontology (GO) analysis. Consistent with the foregoing results, we found an enrichment of biological pathways associated with both metabolic processes and mitochondrial organization in the Bifidobacterium-treated group vs. the control group (Fig. 4B). Gene set enrichment analysis (GSEA) showed that four gene sets directly related with metabolism, including “bile acid metabolism,” “peroxisome,” “fatty acid metabolism,” and “cholesterol homeostasis,” represented an up-regulated gene signature in colon Tregs after Bifidobacterium treatment (Fig. 4C).
Fig. 4.
Bifidobacterium affects the mitochondrial metabolism of colon Tregs. (A) Concentrations of suberic acid in the serum of mice treated with PBS or Bifidobacterium. *P < 0.05. (B) GO analysis of significantly impaired biological processes. (C) GSEA of colon Tregs. The diagram plots GSEA for four gene sets up-regulated in Bifidobacterium-treated groups (left side, Bifidobacterium; right side, PBS). The vertical axis in the upper graph indicates the enrichment score (ES) for genes in each gene set. The barcode plot indicates the position of genes in each gene set. NES, normalized ES; FDR, false discovery rate. (D) Mitochondrial mass of colon LP Tregs (n = 5), as detected by MitoTracker Deep Red FM labeling and flow cytometry. *P < 0.05. (E) Mitochondrial stress of colon LP Tregs (n = 5), as detected by MitoTracker Red CMXROS labeling and flow cytometry. *P < 0.05. (F) Mitochondria-related gene expression pattern in Bifidobacterium-treated vs. PBS-treated LP Tregs.
To investigate whether the mitochondrial volume and function of Tregs are affected by Bifidobacterium, we stained cells with fluorescent probes to monitor mitochondrial volume and membrane potential. The flow cytometry data showed that Tregs in the Bifidobacterium-treated group exhibited increases in both mitochondrial volume and mitochondrial stress (Fig. 4 D and E) compared with those in the PBS group. Consistent with this finding, multiple genes related to mitochondrial function and mitochondrial structural components were significantly up-regulated by Bifidobacterium treatment (Fig. 4F). Collectively, these results demonstrate that Bifidobacterium modulates metabolic processes and enhances mitochondrial activity in gut Tregs.
Discussion
In this study, we used a mouse DSS colitis model under immune checkpoint blockade conditions to examine the effects of Bifidobacterium on both the commensal community and the host immune system. Importantly, we found that Bifidobacterium administration exerts systemic changes in the gut microbiota. Moreover, this modulation is influenced by mucosal immune regulatory environment, as determined by Tregs.
Interestingly, we found that Bifidobacterium administration significantly altered the abundance of Lactobacillus, suggesting that Bifidobacterium contributes to the remission of intestinal inflammation by constituting a favorable gut ecosystem with other taxa associated with probiotic activity. Notably, a low percentage of colonization of Bifidobacterium (∼0.002%) can lead to an enrichment of Lactobacillus, reaching ∼10% of the total commensal bacteria (Fig. 1B), indicating a role of Bifidobacteria as a pioneer species to allow the colonization of other, more abundant probiotic species. Moreover, we found that this probiotics relay from Bifidobacterium to Lactobacillus is also sensitive for the gut inflammation status set by the Tregs. By identifying B. breve and L. rhamnosum as two specific functional strains from the Bifidobacterium and Lactobacillus genera, we further confirmed the positive roles of these bacteria in helping control CTLA-4–induced intestinal toxicity.
In summary, we have found that introducing Bifidobacterium into mice profoundly alters their microbiome, indicating that it is a dynamic and interconnected ecosystem. Relevant to the effect of these bacteria in blunting the harmful effects of anti–CTLA-4/gut injury, we found both increased IL-10Ra expression and increased IL-10 production in intestinal Tregs, suggesting that Bifidobacterium can directly or indirectly enhance the suppressive function of Tregs by stimulating an IL-10/IL10Ra signaling loop in Tregs without altering their abundance. Furthermore, we show that this process is coupled to an enhanced mitochondrial activity in these Tregs, providing a metabolic link to their enhanced function in this system.
Materials and Methods
Mice.
Il-10 KO mice (Il-10−/−; C57BL/6-Il10tm1Cgn) were purchased from The Jackson Laboratory. All experiments were performed using 6- to 14-wk-old female mice. The mice were maintained in a specific pathogen-free facility at Shanghai Jiao Tong University or Stanford University. The mouse experiments were approved by the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University School of Medicine and Stanford University.
DSS Colitis Model under CTLA-4 Blockade Conditions.
Between 2% and 4% DSS (MP Biomedicals) was added to the mouse drinking water for 7 to 12 d. Weight changes were monitored each day. For gut commensal manipulation, the mice received vancomycin (0.5 g/L; Sigma-Aldrich) at least 14 d before DSS administration. The mice were injected with 200 µg of an anti–CTLA-4 mAb (BioXCell, clone 9D9) or an isotype control at the start of DSS administration.
Histological Analysis.
Colon tissues were fixed with 4% paraformaldehyde, embedded in paraffin, sectioned at 3 to 6 µm, and stained with H&E. Each segment was given a score of 0 to 4 based on five criteria: severity of inflammation, percent of area affected by inflammation, degree of hyperplasia, percentage of area affected by hyperplastic changes, and ulceration.
Probiotic Administration.
A mixture of four Bifidobacterium species consisting of B.bifidum, B. longum, B. lactis, and B. breve (Seeking Health) or individual strains of probiotics were resuspended in PBS. Each mouse received 1 × 109 bacterial CFU by oral gavage. For DSS colitis, probiotics were administered before DSS treatment.
Fecal DNA Extraction and 16S Sequence Analysis.
Genomic DNA was isolated from fecal samples (collected 4 d after oral gavage probiotics) using the PowerSoil DNA Isolation Kit (MO BIO Laboratories) following the manufacturer's instructions. The 16S universal eubacterial primers 515F GTGCCAGCMGCCGCGGTAA and 806R GGACTACHVGGGTWTCTAAT were used to evaluate the microbial ecology of each sample on an Illumina HiSeq 2500 sequencing system. The sequence data derived from the sequencing process were processed using a proprietary analysis pipeline (MR DNA). OTUs were then taxonomically classified using BLASTn against a curated GreenGenes/Ribosomal Database Project/National Center for Biotechnology Information-derived database.
Isolation of Intestinal LP Lymphocytes.
LP lymphocytes were isolated as described previously with a simple modification (19). In brief, the mice were killed, and the colons were removed and opened longitudinally. The intestines were thoroughly washed in PBS and cut into 1.5-cm pieces. The intestines were shaken in PBS containing 1 mM DTT, 30 mM EDTA, and 10 mM Hepes at 37 °C for 10 min. Then the intestines were shaken in PBS containing 30 mM EDTA and 10 mM Hepes at 37 °C for 10 min. After washing with complete RPMI 1640 medium, the tissues were digested in RPMI 1640 containing 16% collagenase VIII (Sigma-Aldrich; 50 KU) and DNase I (Sigma Aldrich; 90 mg/mL) at 37 °C for 55 min. The cell suspensions from the enzyme digestion were then applied to a Percoll (GE Healthcare) gradient (for lymphocytes: 40% Percoll on the top, 80% Percoll on the bottom) by centrifugation at 2500 rpm for 25 min at room temperature. Lymphocytes were harvested from the interphase and washed twice with 0.5% BSA-PBS.
Flow Cytometry.
The cells were stained with the indicated fluorochrome-conjugated antibodies in PBS containing 0.5% BSA for surface marker analysis. Flow cytometric analysis was performed using a Fortessa flow cytometer (BD Biosciences) with FlowJo software.
Intracellular Cytokine Staining.
The intracellular expression levels of IL-17A, IL-22, and IL-10 in CD4+ T cells were analyzed with the Foxp3/Transcription Factor Staining Buffer Set (Invitrogen) according to the manufacturer’s instructions. In brief, intestinal LP were incubated with cell stimulation mixture plus protein transport inhibitor (Invitrogen) in complete RPMI 1640 at 37 °C for 5 h. Surface staining was performed at 4 °C for 30 min. After fixation and permeabilization treatment, intracellular staining was performed with anti–IL-17A (eBioscience), anti–IL-22 (eBioscience), anti–IL-10 (BD Biosciences), anti-Foxp3 (eBioscience), and anti-RORγt (eBioscience) antibodies for 1 h. Data were acquired with a BD Biosciences Fortessa flow cytometer and analyzed with FlowJo software.
Serum Cytokine Assay.
Blood samples were collected on days 6 to 7 after colitis induction with DSS. After clotting at room temperature for at least 30 min, the serum was separated at 1,200 RCF for 10 min with a centrifuge. Cytokine measurements were performed following the instructions in the Luminex system manual.
RNA-Sequencing Data Analysis.
Each individual sample had an average of 35 million 75-bp paired-end reads. Fastqc (version 0.11.4) was used to assess sequencing quality. The reads were then aligned to the human (hg19) transcriptome using Bowtie version 2.2.7, with splice junctions defined in a GTF file (obtained from UCSC). An average of 65% of reads were aligned to the reference transcriptome. Expression at the gene level was determined by calculating reads per kilobase per million aligned reads and raw counts using RSEM version 1.2.30. Differentially expressed genes with fold changes were further detected by DEseq2 version 1.10.1 for the two comparable conditions. GO analysis was conducted via David online tool (https://david.ncifcrf.gov). GSEA was carried out with the preranked model and gene sets with FDR lower than 25% were considered as significant enrichment.
Treg Suppression Assay.
Effector T cells (CD4+TCRβ+GFP−CD44+CD62L−) and non-T cells (TCRβ−) were purified by flow cytometry cell sorting from the spleens of C57BL/6 mice. T cells were labeled with CellTrace Violet (Invitrogen; 5 µm). Non-T cells were incubated with mitomycin C (Sigma-Aldrich; 50 µg/mL) at 37 °C for 30 min. Tregs (CD45+CD4+TCRβ+GFP+) were sorted from the colon LP of Foxp3-GFP mice. Tregs and labeled effector T cells (Teffs) were plated in 96-well plates at Treg:Teff ratios of 1:8, 1:16, 1:32, and 1:64, and then non-T cells were added to each well at three times the Teff cell number. The cells were cultured with anti-CD3ε (BD Biosciences; clone 2C11) at 0.1 µg/mL for 3 d. The suppressive activity of the Tregs was assessed as the proliferation of Teff cells based on the dilution of the cytosolic dye CellTrace Violet.
Mitochondrial Analysis.
Colon LP lymphocytes were isolated and then stained with cell surface markers. After two washings with 0.5% BSA-PBS, the cells were incubated with a mitochondrial stain (Invitrogen) (for Mito Tracker Red FM, 50 nM; for Mito Tracker Red CMXRos, 100 nM) at 37 °C for 25 min.
Metabolomics Analysis.
Serum samples were extracted with methanol, and precipitated protein was pelleted by centrifugation. Extracts were then dried down and reconstituted in 50% methanol. Samples were then analyzed by liquid chromatography mass spectrometry (LC-MS) using an Agilent 6545 Q-TOF involving three methods, including positive- and negative-mode reverse-phase LC-MS and hydrophilic interaction chromatography. Peak identification was then performed using a mass spectrometry library of standards and related software from IROA Technologies.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism version 7.00.
Supplementary Material
Acknowledgments
We thank Shashuang Zhang, Lei Ding, Ru Feng, Lei Chen, Guojun Qu, Bing Su, Shuo Han, Yueh-hsiu Chien, William Van Treuren, and Curt Fischer for helpful discussions and technical assistance, as well as the Stanford University ChEM-H Metabolite Chemistry Analysis Center. This study was supported by grants from the National Key Research and Development Program of China (SQ2018YFA090045-01), the National Natural Science Foundation of China (82071852, 81771739), the Program for Professor of Special Appointments (Eastern Scholar) at Shanghai Institutions of Higher Learning and the Technology Committee of Shanghai Municipality (18JC1414100, 20410713800 to F.W.), HHMI, and the Parker Institute for Cancer Immunotherapy (to M.M.D.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921223117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Bates S. E., Refining immunotherapy approvals. Clin. Cancer Res. 23, 4948–4949 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Michot J. M.et al., Immune-related adverse events with immune checkpoint blockade: A comprehensive review. Eur. J. Cancer 54, 139–148 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Soularue E.et al., Enterocolitis due to immune checkpoint inhibitors: A systematic review. Gut 67, 2056–2067 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Roy S., Trinchieri G., Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 17, 271–285 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Matson V.et al., The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopalakrishnan V.et al., Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vétizou M.et al., Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Routy B.et al., Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Wang Y.et al., Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 24, 1804–1808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivan A.et al., Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubin K.et al., Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 7, 10391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaput N.et al., Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 28, 1368–1379 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Wang F., Yin Q., Chen L., Davis M. M., Bifidobacterium can mitigate intestinal immunopathology in the context of CTLA-4 blockade. Proc. Natl. Acad. Sci. U.S.A. 115, 157–161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Baarlen P., Wells J. M., Kleerebezem M., Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 34, 208–215 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Ruiz L., Delgado S., Ruas-Madiedo P., Sánchez B., Margolles A., Bifidobacteria and their molecular communication with the immune system. Front. Microbiol. 8, 2345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamanaka M.et al., Memory/effector (CD45RB(lo)) CD4 T cells are controlled directly by IL-10 and cause IL-22-dependent intestinal pathology. J. Exp. Med. 208, 1027–1040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M. O., Flavell R. A., Contextual regulation of inflammation: A duet by transforming growth factor-beta and interleukin-10. Immunity 28, 468–476 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Shouval D. S.et al., Interleukin 10 receptor signaling: Master regulator of intestinal mucosal homeostasis in mice and humans. Adv. Immunol. 122, 177–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson J. K.et al., Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Nieuwdorp M., Gilijamse P. W., Pai N., Kaplan L. M., Role of the microbiome in energy regulation and metabolism. Gastroenterology 146, 1525–1533 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Dodd D.et al., A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X. D., Gillespie S. K., Borrow J. M., Hersey P., The histone deacetylase inhibitor suberic bishydroxamate: A potential sensitizer of melanoma to TNF-related apoptosis-inducing ligand (TRAIL) induced apoptosis. Biochem. Pharmacol. 66, 1537–1545 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.