Significance
Recent research has shown that intracellular proteinaceous condensates (membraneless organelles [MLOs]) are involved in various processes, ranging from Alzheimer’s disease to RNA processing, and here we demonstrate that this phenomenon governs a mechanism of anhydrobiosis. The protein AfrLEA6 is found in the desiccation-tolerant life stage of the animal extremophile Artemia franciscana, and the protein engages in two distinct molecular mechanisms to confer protection during water loss. AfrLEA6 forms MLOs that may act as protective nodes for desiccation-sensitive proteins, while a cytosolic fraction of the protein promotes structural integrity of cells during anhydrobiosis. These findings significantly advance our understanding of “life without water” and promote transformative advancements in various fields, ranging from cell preservation technology to improvement of crop desiccation tolerance.
Keywords: cryptobiosis, late embryogenesis abundant, membraneless organelle, water stress, liquid-liquid phase separation
Abstract
Proteinaceous liquid-liquid phase separation (LLPS) occurs when a polypeptide coalesces into a dense phase to form a liquid droplet (i.e., condensate) in aqueous solution. In vivo, functional protein-based condensates are often referred to as membraneless organelles (MLOs), which have roles in cellular processes ranging from stress responses to regulation of gene expression. Late embryogenesis abundant (LEA) proteins containing seed maturation protein domains (SMP; PF04927) have been linked to storage tolerance of orthodox seeds. The mechanism by which anhydrobiotic longevity is improved is unknown. Interestingly, the brine shrimp Artemia franciscana is the only animal known to express such a protein (AfrLEA6) in its anhydrobiotic embryos. Ectopic expression of AfrLEA6 (AWM11684) in insect cells improves their desiccation tolerance and a fraction of the protein is sequestered into MLOs, while aqueous AfrLEA6 raises the viscosity of the cytoplasm. LLPS of AfrLEA6 is driven by the SMP domain, while the size of formed MLOs is regulated by a domain predicted to engage in protein binding. AfrLEA6 condensates formed in vitro selectively incorporate target proteins based on their surface charge, while cytoplasmic MLOs formed in AfrLEA6-transfected insect cells behave like stress granules. We suggest that AfrLEA6 promotes desiccation tolerance by engaging in two distinct molecular mechanisms: by raising cytoplasmic viscosity at even modest levels of water loss to promote cell integrity during drying and by forming condensates that may act as protective compartments for desiccation-sensitive proteins. Identifying and understanding the molecular mechanisms that govern anhydrobiosis will lead to significant advancements in preserving biological samples.
It is a biological truism that water on Earth appears to be the only solvent suitable for life to occur, although nonaqueous solvents might be available to support life elsewhere in the universe (1–3). Challenged by variations in water availability, animals have developed a variety of mechanisms to maintain an optimal hydration level, which is considered necessary to maintain organismal homeostasis, ecological competitiveness, and species survival (4, 5). In response to the challenge of severe desiccation in terrestrial biotopes, some remarkable organisms have developed mechanisms to survive virtually complete water loss (reviewed in ref. 6). Anhydrobiosis is the state of life in which an organism has lost virtually all intracellular water but is capable of resuming its biological processes and life cycle on rehydration. However, the molecular mechanisms governing anhydrobiosis are still largely unknown. Insights gained over several decades of research have demonstrated that there is no single mechanism that enables anhydrobiosis, but rather that this phenomenon entails a complex array of mechanisms that are carefully orchestrated by certain proteins and osmolytes (7–12).
Late embryogenesis abundant (LEA) proteins form a large subgroup of anhydrobiosis-related intrinsically disordered (ARID) proteins, and were named for their high expression during late embryogenesis in cotton seeds (13, 14). Since their discovery in plants, LEA proteins have also been found in animals belonging to several phyla, including Rotifera, Nematoda, and Arthropoda (8, 15–20). While classification of LEA proteins via bioinformatics has led to multiple organization systems, we use the grouping scheme proposed by Wise et al. (21). It is noteworthy that all known animal LEA proteins belong to group 3, with the exception of the brine shrimp Artemia franciscana, which also expresses LEA proteins belonging to group 6 (22) and group 1 (23, 24) in their anhydrobiotic embryos. Interestingly, group 6 LEA proteins have been linked to the prolonged desiccation tolerance of orthodox seeds, but the mechanism(s) by which these proteins confer protection warrants further evaluation (10, 25, 26). Here we provide evidence that the seed maturation protein (SMP) domains (PF04927) contained in AfrLEA6 (AWM11684) from A. franciscana promote liquid-liquid phase separation (LLPS) in vivo, and that these condensates can selectively compartmentalize target proteins based on their net surface charge in vitro.
Using bioinformatics, AfrLEA6 can be organized into three regions: a region of two SMP domains, a proline-glycine spacer, and a domain predicted to engage in protein interactions with unknown targets (27). Similar to results obtained for several group 3 LEA proteins from A. franciscana (28), analysis of circular dichroism data predicts that AfrLEA6 compacts into α-helices during desiccation and exists as an intrinsically disordered protein when it is fully hydrated with ∼90% of its residues involved in random coiling (29). A large conformational change and increase in the overall compactness of the protein was also suggested by results obtained in vivo using cellular dielectrophoresis, indicating that ectopically expressed AfrLEA6 undergoes a decrease in its hydrodynamic radius in osmotically dehydrated cells (30). However, previous bioinformatics analysis of AfrLEA6 also identified sequence features associated with proteins known to undergo LLPS to form membraneless organelles (MLOs), which warranted a more thorough evaluation on the possible role of LLPS in animal anhydrobiosis (14, 27, 31, 32).
MLOs are superstructures formed by driver proteins that undergo LLPS via weak multivalent interactions with one another, with scaffolding and client proteins, and/or with nucleic acids (reviewed in refs. 33, 34) and form proteinaceous compartments that are not enclosed by a membrane (35–39). MLOs may be persistent within the cell or arise in response to some signal and perform a variety of cellular functions, including regulating gene expression or partitioning biomolecules. Transient LLPS occurs when the proteins become insoluble in the liquid phase of the cellular milieu or when water becomes unavailable, thereby permitting overall less favorable protein-protein associations. This can occur when the concentration of dissolved ions increases, when other polymers crowd the cellular space, or in response to temperature or pH changes (40). Conditions that may promote LLPS of proteins in the cytoplasm of A. franciscana are plentiful during the cryptobiotic period, when the encysted embryos are released during oviparous reproduction and dehydrate after being washed up at the shoreline of the habitat (41). The cytoplasmic concentrations of ions and protein crowding increase during desiccation of the embryo, which in turn will promote LLPS of AfrLEA6. Consequently, we investigated the behavior of AfrLEA6 when ectopically expressed in Drosophila melanogaster Kc167 cells under desiccation stress and the behavior of the purified protein in solution. We found that the protein promotes desiccation tolerance via two distinct molecular mechanisms: by raising cytoplasmic viscosity at even modest levels of water loss to promote cell integrity during drying and by forming MLOs that may act as protective compartments for desiccation-sensitive proteins.
Methods
Bioinformatics.
Several bioinformatics tools were used to gain insight into the structural propensities of AfrLEA6 and to develop testable hypothesis for wet-bench experiments. SMART EMBL (available at http://smart.embl-heidelberg.de/) was used to identify homologous regions and internal repeats of amino acids in the AfrLEA6 protein sequence (42, 43). The net charge per residue was calculated at a pH of 7.2 with a sliding window of five amino acids using the LocalCIDER program, available at https://pappulab.github.io/localCIDER/ (44). The I-Tasser server was used to model potentially secondary and tertiary structure motifs of AfrLEA6 by comparison to homologous structures in proteins with known crystal structures (45–47).
Protein Cloning, Expression, and Purification.
DNA encoding AfrLEA6 (GenBank accession no. MH351624) and green fluorescent protein (Addgene; 51562) were cloned into the pTXB1 vector (New England BioLabs) using standard techniques and expressed as a fusion protein composed of AfrLEA6 or sGFP (net surface charge of −7) in frame with a chitin-binding protein and self-splicing intein protein spacer. The resulting constructs were used to transform the chemically competent Escherichia coli strain BL21 Star (Thermo Fisher Scientific) and cells were grown on Luria-Bertani (LB) medium-based agarose plates containing 100 µg/mL ampicillin. Antibiotic-resistant colonies were selected at random and grown to an optical density of 0.6 at λ = 595 nm in liquid culture on an orbital shaker at 225 rpm and 37 °C in LB medium containing 100 µg/mL ampicillin. Protein expression was induced by adding isopropyl-β-1-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM, and the bacteria were harvested after 2 h via centrifugation at 5,000 × g for 30 min at 4 °C. The bacterial pellets were then resuspended in buffer A (500 mM NaCl and 50 mM Tris⋅HCl, pH 8.5) containing 1 mM phenylmethylsulfonyl fluoride to inhibit serine protease activity. For protein purification, cells were lysed by sonication (Q500; Qsonica), and bacterial debris was removed by centrifugation for 30 min at 5,000 × g at 4 °C. The supernatant was loaded via gravity flow onto a 15-mL chitin resin (New England BioLabs) containing column. The column was washed with 20 column volumes of buffer A. Proteins were eluted from the column after incubation with 50 mM DTT dissolved in buffer A at 4 °C for 48 h, then dialyzed into 50 mM phosphate buffer at a pH of 7.0 and concentrated using centrifugal filter units (Amicon Ultra 10 kDa; Millipore Sigma). The purity of the protein was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and averaged at least 95%. Purified protein aliquots were snap-frozen in liquid nitrogen and stored at −80 °C until use in experiments. Two additional supercharged GFP constructs both containing a 6XHIS tag for purification purposes, pGFP (surface net charge: +36; Addgene, 62937) and nGFP (surface net charge −30; Addgene, 62936), were also expressed in E. coli, but the induction with 0.4 mM IPTG was performed at ambient temperature for 16 h (48). Bacterial lysates were prepared in binding buffer (20 mM imidazole and 500 mM NaCl, pH 7.4) by sonication, and bacterial debris was removed by centrifugation for 30 min at 5,000 × g at 4 °C. The cleared lysates were then applied to 1-mL HisTrap FF columns (Thermo Fisher Scientific) using a 20-mL syringe. Columns were washed with 20 mL of binding buffer, and the proteins were eluted by raising the imidazole concentration to 500 mM. The GFP-containing elution fraction was dialyzed against 50 mM sodium phosphate buffer at a pH of 7.0, and the proteins were concentrated using centrifugal filter units with a 10-kDa molecular weight cutoff (Amicon Ultra 10 kDa; Millipore Sigma). The purity of the protein was confirmed by SDS-PAGE and averaged >95%. Purified protein aliquots were snap-frozen in liquid N2 and stored at −80 °C until use in experiments.
AfrLEA6 Liquid-Liquid Phase Separation.
Stocks of AfrLEA6 were dialyzed against 20 mM sodium phosphate buffer (pH 6.5), or a solution with a composition designed to resemble the crowded conditions in the cytoplasm and concentrations of major osmolytes measured in A. franciscana cysts (osmosome solution: 32 mM NaCl, 98 mM KCl, 11 mM K2PO4, 5 mM CaCl2, 340 mM trehalose, 2.9% wt/vol glycerol, and 25% Ficoll 400, pH 6.5) (49). Liquid-liquid phase separation was observed by light microscopy after pipetting osmosome solution containing AfrLEA6 (0.17 mg/mL) onto glass plates and allowing water to evaporate from the samples at ambient conditions. To investigate interactions between AfrLEA6 droplets and GFP constructs, the fluorescent proteins (35 μM) were added to 1 mg/mL AfrLEA6 (37 μM) in sodium phosphate buffer and the solutions were allowed to desiccate by convective drying as described above. Samples were imaged every 5 min using a Nikon A1R confocal microscope until no liquid water was observed to ensure that interactions between AfrLEA6 and GFP constructs were consistent throughout the drying process.
Scanning Electron Microscopy and Atomic Force Microscopy.
AfrLEA6 superstructures in the desiccated state were observed after drying the protein (1 mg/mL) in sodium phosphate buffer (pH 6.5) on aluminum stages. The air-dried samples were dried further in a sealed desiccation chamber over anhydrous calcium sulfate (Drierite; WA Hammond) for 1 wk. The dried samples were then sputter-coated with gold and examined using SE2 scanning electron microscopy (SEM) using a Zeiss Supra 35 instrument. AFM samples were dried on mica and observed with tapping AFM using a SCANASYST-AIR_HPI probe (Bruker).
Cell Culture and Transgenic Cell Lines.
Drosophila melanogaster Kc167 cells were purchased from the Drosophila Genomics Research Center. Cells were cultured on 100 mm or 60 mm culture-treated dishes (Corning) in Schneider’s media (Caisson) supplemented with 10% heat-inactivated FBS (Atlanta Biologicals). Transfections were performed as described previously, with the exception that Schneider’s media was used in place of M3+BPYE medium (50). The pAc5-STABLE2-neo vector was purchased from Addgene. Detailed information on the pAc5-STABLE2-neo vector is available elsewhere (51). Corrected orientation and nucleotide sequences of all genetic constructs were confirmed by sequencing (GenScript), and immunoblotting was performed as described previously (30). The following primers were used to amplify and clone AfrLEA6: 5′-CGAGGTACCCAAACATGTCTGAGAATATTGGTCATATTAACATAAATGC-3′ and 5′-ATAGCGGCCGCAGTCCATGCGGACATTCCCAATAGTA-3′. The primers used for a chimeric protein composed of AfrLEA6 in frame with GFP were 5′-CGAGGTACCCAAACATGTCTGAGAATATTGGTCATATTAACATAAATGC-3′ and 5′-ACTGAGAATTCGTCCATGCGGACATTCCCAATAGTA-3′.
The primers used to amplify a truncated AfrLEA6 construct lacking the first SMP domain (Ser2 to Gly56) in frame with mCherry at the C terminus were 5′-CGGCGGTACCATGGCCTATGAATCGTGGAATATCAACC-3′ and 5′-ATGTGAATTCGTCCATGCGGACATTCCCAATAGTACTT-3′. The primers used to amplify a truncated AfrLEA6 construct lacking both SMP domains (Met1 to Glu140) fused to mCherry at the n-terminus were 5′-CGGCGCGGCCGCACAGGCCCATTTCAC-3′ and 5′-ATGTGCGGCCGCAGTCCATGCGGACATTCCCAATAGTA-3′.
The primers used to amplify a truncated AfrLEA6 construct lacking the postulated protein-binding domain (Tyr201 to Asp257) in frame with mCherry at the C terminus were 5′-CGGCGTACCATGTCTGAGAATATTGGTCAT3′ and 5′-ATGTGAATTCTGTGGTGATAGAAGGAGGGA-3′.
Confocal Microscopy.
All images were obtained with a Nikon A1R confocal microscope using Kc167 cells plated at ∼2.0 × 106 cells per compartment in a four-compartment, 35-mm CELLview cell culture dish (Greiner Bio-One). Cells were incubated in 500 µL of Schneider’s medium supplemented with 10% heat-inactivated FBS at 25 °C for 1 h before imaging. In experiments using 1,6-hexanediol (97%; Alfa Aesar), the compound was dissolved in Dulbecco’s PBS (DPBS; VWR). Cells were briefly washed with 500 µL of DPBS, followed by the addition of 200 µL of hexanediol solution. Cells were imaged for ∼40 min in the presence of the organic compound. Cells were then briefly rinsed with 500 µL of DPBS, and an additional 200 µL of DPBS was carefully added. After an ∼40-min recovery period, cells were again subjected to confocal microscopy.
Puromycin (Thermo Fisher Scientific) and cycloheximide (CHX; VWR) stock solutions (100 mg/mL and 2.5 mg/mL, respectively) were prepared fresh in cell culture-grade water (VWR). The working concentration of puromycin was 25 µg/mL, and CHX was added at 100 µg/mL to Schneider's medium supplemented with 10% heat-inactivated FBS. Cells were incubated at 25 °C for 2 to 3 h with the given antibiotic or control medium, followed by gentle washing with 200 µL of DPBS, and an additional 200 µL of antibiotic-free media was carefully added to keep cells hydrated while imaging. Nile Red stock solutions were generated by dissolving 1.0 mg/mL of 9-diethylamino-5H-benzo[a]phenoxazin-5-one (TCI America) in acetone. The working concentration of Nile Red was 0.1 µg/mL dissolved in DPBS. Cells were incubated with 500 µL of dilute Nile Red for 5 min at room temperature. After incubation, the cells were briefly rinsed with 500 µL of DPBS and resuspended in a final volume of 200 µL of DPBS. To observe Nile Red fluorescence during desiccation in control and AfrLEA6-expressing transgenic cells, 150 µL of DPBS was removed, and cells were allowed to air-dry while being imaged at varying intervals.
Cell Desiccation and Hyperosmotic Stress Experiments.
Desiccation experiments were performed as detailed previously (50). In brief, Kc167 cells were harvested and washed once with 2 to 3 mL of Schneider’s medium supplemented with 10% heat-inactivated FBS and 200 mM trehalose (Pfanstiehl) to yield a concentration of 2.0 × 107 cells/mL (±2.0 × 106 cells/mL) and left on ice for 1 h. Cells suspended in drying medium were placed on 60-mm cell culture dishes as 10 droplets of 15 µL each per plate, and samples were dried over salt isotherms or anhydrous calcium sulfate at 0%, 35% (MgCl2), and 75.5% (NaCl) relative humidity (RH). Culture dishes were measured gravimetrically before adding cells, after adding cells, and after desiccation to calculate gram water per gram dry weight using the dry weight determined after 12 h at 60 °C. After desiccation to various moister contents, samples were hydrated with 3 mL of Schneider’s medium supplemented with 10% heat-inactivated FBS and then incubated at 25 °C for 24 h. The cells in samples were subsequently enumerated, and percent membrane integrity relative to nondried control samples was determined by trypan blue exclusion. For statistical analysis, significance was set to P ≤ 0.05, and ANCOVA was performed using XLSTAT version 2018.6 (Addinsoft). Hyperosmotic stress experiments were performed by plating 3 to 4 × 107 cells/mL in Schneider’s medium supplemented with 10% heat-inactivated FBS and either 400 mOsmol/kg of NaCl or sucrose (VWR). The osmolarity of culture medium was verified by an Osmomat 030 freezing point depression osmometer (Gonotec). Cells were enumerated as described above. For statistical analysis, significance was set to P ≤ 0.05, and one-way ANOVA was performed using XLSTAT version 2018.6 (Addinsoft).
Results
Bioinformatics.
The A. franciscana protein AfrLEA6 is composed of three structurally distinct regions that exhibit unique sequence profiles. According to SMART-EMBEL, the N-terminus is dominated by two SMP domains with e-values of 2.6E−6 and 1.6E−4 compared with the PF04927 consensus sequence. Each of these SMP domains overlaps with a larger internal repeat that is enriched in a low-complexity alanine region. I-Tasser predicts that the two N-terminal SMP domains fold into α-helices (Fig. 1 A and B) that are overall negatively charged, with a mean net charge value of −0.2 (Fig. 1C). The N-terminal α-helices are separated from the C-terminal region by a 40- to 60-aa-long intrinsically disordered spacer region starting at amino acid position 140 that is enriched in proline, glycine, and aromatic or hydrophobic residues. At the C terminus, starting at around amino acid position 200, the net charge distribution alternates between highly positive (+0.5) and highly negative (−0.5), which is indicative of a protein-binding site. This region is predicted by I-Tasser to bind transcription-associated proteins, such as β-catenin (1g3jA; C-score 0.05). Overall, proteins with structural homology to AfrLEA6 and topological similarity scores >0.7 are associated with nuclear pores (e.g., Protein Data Bank [PDB] ID codes 4KNH, 4KF7, 5IJN) and DNA repair mechanisms (e.g., PDB ID codes 5YZ0, 5DLQ, 5LOI).
Fig. 1.
AfrLEA6 is predicted to have three distinct regions. (A) The N-terminal SMP repeats (purple) exhibit alpha-helical propensity. The C-terminal domain (green) exhibits fuzzy self-interactions. These two domains are linked by an intrinsically disordered spacer enriched in proline, glycine, and aromatic residues (blue). I-Tasser structural prediction of AfrLEA6 is based on hierarchical stability of known crystal structures, thus associating this structure with a possible conformation in the dried state. (B) SmartEMBLE identifies two N-terminal SMP domains in AfrLEA6 (purple). (C) The protein is overall negatively charged, with alternating charges (green) at the C terminus promoting interactions with other proteins or itself.
Behavior of AfrLEA6 in Solution.
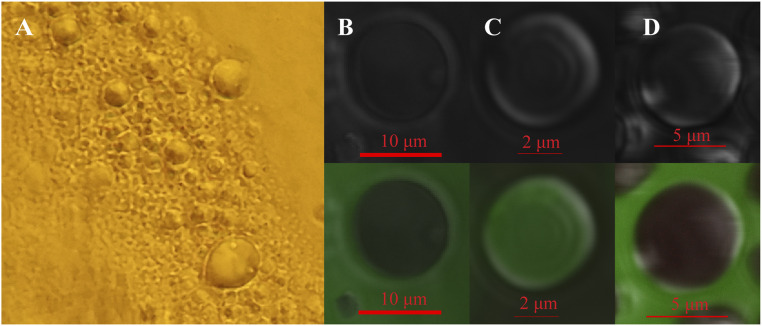
AfrLEA6 dissolves readily at a physiological concentration of ∼0.17 mg/mL (29) in a buffer mimicking the ionic strength and concentrations of the most dominant solutes present in the cytoplasm of hydrated diapausing cysts of A. franciscana (osmosome solution). During evaporative water loss from an osmosome solution containing AfrLEA6 at a starting concentration of 0.17 mg/mL, the protein readily separates from the remaining solvent as a liquid condensate when reaching a concentration of ∼0.25 mg/mL even in the absence of other proteins or nucleic acids (Fig. 2A). The formed droplets fuse on contact with one another and maintain a spherical structure, both during and after any fusion events. In a solution of pure water containing AfrLEA6 at a starting concentration of ∼0.2 mg/mL, droplets do not form during drying, and a thin layer of AfrLEA6 that resembles a glassy state lines the bottom of the sample after complete desiccation (SI Appendix, Fig. S1). The LLPS of AfrLEA6 in osmosome solution is likely to occur in the cytoplasm of desiccating brine shrimp embryos, but the cyst shell does not allow for direct imaging of this process, and cell fixatives used in preparation for electron microscopy may disrupt the detection of these structures.
Fig. 2.
AfrLEA6 undergoes a liquid-liquid phase separation that sequesters GFP based on surface charge. (A) AfrLEA6 (0.17 mg/mL) separates from solution into a liquid phase in a buffer mimicking the intracellular milieu of A. franciscana. (B) Standard GFP (stGFP-7) is partitioned outside of the AfrLEA6 droplet. (C) Positive GFP (pGFP+36) is selectively partitioned and enriched within the droplet, and (D) highly natively charged GFP (nGFP-30) is also excluded.
AfrLEA6 Condensates Partition GFP Constructs According to the Net Surface Charge of these Proteins.
To investigate whether protein surface charge affects intermolecular interactions between AfrLEA6 and other proteins, a 35-μM solution of AfrLEA6 was desiccated in a buffer containing 20 mM sodium phosphate (pH 6.5) to induce droplet formation in the presence of supercharged GFP proteins added at an equimolar ratio. The GFP proteins used have a calculated net surface charge of −7 (sGFP-7), +36 (pGFP+36), and −30 (nGFP-30) (52). When a solution of AfrLEA6 plus sGFP-7 was desiccated, LLPS formed droplets of similar size and shape as droplets formed in a solution of AfrLEA6 only. Interestingly, sGFP-7 is clearly excluded from the interior space of the formed protein droplets (Fig. 2B). In contrast, when AfrLEA6 is desiccated in the presence of pGFP+36, the protein droplets form more readily and are smaller compared with those that form in absence of other proteins, and pGFP+36 is incorporated into the droplet with such high efficiency that the surrounding solution becomes depleted of the fluorescent protein (Fig. 2C). As desiccation proceeds, these protein condensates expand and maintain a seemingly constant ratio of pGFP+36 and AfrLEA6. AfrLEA6 condensates formed in the presence of nGFP-30 also exclude this negatively charged fluorescent protein (Fig. 2D).
AfrLEA6 Enters a Gel Phase During Desiccation.
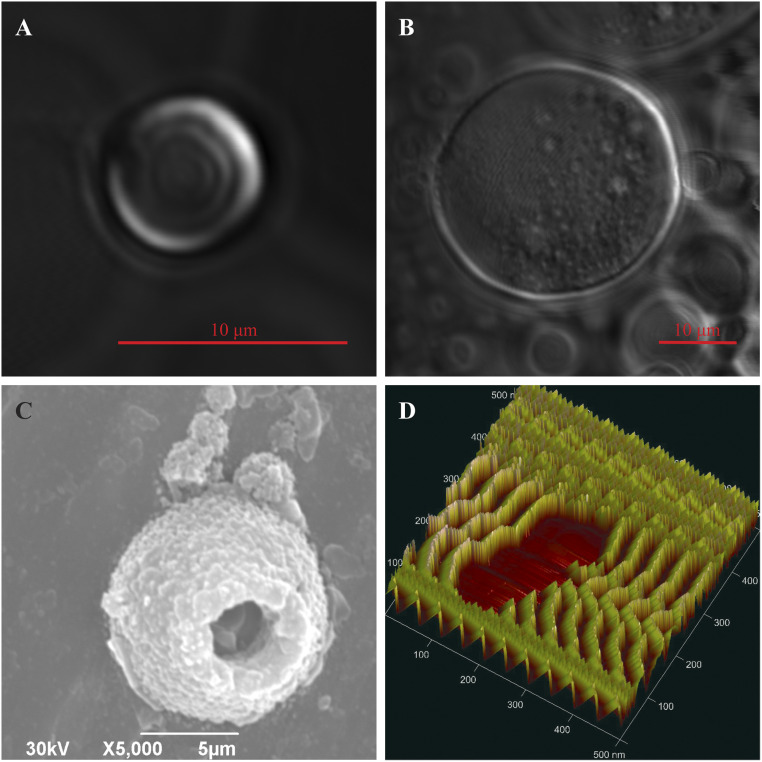
To characterize the behavior of AfrLEA6 during transition from a low-water environment to a fully desiccated state, samples of AfrLEA6 were observed using confocal microscopy during desiccation in 20 mM sodium phosphate buffer (pH 6.5) (Fig. 3A). As evaporation concentrates the proteins, the AfrLEA6 condensates expand in size and lose some of their liquid properties. These larger structures do not completely return to a spherical shape, but encapsulate spherical droplets of media, thus behaving more similarly to a hydrogel than a true liquid (53) (Fig. 3B). The hydrogel structure appears to be preserved in the desiccated state as shown by SEM and atomic force microscopy (Fig. 3 C and D), which is in contrast to the glassy state that AfrLEA6 enters if the protein is dried in absence of added salts (SI Appendix, Fig. S1).
Fig. 3.
AfrLEA6 undergoes phase transitions during desiccation. (A) Confocal microscopy shows that in vitro AfrLEA6 condensates are spherical and heterogenous at low to moderate dehydration. (B) AfrLEA6 condensates in vitro increase in viscosity and form a gel-like matrix at moderate to severe desiccation. (C) SEM imaging shows that some AfrLEA6 condensates maintain a spherical structure in the desiccated state. (D) AFM imaging reveals a series of mobile parallel proteins, aligning into a hydrogel structure.
LLPS of AfrLEA6 is Governed by the SMP Domains.
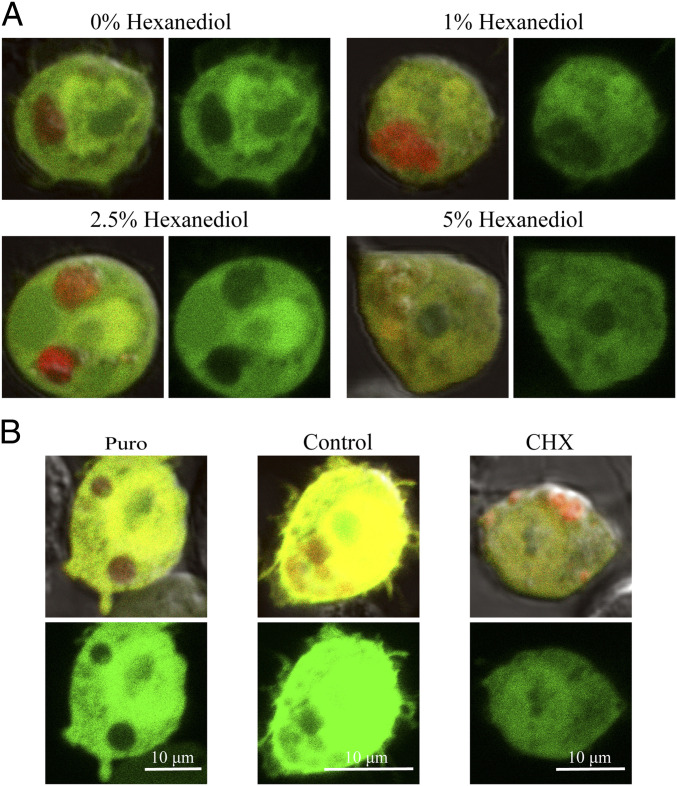
Kc167 cells concurrently expressing AfrLEA6-mCherry and enhanced GFP (eGFP) were imaged using confocal microscopy. The fluorescent proteins eGFP and mCherry both have a mean net surface charge of −5, which is similar to sGFP-7 used in the in vitro experiments described above (54, 55). Ectopic expressed AfrLEA6 concentrates into spherical structures that excluded eGFP and ranged in diameter from ∼1.0 µm to ∼5.0 µm (Fig. 4A and SI Appendix, Fig. S2). In addition, AfrLEA6 condensates imaged with differential interference contrast (DIC) show a sharp change in the refractive index compared to the surrounding cytosol (SI Appendix, Fig. S2, Bottom). A range of 1,6-hexanediol concentrations was used to determine whether ectopic expressed AfrLEA6 undergoes a liquid-liquid phase separation in the cytoplasm of Kc167 cells (Fig. 4A). This compound does not interfere with membrane-surrounded compartments but is able to interfere with weak intermolecular forces governing LLPS in vitro and in vivo (56–60). The addition of 1,6-hexanediol at 1% or 2.5% did not cause significant dispersal of AfrLEA6 condensates, while immediately after exposure to 5%, some cytoplasmic eGFP started to permeate into the AfrLEA6 droplets and prolonged exposure to this concentration (≥40 min) caused AfrLEA6 to fully disperse into the cytosol (Fig. 4A). However, cells exposed to 1,6-hexanediol at 1% and 2.5% regain a morphology similar to control cells in 20 min or less after the compound was removed, while cells treated with 5% remained intact but lost their ability to attach to the glass bottom of the culture dish (SI Appendix, Fig. S3).
Fig. 4.
AfrLEA6 condensates behave like stress granules in vivo. Colocalization of GFP with or exclusion from AfrLEA6 condensates is visualized by comparing images that overlay red (mCherry) and green (GFP) fluorescence with images showing only the green fluorescence signal (Right). (A) Cells expressing AfrLEA6 tagged with mCherry concurrently with GFP were incubated for 1 h with 0 to 5% (wt/vol) of 1,6-hexanediol in DPBS. (Images to the Left show overlay of red and green fluorescence and images to the Right show green fluorescence only.) (B) Cells expressing AfrLEA6-mCherry were subjected to either puromycin (25 µg/mL) or CHX (100 µg/mL) for 2 to 3 h. (Top images shows overlay of red and green fluorescence and Bottom image shows green fluorescence only.)
To further investigate the properties of AfrLEA6 condensates, puromycin and CHX were used to alter the size of the free ribosome pool in the cytoplasm via a decrease or an increase in polysome stability, respectively. After a 1-h incubation, the addition of puromycin caused AfrLEA6 condensates to become more spherical and larger and to exclude cytosolic eGFP more effectively compared with untreated controls. In contrast, CHX treatment caused AfrLEA6 condensates to become less spherical and more dispersed in the cytosol. In the few instances when AfrLEA6 droplets were still visible after 2 to 3 h of exposure to CHX, permeation of eGFP into the MLOs was clearly increased above that in untreated controls (Fig. 4B).
The overall architecture of AfrLEA6 can be bioinformatically described as a composite of three distinct protein regions: two SMP domains, a proline-glycine spacer, and a domain predicted to engage in interactions with other proteins (protein-binding domain). To characterize the protein domain(s) governing LLPS of AfrLEA6, several truncated forms of the protein were expressed in Kc167 cells. A construct lacking both SMP domains did not undergo LLPS, while a construct containing only the protein-binding domain formed many smaller MLOs compared with cells expressing the full-length protein (Fig. 5 A and B). However, the removal of one SMP domain yielded a construct that was still capable of forming condensates in the cytosol similar to the MLOs observed in cells expressing the full-length protein (Fig. 5C).
Fig. 5.
In vitro LLPS of AfrLEA6 is dependent on the SMP domain, while condensates fusion is facilitated by the predicted protein-binding domain. Kc167 cells expressing truncated versions of AfrLEA6 were imaged using confocal microscopy. Colocalization of GFP with or exclusion from AfrLEA6 condensates is visualized by comparing images that overlay red (mCherry) and green (GFP) fluorescence (Left) with images showing only the green fluorescence signal (Right). (A) LLPS of AfrLEA6 was absent when both SMP domains were removed. (B) LLPS of AfrLEA6 occurred when the predicted protein-binding domain was removed, but condensates fusion was hindered. (C) Removing only one of two SMP repeats had no apparent effect on the LLPS of AfrLEA6.
AfrLEA6 Increases Structural Integrity and Intracellular Viscosity of Kc167 Cells during Desiccation.
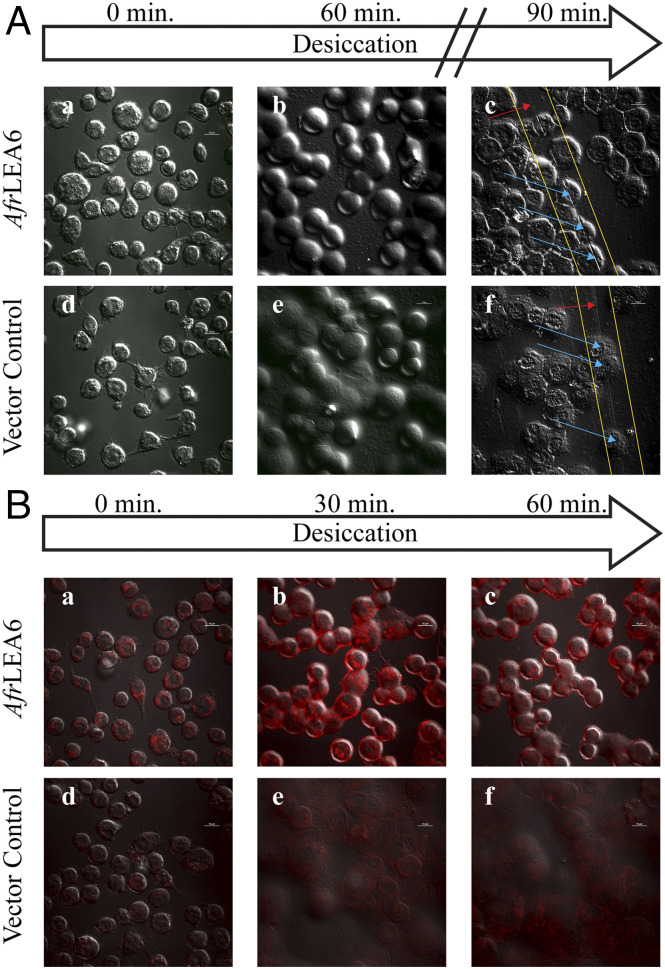
Kc167 cells expressing AfrLEA6 were imaged with DIC confocal microscopy while desiccating on the stage of the microscope (Fig. 6A). There were no apparent morphological differences between cells expressing AfrLEA6 and vector control cells (stably transfected with the corresponding vector construct but lacking AfrLEA6) when fully hydrated (Fig. 6 A, a and d). After 30 min of desiccation, control cells flattened out and plasma membranes tended to fuse among neighboring cells. In contrast, cells expressing AfrLEA6 retained a height closer to the height observed in fully hydrated cells even after a large amount of bulk water had been removed by evaporation. Surprisingly, fusion of membranes among neighboring cells was not as apparent as that in controls (Fig. 6 A, a to c). Furthermore, cells that express AfrLEA6 and were “scratched” after 90 min of desiccation show a sharp contrast in DIC pictures at the points of incisions, whereas control cells lack this response (Fig. 6 A, c, f).
Fig. 6.
AfrLEA6 expression increases the structural integrity and intracellular viscosity of Kc167 cells during desiccation. (A) Kc167 cells expressing AfrLEA6 (A, a to c) were desiccated concurrently with vector control cells (A, d to f). Cells expressing AfrLEA6 retained more of their native spherical morphology (A and B) than vector control cells (A, e). To demonstrate differences in cell height, samples were scratched multiple times with a pipette tip (A, c and f). Yellow lines indicate positions of performed scratches. Red arrows indicate areas where the scratch passed through salt deposits, and blue arrows indicate areas where the scratch passed through cells. (B) Fresh cells from both lines were incubated with Nile Red at a final concentration of 0.1 µg/mL for 5 min. Increasing red fluorescence is indicative of increasing intracellular viscosity. The decrease in red fluorescence between (B, b) and (B, c) may be due to the intracellular environment changing from a viscous gel to a glassy state, where Nile Red starts to precipitate out of solution and become nonfluorescent. Presented images are representative images from one of three separate trials. (Scale bars: 10 µm.)
To investigate the impact of AfrLEA6 on cytoplasmic viscosity during desiccation, control and transgenic Kc167 cells were stained with Nile Red and imaged with confocal microscopy during evaporative water loss (Fig. 6B). Nile Red is a solvatochromatic dye that has been extensively used for staining lipid droplets, but this indicator can also be used to observe viscosity changes in samples (61–64). Spherical structures with low Nile Red fluorescence in nondesiccating control cells are likely lipid droplets, while structures exhibiting bright fluorescence in fully hydrated cells expressing AfrLEA6 are thought to be MLOs composed of AfrLEA6. This assessment was based on the size and distribution of areas with increased fluorescence and the low concentration of Nile Red used, which was ∼50-fold lower than the concentration typically used for staining lipid vesicles in vivo (Fig. 6 B, a and d) (65). After 30 min of desiccation, there was a robust increase in Nile Red fluorescence throughout cells expressing AfrLEA6 but very little change in the fluorescence intensity of control cells (Fig. 6 B, b and e). After 60 min of desiccation, there was a steep decline in red fluorescence in cells expressing AfrLEA6, while fluorescence intensity for control cells remained unchanged (Fig. 6 B, c, f).
AfrLEA6 Increases Desiccation Tolerance of Kc167 Cells.
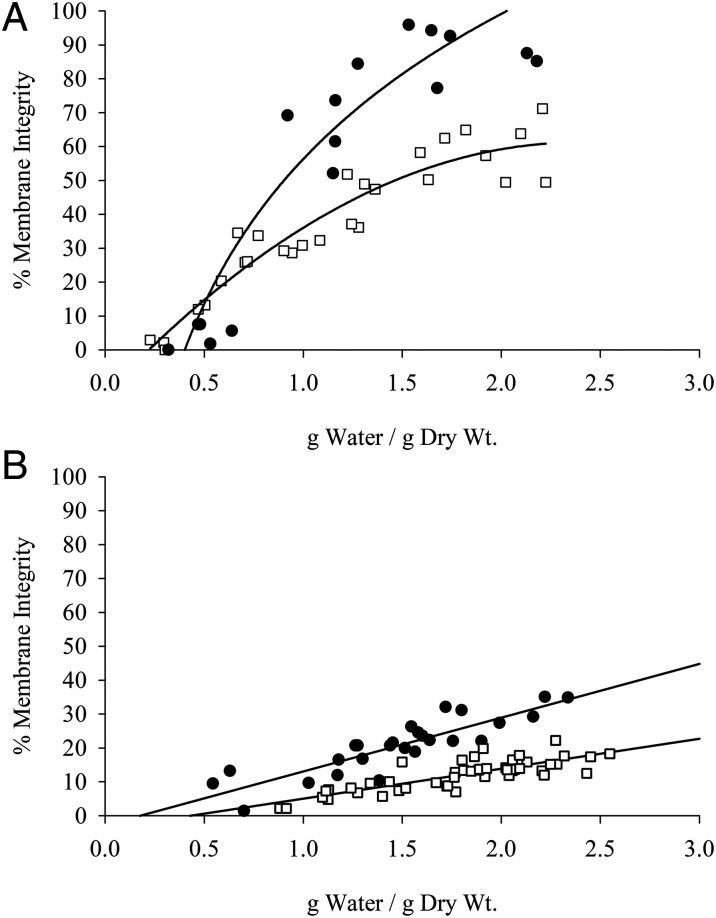
Kc167 cells expressing AfrLEA6 were desiccated at 0% (Fig. 7A) and 75.5% RH (Fig. 7B) to assess the impact of AfrLEA6 on viability after rehydration. Data for nontransfected control cells and cells stably transfected with a vector lacking AfrLEA6 were combined, since the response to desiccation was identical for both cell lines. Cells expressing AfrLEA6 maintained a higher percentage of cells with intact membranes at any given g H2O/g dry weight, and the faster drying rates observed at 0% RH vs. 75.5% RH were positively correlated with postrehydration viability (Fig. 7 A and B). For example, the time for control Kc167 cells to reach a moisture content of ∼0.5 g H2O/g dry weight increased from 2.5 h when desiccated at 0% RH to 8 h when dried at 75.5% RH, and viability dropped by ∼25% (SI Appendix, Fig. S4). In addition to improving desiccation tolerance, expression of AfrLEA6 also increased the osmotic-stress tolerance of Kc167 cells to medium supplemented with either 400 mOsmol/kg sucrose or NaCl (Fig. 8).
Fig. 7.
Desiccation tolerance is improved by AfrLEA6. Control Kc167 cells (open squares) and cells expressing AfrLEA6 (solid circles) were desiccated at an RH of 0% (A) and 75.5% (B). Cells expressing AfrLEA6 were more desiccation-tolerant at either RH level. (A) n = 18 to 28; F-statistic, 68.8 on 2 and 45 df; P < 0.01, ANCOVA. (B) n = 26 to 47; F-statistic, 121.9 on 2 and 72 df; P < 0.01, ANCOVA.
Fig. 8.
AfrLEA6 expression increases osmotic stress tolerance toward NaCl and sucrose. Nontransfected control (NTC), vector control (VC), and cells expressing AfrLEA6 were cultured for 48 h in culture medium supplemented with +400 mOsmol/kg of either NaCl (white bars) or sucrose (gray bars). In both cases, expression of AfrLEA6 significantly increased the osmotic stress tolerance of Kc167 cells. n = 9 to 18; P < 0.05, ANOVA. Different capital letters denote significant differences between cell lines in response to NaCl, while lowercase letters denote significant differences in response to sucrose.
Discussion
This study expands the known functions of proteinaceous condensates to include serving a protective role in anhydrobiosis. In vitro, AfrLEA6 condensates selectively incorporate or exclude GFP variants based on their net surface charge, indicating that in Kc167 cells structures formed that can be classified as MLOs. When ectopically expressed, the response of AfrLEA6 condensates to chemicals known to effect MLO stability, such as 1,6-hexanediol, CHX, and puromycin, is consistent with the behavior of a stress granule. Furthermore, a fraction of AfrLEA6 that remains dissolved in the cytoplasm increases intracellular viscosity significantly above control at modest levels of water loss. A rapid increase in cytoplasmic viscosity during desiccation likely provides physical support to intracellular structures during water loss in anhydrobiotic organisms. AfrLEA6 therefore confers protection during desiccation by engaging in two distinct molecular mechanisms in a previously unknown fashion: by forming target selective MLOs that may serve as alternative solvents for desiccation-sensitive proteins and by reinforcing cytoplasmic integrity.
AfrLEA6 is the only animal protein known to contain SMP domains and a role of this group 6 LEA protein in anhydrobiosis was previously indicated by bioinformatics and the observation that the protein is predominantly expressed in desiccation tolerant embryos of A. franciscana (27, 29). Proteins containing seed maturation domains are common in plants and have been linked to the longevity of orthodox seeds in the desiccated state, but the specific function(s) of the SMP domains remain unclear (10, 25, 66). Bioinformatics further indicated that this group 6 protein was capable of undergoing LLPS (14), and consequently, we investigated how protein architecture governs the LLPS of AfrLEA6 and found that the process was dependent on this domain. This finding may provide further insight into the observation that the group 6 LEA protein Rab28 (At3g22490) from Arabidopsis thaliana is selectively incorporated into the nucleolus of transgenic maize roots (67). Indeed, it would be highly instructive to know whether the SMP domains of other group 6 LEA proteins promote LLPS, and how sequence variations within this family of proteins impact the stability of the formed MLOs, their interactions with other preexisting MLOs (e.g., nucleolus, stress granules, p-granules), and their target selectivity.
Several observations support the hypothesis that the AfrLEA6 condensates behave like stress granules. First, a relatively high concentration of 1,6-hexanediol was required to disperse the in Kc167 cells formed AfrLEA6 condensates or increase their permeability for eGFP. Second, in the presence of puromycin, the structures formed by AfrLEA6 stabilize and become more defined, while the MLOs become destabilized in the presence of CHX. The compound 1,6-hexanediol dissipates membraneless organelles by interfering with the weak interactions that generally govern LLPS (37, 56–58, 60), and a relatively high resistance to being dissolved is characteristic for stress granules, which may not be governed solely by weak interactions (68). Furthermore, the results obtained with truncated AfrLEA6 constructs suggest that core initiation of the condensate is driven by the SMP domains, while the protein-binding domain may be interacting with client proteins at the periphery of the structure allowing the surrounding shells to fuse similar to stress granules (68–71).
A decrease in cytoplasmic conductivity in Kc167 cells expressing AfrLEA6 during hyperosmotic stress has recently been demonstrated via cellular dielectrophoresis (30). This observation adds evidence to the assessment that random coil regions of AfrLEA6 are folding into defined secondary structures when water levels are reduced, thereby decreasing the hydrodynamic radius of the polypeptide chain and subsequent surface area that can interact with free ions. This interpretation is in line with circular dichroism (CD) data on AfrLEA6, where the protein gains substantial amounts of α-helices during desiccation (29). However, CD describes only the ratio of secondary structural motifs for a protein that may exist in a wide ensemble of conformations, which may be especially common for intrinsically disordered proteins (IDPs), such as AfrLEA6 (27, 72, 73). Therefore, it cannot be assumed that all AfrLEA6 polypeptides contain the same secondary structure content. For the fraction of AfrLEA6 that remains in the dilute phase, folding may allow it to become entangled with other AfrLEA6 polypeptides and a wide variety of other proteins, thereby increasing intracellular viscosity by forming weak, cell-wide, promiscuous interactions. A rapid increase in intracellular viscosity via entanglement of AfrLEA6 in the diluted phase may act as prelude to entering a glassy state during severe desiccation and counteracts morphological changes early during water loss, thereby improving viability (74, 75). Structural support conveyed by AfrLEA6 may also contribute to the increase in the osmotic-stress tolerance observed for Kc167, which agrees with other studies that have suggested a role for group 6 LEA proteins in improving water-stress tolerance (76, 77).
Although we cannot exclude the possibility that positively charged proteins may interact with mRNA in vivo to allow for mRNA incorporation, owing to the overall negative charge of AfrLEA6 and the negative charge of the ribose-phosphate backbone, the incorporation of nucleic acids into AfrLEA6 condensates was not explored here. However, AfrLEA6 could significantly hinder cellular development by incorporating various biomolecules, which may explain why the protein is rapidly depleted in the cytoplasm of embryos on termination of diapause in A. franciscana (29). Denaturation of globular proteins with hydrophobic cores during desiccation is often irreversible and molecular shielding by LEA proteins is thought to prevent aggregate formation (78). LLPS may offer an additional protective mechanism to sterically hindering physical interactions among partially denatured proteins. The physicochemical environment of AfrLEA6 condensates might be compatible with the native structure of target proteins, in principle serving as a protective solvent at moderate levels of dehydration and by conformationally locking them through vitrification in the desiccated state. We are just starting to understand the role of LLPS in anhydrobiosis. Furthering our understanding of the mechanisms that govern physiochemical properties of anhydrobiosis-related MLOs offers the exciting possibility to engineer protein-based biomatrices for target-specific biostabilization in the desiccated state.
Supplementary Material
Acknowledgments
We thank Ozgur Yavuzcetin and Scott Janis (University of Wisconsin–Whitewater) for access to their scanning electron microscope and atomic force microscope. This work was supported by the NSF (IOS-1457061 and IOS-1659970).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014463117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Ball P., Water is an active matrix of life for cell and molecular biology. Proc. Natl. Acad. Sci. U.S.A. 114, 13327–13335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benner S. A., Ricardo A., Carrigan M. A., Is there a common chemical model for life in the universe? Curr. Opin. Chem. Biol. 8, 672–689 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Finney J. L., Water? What’s so special about it? Philos. Trans. R Soc. Lond. B Biol. Sci. 359, 1145–1163; discussion 1163–1145, 1323–1148 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yancey P. H., Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208, 2819–2830 (2005). [DOI] [PubMed] [Google Scholar]
- 5.McCluney K. E.et al., Shifting species interactions in terrestrial dryland ecosystems under altered water availability and climate change. Biol. Rev. Camb. Philos. Soc. 87, 563–582 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Alpert P., The constraints of tolerance: Why are desiccation-tolerant organisms small and rare? J. Exp. Biol. 209, 1575–1584 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Crowe J. H.et al., The trehalose myth revisited: Introduction to a symposium on stabilization of cells in the dry state. Cryobiology 43, 89–105 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Tunnacliffe A., Lapinski J., Resurrecting Van Leeuwenhoek’s rotifers: A reappraisal of the role of disaccharides in anhydrobiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1755–1771 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hand S. C., Menze M. A., Toner M., Boswell L., Moore D., LEA proteins during water stress: Not just for plants anymore. Annu. Rev. Physiol. 73, 115–134 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Leprince O., Pellizzaro A., Berriri S., Buitink J., Late seed maturation: Drying without dying. J. Exp. Bot. 68, 827–841 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Clegg J. S., Seitz P., Seitz W., Hazlewood C. F., Cellular responses to extreme water loss: The water-replacement hypothesis. Cryobiology 19, 306–316 (1982). [DOI] [PubMed] [Google Scholar]
- 12.Grelet J.et al., Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 137, 157–167 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dure L., Chlan C., Developmental biochemistry of cottonseed embryogenesis and germination: XII. Purification and properties of principal storage proteins. Plant Physiol. 68, 180–186 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janis B., Belott C., Menze M. A., Role of intrinsic disorder in animal desiccation tolerance. Proteomics 18, e1800067 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Solomon A., Salomon R., Paperna I., Glazer I., Desiccation stress of entomopathogenic nematodes induces the accumulation of a novel heat-stable protein. Parasitology 121, 409–416 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Browne J., Tunnacliffe A., Burnell A., Anhydrobiosis: Plant desiccation gene found in a nematode. Nature 416, 38 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Kikawada T.et al., Dehydration-induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem. Biophys. Res. Commun. 348, 56–61 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Hand S. C., Jones D., Menze M. A., Witt T. L., Life without water: Expression of plant LEA genes by an anhydrobiotic arthropod. J. Exp. Zool. A Ecol. Genet. Physiol. 307, 62–66 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Clark M. S.et al., Surviving extreme polar winters by desiccation: Clues from Arctic springtail (Onychiurus arcticus) EST libraries. BMC Genomics 8, 475 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pouchkina-Stantcheva N. N.et al., Functional divergence of former alleles in an ancient asexual invertebrate. Science 318, 268–271 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Wise M. J., Tunnacliffe A., POPP the question: What do LEA proteins do? Trends Plant Sci. 9, 13–17 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Wu G.et al., Diverse LEA (late embryogenesis abundant) and LEA-like genes and their responses to hypersaline stress in post-diapause embryonic development of Artemia franciscana. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 160, 32–39 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Sharon M. A., Kozarova A., Clegg J. S., Vacratsis P. O., Warner A. H., Characterization of a group 1 late embryogenesis abundant protein in encysted embryos of the brine shrimp Artemia franciscana. Biochem. Cell Biol. 87, 415–430 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Warner A. H.et al., Evidence for multiple group 1 late embryogenesis abundant proteins in encysted embryos of Artemia and their organelles. J. Biochem. 148, 581–592 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Chatelain E.et al., Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 35, 1440–1455 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Leprince O., Buitink J., Introduction to desiccation biology: From old borders to new frontiers. Planta 242, 369–378 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Janis B., Uversky V. N., Menze M. A., Potential functions of LEA proteins from the brine shrimp Artemia franciscana: Anhydrobiosis meets bioinformatics. J. Biomol. Struct. Dyn. 36, 3291–3309 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Boswell L. C., Menze M. A., Hand S. C., Group 3 late embryogenesis abundant proteins from embryos of Artemia franciscana: Structural properties and protective abilities during desiccation. Physiol. Biochem. Zool. 87, 640–651 (2014). [DOI] [PubMed] [Google Scholar]
- 29.LeBlanc B. M., Le M. T., Janis B., Menze M. A., Hand S. C., Structural properties and cellular expression of AfrLEA6, a group 6 late embryogenesis abundant protein from embryos of Artemia franciscana. Cell Stress Chaperones 24, 979–990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashed M. Z., Belott C. J., Janis B. R., Menze M. A., Williams S. J., New insights into anhydrobiosis using cellular dielectrophoresis-based characterization. Biomicrofluidics 13, 064113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dirk L. M. A.et al., Late embryogenesis abundant protein-client protein interactions. Plants 9, 814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuevas-Velazquez C. L., Dinneny J. R., Organization out of disorder: Liquid-liquid phase separation in plants. Curr. Opin. Plant Biol. 45, 68–74 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Gomes E., Shorter J., The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Leeuwen W., Rabouille C., Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 20, 623–638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nott T. J.et al., Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt H. B., Görlich D., Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 41, 46–61 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Sehgal P. B., Westley J., Lerea K. M., DiSenso-Browne S., Etlinger J. D., Biomolecular condensates in cell biology and virology: Phase-separated membraneless organelles (MLOs). Anal. Biochem. 597, 113691 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Uversky V. N., Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18–30 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Brangwynne C. P., Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 203, 875–881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abatzopoulos T. J., Beardmore J., Clegg J. S., Sorgeloos P., Artemia: Basic and Applied Biology, (Springer, ed. 1, 2002). [Google Scholar]
- 42.Letunic I., Bork P., 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I., Doerks T., Bork P., SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holehouse A. S., Das R. K., Ahad J. N., Richardson M. O., Pappu R. V., CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 112, 16–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J.et al., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J., Zhang Y., I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson D. B., Cronican J. J., Liu D. R., Engineering and identifying supercharged proteins for macromolecule delivery into mammalian cells. Methods Enzymol. 503, 293–319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glasheen J. S., Hand S. C., Metabolic heat dissipation and internal solute levels of artemia embryos during changes in cell-associated water. J. Exp. Biol. 145, 263–282 (1989). [Google Scholar]
- 50.Marunde M. R.et al., Improved tolerance to salt and water stress in Drosophila melanogaster cells conferred by late embryogenesis abundant protein. J. Insect Physiol. 59, 377–386 (2013). [DOI] [PubMed] [Google Scholar]
- 51.González M.et al., Generation of stable Drosophila cell lines using multicistronic vectors. Sci. Rep. 1, 75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence M. S., Phillips K. J., Liu D. R., Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–10112 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonker A. M., Löwik D. W. P. M., van Hest J. C. M., Peptide- and protein-based hydrogels. Chem. Mater. 24, 759–773 (2012). [Google Scholar]
- 54.Kim B., Lam C. N., Olsen B. D., Nanopatterned protein films directed by ionic complexation with water-soluble diblock copolymers. Macromolecules 45, 4572–4580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norris D. M.et al., An improved Akt reporter reveals intra- and inter-cellular heterogeneity and oscillations in signal transduction. J. Cell Sci. 130, 2757–2766 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Updike D. L., Hachey S. J., Kreher J., Strome S., P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192, 939–948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kroschwald S.et al., Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molliex A.et al., Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel S. S., Belmont B. J., Sante J. M., Rexach M. F., Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Ribbeck K., Görlich D., The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haidekker M. A., Theodorakis E. A., Environment-sensitive behavior of fluorescent molecular rotors. J. Biol. Eng. 4, 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levitt J. A., Chung P. H., Suhling K., Spectrally resolved fluorescence lifetime imaging of Nile red for measurements of intracellular polarity. J. Biomed. Opt. 20, 096002 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Swain J., Mishra A. K., Nile red fluorescence for quantitative monitoring of micropolarity and microviscosity of pluronic F127 in aqueous media. Photochem. Photobiol. Sci. 15, 1400–1407 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Yablon D. G., Schilowitz A. M., Solvatochromism of nile red in nonpolar solvents. Appl. Spectrosc. 58, 843–847 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Greenspan P., Mayer E. P., Fowler S. D., Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100, 965–973 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verdier J.et al., A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 163, 757–774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amara I., Capellades M., Ludevid M. D., Pagès M., Goday A., Enhanced water stress tolerance of transgenic maize plants over-expressing LEA Rab28 gene. J. Plant Physiol. 170, 864–873 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Wheeler J. R., Matheny T., Jain S., Abrisch R., Parker R., Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain S.et al., ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis D.et al., Human antiviral protein MxA forms novel metastable membraneless cytoplasmic condensates exhibiting rapid reversible tonicity-driven phase transitions. J. Virol. 93, e01014-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shiina N., Liquid- and solid-like RNA granules form through specific scaffold proteins and combine into biphasic granules. J. Biol. Chem. 294, 3532–3548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uversky V. N., Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 11, 739–756 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oldfield C. J., Dunker A. K., Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Chakraborty N.et al., A spin-drying technique for lyopreservation of mammalian cells. Ann. Biomed. Eng. 39, 1582–1591 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Boothby T. C.et al., Tardigrades use intrinsically disordered proteins to survive desiccation. Mol. Cell 65, 975–984.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boudet J.et al., Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol. 140, 1418–1436 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borrell A.et al., Arabidopsis thaliana atrab28: A nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol. 50, 249–259 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Chakrabortee S.et al., Intrinsically disordered proteins as molecular shields. Mol. Biosyst. 8, 210–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.