Significance
Chikungunya virus (CHIKV) is a significant human pathogen that causes debilitating and long-lasting arthritis. Currently, there is no approved vaccine or specific therapeutic. We show that two highly potent anti-CHIKV antibodies—CHK-124 and CHK-263—can inhibit multiple steps of the CHIKV infection cycle and determined their cryogenic electron microscopy structures in complex with CHIKV particles to a 4- to 5-Å resolution. We describe the structural details of the epitopes of CHK-124 and CHK-263 and how they relate to their functional mechanisms of neutralization. Our results provide important information that will advance antibody therapeutics and vaccine development against this emerging pathogen.
Keywords: antibody, chikungunya virus, cryo-EM, epitope
Abstract
Chikungunya virus (CHIKV) is an emerging viral pathogen that causes both acute and chronic debilitating arthritis. Here, we describe the functional and structural basis as to how two anti-CHIKV monoclonal antibodies, CHK-124 and CHK-263, potently inhibit CHIKV infection in vitro and in vivo. Our in vitro studies show that CHK-124 and CHK-263 block CHIKV at multiple stages of viral infection. CHK-124 aggregates virus particles and blocks attachment. Also, due to antibody-induced virus aggregation, fusion with endosomes and egress are inhibited. CHK-263 neutralizes CHIKV infection mainly by blocking virus attachment and fusion. To determine the structural basis of neutralization, we generated cryogenic electron microscopy reconstructions of Fab:CHIKV complexes at 4- to 5-Å resolution. CHK-124 binds to the E2 domain B and overlaps with the Mxra8 receptor-binding site. CHK-263 blocks fusion by binding an epitope that spans across E1 and E2 and locks the heterodimer together, likely preventing structural rearrangements required for fusion. These results provide structural insight as to how neutralizing antibody engagement of CHIKV inhibits different stages of the viral life cycle, which could inform vaccine and therapeutic design.
Chikungunya virus (CHIKV), a single-stranded positive-sense RNA envelope virus, is an emerging alphavirus transmitted to humans by Aedes species mosquitoes (1, 2). CHIKV consists of three related genotypes: Asian, East/Central/South African (ECSA), and West African (3). According to the Centers for Disease Control and Prevention, there have been millions of cases reported in approximately 100 countries. CHIKV infection causes an acute febrile illness accompanied by musculoskeletal disease (4, 5). A subset of cases (∼30%) showed that chronic arthritis can develop and persist for months to years (6, 7).
The 12-kb positive-sense RNA genome is packaged within an icosahedral nuclear capsid core composed of 240 copies of capsid proteins, which is surrounded by a host-derived lipid bilayer. The surface of the mature CHIKV particle (diameter ∼700 Å) has 80 trimeric envelope E1-E2 heterodimer protein spikes anchored on the lipid bilayer membrane (SI Appendix, Fig. S1A) arranged in T = 4 icosahedral symmetry. E1 and E2 protein ectodomains each consist of three domains: E1-DI; E1-DII and E1-DIII; and E2-DA, E2-DB, and E2-DC (SI Appendix, Fig. S1B). The fusion loop on the distal end of E1-DII mediates endosomal membrane fusion. The groove formed by E2-DA and E2-DB shields the fusion loop of E1 protein from premature membrane fusion at neutral pH (8). Multiple attachment factors have been implicated in CHIKV entry of cells (9), and E2-DB reportedly contains receptor-binding sites (10, 11). Mxra8, a recently identified alphavirus receptor (12), recognizes an epitope spanning both the E1 and E2 proteins (13, 14).
The virus infection cycle starts with the E1-E2 proteins binding to the cell-surface receptors (12). The virion is then internalized into the endosome (15, 16). The acidic condition of the endosome causes E1-E2 heterodimers to undergo conformational changes, exposure of the E1 fusion loop for insertion into the endosomal membrane, and subsequent reorganization of the E1 protein into E1 trimers to allow endosomal membrane fusion (17, 18). After fusion, the capsid and RNA genome are released into the cytoplasm (19) to allow translation and replication of the viral genome. The newly synthesized virus buds at the plasma membrane (20).
Currently, there exist no licensed CHIKV vaccine or therapeutics. Neutralizing antibodies have been shown to confer both prophylactic and therapeutic protection in animal models (21–28). Here we show the potencies of two CHIKV antibodies, CHK-124 and CHK-263, in vivo and demonstrate that they inhibit multiple steps in the virus infection cycle in vitro. We also determined the cryogenic electron microscopy (cryo-EM) structures of their Fab fragments complexed with CHIKV to 4- to 5-Å resolution. For CHK-124, the predominant neutralization mechanisms are aggregation of virus particles and inhibition of receptor binding. For CHK-263, the mechanism is the inhibition of fusion by locking E1 and E2 proteins together. Altogether, our study provides a structural understanding as to how potent antibodies block CHIKV infection.
Results
CHK-124 and CHK-263 Are Potently Neutralizing In Vivo.
We previously reported that the mouse monoclonal antibodies (mAbs) CHK-124 and CHK-263 exhibit potent neutralizing activity in vitro using focus reduction neutralization tests (21). We confirmed the neutralization activity of CHK-124 and CHK-263 using a separate assay (a plaque reduction neutralization test [PRNT]50 of 1.8 and 3.8 ng/mL) against a clinical isolate CHIKV East African strain in Vero cells using PRNT (SI Appendix, Fig. S2).
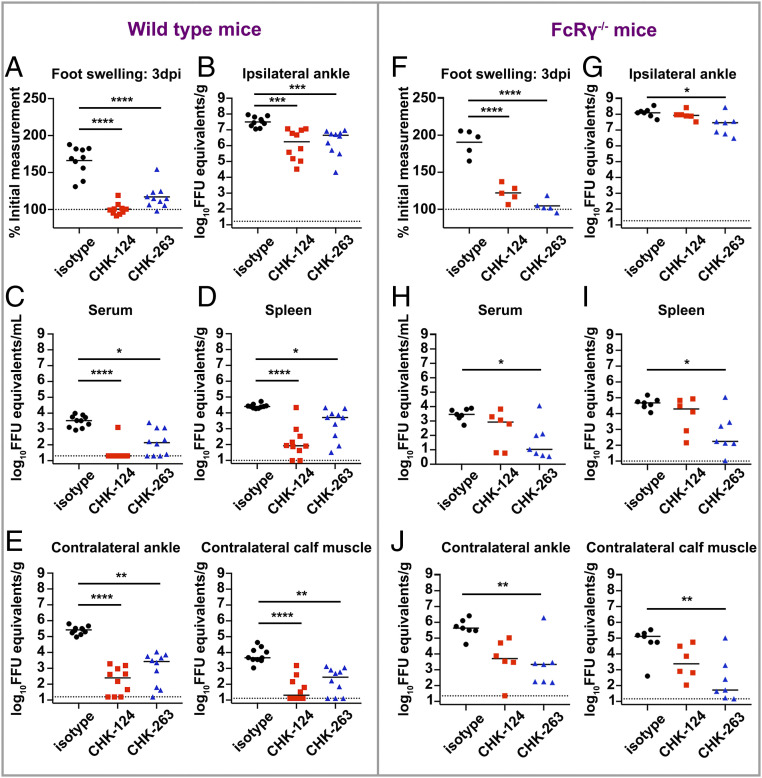
We next evaluated the efficacy of CHK-124 and CHK-263 in vivo using a wild-type (WT) C57BL/6 mouse model of CHIKV arthritis and musculoskeletal disease by subcutaneous inoculation in the foot (29, 30). Treatment with CHK-124 or CHK-263 prior to CHIKV inoculation significantly reduced foot swelling at 3 d post infection (3 dpi) (Fig. 1A). This phenotype was associated with reduced levels of viral RNA in the ipsilateral ankle (Fig. 1B), diminished viremia, reduced infection of the spleen, and decreased spread to contralateral ankle and gastrocnemius (calf) muscle compared to the isotype control mAb (Fig. 1 C–E). Thus, CHK-124 and CHK-263 are potently neutralizing mAbs in vitro and confer protective activity in vivo.
Fig. 1.
CHK-124 and CHK-263 protect mice against CHIKV diseases and dissemination. (A–J) CHK-124 and CHK-263 mAbs reduced foot swelling and the viral RNA levels in ankles, muscles, blood, and spleen of CHIKV-infected wild-type (A–E) or congenic FcRγ−/− (F–J) mice. Four-week-old wild-type or FcRγ−/− C57BL/6 mice were treated with 100 µg of CHK-124, CHK-263, or an isotype control (WNV E60) 1 d before infection with 103 FFU of CHIKV. (A and F) Foot swelling was measured at 3 dpi. Horizontal line indicates mean values (n = 5 to 10/group; two experiments; one-way ANOVA with a Tukey’s posttest; ****P < 0.0001). (B and G) Ipsilateral ankle, (C and H) serum, (D and I) spleen, and (E and J) contralateral ankle and gastrocnemius (calf) muscle were harvested 3 dpi, and viral RNA levels were determined by qRT-PCR. Bars indicate median values (n = 5 to 10/group); two experiments; Kruskal–Wallis with a Dunn’s posttest; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Each symbol in this figure represents data from an individual mouse.
To investigate whether the inhibitory activity of these antibodies requires FcR effector functions, we repeated the same infection experiments in congenic mice lacking the Fc receptor common gamma chain (FcRγ−/−). In WT mice, both antibodies prevented CHIKV-induced infection and joint swelling, with CHK-124 showing slightly better prevention activity than CHK-263. However, this pattern was reversed in FcRγ−/− mice (Fig. 1 F–J). Thus, the in vivo inhibitory activity of CHK-124 is partially FcRγ-dependent whereas the protective effect of CHK-263 is largely independent.
CHK-124 and CHK-263 Block Multiple Pathways in the Virus Infection Cycle.
Antibodies can neutralize virus infection by different mechanisms including aggregation of virus particles, prevention of viral attachment, fusion inhibition, or viral egress blockade.
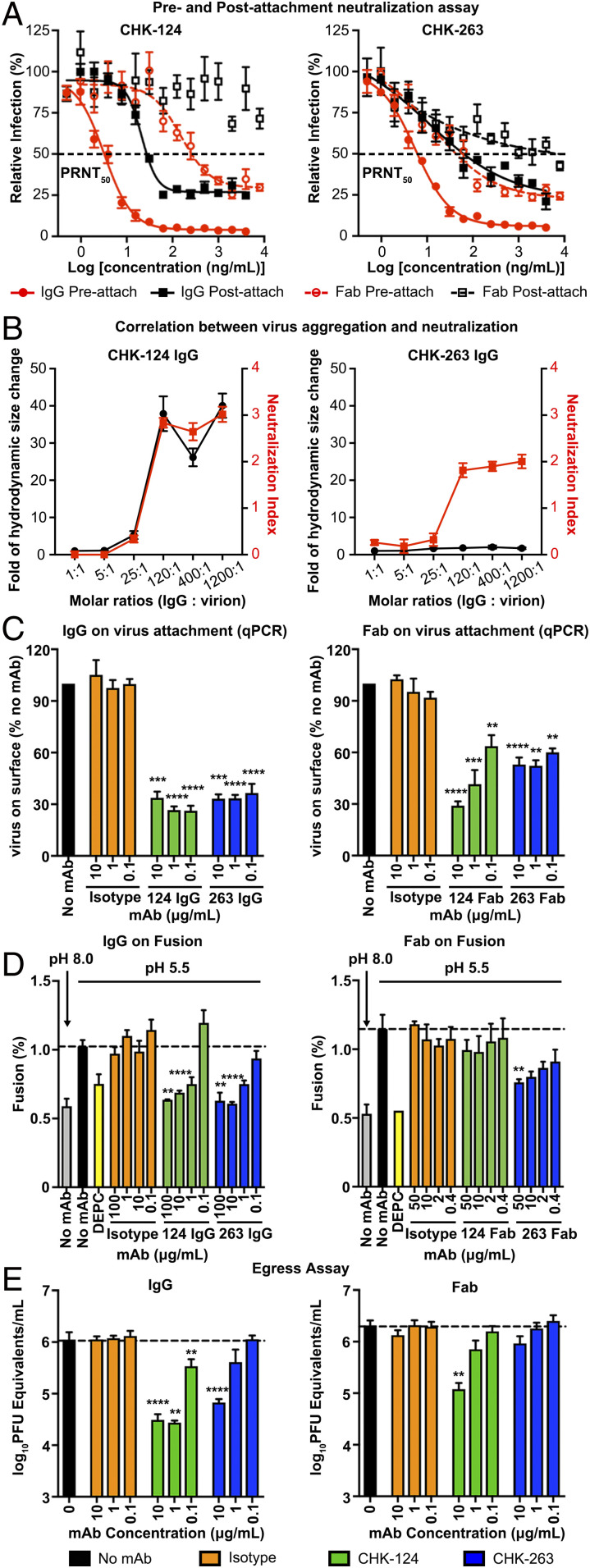
To investigate whether CHK-124 and CHK-263 inhibit viral entry, we conducted preattachment and postattachment neutralization assays (21, 31). In these assays, antibody was added either before or after the virus attachment to cells, respectively. The preattachment neutralization assay determines if antibodies block viral engagement prior to cellular attachment (e.g., through aggregation of virions or inhibition of attachment to cells). Both CHK-124 IgG and CHK-263 IgG neutralize primarily by inhibiting preattachment stages of infection with the PRNT50 value of 3.2 and 5.5 ng/mL, respectively (Fig. 2A), whereas their PRNT50 values are poorer in postattachment neutralization—21.2 and 10.8 ng/mL, respectively. In the postneutralization assay, both CHK-124 IgG and CHK-263 IgG were unable to neutralize 30% of virus even at high antibody concentrations. This phenotype also was observed previously for other antibodies (21, 32, 33). Although the reason is not known, we speculate that, when the virus is first allowed to bind to the cell surface, some particles engaging the cell are less accessible to antibody binding. Because these regions cannot be bound by antibody, the stoichiometry of binding required for virus neutralization may not be achieved (33) and these viral particles still can carry out entry and fusion.
Fig. 2.
CHK-124 and CHK-263 target multiple pathways in the CHIKV infection cycle. In general, CHK-124 inhibitory activities were more dramatically reduced when bivalency of IgG was abolished by the use of Fab fragments compared to CHK-263. (A) CHK-124 and CHK-263 IgGs are highly neutralizing in both pre- and postattachment neutralization assays, whereas their Fabs have weaker activities. PRNT50 values indicating the antibody concentration that neutralized 50% of the plaque-forming units was determined by curve fitting using nonlinear regression in GraphPad Prism v8.0. (B) CHK-124 IgG induced virus aggregation, which correlates with its neutralization profile, whereas CHK-263 does not cause virus aggregation. The hydrodynamic size of CHIKV:CHK-124 IgG and CHIKV:CHK-263 IgG complexes in a different IgG:virion molar ratio was measured by dynamic light scattering (black curve). The neutralization profile of these CHIKV:IgG complexes (red curve) is shown as a neutralization index (right y axis) calculated as the log10 fold reduction of the virus titer compared to the virus-only control. (C) Both CHK-124 and CHK-263 IgG and Fab prevent virus from attaching to cells if antibody:CHIKV complex is formed before addition to cells. Isotype IgG/Fab and no antibody controls were included. (D) Virus:liposomal membrane fusion assays showed that both IgGs of CHK-124 and CHK-263 inhibit virus:liposomal membrane fusion at pH 5.5, similar to the positive control (dethylpyrocarbonate [DEPC]). When Fab fragments were used, the inhibitory effect of CHK-124 was abolished whereas that of CHK-263 Fab was maintained. The extent of fusion was calculated as the percentage of the fluorescence emission before adding Triton X-100 to the full fluorescence emission after adding Triton X-100. (E) CHK-124 has stronger inhibitory activity of viral egress than CHK-263. (A–E) Data are the mean ± SEM from at least three independent experiments. Significance was determined by one-way ANOVA with Dunnett’s posttest compared to isotype control. (**P < 0.01, ***P < 0.001, ****P < 0.0001).
We also conducted preattachment and postattachment neutralization assays with Fab fragments of CHK-124 and CHK-263; they are consistently less neutralizing than their IgG counterparts (Fig. 2A). In the preattachment neutralization, CHK-124 Fab (PRNT50 = 152.9 ng/mL) and CHK-263 Fab (PRNT50 = 13.9 ng/mL) have 47- and 2.5-fold poorer activities than the corresponding IgG. In the postattachment neutralization assay, CHK-124 Fab completely lost its neutralization activity, whereas CHK-263 Fab has 1,000-fold less activity (PRNT50 = ∼10 μg/mL) than its IgG. Thus, bivalent binding and/or virus cross-linking likely enhances the ability of these antibodies to neutralize virus. This is especially important for CHK-124.
To determine if the antibodies aggregate virions in solution, we added different molar ratios of IgG to virion and measured their hydrodynamic size using dynamic light scattering (Fig. 2B). CHK-124 started aggregating virus particles at IgG:virus molar ratios above 25:1, which correlated with its neutralizing activity. In comparison, CHK-263 did not aggregate virus at any IgG:virus molar ratios tested, including molar ratios above 25:1. The cryo-EM images of these complexes at a IgG:E2 (on virus) molar ratio of 1.5:1 also showed CHK-124 IgG severely aggregated viruses whereas CHK-263 did not (SI Appendix, Fig. S3).
We next tested whether CHK-124 and CHK-263 could inhibit CHIKV binding to cells. We preincubated CHIKV with IgG or their Fabs at neutralizing concentrations prior to adding to Vero cells at 4 °C, which prevents virus internalization. The amount of cell-bound virus was then determined by RT-qPCR. Both CHK-124 IgG and CHK-263 IgG at the concentrations tested (0.1 to 10 μg/mL) inhibited virus attachment to cells (Fig. 2 C, Left). Their Fab fragments retained their ability to inhibit virus attachment to cells, although less efficiently (Fig. 2 C, Right). This indicates that bivalency of IgG enhances the inhibitory effect on virus attachment.
Upon viral entry, antibodies can inhibit endosomal membrane fusion and penetration into the cytoplasm. To determine if CHK-124 and CHK-263 inhibited fusion, we used an in vitro system, where 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD)-labeled purified CHIKV particles were tested for their ability to fuse with liposomes in the presence or absence of antibodies. In cases where fusion occurs, the DiD in the viral membrane is diluted within the liposomal membrane, thereby emitting fluorescence. Both CHK-124 and CHK-263 IgG inhibited membrane fusion in a dose-dependent manner (Fig. 2 D, Left). However, CHK-124 Fab was unable to inhibit fusion (Fig. 2 D, Right), possibly due to its inability to aggregate virions in a monovalent form (Fig. 2B). In comparison, CHK-263 Fab moderately inhibited virus fusion with liposomes (Fig. 2 D, Right).
We next evaluated whether both IgG and Fab of CHK-124 and CHK-263 block virus egress. We inoculated cells with CHIKV and then removed unbound virus particles 1 h later through extensive rinsing of monolayers. Subsequently, mAbs were added and, 1 h later, viral RNA was measured in the supernatant; as expected, at this early time point, no antibody inhibitory effect was detected (SI Appendix, Fig. S4). We then measured the amount of viral RNA at 6 h post infection, when the first round of virus secretion started. Compared to the no mAb or isotype control mAb conditions, CHK-124 IgG has a greater ability to inhibit viral egress than CHK-263 IgG (Fig. 2 E, Left). The Fab fragments of CHK-124 and CHK-263 has less and virtually no ability to block egress, respectively (Fig. 2 E, Right).
The cryo-EM Structures of CHIKV Complexed with Fab Fragments of CHK-124 and CHK-263.
To determine CHK-124 and CHK-263 epitopes, we performed cryo-EM studies using Fab fragments of these antibodies complexed with CHIKV. Micrographs showed that preincubation with CHK-124 or CHK-263 Fab fragments did not aggregate virus or substantially alter its morphology (SI Appendix, Fig. S5).
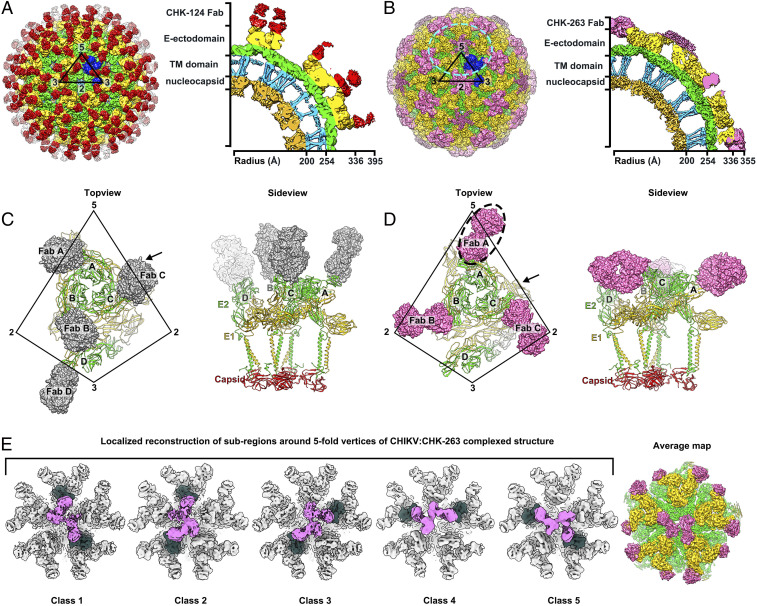
The cryo-EM reconstructions of CHIKV:CHK-124 Fab and CHIKV:CHK-263 Fab were determined to 5.2- and 4.7-Å resolutions, respectively, based on the Fourier Shell Correlation cutoff of 0.143 (SI Appendix, Figs. S6A and S7A). Binding of CHK-124 Fab and CHK-263 Fab to CHIKV did not induce large conformational changes to the overall quaternary virus structure, with the root mean square deviation (rmsd) values of 1.497 and 1.596, respectively, compared to unliganded CHIKV-VLPs (Protein Data Bank [PDB] ID: 6NK5) (13). We observed 240 CHK-124 Fab fragments bound to the CHIKV virion at the top of the spikes, orienting perpendicularly with respect to the viral lipid envelope (Fig. 3A and SI Appendix, Fig. S6 A–C).
Fig. 3.
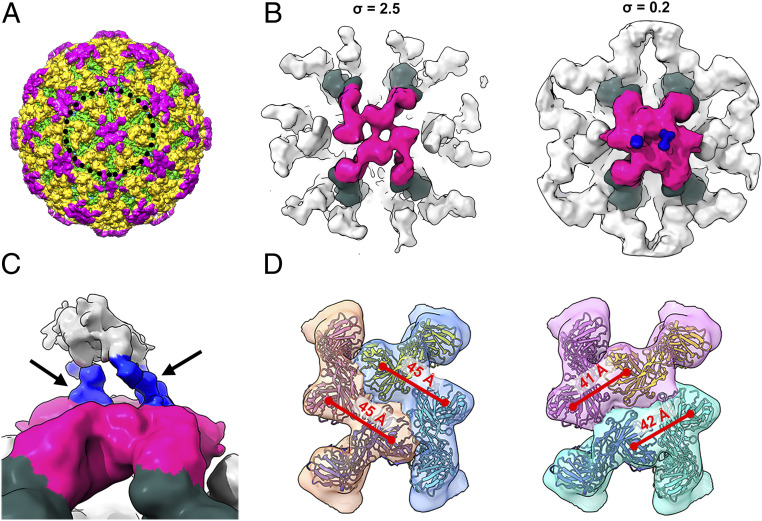
The cryo-EM maps of CHIKV complexed with Fab fragment of CHK-124 or CHK-263. (A and B) The cryo-EM density maps of CHIKV:CHK-124 Fab (A) and CHIKV:CHK-263 Fab (B) were determined to 5.2- and 4.7-Å resolutions, respectively. Their surface and a cross-section of a quarter of cryo-EM density maps are shown in the Left and Right panels, respectively. The black triangles represent an icosahedral asymmetric unit, and their five-, three-, and two-fold vertices are indicated. The E1-E2 heterodimers in one icosahedral asymmetric unit are colored as blue. (C and D) Top (Left) and side (Right) view of the binding of CHK-124 Fab (C) and CHK-263 Fab (D) to an asymmetric unit of CHIKV surface E1-E2 proteins. CHK-124 Fabs bind to all individual E1-E2 heterodimers within an asymmetric unit in an orientation perpendicular to the virus lipid envelope surface. CHK-263 Fabs lie laterally to the virus surface binding to three of the four E1-E2 (molecules A–C but not D). The Fab A (dashed circle) density is poor, suggesting partial occupancy around the five-fold vertex. E1:E2 heterodimers are shown as ribbons and the Fab molecules as surface representations. CHK-124 and CHK-263 Fab molecules are colored in gray and pink, respectively. The CHIKV E1, E2, and capsid proteins are colored in yellow, green and red, respectively. Vertices are indicated. (E) Localized reconstruction of subregions around the five-fold vertices in the CHIKV:CHK-263 Fab-complexed structure (cyan circle in B, Left). Results showed five different classes of the subparticles. All indicate that at most only two Fabs can bind around the five-fold vertex and are unable to bind to two E1-E2 heterodimers located right next to each other. The Fab densities are colored as hot pink, and the E1-E2 heterodimers bound by these Fabs are colored in dark green. (Right) The averaged map of these classes after they have been rotationally aligned to each other at 4.5-Å resolution.
CHK-263 Fab fragments, in contrast, bound laterally to the E1-E2 heterodimers on the virus surface (Fig. 3B). We observed Fab densities at the two- and five-fold vertices but not in the E1-E2 trimeric spikes at the three-fold vertex. Although the Fab densities were observed around the five-fold vertex, weaker electron density likely indicates partial occupancy (Fig. 3B). To understand how CHK-263 Fab binds around the five-fold vertex, we performed localized three-dimensional (3D) reconstruction (34) with classification but no shift or rotation alignment nor symmetry averaging imposed. The result shows five classes of subparticles (Fig. 3E), with different combinations of two Fab molecules binding to two different E1-E2 heterodimers. Consistent among these classes, we observed that the two Fab molecules around the five-fold vertex were unable to bind to their immediate neighboring E1-E2 heterodimers, likely due to the need for spacing to avoid steric hindrance between Fabs. To obtain a higher resolution map, we combined the subregion classes by rotational alignment and then averaged them (35). The structure of the averaged subregions showing two Fabs bound around the five-fold vertex was determined to a resolution of 4.5-Å (Fig. 3E and SI Appendix, Fig. S7B). We also performed localized reconstruction for subregions around the three-fold vertex and two-fold (also known as quasi-six-fold) vertex (SI Appendix, Fig. S7 C and D). Their structures are similar to the icosahedrally averaged CHKV:CHK-263 complex. No Fabs were bound near the three-fold vertex. Around the icosahedral two-fold (or quasi-six-fold) vertex, the occupancy of CHK-263 Fab is not full: only four of the six epitopes are bound (SI Appendix, Fig. S7D). The occupancy here is greater than around the five-fold vertex, which has only two Fabs bound, because there is more space and less steric hindrance around the quasi-six-fold vertex. Overall, CHK-263 Fab showed selective binding to the E1-E2 proteins on the virus surface with only 144 Fabs (60% occupancy) bound to the virion.
We then interpreted the cryo-EM density by fitting the CHK-124 and CHK-263 Fabs and the CHIKV E1-E2 heterodimer into their corresponding densities (SI Appendix, Figs. S6D, S7E, and Table S1). Contrary to the previously determined CHIKV-181/25 cryo-EM structure (13, 32), we did not observe densities corresponding to the E3 protein, a protein present on immature CHIKV. Fitting of the complexed particles and subregions at different vertices showed similar Fab:epitope-interacting interfaces. Due to the resolution, some side-chain densities cannot be placed accurately. We thus identified the likely contact residues between the CHIKV envelope proteins and the Fab molecules using a cutoff of distance <∼8 Å between pairs of Cα atoms. We also considered the local hydrophilic or hydrophobic environment.
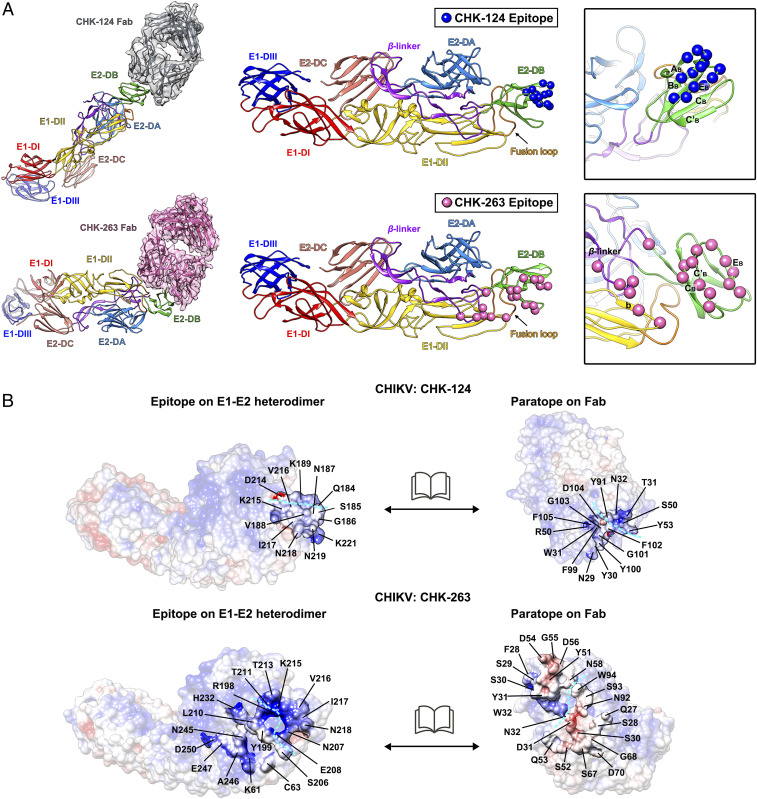
Within an asymmetric unit, all four CHK-124 Fab molecules bind to E2 proteins in a similar conformation (Fig. 3C) with an average rmsd value of 0.813 (SI Appendix, Fig. S8A). The binding interfaces of the CHK-124 Fab molecules on the E2 protein occlude ∼489 Å2 of surface area. The interaction is focused on a small region on the outer surface of the E2-DB, without contacting the E1 protein or adjacent E1-E2 heterodimers (Fig. 3C). CHK-124 Fab primarily binds to two peptides on the E2-DB (Fig. 4A). One peptide fragment is located in the ABBB loop and extends to the BB strand (Fig. 4A and SI Appendix, Fig. S9A). A second peptide fragment stretches from the EB strand to the EBη3 loop. The paratope of the CHK-124 Fab consists of residues from the heavy chain and light chain (Fig. 4 B, Top, and SI Appendix, Table S2). The epitope and paratope side-chain charges are not highly complementary (Fig. 4 B, Top), suggesting that some side chains may interact with the main chain, similar to that found in other virus-antibody studies [e.g., the Zika virus-C8 mAb interaction (36)].
Fig. 4.
Epitopes bound by CHK-124 Fab and CHK-263 Fab. (A) Binding mode of CHK-124 Fab (Top) and CHK-263 Fab (Bottom) to the ectodomains of an E1-E2 heterodimer. Epitopes of CHK-124 and CHK-263 are shown as blue and pink spheres, respectively, on an E1-E2 heterodimer (Middle and Right). (Right) Zoom-in view of the epitope with strands labeled. (B) Open book representation showing the electrostatic potential of the interacting interface between the epitope and paratope of CHIKV:CHK-124 (Top) and CHIKV:CHK-263 Fab (Bottom) structures. Positive, negative, and neutral charged residues are colored in blue, red, and white, respectively. Cyan dashed lines indicate the boundaries of the antibody heavy and light chains in the paratope and also its corresponding binding epitope. Residues at the interacting interface are indicated.
Despite the different chemical environments, the three bound CHK-263 Fabs (A, B, and C) interact with the CHIKV E1-E2 heterodimer in a similar conformation (Fig. 3D) with average rmsd values of 0.896 (SI Appendix, Fig. S8B). In contrast to CHK-124, the CHK-263 Fab molecule has a larger footprint (∼918 Å2). The epitope on the CHIKV envelope proteins spans across E1 and E2 proteins (Fig. 4A), and it has complementary charges to the paratope (Fig. 4 B, Bottom, and SI Appendix, Table S3).
Most of the residues in the epitope of CHK-263 are located in the E2-DB, engaging the CB strand, CBC′B loop, C′B strand, C’BEB loop, and the EB strand (Fig. 4A and SI Appendix, Fig. S9A). The β-linker connecting E2-DB with E2-DC and two residues (K61 and C63) on the E1-DII also contribute to the interaction with the CHK-263 Fab molecule.
In conclusion, the cryo-EM structures of CHIKV complexed with the Fab fragments of CHK-124 and CHK-263 showed that the CHK-124 Fab binds exclusively to E2-DB, whereas CHK-263 Fab binds to an epitope spanning E2-DB, E2 β-linker, and E1-DII. We subsequently used biolayer interferometry to determine the affinity of the CHK-124 and CHK-263 Fabs to CHIKV. Notably, CHK-263 Fab binds to CHIKV with a KD of ∼2.5 nM, which is an ∼20-fold higher affinity than the CHK-124 Fab (45.4 nM) (SI Appendix, Fig. S10). This higher affinity of binding correlates with the footprint area of Fabs on CHIKV, as CHK-263 has a two-fold larger interface (∼918 Å2) than CHK-124 (∼489 Å2).
Cryo-EM Structure of CHK-263 IgG Complexed with CHIKV.
Since CHK-263 IgG does not aggregate CHIKV, we determined the icosahedrally averaged structure of the CHIKV:CHK-263 IgG complex to 5.9-Å resolution (Fig. 5A and SI Appendix, Fig. S11A). The location of the densities of the CHK-263 IgG (Fig. 5A) largely overlap with the Fab:CHIKV complex (Fig. 3B and SI Appendix, Fig. S11B). In addition, there are some additional weak densities above the Fab that likely belong to the Fc region of the IgG around both five- and two- (or quasi-six-) fold vertices (SI Appendix, Fig. S11A). The poor densities of the Fc could be due to icosahedral averaging, so we conducted localized reconstruction of the regions around the five- and two- (or quasi-six-) fold vertices without imposing symmetry. The IgG densities around the five-fold vertex did not improve: the Fab and Fc region densities were weak, likely because the IgG binding is flexible. As for the two-fold (quasi-six-fold) vertex, asymmetric localized reconstruction yielded a 9.4-Å resolution map (SI Appendix, Fig. S11C), clearly showing two hinge densities (Fig. 5 B and C and SI Appendix, Fig. S11C) (hinge region links the Fabs to the Fc region of IgG). This suggests that, around this vertex, two IgG molecules can bind at the same time (Fig. 5 B and C). Although we were unable to determine specifically which two Fab molecules belongs to an IgG, we narrowed down the binding mode of the IgGs to two possibilities (Fig. 5D) by measuring distances between neighboring Fabs (distance between two Fabs in an IgG should be <87 Å [PDB ID: 1IGT]) (37).
Fig. 5.
The cryo-EM structure of CHIKV complexed with CHK-263 IgG. (A) Surface of the cryo-EM structure of CHIKV complexed with CHK-263 IgG. E1, E2, and IgG are colored in green, yellow and magenta, respectively. (B) The cryo-EM map of the localized reconstruction of subregions around the two-fold (quasi-six-fold) vertex of CHIKV:CHK-263 IgG complex, displayed at different contour levels. The densities corresponding to the Fabs of the IgG are colored in magenta, while their bound E1-E2 heterodimers are colored in dark green. Blue densities indicate two hinge densities each from an IgG. We therefore detected two IgGs bound around the two-fold vertex. (C) Zoom in side view of the bound IgGs. Black arrows pointing to the two hinges (in blue) each from an IgG. Weak densities corresponding to the Fc regions are colored in gray. (D) Two possible arrangements of the two IgGs around the two-fold vertex. The densities of two Fabs possibly from an IgG are in the same color. Distance between the adjacent Fab molecules of CHK-263 IgG is indicated in red fonts above the red lines. Measurement of an IgG crystal structure (PDB ID: 1IGT) shows that the distance between two Fabs in an IgG is within 87 Å.
Discussion
A previous study (21) and our current study demonstrate that mAb CHK-124 and CHK-263 are highly neutralizing in vitro across strains from the three CHIKV genotypes (ECSA, Asian, and West African) as well as other alphaviruses. Sequence comparison shows that the CHK-124 epitope on E2-DB is conserved across not only the three CHIKV genotypes but also across other alphaviruses including O’nyong’nyong virus (ONNV), Mayaro virus (MAYV), Ross River virus (RRV), Sagiyama virus, Semliki Forest virus (SFV) and Una virus (SI Appendix, Fig. S9B). This is consistent with the previous study showing that CHK-124 broadly neutralizes CHIKV, MAYV, SFV, and ONNV (26). The CHK-263 epitope across E1 and E2 also showed high conservation across the three CHIKV genotypes (SI Appendix, Fig. S9A), consistent with its cross-neutralization activities.
Most anti-CHIKV antibodies (e.g., 5F10, CHK-265, and RRV-12) that have been characterized structurally (26, 32, 38–40) are similar to CHK-124, as they bind largely to E2-DB. CHK-263 and CHK-166 (21) are interesting as they both bind across E1 and E2 proteins, although only the epitope of CHK-263 is confirmed structurally. A possible reason for why most antibodies bind only to E2 could be due to its higher solvent accessibility compared to E1 protein, which is partially hidden underneath. We also showed that CHK-263 Fab has a 20-fold higher affinity than CHK-124 Fab, which correlates to the size of their footprints.
In the structure of the CHIKV:CHK-263 Fab complex, only three (E1-E2 molecules [mols] A to C) of the four epitopes were engaged by the Fabs within an asymmetric unit even though all epitopes are accessible (mols A to D) (Fig. 3D). Superposition of CHK-263 Fab fragment onto E1-E2 mol D shows that this Fab would clash with the neighboring bound Fabs B and C′ (from the neighboring asymmetric unit) (SI Appendix, Fig. S12). Thus, binding of Fabs to either E1-E2 mols B or C′ could block engagement of Fab to E1-E2 mol D. This suggests that, at low antibody concentrations, if both E1-E2 mols B and C are not occupied, Fab may be able to engage E1-E2 mol D.
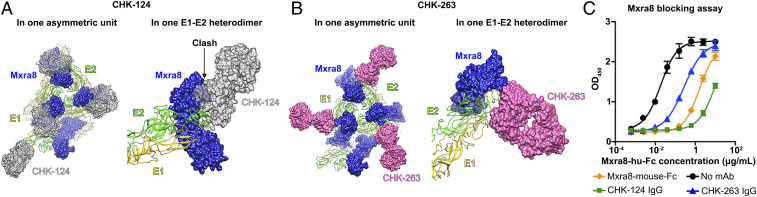
Both CHK-124 and CHK-263 inhibit virus attachment to cells. Mxra8 receptor had been shown to bind across different E1-E2 heterodimers on the CHIKV surface and its epitopes include E2-DA and E2-DB; E2 β-linker; and E1-DII (13, 14). Superposition of Mxra8 and CHK-124 Fab onto the E1-E2 heterodimer shows clashes (Fig. 6A). This suggests that CHK-124 may inhibit CHIKV from binding to the Mxra8 receptor. Indeed, our direct-binding enzyme-linked immunosorbent assay (ELISA) data showed that CHK-124 IgG strongly blocked Mxra8 binding to CHIKV virions (Fig. 6C). Antibody-induced virus aggregation had been shown to reduce the number of infectious units (41), but it may not prevent these aggregated viruses from attaching to cells (42, 43). For CHK-124, we observed that, in addition to causing virus aggregation (Fig. 2B and SI Appendix, Fig. S3), it also prevents viral attachment to cells (Fig. 2 C, Left), likely because Fab specifically blocks Mxra8 receptor binding. This idea is supported by the observation that, when aggregation is abolished using Fab fragments, virus attachment to cells is still inhibited (Fig. 2 C, Right). As the CHK-263 epitope does not overlap with the Mxra8-binding site (Fig. 6B), its virus-attachment–inhibiting activity might target other unknown attachment factors (9).
Fig. 6.
Superposition of the CHIKV receptor Mxra8 onto the cryo-EM structures of the CHIKV:Fab complexes. (A and B) Superposition of the CHIKV receptor Mxra8 to the CHK-124 Fab (A) and CHK-263 Fab (B) structures within an asymmetric unit (Left). Zoom-in side views of E1-E2 heterodimer (Right) show that the binding of CHK-124 may clash with Mxra8, whereas CHK-263 Fab and Mxra8 can bind simultaneously to the E1-E2 heterodimer. E1: yellow ribbons; E2: green ribbons; Mxra8: blue surfaces; CHK-124: gray surfaces; CHK-263: pink surfaces. (C) CHK-124 IgG can strongly inhibit virus from binding to Mxra8 whereas CHK-263 IgG has only moderate activity. Blocking of Mxra8-Fc binding to CHK-124 or CHK-263 complexed CHIKV was determined by competition ELISA. CHIKV VLPs were captured with CHK-152 and CHK-166 before addition of the CHIKV mAbs or Mxra8-mouse-Fc followed by Mxra8-hu-Fc (human Fc). Black circles indicate Mxra8-hu-Fc binding to CHIKV VLP in the absence of anti-CHIKV mAbs. A rightward shift of the curve indicates competition of CHIKV mAbs with Mxra8-mFc for binding to CHIKV. Data are the mean ± SEM from two independent experiments performed in duplicate.
From the aggregation assay data (Fig. 2B) and cryo-EM images (SI Appendix, Fig. S3), we showed that CHK-263 does not cause aggregation of virus. The asymmetric localized reconstruction of the CHK-263 IgG complexed with CHIKV around its two-fold (quasi-six-fold) vertex showed that there are two IgG molecules bound (Fig. 5 B–D). Since the Fab arms of both IgGs are engaged, they are unlikely to cross-link other virus particles, thereby preventing virus aggregation. We were unable to determine the structure of the CHK-124 IgG complexed with CHIKV, as this antibody aggregates virus particles (SI Appendix, Fig. S3). To understand the mechanism of how this antibody causes aggregation, we superimposed a Fab arm of the crystal structure of an IgG (37) onto the Fabs in the cryo-EM structure of the CHIKV:CHK-124 Fab complex (SI Appendix, Fig. S13A). This shows that when one arm of the Fab of an IgG is bound to the virus, another Fab points away to engage other virus particles (SI Appendix, Fig. S13 A and B).
In conclusion, both CHK-124 and CHK-263 potently inhibit different steps of the virus infection cycle. The major neutralization mechanism of CHK-124 IgG is antibody-induced aggregation, which inhibit virus-endosomal membrane fusion and egress. Aggregation of virions in solution or viral antigen on the plasma membrane of cells may more efficiently activate effector functions and immune cells (44, 45). This idea is consistent with the greater dependency of CHK-124 on FcRγs for protection. A more specific inhibitory mechanism of CHK-124 is that it directly blocks virus interaction with the Mxra8 receptor. In contrast, CHK-263 binds laterally to the virus surface and its epitope spans across E1 and E2 proteins. While the mechanism by which CHK-263 partially blocks virus attachment to cells requires further investigation, its ability to bridge E1 and E2 together likely prevents E1 dissociation from E2 during endosomal fusion. The thorough understanding of the neutralization mechanisms and how the antibodies interact with virus particles could inform the development of vaccines and therapeutics to combat CHIKV infection.
Materials and Methods
A detailed description of all materials and methodology is included in SI Appendix, Materials and Methods. This includes the cells and virus, mAb CHK-263 and CHK-124 production and digestion, plaque reduction neutralization test, in vivo experiments, pre- and postattachment neutralization assays, dynamic light scattering assay, attachment RT-qPCR assay, DiD-labeled virus fusion assay, virus egress assay, Mxra8-blocking experiment, biolayer interferometry assay, statistical analysis, cryo-EM sample preparation, data collection, processing, and analysis.
Sample Preparation for Cryo-EM.
CHIKV virus samples were mixed with the CHK-263 Fab or CHK-124 Fab or CHK-263 IgG at a molar ratio of 1.5 Fab/IgG to every E2 protein, incubated at 37 °C for 30 min, and then kept at 4 °C before freezing on cryo-EM grids. About 2 μL of the sample was applied to a precooled cryo-EM grid with lacey carbon covered with a thin carbon film. The grid was then blotted with filter paper at 4 °C with 100% humidity for 1 to 2 s and flash-frozen in liquid ethane by using the Vitrobot Mark IV plunger (FEI).
Data Collection and Image Processing.
The images of the frozen CHIKV-Ross:CHK-124 Fab complex, CHIKV-East African:CHK-263 Fab complex, and CHIKV-East African:CHK-263 IgG complex were taken with the FEI Titan Krios electron microscope at 300 keV with a nominal magnification of 47,000× and a FEI Falcon II direct electron detector. Images were collected using Leginon (46) in movie mode, with a total exposure of 2 s and a total dose of ∼20 e−/Å2. The frames from each movie were aligned using MotionCorr (47) to produce full-dose images used for particle selection and orientation search, and the images from the first several frames amounting to the dose of about 20 e−/Å2 were used in 3D reconstruction. The images were taken at a defocus range of 0.5- to 2.5-μm. cisTEM (48) and Gctf (49) were used to estimate the astigmatic defocus parameters. Particle picking was undertaken using cisTEM. Orientation search and 3D reconstruction of the picked particles were performed using cisTEM and RELION (50, 51). The reconstruction process is detailed in SI Appendix, Materials and Methods and Table S1.
Supplementary Material
Acknowledgments
This work was supported by the Duke-National University of Singapore Signature Research Programme funded by the Ministry of Health, Singapore, and awarded to S.-M.L.; and by NIH Grants R01 AI141436, R01 AI114816, R01 AI127513, U19 AI142790, and contract AI201800001 awarded to M.S.D. We thank the Defense Medical & Environmental Research Institute for the generous gifts of the Chikungunya virus Ross strain and Chikungunya virus East African strain.
Footnotes
Competing interest statement: M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Emergent BioSolutions and serves on the Scientific Advisory Board of Moderna. The M.S.D. laboratory at Washington University School of Medicine has received sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008051117/-/DCSupplemental.
Data Availability.
The cryo-EM map of the CHIKV:CHK-124 Fab complex, CHIKV:CHK-263 Fab complex, CHIKV:CHK-263 IgG complex, subregion around the five-fold vertex of the CHIKV:CHK-263 Fab complex, and subregion around the two-fold vertex of the CHIKV:CHK-263 IgG complex has been deposited in the Electron Microscopy Data Bank under the accession codes EMD-30476, EMD-30477, EMD-30478, EMD-30479, and EMD-30480, respectively. Their atomic models have been deposited in the Protein Data Bank under the accession codes 7CVY, 7CVZ, 7CW0, 7CW2, and 7CW3, respectively.
All study data are included in the article and supporting information.
References
- 1.Tsetsarkin K. A., Vanlandingham D. L., McGee C. E., Higgs S., A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 3, e201 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahid B., Ali A., Rafique S., Idrees M., Global expansion of chikungunya virus: Mapping the 64-year history. Int. J. Infect. Dis. 58, 69–76 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Powers A. M., Brault A. C., Tesh R. B., Weaver S. C., Re-emergence of chikungunya and O’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 81, 471–479 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Pialoux G., Gaüzère B.-A., Jauréguiberry S., Strobel M., Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 7, 319–327 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Couderc T., Lecuit M., Chikungunya virus pathogenesis: From bedside to bench. Antiviral Res. 121, 120–131 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Hoarau J.-J., et al., Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J. Immunol. 184, 5914–5927 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Young A. R., et al., Dermal and muscle fibroblasts and skeletal myofibers survive chikungunya virus infection and harbor persistent RNA. PLoS Pathog. 15, e1007993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss J. E., et al., Glycoprotein organization of chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Schnierle B. S., Cellular attachment and entry factors for chikungunya virus. Viruses 11, 1078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva L. A., et al., A single-amino-acid polymorphism in chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J. Virol. 88, 2385–2397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber C., et al., Identification of functional determinants in the chikungunya virus E2 protein. PLoS Negl. Trop. Dis. 11, e0005318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang R., et al., Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basore K., et al., Cryo-EM structure of chikungunya virus in complex with the Mxra8 receptor. Cell 177, 1725–1737.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song H., et al., Molecular basis of arthritogenic alphavirus receptor MXRA8 binding to chikungunya virus envelope protein. Cell 177, 1714–1724.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Bernard E., et al., Endocytosis of chikungunya virus into mammalian cells: Role of clathrin and early endosomal compartments. PLoS One 5, e11479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoornweg T. E., et al., Dynamics of chikungunya virus cell entry unraveled by single-virus tracking in living cells. J. Virol. 90, 4745–4756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbons D. L., et al., Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Li L., Jose J., Xiang Y., Kuhn R. J., Rossmann M. G., Structural changes of envelope proteins during alphavirus fusion. Nature 468, 705–708 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung J. Y., Ng M. M., Chu J. J., Replication of alphaviruses: A review on the entry process of alphaviruses into cells. Adv. Virol. 2011, 249640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown R. S., Wan J. J., Kielian M., The alphavirus exit pathway: What we know and what we wish we knew. Viruses 10, 89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal P., et al., Development of a highly protective combination monoclonal antibody therapy against chikungunya virus. PLoS Pathog. 9, e1003312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goh L. Y., et al., Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin. Immunol. 149, 487–497 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Fric J., Bertin-Maghit S., Wang C.-I., Nardin A., Warter L., Use of human monoclonal antibodies to treat chikungunya virus infection. J. Infect. Dis. 207, 319–322 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Fong R. H., et al., Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J. Virol. 88, 14364–14379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith S. A., et al., Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against chikungunya virus. Cell Host Microbe 18, 86–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox J. M., et al., Broadly neutralizing alphavirus antibodies bind an epitope on E2 and inhibit entry and egress. Cell 163, 1095–1107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broeckel R., et al., Therapeutic administration of a recombinant human monoclonal antibody reduces the severity of chikungunya virus disease in rhesus macaques. PLoS Negl. Trop. Dis. 11, e0005637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couderc T., et al., Prophylaxis and therapy for chikungunya virus infection. J. Infect. Dis. 200, 516–523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner J., et al., Chikungunya virus arthritis in adult wild-type mice. J. Virol. 84, 8021–8032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison T. E., et al., A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: Evidence of arthritis, tenosynovitis, myositis, and persistence. Am. J. Pathol. 178, 32–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fibriansah G., et al., A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 6, 6341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell L. A., et al., Human mAbs broadly protect against arthritogenic alphaviruses by recognizing conserved elements of the Mxra8 receptor-binding site. Cell Host Microbe S1931-3128, 30404-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierson T. C., et al., The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1, 135–145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilca S. L., et al., Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres S. H. W., Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 579, 125–157 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Barba-Spaeth G., et al., Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Harris L. J., Larson S. B., Hasel K. W., McPherson A., Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Sun S., et al., Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2, e00435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long F., et al., Cryo-EM structures elucidate neutralizing mechanisms of anti-chikungunya human monoclonal antibodies with therapeutic activity. Proc. Natl. Acad. Sci. U.S.A. 112, 13898–13903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porta J., et al., Structural studies of chikungunya virus-like particles complexed with human antibodies: Neutralization and cell-to-cell transmission. J. Virol. 90, 1169–1177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brioen P., Dekegel D., Boeyé A., Neutralization of poliovirus by antibody-mediated polymerization. Virology 127, 463–468 (1983). [DOI] [PubMed] [Google Scholar]
- 42.Wang J., et al., A human bi-specific antibody against Zika virus with high therapeutic potential. Cell 171, 229–241e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumkosit U., et al., Anti-chikungunya virus monoclonal antibody that inhibits viral fusion and release. J. Virol. 94, e00252-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin J., et al., Neutralizing antibodies inhibit chikungunya virus budding at the plasma membrane. Cell Host Microbe 24, 417–428.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klimpel G. R., “Immune defenses” in Medical Microbiology, ed. 4, Baron S, Ed. (University of Texas Medical Branch at Galveston, 1996). [PubMed] [Google Scholar]
- 46.Carragher B., et al., Leginon: An automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 132, 33–45 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Li X., Zheng S., Agard D. A., Cheng Y., Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 192, 174–178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant T., Rohou A., Grigorieff N., cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zivanov J., et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM map of the CHIKV:CHK-124 Fab complex, CHIKV:CHK-263 Fab complex, CHIKV:CHK-263 IgG complex, subregion around the five-fold vertex of the CHIKV:CHK-263 Fab complex, and subregion around the two-fold vertex of the CHIKV:CHK-263 IgG complex has been deposited in the Electron Microscopy Data Bank under the accession codes EMD-30476, EMD-30477, EMD-30478, EMD-30479, and EMD-30480, respectively. Their atomic models have been deposited in the Protein Data Bank under the accession codes 7CVY, 7CVZ, 7CW0, 7CW2, and 7CW3, respectively.
All study data are included in the article and supporting information.