Significance
Marine sediment covers 70% of Earth’s surface and harbors as much biomass as seawater. However, the global taxonomic diversity of marine sedimentary communities, and the spatial distribution of that diversity remain unclear. We investigated microbial composition from 40 globally distributed sampling locations, spanning sediment depths of 0.1 to 678 m. Statistical analysis reveals that oxygen presence or absence and organic carbon concentration are key environmental factors for defining taxonomic composition and diversity of marine sedimentary communities. Global marine sedimentary taxonomic richness predicted by species–area relationship models is 7.85 × 103 to 6.10 × 105 for Archaea and 3.28 × 104 to 2.46 × 106 for Bacteria as amplicon sequence variants, which is comparable to the richness in seawater and that in topsoil.
Keywords: subseafloor life, microbial diversity, marine sediment
Abstract
Microbial life in marine sediment contributes substantially to global biomass and is a crucial component of the Earth system. Subseafloor sediment includes both aerobic and anaerobic microbial ecosystems, which persist on very low fluxes of bioavailable energy over geologic time. However, the taxonomic diversity of the marine sedimentary microbial biome and the spatial distribution of that diversity have been poorly constrained on a global scale. We investigated 299 globally distributed sediment core samples from 40 different sites at depths of 0.1 to 678 m below the seafloor. We obtained ∼47 million 16S ribosomal RNA (rRNA) gene sequences using consistent clean subsampling and experimental procedures, which enabled accurate and unbiased comparison of all samples. Statistical analysis reveals significant correlations between taxonomic composition, sedimentary organic carbon concentration, and presence or absence of dissolved oxygen. Extrapolation with two fitted species–area relationship models indicates taxonomic richness in marine sediment to be 7.85 × 103 to 6.10 × 105 and 3.28 × 104 to 2.46 × 106 amplicon sequence variants for Archaea and Bacteria, respectively. This richness is comparable to the richness in topsoil and the richness in seawater, indicating that Bacteria are more diverse than Archaea in Earth’s global biosphere.
Over the last two decades, scientists have explored the nature and extent of subseafloor life through scientific ocean drilling in various oceanographic settings. The total number of microbial cells in marine sediment is presently estimated as 2.9 × 1029 to 5.4 × 1029 cells, accounting for 0.18 to 3.6% of Earth’s total living biomass (1, 2). The abundance of microbes in marine sediment generally decreases with increasing depth and increasing sediment age (1, 3). Cell concentrations are usually orders of magnitude higher in the organic-rich anoxic sediment of continental margins than in the organic-poor oxic sediment of the open ocean (4). A recent study used microfluidic digital PCR to estimate that archaeal cells constitute 37.3% of all marine sedimentary cells, with notably higher percentages of archaeal cells in ocean-margin sediment than in open-ocean sediment (40.0% and 12.8%, respectively; ref. 5). Another recent study estimates that 2.5 × 1028 to 1.9 × 1029 bacterial endospores (4.6 to 35 Pg of biomass carbon) exist in the uppermost kilometer of marine sediment (6).
Profiles of cell counts and porewater chemistry indicate that microbial activity in subseafloor sediment is usually extraordinarily low, with mean respiration rates ranging from 2.8 × 10−18 to 1.1 × 10−14 moles of electrons per cell per year, depending on the availability of electron donors and acceptors (4, 7, 8). Single cell-targeted, stable isotope-probing incubation and nanometer-scale secondary ion mass spectrometry have shown that most microbial cells in samples of subseafloor sediment can assimilate a diverse range of carbon and nitrogen compounds into cellular biomass, even from 2-km-deep anoxic Miocene sediment (9–12) and 101.5-Ma oxic sediment.
Factors that may limit the deep sedimentary biosphere are not restricted to scarcity of nutrients and scarcity of energy-yielding substrates. Limiting factors may also include temperature, pressure, pH, salinity, water availability, sediment porosity, or permeability. For example, without fluid transport in sediment, dispersal of subseafloor microbial cells may be limited to diffusive transport since it is unlikely that proton pump-driven flagellar motility can occur under such low energy flux (13). In this case, transport may only reach 6 m over 1 million years, even in very porous layers (8, 14, 15). Recent studies of community compositional relationships between shallow sediment and seawater have demonstrated that deep-subseafloor sediment is populated by descendants of seafloor-sediment communities, which become predominant through preferential survival as the communities are buried over thousands to hundreds of thousands of years (16–18). Some deeply buried cells may later be introduced to the ocean through fluid transport (flow through faults, mud volcanism, and hydrocarbon seepage) at plate-convergent margins, being dispersed as “deep-biosphere seeds” (19–22). Based on mathematical models of energy availability and microbial processes, the energy required to simply survive, rather than to grow, comprises a substantial component of the total power consumed by subseafloor-sedimentary communities (23–25).
Despite the ecological and evolutionary significance of marine sedimentary life, the spatial distribution and environmental constraints of microbial diversity in marine sediment are poorly delineated, in part due to limited microbiological sampling of subseafloor sediment and partly due to use of different quality controls and different analytical protocols by different studies (26–28). Despite these limitations, studies of 16S ribosomal RNA (rRNA) gene sequences have demonstrated that 1) diverse bacterial and archaeal taxa are ubiquitous in organic-rich anoxic sediment, 2) microbial communities are stratified by sediment depth, and 3) geochemical and sedimentological properties influence microbial community composition (10, 28–36). In addition, single-cell genomic, metagenomic, and functional gene analyses have demonstrated that predominant bacterial and archaeal taxa (e.g., members of the Atribacteria and Bathyarchaeota) possess metabolic capabilities that can contribute to the heterotrophic subseafloor ecosystem, such as homoacetogenesis and the ability to degrade diverse organic compounds (37–42).
With recent advances in molecular ecological and computational approaches, microbiomes of other habitats, such as seawater and topsoil, have been investigated comprehensively at the global scale (43, 44). For example, the global diversity of marine prokaryotes (Archaea and Bacteria) in the near-surface ocean (0 to 1,000 m below sea level [mbsl]) was estimated (3.75 × 104 operational taxonomic units) using seawater samples from the Tara Oceans project (43). The community composition of these seawater samples covaries with temperature (43). Investigation of the global microbial diversity of topsoil has similarly revealed that bacterial community composition of topsoil is strongly affected by soil pH and climatic variables (44). Recently, Magnabosco et al. (45) compiled published microbial diversity data from diverse groundwater sources and estimated that the continental subsurface biosphere contains 2 × 1029 to 6 × 1029 cells (23 to 31 Pg of biomass carbon), with community composition correlated with sample lithology. However, the authors cautioned that these results are likely influenced by batch effects, since data for proximal sites were generally generated by the same laboratory using the same methodologies (e.g., sampling and storage conditions, DNA-extraction and -purification procedures, primers, and sequencing technologies).
To discover the global diversity of the marine sedimentary microbiome and to exclude potential experimental biases, we established a consistent subsampling and experimental procedure and analyzed all samples at a single laboratory. While paying particular attention to quality control and quality assurance (QA/QC) of sample processing (28), we analyzed a total of 299 deep-frozen samples collected from 40 sites by scientific ocean drilling and coring expeditions, starting with the first deep biosphere-dedicated Ocean Drilling Program (ODP) Leg 201 in 2002 (7). Using this approach, we investigate the distribution of diversity in the global marine sedimentary microbiome and elucidate environmental factors that constrain its community composition at a global scale.
Results and Discussion
Samples and Sequencing.
The 299 sediment samples were collected from core depths of 0.1 to 678 m below the seafloor (mbsf) (Fig. 1 and Dataset S1). The samples were taken aseptically on the ship immediately after core recovery, frozen at −80 °C immediately after sampling, and stored at −80 °C until subsampling. Consistent QA/QC procedures were implemented for subsampling, DNA extraction, PCR amplification, and sequencing. For example, to better understand microbial taxonomic richness and diversity, the same primer sets and sequencing technology were used throughout the study, including negative experimental controls.
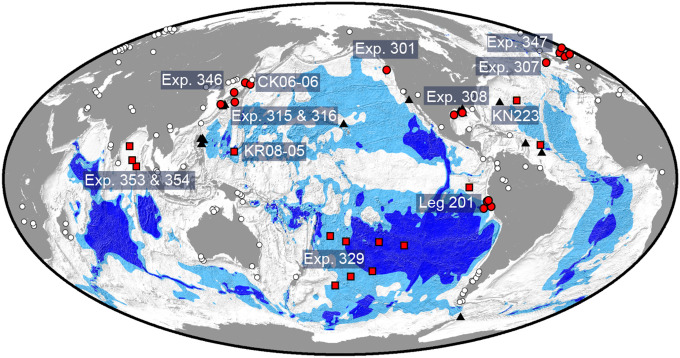
Fig. 1.
Location of sampling sites. Red circles indicate ocean-margin sites, and red squares indicate open-ocean sites, respectively. Light blue and dark blue regions, respectively, show maximum and minimum areas where dissolved oxygen and aerobic activity may occur throughout the sediment (4). A total of 299 sediment samples from many different depths below the seafloor (0.1 to 678 mbsf) were collected from 40 sites during 14 scientific expeditions: Ocean Drilling Program (ODP) Leg 201; IODP Exp. 301; ODP Exp. 307; IODP Exp. 308; IODP Exps. 315 and 316; IODP Exp. 346; IODP Exp. 347; IODP Exp. 353; IODP Exp. 354; drilling vessel Chikyu Exp. CK06-06; research vessel (R/V) Knorr cruise KN223; and R/V Kairei cruise KR08-05. White circles mark topsoil sites (44), and black triangles mark seawater sites (19, 46–49) that are included for comparison between different global biomes.
Archaea-specific, Bacteria-specific, and universal primers were used to amplify 16S rRNA gene sequences from 236, 299, and 287 sediment samples, respectively (Materials and Methods). Total samples amplified with the Archaea-specific primer and with the universal primer are fewer than 299 because those primer sets did not yield amplicons from several samples. After filtering the raw data, totals of 12.5, 16.0, and 18.6 million sequence reads were obtained using the Archaea-specific, Bacteria-specific, and universal primers, respectively. The datasets obtained with these primer sets are referred to in the following sections as “Archaea,” “Bacteria,” and “Universal.”
Taxonomic Composition of Archaeal Communities.
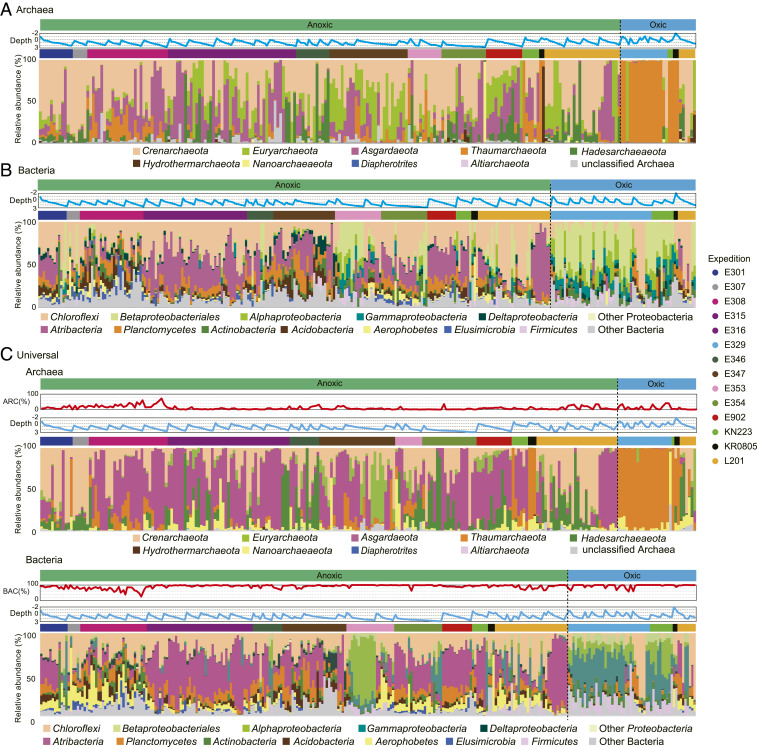
The taxonomic composition of the archaeal community in anoxic subseafloor sediment differs markedly from that in oxic subseafloor sediment (Fig. 2 A and C and SI Appendix, Fig. S1). Fig. 2A shows the results of the Archaea-sequencing library. It illustrates the microbial community composition for 235 sediment samples. Members of Crenarchaeota, including Bathyarchaeia, are prevalent within the anaerobic communities. Euryarchaeota and Asgardaeota are highly abundant in anaerobic communities from some regions, such as the Baltic Sea (Expedition [Exp.] 347) and off the Shimokita Peninsula (Exp. 902). In some samples from the Japan Sea (Exp. 346), the Peru margin (Leg 201), and off the Shimokita Peninsula, members of the Hadesarchaeaeota account for more than 70% of the archaeal community, and their relative abundance increases with increasing sediment depth (Fig. 2A). Members of Nanoarchaeaeota constitute more than 95% of the archaeal community in the deepest sediment sample (678 mbsf) from the Bay of Bengal (Exp. 354). In contrast to the anaerobic communities, the aerobic archaeal communities are dominated by members of the Thaumarchaeota (e.g., Nitrososphaera). This dominance by Thaumarchaeota suggests the possibility that seawater microorganisms were incorporated into the oxic sediment and subsist there (15). Of the 3,892 total archaeal amplicon sequence variants (ASVs) detected in the Archaea dataset, only 260 ASVs are common. These 260 ASVs account for roughly 70% of the total archaeal sequences (SI Appendix, Fig. S1).
Fig. 2.
Taxonomic composition of marine sedimentary microbial communities. (A) Archaeal community composition obtained for 235 samples by using Archaea-specific primers. (B) Bacterial community composition obtained for 272 samples by using Bacteria-specific primers. (C) Archaeal community composition for 245 samples and bacterial community composition for 281 samples obtained by using the universal primers. For A–C, samples containing fewer than 1,000 sequences are not shown. The upper light blue line charts in each panel show sediment depth on a logarithmic scale. Colored bars above the bar charts in A–C indicate expeditions. The upper red line charts in C indicate relative abundances of Archaea (ARC) or Bacteria (BAC) in the Universal sequence library. Classification is based on the SILVA 132 SSU database (https://www.arb-silva.de/). Gammaproteobacteria in the charts do not include Betaproteobacteriales, which are enumerated separately.
The archaeal community compositions obtained using the universal primer set are slightly different from those obtained using the archaea-specific primers. For many sediment samples, Asgardaeota is highly dominant in the Universal dataset, whereas Crenarchaeota and Euryarchaeota are dominant in the Archaea dataset. In particular, Asgardaeota is highly dominant in the Universal dataset for deep sediment from the Gulf of Mexico (Exp. 308), Nankai Trough (Exps. 315 and 316), and off the Shimokita Peninsula (Exp. 902), with ∼80%. These differences confirm that primer coverage influences apparent community composition. Despite these slight differences between the Archaea and Universal libraries, both libraries show that archaeal communities in anoxic sediment are distinct from those in oxic sediment. For example, both the Archaea dataset and the Universal dataset show Thaumarchaeota to be dominant only in samples from oxic sediment.
The relative abundance of Archaea in the Universal dataset is below 10% for most samples (Fig. 2C), although this result is not quantitative due to the difference in PCR efficiency. A previous study used digital PCR quantification to identify percentages of Archaea as 5.6 and 22.6% in open-ocean sediment and ocean-margin sediment, respectively (5). The percentages in the present study are 4.3 and 12.2%. Although these percentages are slightly lower than in the previous study, they are consistent in showing Bacteria to dominate significantly over Archaea in subseafloor sediment (SI Appendix, Fig. S2).
Taxonomic Composition of Bacterial Communities.
As with the archaeal communities, bacterial communities in anoxic sediment are very different from those in oxic sediment (Fig. 2 B and C and SI Appendix, Fig. S1). The bacterial community compositions obtained from the Bacteria-specific primers and the Universal primers were in agreement, except for differences in composition within Proteobacteria. Members of Proteobacteria, including Alphaproteobacteria and Betaproteobacteriales (Gammaproteobacteria), predominate, along with members of the Firmicutes, in oxic subseafloor sediment. In contrast, members of Atribacteria, Chloroflexi, and Planctomycetes are prevalent in anoxic sediment, as previously reported by more geographically limited studies (e.g., refs. 40 and 50). However, some anoxic sediment, such as samples collected from the Bay of Bengal (Exp. 353), is inhabited by bacterial communities typical of oxic sediment. Despite the consistent differences in dominant members of anaerobic and aerobic bacterial communities, 5,212 of the 30,874 ASVs are shared by both anaerobic and aerobic communities. These 5,212 ASVs collectively constitute about 80% of the 16S rRNA sequences in our dataset (SI Appendix, Fig. S1).
Comparison of the Marine Sedimentary Biome with the Seawater and Topsoil Biomes.
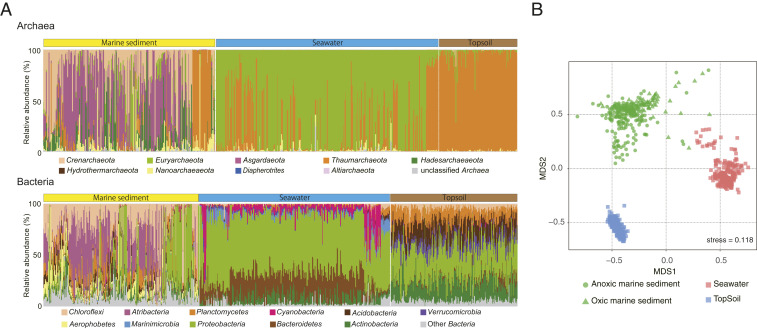
To assess the relationship of the marine sedimentary biome to other major biomes, we compare our Universal library for marine sediment to published sequence libraries from seawater and topsoil (references in Fig. 1) that use the same primers (or slightly modified) as the universal primer used in this study (Fig. 3). Bacterial and archaeal community compositions of the three biomes are distinctly different. As shown in Fig. 2, Crenarchaeota and Asgardaeota are dominant in marine sediment, whereas Euryarchaeota and Thaumarchaeota are dominant in seawater and Thaumarchaeota in topsoil. Bacterial community composition shows dominance of Proteobacteria in the seawater and topsoil data. In addition, Cyanobacteria, Marinimicrobia, and Bacteroidetes are dominant in seawater, whereas Acidobacteria and Verrucomicrobia are particularly dominant in topsoil. Atribacteria and Aerophobetes are prevalent in marine sediment but absent from the other two biomes, suggesting that marine sedimentary microbes constitute a unique biome. Intersample similarities in ASV composition indicate distinct communities in the three biomes (Fig. 3B) and higher intersample diversity in marine sediment. This high diversity may reflect more drastic variation in habitat conditions of marine sediment (e.g., with depth below seafloor, from oxic to anoxic or from energy-rich to energy-poor) than in the other biomes.
Fig. 3.
Comparison of the microbial community compositions of marine sediment, seawater, and topsoil. (A) Archaeal and bacterial community compositions obtained by using the 16S rRNA universal primers. The gene sequences of seawater and topsoil samples were compiled from previous publications (19, 44, 46–49). (B) NMDS ordination plots generated using Jaccard similarity index values derived from the rarefied ASV populations.
Environmental Factors Constraining Marine Sedimentary Community Compositions.
To understand which environmental factors constrain microbial-community composition at a global scale and across a broad range of sediment depths, we performed nonmetric multidimensional scaling (NMDS) analysis of community composition and diverse environmental properties. These properties included water depth, sediment depth, sulfate concentration, total organic carbon concentration, and the presence or absence of dissolved oxygen (Fig. 4 A–C and Dataset S1). For this analysis, we excluded ASVs that were statistically determined to be contaminants. We also analyzed the data after subtracting all ASVs detected in the negative control to confirm that the NMDS coordination does not result from contamination (SI Appendix, Fig. S3).
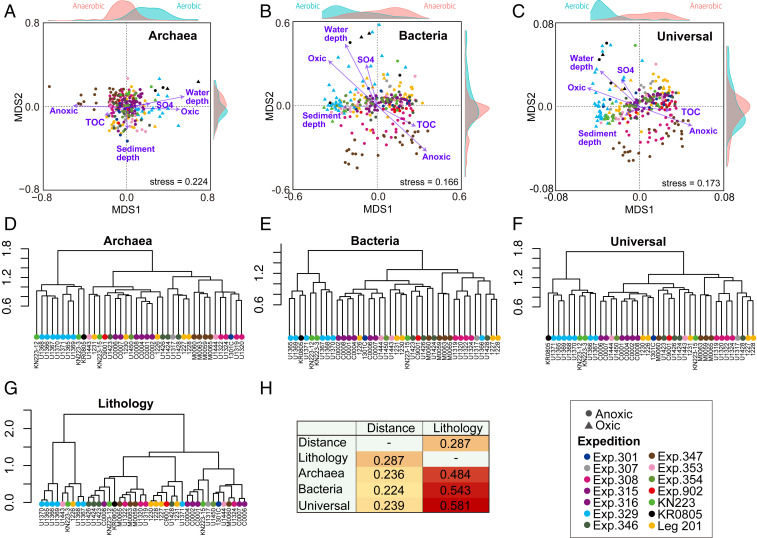
Fig. 4.
Beta diversity of microbial communities in marine sediment. (A–C) NMDS ordination plots for the Archaea (A), Bacteria (B), and Universal (C) libraries, generated using Jaccard similarity index values. The Jaccard values were derived from the rarefied ASV populations in 235, 272, and 281 sediment samples for the Archaea, Bacteria, and Universal libraries, respectively. Purple arrows represent vectors that were fitted with environmental factors characterized by significance values of P < 0.01. (D–F) Unweighted pair-group method with arithmetic mean (UPGMA) dendrograms of the archaeal (D), bacterial (E), and universal (F) communities, respectively, from the 40 sites, based on Jaccard similarity index values. (G) UPGMA dendrogram of lithology, based on mineral composition of sediment at the 40 sites (Dataset S2). (H) Mantel test results used to determine if geographic distance or lithology is correlated with microbial community composition.
The global-scale compilation of marine sedimentary archaeal communities can be categorized into two broad groups: an aerobic open-ocean group and an anaerobic group inhabiting organic-rich habitats. The bacterial communities similarly cluster into an aerobic open-ocean group and an anaerobic group inhabiting continental margins and other upwelling regions (Fig. 4B). The respective associations of these broadly defined groups, with 1) the presence of dissolved oxygen and low total organic abundance and 2) the absence of dissolved oxygen and high organic abundance, respectively, indicate that these environmental factors significantly contribute to the taxonomic composition of marine sedimentary microbiomes and, hence, the ecological functions of aerobic and anaerobic marine sedimentary microbial ecosystems. The NMDS ordination for the universal primers (both bacterial and archaeal 16S rRNA gene sequences) more closely resembles the ordination for the Bacteria-specific primers than the ordination for the Archaea-specific primers (Fig. 4 A–C), most likely due to the greater diversity of bacterial taxa, when compared with the diversity of archaeal taxa, in the marine sedimentary biome (5). Among the examined environmental factors, the sediment-depth vector is perpendicular to the other factor vectors, regardless of NMDS analysis, suggesting that sediment depth may be independently correlated with community composition. Such a relationship might result from decreasing availability of nutrient and energy substrates with increasing sediment depth (51).
To examine the correlation of geographic distance and sediment lithology to microbial-community composition, we performed a Mantel test for the compiled community assemblage for the individual sites. The test yielded a highly significant (P < 0.001) but relatively weak positive correlation between geographic distance and community composition (r = 0.236, 0.224, and 0.239 for the Archaea, Bacteria, and Universal datasets, respectively; Fig. 4H). This analysis shows that sediment lithology (Dataset S2) is strongly correlated with community composition (r = 0.484, 0.543, and 0.581 for the Archaea, Bacteria, and Universal datasets, respectively). This strong correlation may be due to different quantities and compositions of organic matter in the different sedimentary lithologies, resulting from different sediment sources and sedimentation rates. For individual sediment samples, sedimentation rate and sediment age may also be important determinants of community composition (52); however, we did not include these variables for NMDS analysis because sedimentation rates are not available for more than half of the sediment samples used in this study.
Network Analysis of Marine Sedimentary Microbial Communities.
To determine cooccurrence patterns, we performed network analysis based on Spearman’s rank correlation of ASVs obtained using the universal primers. The networks were constructed using only strong, positive, and significant correlations (r > 0.6, P < 0.05; Fig. 5).
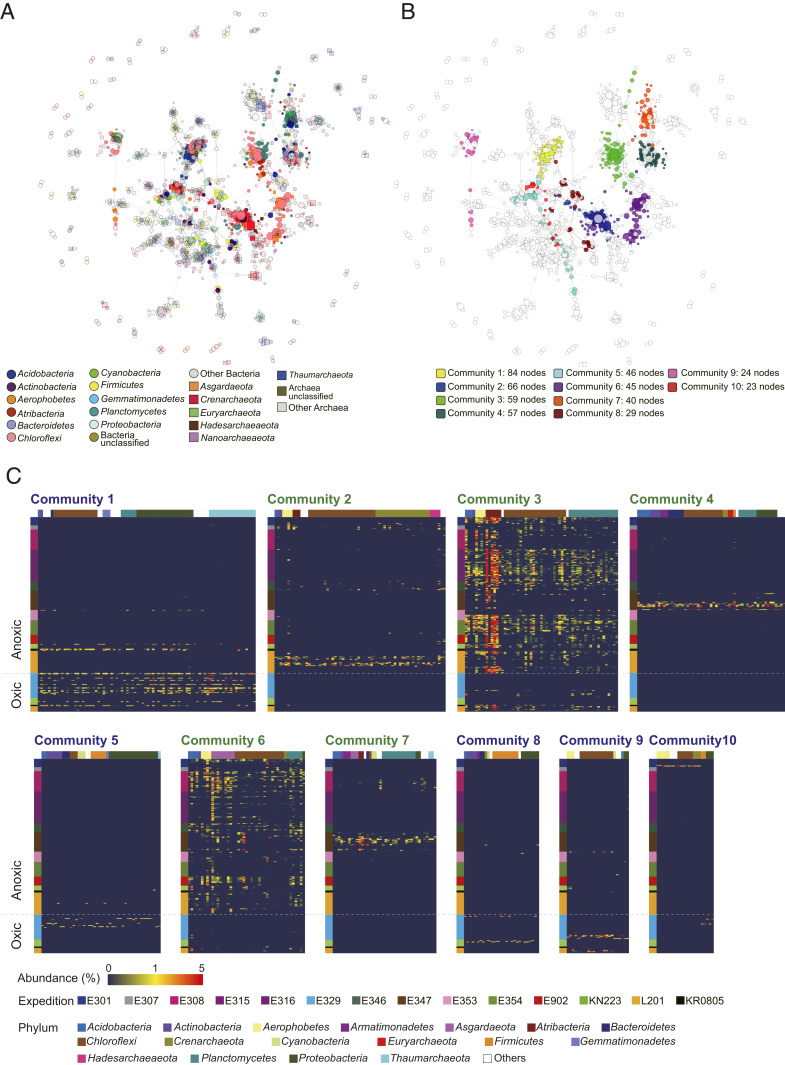
Fig. 5.
Network analysis of co-occurring ASVs. Node sizes are proportional to PageRank. (A) Node colors indicate phylogenetic groups. Circles and squares respectively represent bacterial and archaeal groups. Lightly tinted nodes indicate ASVs not in the top 10 largest communities. (B) Node colors indicate the 10 largest communities in the network shown in A. Nodes not in the 10 largest communities are shown as white dots with light gray outlines. (C) Heat maps of the relative proportions of ASVs in each sample. The color bar on the left represents expeditions, and the color bar on the top represents phylum-level taxonomies. The names of communities mainly consisting of ASVs from anoxic sediment are in green, whereas communities mainly consisting of ASVs from oxic sediment are in blue.
The resulting network graph contains 1,533 nodes (ASVs). Fifty communities were detected, based on edge-betweenness clustering (53). The modularity of the network is high (0.84). In Fig. 5B, only the 10 largest communities are highlighted. These 10 communities reflect the sedimentary redox environment, with five communities consisting of ASVs prevalent in oxic sediment (Communities 1, 5, 8, 9, and 10) and five communities consisting of ASVs prevalent in anoxic sediment (Communities 2, 3, 4, 6, and 7).
Of these 10 communities, Community 1 contains the largest number of ASVs (84 ASVs), which consist mainly of Chloroflexi, Proteobacteria, and Thaumarchaeota (Fig. 5C). The network diagram shows that Chloroflexi and Thaumarchaeota are important members of the cooccurrence network (Fig. 5A) based on PageRank. These Thaumarchaeota were detected primarily in surface sediment from the South Pacific Gyre (Exp. 329). They belong to the family Nitrosopumilaceae (Dataset S3). An isolate from this Thaumarchaeota group is an autotropic ammonia oxidizer. Nitrosopumilaceae are widespread in the ocean and may be deposited on the seafloor from seawater. They have been reported to survive in oceanic crust using ammonia produced by aerobic mineralization of organic matter and may be sustained similarly in the sediment (54). The members of Chloroflexi in this community belong to the families Dehalococcoidia and Anaerolineae. Although Dehalococcoidia are generally considered as strictly anaerobic bacteria, they also appear in aerobic Community 10. The Anaerolineae includes isolates capable of aerobic growth (55), which is in good agreement with the potential for catabolic O2 reduction inferred for related taxa from metagenomes (56–58). The members of Proteobacteria in Community 1 include taxa assigned to families with members involved in metal cycling in manganese nodules and manganese crusts, such as Magnetospiraceae (microaerobic heterotrophs) and Kiloniellaceae (59) (reported to be chemoheterotrophic aerobes; refs. 60 and 61). The other communities detected in oxic sediment (Communities 5, 8, 9, and 10) are mostly composed of ASVs assigned to higher taxa associated with anaerobic metabolisms. The extent to which these ASVs are capable of aerobic activities is generally unknown. In addition to the Thaumarchaeota, seawater-associated taxa in the oxic-sediment communities include Proteobacteria, such as the SAR86 clade of Gammaproteobacteria (in Community 5), and phylum Cyanobacteria (in Communities 5, 8, and 9). Cyanobacteria have been reported to occur deep beneath the continental surface, where they are hypothesized to survive by hydrogenotrophy in the absence of light (62). They may similarly rely on hydrogenotrophy in subseafloor sediment.
Of the five anoxic-sediment communities, Community 3 consists primarily of Atribacteria, Chloroflexi, and Planctomyces that are typically found in anoxic subseafloor sediment. The network diagram confirms that Chloroflexi is a prominent component. However, almost all of the Chloroflexi detected are affiliated with MSBL9, a candidate order with little-known properties. The reported genome of Phycisphaerae in MSBL9 suggests that the bacterium is adapted to utilize a variety of complex sugar polymers (63). Atribacteria are prevalent in organic-rich subseafloor sediment (30, 64). They are heterotrophic anaerobic bacteria that ferment organic acids to produce acetate and carbon dioxide (40). Dehaloccoidia, belonging to Chloroflexi, are likewise anaerobic heterotrophic bacteria and may compete with the MSBL9-affiliated bacteria and/or the Atribacteria for a common substrate. Alternatively, since Dehaloccoidia can utilize aromatic and halogenated organic compounds (65), they may coexist by utilizing a different substrate. The network may also represent the use of organic acids, a metabolite of Chloroflexi, by Atribacteria and Planctomyces. In Community 6, the network diagram shows that Asgardaeota and Chloroflexi are the main members. Almost all Asgardaeota in this community are Lokiarchaeia (Dataset S3). Metagenome-assembled genomes of Lokiarchaeia from Costa Rica margin sediment suggest that they live heterotrophically by utilizing aromatic hydrocarbons but thermodynamically require symbiosis with sulfate-reducing bacteria (66). Another Chloroflexi clade in Community 6 is the Anaerolineae, which is generally known to be anaerobic and heterotrophic. The other three anaerobic communities were distinct to their sampling locations. Communities 4 and 7 were characteristic of the Baltic Sea (Exp. 347) samples. Community 2 was characteristic of the Peru Margin (Leg 201) samples. Exp. 347 drilled glacial and interglacial sediments, in which Community 4 consisted of members derived from glacial sediment with low dissolved alkalinity.
In short, the cooccurrence network analysis indicates that marine sedimentary, microbial-community composition is shaped by the redox state and electron donors available in the sediment. The results of this analysis also suggest that subseafloor-sedimentary microbes interact with each other to make effective use of the limited substrates available in this extreme environment.
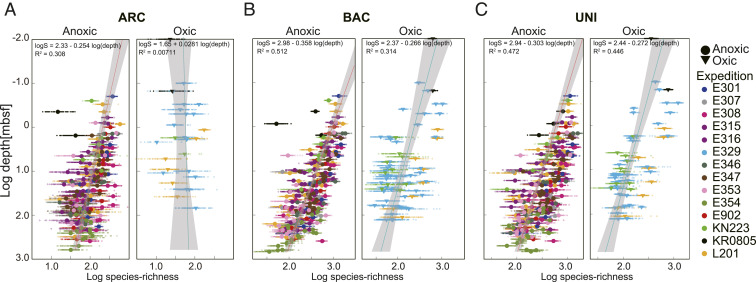
Depth Profile of Taxonomic Richness.
To estimate taxonomic richness for each sample, 1,000 sequences were randomly selected from the sequence data for the sample and used to calculate ASV richness (Chao-1) for the sample. To ensure the robustness of our richness estimates, this process was repeated 100 times for each sample. Bacterial ASV richness and archaeal ASV richness both generally decrease with increasing depth in anoxic sediment (Fig. 6). However, in oxic sediment, bacterial richness generally decreases with increasing depth, whereas archaeal richness remains relatively constant throughout the sediment column. Bacterial richness is 10-fold higher than archaeal richness and is also higher in anoxic sediment than in oxic sediment (Fig. 6B), possibly because organic-rich anoxic sediment generally harbors higher cell abundance than organic-poor oxic sediment (1, 5). In general, the steepness of depth-related declines in richness is positively related to the richness of the communities in shallow sediment. For example, based on the steeper slope of the regression lines for ASV richness and sediment depth in anoxic sediment than in oxic sediment (Fig. 6 A–C), a smaller fraction of initial richness survives subseafloor selection pressure in anoxic sediment than in oxic sediment. Similarly, in both anoxic and oxic sediment, archaeal richness decreases less dramatically than bacterial richness with increasing sediment depth. The lower decrease in archaeal richness with depth is consistent with suggestions that archaeal taxa are generally more tolerant of very low energy availability than bacterial taxa (8). However, despite this lower rate of decrease in archaeal richness with sediment depth, archaeal richness is generally lower than bacterial richness at all sediment depths.
Fig. 6.
Depth profiles of microbial richness in oxic marine sediment and anoxic marine sediment. (A–C) Archaeal (ARC), bacterial (BAC), and universal (UNI) ASV richness (Chao-1 estimator), calculated using data rarified to 1,000 reads per sample. Small, pale points represent the 100 resamplings per sample, and large points represent the mean values of the resamplings.
Total Taxonomic Richness of the Marine Sedimentary Biome.
We used the three sets of 16S rRNA gene sequence libraries (Archaea, Bacteria, and Universal) to estimate the global number of ASVs in marine sediment (Materials and Methods). To ensure the robustness of our results, we used five different species–area relationship (SAR) models for independent estimates of taxonomic richness (SI Appendix, Fig. S4). Of these five models (Arrhenius, Gleason, asymptotic, logistic, and Gitay) (SI Appendix, Table S1), the Akaike information criterion (AIC) scores indicate that the asymptotic and Gitay models are the best and second-best for the three datasets, respectively (Table 1). When extrapolated to the global volume of marine sediment (67), the total number of archaeal ASVs is 7.85 × 103 using the asymptotic model and 6.10 × 105 using the Gitay model. The total number of bacterial ASVs in global marine sediment is 3.28 × 104 using the asymptotic model and 2.46 × 106 using the Gitay model. With both models, global bacterial richness is about fourfold higher than global archaeal richness (Table 1). The ranges of model extrapolations are 6.18 × 103 to 7.90 × 1015 for archaeal and 2.88 × 104 to 1.32 × 1015 for bacterial ASVs (Table 1 and SI Appendix, Fig. S4). Using the Universal dataset to estimate global diversity, we again found the asymptotic model to be the best model, with archaeal and bacterial diversity of 1.98 × 103 and 3.90 × 104, respectively (SI Appendix, Table S2 and Fig. S5). This result is consistent with the results described above for the Bacteria and Archaea datasets. These estimates using the asymptotic and Gitay models are far below previous estimates of microbial taxonomic richness in the terrestrial subsurface biome (i.e., 109 to 1012) (45). These previous terrestrial subsurface estimates were based on Arrhenius scaling relationships (R = aNb, where a is a constant, b is a scaling factor, R is taxonomic richness, and N is the number of sequence reads) (45). With the Arrhenius model, where N is the number of samples instead of the number of sequence reads, total ASV richness of the marine sedimentary microbiome is 8.71 × 1013. This value is similar to the Arrhenius-based estimate for the terrestrial subsurface biome (45). However, the Arrhenius model consistently yielded a higher AIC score than the asymptotic model for marine sediment (Table 1). It also yielded a much higher richness estimate (5 to 12 orders of magnitude) than the other models (Table 1). Consequently, we consider the global ASV value calculated using the Arrhenius model to be a very large overestimate (Table 1; cf. ref. 68; SI Appendix, Supplementary Text).
Table 1.
Global number of microbial taxa (ASVs) predicted using different SAR models
| Model | Mean | Minimum | Maximum | AIC | Source | |
| Archaea | Asymptotic | 7.85 × 103 | 6.58 × 103 | 1.13 × 104 | 2,988 | This study |
| Marine sediment | Gitay | 6.10 × 105 | 3.63 × 105 | 8.82 × 105 | 3,010 | |
| Logistic | 6.68 × 103 | 6.18 × 103 | 7.38 × 103 | 3,159 | ||
| Arrhenius | 2.18 × 1013 | 3.18 × 1010 | 7.90 × 1015 | 3,163 | ||
| Gleason | 7.83 × 105 | 6.90 × 104 | 8.99 × 104 | 3,372 | ||
| Bacteria | Asymptotic | 3.28 × 104 | 3.07 × 104 | 3.72 × 104 | 4,366 | This study |
| Marine sediment | Gitay | 2.46 × 106 | 1.66 × 106 | 3.50 × 106 | 4,378 | |
| Logistic | 2.98 × 104 | 2.88 × 104 | 3.09 × 104 | 4,658 | ||
| Arrhenius | 1.41 × 1013 | 1.98 × 1011 | 1.32 × 1015 | 4,884 | ||
| Gleason | 3.52 × 105 | 3.16 × 105 | 3.94 × 106 | 4,883 | ||
| Archaea and Bacteria | Asymptotic | 4.03 × 104 | 3.67× 104 | 4.50 × 104 | 4,791 | This study |
| Marine sediment | Gitay | 3.30 × 106 | 2.22 × 106 | 4.56 × 106 | 4,898 | |
| Arrhenius | 8.71 × 1013 | 1.07 × 1012 | 1.31 × 1016 | 5,056 | ||
| Logistic | 3.55 × 104 | 3.41 × 104 | 3.71 × 104 | 5,063 | ||
| Gleason | 4.27 × 105 | 3.77 × 105 | 4.75 × 105 | 5,332 | ||
| Archaea and Bacteria | Asymptotic | 7.88 × 104 | 7.73 × 104 | 8.05 × 104 | 4,134 | Ref. 40 |
| Topsoil | Gitay | 1.69 × 107 | 3.65 × 107 | 5.10 × 107 | 4,188 | |
| Gleason | 8.33 × 106 | 7.94 × 106 | 8.88 × 106 | 4,281 | ||
| Logistic | 7.58 × 104 | 7.46 × 104 | 7.68 × 104 | 4,382 | ||
| Arrhenius | 2.91 × 1011 | 6.65 × 1010 | 1.87 × 1012 | 4,338 | ||
| Archaea and Bacteria | Asymptotic | 3.00 × 104 | 2.92 × 104 | 3.17 × 104 | 5,464 | Refs. 18 and 42–45 |
| Seawater | Gitay | 1.69 × 106 | 1.27 × 106 | 2.07 × 106 | 5,589 | |
| Arrhenius | 2.80 × 1011 | 1.50 × 1010 | 4.93 × 1012 | 5,963 | ||
| Logistic | 2.85 × 104 | 2.80 × 104 | 2.92 × 104 | 6,019 | ||
| Gleason | 3.08 × 105 | 2.80 × 105 | 3.33 × 105 | 6,141 |
The results are ordered by AIC, from lowest to highest, with AIC calculated as the highest AIC value for 100 model permutations.
To compare the global bacterial and archaeal richness of the marine sedimentary biome with the global richness of other major biomes, we compiled published 16S rRNA gene sequence reads from topsoil and seawater (topsoil: 190 sites, 234 samples; seawater: 20 sites, 352 samples; ref. 44 and references in Fig. 1). The same 16S region was analyzed for all three biomes. However, the data for the different biomes were obtained using different DNA-extraction methods, different PCR conditions, and slight differences in primer sequences. After fitting the five SAR models to the topsoil data and the seawater data, the AIC scores indicate the asymptotic and Gitay models to be better for both topsoil and seawater, as with the marine sediment. The global ASV richness of topsoil and the global ASV richness of seawater are, respectively, 7.88 × 104 and 3.00 × 104 using the asymptotic model (Table 1). They are 1.69 × 107 and 1.69 × 106 using the Gitay model (Table 1). The combined global total of the three biomes (the sum of all AVS richness for topsoil, sweater, and marine sediment) is 1.49 × 105 using the asymptotic model and 2.19 × 107 using the Gitay model (Table 1). As in marine sediment, global bacterial diversity exceeds global archaeal-diversity topsoil and seawater (SI Appendix, Table S2).
Conclusions
In this study, we analyzed microbial diversity in 299 marine sediment samples from throughout the world, using a precisely controlled and consistent method. Our results demonstrate that microbial communities in marine sediment comprise two major microbial groups, one anaerobic and the other aerobic. Diversity in marine sediment decreases with increasing sediment depth. The rate of decrease is generally lower for Archaea than Bacteria. Despite the lower rate of decrease for archaeal diversity with depth, bacterial diversity exceeds archaeal diversity at every depth in the sediment. Of five ASV–area relationship models, the asymptotic model best describes our data, predicting total ASVs in marine sediment to be 7.85 × 103 for Archaea and 3.28 × 104 for Bacteria. Comparative analysis of marine sediment, topsoil, and seawater microbiomes showed that each biome contains a distinctly different community but roughly similar levels of global microbial richness. This result indicates that Bacteria are more diverse than Archaea in Earth’s global biosphere.
Materials and Methods
Sediment Core Samples.
The 299 sediment samples used in the present study (0.1 to 678 mbsf) are from 40 sites sampled over the course of 14 drilling or coring expeditions, including Integrated Ocean Drilling Program (IODP) expeditions in 2002 to 2015 (Fig. 1 and Dataset S1). Sediment cores were sampled aseptically on board, and the samples were frozen at −80 °C immediately after sampling. The frozen sediment samples were stored at −80 °C until postexpedition subsampling in a cleanroom. The environmental metadata (sediment depth, age, porewater chemistry) for each sediment sample were obtained from published expedition reports (Dataset S1). Mineral composition was determined based on the observation of smear slides of frozen samples under a polarized microscope (Dataset S2).
DNA Extraction.
From each frozen core sample, 5 g of frozen sediment was aseptically subsampled for DNA extraction, as described previously (19). To diminish biases that could be caused by using different procedures, subsampling, DNA extraction, 16S rRNA gene amplification, and sequencing were performed using the same procedures, in the same laboratory, and at the same time. In addition, a negative control (5 mL of water) was included with each batch of subsamples and subject to DNA extraction. DNA was extracted from each subsample using the PowerMax Soil DNA Isolation kit (Qiagen), according to the manufacturer’s instructions. Each of the resulting 5-mL DNA solutions was concentrated by ethanol precipitation and then stored at –20 °C.
Sequencing and Analysis.
The V3–V4 hypervariable region of the 16S rRNA gene was amplified by PCR using universal primers (U515F and U806R; ref. 69). In addition, the V4–V5 region of the 16S rRNA gene was amplified using domain-specific primer sets (518F/926R and 517F/958R; https://vamps.mbl.edu/resources/primers.php) that target Bacteria and Archaea, respectively.
The volume of the DNA extracts used for all PCRs was 1 µL. To avoid overamplification, the number of PCR cycles was set between 25 and 40 based on the results of preliminary PCR tests monitoring amplification curves. After the PCR products were purified, index and adapter sequences were added by eight cycles of PCR, followed by purification by using AMPure XP (Beckman Coulter). The PCR products were then sequenced using the MiSeq platform with either the MiSeq Reagent Kit v3 (600 cycles) or MiSeq Reagent Kit v2 (500 cycles; Illumina).
After quality trimming, the demultiplexed sequence reads were processed using Mothur (version 1.35.0) (70) and USEARCH (64-bit version; www.drive5.com/usearch/), in order to merge pair-end reads and to cluster sequence reads into taxonomic units (ASVs). ASVs with abundance of 3 or less were discarded. Qiime (71) was used to determine which ASVs were significantly more abundant (P < 0.20; ANOVA) in the negative-control sequences than in the sediment-sample sequences. For this comparison, we used 18 negative controls. ASVs inferred to be potential contaminants were removed from all samples and excluded from further analysis. After this removal of potential contaminants, samples with fewer than 1,000 sequences for Archaea and fewer than 10,000 sequences for Bacteria and Universal were removed. As a result, 235, 272, and 281 samples for Archaea, Bacteria, and Universal, respectively, remained for further analyses.
For analyses of community composition using each dataset, the number of sequences in each sample was rarefied to the lowest number of sequences in any sample in that dataset. Using these rarefied data, NMDS analysis was performed using the Jaccard index and the “vegan” package in R, as described elsewhere (72, 73). In addition, a permutation test was conducted using the envfit function of “vegan” to fit the vectors of the environmental metadata to the NMDS ordination plots, and the taxonomic (ASV) richness (Chao estimator) of each sediment sample was calculated using the “iNEXT” package (74) in R. Cytoscape (https://cytoscape.org) was used to generate cooccurrence networks. These networks were used to visualize relationships between ASVs in the sediment samples. Only abundant genera, defined as genera that comprise >1.0% of reads at least in one sediment sample (2,246 ASVs in total), were selected from the Universal sequence library to construct the network. Spearman’s correlation coefficients were calculated, and strong, positive, and significant correlations (Spearman’s ρ > 0.6; P < 0.05) were used for network construction. Calculation of PageRank of the nodes and identification of communities by edge-betweenness (75) was performed using the “igraph” package in R.
Modeling of Global Richness.
The number of global ASVs was evaluated using five SAR models (SI Appendix, Supplementary Text). Various SAR models have been used over the decades (76). Power-law functions, such as the Arrhenius model, have been mainly used classically, but there are many SAR models with different expression functions (e.g., saturation, linear, and quadratic logarithmic). To increase confidence in the results, we used five models to evaluate the relationship between numbers of samples and ASVs, as well as to predict the number of global ASVs (SI Appendix, Table S1 and Supplementary Text). For this analysis, the ASV composition data of each sediment sample was rarefied using a coverage-based method in the “rtk” package of R (77) to mitigate bias caused by sequencing efforts. We separately applied the SAR models to the Archaea, Bacteria, and Universal datasets (Fig. 1) (SI Appendix, Fig. S5 and Table S2). Prior to performing SAR analysis, we first excluded samples with fewer than 10,000 sequences from the datasets. As a result, 213, 272, and 281 samples from the Archaea, Bacteria, and Universal datasets were used for analysis, respectively. The ocean and topsoil datasets compiled from the previous studies (references in Fig. 1) yielded 349 and 227 samples, respectively. The sampling depth for rarefying was set to the minimum total reads of any sample in each dataset according to Saary et al. (77). These sampling depths were 10,183, 10,092, and 10,546 sequences for the Archaea, Bacteria, and Universal datasets, respectively. For the ocean and topsoil datasets, the minimum reads were 11,030 and 11,165, respectively.
The “rtk” algorithm works by transforming input counts into a vector of feature occurrences and shuffles it using Mersenne Twister random number. A subset of this shuffled vector of length equal to the sampling depth was used to construct the rarefied sample and to estimate diversity (77). The five SAR models were run using the rarefied ASV datasets and environmental metadata (SI Appendix, Table S1), with 1,000 random iterations, using the specaccum and fitspecaccum functions in of the “vegan” package in R. The global ASV richness of marine sediment was estimated using each of the SAR models. Volume was used, instead of area, to predict the ASV richness of global marine sediment and seawater (78). Global ASV richness in marine sediment was extrapolated for a global marine sediment volume of 3.01 × 108 km3 (67). The ASV richness of topsoil and ocean were extrapolated for volumes of 14.7 × 108 km2 (79) and 1.33 × 109 km3 (80), respectively.
To test the effect of sequencing depth on our global richness estimates, we preliminarily performed the SAR model analysis with the samples rarefied to 1,000 sequences and with the samples rarified to 10,000 sequences (SI Appendix, Figs. S6 and S7 and Tables S3 and S4). To check the potential effect of contaminants, we also did the SAR analysis using the datasets with and without potential contaminant removal (SI Appendix, Fig. S8 and Table S5). The estimates from these tests differed by less than 1.6-fold, confirming that sequence depth and contamination removal had little effect on the estimates of species richness by SAR models.
Supplementary Material
Acknowledgments
We are grateful to the crew, drilling or coring team members, and shipboard scientists of ODP Leg 201; IODP Exps. 301, 307, 308, 315, 316, 329, 346, and 347; International Ocean Discovery Program Exps. 353 and 354; Chikyu shakedown cruise CK06-06; and R/V Knorr cruise KN223. This research used samples and data that were provided by ODP and IODP. We are also grateful for the technical assistance of S. Hashimoto and M. Tsutsumi (Kochi Institute for Core Sample Research, JAMSTEC) and T. Terada (Marine Works Japan, Ltd.). This is a contribution to the Deep Carbon Observatory (DCO) and Earth 4D: Subsurface Science and Exploration, Canadian Institute for Advanced Research (CIFAR). This is Center for Dark Energy Biosphere Investigations publication 544. This work was partially supported by the Japan Society for the Promotion of Science Grant-in-Aid for Science Research (JSPS) Strategic Fund for Strengthening Leading-Edge Research and Development (to JAMSTEC and F.I.), the JSPS Funding Program for Next Generation World-Leading Researchers (GR102 [to F.I.]); JSPS Grants-in-Aid for Scientific Research 26251041 (to F.I.), 17H03956 (to T.H.), and 19H05503 and 20K20429 (to F.I and T.H.); the DCO Deep Life Community pilot project (T.H.), supported by the Alfred P. Sloan Foundation; the US NSF through Center for Deep Dark Energy Biosphere Investigations Grant NSF-OCE-0939564 (to S.D.), Deutsche Forschungsgemeinschaft Grant AD 511/2-1 (to R.R.A.); and the Deutsche Forschungsgemeinschaft through Center of Excellence “The Ocean Floor—Earth’s Uncharted Interface” Project 390741603 (K.-U.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919139117/-/DCSupplemental.
Data Availability.
The 16S rRNA gene sequences, including the negative controls generated by this study, have been deposited in the DNA Data Bank of Japan nucleotide sequence database (accession nos. DRA005492, DRA008420, DRA008547, and DRA008548).
References
- 1.Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S., Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes R. J.et al., A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar. Geol. 352, 409–425 (2014). [Google Scholar]
- 3.Parkes R. J.et al., Deep bacterial biosphere in Pacific Ocean sediments. Nature 371, 410–413 (1994). [Google Scholar]
- 4.D’Hondt S.et al., Presence of oxygen and aerobic communities from seafloor to basement in deep-sea sediment. Nat. Geosci. 8, 299–304 (2015). [Google Scholar]
- 5.Hoshino T., Inagaki F., Abundance and distribution of Archaea in the subseafloor sedimentary biosphere. ISME J. 13, 227–231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wörmer L.et al., Microbial dormancy in the marine subsurface: Global endospore abundance and response to burial. Sci. Adv. 5, eaav1024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Hondt S.et al., Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2221 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Hoehler T. M., Jørgensen B. B., Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Morono Y.et al., Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18295–18300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inagaki F.et al., Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science 349, 420–424 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Trembath-Reichert E.et al., Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc. Natl. Acad. Sci. U.S.A. 114, E9206–E9215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morono Y.et al., Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years. Nat. Commun. 11, 3626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg H. C., The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Fenchel T., Motility of bacteria in sediments. Aquat. Microb. Ecol. 51, 23–30 (2008). [Google Scholar]
- 15.Jørgensen B. B., Marshall I. P., Slow microbial life in the seabed. Annu. Rev. Mar. Sci. 8, 311–332 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Walsh E. A.et al., Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10, 979–989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starnawski P.et al., Microbial community assembly and evolution in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 114, 2940–2945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkpatrick J. B., Walsh E. A., D’Hondt S., Microbial selection and survival in subseafloor sediment. Front. Microbiol. 10, 956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshino T.et al., Atribacteria from the subseafloor sedimentary biosphere disperse to the hydrosphere through submarine mud volcanoes. Front. Microbiol. 8, 1135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ijiri A.et al., Deep-biosphere methane production stimulated by geofluids in the Nankai accretionary complex. Sci. Adv. 4, eaao4631 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty A.et al., Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proc. Natl. Acad. Sci. U.S.A. 117, 11029–11037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruff S. E.et al., In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J. 13, 197–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Hondt S., Wang G., Spivack A. J., . “The underground economy (energetic constraints of subseafloor life)” in Earth and Life Processes Discovered from Subseafloor Environment: A Decade of Science Achieved by the Integrated Ocean Drilling Program (IODP), Stein R., Blackman D., Inagaki F., Larsen H.-C., Eds. (Series Developments in Marine Geology, Elsevier, Amsterdam, The Netherland, New York, NY, 2014), Vol. chap. 2.3, pp. 127–148. [Google Scholar]
- 24.LaRowe D. E., Amend J. P., Power limits for microbial life. Front. Microbiol. 6, 718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley J. A., Amend J. P., LaRowe D. E., Survival of the fewest: Microbial dormancy and maintenance in marine sediments through deep time. Geobiology 17, 43–59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teske A., Sørensen K. B., Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2, 3–18 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Lloyd K. G., May M. K., Kevorkian R. T., Steen A. D., Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morono Y., Inagaki F., Analysis of low-biomass microbial communities in the deep biosphere. Adv. Appl. Microbiol. 95, 149–178 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Inagaki F.et al., Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69, 7224–7235 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inagaki F.et al., Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 103, 2815–2820 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkes R. J.et al., Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature 436, 390–394 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Biddle J. F., Fitz-Gibbon S., Schuster S. C., Brenchley J. E., House C. H., Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U.S.A. 105, 10583–10588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fry J. C., Parkes R. J., Cragg B. A., Weightman A. J., Webster G., Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol. Ecol. 66, 181–196 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Teske A., . “Marine deep sediment microbial communities” in The Prokaryotes, Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F., Eds. (Springer, Berlin, Heidelberg, Germany, 2013), pp. 123–138. [Google Scholar]
- 35.Ciobanu M.-C.et al., Microorganisms persist at record depths in the subseafloor of the Canterbury Basin. ISME J. 8, 1370–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X.et al., Bioturbation as a key driver behind the dominance of Bacteria over Archaea in near-surface sediment. Sci. Rep. 7, 2400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lloyd K. G.et al., Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Baker B. J.et al., Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat. Microbiol. 1, 16002 (2016). [DOI] [PubMed] [Google Scholar]
- 39.He Y.et al., Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1, 16035 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Nobu M. K.et al., Phylogeny and physiology of candidate phylum “Atribacteria” (OP9/JS1) inferred from cultivation-independent genomics. ISME J. 10, 273–286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T.et al., Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc. Natl. Acad. Sci. U.S.A. 115, 6022–6027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuillemin A.et al., Archaea dominate oxic subseafloor communities for 15 million years. Sci. Adv. 5, eaaw4108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunagawa S.et al., Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Bahram M.et al., Structure and function of the global topsoil microbiome. Nature 560, 233–237 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Magnabosco C.et al., The biomass and biodiversity of the continental subsurface. Nat. Geosci. 11, 707–717 (2018). [Google Scholar]
- 46.Gajigan A. P.et al., Diversity and community structure of marine microbes around the Benham Rise underwater plateau, northeastern Philippines. PeerJ 6, e4781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mason O. U., Canter E. J., Gillies L. E., Paisie T. K., Roberts B. J., Mississippi River plume enriches microbial diversity in the Northern Gulf of Mexico. Front. Microbiol. 7, 1048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parada A. E., Needham D. M., Fuhrman J. A., Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Teeling H.et al., Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5, e11888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster G.et al., A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ. Microbiol. 8, 1575–1589 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Oni O. E.et al., Microbial communities and organic matter composition in surface and subsurface sediments of the Helgoland mud area, North Sea. Front. Microbiol. 6, 1290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh E. A.et al., Relationship of bacterial richness to organic degradation rate and sediment age in subseafloor sediment. Appl. Environ. Microbiol. 82, 4994–4999 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Girvan M., Newman M. E. J., Community structure in social and biological networks. Proc. Natl. Acad. Sci. U.S.A. 99, 7821–7826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao R., Dahle H., Ramírez G. A., Jørgensen S. L., Indigenous ammonia-oxidizing archaea in oxic subseafloor oceanic crust. mSystems 5, e00758-e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakahara N.et al., Aggregatilinea lenta gen. nov., sp. nov., a slow-growing, facultatively anaerobic bacterium isolated from subseafloor sediment, and proposal of the new order Aggregatilineales ord. nov. within the class Anaerolineae of the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 69, 1185–1194 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Ward L. M., Hemp J., Pace L. A., Fischer W. W., Draft genome sequence of Leptolinea tardivitalis YMTK-2, a mesophilic anaerobe from the Chloroflexi class Anaerolineae. Genome Announc. 3, e01356-e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward L. M., McGlynn S. E., Fischer W. W., Draft genome sequences of two basal members of the Anaerolineae class of Chloroflexi from a sulfidic hot spring. Genome Announc. 6, e00570-e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fincker M.et al., Metabolic strategies of marine subseafloor Chloroflexi inferred from genome reconstructions. Environ. Microbiol. 22, 3188–3204 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Wang L., Li X., Lai Q., Shao Z., Kiloniella litopenaei sp. nov., isolated from the gut microflora of Pacific white shrimp, Litopenaeus vannamei. Antonie Van Leeuwenhoek 108, 1293–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Santelli C. M.et al., Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Blöthe M.et al., Manganese-cycling microbial communities inside deep-sea manganese nodules. Environ. Sci. Technol. 49, 7692–7700 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Puente-Sánchez F.et al., Viable cyanobacteria in the deep continental subsurface. Proc. Natl. Acad. Sci. U.S.A. 115, 10702–10707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robbins S. J., Evans P. N., Parks D. H., Golding S. D., Tyson G. W., Genome-centric analysis of microbial populations enriched by hydraulic fracture fluid additives in a coal bed methane production well. Front. Microbiol. 7, 731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blazejak A., Schippers A., High abundance of JS-1- and Chloroflexi-related bacteria in deeply buried marine sediments revealed by quantitative, real-time PCR. FEMS Microbiol. Ecol. 72, 198–207 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Kaster A. K., Mayer-Blackwell K., Pasarelli B., Spormann A. M., Single cell genomic study of Dehalococcoidetes species from deep-sea sediments of the Peruvian Margin. ISME J. 8, 1831–1842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farag I. F.et al., Metabolic potentials of archaeal lineages resolved from metagenomes of deep Costa Rica sediments. ISME J. 14, 1345–1358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.LaRowe D. E., Burwicz E., Arndt S., Dale A. W., Amend J. P., Temperature and volume of global marine sediments. Geology 45, 275–278 (2017). [Google Scholar]
- 68.DeMalach N., Saiz H., Zaady E., Maestre F. T., Plant species-area relationships are determined by evenness, cover and aggregation in drylands worldwide. Glob. Ecol. Biogeogr. 28, 290–299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caporaso J. G.et al., Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schloss P. D.et al., Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso J. G.et al., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dixon P., VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 73.Doi H., Okamura H., Similarity indices, ordination, and community analysis tests using the software R. Jap. J. Ecol. 61, 3–20 (2011). [Google Scholar]
- 74.Hsieh T. C., Ma K. H., Chao A., A. iNEXT: An R package for rarefaction and extrapolation of species diversity (H ill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016). [Google Scholar]
- 75.Newman M. E. J., Girvan M., Finding and evaluating community structure in networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 69, 026113 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Dengler J., Which function describes the species–area relationship best? A review and empirical evaluation. J. Biogeogr. 36, 728–744 (2009). [Google Scholar]
- 77.Saary P., Forslund K., Bork P., Hildebrand F., RTK: Efficient rarefaction analysis of large datasets. Bioinformatics 33, 2594–2595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taylor C. M., Fish species richness and incidence patterns in isolated and connected stream pools: Effects of pool volume and spatial position. Oecologia 110, 560–566 (1997). [DOI] [PubMed] [Google Scholar]
- 79.Gaston K. J., Blackburn T. M., Klein Goldewijk K., Habitat conversion and global avian biodiversity loss. Proc. Biol. Sci. 270, 1293–1300 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Charette M. A., Smith W. H., The volume of Earth’s ocean. Oceanography 23, 112–114 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequences, including the negative controls generated by this study, have been deposited in the DNA Data Bank of Japan nucleotide sequence database (accession nos. DRA005492, DRA008420, DRA008547, and DRA008548).