SUMMARY
Proteins are dynamic molecules that can undergo rapid conformational rearrangements in response to stimuli. These structural changes are often critical to protein function, and thus elucidating time-dependent conformational landscapes has been a long-standing goal of structural biology. To harness the power of single particle cryo-EM methods to enable ‘time-resolved’ structure determination, we have developed a light-coupled cryo-plunger that pairs flash-photolysis of caged ligands with rapid sample vitrification. The ‘flash-plunger’ consists of a high-power ultraviolet LED coupled with focusing optics and a motorized linear actuator, enabling the user to immobilize protein targets in vitreous ice within a programmable time window – as short as tens of milliseconds – after stimulus delivery. The flash-plunger is a simple, inexpensive and flexible tool to explore short-lived conformational states previously unobtainable by conventional sample preparation methods.
INTRODUCTION
Proteins are inherently dynamic and often undergo large-scale conformational rearrangements in response to stimuli. Occupancy of individual functional states and the kinetics by which proteins move from one state to another vary widely and, given the critical nature of protein dynamics to cellular function, the elucidation of molecular structures associated with specific functional states is an area of scrutiny. Ion channels, for example, exhibit a rich landscape of conformational dynamics, opening on a micro- to millisecond time scale and, in many instances, transitioning to non-conducting inactive/desensitized conformations nearly as rapidly (Hille, 2001). Historically, a variety of approaches including mutagenesis (Hou et al., 2018; Pau et al., 2017) and the inclusion of biological toxins (Baconguis et al., 2014; Cao et al., 2013; Chen et al., 2014; Gao et al., 2016; Shen et al., 2018; Xu et al., 2019) have been deployed in order to stabilize specific conformational states. Nevertheless, developing ‘time-resolved’ structural methods to capture specific conformational states, without the use of genetic or molecular tools, remains a long-sought goal of the structural biology field (Frank, 2017; Orville, 2018).
Despite their information rich nature, macromolecular structures typically provide static ‘snap-shots’ of protein conformations and are devoid of biologically-relevant kinetic details. Furthermore, the successful formation of well-ordered crystals or the determination of high-resolution cryo-electron microscopy (cryo-EM) reconstructions often precludes the ability to capture transient conformational states via conventional structural means and instead selects for long-lived states (Palamini et al., 2016). Therefore, while the wealth of information gained from the pursuit of high-resolution protein structures has contributed immensely to our understanding of biology, the lack of a meaningful correlation to kinetics represents a substantial gap in our understanding of protein structure/function relationships (Frank, 2017).
Given the inherent limitations of x-ray crystallography imposed by the crystal lattice, cryo-EM is a powerful technique for exploring time-resolved macromolecular structural changes, particularly in light of advancements in data processing algorithms aimed at parsing conformational heterogeneity (Grant et al., 2018; Nakane et al., 2018; Punjani et al., 2017; Zhang et al., 2019). Indeed, no special equipment is required to study slower structural transitions – such as those that occur on the order of tens of seconds or more – via cryo-EM. Rather, the user simply freezes grids at defined timepoints following the initiation of a reaction (Fischer et al., 2010). However, to study conformational changes within a time window of seconds or faster, such as those associated with activation and inactivation of ion channels, special techniques are needed to overcome the temporal limitations unavoidable in conventional cryo-EM sample preparation, including both blotting and sample handling delays.
A notable example of applying time-resolved cryo-EM to study rapid conformational changes in macromolecular structures was the landmark study of nicotinic acetylcholine receptors (nAChRs) by Nigel Unwin and John Berriman (Berriman and Unwin, 1994; Unwin, 1995, 2003). Even though nAChRs open in microseconds and undergo desensitization within tens of milliseconds in the presence of acetylcholine (Matsubara et al., 1992), Unwin and Berriman captured the receptor in both closed and open states by spraying acetylcholine on cryo-EM grids immediately prior to vitrification. The spray-plunge technique has since evolved to encompass multiple approaches and apparatuses, from designs expanding upon Unwin and Berriman’s initial method of spraying ligand onto a sample suspended on a cryo-EM grid (Berriman and Unwin, 1994), to recent developments aimed at mixing reactants prior to spray application (Chen et al., 2015; Feng et al., 2017; Fu et al., 2019; Kaledhonkar et al., 2018; Kharlamova et al., 2010; Lu et al., 2014). Depending on the method used and the details of the experimental setup, temporal resolution on the single digit milliseconds timescale is theoretically attainable. However, spray-plunging methods are not without downsides, as control over ice thickness, an increase in the air-water interface due to the formation of liquid droplets via aerosolization, and specialized equipment, present challenges.
Light has also been explored as a method to catalyze conformational changes for cryo-EM studies (Shaikh et al., 2009; White et al., 2003; White et al., 1998) and has been successful in electron diffraction experiments (Subramaniam et al., 1993; Subramaniam and Henderson, 1999). These early ‘flash-plunge’ setups employed high-output flash-lamps coupled to liquid light guides and custom manual plunge apparatuses to vitrify samples following light exposure. However, despite the initial promise of light-based time-resolved cryo-EM methods and the aforementioned success when used in conjunction with electron crystallography, the approach has seen comparatively little development.
Light is an attractive mechanism with which to initiate chemical reactions due to its essentially instantaneous delivery and minimal sample perturbation. Furthermore, leveraging photolabile compounds has been instrumental in the study of many biological systems and has met with particular success in the field of neuroscience research (Callaway and Yuste, 2002; Eder et al., 2004; Korkotian et al., 2004; Watanabe et al., 2013; Watanabe et al., 2014). As such, the chemistry of photolabile neuroactive compounds is well developed and there are many commercially available caged neurotransmitters, including ATP (Kaplan et al., 1978), GABA (Wieboldt et al., 1994b), glutamate (Wieboldt et al., 1994a), serotonin (Breitinger et al., 2000), and dopamine (Araya et al., 2013).
Here, we present a light-coupled cryo-plunger for time-resolved cryo-EM that utilizes light to initiate photochemical reactions in a sample suspended on a cryo-EM grid, immediately prior to sample vitrification. Furthermore, we demonstrate that LED-based UV irradiation is sufficient to uncage protons on a tens of millisecond time scale, driving large-scale conformational changes in chicken acid-sensing ion channel 1a (cASIC1a) on a cryo-EM grid. This simple but robust system is easy to assemble and use, requires less than a square meter of bench space and is fully adaptable to fill a variety of unique experimental requirements. As such, our flash-plunger represents an accessible and broadly applicable platform for time-resolved cryo-EM studies.
RESULTS
Design of the flash-plunge apparatus
The flash-plunger consists of two primary components: a motorized linear actuator mounted vertically above a foam cryogen container, and a high-power LED light source mounted perpendicular to the motion axis of the actuator (Figure 1). To maintain alignment of the components and facilitate transport into a variety of environments, such as into a laboratory cold room, the flash-plunger is housed on an aluminum breadboard. The LED and linear actuator are interfaced to a motion control board, which is programmed by and under control of the user via a graphical user interface (GUI) (Supplementary Data Figure 1).
Figure 1. Instrument design and schematic.
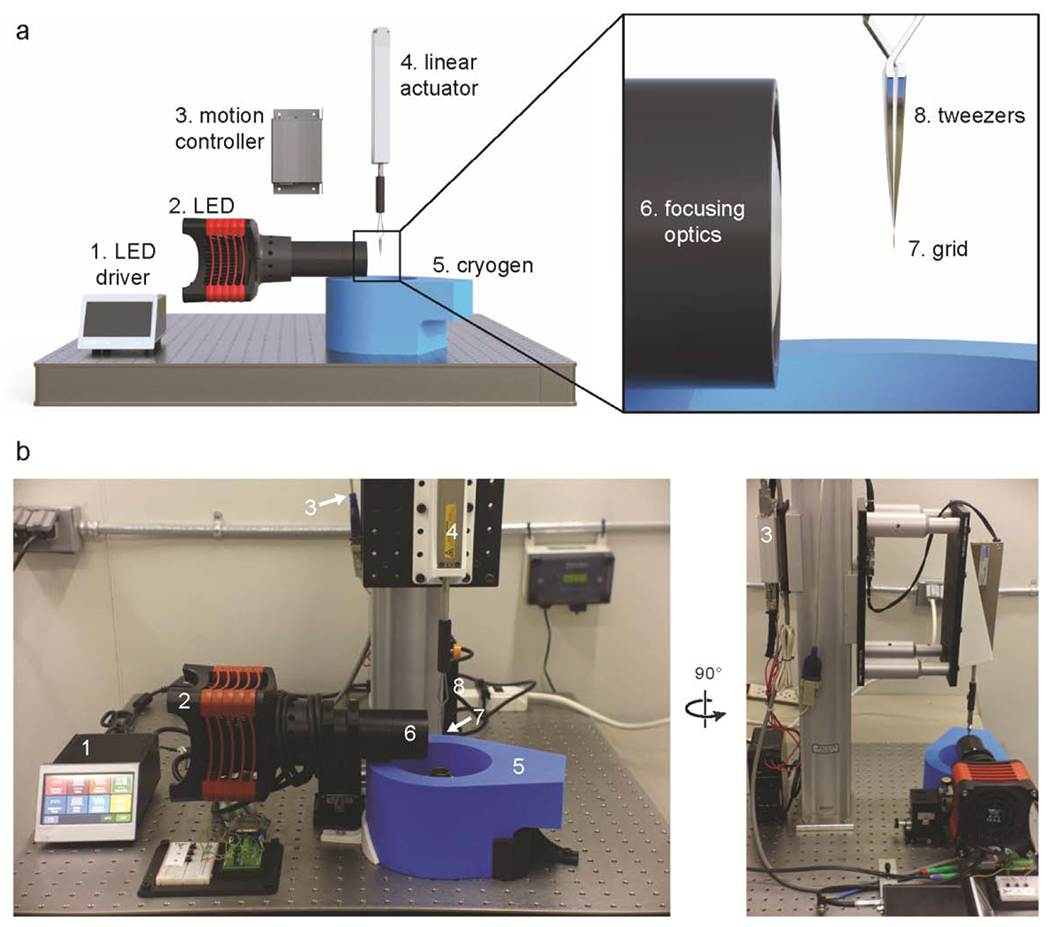
a, Cartoon representation of the flash-plunge device with inset showing close-up view of focusing optics and grid. b, Picture of the actual flash-plunge device with a side-view shown on the right. Numbers in white correlate to components outlined in panel (a).
Three pushbuttons and a foot switch allow the user to select a desired vitrification protocol and to proceed through the selected protocol, respectively (Figure 2). Programmable digital input pins detect user input via pushbutton during the experimental session, allowing for up to three independent protocols to be selected on a grid-by-grid basis without re-programming or re-initializing the system. Such control enables the user to explore multiple timepoints, postphotochemical reaction, within a single grid preparation session. Moreover, the motion control board provides a central node of communication for the entire system, allowing the apparatus to be operated independently of a computer.
Figure 2. Sample workflow.

Overview of the general workflow for a typical flash-plunge experiment.
LED light source
To catalyze photochemical reactions within a short time window we use a high-power UV LED emitter capable of providing at least three watts of output power, measured at the grid, with a dominant wavelength of 365 nm and a bandwidth of 10 nm (full width at half maximum) according to manufacturer specifications. Photons exit the UV LED as a 2-inch diameter collimated beam, passing through a 50 mm wide aspheric condenser lens that focuses the photons onto the cryo-EM grid (Figure 3A–B). Power readings taken with and without a grid in place (carbon side facing light source) indicate roughly 30% of the incident light is absorbed by the grid. Importantly, this data serves only to illustrate the amount of light the sample is exposed to, as we are unable to separate light blocked by the grid from light absorbed by the sample. Given the low absorptivity of water in the UV range (Jaffe et al., 1994), however, the energy absorbed by the sample is likely to be much lower than the estimated 30% total incident light. Further, though we attempted to minimize reflectance by orienting the grid with the carbon side facing the light source, we are unable to empirically determine the ratio of photons reflected vs absorbed by the grid. While optimizing beam dimensions and maximizing power delivery to the grid, this ‘high-power’ setup necessitates short working distances between lens and grid. For photochemical reactions requiring fewer photons, an alternate lens system may be used which provides a more diffuse beam and an increased working distance (Figure 3C–D).
Figure 3. Light pathway.

a-b, Ray-trace diagram (a) and irradiance distribution plot (b) for the flash-plunger equipped with an aspheric condenser lens. c-d, Ray-trace diagram (c) and irradiance distribution plot (d) for the long working distance configuration equipped with a plano convex lens. Ray-trace diagrams were generated using FRED optical engineering software (see Materials and Methods).
Caged compounds are designed to be sensitive to specific wavelengths of light and given differences in quantum yields and compound concentration requirements, different experiments will likely have unique illumination needs. To accommodate a wide array of potential experiments we have designed the LED component of the flash-plunge system to be modular, allowing the user to easily swap LEDs without making substantial adjustments to the rest of the platform, thus providing access to a wide variety of commercially available light sources. Furthermore, while transistor-transistor logic (TTL) signals from the motion control board switch the LED on or off, power output is fully scalable by varying the LED current via the LED driver, allowing the user to tailor the LED output to experimental requirements (Figure 2, Supplementary Data Figure 1).
Protocol design for photo-uncaging and vitrification
Control over the interval separating sample vitrification from the initiation of a conformational change is an essential aspect of time-resolved cryo-EM. In our flash-plunge system, the user sets the desired irradiation time by programming the behavior of a general-purpose digital output pin which switches the LED on or off via a TTL signal. Additionally, the motion profile of the linear actuator, including but not limited to piston start/stop position, timing, acceleration/deceleration and velocity is fully programmable via the GUI. Therefore, both the length of the light exposure and the time between exposure and vitrification are under the user’s control and can be tailored to experimental requirements.
In addition to programmable light exposure and plunge times, the system can accommodate multiple light exposure and vitrification strategies. For projects requiring maximal irradiation, a sequential ‘flash-then-plunge’ protocol (Figure 4A, Supplementary Data Video 1) will increase light exposure at the expense of temporal resolution, which, with our current hardware configuration, is limited to tens of milliseconds. Though there is no appreciable delay between irradiation and plunge initiation, considerations impacting temporal resolution include the large diameter of the optical components, which require the grid to be held for irradiation at least 25 mm above the cryogen, as well as the maximum average speed of the linear actuator which, over a short plunge path, is limited by acceleration. In our tests, the linear actuator moved a load consisting of tweezers, 3D printed tweezer adapter and a grid 50 mm in ~ 45 ms.
Figure 4. Flash-plunge protocols.
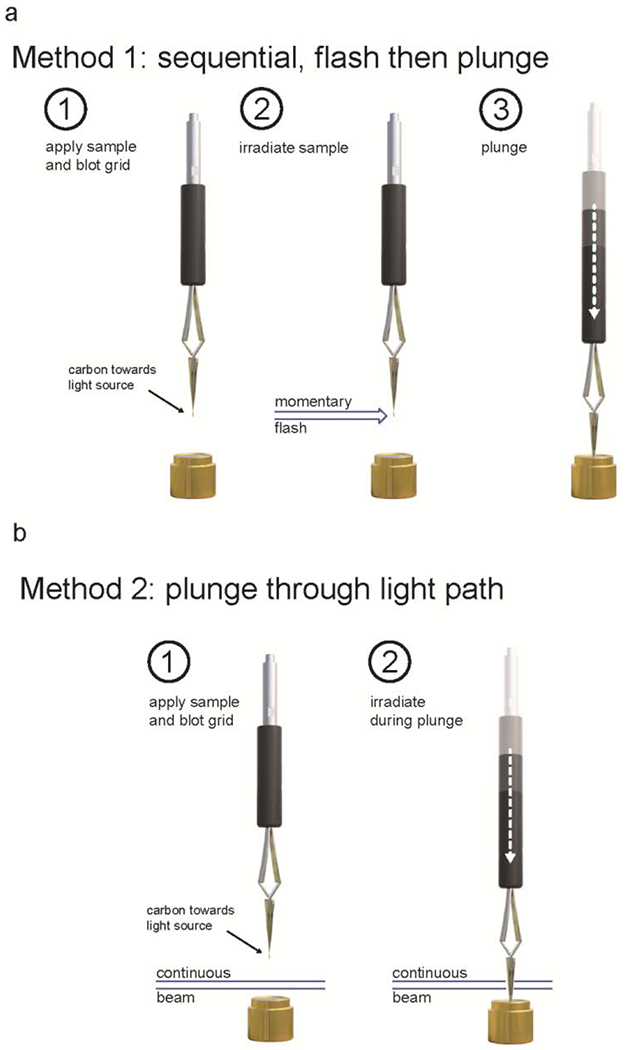
a-b, Graphical representations of the sequential ‘flash-then-plunge’ protocol (a) and the alternate protocol (b) wherein a blotted grid is moved through a continuous beam path.
Alternatively, the desired photochemistry may be achieved by simply plunging a blotted grid through the path of the UV light beam (Figure 4B). Depending on the distance between the beam path and the cryogen, as well as the user-defined motion profile of the linear actuator, adopting this exposure method improves temporal resolution to single milliseconds, thus approaching the limitations set by the formation of vitreous ice (Brüggeller and Mayer, 1980; Kasas et al., 2003). Notably, this approach will provide less irradiation than the aforementioned ‘flash-then-plunge’ approach, limiting its usefulness to projects that require fewer photochemical events to achieve a desired conformational change.
Photolysis of caged compounds
To demonstrate the capabilities of the flash-plunge apparatus to facilitate photochemical reactions on a cryo-EM grid, we used UV radiation to release protons from the sodium salt of 2-methoxy-5-nitrophenyl sulfate (MNPS·Na) (Abbruzzetti et al., 2005) (Figure 5A) in Tris-buffered saline (TBS). We began the ‘pH drop’ experiments by first measuring the capacity of the photolyzed caged proton source to decrease the pH of a droplet of mildly buffered solution on an EM grid. To do this, we employed 20 mM of MNPS·Na (final concentration) in a solution of 5 mM Tris pH 8.0, 150 mM NaCl and exposed 2 μl of the solution deposited on a holey carbon cryo-EM grid to 365 nm radiation. The pH was then estimated by using a UV-VIS spectrofluorophotometer and the pH-dependent probe SNARF-4F, which harbors a characteristic ratiometric and pH-dependent emission spectrum when excited with 543 nm light (Supplementary Data Figure 2). At the maximum current supported by our LED (4500 mA), exposure to UV radiation was sufficient to decrease the pH of the sample from ~ 8.0 to near 6.0 in 15 ms, and to pH 5 in 25 ms (Figure 5B). Alternatively, we were also able to titrate the pH drop by varying the LED current while maintaining a constant exposure time of 25 ms (Figure 5C).
Figure 5. Uncaging protons via light exposure.
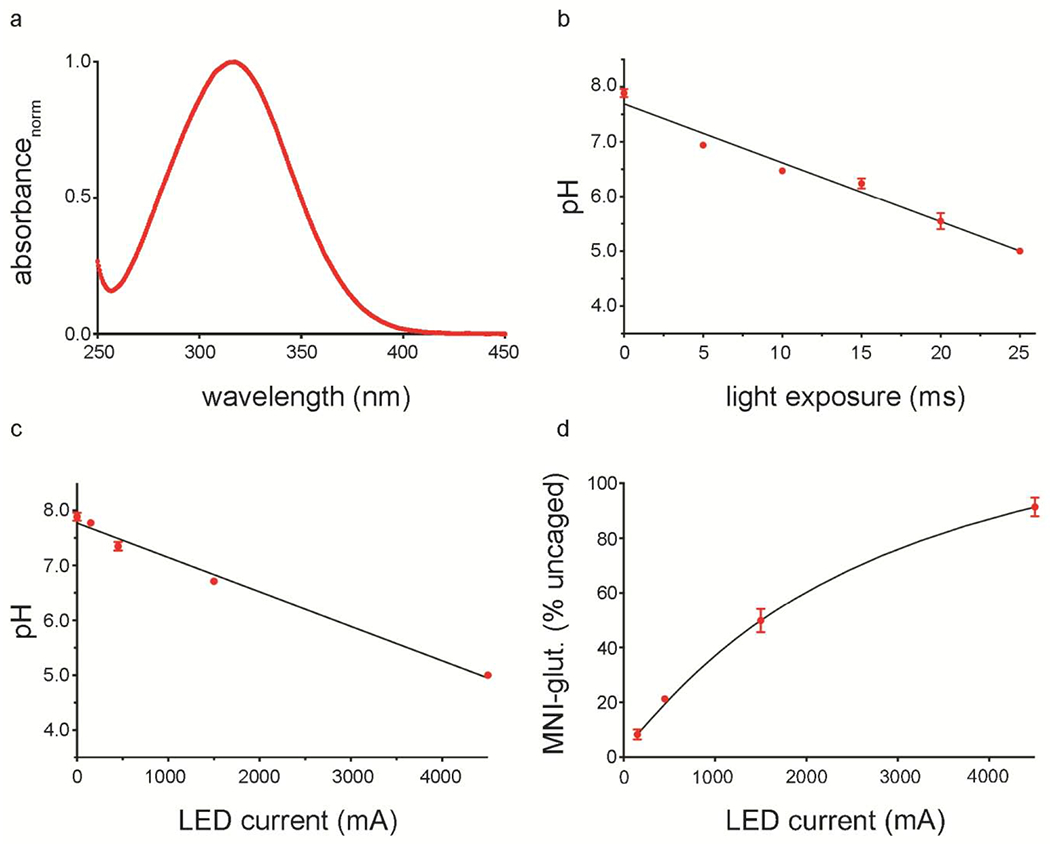
a, Absorbance spectrum for MNPS·Na. b-c, Plot of pH vs light exposure time at 4500 mA LED current (b) or pH vs LED current for a 25 ms exposure (c) for MNPS·Na. d, Plot of uncaged glutamate vs LED current for a 25 ms exposure. For b-d, n=3, data represent mean, error bars represent SEM and black lines represent interpolated linear (b-c) and sigmoidal (d) fits.
To assess the capacity of our apparatus to uncage compounds more commonly used in biochemistry or neuroscience experiments, we tested the flash-plunge device with 4-methoxy-7-nitroindolinyl caged l-glutamate (MNI-glutamate) (Matsuzaki et al., 2001), a caged compound routinely used for photo-activation of N-methyl-d-aspartate (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) glutamate receptors (Araya et al., 2006; Carter and Jahr, 2016; Digregorio et al., 2007; Pugh and Jahr, 2011). We exposed solutions containing 1 mM MNI-glutamate to UV radiation for 25 ms while varying the LED current (Figure 5D). Photolysis efficiency was assessed via HPLC analysis of the resulting grids and the extent of glutamate release was ~ 50% for a 1500 mA flash and over 90% for a 4500 mA flash. Because the EC50 values for glutamate activation of NMDA and AMPA receptors are in the low micromolar range, the initiation of receptor activation may require less illumination than the values tested here.
To determine the impact of UV irradiation on biological samples immobilized on a cryo-EM grid, we flashed grids containing apo ferritin in TBS with UV light immediately prior to vitrification. To mimic the conditions tested in our photolysis experiments, we irradiated grids for 25 ms with the LED set to either 10% (450 mA), 30% (1350 mA) or 100% (4500 mA) power. Ice quality and particle distribution were not noticeably impacted for 10% or 30% power flashes when compared to control grids (Supplementary Data Figure 3A–F). Grids exposed to full high-power irradiation from the UV LED (4500 mA), however, showed thinned ice, higher background signal likely due to evaporation-induced increases in salt and buffer concentrations, and fewer particles (Supplementary Data Figure 3G–H). While grids were frozen in a cold room environment with moderate humidity levels 60-70%, increasing room humidity or engineering a high-humidity chamber (Bellare et al., 1988) may further mitigate evaporation-induced effects, improving the consistency of results and potentially increasing the maximum tolerable irradiation levels.
The flash-plunge system described here is capable of releasing sufficient protons from MNPS·Na to substantially alter the pH of a buffered system by ~ 3 pH units at high power, and moderate irradiation is capable of releasing ~ 500 μM glutamate in 25 ms. Thus, we propose that the apparatus is capable of photochemically modifying many commonly available caged and photo-switchable compounds and thus will be applicable to a wide variety of projects. Moreover, ‘flashed’ apo ferritin grids confirmed that sample and grid quality did not suffer when exposed to low to moderate levels of light from the UV LED.
Photo-uncaging protons drives conformational changes in a ligand-gated ion channel
Acid-sensing ion channels (ASICs) are proton-gated members of the epithelial sodium channel/degenerin superfamily of ion channels (Kellenberger and Schild, 2002; Waldmann and Lazdunski, 1998). ASICs occupy a non-conducting resting state at high pH, open in response to low pH exposure, and enter a long-lived proton-bound but non-conducting desensitized state tens to hundreds of milliseconds following channel activation (Zhang et al., 2006). Proton-dependent structural rearrangements in cASIC1a are well documented and have been captured previously by both single-particle cryo-EM (Yoder et al., 2018) and by x-ray crystallography (Baconguis et al., 2014; Baconguis and Gouaux, 2012; Dawson et al., 2012; Gonzales et al., 2009; Jasti et al., 2007). Therefore, cASIC1a is an ideal model system to test photo-uncaging of protons and the applicability of the flash-plunge system.
We thus added 20 mM MNPS·Na to cASIC1a purified at pH 8.0 and exposed the sample to UV radiation prior to vitrification on holey carbon cryo-EM grids. Each sample was irradiated following the aforementioned ‘flash then plunge’ technique (Figure 4A, Supplementary Data Video 1). For these experiments, LED current was set to either 0, 1000 or 3000 mA and exposure time was kept constant at 25 ms, yielding a final pH of either ~ 8.0, 7.0 or 6.0, respectively (Figure 5C). For all experiments there was a ~ 70 ms interval between initial UV exposure and vitrification composed of irradiation (25 ms) and plunge (45 ms) steps.
cASIC1a particles not exposed to UV radiation (Yoder and Gouaux, 2020) occupy a high pH resting conformation (Figure 6A–B) characterized by an expanded acidic pocket and a gate closed to ion permeation, consistent with existing x-ray and cryo-EM structures of cASIC1a in a high pH resting state (Yoder et al., 2018). Similarly, channels that were subjected to a 1000 mA flash – ostensibly reducing the pH to 7.0 (Figure 5C) – also retained a high pH resting state conformation (Figure 6C–D, Supplementary Data Figure 4, Supplementary Data Table 1), indicative of insufficient channel activation and unsurprising given a pH50 of ~ 6.7 for cASIC1a activation (Jasti et al., 2007). In contrast, channels exposed to a 3000 mA flash – bringing the pH to an estimated value of 6.0 (Figure 5C) – primarily occupy a low pH desensitized conformation (Figure 6E–G, Supplementary Data Figure 5, Supplementary Data Table 1), characterized by a proton-bound collapsed acidic pocket (Figure 6H) and a characteristic arrangement of the channel’s β11-β12/β1-β2 linkers unique to the desensitized conformation (Figure 6I) (Baconguis and Gouaux, 2012; Gonzales et al., 2009).
Figure 6. A photochemically-initiated conformational change in cASIC1a.

a-b, Representative micrograph with ten selected top-down views shown in white circles (a) and cryo-EM reconstruction (b) of cASIC1a not exposed to UV radiation, occupying the resting state (Yoder and Gouaux, 2020). Inset shows close-up view of the acidic pocket, c-d, Example micrograph with ten selected top-down views shown in white circles (c) and cryo-EM reconstruction (d) of cASIC1a exposed to a 1000 mA UV flash for 25 ms with coordinates for the resting channel (PDB 5WKU (Yoder et al., 2018) superimposed). Inset shows close-up view of the acidic pocket, e-f, Example micrograph with ten selected top-down views shown in white circles (e) and cryo-EM reconstruction (f) of cASIC1a exposed to a 3000 mA UV flash for 25 ms with coordinates for the desensitized channel (PDB 4NYK (Baconguis et al., 2014; Gonzales et al., 2009) superimposed). Inset shows close-up view of the acidic pocket, g-h, Locally-refined map of the ECD from the 3000 mA flash dataset (g) demonstrating conformational changes at the acidic pocket (h) and β1-β2/β11-β12 linkers (i) associated with the desensitized channel conformation. Coordinates for the x-ray structure of a desensitized channel (PDB 4NYK (Baconguis et al., 2014; Gonzales et al., 2009), red) were rigid body fit into the ECD-map. For structural comparisons in h-i, coordinates for the resting (PDB 5WKU (Yoder et al., 2018), blue) and open (PDB 4NTW (Baconguis et al., 2014), grey) channels were superposed.
Ultimately, particle distribution deteriorated when LED current was raised to 3000 mA, resulting in densely packed regions and clusters at the hole edges rather than an even distribution (Figure 6E). Furthermore, orientation distribution skewed towards top-down views, likely due to the thinner ice associated with the 3000 mA flash. Accordingly, map quality suffered, and the transmembrane domains were not well resolved in our reconstructions of the entire channel (Figure 6F). Interestingly, local refinement including only the extracellular domain (ECD) showed improved map quality (Figure 6G, Supplementary Data Figure 5) and confirmed the unique structural features at the ECD that correspond to proton-bound (Figure 6H) and desensitized (Figure 6I) channels solved previously (Baconguis et al., 2014; Baconguis and Gouaux, 2012; Gonzales et al., 2009). Unfortunately, we were unable to capture a proton-bound open channel conformation with these experiments, demonstrating a necessity for greater temporal resolution to capture the channel in a proton-activated state.
These results demonstrate that our flash-plunge system is capable of generating meaningful conformational changes in samples immobilized on cryo-EM grids and that moderate light exposure does not prohibit structure determination for even challenging membrane proteins. Of note, at high irradiation levels – such as those provided by a 3000 mA flash – we observed a degradation of particle quality that hampered, but did not prevent, structure determination. Therefore, while the amount of light applied to a grid to facilitate photochemical reactions may result in undesirable side effects, neither formation of vitreous ice nor structure determination is precluded by exposing blotted grids to substantial UV radiation.
DISCUSSION
Here we present an apparatus that couples light delivery and sample vitrification to observe time-resolved structural changes by cryo-EM. In our system, delivery of photons from a high-power LED light source to a sample immobilized on a cryo-EM grid initiates photochemical reactions that catalyze protein conformational changes captured by rapid vitrification. Most of the components that comprise the flash-plunge apparatus are inexpensive, commercially available as well as easily assembled, programed and operated. Moreover, the inherent flexibility of the system allows the plunger to be readily adapted to a variety of projects. Finally, we demonstrate the efficacy of LED-based light sources for catalyzing photochemical reactions on grids by reducing the pH of buffered solutions by photo-uncaging protons and, in doing so, driving large-scale conformational changes in recombinant proton-sensitive cASIC1a channels.
Importantly, large amounts of light, such as the estimated ~ 1 Watt(1 mJ/ms) blocked by the grid during maximal irradiation, may have detrimental effects on the sample. Despite the low absorption of water in the UV range (Buiteveld et al., 1994), the deposition of energy via photons onto the cryo-EM grid will nevertheless drive an increase in sample temperature, as demonstrated in earlier iterations of a flash-plunge device (Shaikh et al., 2009). While the factors governing the severity of the temperature rise are complex and difficult to predict or measure accurately, our basic understanding of common grid materials as well as the relationship between heat transfer and temperature (Q=mcΔT) allow us to estimate the maximum temperature increases. Specifically, for a grid (1 mg) composed entirely of gold, copper or carbon, 1 mJ of energy absorbed during 1 ms of full-power irradiation would result in an increase in temperature of 7.7, 2.6 or 1.4°C, respectively (specific heats, c = 0.13, 0.39 and 0.72 J/g°C for gold, copper and carbon, respectively). Furthermore, though we submit that absorption of UV photons by the sample is minimal when compared to that of the grid, it is worth noting that a 1 mJ increase in energy would raise the temperature of an equivalent mass of water (c = 4.18 J/g°C) by 0.24 °C.
Notably, the above calculations omit numerous relevant factors that will directly impact the magnitude of temperature jumps including reflected photons, heat transfer to the grid tweezers and evaporative cooling effects. Regardless, the predicted temperature shifts, especially for gold grids, are striking and illustrate the potential for thermal denaturation (Chick, 1910) in cases of extreme irradiation. Additionally, photolabile compounds requiring UV irradiation, such as those used in these studies, generate the potential for UV-induced protein denaturation (Clark, 1935). While the extent of the negative impact of irradiation is likely to be influenced by a variety of factors including exposure time, irradiance and wavelength, care must be taken in experimental design to minimize irradiation requirements whenever possible.
As demonstrated in the reconstructions of cASIC1a exposed to a 1000 mA flash, we did not observe significant impacts on particle quality or structure determination following ‘moderate’ UV irradiation. Therefore, photochemically-demanding projects, such as those requiring unusually high concentrations of photo-uncaged ligand or for whom only poorly photosensitive ligands – i.e. those with a low quantum yield – are available, will be more prone to the detrimental effects of light exposure. However, these effects are less likely to be observed at the levels of irradiation required for common flash-plunge applications and may be further mitigated through the use of non-UV sensitive photolabile compounds.
We acknowledge that a photochemical approach is not a ‘one size fits all’ solution to time-resolved cryo-EM, and that alternative methods, such as mixing-spraying techniques, are effective in certain situations (Fu et al., 2019; Kaledhonkar et al., 2019). Rather, we propose that our light-based apparatus mitigates the ongoing challenge of ice variability associated with a mixing-spraying approach (Feng et al., 2017) and avoids the possibility of local concentration differences inherent to a solution mixing step. Furthermore, given the low cost and simple setup, our light-coupled cryo-plunger improves upon the accessibility of implementing a time-resolved system in most laboratory environments. As such, the flash-plunge system complements existing methods of sample preparation and provides the cryo-EM community with a platform for time-resolved cryo-EM.
MATERIALS AND METHODS
Flash-plunger components:
The major components are as follows: (1) 100 mm stroke multi-phase 6 coil linear actuator with built in 5 μm encoder (LCA25-100-35-6, SMAC); (2) single axis brushless motion controller with built in amplifier (LCC-10, SMAC), an RS232 Kit (SMAC) and DB26HD breakout board (CZH Labs); (3) 240 W power supply (S8VK-G24048, Omron Automation); (4) high-power 365 nm UV LED (SOLIS-365C, Thor Labs) controlled by an LED driver (DC2200, Thor Labs) and equipped with an aspheric condenser lens AR-coated for 350-700 nm (ACL50832U-A, Thor Labs) housed in 2-inch lens tubes (Thor Labs). The linear actuator was affixed to a stainless-steel breadboard via a large diameter aluminum post (Spindle and Hoyer) and the LED and optical components were affixed to the breadboard via a three-axis manual micromanipulator (1680 XYZL, Siskiyou). The ethane/propane mixture was contained in a brass cup from a Vitrobot Mark III (ThermoFisher Scientific) and housed within a small foam vessel (FD-500, SpearLab). Additional electrical components included a breadboard connected to a foot switch and affixed with three pushbuttons as well as a circular white LED for postvitrification illumination of the working space (144W-ZK, AmScope). All programming, tuning and testing of the motion parameters was done in LCC Control Center V2.0.1 (SMAC) or MotionLab V1.8.1.0 (SMAC).
Several components of the flash-plunge apparatus were 3D printed ‘in house’ on a large-format FDM 3D printer (N2Plus, Raise3D) using PLA or ABS filament. The 3D printed parts include the adapter connecting the tweezers to the actuator piston, as well as mounting brackets for the ethane/propane mixture, motion control board, linear actuator, breadboard/breakout board and foam cryogen container.
Flash-tests with caged compounds:
A 2 μl sample containing 20 mM MNPS·Na, 5 mM Tris pH 8.0, 150 mM NaCl, 10 μM SNARF-4F was applied to a holey carbon grid held in selfclosing tweezers (5376-NM, Pelco) with the carbon side facing the light source and irradiated via the UV LED. LED power was set by adjusting LED current via the LED driver and irradiation time was programmed via the GUI by adjusting the length of a TTL pulse from the motion controller to the LED driver. The sample was diluted to 10 μl total volume in a sub-micro fluorometer cuvette (701MFLUV10.10B, FireflySci) and emission spectra were recorded (Ex. 543 nm) on a UV-VIS spectrofluorophotometer (RF-5301PC, Shimadzu). The ratio of emission peaks at 642 and 586 nm (R642/586) was calculated and final pH values interpolated using a calibration curve generated using pH-adjusted buffer containing 80 mM Tris, 40 mM MES and 10 μM SNARF-4F. Experiments for standard curve data as well as all flash tests were performed in triplicate. The caged proton source MNPS Na was synthesized (Fibich et al., 2007) by the medicinal chemistry core at Oregon Health and Science University (OHSU) and stored at −80°C. Solutions of MNPS Na in water were stable for months at −20°C and through multiple freeze thaw cycles.
For glutamate uncaging, 1 mM MNI-glutamate samples in water were prepared in a dark room. A 4 μl sample was applied to a holey carbon grid as described above and irradiated via the UV LED. Following irradiation, a 2 μl sample was removed and diluted to 98 μl for HPLC-based measurement of uncaged glutamate.
Motion and illumination analysis:
Given the short duration of irradiation as well as the velocity profile of the plunging grid, we used a camera capable of collecting 1000 frames per second (fps) modified to accept aftermarket lens systems (RibCage RX0 camera, Sony; Voightlander Nokton 17.5 mm f/0.95 lens). Recordings were collected at 1080p/1000 fps and movies were analyzed frame-by-frame in Premier Pro (Adobe).
Ray trace analysis for design and optimization of the lens system was performed using FRED optical engineering software (Photon Engineering). Briefly, 3D models corresponding to the UV LED and associated optics and lens tubes were imported into the software and simulations were conducted for an incoherent source (3 W output at 365 nm) of roughly the same size as the LED emitter (4 mm x 4 mm square). LED power was measured at the grid with a wireless power meter (PM160T-HP, Thor Labs) set to 365 nm.
Expression and purification of cASIC1a:
Isolated membrane fractions containing recombinant cASIC1a (Gallus gallus) containing an N-terminal 8x His EGFP tag were prepared as previously described (Yoder et al., 2018). For control and 1000 mA flash experiments, membrane pellets were suspended using a Dounce homogenizer and solubilized in Tris-buffered saline (TBS) containing 2% (w/v) SL300010 (Polyscope) styrene-maleic acid copolymer. Membrane debris was removed via centrifugation at 125,171 ref and SMA-cASIC1a particles were bound, in batch mode, to Ni-NTA beads overnight at 4°C, in the presence of 10 mM imidazole.
The following day, the SMA-cASIC1a protein was eluted from the Ni-NTA beads packed into an XK-16 column with TBS containing 250 mM imidazole. Peak fractions were pooled and concentrated to ~ 5 mg/ml and the 8x-His EGFP tag was removed via thrombin digestion (1:25) overnight at room temperature (RT). Following tag removal, the SMA-cASIC1a protein was subjected to size-exclusion chromatography (Superose 6 Increase) with a mobile phase containing TBS supplemented with 1 mM DTT. Peak fractions were concentrated to 1 mg/ml for cryo-EM sample preparation.
For samples flashed at 3000 mA LED current, cASIC1a membranes were suspended using a Dounce homogenizer in TBS with 1% (w/v) digitonin and membrane debris was removed via centrifugation. Subsequent purification of digitoninn-cASIC1a occurred as described above but with some exceptions. First, the digitonin-cASIC1a protein was bound to Ni-NTA beads in batch for 1.5 hours at 4°C. Second, the 8x-His EGFP tag was removed from the digitonin-cASIC1a protein via thrombin digestion (1:50) for 1 hour at RT. Finally, purification buffers for digitonin-cASIC1a were identical to those detailed for purification of SMA-cASIC1a but contained 0.1% (w/v) digitonin. Purified digitonin-cASIC1a sample was used for flash-plunge grid preparation at 5 mg/ml.
Sample preparation for cryo-EM flash-plunge experiments:
All flash-plunge experiments took place in an environmentally controlled room maintained 4°C and 60-70% humidity. MNPS Na was added to each sample to achieve a final concentration of 20 mM. Samples were applied to 200 mesh Au Quantifoil holey carbon grids – R1.2/1.3 for un-flashed cASIC1a, R2/2 for 1000 mA-flashed cASIC1a, and R2/1 for 3000 mA-flashed cASIC1a. The grids were glow-discharged (PELCO easiGlow) for 1 min at 15 mA, carbon side up, and pre-cooled to 4°C.
A 4 μl sample was applied to the carbon side of a glow-discharged grid and blotted by hand using pre-cooled filter paper (Whatman, grade 1). The sequential ‘flash-then-plunge’ method described above was used to vitrify all samples in a mixture of 35% ethane and 65% propane (Tivol et al., 2008). A 25 ms flash was used for all grids and the LED current was varied to defined irradiation levels, resulting in a total time of ~ 70 ms (25 ms flash, 45 ms plunge) between blot and vitrification. For all samples, the carbon side of the grid was oriented towards the light source. For control grids that were not irradiated, the LED current was set to 0 mA to maintain a constant time interval.
Cryo-EM data collection:
Cryo-EM data were collected on a Titan Krios cryo-electron microscope (ThermoFisher) operated at 300 kV. Images were recorded on a Gatan K3 camera positioned after an energy filter (20-eV slit width) operating in super-resolution mode with a binned pixel size of 0.83 Å. Data were collected using SerialEM (Mastronarde, 2003) and dose-fractionated to 50 frames for a total exposure time of 2-3 s and a total dose of 40-50 e− Å−2.
Cryo-EM data processing for cASIC1a:
Images were motion-corrected with UCSF MotionCor2 (Zheng et al., 2017) and defocus values were estimated with Gctf (Zhang, 2016) for 1000 mA-flashed cASIC1a or patch CTF estimation in cryoSPARC V2 (Punjani et al., 2017) for 3000 mA-flashed cASIC1a. Particles were picked using DoGPicker (Voss et al., 2009) and reference-free 2D classification was performed in cryoSPARC. For 3000 mA-flashed particles a second round of particle picking was performed using initial 2D classes as templates. Following 2D classification, an ab-initio model was generated in cryoSPARC and particles were subjected to iterative rounds of 3D classification and refinement, also in cryoSPARC. Of note, extensive 3D classification and refinement did not indicate the presence of additional classes in any of the flashed datasets. Final reconstructions were obtained via non-uniform refinement (C3 symmetry) in cryoSPARC for control particles or in RELION 3.0 (Zivanov et al., 2018) for 1000 mA-flashed particles.
For 3000 mA-flashed particles, particle subtraction was performed in cryoSPARC V2 using a mask that removed signal from density corresponding to either the micelle or to the micelle and the TMD. Local refinement (C1 symmetry) was then performed on the subtracted particle stack to obtain reconstructions of the ECD alone or of the entire channel.
Supplementary Material
Supplementary Data Video 1. ‘Flash-then-plunge’ method. Video recorded at 1000 frames per second showing a cryo-EM grid irradiated for 25 ms prior to plunge into ‘cryogen’ (water). Time from initial UV exposure to ‘cryogen’ immersion ~ 70 ms. Video playback is 25 frames per second.
Supplementary Data Figure 1. Instrument control diagram. a, A schematic showing how the instrument is controlled and programmed. Red denotes input required from the user to initiate the experiment; blue denotes information programmed by the user prior to the experiment. b, Picture of the actual flash-plunge device with a side-view shown on the right. Numbers in white correlate to components outlined in (a).
Supplementary Data Figure 2. Fluorescence changes of SNARF-4F probe as a function of pH. a, Normalized fluorescence emission spectra (excitation at 543 nm) of pH-adjusted buffer containing 8 mM Tris, 4 mM MES, 150 mM NaCl and 10 μM SNARF-4F measured between 565 nm and 750 nm. Data represent the mean, n=3. b, Plot of pH vs ratio of fluorescence at 642nm (high pH peak) and 586 nm (low pH peak). Data represent mean (black circles), n=3, and interpolated standard curve is shown as solid green line with 95% confidence internal shown in dashed green.
Supplementary Data Figure 3. Flashed apo ferritin grids. a-h, Representative micrograph and Gctf power spectrum for apo ferritin grids not exposed to UV radiation (a and b, respectively), or for grids exposed to a 25 ms 450 mA (c and d, respectively), 1350 mA (e and f, respectively) or 4500 mA (g and h, respectively) UV LED flash.
Supplementary Data Figure 4. Cryo-EM data processing of 1000 mA flashed cASIC1a. a, Selected 2D classes. b, Schematic of cryo-EM data processing workflow. c-e, local resolution estimation (c) particle distribution (d) and resolution estimation (e) from final non-uniform refinement in cryoSPARC V2.
Supplementary Data Figure 5. Cryo-EM data processing of 3000 mA flashed cASIC1a. a, Selected 2D classes, b, Schematic of cryo-EM data processing workflow, c-e, local resolution estimation (c) particle distribution (d) and resolution estimation (e) from final local refinement using a map containing the whole channel, f-h, local resolution estimation (c) particle distribution (d) and resolution estimation (e) from final local refinement using a map containing the only the ECD.
HIGHLIGHTS.
A light-coupled cryo-plunger provides a flexible platform for time-resolved cryo-EM.
Coupling irradiation with high-speed mechanical plunge freezing enables temporal resolutions in the range of tens of milliseconds.
Ultraviolet irradiation from a light emitting diode robustly uncages glutamate and protons.
On-grid photolysis of ‘caged protons’ activates acid-sensing ion channels and enables structure elucidation by single-particle cryo-EM.
ACKNOWLEDGEMENTS
We thank L. Vaskalis for help with figures, H. Owen for manuscript preparation and all Gouaux lab members for their support. Additionally, we thank Thomas Braun and Lucas Rima for helpful instruction regarding manual plunger design and Hans-Jürgen Apell for advice concerning the selection, synthesis and use of caged proton compounds. This research was supported by the National Institute of Diabetes and Digestive Kidney Diseases (5T32DK007680), the National Institute of Neurological Disorders and Stroke (5F31NS096782 to N.Y., 5F32MH115595 to F.J.-Y, and 5R01NS038631 to E.G.), the American Heart Association (18PRE33990205 to S.N.), the National Science Foundation (DGE-1937961 to A.H.) and the National Institutes of Health (DP5OD017871 to I.B.). Initial electron microscopy work was performed at the Multiscale Microscopy Core at OHSU. A portion of this research was performed at the National Center for Cryo-EM Access and Training and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24GM129539) and by grants from the Simons Foundation (SF349247) and New York State. Subsequent research was supported by NIH grant U24GM129547 and performed at the Pacific Northwest Cryo-EM Center at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Additional support was provided by ARCS Foundation and Tartar Trust fellowships. E.G. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
DATA AVAILABILITY
All cryo-EM maps have been deposited in the Electron Microscopy Data Bank under the accession codes EMD-21385 for 1000 mA-flashed cASIC1a and EMD-21384 for 3000 mA-flashed cASIC1a. Locally refined ECD-only map deposited as an additional map under entry EMD-21384. Technical drawings for most commercial components are available directly from the manufacturer and drawings for custom 3D printed parts will be made available upon request.
REFERENCES
- Abbruzzetti S, Sottini S, Viappiani C, and Corrie JE (2005). Kinetics of proton release after flash photolysis of 1-(2-nitrophenyl)ethyl sulfate (caged sulfate) in aqueous solution. J Am Chem Soc 127, 9865–9874. [DOI] [PubMed] [Google Scholar]
- Araya R, Andino-Pavlovsky V, Yuste R, and Etchenique R (2013). Two-Photon Optical Interrogation of Individual Dendritic Spines with Caged Dopamine. ACS Chemical Neuroscience 4, 1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya R, Eisenthal KB, and Yuste R (2006). Dendritic spines linearize the summation of excitatory potentials. Proceedings of the National Academy of Sciences 103, 18799–18804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I, Bohlen CJ, Goehring A, Julius D, and Gouaux E (2014). X-ray structure of acid sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell 156, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baconguis I, and Gouaux E (2012). Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare JR, Davis HT, Scriven LE, and Talmon Y (1988). Controlled environment vitrification system: An improved sample preparation technique. Journal of Electron Microscopy Technique 10, 87–111. [DOI] [PubMed] [Google Scholar]
- Berriman J, and Unwin N (1994). Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy 56, 241–252. [DOI] [PubMed] [Google Scholar]
- Breitinger H-GA, Wieboldt R, Ramesh D, Carpenter BK, and Hess GP (2000). Synthesis and Characterization of Photolabile Derivatives of Serotonin for Chemical Kinetic Investigations of the Serotonin 5-HT3Receptor†. Biochemistry 39, 5500–5508. [DOI] [PubMed] [Google Scholar]
- Brüggeller P, and Mayer E (1980). Complete vitrification in pure liquid water and dilute aqueous solutions. Nature 288, 569–571. [Google Scholar]
- Buiteveld H, Hakvoort J, and Donze M (1994). Optical properties of pure water, Vol 2258 (SPIE). [Google Scholar]
- Callaway EM, and Yuste R (2002). Stimulating neurons with light. Current Opinion in Neurobiology 12, 587–592. [DOI] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, and Julius D (2013). TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BC, and Jahr CE (2016). Postsynaptic, not presynaptic NMDA receptors are required for spike-timing-dependent LTD induction. Nature Neuroscience 19, 1218–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu T-M, Ruben, and Frank J (2015). Structural Dynamics of Ribosome Subunit Association Studied by Mixing-Spraying Time-Resolved Cryogenic Electron Microscopy. 23, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Durr KL, and Gouaux E (2014). X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science 345, 1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick H (1910). On the ‘heat coagulation’ of proteins. The Journal of Physiology 40, 404–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JH (1935). The denaturation of egg albumin by ultra-violet radiation. The Journal of General Physiology 19, 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson RJ, Benz J, Stohler P, Tetaz T, Joseph C, Huber S, Schmid G, Hugin D, Pflimlin P, Trube G, et al. (2012). Structure of the acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nat Commun 3, 936. [DOI] [PubMed] [Google Scholar]
- Digregorio DA, Rothman JS, Nielsen TA, and Silver RA (2007). Desensitization Properties of AMPA Receptors at the Cerebellar Mossy Fiber Granule Cell Synapse. Journal of Neuroscience 27, 8344–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder M, Zieglgänsberger W, and Dodt H-U (2004). Shining Light on Neurons - Elucidation of Neuronal Functions by Photostimulation. In Reviews in the Neurosciences, pp. 167. [DOI] [PubMed] [Google Scholar]
- Feng X, Fu Z, Kaledhonkar S, Jia Y, Shah B, Jin A, Liu Z, Sun M, Chen B, Grassucci RA, et al. (2017). A Fast and Effective Microfluidic Spraying-Plunging Method for High-Resolution Single-Particle Cryo-EM. Structure 25, 663–670 e663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibich A, Janko K, and Apell HJ (2007). Kinetics of proton binding to the sarcoplasmic reticulum Ca-ATPase in the E1 state. Biophys J 93, 3092–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, and Stark H (2010). Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 466, 329–333. [DOI] [PubMed] [Google Scholar]
- Frank J (2017). Time-resolved cryo-electron microscopy: Recent progress. J Struct Biol 200, 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Indrisiunaite G, Kaledhonkar S, Shah B, Sun M, Chen B, Grassucci RA, Ehrenberg M, and Frank J (2019). The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy. Nature Communications 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cao E, Julius D, and Cheng Y (2016). TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, and Gouaux E (2009). Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). cisTEM, user-friendly software for single-particle image processing. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B (2001). Ion channels of excitable membranes, 3rd edn (Sunderland, Mass.: Sinauer; ). [Google Scholar]
- Hou X, Burstein SR, and Long SB (2018). Structures reveal opening of the store-operated calcium channel Orai. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe JS, Buiteveld H, Hakvoort JHM, and Donze M (1994). Optical properties of pure water. In Ocean Optics XII, pp. 174–183. [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, and Gouaux E (2007). Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449, 316–323. [DOI] [PubMed] [Google Scholar]
- Kaledhonkar S, Fu Z, Caban K, Li W, Chen B, Sun M, Gonzalez RL, and Frank J (2019). Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaledhonkar S, Fu Z, White H, and Frank J (2018). Time-Resolved Cryo-electron Microscopy Using a Microfluidic Chip. Methods Mol Biol 1764, 59–71. [DOI] [PubMed] [Google Scholar]
- Kaplan JH, Forbush B, and Hoffman JF (1978). Rapid photolytic release of adenosine 5’-triphosphate from a protected analog: utilization by the sodium:potassium pump of human red blood cell ghosts. Biochemistry 17, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Kasas S, Dumas G, Dietler G, Catsicas S, and Adrian M (2003). Vitrification of cryoelectron microscopy specimens revealed by high-speed photographic imaging. Journal of Microscopy 211, 48–53. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, and Schild L (2002). Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev 82, 735–767. [DOI] [PubMed] [Google Scholar]
- Kharlamova A, Prentice BM, Huang TY, and McLuckey SA (2010). Electrospray droplet exposure to gaseous acids for the manipulation of protein charge state distributions. Anal Chem 82, 7422–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkotian E, Oron D, Silberberg Y, and Segal M (2004). Confocal microscopic imaging of fast UV-laser photolysis of caged compounds. Journal of Neuroscience Methods 133, 153–159. [DOI] [PubMed] [Google Scholar]
- Lu Z, Barnard D, Shaikh TR, Meng X, Mannella CA, Yassin A, Agrawal R, Wagenknecht T, and Lu TM (2014). Gas-Assisted Annular Microsprayer for Sample Preparation for Time-Resolved Cryo-Electron Microscopy. J Micromech Microeng 24, 115001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2003). SerialEM: A program for automated tilt series acquisition on Tecnai microscopes using prediction of speciment position. Microscopy and Microanalysis 9, 1182–1183. [Google Scholar]
- Matsubara N, Billington AP, and Hess GP (1992). How fast does an acetylcholine receptor channel open? Laser-pulse photolysis of an inactive precursor of carbamoylcholine in the microsecond time region with BC3H1 cells. Biochemistry 31, 5507–5514. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Ellis-Davies GCR, Nemoto T, Miyashita Y, Iino M, and Kasai H (2001). Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nature Neuroscience 4, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kimanius D, Lindahl E, and Scheres SH (2018). Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orville AM (2018). Entering an era of dynamic structural biology…. BMC Biology 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamini M, Canciani A, and Forneris F (2016). Identifying and Visualizing Macromolecular Flexibility in Structural Biology. Frontiers in Molecular Biosciences 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau V, Zhou Y, Ramu Y, Xu Y, and Lu Z (2017). Crystal structure of an inactivated mutant mammalian voltage-gated K(+) channel. Nat Struct Mol Biol 24, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh JR, and Jahr CE (2011). NMDA Receptor Agonists Fail To Alter Release from Cerebellar Basket Cells. Journal of Neuroscience 31, 16550–16555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Barnard D, Meng X, and Wagenknecht T (2009). Implementation of a flash-photolysis system for time-resolved cryo-electron microscopy. J Struct Biol 165, 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li Z, Jiang Y, Pan X, Wu J, Cristofori-Armstrong B, Smith JJ, Chin YKY, Lei J, Zhou Q, et al. (2018). Structural basis for the modulation of voltage-gated sodium channels by animal toxins. Science 362, eaau2596. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Gerstein M, Oesterhelt D, and Henderson R (1993). Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, and Henderson R (1999). Electron crystallography of bacteriorhodopsin with millisecond time resolution. J Struct Biol 128, 19–25. [DOI] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, and Jensen GJ (2008). An Improved Cryogen for Plunge Freezing. Microscopy and Microanalysis 14, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N (1995). Acetylcholine receptor channel imaged in the open state. Nature 373, 37–43. [DOI] [PubMed] [Google Scholar]
- Unwin N (2003). Structure and action of the nicotinic acetylcholine receptor explored by electron microscopy. FEBS Lett 555, 91–95. [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, and Carragher B (2009). DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. Journal of Structural Biology 166, 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann R, and Lazdunski M (1998). H(+)-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol 8, 418–424. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Rost BR, Camacho-Pérez M, Davis MW, Söhl-Kielczynski B, Rosenmund C, and Jorgensen EM (2013). Ultrafast endocytosis at mouse hippocampal synapses. 504, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Trimbuch T, Camacho-Pérez M, Rost BR, Brokowski B, Söhl-Kielczynski B, Felies A, Davis MW, Rosenmund C, and Jorgensen EM (2014). Clathrin regenerates synaptic vesicles from endosomes. Nature 515, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HD, Thirumurugan K, Walker ML, and Trinick J (2003). A second generation apparatus for time-resolved electron cryo-microscopy using stepper motors and electrospray. J Struct Biol 144, 246–252. [DOI] [PubMed] [Google Scholar]
- White HD, Walker ML, and Trinick J (1998). A computer-controlled spraying-freezing apparatus for millisecond time-resolution electron cryomicroscopy. J Struct Biol 121, 306–313. [DOI] [PubMed] [Google Scholar]
- Wieboldt R, Gee KR, Niu L, Ramesh D, Carpenter BK, and Hess GP (1994a). Photolabile precursors of glutamate: synthesis, photochemical properties, and activation of glutamate receptors on a microsecond time scale. 91, 8752–8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieboldt R, Ramesh D, Carpenter BK, and Hess GP (1994b). Synthesis and Photochemistry of Photolabile Derivatives of.gamma.-Aminobutyric Acid for Chemical Kinetic Investigations of the .gamma.-Aminobutyric Acid Receptor in the Millisecond Time Region. Biochemistry 33, 1526–1533. [DOI] [PubMed] [Google Scholar]
- Xu H, Li T, Rohou A, Arthur CP, Tzakoniati F, Wong E, Estevez A, Kugel C, Franke Y, Chen J, et al. (2019). Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Cell 176, 702–715.e714. [DOI] [PubMed] [Google Scholar]
- Yoder N, and Gouaux E (2020). Conserved His-Gly motif of acid-sensing ion channels resides in a reentrant ‘loop’ implicated in gating and ion selectivity. eLife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder N, Yoshioka C, and Gouaux E (2018). Gating mechanisms of acid-sensing ion channels. Nature 555, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cantara W, Jeon Y, Musier-Forsyth K, Grigorieff N, and Lyumkis D (2019). Analysis of discrete local variability and structural covariance in macromolecular assemblies using Cryo-EM and focused classification. Ultramicroscopy 203, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. Journal of Structural Biology 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Sigworth FJ, and Canessa CM (2006). Gating of acid-sensitive ion channel-1: release of Ca2+ block vs. allosteric mechanism. Journal of General Physiology 127, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Video 1. ‘Flash-then-plunge’ method. Video recorded at 1000 frames per second showing a cryo-EM grid irradiated for 25 ms prior to plunge into ‘cryogen’ (water). Time from initial UV exposure to ‘cryogen’ immersion ~ 70 ms. Video playback is 25 frames per second.
Supplementary Data Figure 1. Instrument control diagram. a, A schematic showing how the instrument is controlled and programmed. Red denotes input required from the user to initiate the experiment; blue denotes information programmed by the user prior to the experiment. b, Picture of the actual flash-plunge device with a side-view shown on the right. Numbers in white correlate to components outlined in (a).
Supplementary Data Figure 2. Fluorescence changes of SNARF-4F probe as a function of pH. a, Normalized fluorescence emission spectra (excitation at 543 nm) of pH-adjusted buffer containing 8 mM Tris, 4 mM MES, 150 mM NaCl and 10 μM SNARF-4F measured between 565 nm and 750 nm. Data represent the mean, n=3. b, Plot of pH vs ratio of fluorescence at 642nm (high pH peak) and 586 nm (low pH peak). Data represent mean (black circles), n=3, and interpolated standard curve is shown as solid green line with 95% confidence internal shown in dashed green.
Supplementary Data Figure 3. Flashed apo ferritin grids. a-h, Representative micrograph and Gctf power spectrum for apo ferritin grids not exposed to UV radiation (a and b, respectively), or for grids exposed to a 25 ms 450 mA (c and d, respectively), 1350 mA (e and f, respectively) or 4500 mA (g and h, respectively) UV LED flash.
Supplementary Data Figure 4. Cryo-EM data processing of 1000 mA flashed cASIC1a. a, Selected 2D classes. b, Schematic of cryo-EM data processing workflow. c-e, local resolution estimation (c) particle distribution (d) and resolution estimation (e) from final non-uniform refinement in cryoSPARC V2.
Supplementary Data Figure 5. Cryo-EM data processing of 3000 mA flashed cASIC1a. a, Selected 2D classes, b, Schematic of cryo-EM data processing workflow, c-e, local resolution estimation (c) particle distribution (d) and resolution estimation (e) from final local refinement using a map containing the whole channel, f-h, local resolution estimation (c) particle distribution (d) and resolution estimation (e) from final local refinement using a map containing the only the ECD.


