Abstract
Disease surveillance systems are a cornerstone of public health tracking and prevention. This review addresses the use, promise, perils, and ethics of social media and internet-based data collection for public health surveillance. Our review highlights untapped opportunities for integrating digital surveillance in public health, and current applications that could be improved through better integration, validation and clarity on rules surrounding ethical considerations. Promising developments include hybrid systems that couple traditional surveillance data with data from search queries, social media posts, and crowdsourcing. In the future, it will be important to identify opportunities for public and private partnerships, train public health experts in data science, reduce biases related to digital data (gathered from internet use, wearable devices, etc.), and address privacy. We are on the precipice of an unprecedented opportunity to track, predict, and prevent global disease burdens in the population using digital data.
Keywords: Social media, digital health, mhealth, big data, infectious diseases, surveillance
INTRODUCTION
Disease surveillance in the community setting is a cornerstone of public health tracking and prevention. The World Health Organization defines public health surveillance as “the continuous, systematic collection, analysis, and interpretation of health-related data needed for the planning, implementation, and evaluation of public health practice”(4). Public health surveillance acts as a sentinel for identifying trends in disease and emerging public health concerns and can help to identify potential points of intervention. Furthermore, surveillance data can provide benchmarks for evaluating intervention measures for curbing disease spread in populations and allow health experts set priorities and policies.
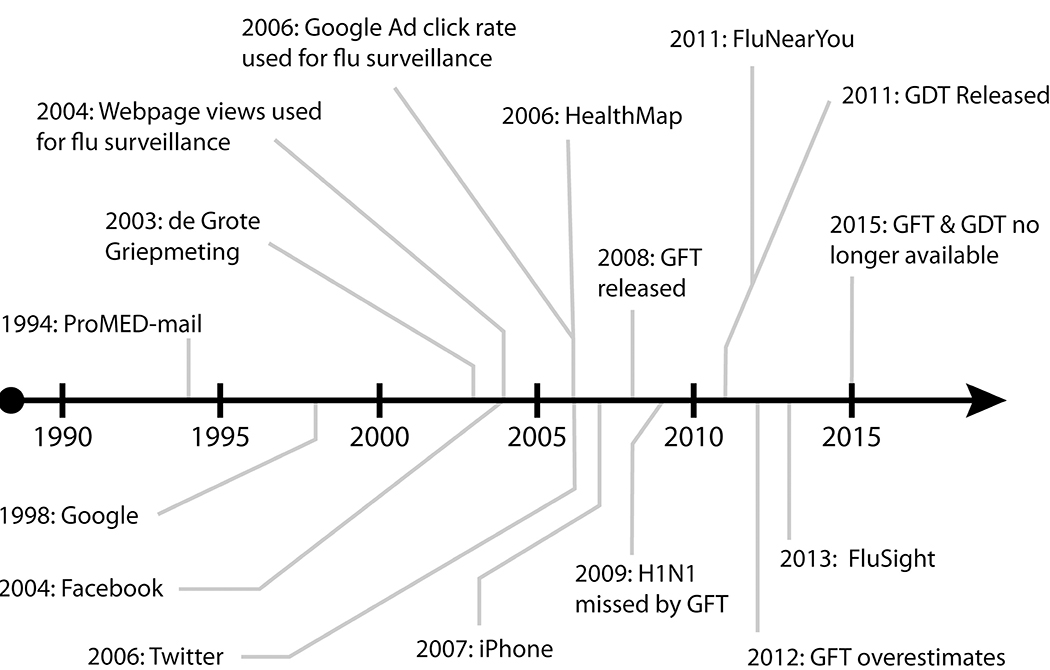
Surveillance has undergone numerous changes over the years (26) and will likely continue to evolve. Recent changes in disease surveillance systems are a result of technological advances for data collection related to access to the internet and improved computational power. Digital disease surveillance can be defined as the use of internet-based data in the explicit development or application of systems aimed at nowcasting or forecasting of disease incidence or prevalence (78). In recent years, internet-based search tools and social media have created exciting new prospects for expanding disease surveillance by capturing real time data and trends for health outcomes. Figure 1 highlights some of the key developments related to digital public health surveillance, including the introduction of Google Flu Trends.
Figure 1:
Major events in digital public health surveillance. Abbreviations: GFT: Google Flu Trends, GDT: Google Dengue Trends. ProMED-mail (69), de Grote Griepmeting (63), Web page views used for flu surveillance (59), Google ad click rate used for surveillance (41), HealthMap (20), H1N1 missed by GFT (31), FluNearYou (102), GFT overestimates (77), FluSight (6), GFT and GDT no longer available (2).
Data collected via the internet and social-media sites has been typically used as a complementary source of surveillance data for existing outpatient, hospital, and laboratory-based systems. This complement can be extremely useful due to traditional surveillance systems reliance on individuals seeking care, and therefore underestimate the total disease burden from a lack of representativeness (105). One example of the complementary approach is the inclusion of electronic health records and historical influenza like illness data in models built on Google search terms, which increased accuracy over Google search trend data alone (119; 120).
However, some digital surveillance arises solely from digital data without necessarily complementing traditional public health surveillance systems. For example, Google searches for “diarrhea”, “food poisoning” and other related terms were shown to coincide independently with a peanut butter associated outbreak of Salmonella enterica (19). While this example of digital data surveillance was never directly integrated with traditional passive public health surveillance, it is possible to link these types of data with traditional food-borne illness surveillance data. Examples where the link with traditional public health surveillance is less clear, but may still provide important public health surveillance information, include the internet collection of data on public attitudes towards vaccination (93), health behaviors such as tracking diet success (43), and smoking cessation (10).
Digital surveillance has been conducted for multiple health events using various sources of search query data, including dengue virus incidence using Google search queries (8), queries from Baidu (a large Chinese search engine similar to Google) (67), and vaccine effectiveness using Google search queries.(98) Online restaurant reservation and review logs have been used to identify foodborne disease and influenza like illness outbreaks (54; 75). While there are a variety of new applications in digital public health surveillance, the most utilized has been within the area of influenza surveillance and tracking.
There are several digital surveillance systems that have been used for influenza, including Google Flu Trends, Influenzanet, and FluNearYou. These systems broadly aim to provide timely reports of influenza incidence in local areas (24; 29; 102). While these influenza examples and other digital health surveillance demonstrate promise for expanding surveillance through digital means, there are also some key trade-offs, such as concerns related to accuracy, privacy and navigating public/private partnerships for connecting technology companies with government surveillance efforts.
This review will address the use, promise, potential perils, and ethics of the use of social media and internet-based data collection for public health surveillance. Within this review, we present a range of examples with a larger focus on influenza, given the high utility of surveillance for this critical public health concern. We also discuss next steps and potential for future application of surveillance through digital sources.
DIGITAL HEALTH SURVEILLANCE
Public health surveillance is the “systematic and continuous collection, analysis, and interpretation of data, closely integrated with the timely and coherent dissemination of the results and assessment to those who have the right to know so that action can be taken” (82). Digital public health surveillance, which we will refer to as digital surveillance hereafter, is the inclusion of digital data, particularly from social media or other internet-based sources, for this same purpose. Others have further distinguished digital surveillance as data collected outside the public health system (90). Moreover, digital health surveillance data is often linked to a non-health data source (e.g. pulling health data from a twitter user who posts on a range of topics which may also include mentions of their health related information), unlike more traditional public health surveillance systems.
Since the early 1990s, digital health surveillance has evolved closely in tandem with the internet itself. Early systems such as ProMED mail, an expert-moderated list of email messages related to the spread of emerging infectious diseases, helped galvanize interest in the internet-based public health through the promise of early widespread outbreak notification (69). Beginning in the early 2000s, digital surveillance efforts have largely encompassed three major types of web-based activity: 1) aggregate trends derived from searches (e.g. Google search trends, Wikipedia page views), 2) social media postings (e.g. Facebook posts, tweets), and 3) participatory surveillance efforts (e.g. FluNearYou, Influenzanet). It is important to note that each of these use activities rely on differing online interactions for input. Some require individuals to be actively seeking health information online, thereby providing relevant public health surveillance data indirectly. Conversely, individuals may opt, passively or purposefully, to share health-relevant information on social media for a variety of reasons. Distinguishing between these different data types of interaction with digital sources of surveillance data, may be important because active versus passive information provides different levels of confidence in the specificity of the data - i.e., there are many reasons to visit the Wikipedia page on influenza, but a tweet describing one’s symptoms may be a more accurate reflection of actual illness. Additionally, the population captured by these approaches likely differs, as certain types of individuals may be more likely to engage in providing participatory data on their health or tweet about symptoms to get input about their health from others, versus a simple query to search the web about symptoms or a disease.
Digital data has primarily been processed to either “forecast” or “nowcast” infectious disease outbreaks. Nowcasting is short-term prediction that attempts to track the present state of incidence in near real-time, whereas forecasting aims to predict the future. The way in which the data are prepared for these purposes often requires substantial pre-processing of raw data. To use aggregate trend data from searches, data need to be filtered by keywords that correspond with disease incidence. The selection of search terms to build these prediction algorithms requires careful consideration, since the choice of words can have a significant impact on the accuracy of the surveillance predictions (31). With social media data, natural language processing and image analysis applied to posts may be used to further extract features of users posts. Due to the volume and types of data in digital sources, surveillance projects often utilize machine learning algorithms (8; 85; 89). Machine learning is a broad term that refers to approaches that adapt to patterns in data without explicitly programming of the prediction task (74). Some examples of algorithms include decision trees, neural networks, and support vector machines. Digital surveillance systems generally use supervised learning, where data patterns are compared to a user-specified outcome. Supervised machine learning has seen success in with problems of prediction, particularly with image analysis, including classification of skin lesions (40), identification of early indicators of breast cancer (111), and detection of diabetic retinopathy (51). However, more data with more advanced prediction algorithms does not necessarily imply improvements to prediction (28). Merely the use of digital surveillance and associated tools, like machine learning, do not necessarily mean these systems will be better than traditional surveillance (72).
Examples of Digital Surveillance.
There are many ways in which digital surveillance can be used to monitor trends and detect disease outbreaks, including through enhancing other data sources, identifying geographic spread, and optimizing existing surveillance systems. An example of digital surveillance being used to enhance other data sources is the incorporation of Google data as a “virtual provider” to enhance accuracy of an existing influenza like illness surveillance system based on a network of outpatient providers (97), using tweets to identify restaurants potentially responsible for food borne infections and subsequently target inspections (53), and using Google to nowcast a plague outbreak in Madagascar, ground-truthed against healthcare-based statistics.(16) Digital data has also been used to identify geographical spread of infectious diseases; in particular Twitter geolocation data has been used along with air traffic data to track the spread of Chikungunya virus,(86) and incorporated into mechanistic models of the flu to forecast peak time and intensity.(123) Further, digital data has optimized traditional surveillance through various means. For influenza surveillance, digital data has been directly integrated with existing surveillance systems on influenza-like illness (1; 6; 97). Possible cases of foodborne illness have been identified through tweets with natural language processing and users were provided information on how to report foodborne illnesses (52; 53). Lastly, digital data has also aided identification of foodborne outbreak point sources through tweets (53), Google search and location logs (89), and Yelp reviews (76). Other examples of digital data use for surveillance abound, including mosquito-borne infectious diseases (8; 24; 25; 86; 113), foodborne infectious diseases (35; 52; 76; 89), and attitudes/behaviors, including those related to vaccination (10; 43).
Search Query Examples.
The most prominent examples of digital surveillance have been related to efforts for tracking influenza, where initial strategies focused on search query data. An early example of this approach was demonstrating the correlation between Google ad click rates and influenza incidence in Canada (41), and Yahoo search trends correlation with influenza incidence (81). Inspired by these approaches, Google Flu Trends was a publicly-available platform communicating predicted influenza incidence from a model based on Google search query volumes. CDC’s weekly influenza-likeillness reports typically contain 1–2 week lag (17), so the goal of Google Flu Trends was to predict influenza approximately 1 week ahead of CDC. Google Flu Trends used a linear regression model of 45 unique search queries highly correlated with the influenza time series, manually pruned for feature relevance (e.g., terms related to basketball, which seasonally correlate with flu, were manually removed) (47). After Google Flu Trends’s release in 2008, it was found to highly correlate with traditional surveillancesystems in several countries and reliably predict influenza-like illness incidence one to two weeks in advance for the 2007–2008 and 2008–2009 seasons (47; 61; 115). However, failure to detect the 2009 A/H1N1 pandemic led to an initial update to the model fit that correctly predicted the pandemic in retrospect (31). Despite the update, both the original and the updated Google Flu Trends vastly overestimated the peak intensity for the 2012–2013 influenza season (77). Ultimately these failures led to the removal of the public facing site (although Google still makes its Google Flu Trends data available to researchers who request it directly (2)). Current efforts to predict infectious diseases using Google typically rely on aggregate search volume data available through Google Trends (time series of relative volumes of specific search terms) and Google Correlate (correlations in Google Trends for different terms, and comparative Google Trends between US states).
Another example in the influenza digital surveillance field was CDC’s “Forecast the Influenza Season Collaborative Challenge” (a.k.a. FluSight), an annual competition in which teams of researchers compete to develop the most accurate weekly regional-level influenza-like illness predictions, with a requirement that teams use some form of digital data - whether it is search query, social media, or other internet-based data (6; 12). Teams were also allowed to incorporate traditional data sources for influenza surveillance, such as ILINet, which provides data on outpatient reports of influenza-like illness rates in a timely manner (17) This effort spurred broad interest in influenza modeling using digital data, with more teams competing every year (12; 13; 71). A major contribution of the FluSight competition has been the development of useful targets for prediction; in the first season these were timing of season onset, peak week, peak intensity, and duration (12). Compared to purely statistical targets such as correlation or mean squared error with the observed trend, which are frequently used (27; 118), these targets provide information about the public health impact of a given model.(12; 27). A detailed summary of the performance of different FluSight predictive approaches was recently published (85). Performance of algorithms was assessed by onset of the influenza season, peak of the influenza season, and forecast incidence at 1-, 2-, 3-, and 4-weeks in advance. All models were compared to the predictions based on the historical average of past seasons. Most models outperformed the historical average approach. As theory regarding ensemble approaches would suggest (36; 106), an ensemble of all submitted FluSight models outperformed each of the individual models (85). Similar competitions to FluSight have since been developed for other diseases, such as dengue (21), chikungunya (33) and Ebola (109).
Social Media Examples.
In addition to search queries, social media data have formed a vital part of digital surveillance efforts, with Twitter being a highly used platform. This is facilitated by Twitter’s relatively open data policy, allowing public access to a 1% random sample of raw tweets (3). For this reason, Twitter has bec ome a “model organism” for digital research data (104). Six of nine teams in the first season of FluSight used Twitter alone or in addition to other digital sources.(12) The most typical use of Twitter data involves content identification, either through keyword search or natural language processing, to identify tweets related to health conditions, such as the flu. Epidemic levels are then modeled as a function of tweet frequencies.(87) However, an additional benefit of Twitter data is the availability of geolocated tweets, which can be used to model disease spread as a function of human geographic movement, potentially offering greater accuracy (86). While Twitter is by far the most frequently used platform in digital surveillance, many others have been used as well. For example, Facebook “like” patterns have been shown to correlatestrongly with a wide range of health conditions and behaviors (48), and Instagram timelines have been used to identify adverse drug reactions (32).
Crowd Sourced Data.
Besides social media and search query data, large-scale, crowd-sourced participatory surveillance systems such as Flu Near You (102) and Influenzanet (63) represent major innovations in the digital sphere. These systems recruit users through online and traditional media to participate in repeated web-based surveys including detailed symptom reports, and report on observed disease distribution through online maps and newsletters (116). For example, Influenzanet was established in 2009 and includes 10 European countries (although it was built on the Dutch and Belgian platform, de Grote Griepmeting, launched in 2003–2004) (50). The standardized web survey in Influenzanet collects detailed flu symptom data chosen to allow multiple influenza-like illness case definitions adopted by different European health agencies. FluNearYou is a similar system in the United States (102), and Dengue-na Web extended this model to monitoring dengue in Salvador de Bahia, Brazil (116). In addition to providing prevalence estimates based on standardized case definitions, a major advantage of participatory surveillance systems is that they provide individual-level demographic and risk factor data, allowing investigators to define the variables of interest and ultimately address research questions of interest (23; 38). Over traditional surveillance systems, clear advantages of these crow sourced approaches, include lower cost and greater flexibility, as they allow integration of additional questions and varying case definitions.(50)
Hybrid Digital/Traditional public health data.
Given the complicated biases present in internet and social media data (covered below), digital data are often best used to supplement rather than replace non-digital public health surveillance data sources. Indeed, the greatest potential use for digital data has been described as the development of hybrid systems (65; 101; 115). Outside of the FluSight competition (12), a few authors have attempted to formally integrate web-based surveillance with more traditional sources. Santillana et al. (95) combined Google, Twitter, and FluNearYou data with influenza-like illness percentages from a private healthcare insurer, showing improvement over Google Flu Trends in terms of root mean square error and forecasting horizon (4-weeks), but do not report on more public health-relevant performance metrics (e.g., start week, peak week, peak percentage, and duration). Incorporation of humidity (a known weather-related determinant of influenza transmission) data with Google Flu Trends data into mechanistic models has shown promising results (60; 99; 100). Other sources of “big data” can complement as well. Cloud-based electronic health record data is increasingly available in near real-time; a study combining electronic health record influenza-like illness estimates with Google search data reduced the root mean squared error of predicting the flu intensity 4 weeks in advance of the CDC relative to HealthMap (119). Another example is the use of air travel volume data combined with Twitter geolocation data to predict the spread of Chikungunya virus(86).
VALIDITY AND BIASES OF SOCIAL MEDIA AND INTERNET DATA
Two key distinctions for digital surveillance with regards to bias are that (a) digital data are not owned by the public, and (b) except for participatory surveillance, the data are capturing public awareness rather than actual occurrence of disease. An early example of these issues is the inaccurate predictions by Google Flu Trends that sparked criticism from researchers and led to the removal of the publicly available Google Flu Trends site in 2015 (2). Google Flu Trends was unable to detect the influenza A/H1N1 pandemic in 2009, and greatly overestimated the peak intensity of the 2012–2013 season (77). Proposed explanations for divergent predictions include changes in search behaviors over time - since the 2009 H1N1 pandemic occurred in the spring / summer, the terms used in influenza-related internet searches may have deviated from the terms more commonly used in the winter (31). For the 2012–2013 season, media coverage is believed to have caused exaggerated public awareness of influenza, leading to a higher volume of searches and inflating predictions (22). The inaccurate predictions by Google Flu Trends in multiple seasons raised awareness of potential biases inherent in digital surveillance (65), including changes to search algorithms (stability), non-independence of data sources (posting and searching can be influenced by others), confounding (of search terms), representativeness (access to internet), and lack of case validation (no clinical ascertainment). We highlight each of these potential sources of bias and present some ideas for mitigating them.
Stability of Digital Data Sources.
Search engine companies make frequent changes to query algorithms without notifying the public. An almost automatic result of this is that predictive models degrade in accuracy over time. Therefore, relying on these sets of data over time can lead to a bias, referred to as “concept drift” (114). Forecasting is especially challenging - across multiple infectious diseases, prediction accuracy degrades rapidly with forecast horizon, typically only extending reasonably to 2–4 weeks (110). Models with excellent nowcasting skill can have zero forecasting value (83), since digital data may only indicate public awareness and coincide with outbreak peaks (86). However, even with nowcasting, degradation is cumulative. Priedhorsky et al. (83) found that an influenza model trained on Wikipedia traffic showed “staleness” after four months, meaning models were no longer effective and needed to be retrained. This problem was largely unaided by adding more data over a longer period. Such “staleness” does not just cause noisy predictions but can create erroneous spikes.(83) Further, a website can be deactivated at any time or change owners and therefore, the persistence of data is not guaranteed. The concerns related to disappearing or changing data on websites, and algorithm changes, makes the process of replication of digital surveillance highly tenuous.
Non-Independent Digital Data.
Internet data are fundamentally non-independent and contain self-perpetuating feedback loops of multiple kinds. When predictors are trained on such data, their sensitivity/specificity and predictive values can be expected to change over time, leading to erroneous conclusions. Recommendation systems generate suggestions based in part on the popularity of a particular search term, creating dependence between observations. In social media, “trending” topics are self-perpetuating and can be manipulated (104). Media attention on an epidemic causes spikes in topic frequency not related to disease rates. For instance, CDC conducted a press conference in April of 2013 regarding the H7N9 “bird flu” pandemic, which led to a spike in flu-related tweets not corresponding to incidence.(18)
Digital Search Confounding.
Confounding of search terms is also present in correlation-based feature selection, particularly when relying on simple terms without sophisticated filtering. For example, terms related to basketball correlate strongly with influenza due to seasonal overlap (47; 83), and Google trends for the word “cholera” revealed a cholera “epidemic” in the US in 2007 related to Oprah Winfrey selecting Love in the Time of Cholera for her book of the month club (101). Prediction accuracy has also been observed to drop off precipitously during holidays (121). These examples illustrate the necessity of semantic filtering, which was performed by hand in the initial Google Flu Trends algorithm (47), but is possible to automate the process (83). Related to confounding, there may be social desirability biases related to the use of certain search terms. If individuals are concerned that their searches are being tracked or can be identified through their computer searches, they may be less likely to use terms that relate to certain diseases and conditions that may be stigmatized. This bias may reduce the accuracy of digital data surveillance for certain conditions.
Digital Representativeness.
Representativeness is a key characteristic of an ideal surveillance system. Well-known sampling biases exist in internet and social media use. Heavily discussed is the so-called “digital divide” – the fact that socioeconomic inequality exists in internet access and usage (37), and that internet access is substantially less dense in developing countries (49). While roughly 22% of the US adult population uses Twitter, there is overrepresentation of individuals of higher SES, ages 30–44, and those living in urban areas (80), reflecting a notable underrepresentation of groups at highest risk for infectious disease morbidity and mortality. Participatory surveillance systems, though they may mitigate the confounding and non-independence discussed above, suffer similar representativeness issues, with individuals needing to sign up and manually provide their information. For example, Influenzanet shows underrepresentation of males and the youngest and oldest age groups, as well as a higher influenza vaccination rate among older participants compared to the same age group in the target population (50). As expected, the population participating in Influenzanet may be healthier and more advantaged, showing lower prevalence of asthma and diabetes and higher income and education, which are predictors of lower influenza risk (63). To our knowledge, no study has specifically aimed to explore the effect of underrepresentation of those most susceptible to flu, the very young and old, on population-level flu estimates based on internet data. One promise of big data is the potential for highly granular predictions in terms of geography; hence, some Twitter-based studies rely on geolocation data. However, geolocation of tweets is turned off by default, with roughly 1–2% of users turning it on, likely introducing additional sampling biases (66). In particular, geolocation users have measurable differences in language use and are more likely to be men over 40 (79). To our knowledge, no investigation has examined the relationship between use of geolocation tags and disease, but it might reasonably correlate negatively with influenza risk (particularly by age) and with HIV risk behaviors, many of which are stigmatized or illegal (103).
Digital Validation.
Much of traditional public health surveillance relies on validation through clinical case ascertainment, which is not possible with digital surveillance (perhaps except in cases where participatory surveillance is required). To properly evaluate the performance of digital surveillance, appropriate evaluation metrics are needed. A suitable comparator data source for the surveillance system must be chosen, since prediction will also replicate any biases present in the ground truth data. Using influenza as an example, diagnosed case reports (which are used in most studies of the flu) underestimate prevalence, especially among persons with lower access to regular health care. CDC influenza-like illness percentages are the most common ground truth data used for the digital flu surveillance; these are based on weekly reports from approximately 2,200 outpatient providers throughout the US reflecting the total number of patients meeting the CDC’s syndromic case definition (7). Because some providers may exhibit delays in case reporting, influenza-like illness reports are often revised weeks or months after their initial release, which has been shown to negatively impact forecasting ability (85). Also, influenza-like illness reports reflect incidence in the general population, which may not accurately reflect the impact of the flu on those who are most physiologically vulnerable, such as the very young and old. In order to better capture such impacts, digital surveillance efforts should also consider incorporating other routine indicators, such as age-stratified influenza-like illness, and influenza-related hospitalization and mortality (14; 60).
Possible solutions to biases.
There are several potential solutions to the biases described above. Addressing both concept drift and non-independence have been explored previously. Concept drift and resulting model staleness can likely be addressed by including a plan to dynamically retrain models to account for always-changing online behavior and disease dynamics. For example, Santillana et al. (96) were able to dramatically improve Google Flu Trends’s accuracy by automatically updating the model when weekly CDC estimates were released, highlighting that language used in searches changes over time and so should independent contributions of search terms used to estimate flu trends. Future work could examine precisely how frequently models need to be recalibrated.
Non-independence of data is a more challenging problem, since self-perpetuating public awareness potentially impacts all forms of health-related online behavior, but can be addressed at least in part by improving text classification accuracy. For example, Broniatowski et al. (18) used a multi-stage filtering approach to distinguish media-related “chatter” from tweets that are true markers of influenza incidence that doubled the accuracy in predicting the weekly direction of change in incidence (ie, up or down).
Solutions to sampling bias and non-representativeness have been less well explored. A classical approach is to use weighting to adjust samples to be population representative, either through post-stratification weights, inverse probability weights, or raking (122). These are standard practices in national surveys with known sampling probabilities, but may also be reasonable to apply in a convenience sample such as a selection of Google search queries or tweets. Wang et al. (112) used data from a series of daily voter intention polls conducted on the Xbox gaming platform, a sample not unlike social media or search query data, to predict successfully the 2012 US presidential election using post-stratification weights based on simple demographics like age, sex, race, and political party affiliation. Although these variables are not often available from Google or Twitter data, they may be straightforward to learn from the available data - reasonably accurate models exist to predict demographics on Twitter from names alone (117). Though population rates of internet usage have been increasing for all groups, it is worthwhile to note that sampling bias is not necessarily mitigated by high usage rates overall-ironically, as innovations spread through social networks, holdouts can represent a more and more unusual group (88).
ETHICAL CONSIDERATIONS FOR DIGITAL DATA SURVEILLANCE
The primary ethical challenge of public health is appropriately balancing risks and harms to individuals, while protecting and promoting population health (42; 57). This challenge remains for digital surveillance as the primary ethical issue (56; 90), but the non-health purpose of the data raises ethical concerns in a different light. To frame our discussion of ethics of digital surveillance, we focus on the following five principles; beneficence, non-maleficence, respect for autonomy, equity, and efficiency. These principles have been previously used to frame ethical considerations in public health (70) and to examine the ethics of public health surveillance more generally (62). More broadly, consideration of these five principles can help in the decision of public health actions and the ethics of that decision (e.g. harm minimization or precautionary principle)
Beneficence.
The concept of beneficence is the principle that public health surveillance should improve the health of the target population (70). While digital surveillance has the potential to improve infectious disease surveillance systems, part of this principle is clearly defining the target population of the surveillance system. As such, careful consideration should be directed at identifying who is and who is not captured by digital surveillance systems. Identification of blind spots is necessary to ensure that public health surveillance works towards improving the health of the target population it purports. Digital surveillance is promising since members of the target population that do not come into contact with more traditional medical-based surveillance systems can be captured. Additionally, these systems can capture events that may be missed by traditional surveillance. One example is foodborne illness, where only a fraction of cases are captured by traditional surveillance (75). Under this view, the additional coverage of digital surveillance, while still a biased sample, can improve the overall coverage of the target population relative to traditional epidemiologic surveillance approaches alone.
Beneficence also charges us with considering how health would be improved under a digital surveillance system. Merely monitoring a health outcome does not necessarily improve population health. Improving population health depends on effective communication and interventions. Previous work has made the argument that digital surveillance allows for earlier outbreak response (25; 86; 91; 92; 107), but few have actually described interventions that resulted from the earlier detection via digital surveillance and its effectiveness. In part due to forecast windows offered by existing models (2–4 weeks in advance) offer insufficient time to plan meaningful responses, such as adjustments to hospital surge capacity and vaccine manufacturing (110).
Non-maleficence.
The concept of non-maleficence is defined by actions that reduce the potential harms and burdens of collecting data and promote the benefits of doing so - to the greatest degree possible. Several threats to non-maleficence include; use of non-health data and stigmatization of risk factors, violation of privacy, and mistrust in public health information and intentions. Digital surveillance primarily relies on systems not primarily built for explicit collection of health-related data but a substantial amount of non-health data is also often collected during queries. If these non-health data are subsequently labelled as “risk factors,” it may attach stigma to certain behaviors or groups that are a proxy for the true underlying risk factors. There are many examples of stigma manifestation that occurred before the internet-including the HIV epidemic - with groups being stigmatized based on sexual orientation, country of origin, and race/ethnicity (46; 94). Collection of non-health data for digital surveillance and the speed of nowcasting sets up similar misattribution of risk and stigma. What is communicated from digital surveillance and how it is presented to the public should be considered thoughtfully and cautiously.
A core tenet of public health is building and maintaining public trust. A major barrier to public trust is false detections and missed outbreaks by surveillance systems. False detections can erode public trust in these surveillance systems, lead to misuse of limited resources, and result in poor risk communication to the public (15; 39). False detections and missed outbreaks can occur for any surveillance system but the use of non-health data, black-box machine learning algorithms, and rapidly declining performance, open up more possibilities for false detections to occur. Furthermore, mistrust may result from social media sites themselves through data breaches or sites using public health outbreak detection to market products to users. Lastly, users may knowingly publicly share information but may not expect its continual collection and analysis. While aggregate data may alleviate some concerns over violations of privacy, the same cannot be said of individual-specific internet data. Therefore, digital surveillance data may warrant the same privacy protections as data from more traditional surveillance systems. Public and private data use must be implemented cautiously and the possibility for misuse should be examined.
Autonomy.
The concept of respect for autonomy involves recognizing the right to self-determination of individuals and minimizing subsequent violations. A fundamental concern is informed consent within surveillance systems (62), which is also often cited as a major ethical concern of digital surveillance (55; 101). Informed consent is forgone in surveillance systems to improve the accuracy, with various justifications making the lack of informed consent more acceptable (62). In particular, the anonymization or aggregation of data help to justify the lack of informed consent for medical institution data (73). For digital surveillance, monitoring of aggregate search trends or page views is similarly defensible. Issues of informed consent for digital surveillance on individual-level data lead to unique complications. Three distinguishing features of digital epidemiology with regards to informed consent are: 1) data is not sourced from formal medical institutions, 2) data come from proprietary online platforms, and 3) data are not limited to health but often include personal attributes (73).
In the clinical setting, there is often a legal mandate to report certain diseases, medical professionals have their own long-standing code of ethics, and data are collected explicitly for health purposes. Health data from medical institutions are covered under the Health Insurance Portability and Accountability Act. While a patient at a medical institution may reasonably believe that their medical data will be used by health departments to monitor public health, the same is not true for social media. Social media users may not expect their data to be used towards monitoring public health trends, since there is no legal mandate for public health reporting of social media data, there are no standard ethics for social media creators, and data are primarily non-health related. Recently, social media sites and other internet platforms have been under scrutiny regarding privacy concerns (30). While the user agreements (the ‘Terms and Conditions’) cover consent from a legal aspect, the “informed” part has been a concern due to the complexity of the language used and the volume of conditions. While efforts like the European Union General Data Protection Regulation (GDPR) seeks to give internet users greater autonomy over their personal data collected by social media companies and reduce the complexity of user-agreements (5), we remain in a tumultuous time regarding privacy and consent to online data collection. Since digital data consists of largely non-health data, it becomes less clear that informed consent can be broken for individual-level health data.
As public health surveillance continues to rely on these methods, we need to consider and openly discuss the lack of informed consent, and whether there is sufficient justification to warrant digital surveillance without it. Whether the lack of informed consent for digital surveillance is justified depends on the specific scenario and the corresponding risk to public health. With GDPR giving users more control over their data, informed consent could be obtained by directly requesting individuals to share their social media with public health authorities (90). This approach has the additional benefit of avoiding the issue of social media companies retracting access to data. However, this approach has its own ethical concerns (44).
Equity.
Individuals in the target population should have equal opportunity to receive a given public health intervention and a just distribution of benefits. As stated for beneficence, digital surveillance offers the opportunity to identify problems of individuals not in contact with the medical system and thereby may increase opportunities for enhancing equity in who is included in surveillance data. However, as described above in biases, there still remains segments of the population that do not have access to the internet or digital technology, leaving open the potential for loss of equity in digital surveillance. Con sidering the limitations in representativeness (and potentially, heterogeneous predictive power (58)) of most internet data sources, it is unlikely that surveillance data and associated interventions will be justly distributed without taking explicit account of these biases.
Efficiency.
The concept of efficiency is related to the cost-benefit of a surveillance system. For digital surveillance to produce benefits, it needs to overcome the biases previously discussed and provide improved tracking of health issues. For these systems to be cost-efficient, they require automated programs to manage and analyze the data. These automated systems may require substantial start-up cost and require regular maintenance to prevent algorithms from becoming stale. Since the data collected by these private companies is proprietary and there is no legal mandate for data provisions to public health, underlying algorithms by the companies require continuous update, and access could be discontinued at any time without warning (55). If access is revoked, the resources spent on creating the digital surveillance system become poorly spent. Furthermore, proprietary algorithms to detect outbreaks can also be revoked at any time and may not be reproducible (31; 64; 90; 108). An alternative that avoids platforms revoking access is to build legal mandates to allow for access, or allow users control over their data and to ask them for permission to share their data with public health professionals (90). Legal mandates for data provision may violate the autonomy of these companies. Some digital surveillance systems use a variation of the later approach, like FluNearYou and Influenzanet, where users directly provide their data through web-based surveys.
In summary, the use of internet-based data collection offers new opportunities but raises several ethical new concerns compared to traditional public health surveillance. These issues should continue to be addressed and require communication with the public. As a starting point Mettelstadt et al. (73) have provided conditions to consider and several case-studies related to the ethics of digital surveillance.
DISCUSSION
Digital public health surveillance affords the opportunity to revolutionize existing public health surveillance infrastructure. While some public health officials report monitoring digital data sources such as Google Flu Trends for contextual information (22), our review indicates that public health, in any official capacity, has yet to embrace and build upon existing opportunities that have arisen because of new digital data sources. While a variety of conditions examined in public health surveillance have been explored to some degree using digital data sources, the biggest focus has been on influenza. Moreover, road maps describing how public/private partnerships could advance and work together towards creating sustainable, efficient, and ethical digital surveillance systems have yet to be fully developed. In addition, clear standards by which digital public health surveillance can be compared to traditional surveillance systems have yet to be established, making it difficult to assess whether these sources truly amplify the benefits of traditional public health for disease tracking and prevention. While we cover applied uses related to search queries, social media posts, and crowdsourcing, another potentially promising area that has yet to be explored is the use of data from existing digital health collection platforms based on smartphones and wearable devices, such as Apple HealthKit. These sources could offer highly precise information related to both infectious and non-infectious conditions and risk factors at a broad scale, but with many of the biases and representation mentioned above.
Ultimately, digital surveillance systems will need to be developed in ways that avoid the numerous potential pitfalls associated with biases and ethical considerations described in this review. As with all surveillance systems, experts will need to determine whether digital surveillance is necessary and ethically justifiable. In future applications, digital surveillance is likely to have the largest public health impact when integrated with traditional surveillance systems, like traditional laboratory data, case reports, and electronic health records (101). While digital surveillance offers the ability to build novel surveillance systems where no existing surveillance system exists, appropriate and accurately measured training data sets are needed.
In the future, it will be important to identify the most beneficial ways to use digital data sources through hybrid or completely new independent systems. The prospect of a new system, solely driven by digital technology, seems unlikely at present, but the continued advancement of machine learning, and related technologies for managing and deriving meaning from large sets of data, may bring this idea closer to reality in the future. A challenge will be in the training of public health experts in computer science, big data, and machine learning, to harness novel sources of digital data and support innovation in digital surveillance while reducing possible harms. In conclusion, we are on the precipice of an unprecedented opportunity to track, predict, and prevent global disease burdens in the population using digital data, and it is of great importance that public health institutions receive the training and resources to provide the right input and help to build new systems to move traditional public health surveillance into the future.
Table 1:
Glossary of terms
| Public health surveillance | “Systematic and continuous collection, analysis, and interpretation of data, closely integrated with the timely and coherent dissemination of results and assessment to those who have the right to know so that action can be taken”* |
| Digital public health surveillance | Public health surveillance with the inclusion of digital data, particularly from social media or other internet-based sources. |
| Nowcast | Short-term forecasting meant to provide near real-time information. |
| Cloud-based | Adjective describing data that is stored, processed, or analyzed on-demand via remote servers hosted made available through the Internet. |
| Machine learning | Algorithmic approaches that adapt to patterns in data without explicitly programming the prediction task.† |
| Supervised machine learning | Machine learning algorithms where the outcome variable for prediction is explicitly observed and focus is on accurate predictions of that outcome.† |
| Search Query-based digital surveillance | Digital public health surveillance systems that use aggregate search query data to monitor disease trends. Examples include Google Flu Trends and Google Dengue Trends. |
| Social Media-based digital surveillance | Digital public health surveillance systems that use social media posts to monitor disease trends. Social media-based surveillance requires pre-processing of the data, such as keyword searches or natural language processing. |
| Crowd-based digital surveillance | Digital public health surveillance systems where participants voluntarily provide health-relevant information through potentially repeated web-based surveys to monitor disease trends. Examples include Flu Near You and Influenzanet. |
| Hybrid digital surveillance | Integration of digital surveillance data with traditional public health surveillance data, or multiple sources of digital along with traditional public health surveillance data to monitor disease trends. |
| Google Flu Trends | Publicly available site that used aggregate Google search trend data to forecast influenza-like illness incidence. After failure to predict the 2009 pandemic and overestimating the peak intensity of 2012–2013 influenza season, the publicly available site was removed.‡ |
| Flu Near You | Crowd-sourced participatory surveillance system to track influenza-like illness in the United States via weekly surveys completed by participants.# |
| FluSight | The United States Centers for Disease Control and Prevention’s “Forecast the Influenza Season Collaborative Challenge”. An annual competition in which researchers compete to develop the most accurate weekly influenza-like illness predictions with some form of digital data.** |
Porta M. 2008. A Dictionary of Epidemiology. Oxford University Press. 320 pp
Mooney SJ, Pejaver V. 2018. Big Data in Public Health: Terminology, Machine Learning, and Privacy. Annu. Rev. Public Health 39:95–112
Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. 2009. Detecting influenza epidemics using search engine query data. Nature 457:1012–4
The Next Chapter for Flu Trends. https://ai.googleblog.com/2015/08/the-next-chapter-for-flu-trends.html
Smolinski MS, Crawley AW, Baltrusaitis K, Chunara R, Olsen JM, et al. 2015. Flu Near You: Crowdsourced Symptom Reporting Spanning 2 Influenza Seasons. Am. J. Public Health 105:2124–30
Biggerstaff M, Alper D, Dredze M, Fox S, Fung IC-H, et al. 2016. Results from the Centers for Disease Control and Prevention’s predict the 2013–2014 Influenza Season Challenge. BMC Infect. Dis. 16:357
Acknowledgements:
We are grateful to the Carolina Population Center and the NIH/NICHD center grant (P2C HD50924). AR and PZ received funding from the Carolina Population Center Biosocial Training Grant T32 HD091058 (MPI Aiello).
Footnotes
Disclosures: AEA has consulted for Kinsa Inc. on their research studies of Kinsa Smart Thermometer and received an unrestricted gift from Gojo Industries, Inc for research on hand hygiene.
LITERATURE CITED
- 1.Epidemic Prediction Initiative, https://predict.cdc.gov/post/5bal504e5619f003acb7el8f
- 2.The Next Chapter for Flu Trends, https://ai.googleblog.com/2015/08/the-next-chapter-for-flu-trends.html
- 3.Overview: Sample realtime Tweets, https://developer.twitter.com/en/docs/tweets/sample-realtime/overview.html
- 4.2017. WHO ǀ Public health surveillance https://www.who.int/topics/public_health_surveillance/en/
- 5.2018. EU data protection rules
- 6.2019. CDC Competition Encourages Use of Social Media to Predict Flu ǀ CDC https://www.cdc.gov/flu/news/predict-flu-challenge.htm
- 7.2019. Overview of Influenza Surveillance in the United States ǀ CDC https://www.cdc.gov/flu/weekly/overview.htm
- 8.Althouse BM, Ng YY, Cummings DAT. 2011. Prediction of dengue incidence using search query surveillance. PLoSNegl. Trop. Dis 5:e1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Althouse BM, Scarpino SV, Meyers LA, Ayers JW, Bargsten M, et al. 2015. Enhancing disease surveillance with novel data streams: challenges and opportunities. EPJ Data Sci 4. [DOI] [PMC free article] [PubMed]
- 10.Aphinyanaphongs Y, Lulejian A, Brown DP, Bonneau R, Krebs P. 2016. Text classification for automatic detection of e-cigarette use and use for smoking cessation from Twitter: a feasibility pilot. Pac. Symp. Biocomput 21:480–91 [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal S, Chowell G, Simonsen L, Vespignani A, Viboud C. 2016. Big Datafor Infectious Disease Surveillance and Modeling. J. Infect. Dis 214:S375–S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biggerstaff M, Alper D, Dredze M, Fox S, Fung IC-H, et al. 2016. Results from the centers for disease control and prevention’s predict the 2013–2014 Influenza Season Challenge. BMC Infect. Dis 16:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biggerstaff M, Johansson M, Alper D, Brooks LC, Chakraborty P, et al. 2018. Results from the second year of a collaborative effort to forecast influenza seasons in the United States. Epidemics 24:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biggerstaff M, Kniss K, Jemigan DB, Brammer L, Bresee J, et al. 2018. Systematic Assessment of Multiple Routine and Near Real-Time Indicators to Classify the Severity of Influenza Seasons and Pandemics in the United States, 2003–2004 Through 2015–2016. Am. J. Epidemiol 187:1040–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Böl G-F. 2016. Risk communication in times of crisis: Pitfalls and challenges in ensuring preparedness instead of hysterics. EMBO Rep 17:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bragazzi NL, Mahroum N. 2019. Google Trends Predicts Present and Future Plague Cases During the Plague Outbreak in Madagascar: Infodemiological Study. JMIR Public Health Surveill 5:e13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brammer L, Blanton L, Epperson S, Mustaquim D, Bishop A, et al. 2011. Surveillance for Influenza during the 2009 Influenza A (H INI) Pandemic-United States, April 2009-March 2010. Clinical Infectious Diseases 52:S27–S35 [DOI] [PubMed] [Google Scholar]
- 18.Broniatowski DA, Paul MJ, Dredze M. 2013. National and local influenza surveillance through Twitter: an analysis of the 2012–2013 influenza epidemic. PLoS One 8:e83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brownstein JS, Freifeld CC, Madoff LC. 2009. Digital disease detection—harnessing the Web for public health surveillance. N. Engl. J. Med 360:2153–5, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brownstein JS, Freifeld CC, Reis BY, MandlKD. 2008. Surveillance Sans Frontières: Internet-based emerging infectious disease intelligence and the HealthMap project. PLoS Med 5:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buczak AL, Baugher B, Moniz LJ, Bagley T, Babin SM, Guven E. 2018. Ensemble method for dengue prediction. PLoS One 13:e0189988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler D. 2013. When Google got flu wrong. Nature 494:155–6 [DOI] [PubMed] [Google Scholar]
- 23.Carlson SJ, Durrheim DN, Dalton CB. 2010. Flutracking provides a measure of field influenza vaccine effectiveness, Australia, 2007–2009. Vaccine 28:6809–10 [DOI] [PubMed] [Google Scholar]
- 24.Cameiro HA, Mylonakis E. 2009. Google trends: a web-based tool for real-time surveillance of disease outbreaks. Clin. Infect. Dis 49:1557–64 [DOI] [PubMed] [Google Scholar]
- 25.Chan EH, Sahai V, Conrad C, Brownstein JS. 2011. Using web search query data to monitor dengue epidemics: a new model for neglected tropical disease surveillance. PLoS Negl. Trop. Dis 5:e1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi BCK. 2012. The past, present, and future of public health surveillance. Scientifica 2012:875253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chretien J-P, George D, Shaman J, Chitale RA, McKenzie FE. 2014. Influenza forecasting in human populations: a scoping review. PLoS One 9:e94130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. 2019. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol 110:12–22 [DOI] [PubMed] [Google Scholar]
- 29.Chunara R, Aman S, Smolinski M, Brownstein JS. 2013. Flu Near You: An Online Self-reported Influenza Surveillance System in the USA. Online J. Public Health Inform 5
- 30.Confessore N. 2018. Cambridge Analytica and Facebook: The Scandal and the Fallout So Far. The New York Times 2018/4/4
- 31.Cook S, Conrad C, Fowlkes AL, Mohebbi MH. 2011. Assessing Google flu trends performance in the United States during the 2009 influenza virus A (H1N1) pandemic. PLoS One 6:e23610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Correia RB, Li L, Rocha LM. 2016. Monitoring potential drug interactions and reactions via network analysis of Instagram user timelines. Pac. Symp. Biocomput 21:492–503 [PMC free article] [PubMed] [Google Scholar]
- 33.Del Valle SY, McMahon BH, Asher J, Hatchett R, Lega JC, et al. 2018. Summary results of the 2014–2015 DARPA Chikungunya challenge. BMC Infect. Dis 18:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denecke K. 2017. An ethical assessment model for digital disease detection technologies. Life Sciences, Society and Policy 13:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desai R, Hall AJ, Lopman BA, Shimshoni Y, Rennick M, et al. 2012. Norovirus disease surveillance using Google Internet query share data. Clin. Infect. Dis 55:e75–8 [DOI] [PubMed] [Google Scholar]
- 36.Dietterich TG. 2000. Ensemble Methods in Machine Learning. In Multiple Classifier Systems, pp. 1–15: Springer; Berlin Heidelberg [Google Scholar]
- 37.DiMaggio P, Hargittai E, Neuman WR, Robinson JP. 2001. Social Implications of the Internet. Annu. Rev. Sociol 27:307–36 [Google Scholar]
- 38.Eames KTD, Brooks-Pollock E, Paolotti D, Perosa M, Gioannini C, Edmunds WJ. 2012. Rapid assessment of influenza vaccine effectiveness: analysis of an internet-based cohort. Epidemiol. Infect 140:1309–15 [DOI] [PubMed] [Google Scholar]
- 39.Eckmanns T, Fuller H, Roberts SL. 2019. Digital epidemiology and global health security; an interdisciplinary conversation. Life Sci Soc Policy 15:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, et al. 2017. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eysenbach G. 2006. Infodemiology: tracking flu-related searches on the web for syndromic surveillance. AMIA Annu. Symp. Proc.:244–8 [PMC free article] [PubMed] [Google Scholar]
- 42.Fairchild AL, Haghdoost AA, Bayer R, Selgelid MJ, Dawson A, et al. 2017. Ethics of public health surveillance: new guidelines. Lancet Public Health 2:e348–e9 [DOI] [PubMed] [Google Scholar]
- 43.Fried D, Surdeanu M, Kobourov S, Hingle M, Bell D. 2014. Analyzing the language of food on social media. arXiv
- 44.Geneviève LD, Martani A, Wangmo T, Paolotti D, Koppeschaar C, et al. 2019. Participatory Disease Surveillance Systems: Ethical Framework. J. Med. Internet Res 21:e12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.German RR, Horan JM, Lee LM, Milstein B, Pertowski CA. 2001. Updated guidelines for evaluating public health surveillance systems; recommendations from the Guidelines Working Group [PubMed]
- 46.Gilman SL. 1987. AIDS and Syphilis: The Iconography of Disease. October 43:87–107 [Google Scholar]
- 47.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. 2009. Detecting influenza epidemics using search engine query data. Nature 457:1012–4 [DOI] [PubMed] [Google Scholar]
- 48.Gittelman S, Lange V, Gotway Crawford CA, Okoro CA, Lieb E, et al. 2015. A new source of data for public health surveillance: Facebook likes. J. Med. Internet Res 17:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graham M, Hale S, Stephens M. 2012. Featured Graphic: Digital Divide: The Geography of Internet Access. Environ. Plan. A 44:1009–10 [Google Scholar]
- 50.Guerrisi C, Turbelin C, Blanchon T, Hanslik T, Bonmarin I, et al. 2016. Participatory Syndromic Surveillance of Influenza in Europe. J. Infect. Dis 214:S386–S92 [DOI] [PubMed] [Google Scholar]
- 51.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, et al. 2016. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 316:2402–10 [DOI] [PubMed] [Google Scholar]
- 52.Harris JK, Hawkins JB, Nguyen L, Nsoesie EO, Tuli G, et al. 2017. Using Twitter to Identify and Respond to Food Poisoning: The Food Safety STL Project. J. Public Health Manag. Pract 23:577–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris JK, Mansour R, Choucair B, Olson J, Nissen C, et al. 2014. Health department use of social media to identify foodbome illness - Chicago, Illinois, 2013–2014. MMWR Morb. Mortal. Wkly. Rep 63:681–5 [PMC free article] [PubMed] [Google Scholar]
- 54.Harrison C, Jorder M, Stem H, Stavinsky F, Reddy V, et al. 2014. Using online reviews by restaurant patrons to identify unreported cases of foodbome illness - New York City, 2012–2013. MMWR Morb. Mortal. Wkly. Rep 63:441–5 [PMC free article] [PubMed] [Google Scholar]
- 55.Heitmueller A, Henderson S, Warburton W, Elmagarmid A, Pentland AS, Darzi A. 2014. Developing public policy to advance the use of big data in health care. Health Aff 33:1523–30 [DOI] [PubMed] [Google Scholar]
- 56.Höhle M. 2017. A statistician’s perspective on digital epidemiology. Life Sci Soc Policy 13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holland S. 2015. Public Health Ethics John Wiley & Sons. 288 pp. [Google Scholar]
- 58.Huang D-C, Wang J-F, Huang J-X, Sui DZ, Zhang H-Y, et al. 2016. Towards Identifying and Reducing the Bias of Disease Information Extracted from Search Engine Data. PLoS Comput. Biol 12:e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson HA, Wagner MM, Hogan WR, Chapman W, Olszewski RT, et al. 2004. Analysis of Web access logs for surveillance of influenza. Stud. Health Technol. Inform 107:1202–6 [PubMed] [Google Scholar]
- 60.Kandula S, Pei S, Shaman J. 2019. Improved forecasts of influenza-associated hospitalization rates with Google Search Trends. J. R. Soc. Interface 16:20190080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelly H, Grant K. 2009. Interim analysis of pandemic influenza (H1N1) 2009 in Australia: surveillance trends, age of infection and effectiveness of seasonal vaccination. Euro Surveill 14. [DOI] [PubMed]
- 62.Klingler C, Silva DS, Schuermann C, Reis AA, Saxena A, Strech D. 2017. Ethical issues in public health surveillance: a systematic qualitative review. BMC Public Health 17:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koppeschaar CE, Colizza V, Guerrisi C, Turbelin C, Duggan J, et al. 2017. Influenzanet: Citizens Among 10 Countries Collaborating to Monitor Influenza in Europe. JMIR Public Health Surveill 3:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kostkova P. 2018. Disease surveillance data sharing for public health: the next ethical frontiers. Life Sci Soc Policy 14:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazer D, Kennedy R, King G, Vespignani A. 2014. The Parable of Google Flu: Traps in Big Data Analysis. Science 343:1203–5 [DOI] [PubMed] [Google Scholar]
- 66.Leetaru K, Wang S, Cao G, Padmanabhan A, Shook E. 2013. Mapping the global Twitter heartbeat: The geography of Twitter. First Monday 18
- 67.Li Z, Liu T, Zhu G, Lin H, Zhang Y, et al. 2017. Dengue Baidu Search Index data can improve the prediction of local dengue epidemic: A case study in Guangzhou, China. PLoS Negl. Trop. Dis 11:e0005354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu D. 2019. Creating an AI can be five times worse forthe planet than a car. New Scientist
- 69.Madoff LC, Woodall JP. 2005. The internet and the global monitoring of emerging diseases: lessons from the first 10 years of ProMED-mail. Arch. Med. Res 36:724–30 [DOI] [PubMed] [Google Scholar]
- 70.Marckmann G, Schmidt H, Sofaer N, Strech D. 2015. Putting public health ethics into practice: a systematic framework. Front Public Health 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McGowan CJ, Biggerstaff M, Johansson M, Apfeldorf KM, Ben-Nun M, et al. 2019. Collaborative efforts to forecast seasonal influenza in the United States, 2015–2016. Sci. Rep 9:683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mignan A, Broccardo M. 2019. One neuron is more informative than a deep neural network for aftershock pattern forecasting. arXiv [physics.geo-ph]
- 73.Mittelstadt B, Benzler J, Engelmann L, Prainsack B, Vayena E. 2018. Is there a duty to participate in digital epidemiology? Life Sci Soc Policy 14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mooney SJ, Pejaver V. 2018. Big Data in Public Health: Terminology, Machine Learning, and Privacy. Annu. Rev. Public Health 39:95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nsoesie EO, Buckeridge DL, Brownstein JS. 2014. Guess who’s not coming to dinner? Evaluating online restaurant reservations for disease surveillance. J. Med. Internet Res 16:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nsoesie EO, Kluberg SA, Brownstein JS. 2014. Online reports of foodbome illness capture foods implicated in official foodbome outbreak reports. Prev. Med 67:264–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olson DR, Konty KJ, Paladini M, Viboud C, Simonsen L. 2013. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: a comparative epidemiological study at three geographic scales. PLoS Comput. Biol 9:e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park H-A, Jung H, On J, Park SK, Kang H. 2018. Digital Epidemiology: Use of Digital Data Collected for Non-epidemiological Purposes in Epidemiological Studies. Healthc. Inform. Res 24:253–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavalanathan U, Eisenstein J. 2015. Confounds and Consequences in Geotagged Twitter Data. arXiv [cs.CL]
- 80.Perrin A, Anderson M. Share of U.S. adults using social media, including Facebook, is mostly unchanged since 2018
- 81.Polgreen PM, Chen Y, Pennock DM, Nelson FD. 2008. Using internet searches for influenza surveillance. Clin. Infect. Dis 47:1443–8 [DOI] [PubMed] [Google Scholar]
- 82.Porta M. 2008. A Dictionary of Epidemiology Oxford University Press. 320 pp. [Google Scholar]
- 83.Priedhorsky R, Osthus D, Daughton AR, Moran KR, Generous N, et al. 2017. Measuring Global Disease with Wikipedia. Proceedings of the 2017 ACM Conference on Computer Supported Cooperative Work and Social Computing - CSCW’17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quinn P. 2018. Crisis Communication in Public Health Emergencies: The Limits of Ľegal Control’ and the Risks for Harmful Outcomes in a Digital Age. Life Sci Soc Policy 14:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reich NG, Brooks LC, Fox SJ, Kandula S, McGowan CJ, et al. 2019. A collaborative multiyear, multimodel assessment of seasonal influenza forecasting in the United States. Proc. Natl. Acad. Sci. U. S. A 116:3146–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rocklöv J, Tozan Y, Ramadona A, Sewe MO, Sudre B, et al. 2019. Using Big Data to Monitor the Introduction and Spread of Chikungunya, Europe, 2017. Emerging Infectious Disease journal 25:1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodríguez-Martínez M, Garzón-Alfonso CC. 2018. Twitter Health Surveillance (THS) System. Proc IEEE Int Conf Big Data 2018:1647–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogers EM. 2003. Diffusion of Innovations, 5th Edition. Simon and Schuster. 576 pp. [Google Scholar]
- 89.Sadilek A, Caty S, DiPrete L, Mansour R, Schenk T Jr, et al. 2018. Machine-learned epidemiology: real-time detection of foodbome illness at scale. NPJ Digit Med 1:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salathé M. 2018. Digital epidemiology: what is it, and where is it going? Life sciences, society and policy 14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salathé M, Bengtsson L, Bodnar TJ, Brewer DD, Brownstein JS, et al. 2012. Digital epidemiology. PLoS Comput. Biol 8:e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salathé M, Freifeld CC, Mekaru SR, Tomasulo AF, Brownstein JS. 2013. Influenza A (H7N9) and the importance of digital epidemiology. N. Engl. J. Med 369:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salathé M, Khandelwal S. 2011. Assessing vaccination sentiments with online social media: implications for infectious disease dynamics and control. PLoS Comput. Biol 7:e1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santana MA, Dancy BL. 2000. The stigma of being named "AIDS carriers" on Haitian-American women. Health Care Women Int 21:161–71 [DOI] [PubMed] [Google Scholar]
- 95.Santillana M, Nguyen AT, Dredze M, Paul MJ, Nsoesie EO, Brownstein JS. 2015. Combining Search, Social Media, and Traditional Data Sources to Improve Influenza Surveillance. PLoS Comput. Biol 11:e1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santillana M, Zhang DW, Althouse BM, Ayers JW. 2014. What can digital disease detection learn from (an external revision to) Google Flu Trends? Am. J. Prev. Med 47:341–7 [DOI] [PubMed] [Google Scholar]
- 97.Scarpino SV, Dimitrov NB, Meyers LA. 2012. Optimizing provider recmitment for influenza surveillance networks. PLoS Comput. Biol 8:e1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shah MP, Lopman BA, Tate JE, Harris J, Esparza-Aguilar M, et al. 2018. Use of Internet Search Data to Monitor Rotavirus Vaccine Impact in the United States, United Kingdom, and Mexico. J Pediatric Infect Dis Soc 7:56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shaman J, Karspeck A. 2012. Forecasting seasonal outbreaks of influenza. Proc. Natl. Acad. Sci. U. S. A 109:20425–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shaman J, Karspeck A, Yang W, Tamerius J, Lipsitch M. 2013. Real-time influenza forecasts during the 2012–2013 season. Nat. Commun 4:2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simonsen L, Gog JR, Olson D, Viboud C. 2016. Infectious Disease Surveillance in the Big DataEra: Towards Faster and Locally Relevant Systems. J. Infect. Dis 214:S380–S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smolinski MS, Crawley AW, Baltmsaitis K, Chunara R, Olsen JM, et al. 2015. Flu Near You: Crowdsourced Symptom Reporting Spanning 2 Influenza Seasons. Am. J. Public Health 105:2124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stoové MA, Pedrana AE. 2014. Making the most of a brave new world: opportunities and considerations for using Twitter as a public health monitoring tool. Prev. Med 63:109–11 [DOI] [PubMed] [Google Scholar]
- 104.Tufekci Z. 2014. Big Questions for Social Media Big Data: Representativeness, Validity and Other Methodological Pitfalls. arXiv [cs.SI]
- 105.Uiters E, Devillé W, Foets M, Spreeuwenberg P, Groenewegen PP. 2009. Differences between immigrant and non-immigrant groups in the use of primary medical care; a systematic review. BMC Health Serv. Res 9:76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Laan MJ, Polley EC, Hubbard AE. 2007. Super learner. Stat. Appl. Genet. Mol. Biol 6:Article 25 [DOI] [PubMed]
- 107.Vayena E, Salathé M, Madoff LC, Brownstein JS. 2015. Ethical challenges of big data in public health. PLoS Comput. Biol 11:e1003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Velasco E. 2018. Disease detection, epidemiology and outbreak response: the digital future of public health practice. Life Sci Soc Policy 14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Viboud C, Sun K, Gaffey R, Ajelli M, Fumanelli L, et al. 2018. The RAPIDD ebola forecasting challenge: Synthesis and lessons learnt. Epidemics 22:13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Viboud C, Vespignani A. 2019. The future of influenza forecasts. Proc. Natl. Acad. Sci. U. S. A 116:2802–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang J, Yang X, Cai H, Tan W, Jin C, Li L. 2016. Discrimination of Breast Cancer with Microcalcifications on Mammography by Deep Learning. Sci. Rep 6:27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang W, Rothschild D, Goel S, Gelman A. 2015. Forecasting elections with non-representative polls. Int. J. Forecast 31:980–91 [Google Scholar]
- 113.Wesolowski A, Eagle N, Tatem AJ, Smith DL, Noor AM, et al. 2012. Quantifying the impact of human mobility on malaria. Science 338:267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Widmer G, Kubat M. 1996. Learning in the presence of concept drift and hidden contexts. Mach. Learn 23:69–101 [Google Scholar]
- 115.Wilson N, Mason K, Tobias M, Peacey M, Huang QS, Baker M. 2009. Interpreting “Google Flu Trends” data for pandemic H1N1 influenza: The New Zealand experience. Eurosurveillance 14:19386. [PubMed] [Google Scholar]
- 116.Woójcik OP, Brownstein JS, Chunara R, Johansson MA. 2014. Public health for the people: participatory infectious disease surveillance in the digital age. Emerg. Themes Epidemiol 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wood-Doughty Z, Andrews N, Marvin R, Dredze M. 2018. Predicting Twitter User Demographics from Names Alone. Proceedings of the Second Workshop on Computational Modeling of People’s Opinions, Personality, and Emotions in Social Media:105–11
- 118.Yan SJ, Chughtai AA, Macintyre CR. 2017. Utility and potential of rapid epidemic intelligence from internet-based sources. Int. J. Infect. Dis 63:77–87 [DOI] [PubMed] [Google Scholar]
- 119.Yang S, Santillana M, Brownstein JS, Gray J, Richardson S, Kou SC. 2017. Using electronic health records and Internet search information for accurate influenza forecasting. BMC Infect. Dis 17:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang S, Santillana M, Kou SC. 2015. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc. Natl. Acad. Sci. U. S. A 112:14473–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Young SD, Torrone EA, Urata J, Aral SO. 2018. Using Search Engine Data as a Tool to Predict Syphilis. Epidemiology 29:574–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang L-C. 2000. Post-Stratification and Calibration—A Synthesis. Am. Stat 54:178-? [Google Scholar]
- 123.Zhang Q, Perra N, Perrotta D, Tizzoni M, Paolotti D, Vespignani A. 2017. Forecasting Seasonal Influenza Fusing Digital Indicators and a Mechanistic Disease Model. Proceedings of the 26th International Conference on World Wide Web:311–9