Significance
The latent reservoir for HIV-1 is a major barrier to curing the infection. Current cure strategies are focused on eliminating the reservoir through a two-step “shock and kill” approach involving the use of small-molecule latency-reversing agents (LRAs) followed by immune interventions to kill infected cells. Numerous candidate LRAs have been identified in studies using in vitro models of latency, but none have been shown to reduce the reservoir in vivo. We show here that many candidate LRAs work through an unexpected pathway involving the transcription factor HSF1, a result with implications for developing effective combinations of LRAs for use in HIV cure strategies.
Keywords: HIV, latency, reservoir, LRA, HSF1
Abstract
HIV-1 latency is a major barrier to cure. Identification of small molecules that destabilize latency and allow immune clearance of infected cells could lead to treatment-free remission. In vitro models of HIV-1 latency involving cell lines or primary cells have been developed for characterization of HIV-1 latency and high-throughput screening for latency-reversing agents (LRAs). We have shown that the majority of LRAs identified to date are relatively ineffective in cells from infected individuals despite activity in model systems. We show here that, for diverse LRAs, latency reversal observed in model systems involves a heat shock factor 1 (HSF1)-mediated stress pathway. Small-molecule inhibition of HSF1 attenuated HIV-1 latency reversal by histone deactylase inhibitors, protein kinase C agonists, and proteasome inhibitors without interfering with the known mechanism of action of these LRAs. However, latency reversal by second mitochondria-derived activator of caspase (SMAC) mimetics was not affected by inhibition of HSF1. In cells from infected individuals, inhibition of HSF1 attenuated latency reversal by phorbol ester+ionomycin but not by anti-CD3+anti-CD28. HSF1 promotes elongation of HIV-1 RNA by recruiting P-TEFb to the HIV-1 long terminal repeat (LTR), and we show that inhibition of HSF1 attenuates the formation of elongated HIV-1 transcripts. We demonstrate that in vitro models of latency have higher levels of the P-TEFb subunit cyclin T1 than primary cells, which may explain why many LRAs are functional in model systems but relatively ineffective in primary cells. Together, these studies provide insights into why particular LRA combinations are effective in reversing latency in cells from infected individuals.
Despite advances in antiretroviral therapy (ART), HIV-1 remains incurable due to a stable reservoir of latent but replication-competent proviruses (1–3). An important cure strategy, termed “shock-and-kill,” envisions stimuli that induce viral transcription, resulting in clearance of the latent reservoir (4). Diverse latency-reversing agents (LRAs) targeting signaling pathways (5–8), transcriptional regulation (9), and epigenetic modifications (10–14) have been identified using in vitro models of HIV-1 latency. However, most LRAs are less effective in reversing latency in cells from patients on ART (15). We show here that latency reversal by many candidate LRAs with different proposed mechanisms of action is dependent on a heat shock factor 1 (HSF1)-mediated stress response which increases elongation of HIV-1 transcripts. Our results explain why model systems overreport LRA activity. The finding that many LRAs identified in model systems function through generalized cellular stress will aid in the search for effective shock agents in cure strategies.
The low frequency of CD4+ T lymphocytes harboring intact latent proviruses (∼60 to 100/106 cells) (16) complicates ex vivo analysis of latency reversal, and thus in vitro latency models are used to identify LRAs. These model systems implicate epigenetic restriction as central to HIV-1 latency, through both DNA methylation (11, 12) and histone modifications (13, 17–19). In addition, latency is promoted in resting CD4+ T cells by the absence or sequestration of critical host factors involved in the initiation of transcription such as NF-κB (5–8) and NFAT (20) or in transcriptional elongation such as pTEFb (9, 21, 22). Classes of LRAs that may reverse these blocks include cytokines (23, 24), histone deacytelase (HDAC) inhibitors (17, 25, 26), DNA methylase inhibitors (27), protein kinase C (PKC) activators (28–31), and proteasome inhibitors (32). Although a wide variety of pathways have been implicated in latency reversal in model systems, individual LRAs targeting specific pathways can reverse latency in these systems. We therefore hypothesized that many candidate LRAs identified in model systems function through a common mechanism that reverses latency in these systems but is less effective in primary cells.
In addition to LRAs targeting specific pathways, certain heat shock proteins have been shown to regulate HIV-1 latency in model systems (33, 34), and previous work in a cell line model of latency has shown that HSF1 can recruit elongation factors to the HIV-1 promoter (35). The potential involvement of HSF1 in HIV-1 transcription is compelling, given that HSF1 responds to a wide variety of stimuli and functions as a hub in a complex signaling network involved in cellular response to thermal and chemical stress. Here, we analyzed the involvement of HSF1 signaling in LRA-induced HIV-1 transcription in primary cells both in vitro and ex vivo.
Results
Stress Responses Trigger Latency Reversal in a Primary Cell Model.
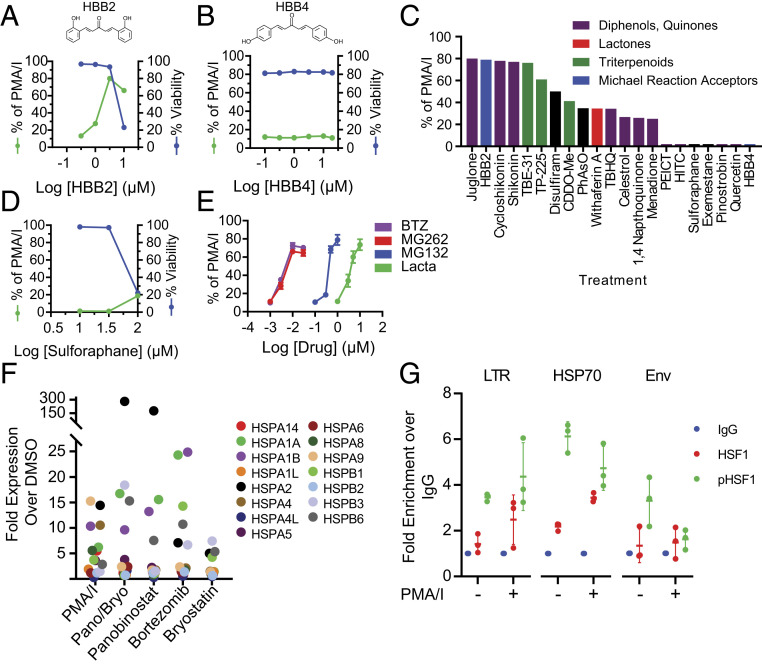
An initial screen of stress-inducing compounds in a primary cell model of HIV-1 latency (36) revealed impressive latency reversal by some compounds that can modify thiols. Thiol adduct formation potently induces chemical stress in mammalian cells (37) and initiates HSF1 nuclear localization (38). To probe the importance of thiol modification, we compared latency reversal by the thiol adduct forming compound HBB2 with that of a closely related isomer HBB4, which has a substantially lower propensity to form thiol adducts (39). HBB2 reversed latency in the primary cell model as evidenced by expression of green fluorescent protein (GFP) from latent proviruses with GFP in the env open reading frame (Fig. 1A). The isomer HBB4, which forms adducts less readily, did not reverse latency (Fig. 1B). Interestingly, latency reversal by HBB2 occurred at concentrations slightly below those that were toxic. HBB2 belongs to a diverse family of compounds that activate an adaptive redox stress response involving the nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf2) (40). Several other Nrf2 activators reversed latency in model cells, typically at concentrations slightly below those that were toxic (SI Appendix, Fig. S1). However, a large-scale screen of Nrf2 activators revealed that only a subset of these compounds reversed HIV-1 latency (Fig. 1C). Specifically, the canonical Nrf2 activator sulforaphane (41) did not reverse latency even at high concentrations (Fig. 1D). Therefore, we analyzed the role in latency reversal of a distinct but related stress pathway involving the transcription factor HSF1. We used proteasome inhibitors, which activate the HSF1 pathway independently of thiol modification by promoting the accumulation of denatured proteins as in the case of thermal stress (42). We observed pronounced latency reversal by all proteasome inhibitors tested at concentrations reflecting their known potency in inhibiting the proteasome (Fig. 1E), consistent with previous studies (32, 43). These results suggest that accumulation of denatured proteins that are normally degraded by the proteasome triggers latency reversal in this model.
Fig. 1.
A primary cell model of HIV-1 latency (61) is highly responsive to cellular stress. Primary model cells were treated with varying concentrations of HBB2 (A) or HBB4 (B) for 18 h. GFP expression was measured by flow cytometry and expressed as a percentage of the response to the positive control response to the PMA and I, a combination that mimics T cell activation. Viability was measured by 7AAD staining. (C) Compounds known to induce chemical stress (37, 40) were assayed for latency reversal in primary model cells. Model cells were treated with optimal concentrations of the indicated compounds for 18 h before analysis by flow cytometry. Optimal concentrations were predetermined through dose–response analysis with each compound. Colors indicate distinct chemical classes. Results are presented as a percentage of GFP expression observed with PMA/I stimulation. (D) Dose–response curve for reactivation of latent HIV-1 by sulforaphane in primary model cells. Latency reversal and viability were assessed as described in A. (E) Dose responses for reactivation of latent HIV-1 by four proteasome inhibitors in primary model cells. (F) Effect of LRAs on expression of HSP70 gene family members in primary resting CD4+ T cells. Cells were incubated with LRAs for 24 h at concentrations used to reverse latency, and HSP70 family mRNA levels were quantified by RT-qPCR. Results for each individual gene is shown as fold change over the DMSO control. (G) HSF1 and phospho-HSF1 antibodies enrich LTR and HSP70 promoter sequences relative to nontargeted IgG controls in J-Lat cells. Data are shown as fold change over IgG control for each individual experimental replicate.
Common LRAs Induce HSF1-Mediated Signaling.
Given the possible relationship between latency reversal and the HSF1-mediated stress pathway, we tested a panel of known LRAs for ability to induce HSF1-mediated signaling. We treated resting CD4+ T cells from healthy donors with LRAs and LRA combinations at concentrations known to reverse HIV-1 latency and measured the expression of the HSP70 family of protein chaperones, which are canonical targets of HSF1 signaling (44). Although these agents are thought to act through different mechanisms, we observed potent up-regulation of multiple genes within the HSP70 family, consistent with the hypothesis that these LRAs initiate an HSF1-mediated stress response (Fig. 1F). The combination of the PKC agonist phorbol 12-myristate 13-acetate (PMA) and the calcium ionophore ionomycin (I), which most robustly promotes latency reversal in primary cells and model systems (45), increased transcription of the largest number of chaperone genes tested. The combination of the HDAC inhibitor panobinostat and the PKC agonist bryostatin, which also reverses latency ex vivo in cells from treated patients (15, 46), also activated transcription of HSP70 family genes as did panobinostat alone. As single agents, bortezomib and bryostatin also induced expression of HSP70 family genes. We observed significant overlap between the HSP genes up-regulated by PMA/I and those up-regulated by LRAs, suggesting a shared stress response. These results led us to hypothesize that these LRAs may reverse latency in model systems primarily through off-target stress pathways rather than through their expected pharmacological targets.
A possible mechanism by which off-target stress pathways induce HIV-1 expression is through HSF1 association with the HIV-1 5′ long terminal repeat (LTR). Evidence of HSF1 associating with the HIV-1 5′ LTR has been shown previously and has been proposed as a targetable mechanism to achieve latency reversal (35, 47). To confirm the interaction of HSF1 with the HIV-1 LTR, we performed a series of chromatin immunoprecipitation (ChIP) assays with antibodies targeting bulk HSF1 and activated phospho-HSF1. We used the best characterized in vitro model of HIV-1 latency, J-Lat cells, which were generated through transduction of Jurkat cells with a recombinant HIV-1 vector carrying GFP (48). We observed that immunoprecipitations of HSF1 and phospho-HSF1 substantially enriched both LTR sequences and canonical HSP70 promoter regions relative to nontargeted IgG controls (Fig. 1G). This suggests that HSF1, particularly the active phosphorylated form, associates with the HIV LTR. We believe enrichment of LTR sequences in the vehicle control is due to basal levels of HSF1 stimulation in J-Lat cells related to their continuous proliferation and high metabolic activity.
Inhibition of HSF1-Mediated Signaling Attenuates HIV-1 Latency Reversal.
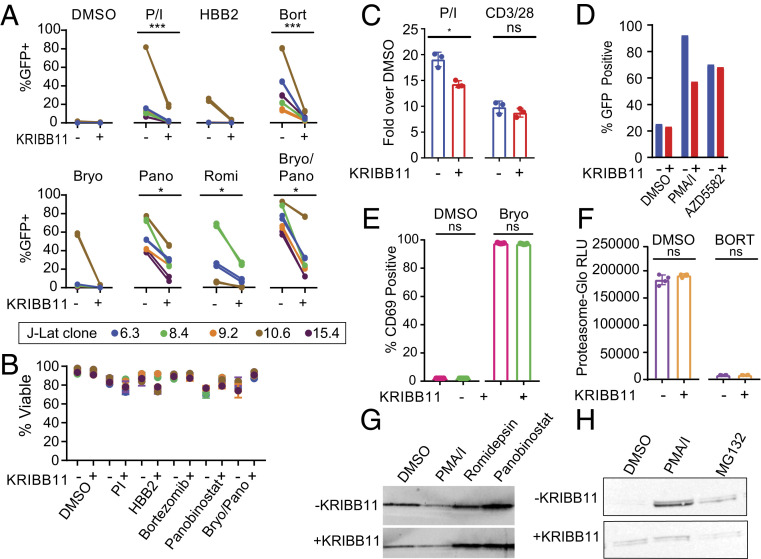
To directly test the involvement of the HSF1 pathway in HIV-1 latency reversal, we tested several clones of J-Lat cells that have different proviral integration sites and differences in response to some LRAs (31, 45). We stimulated J-Lat clones with LRAs in the presence or absence of the specific HSF1 inhibitor KRIBB11, which blocks HSF1-mediated recruitment of P-TEFb to heat shock promoters (49). For LRAs thought to act through different mechanisms, we observed uniform inhibition of latency reversal in the presence of KRIBB11 (Fig. 2A). The combination PMA/I strongly reverses latency in the J-Lat 10.6 clone, but the effect was almost completely blocked by KRIBB11. Latency reversal by HBB2 and bortezomib, which activate stress pathways, was also blocked by KRIBB11. More surprising was the finding that latency reversal by the PKC agonist bryostatin, by the HDAC inhibitors panobinostat and romidepsin, and by combinations of these agents were all blocked by KRIBB11 (Fig. 2A). Inhibition by KRIBB11 occurred without significant effects on cell viability (Fig. 2B). These results suggest that LRAs active in model systems may reverse HIV-1 latency primarily through an HSF1-mediated pathway rather than through pathways that the LRAs are known to target, such as the PKC-signaling cascade or histone deacetylation.
Fig. 2.
Reversal of HIV-1 latency by certain LRAs is blocked by KRIBB11. (A) Effect of KRIBB11 on latency reversal by candidate LRAs in five J-Lat clones. Cells were treated with optimal concentrations of the indicated LRAs in the presence or absence of 5 µM KRIBB11. Results are expressed as the percentage of GFP+ cells after a 24-h stimulation. Statistical analysis was performed with a ratio-paired t test. (B) Viability of J-Lat cells after 24-h LRA treatment in the presence or absence of 5 µM KRIBB11. Viability was measured by flow cytometry using a vital dye. (C) Effect of KRIBB11 on the NF-κB–signaling pathway. An NF-κB–driven luciferase reporter was transfected into Jurkat cells and used to detect NF-κB signaling after activation with PMA/I or anti-CD3/CD28 in the presence or absence of 5 µM KRIBB11. Three independent replicates were performed and compared with a ratio-paired t test. (D) Effect of KRIBB11 on latency reversal mediated by the SMAC mimetic AZD-5582 (1 µM) in a polyclonal population of J-Lat cells. (E) Effect of KRIBB11 on induction of surface expression of CD69 by the PKC agonist bryostatin. CD69 expression on resting CD4+ T cells from infected individuals was measured with flow cytometry after treatment with 10 nM bryostatin for 24 h in the presence or absence of 5 µM KRIBB11. (F) Effect of KRIBB11 on inhibition of 20S proteasome activity by bortezomib. Resting CD4+ T cells were treated with bortezomib in the presence or absence of 5 µM KRIBB11. Proteasome activity was measured using a labeled proteasome substrate. Four independent replicates were performed and analyzed with a ratio-paired t test. (G) Western blot analysis of histone H3 acetylated at the N terminus after treatment of resting CD4+ T cells from healthy donors with the vehicle control (DMSO), PMA/I, or the HDAC inhibitors romidepsin or panobinostat. (H) Western blot analysis of the effect of KRIBB11 on induction of HSP70 by the vehicle control (DMSO), PMA/I, and MG132. KRIBB11 was used at a concentration of 5 µM.
The KRIBB11-mediated inhibition of latency reversal by several different classes of LRAs could reflect the involvement of HSF1 signaling in latency reversal or the off-target effects of KRIBB11 on diverse regulatory pathways. Because many candidate LRAs, particularly PKC agonists, are thought to function through NF-κB–dependent pathways (5), we confirmed that the inhibitory effect of KRIBB11 was not due to off-target inhibition of NF-κB–mediated signaling. Jurkat cells were transfected with Igκ2-IFN-LUC, a plasmid in which luciferase expression is controlled by two upstream NF-κB sites (50). Stimulation with PMA/I or anti-CD3/CD28 caused dramatic increases in luciferase activity that were only marginally inhibited by KRIBB11 (Fig. 2C). Recently described SMAC mimetics have also been proposed to reverse HIV-1 latency through a noncanonical NF-κB mechanism (51). While we did not observe latency reversal mediated by the SMAC mimetic AZD5582 in clonal populations of J-Lats, we observed substantial latency reversal in a polyclonal population of J-Lat cells (Fig. 2D). We further found that latency reversal by AZD5582 was not inhibited by the inclusion of KRIBB11, suggesting that AZD5582 does not function primarily through a HSF1-mediated mechanism (Fig. 2D). Similarly, up-regulation of the activation marker CD69 on resting CD4+ T cells by the PKC agonist bryostatin was not inhibited by KRIBB11 (Fig. 2E). Additionally, the proteasome inhibitor bortezomib blocked the 20S proteasome-mediated cleavage of a luminescent reporter even in the presence of KRIBB11 (Fig. 2F). We also tested whether KRIBB11 blocked the canonical activity of histone deacetylase inhibitors, which have been widely studied as LRAs (13, 19). Through Western blot analysis of acetylated histone H3, we showed that the potent histone deacetylase inhibitors panobinostat and romidepsin induced a dramatic increase in acetylated histone H3 levels in resting CD4+ T cells that was not blocked by KRIBB11 (Fig. 2G). These results demonstrate that KRIBB11 inhibits latency reversal by multiple distinct mechanistic classes of LRAs without interfering with the known targeted activity of each LRA. We confirmed that KRIBB11 did prevent up-regulation of HSP70 in response to PMA/I treatment, in line with the known function of KRIBB11 (Fig. 2H). For the majority of LRAs tested, KRIBB11 did not interfere with the proposed LRA mechanism of action despite substantially attenuating HIV-1 latency reversal. Together, these findings strongly suggest that HSF1 signaling significantly contributes to latency reversal by the best-known LRAs including romidepsin and bryostatin.
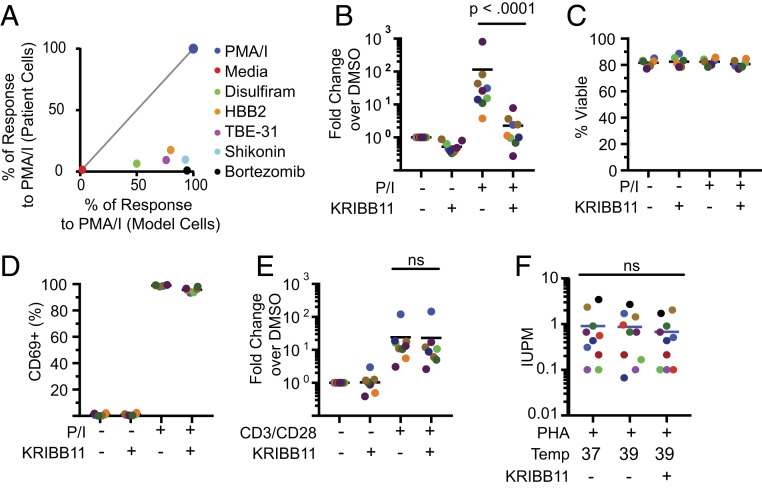
We next determined whether compounds that activate stress pathways and reverse HIV-1 latency in primary cell models activated HIV-1 gene expression ex vivo in cells from patients on ART. Diverse compounds meeting these criteria were tested. For each, a dose–response curve was obtained to identify optimal concentrations for latency reversal in model cells (SI Appendix, Table S1). Resting CD4+ T cells from patients on suppressive ART were stimulated with optimal concentrations of these compounds for 24 h, and up-regulation of HIV-1 gene expression was quantified by RT-qPCR detection of correctly terminated HIV-1 transcripts as previously described (52). Despite efficacy in the primary cell model, none of these compounds induced significant up-regulation of HIV-1 gene expression in patient cells relative to T cell activation (Fig. 3A). These results are consistent with previous studies indicating that many LRAs active in model systems do not effectively reverse latency ex vivo in cells from infected individuals (15) and demonstrate that induction of a stress response is insufficient to reverse latency ex vivo.
Fig. 3.
Analysis of latency reversal ex vivo in cells from patients on ART. (A) Compounds that induce cellular stress responses reverse latency in a primary cell model but not in resting CD4+ T cells from treated patients. Identical treatment conditions were used for the primary cell model and for resting CD4+ T cells from infected individuals. Graph shows a correlation plot for responses of the two cell types to each stimulus. Latency reversal in the primary cell model was measured using GFP expression. Latency reversal in patient cells was measured by RT-qPCR as described above. In both cases, responses are expressed as the percentage of the positive control response to PMA/I. (B) Effect of KRIBB11 on latency reversal by PMA/I in resting CD4+ T cells from patients on ART (n = 9). Cells were stimulated with PMA/I for 24 h in the presence or absence of 5 µM KRIBB11 and then assayed for HIV-1 transcripts by RT-qPCR. Colored symbols indicate different patients. Statistical significance was determined using ratio-paired t tests. (C) Effect of KRIBB11 on cell viability. Viability of treated primary cells from B was measured with flow cytometry using a vital dye. (D) Effect of KRIBB11 on induction of CD69 expression by PMA/I. Levels of CD69 on cells from B were measured by flow cytometry. (E) Effect of KRIBB11 on latency reversal by anti-CD3/CD28 in resting CD4+ T cells from patients on ART (n = 9). Cells were stimulated with CD3/CD28 stimulation for 72 h in the presence or absence of 5 µM KRIBB11 and then assayed for production of HIV-1 transcripts by RT-qPCR. Statistical significance was determined using ratio-paired t tests. (F) Effect of elevated temperature and KRIBB11 on latency reversal by PHA and allogenic stimulation. Primary resting CD4+ T cells were assayed with a 21-d quantitative viral outgrowth assay at different temperatures in the presence or absence of 5 µM KRIBB11. Data are presented as IUPM.
One small-molecule stimulus that does induce a high level of latency reversal in cells from infected individuals is the combination PMA/I (15). We therefore tested whether HSF1-mediated signaling plays a role in PMA/I-induced HIV-1 latency reversal in resting CD4+ T cells from treated patients. We activated resting CD4+ T cells with PMA/I for 24 h in the presence or absence of KRIBB11 and measured the production of correctly terminated HIV-1 transcripts. In the presence of KRIBB11, we observed a >90% reduction in the number of HIV-1 transcripts (Fig. 3B). Decreases in PMA/I-driven HIV-1 gene expression by KRIBB11 were not due to generalized cell death as no effect on cell viability was observed (Fig. 3C). Furthermore, staining for the activation marker CD69 revealed that KRIBB11 does not repress PMA/I-mediated T cell activation (Fig. 3D). To test whether KRIBB11 inhibits latency reversal by a stimulus that more closely approximates physiologic T cell activation, we carried out similar experiments using anti-CD3/CD28 as the activating stimulus. In this case, KRIBB11 caused only a minimal reduction in the quantity of HIV-1 transcripts produced, suggesting that latency reversal following physiologic stimulation of resting CD4+ cells occurs through mechanisms largely independent of the HSF1 pathway (Fig. 3E). To further test the involvement of the HSF1 pathway in the context of physiologic stimulus, we performed a series of viral outgrowth assays at elevated temperatures (Fig. 3F). When stimulated with the T cell-activating lectin phytohemagglutinin (PHA) and cocultured with irradiated allogenic peripheral blood mononuclear cells (PBMCs), increases in temperature did not increase latency reversal as measured in the quantitative viral outgrowth assay (53). Furthermore, inclusion of KRIBB11 did not inhibit latency reversal by PHA and allogeneic PBMC. Taken together, these results demonstrate that HSF1 signaling plays a role in pharmacologic HIV-1 latency reversal and is minimally involved in physiological stimuli that activate T cells through surface receptors. These results also show that KRIBB11 does not generally inhibit HIV-1 gene expression. Rather, it blocks HIV-1 transcription induced by stimuli that act through HSF1. Taken together, these data demonstrate that inhibition of the HSF1-mediated stress response substantially decreases HIV-1 transcription induced by several classes of LRAs in model cells and by PMA/I in cells from infected individuals.
Inhibition of HSF1-Mediated Signaling Attenuates Transcriptional Elongation.
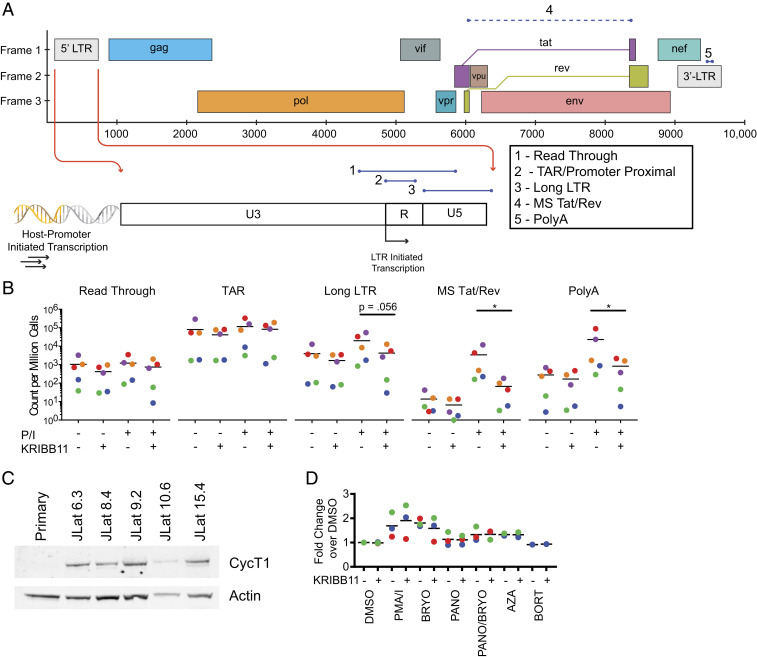
HSF1 increases transcription of heat shock genes through recruitment of P-TEFb, which promotes transcriptional elongation (54). Given that HSF1 functions through a P-TEFb–dependent mechanism in the induction of heat shock genes, we hypothesized that a similar mechanism may be involved in the reactivation of latent HIV-1 by stimuli that activate the HSF1 pathway. If HSF1 functions in HIV-1 latency reversal by recruiting P-TEFb to the LTR, then inhibiting HSF1 with KRIBB11 should specifically inhibit elongation but not initiation of HIV-1 transcripts or read-through transcripts initiating at upstream promoters of genes into which the provirus has integrated (Fig. 4A). To measure the effects of KRIBB11 on different steps in the HIV-1 transcription process, we used a panel of reverse transcription droplet digital PCR assays (55) to quantify the abundance of transcripts having proceeded through various phases of transcription (Fig. 4A). This analysis was carried out using resting CD4+ T cells from patients on ART. PMA/I was used to reverse latency since other LRAs are relatively ineffective ex vivo (Fig. 3A).
Fig. 4.
Analysis of transcriptional elongation in HIV-1 latency reversal. (A) Evaluation of steps in HIV-1 transcription using a panel of RT-ddPCR assays (55) that target five different regions of HIV-1 transcripts. The read-through assay includes HIV-1 sequences upstream of the HIV-1 transcription start site (TSS) and quantifies transcripts resulting from host-promoter–initiated transcription. The TAR assay targets the TAR region directly downstream from the HIV-1 TSS. The Long LTR assay starts downstream of the HIV-1 TSS and ends just past the 5′ LTR. The MS Tat/Rev and PolyA assays target distal sites in HIV-1 transcripts and require correct splicing and termination, respectively. (B) Analysis of HIV-1 transcript levels in resting CD4+ T cells from treated patients with or without PMA/I stimulation in the presence or absence of KRIBB11. Counts of various transcript types are quantified and normalized per million cells as determined by TERT cell equivalents as described (55). (C) Western blot analysis for CycT1 in whole-cell lysates of resting CD4+ T cells and clones of J-Lat cells. (D) Intracellular flow cytometry analysis of CycT1 levels in resting CD4+ T cells from treated patients after stimulation with candidate LRAs. Median fluorescence intensity is plotted as a fold change over DMSO control.
In line with the above hypothesis, KRIBB11 caused no reduction in the quantity of read-through transcripts initiating at upstream promoters of genes into which the provirus has integrated (Fig. 4B). Promoter-proximal HIV-1 transcripts encompassing the transactivation response (TAR) region were present at high levels even in the absence of activation and were unchanged by KRIBB11 treatment. In contrast, levels of more 5′ elongated HIV-1 transcripts were dramatically increased by activation with PMA/I, and the increases were blocked by KRIBB11. We observed significant KRIBB11-induced reductions in elongated HIV-1 transcripts including correctly terminated transcripts detected with amplicons overlapping the poly-A tail and multiply spliced HIV-1 (Tat/Rev) transcripts. Overall, these data demonstrate that KRIBB11 exerts an effect prominently on elongated transcripts, which supports the hypothesis that HSF1-mediated signaling plays a role in HIV-1 transcriptional elongation following PMA/I stimulation.
Our data demonstrate that a substantial component of HIV-1 latency reversal in J-Lat cells is controlled by HSF1 signaling and is inhibited by KRIBB11 (Fig. 2A). Previous work has demonstrated that components of the heat shock pathway, namely HSP90, promote viral replication by increasing the expression of transcription factors necessary for HIV replication (34). In this work we show that HSF1-mediated signaling is induced by LRAs and has demonstrable effects on the HIV-1 transcriptional process. Further study of how LRAs impact other components of the heat shock pathway will further clarify the mechanisms contributing to LRA-mediated latency reversal.
In primary cells from infected individuals, an HSF1-mediated stress response in itself is not sufficient to reverse HIV-1 latency. Because HSF1 is known to enhance recruitment of free PTEF-b to the HIV-1 LTR (35), we hypothesized that differences in basal P-TEFb levels could explain this discrepancy. The regulation of PTEF-b in T cells is complex and multifactorial, involving microRNA-based regulation of transcript levels of Cyclin T1 (CycT1), a component of P-TEFb (56, 57). Indeed, we observed pronounced differences in basal levels of CycT1 protein in primary cells as compared to J-Lat cells (Fig. 4C) by Western blot analysis. Resting CD4+ T cells from both healthy controls and infected individuals had low or undetectable levels of CycT1, while there were readily detectable levels of CycT1 in all clones of J-Lat cells, even in the absence of stimulation (Fig. 4C). Furthermore, intracellular flow cytometry analysis revealed very low levels of CycT1 in primary resting CD4+ T cells relative to J-Lat cells (SI Appendix, Fig. S2). Importantly, the majority of LRAs did not increase CycT1 levels in resting CD4+ T cells (Fig. 4D). However, agents that show ex vivo latency reversal, such as PMA/I (58), did cause slight increases in protein levels of CycT1. Given the pronounced differences in basal P-TEFb levels between J-Lat cells and resting CD4+ T cells, we conclude that insufficient levels of P-TEFb in primary cells explain the inability of HSF1-mediated signaling alone to reverse HIV-1 latency in primary cells from infected individuals.
We hypothesize that different levels of CycT1 in cell lines and resting CD4+ T cells ex vivo represent a central difference between the form of latency present in in vitro models and the latency occurring in primary cells from infected individuals. Furthermore, we hypothesize that CycT1 present in cell line models of latency is recruited to the HIV-1 LTR by a HSF1-dependent mechanism following stimulation with many LRAs, resulting in HSF1-mediated latency reversal in these models of latency. The observation that a significant component of latency reversal mediated by many LRAs occurs through cell stress mechanisms involving HSF1 represents a significant difference relative to more physiologic latency-reversing stimuli and warrants further study of LRAs and their capacity to induce HSF1-mediated signaling. Additionally, dissection of T cell activation pathways to identify mechanisms governing CDK9/CycT1 protein levels would be highly beneficial to the study of HIV latency reversal and could potentially result in much more efficacious and clinically tolerable latency reversal strategies.
Discussion
HIV-1 infection remains incurable with ART alone due to the persistence of a highly stable latent reservoir of intact HIV-1 proviruses. Substantial effort has been devoted to identifying small molecules that reverse latency, allowing for immune recognition and clearance of infected cells (59). Due to the low frequency of HIV-1–infected cells in the blood of infected individuals, several model systems have been developed to facilitate the study of latency and the identification of LRAs. Several LRAs have been identified using in vitro models of latency. These target diverse molecular mechanisms involved in the regulation of HIV-1 gene expression, including signaling pathways involved in T cell activation and epigenetic mechanisms that may enforce latency (5–7, 11, 20, 27). Despite the large number of LRAs identified in in vitro studies, no LRA has achieved clinical success in reducing the size of the HIV-1 reservoir. It has been unclear why model systems overreport LRA activity. Certain combinations of LRAs show good ex vivo activity in cells from infection individuals (15, 46), but there has been no clear molecular explanation of why only certain LRA combinations are active.
By directly measuring the production of properly terminated HIV-1 messenger RNAs (mRNAs), we demonstrated that the majority of LRAs identified in vitro are ineffective at reversing latency in primary cells from infected patients. In this study, we provide evidence that several LRAs targeting diverse pathways stimulate HIV-1 latency reversal in model systems through a common cellular stress pathway involving the transcription factor HSF1. Cotreatment with the HSF1 inhibitor KRIBB11 substantially reduced the HIV-1 latency reversal potential of HDAC inhibitors, PKC agonists, and proteasome inhibitors. In a series of orthogonal assays, we demonstrate that despite significant attenuation of HIV-1 latency reversal, KRIBB11 did not block the expected normal activity of these agents. Interestingly, we observed that cotreatment with KRIBB11 did not substantially attenuate latency reversal mediated by the SMAC mimetic AZD5582, suggesting that AZD5582 does not function through an HSF1-mediated stress pathway. When analyzing resting CD4+ T cells from infected individuals, we observed that KRIBB11 substantially attenuated PMA/I-induced HIV-1 latency reversal, but not latency reversal mediated by αCD3/αCD28 antibodies. Our results differ from Pan et al. (35) who have suggested that HSF1 plays a central role in HIV-1 latency reversal. Here, we show that in primary CD4+ T cells from infected patients, the involvement of HSF1 in HIV-1 latency reversal differs given the type of stimulus. Pharmacologic latency reversal mediated by several well-known LRAs is readily inhibited by the HSF1 inhibitor KRIBB11. In contrast, reversal of HIV-1 latency by physiological activation of T cells through surface receptors is minimally impacted by KRIBB11.
The effects of HSF1 on transcriptional elongation strongly suggest that HSF1 is involved in the HIV-1 transcriptional process in primary cells in response to LRAs, many of which induce a cellular stress response. P-TEFb, a complex of Cyclin T1 and CDK9, plays a central role in activating Pol-II to a fully processive form. Previous work has shown that HSF1 forms a complex with P-TEFb, which is then recruited to the promoters of heat shock genes, allowing transcriptional elongation (54). In a series of chromatin immunoprecipitation assays, we confirm that observations by Pan et al. (35) that HSF1 associates with the HIV-1 LTR and that this association is enhanced upon stimulation with PMA/I. Using a series of droplet digital PCR assays, we demonstrate that inhibition of HSF1 inhibits the formation of elongated and properly terminated HIV-1 transcripts, but not the formation of short, abortive transcripts that can form in the absence of the P-TEFb elongation complex.
Posttranslational inhibition of P-TEFb has been identified as the primary promoter of HIV-1 latency in model systems (21). We show here that, despite posttranslational inhibition, the J-Lat cell line had markedly higher levels of cyclin T1 compared to primary cells. Different levels of Cyclin T1 in primary cells and J-Lat cells may explain differing responses to LRAs, particularly those that may promote the release of P-TEFb from inhibitory complexes such as HDAC inhibitors (22). Furthermore, given the potential for HSF1 to recruit the P-TEFb complex to the HIV-1 LTR, higher levels of cyclin T1 in vitro may predispose in vitro models to be more sensitive to HSF1 stimulation in terms of latency reversal.
Our results help to explain why only certain LRAs and LRA combinations activate HIV-1 gene expression in resting CD4+ T cells from infected individuals on ART. For many LRAs (but not physiologic T cell activation), latency reversal in infected resting cells requires both reversing the restrictions on P-TEFb and promoting HSF1-mediated recruitment of P-TEFb to the LTR. For example, we have shown that the PKC agonist bryostatin can increase CycT1 levels and the HDAC inhibitor panobinostat can induce an HSF1 response. Together, these agents reverse latency ex vivo to a degree that approaches that seen with PMA/I. Thus, these studies provide a framework for understanding which LRA combinations are more likely to effective in vivo. In future studies, it will be important to test other promising LRAs such as recently described analogs of bryostatin that appear to have increased efficacy and tolerability (60).
Further characterization of model systems of latency is essential for progress toward identifying effective LRAs. Understanding key differences between model systems and primary cells is central to designing more informative latency models. We have demonstrated that several LRAs identified using in vitro models appear at least in part through a common mechanism that is distinct from their predicted mechanisms of action. We have shown that many LRAs are relatively ineffective in primary cells and have highlighted a key difference between a common HIV-1 model system and primary cells that may explain why certain LRAs function in model systems but not primary cells. Together, these studies enhance our understanding of molecular mechanisms of HIV-1 latency reversal in both primary and model systems and help to resolve why LRAs identified in model systems may be ineffective in vivo.
Materials and Methods
Study Participants.
Primary, untouched resting CD4+ T cells were isolated from either HIV+ or HIV− individuals (depending on the experiment) through negative selection as previously described (15). For HIV+ individuals, participants were enrolled under the criteria of suppression of viremia <50 copies/mL for at least 6 mo. All participants provided written informed consent. This study was approved by the Johns Hopkins Institutional Review Board.
Cell Isolations.
Peripheral blood mononuclear cells were isolated by density gradient centrifugation of whole blood or leukapheresis product. CD4+ T lymphocytes were isolated from PBMCs using a negative magnetic separation T cell enrichment kit (Stemcell). From bulk populations of CD4+ T cells, activated cells were removed with magnetic beads carrying antibodies against CD25, CD69, and HLA-DR (Miltenyi Biotech).
Latency Reversal Assays.
Primary model cells, J-Lat cells, and cells from infected individuals were treated with identical doses of PMA/I or candidate LRAs for 24 h. Doses known to give optimal latency reversal were identified with dose–response curves in primary model cells. PMA (Sigma) was used at a standard concentration of 50 ng/mL, and ionomycin (Sigma) was used at 1 μM. Both bryostatin and bortezomib were dosed at 10 nM, and panobinostat was dosed at 10 μM. Both KRIBB11 and HBB2 were used at 5 μM. For CD3/CD28 stimulations, antibody-coated magnetic beads (Thermo) were used at a 1:1 cell:bead ratio in the presence of 100 U/mL IL2 (Promega). For treatments involving HIV+ samples, 10 μM enfurvitide (T20) was included to prevent infection events within the culture.
Generation and Treatment of Bcl2-Transduced Model Cells.
Primary model cells were generated as previously described (61). Briefly, untouched primary CD4+ T cells were isolated from healthy donors as described above. Cells were activated with CD3/CD28 cross-linking for 72 h. Activated cells were transduced with the EB-FLV plasmid to overexpress Bcl2. Cells were cultured for ∼4 wk in nonstimulatory conditions to allow contraction and apoptosis of nontransduced cells. A second activation was performed, and cells were expanded for 3 wk in IL-2–supplemented media and then infected with the CM6 reporter virus which contains GFP substituted for a segment of the env gene. A second incubation in the absence of activating stimuli was performed to allow down-regulation of GFP expression as the cells entered latency. To assay for latency reversal, 100,000 GFP primary model cells were plated in a 96-well plate and treated with LRAs for 18 h unless otherwise stated. Following treatment, cells were assessed for GFP expression on a FACSCanto flow cytometer. Viability was assessed by staining with 7AAD. Each condition was normalized to a PMA/I control.
HSP70 Gene Analysis.
Expression of the HSP70 family of genes was measured with an array of qPCR assays. Cell-associated RNA was isolated using phenol-acid-chloroform precipitation (TRIzol Reagent) and then treated with DNase. Relative abundances were calculated using the ΔΔCT method and normalized to expression of DNAJC5, which was the least-perturbed gene between samples. All data are shown as fold over dimethylsulfoxide (DMSO) control.
J-Lat Experiments.
Five clones of J-Lats (6.3, 8.4, 9.2, 10.6, 15.4) were seeded in a 96-well plate at 200,000 cells per well. Treatments were then performed with two technical replicates for each condition. All treatments were performed for 24 h, and then J-Lat cells were analyzed for GFP expression using an Intellicyt I-Que plus flow cytometer. All data are presented as a percentage of GFP-gated cells compared to the total number of cells assayed. In addition to GFP expression, viability was assessed using Zombie Violet viability dye (Biolegend) following standard protocols.
Ex Vivo Cell Stimulations.
Cells from HIV+ individuals were isolated as described and plated in aliquots of 5 million cells per treatment for 24 h unless otherwise stated. Cell-associated RNA was isolated using phenol-acid-chloroform precipitation (TRIzol Reagent; Thermo). RNA was converted to complementary DNA using the SuperScript RT kit (Thermo) and random hexamer primers. Mature, polyadenylated HIV mRNA transcripts were analyzed by qPCR using an Applied Biosystems Viia-7 qPCR thermocycler as described (52). Primers and probes are listed in SI Appendix, Table S2. Cycle thresholds were compared to a plasmid standard to calculate objective transcript counts. Each experimental condition was reported as a fold change over negative control. For treatments involving HSF1 blockade, KRIBB11 was included at 5 µM as a cotreatment for the duration of the experimental stimulus.
Quantitative Viral Outgrowth Assay.
Resting CD4+ T cells were isolated and tested with a quantitative viral outgrowth as described (53). Briefly, cells were then plated at 200,000 cells/well and subjected to stimulation with PHA and irradiated, allogenic PBMCs for 18 h. Stimulations were performed at either 37 °C or 39 °C as stated in the presence or absence of 5 µM KRIBB11. After the 18-h stimulation, cells were washed to remove residual PHA/KRIBB11 and cocultured with MOLT4 cells for 21 d before quantification of p24 protein with an enzyme-linked immunosorbent assay. Infectious units per million (IUPM) were calculated using IUPMStats V1.0 (62).
Measurement of NF-κB Activity.
The NF-κB luciferase construct was a gift from J. Pomerantz, Johns Hopkins University, Baltimore. The plasmid was transfected into Jurkat cells using calcium-phosphate–mediated transfection. After culture for 42 h, cells were stimulated for 5 h with PMA/I or CD3/CD28 dynabeads in the presence and absence of KRIBB11. Luciferase production was measured using a Centro LB 960 Luminometer.
Proteasome Activity.
Proteasome activity was measured in Jurkat cells using the Proteasome-Glo kit (Promega) according to the manufacturer’s instructions. Briefly, Jurkat cells were treated with either media or KRIBB11 for 24 h. Proteasome-Glo was added directly to cells, which emit luminescence when exposed to the 20S proteasome. Luminescence was detected using a Centro LB 960 Luminometer.
Elongation Analysis.
Droplet digital PCR was performed and analyzed as previously described (55). Briefly, resting CD4+ cells were isolated and treated with PMA/I in the presence or absence of KRIBB11 for 24 h. Read-through, total (TAR), elongated (long LTR), completed (polyA), and multiply spliced (Tat-Rev) transcripts in unstimulated and activated rCD4+ T cells were measured by reverse transcription droplet digital PCR (RT-ddPCR) and normalized to cell numbers via ddPCR quantification of the gene Telomere Reverse Transcriptase (TERT) in DNA extracted at the same time as the RNA. All five HIV transcripts were quantified in five patients. For all assays, primer and probe sets are listed in SI Appendix, Table S2.
Intracellular Flow Cytometry Staining.
Cells from three HIV+ individuals were isolated as described and plated in aliquots of 2 million cells per treatment for 24 h. Cells were stained with vital dye and then fixed and permeabilized with paraformaldehyde. Cells were stained with a fluorescent α-CycT1 antibody (Santa Cruz Biotechnology) and analyzed on a FACS Canto flow cytometer. Median fluorescence intensity in live cells was normalized to a fold change over vehicle control.
Western Blot Analysis.
Western blots were performed using 15 µg of bulk cellular lysate obtained through standard radioimmunoprecipitation assay cell lysis. Protein lysates were run on NuPAGE 4 to 12% Bis/Tris gradient gels and transferred to polyvinylidene difluoride membranes using the IBlot2 transfer device (Thermo). Membranes were blocked in a 5% solution of nonfat milk powder and TBS-Tween buffer (Thermo). Western blotting with primary antibodies was performed in blocking buffer. Acetylated histone H3 (ab47813), HSP70 (ab79852), and CycT1 (ab226851) Western blots were performed with rabbit antibodies (Abcam). AF488-conjugated anti-rabbit antibodies raised in goat were used as secondary antibodies. Blots were detected using the iBright digital imager (Thermo).
Chromatin Immunoprecipitation.
J-Lat clone 6.3 cells were stimulated with PMA/I or vehicle control for 24 h. Cells were then cross-linked with paraformaldehyde and sheared using a Covaris instrument into fragments ∼1 kb in size. Immunoprecipitations with HSF1, phospho-HSF1, and nontargeted IgG control antibodies were performed using the Pierce magnetic ChIP kit (Thermo) on equal volumes of cross-linked and sheared genomic DNA. Target genomic sequences were quantified with droplet digital PCR. Each ddPCR assay contained 2x BioRad ddPCR mastermix, 600 nM of forward and reverse primers, 200 nM fluorescent probes, and 4 µL eluted DNA product. Cycling conditions were 95 ºC for 10 min, and then 50 cycles of 95 ºC for 30 s, 56 ºC annealing and extension for 2 min. Post cycling, a final enzyme deactivation was performed at 98 ºC for 10 min. For each antibody pulldown, recovery of genomic targets was normalized to a fold change over the nontargeted IgG control. For all ChIP assays, primers and probes are provided in SI Appendix, Table S2.
Data Availability.
All experimental results described in the paper are presented in the figures and supplementary figures.
Supplementary Material
Acknowledgments
This work was supported by NIH Martin Delaney I4C Grant UM1 AI126603; Beat-HIV Grant UM1 AI126620; and the Delaney AIDS Research Enterprise (DARE) Grant UM1 AI12661 collaboratories; the Johns Hopkins Center for AIDS Research (P30AI094189); NIH Grant 43222; and Howard Hughes Medical Institute and Bill and Melinda Gates Foundation Grant OPP1115715. E.F. was supported by NIH T32 GM007445
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916290117/-/DCSupplemental.
References
- 1.Finzi D.et al., Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Chun T. W.et al., Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387, 183–188 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Wong J. K.et al., Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Deeks S. G.et al.; International AIDS Society Scientific Working Group on HIV Cure , Towards an HIV cure: A global scientific strategy. Nat. Rev. Immunol. 12, 607–614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nabel G., Baltimore D., An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326, 711–713 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Böhnlein E.et al., The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell 53, 827–836 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Duh E. J., Maury W. J., Folks T. M., Fauci A. S., Rabson A. B., Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc. Natl. Acad. Sci. U.S.A. 86, 5974–5978 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks D. G., Arlen P. A., Gao L., Kitchen C. M. R., Zack J. A., Identification of T cell-signaling pathways that stimulate latent HIV in primary cells. Proc. Natl. Acad. Sci. U.S.A. 100, 12955–12960 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterlin B. M., Price D. H., Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Van Lint C., Emiliani S., Ott M., Verdin E., Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15, 1112–1120 (1996). [PMC free article] [PubMed] [Google Scholar]
- 11.Blazkova J.et al., CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5, e1000554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blazkova J.et al., Paucity of HIV DNA methylation in latently infected, resting CD4+ T cells from infected individuals receiving antiretroviral therapy. J. Virol. 86, 5390–5392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman J.et al., Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85, 9078–9089 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott J. H.et al., Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 10, e1004473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laird G. M.et al., Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 125, 1901–1912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruner K. M.et al., Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archin N. M.et al., Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coull J. J.et al., The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74, 6790–6799 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keedy K. S.et al., A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J. Virol. 83, 4749–4756 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinoshita S., Chen B. K., Kaneshima H., Nolan G. P., Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell 95, 595–604 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Tyagi M., Pearson R. J., Karn J., Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84, 6425–6437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartholomeeusen K., Fujinaga K., Xiang Y., Peterlin B. M., Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J. Biol. Chem. 288, 14400–14407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scripture-Adams D. D., Brooks D. G., Korin Y. D., Zack J. A., Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J. Virol. 76, 13077–13082 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehrman G., Ylisastigui L., Bosch R. J., Margolis D. M., Interleukin-7 induces HIV type 1 outgrowth from peripheral resting CD4+ T cells. J. Acquir. Immune Defic. Syndr. 36, 1103–1104 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Contreras X.et al., Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284, 6782–6789 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei D. G.et al., Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 10, e1004071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauder S. E., Bosque A., Lindqvist A., Planelles V., Verdin E., Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5, e1000495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinter A. L., Poli G., Maury W., Folks T. M., Fauci A. S., Direct and cytokine-mediated activation of protein kinase C induces human immunodeficiency virus expression in chronically infected promonocytic cells. J. Virol. 64, 4306–4312 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korin Y. D., Brooks D. G., Brown S., Korotzer A., Zack J. A., Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 76, 8118–8123 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkosky J.et al., Prostratin: Activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98, 3006–3015 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Williams S. A.et al., Prostratin antagonizes HIV latency by activating NF-kappaB. J. Biol. Chem. 279, 42008–42017 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Miller L. K.et al., Proteasome inhibitors act as bifunctional antagonists of human immunodeficiency virus type 1 latency and replication. Retrovirology 10, 120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson I.et al., Heat shock protein 90 controls HIV-1 reactivation from latency. Proc. Natl. Acad. Sci. U.S.A. 111, E1528–E1537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joshi P., Maidji E., Stoddart C. A., Inhibition of heat shock protein 90 prevents HIV rebound. J. Biol. Chem. 291, 10332–10346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan X.-Y.et al., Heat shock factor 1 mediates latent HIV reactivation. Sci. Rep. 6, 26294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H.-C.et al., Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 119, 3473–3486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinkova-Kostova A. T., The role of sulfhydryl reactivity of small molecules for the activation of the KEAP1/NRF2 pathway and the heat shock response. Scientifica (Cairo) 2012, 606104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y.et al., HSF1-dependent upregulation of Hsp70 by sulfhydryl-reactive inducers of the KEAP1/NRF2/ARE pathway. Chem. Biol. 18, 1355–1361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinkova-Kostova A. T., Massiah M. A., Bozak R. E., Hicks R. J., Talalay P., Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc. Natl. Acad. Sci. U.S.A. 98, 3404–3409 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinkova-Kostova A. T.et al., Extremely potent triterpenoid inducers of the phase 2 response: Correlations of protection against oxidant and inflammatory stress. Proc. Natl. Acad. Sci. U.S.A. 102, 4584–4589 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinkova-Kostova A. T.et al., Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi A.et al., The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: Role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. J. Biol. Chem. 289, 12705–12715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z.et al., Reiterative Enrichment and Authentication of CRISPRi Targets (REACT) identifies the proteasome as a key contributor to HIV-1 latency. PLoS Pathog. 15, e1007498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theodorakis N. G., Zand D. J., Kotzbauer P. T., Williams G. T., Morimoto R. I., Hemin-induced transcriptional activation of the HSP70 gene during erythroid maturation in K562 cells is due to a heat shock factor-mediated stress response. Mol. Cell. Biol. 9, 3166–3173 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spina C. A.et al., An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 9, e1003834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darcis G.et al., An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLoS Pathog. 11, e1005063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawat P., Mitra D., Cellular heat shock factor 1 positively regulates human immunodeficiency virus-1 gene expression and replication by two distinct pathways. Nucleic Acids Res. 39, 5879–5892 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jordan A., Bisgrove D., Verdin E., HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22, 1868–1877 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon Y. J.et al., KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J. Biol. Chem. 286, 1737–1747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y.-K.et al., Molecular determinants of scaffold-induced linear ubiquitinylation of B cell lymphoma/leukemia 10 (Bcl10) during T cell receptor and oncogenic caspase recruitment domain-containing protein 11 (CARD11) signaling. J. Biol. Chem. 291, 25921–25936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nixon C. C.et al., Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 578, 160–165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shan L.et al., A novel PCR assay for quantification of HIV-1 RNA. J. Virol. 87, 6521–6525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laird G. M.et al., Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 9, e1003398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lis J. T., Mason P., Peng J., Price D. H., Werner J., P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14, 792–803 (2000). [PMC free article] [PubMed] [Google Scholar]
- 55.Yukl S. A.et al., HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 10, eaap9927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiang K., Sung T.-L., Rice A. P., Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J. Virol. 86, 3244–3252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoque M., Shamanna R. A., Guan D., Pe’ery T., Mathews M. B., HIV-1 replication and latency are regulated by translational control of cyclin T1. J. Mol. Biol. 410, 917–932 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bullen C. K., Laird G. M., Durand C. M., Siliciano J. D., Siliciano R. F., New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shan L.et al., Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36, 491–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marsden M. D.et al., Characterization of designed, synthetically accessible bryostatin analog HIV latency reversing agents. Virology 520, 83–93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim M.et al., A primary cell model of HIV-1 latency that uses activation through the T cell receptor and return to quiescence to establish latent infection. Nat. Protoc. 9, 2755–2770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosenbloom D. I. S.et al., Designing and interpreting limiting dilution assays: General principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect. Dis. 2, ofv123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental results described in the paper are presented in the figures and supplementary figures.