Abstract
Objective
The aim of this study was to carry out whole-genome sequencing (WGS) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), using samples collected from Congolese individuals between April and July 2020.
Methods
Ninety-six samples were screened for SARS-CoV-2 using RT-PCR, and 19 samples with Ct values <30 were sequenced using Illumina Next-Generation Sequencing (NGS). The genomes were annotated and screened for mutations using the web tool ‘coronapp’. Subsequently, different SARS-CoV-2 lineages were assigned using PANGOLIN and Nextclade.
Results
Eleven SARS-CoV-2 genomes were successfully sequenced and submitted to the GSAID database. All genomes carried the spike mutation D614G and were classified as part of the GH clade. The Congolese SARS-CoV-2 sequences were shown to belong to lineage B1 and Nextclade 20A and 20C, which split them into distinct clusters, indicating two separate introductions of the virus into the Republic of Congo.
Conclusion
This first study provides valuable information on SARS CoV-2 transmission in the central African region, contributing to SARS CoV-2 surveillance on a temporal and spatial scale.
Keywords: Republic of Congo, SARS-CoV-2, Whole genome sequencing, SARS-CoV-2 variants, D614G, Lineage B1
Introduction
With the first cases reported on March 14, 2020 (Ntoumi and Velavan, 2020), the Republic of Congo reported a total of 7794 cases with 117 deaths as of January 25, 2021, with transmission driven by the community (WHO, 2021).
The first SARS-CoV-2 genome was described in January 2020, and since then several studies have tracked its evolution worldwide. Mutations found in SARS-CoV-2, using next-generation sequencing methods, are increasingly being studied in order to understand potential associations among transmission dynamics, pathogenicity, diagnostic performance, vaccine efficacy, and immune evasion (ECDC, 2020). There is a paucity of data on SARS-CoV-2 sequences from Central Africa, despite increasing submissions to databases from other regions of Africa.
To date, no studies have reported on SARS-CoV-2 genomic lineages/strains in the Republic of Congo. This first study used samples collected between April and July 2020, and performed next-generation sequencing to understand the SARS-CoV-2 genomes that circulated during the early phases of the outbreak.
Materials and methods
Ethics statement
Informed written consent was obtained from all participants. The study was approved by the ministry of scientific research and technological innovation, Republic of Congo (Approval No. 049/MRSIT-CAB) and by the institutional ethics committee of the Congolese Foundation for Medical Research (Approval No. 027/CIE/FCRM/2020).
Sampling procedures
An epidemiological survey was conducted between April and July 2020 to assess the spread of SARS-COV-2 in the general population of Brazzaville (Batchi-Bouyou et al., 2020). A total of 96 positive samples were randomly selected, and 19 samples (with Ct values <30) were subsequently sequenced using Illumina Next-Generation Sequencing (NGS) methodology.
RT-PCR testing
Viral RNA was extracted from nasopharyngeal samples using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RealStar® SARS-CoV-2 real-time PCR targeting the S gene of SARS-CoV-2 (Altona Diagnostics, Hamburg, Germany) was performed using a LightCycler® 480 Instrument II (Roche Diagnostics, Mannheim, Germany), according to the manufacturer’s protocol. An in-vitro transcribed RNA of the SARS-CoV-2 ‘S’ gene was integrated in each run to determine the number of viral copies with respective Ct values.
NGS sequencing and mutational analysis
Libraries were prepared according to the COVID-19 ARTIC v3 Illumina library construction and sequencing protocol V.5 (DNA Pipelines et al., 2020), using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs). The libraries were quantified (Qubit DNA BR, Thermo Scientific), normalized, and pooled, and sequencing was performed using an Illumina MiSeq v2 with 2 × 250 bp cycles. Viral genome assembly and variant calling was performed using nf-core/viralrecon pipeline (https://nf-co.re/viralrecon/1.1.0) (Ewels et al., 2020). The SARS-CoV-2 genomes were screened for distinct mutations using the online COVID-19 genome annotator ‘coronapp’ (Mercatelli et al., 2020).
Phylogenetic analysis
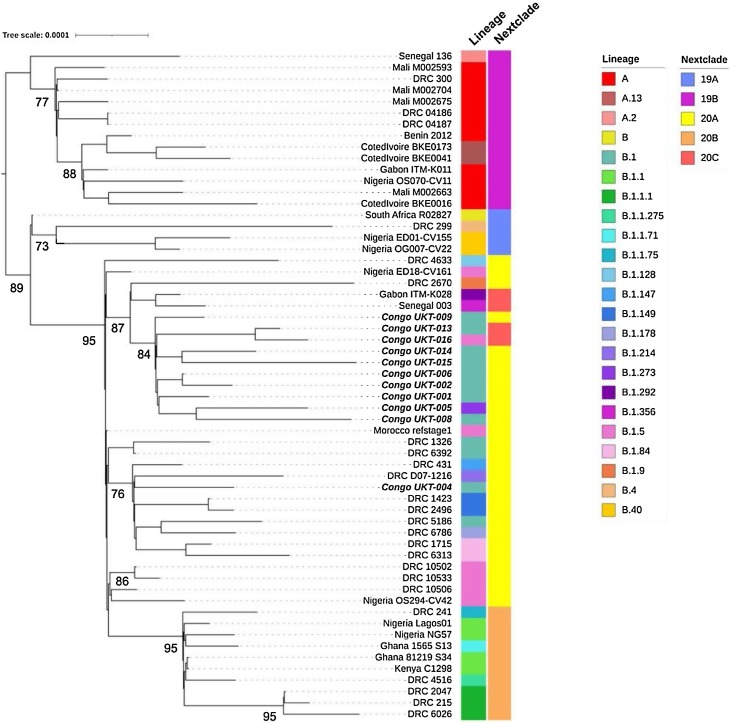
SARS-CoV-2 genomes from African countries were retrieved from GSAID (Shu and McCauley, 2017). The eleven SARS-CoV-2 genomes were comparatively evaluated against reference NC_045512.2-Wuhan-Hu-1 and available SARS-CoV-2 genome sequences collected between April and July 2020, representing countries such as Ghana, Nigeria, Benin, Mali, Senegal, Côte d'Ivoire, Gabon, Democratic Republic of Congo, South Africa, and Kenya. Based on this comparative analysis, a maximum likelihood and phylogenetic tree were reconstructed. All the different SARS-CoV-2 genomes can be accessed using the GSAID database, with the respective IDs provided in Figure 1 .
Figure 1.
Maximum likelihood phylogenetic tree of currently available SARS-CoV-2 genomes from the Republic of Congo (bold text). The genomes were classified into lineages using PANGOLIN and into clades using Nextclade. The tree was rooted between lineages A and B. Branch lengths are drawn according to the number of nucleotide substitutions per site. DRC stands for Democratic Republic of Congo.
The non-coding 3′ and 5′ regions were trimmed using Geneious Prime software, and coding regions were aligned using a multiple sequence alignment (MAFTT) algorithm (Katoh et al., 2002). A maximum likelihood tree was reconstructed with the IQ-TREE server (Trifinopoulos et al., 2016) using the general time-reversible (GTR) model with rate heterogeneity (GTR + G). Branch support was calculated by ultrafast bootstrap, consisting of 1000 alignments (Hoang et al., 2018). SARS-CoV-2 genomes were classified into lineages using Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN) (Rambaut et al., 2020) and viral clades were assigned by Nextclade Beta (https://clades.nextstrain.org/) and Nextstrain (Hadfield et al., 2018). The final dataset was displayed using Interactive Tree of Life (iTOL) v4 (Letunic and Bork, 2019).
Results
Of the 19 samples, 11 met the quality criteria for submission. All SARS-CoV-2 genome sequences were reported from different districts within Brazzaville, the capital of the Republic of Congo. All were symptomatic individuals with a median age of 45 years, and seven were male. Eleven SARS-CoV-2 genomes were deposited on the GISAID platform (Shu and McCauley, 2017) (https://www.gisaid.org/) (Accession numbers EPI_ISL_581455, 581462, 581472, and 581486–581493). The annotated mutations are summarized in Table 1 . The resulting SARS-CoV-2 genomes were compared with the reference NC_045512.2-Wuhan-Hu-1. The amino acid substitutions S (D614G) in the spike protein and NSP12b in the non-structural protein (NSP) occurred in all SARS-CoV-2 isolates. In addition, 10 of the 11 SARS-CoV-2 isolates carried the NSP2 (Y537Y), NSP3 (F106F), and ORF3a (Q57H) substitutions (Table 1). The SARS-CoV-2 genomes analyzed belonged to the B lineage (B.1 and B.1.1), with almost all Congolese sequences clustered together (Figure 1). The Nextclade analysis revealed that nine of the 11 SARS-CoV-2 genomes belonged to clade 20A, with the rest being part of clade 20C (Figure 1).
Table 1.
SARS-CoV-2 genomes annotated in this study.
| No. | ID | Lineage/Nextclade | Mutations observed |
|---|---|---|---|
| 1 | Congo/UKT-001 | B.1/20A | (n = 8) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP3 (F430S), NSP12b (P314L), NSP16 (N297D), S (D614G), ORF3a (Q57H) |
| 2 | Congo/UKT-002 | B.1/20A | (n = 9) – 5'UTR (241), NSP2 (N9N), NSP2 (Y537Y), NSP3 (F106F), NSP12b (P314L), NSP15 (Y88Y), S (D614G), ORF3a (Q57H), N (P6S) |
| 3 | Congo/UKT-004 | B.1/20A | (n = 9) – 5'UTR (241), NSP3 (F106F), NSP5 (D176D), NSP8 (I156I), NSP12b (P314L), NSP12b (N619N), S (S71F), S (S116S), S (D614G) |
| 4 | Congo/UKT-005 | B.1.273/20A | (n = 12) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP3 (T763T), NSP12a (S6L), NSP12b (P314L), NSP14 (T113I), NSP15 (E145E), NSP15 (M271K), S (D614G), ORF3a (Q57H), ORF3a (V228A) |
| 5 | Congo/UKT-006 | B.1/20A | (n = 7) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP12b (P314L), S (D614G), ORF3a (Q57H), N (P6S) |
| 6 | Congo/UKT-008 | B.1/20A | (n = 13) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP3 (T763T), NSP3 (N1337N), NSP3 (T1607T), NSP12b (Y285Y), NSP12b (P314L), NSP13 (L297L), S (D614G), S (K1191N), ORF3a (Q57H), N (F274F) |
| 7 | Congo/UKT-009 | B.1/20A | (n = 8) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP12b (P314L), S (D614G), ORF3a (Q57H), ORF8 (E19D), N (P365L) |
| 8 | Congo/UKT-013 | B.1/20C | (n = 11) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP3 (K1804N), NSP4 (Y451Y), NSP12b (P314L), NSP13 (E261D), NSP13 (A454V), S (D614G), ORF3a (Q57H), N (T205I) |
| 9 | Congo/UKT-014 | B.1/20A | (n = 10) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP5 (A94V), NSP10 (V108V), NSP12b (P314L), S (A348S), S (D614G), ORF3a (Q57H), N (S183Y) |
| 10 | Congo/UKT-015 | B.1/20A | (n = 14) – 5'UTR (241), NSP2 (Y537Y), NSP3 (F106F), NSP3 (S1038S), NSP6 (Y80Y), NSP6 (K270R), NSP10 (V108V), NSP12b (P314L), NSP15 (A81V), S (D614G), S (L828I), ORF3a (Q57H), N (P13L), N (Q181K) |
| 11 | Congo/UKT-016 | B.1.5/20C | (n = 10) – 5'UTR (241), NSP2 (Y537Y), NSP3 (K1804N), NSP5 (R217M), NSP12b (P314L), NSP13 (E261D), NSP13 (A454V), S (D614G), ORF3a (Q57H), N (T205I) |
Figures in parentheses: amino acid substitutions/position aligned with reference NC_045512.2- Wuhan-Hu-1.
NSP: non-structural protein; ORF: open reading frame; S: spike protein; N: nucleoprotein.
Discussion
Genomics-based surveillance has helped researchers to assess the transmission and evolutionary dynamics of the SARS-CoV-2 virus. This first study from the Republic of Congo aimed to provide crucial information on circulating strains during the early pandemic period (between April and July 2020), when sufficient diagnostic capacity was not yet available (Batchi-Bouyou et al., 2020).
All SARS-CoV-2 genomes carried the spike mutation D614G, which is associated with efficient replication ex vivo and transmission in vivo (Hou et al., 2020). The B.1/B.1.1 lineage, as observed in this study, is also widespread in other African regions (Simulundu et al., 2021). Phylogenetic analysis revealed two distinct clusters, suggesting two separate introductions of the virus into the country, although the origins of these introductions remain unclear.
The second phylogenetic cluster with an isolate of lineage B.1 (Congo UKT-004) suggests that this variant may have been introduced from the neighboring country Democratic Republic of Congo. Furthermore, the clustering pattern, with rapid branching and diversification, suggests community transmission. The Congolese SARS-CoV-2 genomes representing clades 20A and 20C are predominantly seen in other central African countries, such as Gabon and in Democratic Republic of Congo.
Taken together, this study provides primary information on SARS CoV-2 transmission in the central African region, contributing to SARS CoV-2 surveillance data on a temporal and spatial scale.
Author contributions
FN and TPV designed the study. FN supervised the overall study in Brazzaville and TPV supervised the study in Germany. FN and CCMM recruited the patients and collected all data. AT performed the phylogenetic analysis, SRP performed the mutational analysis, LTKL performed experimental procedures, NC, AA, MS, and SP were involved in whole-genome sequencing, PGK contributed to the materials, and TPV and SRP wrote the manuscript.
Funding
The field study was supported by PANDORA-ID-NET network (EDCTP-RIA2016E-1609). The authors TPV and FN acknowledge the European and Developing Countries Clinical Trials Partnership (EDCTP) Central African Network for Clinical Research (CANTAM) (EDCTP-RegNet 2015-1045), and the Pan-African Network for Rapid Research, Response, and Preparedness for Infectious Diseases Epidemics Consortium (PANDORA-ID-NET) (EDCTP-RIA2016E-1609). NGS sequencing was performed with the support of the DFG-funded NGS Competence Center Tübingen (INST 37/1049-1). The author MS acknowledges the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 286/2020B01 – 428994620.
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Data availability statement
Data supporting the reported results are available on request.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgement
We acknowledge Marie Gauder from the Quantitative Biology Center (QBiC), Tübingen, for her support in data management. We also thank all study subjects for their participation.
References
- Batchi-Bouyou A.L., Lobaloba L., Ndounga M., Vouvoungui J.C., Mfoutou C.M., Boumpoutou K.R. High SARS-COV2 IgG/IGM seroprevalence in asymptomatic Congolese in Brazzaville, the Republic of Congo. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.12.065. S1201-9712(20)32589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DNA Pipelines R&D, Rajan Diana, Betteridge Emma, Shirley Lesley, Quail Michael, Park Naomi. 2020. COVID-19 ARTIC v3 Illumina library construction and sequencing protocol V.5. [Google Scholar]
- ECDC . ECDC; Stockholm: 2020. European centre for disease prevention and control. Sequencing of SARS-CoV-2 2. 2020. [Google Scholar]
- Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38(3):276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35(2):518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Triboli L., Fornasari E., Ray F., Giorgi F.M. Coronapp: a web application to annotate and monitor SARS-CoV-2 mutations. J Med Virol. 2020 doi: 10.1002/jmv.26678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoumi F., Velavan T.P. COVID-19 in Africa: between hope and reality. Lancet Infect Dis. 2021;21(3):315. doi: 10.1016/S1473-3099(20)30465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data — from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simulundu E., Mupeta F., Chanda-Kapata P., Saasa N., Changula K., Muleya W. First COVID-19 case in Zambia – comparative phylogenomic analyses of SARS-CoV-2 detected in African countries. Int J Infect Dis. 2021;102:455–459. doi: 10.1016/j.ijid.2020.09.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232–5. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. WHO coronavirus disease (COVID-19) dashboard. updated on 25.01.2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the reported results are available on request.