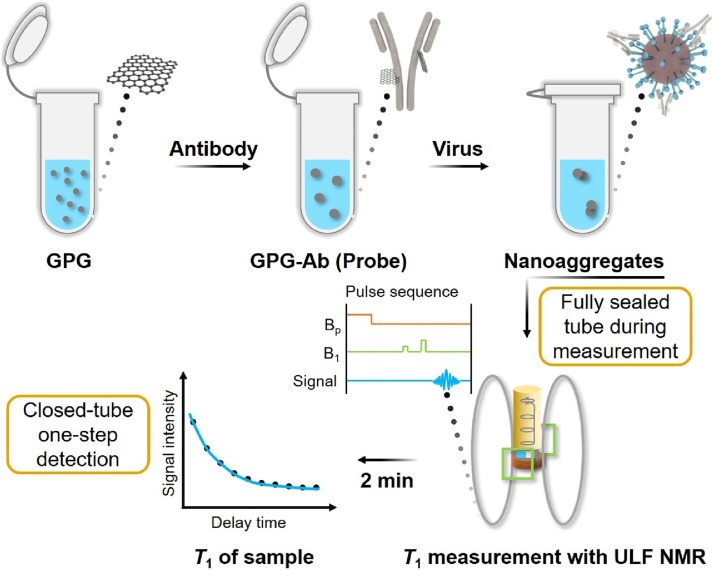
Graphical abstract
Abbreviations: COVID-19, coronavirus disease 2019; MRSw, magnetic relaxation switch; MNPs, magnetic nanoparticles; GQDs, graphene quantum dots; PEG, polyethylene glycol; PEG6, hexaethylene glycol; GPG, Gd3+ loaded PEG modified GQDs; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; S protein, spike protein; Ab, specific antibody against SARS-CoV-2 antigen S protein; RT-PCR, reverse transcription-polymerase chain reaction; LOD, limit of detection; NMR, nuclear magnetic resonance; ULF NMR, ultra-low field NMR; Fe3O4, ferrosoferric oxide; T1, longitudinal relaxation time; PBS, phosphate buffer saline; TEM, transmission electron microscopy; HR-TEM, high resolution TEM; AFM, atomic force microscopy; XPS, X-ray photoelectron spectroscopy; OSR, outer sphere relaxation theory; ELISA, enzyme-linked immune-sorbent assay; BSA, bull serum albumin; SQUID, superconducting quantum interface device; SD, standard deviation
Keywords: SARS-CoV-2, Spike, Graphene quantum dots, Ultra-low field nuclear magnetic resonance, Magnetic relaxation switch
Abstract
The rapid and sensitive diagnosis of the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is one of the crucial issues at the outbreak of the ongoing global pandemic that has no valid cure. Here, we propose a SARS-CoV-2 antibody conjugated magnetic graphene quantum dots (GQDs)-based magnetic relaxation switch (MRSw) that specifically recognizes the SARS-CoV-2. The probe of MRSw can be directly mixed with the test sample in a fully sealed vial without sample pretreatment, which largely reduces the testers’ risk of infection during the operation. The closed-tube one-step strategy to detect SARS-CoV-2 is developed with home-made ultra-low field nuclear magnetic resonance (ULF NMR) relaxometry working at 118 μT. The magnetic GQDs-based probe shows ultra-high sensitivity in the detection of SARS-CoV-2 due to its high magnetic relaxivity, and the limit of detection is optimized to 248 Particles mL‒1. Meanwhile, the detection time in ULF NMR system is only 2 min, which can significantly improve the efficiency of detection. In short, the magnetic GQDs-based MRSw coupled with ULF NMR can realize a rapid, safe, and sensitive detection of SARS-CoV-2.
1. Introduction
As the third coronavirus causing deadly pneumonia to humans in the 21st century [1,2], severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused the global pandemic of coronavirus disease 2019 (COVID-19) [3]. Till December of 2020, the death cases have exceeded 1.8 million globally (World Health Organization COVID-19 dashboard; https://covid19.who.int/). Since currently no specific antiviral drugs are commercially available, the rapid, safe, and sensitive detection of the highly contagious SARS-CoV-2 is still of vital importance for controlling the pandemic. The so-called spike (S) protein, which is the key glycoprotein for the entry of coronavirus into host cells [4], is the most prominent biomarker on the viral surface of SARS-CoV-2. Compared with the RNA-based detection with reverse transcription-polymerase chain reaction (RT-PCR), the detection of S protein could directly identify the coronavirus without sample pretreatment (e.g., nucleic acid extraction and amplification). Recently, several attempts have been made to directly detect coronavirus utilizing S protein via electrochemical sensors [5] and field-effect transistor-based sensors [6]. However, in these methods, the test-tubes still have to be opened during the detection, which could generate aerosol contamination. SARS-CoV-2 was found to be stable [7] and transmitted in the aerosol [8]. Therefore, avoiding opening the tube and preventing the generation of aerosol contamination during the detection could effectively reduce the risk of infection for the testers.
Based on the nuclear magnetic resonance (NMR) phenomenon, magnetic relaxation switches (MRSw) can be applied in biomarker detection [[9], [10], [11], [12]], which can significantly simplify the assay steps [13], achieve closed-loop detection, and enable near background-free sensing [14]. Consequently, MRSw are a class of promising approaches for the rapid and sensitive detection of S protein and thus SARS-CoV-2. Ferriferous oxide (Fe3O4) particles [10,11,15,16], commonly known as magnetite, have been widely used as the probe of MRSw in biomarker detection. In order to enhance the sensitivity of MRSw by 1 or 2 orders of magnitude, many works have focused on signal amplification including integrating the magnetic separation into MRSw [11] and increasing the amount of Fe3O4 [10]. Recent studies show that introducing paramagnetic Gd3+-based probes could also provide high sensitivity [17,18]. For example, magnetic nanoparticles (MNPs) that consist of Gd3+ and graphene quantum dots (GQDs) have numerous applications in developing MRI contrast agents in recent years with guaranteed biocompatibility and high relaxivity [[19], [20], [21]], which would be a promising probe for biomarker detection with high sensitivity. Meanwhile, ultra-low field (ULF) NMR (typical static field less than 250 μT) can realize a two-fold increase of relaxivity of Gd3+-based complexes compared to that measured at 1.5 T [22], which could be beneficial for the biomarker detection. Besides, ULF NMR exhibits a series of advantages including pure heteronuclear J-coupling detection [23,24], wide bandwidth for simultaneous detection of several different nuclei [25,26], low cost [27,28], and portability. Therefore, it has got particular attention to serve as a complementary supplement to high field NMR.
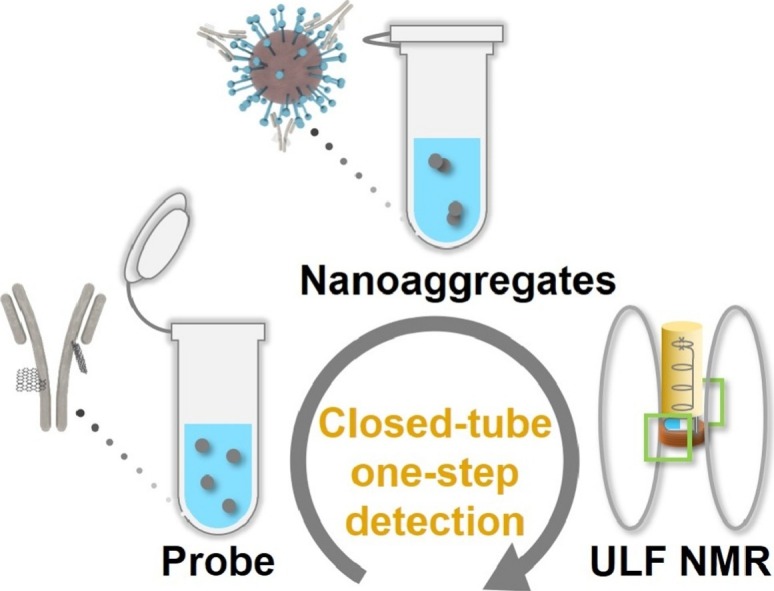
Herein, we developed Gd3+-based MRSw consisting of Gd3+ loaded polyethylene glycol (PEG) modified GQDs (GPG) [21] and specific antibody (Ab) against SARS-CoV-2 antigen S protein. The rapid and closed-tube detection of SARS-CoV-2 pseudovirus in a home-made ULF NMR system was realized with ultra-high sensitivity by measuring the change of longitudinal relaxation times (T 1s). For safety reasons, the SARS-CoV-2 pseudovirus was used in simulated viral samples. The test sample requires no pretreatment before mixing with the MRSw probe, and no additional operation like re-opening the sample vial is needed during the specific combination between the probe and pseudovirus as well as during the detection process, which prevents aerosol contamination and immensely reduces the testers’ risk of infection. By comparing the longitudinal relaxation times measured by home-made ULF NMR relaxometry before and after the specific combination, whether the sample contains SARS-CoV-2 pseudovirus can be distinguished (Scheme 1 ). Thanks to the high magnetic relaxivity of the probe and the high sensitivity of the ULF NMR system, the MRSw can sensitively detect the virus. This approach represents an innovative alternative for rapid, safe, and sensitive diagnosis of COVID-19 without sample pretreatment, and would have wide application for the detection of other viruses by changing the antibody, especially for coronaviruses with S proteins on their surface.
Scheme 1.
The detection process of the MRSw assay with ULF NMR.
2. Materials and methods
2.1. Materials
The GQDs were purchased from CASYUEDA Materials Technology Co., Ltd. (Shanghai, China) and used as received. Hexaethylene glycol (PEG6, 97.0 %), Gd(NO3)3⋅6H2O (99.9 %), and PBS buffer (pH = 7.2) were purchased from Aladdin Co., Ltd. (Shanghai, China) and used without further purification. Deionized water (resistivity ∼18.2 MΩ cm at 25 °C) was obtained using a Milli-Q system and used throughout all the experiments. The SARS-CoV-2 antigen S protein was purchased from Sino Biological Inc. (Beijing, China). The specific Ab against SARS-CoV-2 antigen S protein was acquired from Sino Biological Inc. (Beijing, China). SARS-CoV-2 pseudovirus was obtained from Genomeditech Co., Ltd. (Shanghai, China). Above biomolecules were used according to the manufacturer’s protocols.
2.2. Synthesis of GPG-Ab probe
The GPG was prepared in accordance with the previously proposed method [21]. Briefly, the procedure can be divided into four steps. At first, 15 mg of GQDs and 0.05 mmol of PEG6 were added into 15 mL of deionized water, and then transferred into para-polystyrene lined autoclave heating for 48 h at 240 ℃. Next, 225.7 μg, 0.5 μmol of Gd(NO3)3⋅6H2O was added to the product of the last step and then heated for 24 h at 240 ℃. After that, the obtained solution was dialyzed in a 3500 Da dialysis bag against deionized water for the removal of dissociative Gd3+. At last, a lyophilizer was employed to obtain the GPG powder.
For the purpose of high probe specificity, a highly specific antibody (Cat.# 40592-MM57, Sino Biological Inc, Beijing, China) was selected for the probe. According to the manufacturer’s data, it specifically recognizes SARS-CoV-2, with no cross-reaction with SARS-CoV. In order to obtain the magnetic probe, GPG and Ab were dispersed into PBS buffer with the pH of 7.2 separately. Then, 5 mL of GPG with the Gd3+ concentration of 0.1 mM was mixed with 5 mL of 2 μg mL–1 Ab for dozens of minutes at room temperature, and the pH of the mixture was kept at 7.2 throughout the operation. After sufficient conjugation of the two materials, the GPG-Ab probe was prepared. The probe was stored in PBS buffer and at 4 ℃ for further use.
2.3. Characterization methods
Transmission electron microscopy (TEM) images were captured using a Hitachi H-8100 electron microscope (Hitachi, Tokyo, Japan) with a voltage of 80 kV. Atomic force microscopy (AFM) experiments were carried out using a Bruker Dimension Icon system. X-ray photoelectron spectroscopy (XPS) data was obtained using a PHI Quantera II system (Ulvac-PHI, INC, Japan). Raman spectrum was acquired with an inVia Raman system (Renishaw, UK). Malvern Zetasizer Nano-ZS90 was used to measure the zeta potentials.
2.4. T1 measurement with ULF NMR
ULF NMR was employed to measure the relaxation time T 1. As shown in Fig. S1, the home-made ULF NMR system consists of liquid helium cryostat, static field (B0) coils, earth’s field cancellation coils (Bc), excitation field (B1) coils, per-polarization field (Bp) coil, signal readout and data acquisition module, and pulse controller (not shown here). To date, the measurement field B0 can be adjusted from 47.0 to 234.9 μT, corresponding to a proton Larmor frequency (f L) ranging from 2 to 10 kHz. In this work, B0 was 118 μT, corresponding to f L = 5030 Hz. Before the T 1 measuring sequence, the Bp field of 87 m T, which was more than 2 orders of magnitude stronger than that of B0, was applied to enhance the signal amplitude. The NMR signals were acquired by a superconducting 2nd-order gradiometer inductively coupled to the ultra-sensitive superconducting quantum interface device (SQUID) immersed in liquid helium [29].
During the measurement, the sample was placed beneath the cryostat. The pulse sequence for the T 1 measurement is depicted in Fig. S2. The sample was first pre-polarized by Bp field for 500 ms (T p). After the Bp field was switched-off adiabatically, the sample magnetization freely relaxed in the B0 field for an evolution time . Then, π/2 and π pulses were applied to excite the spin-echo signals. Ten values were chosen to derive the T 1 values based on single-exponential decay fits of signal amplitudes vs. values.
3. Results and discussion
3.1. Strategy of the MRSw assay
In MRSw assays, the amount of targeted molecule can be distinguished by measuring the relaxation time changes due to the target-induced aggregation or disaggregation of MNPs [30,31]. Scheme 1 schematically shows the process of MRSw assay for SARS-CoV-2 pseudovirus detection. Firstly, the probe is formed by the connection of magnetic GPG to Ab via amidation, thus GPG is assembled into nanoaggregates. The prepared magnetic probe (GPG-Ab) can then specifically recognize the S protein on the surface of SARS-CoV-2 through antibody-antigen interaction. Considering the fact that the quantity of S protein on the surface of coronavirus is about 67 [32], GPG-Ab will bind to the viral surface and present in an aggregation state, which can lead to the change of T 1. During the entire testing process, no pretreatment is required. After the sample is mixed with the GPG-Ab, it is kept sealed throughout the whole measurement. The home-made ULF NMR relaxometry acquires the T 1 of a sample in 2 min by running the pulse sequence of T 1 measurement (Figs. S1 and 2, detailed description in Materials and methods section). By comparing the T 1s of the blank sample and the testing sample, the virus in the sample can be detected. It is worth noting that, when dozens of samples are in the queue for the test, the mixing of samples and probes can be initialized in parallel, which means the average detection time for each sample is approximately 2 min.
3.2. Characterization and magnetic dynamics of GPG-Ab
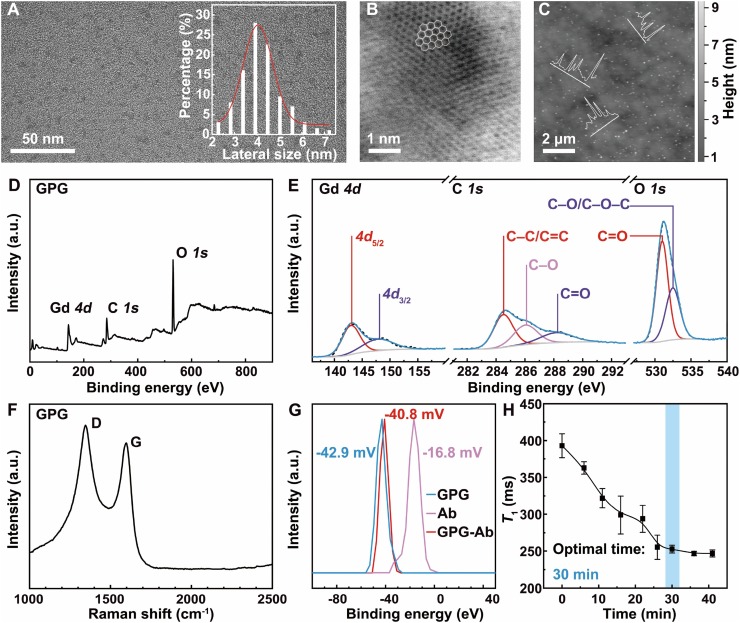
The GPG was prepared in accordance with the previously proposed method by modifing GQDs with PEG and Gd3+ via hydrothermal treatment [21]. As shown in Fig. 1 A, GPG has a uniform lateral size with an average diameter of 4.1 nm. No obvious change in lateral size can be found after the surface modification of GQDs (Figs. S3 and 4). Meanwhile, the image with atomic resolution (Fig. 1B) shows the typical honeycomb lattice structure of graphene, which indicates the excellent crystallinity of GPG [33]. The height of GPG ranging from 0.5 to 1.5 nm corresponds to the 1–4 layered structure (Fig. 1C).
Fig. 1.
(A) TEM image and size distribution histogram of GPG. (B) High-resolution TEM (HR-TEM) image of GPG. (C) AFM topography image of GPG on a 300 nm SiO2/Si substrate. Inset: height profile analysis along the line shown in the image. (D) XPS survey spectrum of GPG. (E) High-resolution XPS C 1s, O 1s, and Gd 4d spectra of GPG. (F) Raman spectrum of GPG. Two intense defect-related D and G peaks are found, centered at 1345.2 and 1597.0 cm–1, respectively. (G) Zeta potentials of GPG, Ab, and GPG-Ab. (H) T1s measured during the conjugation between GPG and Ab. T1 acquired at 0 min is measured from the pure GPG solution. Error bars indicate standard deviation (SD).
XPS survey spectrum shows that the oxygen-rich structure of GPG (Fig. 1D) exhibits a stoichiometry of C to O with the value of 1.1. Peaks located at 285.1, 531.1, and 141.1 eV can be attributed to the C 1s, O 1s, and Gd 4d signals, respectively. High-resolution XPS spectra of GPG are shown in Fig. 1E. In the C 1 s spectrum of GPG, peaks located at 284.5, 286.0, and 288.2 eV are found to be C−C/C=C, C−O, and C=O bonds, respectively [34]. Compared with the C 1s spectrum of GQDs (Fig. S5), the content of C−O bond increases due to the introduction of PEG6. Both the O 1s spectra of GQDs (Fig. S5) and GPG (Fig. 1E) show peaks located at 531.0 and 532.5 eV which can be attributed to C=O and C−O/C−O−C bonds, respectively [35]. The vanishing of O−C=O at 535.5 eV in GQDs is caused by the combination of GQDs and PEG6 via the esterification. These results also indicate the GPG has abundant oxygen-containing groups (i.e., oxhydryl, –OH; carboxyl, –COOH) [21]. Given that there are many amino (–NH2) groups on the surface of Ab, the abundant –COOH groups in the structure of GPG [36] could easily react with the –NH2 groups via amidation. The Gd 4d spectrum of GPG is illustrated in Fig. 1E. The peaks located at 142.9 and 147.8 eV indicate the presence of Gd3+ in the GPG, which brings magnetism into the structure. The Gd content is found to be 2.6 at. %, which means that magnetism is introduced into GPG. Raman spectroscopy reveals that the relative intensity of the D band to that of the G band (I D/I G) for GPG is 0.97 (Fig. 1F), which can be attributed to the relatively high quantity of GPG [37].
The connection of GPG to Ab is further confirmed by zeta potential measurement. As given in Fig. 1G, the zeta potentials of GPG and Ab are –42.9 and –16.8 mV, respectively. The zeta potential of GPG-Ab is –40.8 mV, indicating the stable composite structure between GPG and Ab. Moreover, such low zeta potential of GPG-Ab reveals the excellent water dispersibility of GPG-Ab [38].
In the magnetic dynamic study, the T 1 of GPG in PBS buffer was firstly measured to be 392.9 ± 16.3 ms (Fig. S6). After GPG was mixed with Ab for the formation of the probe, T 1 of the mixture was measured every 5 min. As plotted in Fig. 1H, T 1 decreases as the mixing time increases. After mixing for 30 min, T 1 remains at around 250.0 ms, which reveals that the conjugation between GPG and Ab is finished. Consequently, the T 1 of GPG-Ab is largely reduced after the combination, compared with that of GPG with the same Gd3+ concentration.
The T 1 change after the aggregation of magnetic nanoparticles is determined by two key parameters according to the outer sphere relaxation (OSR) theory: [39] the root-mean-square angular frequency shift Δωr at the particle surface, and the diffusion time τD=r2/D required for a water molecule to diffuse a distance 1.414 r in any specified direction, where r is the particle radius and D is the water diffusion coefficient. When the product of Δωr and τD is less than 1, the relaxation time will decrease after the aggregation of magnetic nanoparticles. Conversely, the relaxation time will exhibit an increase after the aggregation of the magnetic nanoparticles with the condition of τD⋅Δωr>1 [39,40]. Specifically, the relaxation time of the magnetic nanoparticles with a diameter of less than 10 nm will decrease after the aggregation, and the aggregation of the particles larger than 10 nm will lead to the increase of relaxation time after aggregation [41]. In the probe preparation of this MRSw assay, the magnetic GPG has a size smaller than 10 nm. After its conjugation with Ab, the T 1 of GPG-Ab nanoaggregate is thus reduced compared to that of GPG. The averaged diameter of GPG-Ab is 14.2 nm (Figs. S7‒9).
3.3. Performance evaluation of GPG-Ab in S protein detection
The stability of GPG-Ab was investigated by measuring the T 1 over 14 days. The probe was stored at 4 ℃ when it was not measured in the ULF NMR system. No obvious change of T 1 can be found in Fig. S10, which conveys that GPG-Ab stays stable after the storage for 14 days.
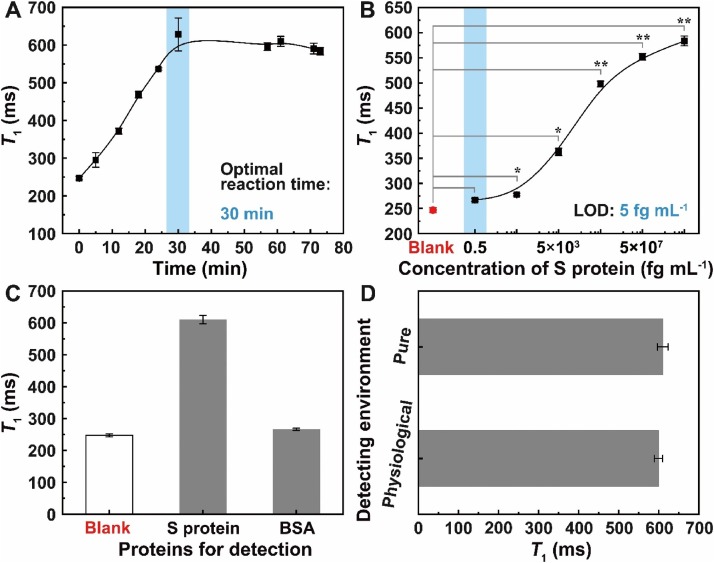
The optimal antibody-antigen reaction time was studied by detecting S protein with a concentration of 5 μg mL–1. As can be seen from Fig. 2 A, T 1 keeps increasing from 246.8 ± 4.7 ms to 609.8 ± 13.2 ms in the first 30 min because of the antibody-antigen binding kinetics. After that, T 1 maintains a constant value around 600 ms, which means that the interaction between Ab and S protein is sufficient. The significant difference in T 1 with and without S protein makes the as-designed MRSw a promising alternative for the rapid detection of S protein and SARS-CoV-2.
Fig. 2.
(A) T1s measured during the reaction procedure between GPG-Ab and S protein. (B) T1s measured in the detection of S protein with different concentrations. Error bars indicate SD. *P < 0.05 and **P < 0.01 are determined by Student’s T-test. (C) T1s measured in the detection of different proteins with the same concentration of 5 μg mL–1. (D) T1 comparison between the detection of S protein in the pure sample and the physiological sample.
The sensitivity, which is crucial for the as-designed MRSw, can be described in terms of the limit of detection (LOD). In this case, S protein with concentrations ranging from 0.5 fg mL–1 to 5 μg mL–1 was measured using the optimal antibody-antigen reaction time of 30 min. The LOD is evaluated from Fig. 2B at which concentration it has a longer T 1 than that of the blank sample (no S protein) plus 3 times the standard deviation, i.e., 0.5 fg mL–1, which is 7 orders of magnitude lower than that of the enzyme-linked immune-sorbent assay (ELISA) platform [6]. This result shows that the MRSw has high sensitivity for S protein detection.
The specificity of the MRSw was performed by detecting different proteins, i.e., S protein and bovine serum albumin (BSA) with the same concentration of 5 μg mL–1. Fig. 2C gives the comparison of T 1s acquired with the presence of different proteins. As a blank control, T 1 is fitted to be 246.8 ± 4.7 ms with no protein in the GPG-Ab solution. T 1 has increased to 609.8 ± 13.2 ms with 5 μg mL–1 of S protein. By contrast, with the addition of BSA to GPG-Ab solution, T 1 is found to be 266.1 ± 3.3 ms. The above data clearly demonstrate that the probe has high specificity towards S protein, in which the Ab specifically recognizes SARS-CoV-2.
To evaluate the interference immunity of GPG-Ab, the S protein was then dissolved in physiological (diluted human saliva) sample with a concentration of 5 μg mL–1 at room temperature. The measuring results in Fig. 2D show that even in a relatively complex environment, the T 1 value is still similar to that in the pure sample, which reveals that the detection can be performed in a nearly background-free manner.
3.4. Detection of SARS-CoV-2 pseudovirus with GPG-Ab
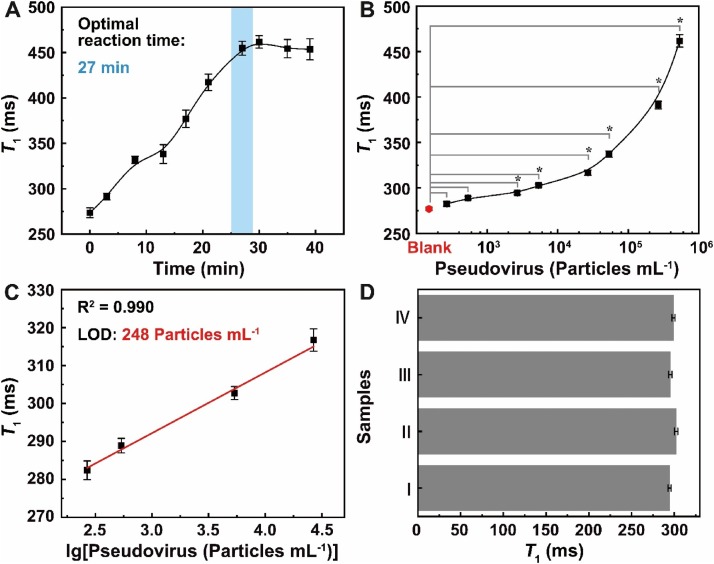
In order to verify that the GPG-Ab can be used to detect the virus, SARS-CoV-2 pseudovirus was introduced. The pseudoviruses have the essential components of spike protein for cell entry and viral infection, but lack nucleic acid and lose self-replication ability. Before the detection, the sample that contained SARS-CoV-2 pseudovirus was mixed with the GPG-Ab probe in a sample vial. As plotted in Fig. 3 A, T 1 increases with the growth of the reaction time in the first 27 min and then settles at 454.8 ± 7.6 ms at room temperature, which indicates that the reaction is completed within 27 min. Note that no sample-tube re-opening is required during the reaction and detection process, which significantly reduces the testers’ risk of infection. The simple operation allows finishing the virus detection in one step.
Fig. 3.
Detection of SARS-CoV-2 pseudovirus. (A) T1s that measured during the reaction procedure between GPG-Ab and pseudovirus. (B) Detection sensitivity for pseudovirus. Error bars indicate SD. *P < 0.05 is determined by Student’s T-test. (C) The linear relationship between T1 and the logarithm of pseudovirus concentration within the low concentration range. (D) Interference immunity study of GPG-Ab by measuring T1s at different detecting environments which have the same pseudovirus concentration of 2.7 × 103 Particles mL–1 (Ⅰ, in pure sample; Ⅱ, in diluted human saliva; Ⅲ, in tap water; Ⅳ, in sanitary sewage).
Using the optimal reaction time of 27 min, the sensitivity of GPG-Ab in detecting SARS-CoV-2 pseudovirus was investigated. Different concentrations of pseudovirus ranging from 2.7 × 102 to 5.4 × 105 Particles mL–1 were tested (Fig. 3B). With the increased concentration of pseudovirus, the difference between T 1s of pseudovirus-contained sample and blank sample becomes greater. In Fig. 3C, a linear fitting between T 1 and the logarithm of the concentration of SARS-CoV-2 pseudovirus is realized in the low range between 2.7 × 102 and 2.7 × 104 Particles mL–1 with R2 of 0.990. The LOD of GPG-Ab in SARS-CoV-2 pseudovirus detection is calculated from the calibration curve in Fig. 3C when T 1 equals to the T 1 of the blank sample plus 3 times the standard deviation [10] (282.7 ms) and corresponds to 248 Particles mL–1. Therefore, sensitive detection of SARS-CoV-2 pseudovirus using GPG-Ab without any sample pretreatment has been demonstrated. As a comparison (Table S1), the performance of the MRSw assay is outstanding compared to that of the newly developed methods for SARS-CoV-2 detection and is comparable to the one that have conducted the virus detection [6].
The interference immunity of the MRSw assay was performed by comparing the T 1s in different detecting environments, including the pure viral sample (Ⅰ), diluted human saliva (Ⅱ), tap water (Ⅲ), and sanitary sewage (Ⅳ). Samples Ⅰ‒Ⅳ have the same SARS-CoV-2 pseudovirus concentration of 2.7 × 103 Particles mL–1. As shown in Fig. 3D, T 1s of samples Ⅱ‒Ⅳ are 302.1 ± 1.9 ms, 295.3 ± 2.0 ms, and 299.1 ± 1.8 ms, respectively, which are of strong similarity with that of sample Ⅰ (294.41 ± 1.8 ms). The result reveals that GPG-Ab has interference immunity in different environments, which can significantly improve the applicability of the assay.
4. Conclusion
In summary, we have utilized magnetic GQDs with high relaxivity as the detection probe, and developed a GQDs-based MRSw for the rapid closed-tube one-step detection of SARS-CoV-2 pseudovirus based on ULF NMR relaxometry. With the help of magnetic GQDs and ULF NMR detection, the assay steps could be significantly simplified to one step. The MRSw-based SARS-CoV-2 detection can detect pseudovirus with a concentration as low as 248 Particles mL–1 within 2 min. The whole procedure does not require sample pretreatment and reopening of the test-tube, so that aerosol pollution is avoided, thus reducing the risk of infection for the clinical testers.
Considering the possible industrialization of this technique, the cost of the probe and the ULF NMR system should not be ignored. As listed in Table S2, the total cost of the GPG for a single test is only USD 1.25. Despite the relatively high cost of the system at current time, ULF NMR with portability can be installed on a truck, which is advantageous for collecting and testing samples in some rural areas lacking well-equipped hospitals and could bring more convenience to the public in the future. Besides, this newly developed assay methodology can be used for virus detection by NMR relaxometry with different static magnetic fields and easily transferred to the detection of other emerging viruses by replacing the antibody.
CRediT authorship contribution statement
Yongqiang Li: Conceptualization, Methodology, Validation, Investigation, Writing - original draft, Visualization. Peixiang Ma: Conceptualization, Investigation, Resources, Writing - review & editing, Funding acquisition. Quan Tao: Software, Validation, Resources. Hans-Joachim Krause: Writing - review & editing, Funding acquisition. Siwei Yang: Conceptualization, Methodology, Resources, Writing - review & editing, Funding acquisition. Guqiao Ding: Resources, Funding acquisition. Hui Dong: Conceptualization, Writing - review & editing, Supervision, Funding acquisition. Xiaoming Xie: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (11874378, 11804353, 11774368), Science and Technology Commission of Shanghai Municipality (19511107100), Guangzhou Institute of Respiratory Health Open Project (Funds provided by China Evergrande Group, 2020GIRHHMS05), Shanghai Local Grant (ZJ2020-ZD-004), and Mobility Programme of the Sino-German Center for Research Promotion (M-0022). The authors sincerely hope the world can soon get over the impact of COVID-19.
Biographies
Yongqiang Li is now a PhD student of Shanghai Institute of Microsystem and Information Technology (SIMIT), Chinese Academy of Science. He received his Bachelor’s degree in 2016 from Jilin University. His research interests are the synthsis, mechanism, and applications (biosensing, bioimaging) of magnetic graphene quantum dots.
Peixiang Ma works as research associated professor at Shanghai Institute for Advanced Immunochemical Studies (SIAIS) in ShanghaiTech University. He received his PhD from Heinrich-Heine-Universität Düsseldorf in 2011. His research focuses on the characterization of protein-ligand interaction, including protein-protein and protein-chemical.
Siwei Yang works as assistant research fellow at Shanghai Institute of Microsystem and Information Technology (SIMIT), Chinese Academy of Science. He received his PhD from SIMIT in 2017. Dr. Siwei Yang first prepared new 2D carbon-based semiconductor C3N in 2017. His research interests include controllable preparation, application (nanoelectronics, nanophotonics, and nanomedicine) of 2D carbon-based semiconductors, graphene quantum dots and other graphene derivatives.
Guqiao Ding is a professor at Shanghai Institute of Microsystem and Information Technology (SIMIT), Chinese Academy of Science. He received his BSc and MS from the Suzhou University and PhD from Shanghai Jiao Tong University. Prof. Dr. Guqiao Ding joined SIMIT in 2010. Research in his laboratory currently includes preparation and application of graphene, graphene oxide, graphene quantum dots and new 2D carbon-based materials.
Hui Dong is now a professor at Shanghai Institute of Microsystem and Information Technology (SIMIT), Chinese Academy of Science. She received her PhD degree from SIMIT in 2011. She worked as visiting scholar in Forschungszentrum Jülich (2008–2010) and University of California at Berkeley (2013–2014). Prof. Dr. Hui Dong mainly works on the development of superconducting quantum interface device (SQUID)-based ultra-low field (ULF) NMR/MRI system and its applications in biosensing/bioimaging and spectroscopic analysis.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2021.129786.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortorici M.A., Veesler D. Structural insights into coronavirus entry, complementary strategies to understand virus structure and function. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 7.Smither S.J., Eastaugh L.S., Findlay J.S., Lever M.S. Experimental aerosol survival of SARS-CoV-2 in artificial saliva and tissue culture media at medium and high humidity. Emerg. Microbes Infec. 2020;9:1415–1417. doi: 10.1080/22221751.2020.1777906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo Y.Y., Uspal W.E., Wei T. Airborne transmission of COVID-19: aerosol dispersion, lung deposition, and virus-receptor interactions. ACS Nano. 2020;14:16502–16524. doi: 10.1021/acsnano.0c08484. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y., Li Y.X., Jiang K., Wang J., White W.L., Yang S.P., et al. Rapid detection of listeria monocytogenes in food by biofunctionalized magnetic nanoparticle based on nuclear magnetic resonance. Food Control. 2017;71:110–116. [Google Scholar]
- 10.Lu W.J., Chen Y.P., Liu Z., Tang W.B., Feng Q., Sun J.S., et al. Quantitative detection of MicroRNA in one step via next generation magnetic relaxation switch sensing. ACS Nano. 2016;10:6685–6692. doi: 10.1021/acsnano.6b01903. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y.P., Xianyu Y.L., Wang Y., Zhang X.Q., Cha R.T., Sun J.S., et al. One-step detection of pathogens and viruses: combining magnetic relaxation switching and magnetic separation. ACS Nano. 2015;9:3184–3191. doi: 10.1021/acsnano.5b00240. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y.T., Medhi R., Nekrashevich I., Litvinov D., Xu S.J., Lee T.R. Specific detection of proteins using exceptionally responsive magnetic particles. Anal. Chem. 2018;90:6749–6756. doi: 10.1021/acs.analchem.8b00593. [DOI] [PubMed] [Google Scholar]
- 13.Chung H.J., Castro C.M., Im H., Lee H., Weissleder R. A magneto-DNA nanoparticle system for rapid detection and phenotyping of Bacteria. Nat. Nanotechnol. 2013;8:369–375. doi: 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bamrungsap S., Shukoor M.I., Chen T., Sefah K., Tan W.H. Detection of lysozyme magnetic relaxation switches based on aptamer-functionalized superparamagnetic nanoparticles. Anal. Chem. 2011;83:7795–7799. doi: 10.1021/ac201442a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M.-H., Son H.-Y., Kim G.-Y., Park K., Huh Y.-M., Haam S. Redoxable heteronanocrystals functioning magnetic relaxation switch for activatable T1 and T2 dual-mode magnetic resonance imaging. Biomaterials. 2016;101:121–130. doi: 10.1016/j.biomaterials.2016.05.054. [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Ma P.X., Dong H., Krause H.J., Zhang Y., Willbold D., et al. A magnetic nanoparticles relaxation sensor for protein-protein interaction detection at ultra-low magnetic field. Biosens. Bioelectron. 2016;80:661–665. doi: 10.1016/j.bios.2016.02.037. [DOI] [PubMed] [Google Scholar]
- 17.Xianyu Y.L., Dong Y.Z., Zhang Z., Wang Z.H., Yu W.B., Wang Z.L., et al. Gd3+-nanoparticle-enhanced multivalent biosensing that combines magnetic relaxation switching and magnetic separation. Biosens. Bioelectron. 2020;155 doi: 10.1016/j.bios.2020.112106. [DOI] [PubMed] [Google Scholar]
- 18.Li J.J., Wang S., Wu C., Dai Y., Hou P.F., Han C.P., et al. Activatable molecular MRI nanoprobe for tumor cell imaging based on gadolinium oxide and Iron oxide nanoparticle. Biosens. Bioelectron. 2016;86:1047–1053. doi: 10.1016/j.bios.2016.07.044. [DOI] [PubMed] [Google Scholar]
- 19.Wang F.H., Bae K., Huang Z.W., Xue J.M. Two-photon graphene quantum dot modified Gd2O3 nanocomposites as a dual-mode MRI contrast agent and cell labelling agent. Nanoscale. 2018;10:5642–5649. doi: 10.1039/c7nr08068a. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y.Q., Chen S.Z., Li H.D., Yuan Y.P., Zhang Z.Y., Xie J.S., et al. Engineered paramagnetic graphene quantum dots with enhanced relaxivity for tumor imaging. Nano Lett. 2019;19:441–448. doi: 10.1021/acs.nanolett.8b04252. [DOI] [PubMed] [Google Scholar]
- 21.Li Y.Q., Dong H., Tao Q., Ye C.C., Yu M.M., Li J.P., et al. Enhancing the magnetic relaxivity of MRI contrast agents via the localized superacid microenvironment of graphene quantum dots. Biomaterials. 2020;250 doi: 10.1016/j.biomaterials.2020.120056. [DOI] [PubMed] [Google Scholar]
- 22.Huang R.Q., Tao Q., Chang B.L., Dong H. Field dependence study of commercial Gd chelates with SQUID detection. IEEE Trans. Appl. Supercond. 2016;26 [Google Scholar]
- 23.Theis T., Blanchard J.W., Butler M.C., Ledbetter M.P., Budker D., Pines A. Chemical analysis using J-coupling multiplets in zero-field NMR. Chem. Phys. Lett. 2013;580:160–165. doi: 10.1063/1.4803144. [DOI] [PubMed] [Google Scholar]
- 24.Elliott D.F., Schumacher R.T. Proton resonance of fluorobenzene in the earth’s magnetic field. J. Chem. Phys. 1957;26:1350. [Google Scholar]
- 25.Huang X.L., Dong H., Tao Q., Yu M.M., Li Y.Q., Rong L.L., et al. Sensor configuration and algorithms for power-line interference suppression in low field nuclear magnetic resonance. Sensors. 2019;19:3566. doi: 10.3390/s19163566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halse M.E., Callaghan P.T. A dynamic nuclear polarization strategy for multi-dimensional earth’s field NMR spectroscopy. J. Magn. Reson. 2008;195:162–168. doi: 10.1016/j.jmr.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Macovski A., Conolly S. Novel approaches to low-cost MRI. Magn. Reson. Med. 1993;30:221–230. doi: 10.1002/mrm.1910300211. [DOI] [PubMed] [Google Scholar]
- 28.Lee S.K., Möβle M., Myers W., Kelso N., Trabesinger A.H., Pines A., et al. SQUID-detected MRI at 132 μT with T1-weighted contrast established at 10 μT–300 mT. Magn. Reson. Med. 2005;53:9–14. doi: 10.1002/mrm.20316. [DOI] [PubMed] [Google Scholar]
- 29.Yu M.M., Tao Q., Dong H., Huang T., Li Y.Q., Xiao Y., et al. Ultra-low noise graphene/copper/nylon fabric for electromagnetic interference shielding in ultra-low field magnetic resonance imaging. J. Magn. Reson. 2020;317 doi: 10.1016/j.jmr.2020.106775. [DOI] [PubMed] [Google Scholar]
- 30.Ling Y.B., Pong T., Vassiliou C.C., Huang P.L., Cima M.J. Implantable magnetic relaxation sensors measure cumulative exposure to cardiac biomarkers. Nat. Biotechnol. 2011;29:273–277. doi: 10.1038/nbt.1780. [DOI] [PubMed] [Google Scholar]
- 31.Perez J.M., Simeone F.J., Saeki Y., Josephson L., Weissleder R. Viral-induced self-assembly of magnetic nanoparticles allows the detection of viral particles in biological media. J. Am. Chem. Soc. 2003;125:10192–10193. doi: 10.1021/ja036409g. [DOI] [PubMed] [Google Scholar]
- 32.Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W.T., Guo H.Z., Li G., Chi Z., Chen H.L., Wang L., et al. White luminescent single-crystalline chlorinated graphene quantum dots. Nanoscale Horiz. 2020;5:928–933. doi: 10.1039/d0nh00053a. [DOI] [PubMed] [Google Scholar]
- 34.Guo H.Z., Wen S.K., Li W.T., Li M., Wang L., Chang Q., et al. A universal strategy to separate hydrophilic hybrid-light carbon quantum dots using pure water as eluent. Appl. Mater. Today. 2020;18 [Google Scholar]
- 35.Han Y., Tang B.J., Wang L., Bao H., Lu Y.H., Guan C.T., et al. Machine-learning-driven synthesis of carbon dots with enhanced quantum yields. ACS Nano. 2020;14:14761–14768. doi: 10.1021/acsnano.0c01899. [DOI] [PubMed] [Google Scholar]
- 36.Huang H., Yang S.W., Liu Y., Yang Y.C., Li H., McLeod J.A., et al. Photocatalytic polymerization from amino acid to protein by carbon dots at room temperature. ACS Appl. Bio Mater. 2019;2:5144–5153. doi: 10.1021/acsabm.9b00805. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., Li W.T., Yin L.Q., Liu Y.J., Guo H.Z., Lai J.W., et al. Full-color fluorescent carbon quantum dots. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J.P., Yang S.W., Deng Y., Chai P.W., Yang Y.C., He X.Y., et al. Emancipating target-functionalized carbon dots from autophagy vesicles for a novel visualized tumor therapy. Adv. Funct. Mater. 2018;28 [Google Scholar]
- 39.Gillis P., Moiny F., Brooks R.A. On T2-shortening by strongly magnetized spheres: a partial refocusing model. Magn. Reson. Med. 2002;47:257–263. doi: 10.1002/mrm.10059. [DOI] [PubMed] [Google Scholar]
- 40.Jun Y.W., Huh Y.M., Choi J.S., Lee J.H., Song H.T., Kim S., et al. Nanoscale size effect of magnetic nanocrystals and their utilization for cancer diagnosis via magnetic resonance imaging. J. Am. Chem. Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 41.Min C., Shao H.L., Liong M., Yoon T.J., Weissleder R., Lee H. Mechanism of magnetic relaxation switching sensing. ACS Nano. 2012;6:6821–6828. doi: 10.1021/nn301615b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.