Abstract
Approximately 20 Leishmania species are known to cause cutaneous, mucocutaneous, and visceral disorders in humans. Identification of the causative species in infected individuals is important for appropriate treatment and a favorable prognosis because infecting species are known to be the major determinant of clinical manifestations and may affect treatments for leishmaniasis. Although Leishmania species have been conventionally identified by multilocus enzyme electrophoresis, genetic analysis targeting kinetoplast and nuclear DNA (kDNA and nDNA, respectively) is now widely used for this purpose. Recently, we conducted countrywide epidemiological studies of leishmaniasis in Ecuador and Peru to reveal prevalent species using PCR-RFLP targeting nDNA, and identified unknown hybrid parasites in these countries together with species reported previously. Furthermore, comparative analyses of kDNA and nDNA revealed the distribution of parasites with mismatches between these genes, representing the first report of mito-nuclear discordance in protozoa. The prevalence of an unexpectedly high rate (~10%) of genetically complex strains including hybrid strains, in conjunction with the observation of mito-nuclear discordance, suggests that genetic exchange may occur more frequently than previously thought in natural Leishmania populations. Hybrid Leishmania strains resulting from genetic exchanges are suggested to cause more severe clinical symptoms when compared with parental strains, and to have increased transmissibility by vectors of the parental parasite species. Therefore, it is important to clarify how such genetic exchange influences disease progression and transmissibility by sand flies in nature. In addition, our aim was to identify where and how the genetic exchange resulting in the formation of hybrid and mito-nuclear discordance occurs.
Keywords: Leishmania, hybrid, mito-nuclear discordance, genetic exchange, Ecuador, Peru
Introduction
Human leishmaniasis is caused by approximately 20 species of the genus Leishmania belonging to the subgenera Leishmania (Leishmania), Leishmania (Viannia), and Leishmania (Mundinia) (Paranaiba et al., 2017; Ruiz-Postigo et al., 2020). The clinical presentation is varied, ranging from a localized cutaneous lesion to a potentially fatal visceral disorder, and the infecting parasite species is the major determinant of the outcome (Ruiz-Postigo et al., 2020). Importantly, several L. braziliensis complex species, such as Leishmania (Viannia) braziliensis and L. (V.) guyanensis, are associated with a risk of metastasizing destructive mucosal lesions after healing of the primary cutaneous lesion (Ruiz-Postigo et al., 2020). In addition, for cutaneous leishmaniasis, variability in disease severity and susceptibility to treatment may be as sociated with the infecting parasite species, although the characteristic cutaneous lesions caused by each infecting species have yet to be determined. Therefore, identification of the causative Leishmania species is important for appropriate treatment and a favorable prognosis.
Leishmania species have been classified by multilocus enzyme electrophoresis (MLEE) as the reference protocol (Rioux et al., 1990; Cupolillo et al., 1994). This method requires parasite isolation in culture, which is time-consuming and associated with risks of contamination with bacteria and fungi on sample collection, and interfusion of other cultures during long-term cultivation. Recently, the application of molecular biological techniques using samples directly obtained from patients’ lesions has facilitated rapid and efficient identification of the parasite species. Kinetoplast DNA (kDNA) is a unique mitochondrial structure found in trypanosomatid parasites, containing 20–50 copies of maxicircle DNA and approximately 10,000 copies of minicircle DNA (Simpson, 1986). Because of the multicopy property, kDNA is widely used as a target for detection and identification of Leishmania species. Although minicircle DNA is more sensitive for detection, it is heterogeneous in sequence. Therefore, maxicircle genes, such as cytochrome b (cyt b), cytochrome c oxidase subunits, and NADH dehydrogenase subunits, are preferentially used as targets for species identification; cyt b gene sequence analysis is widely used and accepted as a reliable marker for this purpose (Luyo-Acero et al., 2004; Asato et al., 2009; Kato et al., 2010; Kato et al., 2011; Leelayoova et al., 2013; Kato et al., 2016a; Kato et al., 2019a). Similarly, among nuclear DNA (nDNA) targets, internal transcribed spacer (ITS) regions of ribosomal RNA and heat shock protein 70 (hsp70) are commonly used for species identification, due to their sensitivity for detection of interspecific sequence divergence (da Silva et al., 2010; Talmi-Frank et al., 2010; de Almeida et al., 2011; Fraga et al., 2012; Montalvo et al., 2012). Generally, genetic analysis of a single target is considered acceptable for reliable identification at the species level; however, analysis of multiple targets may increase accuracy. Furthermore, analysis of single targets may not detect strains that are the product of recombination between different species.
Recently, countrywide surveillances were performed in Ecuador and Peru using cyt b gene analysis, and geographic distributions of Leishmania species were identified (Kato et al., 2010; Kato et al., 2016a; Kato et al., 2019a). Furthermore, comparative analyses of kDNA and nDNA revealed the prevalence of genetically complex Leishmania including hybrids and strains with mismatches between these genes, known as mito-nuclear discordance, at an unexpectedly high rate (~10%) in these countries (Kato et al., 2019b; Tabbabi et al., 2020). In this review, we describe the genetic complexity of Leishmania strains found in Ecuador and Peru that showed hybrid and mito-nuclear discordance characteristics, and discuss where and how such genetic exchange occurs, and its influence on disease severity and expansion of potential vector species.
Leishmaniasis in Ecuador and Perú, and Identification of Causative Species Based on Cytochrome B Gene Analysis
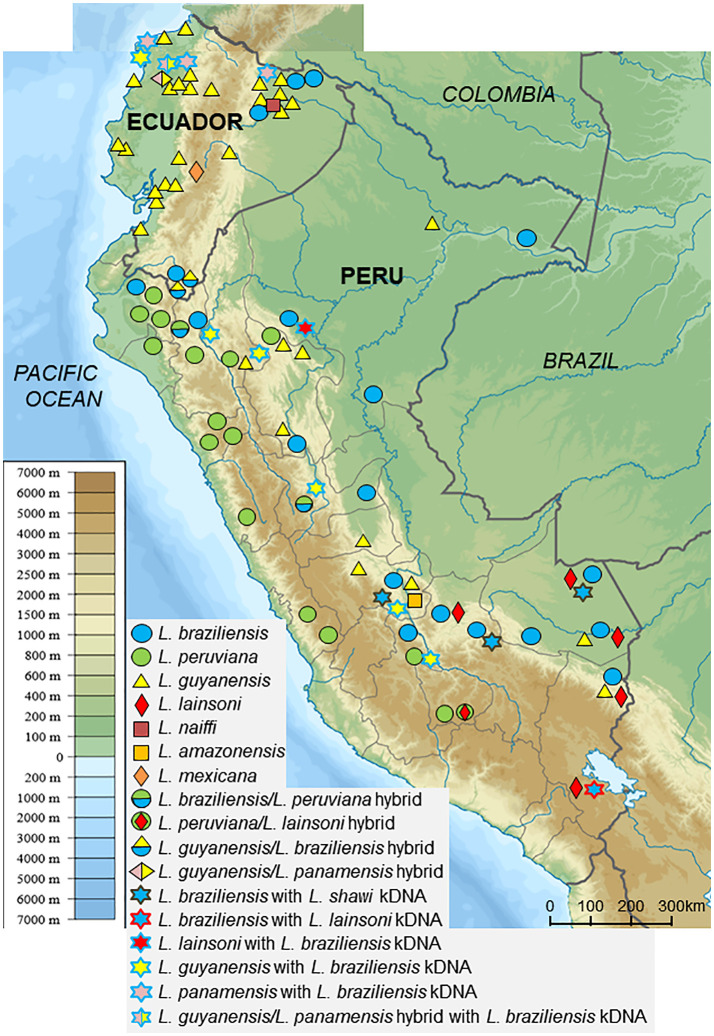
Ecuador is a relatively small country located on the Equator in northwestern South America. The country includes four ecological regions, each with a unique biodiversity and ecosystem: the Pacific coast subtropical areas, Andean highlands, Amazonian rainforest, and Galapagos Islands. Leishmaniasis is endemic in the first three regions (Hashiguchi et al., 2017). Up to the present, eight Leishmania species: L. (V.) guyanensis, L. (V.) panamensis, L. (V.) braziliensis, L. (V.) naiffi, L. (V.) lainsoni, L. (L.) mexicana, L. (L.) amazonensis, and L. (L.) major-like, have been recorded as responsible for cutaneous leishmaniasis (CL) and mucocutaneous leishmaniasis (MCL) (Kato et al., 2016a; Hashiguchi et al., 2017; Kato et al., 2019b) ( Figure 1 ). On the Pacific coast, L. (V.) guyanensis is the dominant causative agent, and infections by L. (V.) panamensis and L. (V.) braziliensis also have been reported. In addition, the distribution of L. (L.) amazonensis has been recorded in certain areas, although infection by it has not been reported recently (Kato et al., 2016a; Hashiguchi et al., 2017). In Amazonian areas, CL and MCL caused by L. (V.) guyanensis and L. (V.) braziliensis have been widely recorded, and CL caused by L. (V.) naiffi and L. (V.) lainsoni was recently reported in several areas (Kato et al., 2013; Kato et al., 2016a; Kato et al., 2016b). In the Andean highlands, areas endemic for CL are limited to the mid-southwestern part of Ecuador, and L. (L.) mexicana is currently the major causative species, whereas infection by L. (L.) major-like was reported previously (Kato et al., 2016a; Hashiguchi et al., 2017; Hashiguchi et al., 2018) ( Figure 1 ). In addition, a hybrid of L. (V.) guyanensis and L. (V.) braziliensis was recorded in southern parts (Bañuls et al., 1997). The observed variety in species and hybrids that cause CL in this relatively small country may reflect the extensive ecological and biological diversities, including among sand fly vectors and reservoir animals. In Ecuador, CL is the most frequently observed form of leishmaniasis, most commonly presenting as an ulcer, followed by popular, nodular, and atypical forms, including diffuse and disseminated lesions, and recidiva cutis (Hashiguchi et al., 2016; Hashiguchi et al., 2017). The characteristic presenting symptoms have not been defined for all infecting species, however, diffuse and disseminated forms of leishmaniasis are caused by L. (L.) mexicana and L. (V.) guyanensis, respectively (Hashiguchi et al., 2017).
Figure 1.
Geographic distribution of Leishmania species in Ecuador and Peru. Leishmania species in clinical samples were identified by sequence analysis of kinetoplast DNA and PCR-RFLP and sequence analyses of nuclear DNA. (Adapted from maps available at https://commons.wikimedia.org/wiki/File:Peru_physical_map.svg and https://commons.wikimedia.org/wiki/File:Ecuador_relief_location_map.svg).
Peru, a larger country located to the south of Ecuador, similarly includes Pacific coast, Andean highland, and Amazonian rainforest regions. Peru is one of the most highly endemic countries for CL, which occurs throughout the country from highlands to lowlands. In contrast, MCL is endemic to Amazonian areas (Kato et al., 2010; Kato et al., 2019a). Six Leishmania species and several hybrids have been recorded as responsible for leishmaniasis (Kato et al., 2010; Kato et al., 2019a; Tabbabi et al., 2020) ( Figure 1 ). Of these, the main causative agents are L. (V.) peruviana, L. (V.) braziliensis, and L. (V.) guyanensis, mainly circulating in the Andean highlands, tropical rainforest, and northern to central rainforest areas, respectively (Kato et al., 2010; Kato et al., 2019a) ( Figure 1 ). In addition to the three dominant species, L. (V.) lainsoni and L. (L.) amazonensis have been reported to be cause disease in lower rainforest areas, and L. (V.) shawi was recently identified as a rare and sporadic species responsible for CL based on cyt b gene sequence analysis (Kato et al., 2010; Kato et al., 2019a) ( Figure 1 ). Furthermore, a hybrid of L. (V.) braziliensis and L. (V.) peruviana first recorded in 1995 in a central area, was recently reported in northern Peru (Dujardin et al., 1995; Koarashi et al., 2016; Kato et al., 2019a) ( Figure 1 ). Unlike Ecuador, CL is highly endemic throughout Andean areas in Peru. Additionally, the cutaneous lesions of patients in the Peruvian Andes were commonly larger and more severe when compared with those of patients in the Ecuadorian Andes (Hashiguchi et al., 2018).
Comparative Nuclear and Kinetoplast DNA Analyses Reveal Genetically Complex Leishmania Strains With Hybrid and Mito-Nuclear Discordance
Sequence analysis targeting kDNA and nDNA is a powerful and reliable tool for the identification of infecting Leishmania species; however, it requires costly reagents and equipment. Therefore, cost-effective alternatives are preferable for more practical use applicable in less-equipped laboratories/countries. Of these, PCR-restriction fragment length polymorphism (RFLP) analysis is a promising candidate. In addition, PCR-RFLP allows analysis of heterozygous multicopy regions. For this purpose, nDNA is considered to be a more suitable target than kDNA, due to the potential effect of polymorphisms in the kDNA sequences of both minicircle and maxicircle genes on restriction fragment patterns. To date, ITS regions of ribosomal RNA and the hsp70 gene have been widely used as targets due to their sensitivity, specificity, and reliability (Garcia et al., 2004; Rotureau et al., 2006; Spanakos et al., 2008; Fraga et al., 2012; Khanra et al., 2012; Fraga et al., 2013; Mouttaki et al., 2014). In addition, PCR-RFLP analyses targeting leishmanial mannose phosphate isomerase (mpi) and 6-phosphogluconate dehydrogenase (6pgd) genes, both of which have been used for multilocus sequence typing (MLST), were recently established, and their reliability for species identification was confirmed (Kato et al., 2019b).
In recent studies, PCR-RFLP analyses of nDNA were applied to 92 and 134 clinical samples from Ecuador and Peru, respectively, and the results were compared with those obtained by kDNA sequence analyses (Kato et al., 2019b; Tabbabi et al., 2020). Interestingly, results that were consistent between the two analyses were obtained only for about 90% of samples each, from Ecuador and Peru (90.2 and 87.3%, respectively). On the other hand, five Ecuadorian samples showed hybrid patterns by PCR-RFLP, and were identified as hybrid strains of L. (V.) guyanensis/L. (V.) braziliensis and L. (V.) guyanensis/L. (V.) panamensis (Kato et al., 2019b). Similarly, six Peruvian samples showing hybrid RFLP patterns were identified as hybrids of L. (V.) braziliensis/L. (V.) peruviana and L. (V.) peruviana/L. (V.) lainsoni (Tabbabi et al., 2020) ( Figure 1 ). Furthermore, these studies unexpectedly identified strains showing incompatibility between kDNA and nDNA, known as mito-nuclear discordance, which had not been reported previously in protozoa, in five Ecuadorian (5.4%) and eleven Peruvian (8.2%) samples (Kato et al., 2019b; Tabbabi et al., 2020). These samples were identified as hybrids of L. (V.) guyanensis/L. (V.) panamensis with L. (V.) braziliensis kDNA, L. (V.) guyanensis with L. (V.) braziliensis kDNA, and L. (V.) panamensis with L. (V.) braziliensis kDNA in Ecuador, and of L. (V.) guyanensis with L. (V.) braziliensis kDNA, L. (V.) braziliensis with L. (V.) shawi kDNA, L. (V.) braziliensis with L. (V.) lainsoni kDNA, and L. (V.) lainsoni with L. (V.) braziliensis kDNA, in Peru. Interestingly, strains with mito-nuclear discordance were detected from geographically separate areas, rather than from delimited areas in both countries ( Figures 1 and 2 ). The distribution of unexpectedly high rates of hybrid or mito-nuclear discordance strains in both Ecuador and Peru indicates that the genetic structure of Leishmania is more complex than expected. In addition, these results suggest that interspecific genetic exchange occurs at a certain frequency in nature. It is important to note that all strains with mito-nuclear discordance are associated with L. (V.) braziliensis, suggesting that the species may have characteristics promoting genetic exchange with other species.
Figure 2.
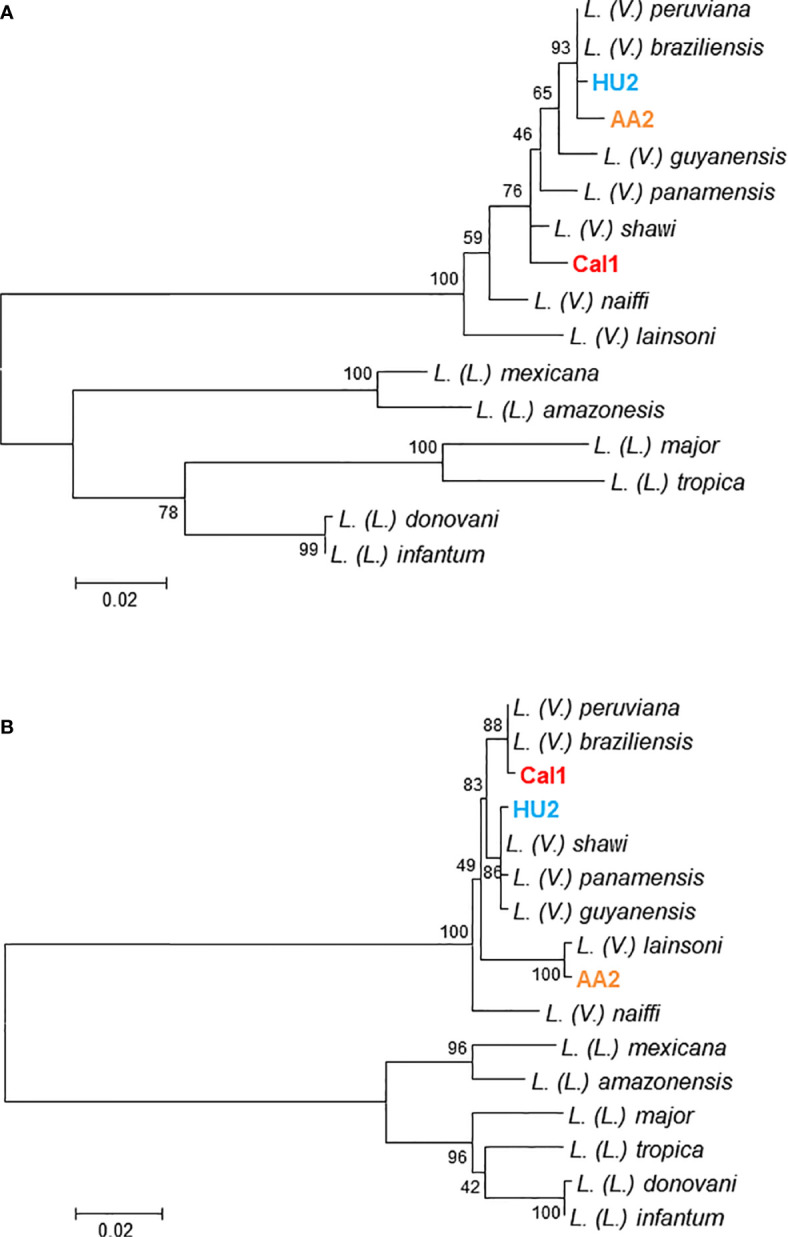
Discordance between cytochrome b and mannose phosphate isomerase gene sequences in clinical samples. Leishmanial cytochrome b (A) and mannose phosphate isomerase (B) genes were determined from clinical samples of patients with cutaneous leishmaniasis (HU2, AA2, and Cal1), and phylogenetic analyses were performed by the maximum likelihood method together with those sequences from 13 Leishmania species. The scale bar represents 0.02% divergence. Bootstrap values are shown above or below branches. Parasites were identified as Leishmania (Viannia) guyanensis with L. (V.) braziliensis kDNA, L. (V.) lainsoni with L. (V.) braziliensis kDNA, and L. (V.) shawi with L. (V.) braziliensis kDNA in clinical samples, HU2, AA2, and Cal1, respectively.
Where and How Does Genetic Exchange Occur?
Hybrids of Leishmania species such as L. (V.) braziliensis/L. (V.) guyanensis, L. (L.) infantum/L. (L.) major, and L. (L.) donovani/L. (L.) aethiopica have been reported in other countries (Delgado et al., 1997; Ravel et al., 2006; Odiwuor et al., 2011). Recently, genome-scale analyses provided evidence of meiotic-like recombination between Leishmania species, resulting in full-genome hybrids (Van den Broeck et al., 2020). Interestingly, this study also showed that the mitochondrial genome of hybrid strains consisted of homogeneous uniparental maxicircles, whereas minicircles originated from both parental species (Van den Broeck et al., 2020).
The mechanisms of genetic exchange in Leishmania resulting in the formation of hybrid and mito-nuclear discordance, and where and how they occur, are still unclear. In Peru, the natural hybridization between L. (V.) braziliensis and L. (V.) peruviana is hypothesized to be associated with a massive migration of people and animals between highland and lowland areas, due to the deterioration and recovery of the political and security situation (Kato et al., 2016c; Van den Broeck et al., 2020). A resulting increased risk for infection by multiple Leishmania species in humans and animals, is thought to give rise to the emergence and establishment of hybrid strains. A hybrid of Leishmania can be generated experimentally in sand fly vectors by co-infecting them with two different strains of the same Leishmania species (Akopyants et al., 2009; Sadlova et al., 2011; Calvo-Álvarez et al., 2014), and in vitro by co-culture of different strains of L. (L.) tropica promastigotes, a stage in the sand fly vector lifecycle (Louradour et al., 2020). In addition, direct evidence of sexual recombination in natural populations was provided by whole genome sequencing of Leishmania isolated from sand flies (Rogers et al., 2014). The midgut adhesion molecule of sand flies is believed to be a major determinant of parasite-vector specificity by supporting species-specific parasite attachment and their growth (Kamhawi et al., 2004). Therefore, interspecific hybrid formation is considered to occur within sand flies if the two parasite species share the midgut molecule for their attachment, which may be possible between closely-related species such as L. (V.) braziliensis and L. (V.) peruviana, and L. (V.) guyanensis and L. (V.) panamensis. However, genetic exchange will not occur between more distantly-related Leishmania species within a sand fly, when its affinities differ for those Leishmania species. Therefore, the potential for genetic exchange within reservoirs with subsequent hybrid formation should be considered.
Mito-Nuclear Discordance
In addition to hybrid species, recent studies have reported the presence of Leishmania strains showing discordance between kDNA and nDNA at a relatively high rate; this represents the first report of mito-nuclear discordance in protozoan parasites (Kato et al., 2019b; Tabbabi et al., 2020). It is not known where and how such incompatibility occurs, or whether the mechanism is the same as for the hybrid formation of nDNA.
Mitochondrial and nuclear genomes co-exist in each cell; however, the rate of evolution of mitochondrial DNA (mtDNA) is more rapid than that of nDNA (Toews and Brelsford, 2012). Incompatibility between mtDNA and nDNA has been reported in various organisms including mammals, birds, reptiles, amphibians, fish, insects, and yeasts (Toews and Brelsford, 2012). It has also been reported in helminth parasites, between Schistosoma turkestanicum populations (Lawton et al., 2017), between Taenia solium lineages (Yanagida et al., 2014), and between T. saginata and T. asiatica (Yamane et al., 2012; Yamane et al., 2013; Sato et al., 2018). Mito-nuclear discordance is considered to result from various processes such as adaptive introgression of mtDNA, demographic disparities, sex-biased asymmetries, hybrid zone movement, an intracellular bacteria, Wolbachia infection in insects, and human actions (Toews and Brelsford, 2012). It is well known that mitochondria play essential roles in cellular energy production, cellular proliferation, and many other metabolic functions (McBride et al., 2006). Although mitochondria contain their own DNA independently, the interaction between mitochondrial and nuclear genomes is important for biological functions of the cell (McBride et al., 2006; Ali et al., 2019). Therefore, it is considered that exchange of kDNA possibly affects pathogenicity and transmission potential by sand flies of Leishmania protozoa, as suggested in a hybrid strain (Cortes et al., 2012). Further studies that involve isolating parasite strains with mito-nuclear discordance are expected to elucidate these issues and provide further insight into the mechanism of genetic exchange between Leishmania protozoa.
Concluding Remarks
This review describes the genetic exchange that results in the establishment of hybrid strains and mito-nuclear discordance in Leishmania under natural conditions. Formation of a hybrid strain was suggested to increase the severity of disease when compared with parental species in an experimental animal model (Cortes et al., 2012). In addition, hybrid strains can increase the potential for sand fly transmission (Volf et al., 2007; Seblova et al., 2015). It is not yet well-established whether strains showing mito-nuclear discordance have increased pathogenicity or vector range. Isolation of strains with mito-nuclear discordance and further studies on the infection in animals and sand flies will be necessary to clarify these issues. Since mitochondria are organelles that are essential for cell energy supply, differentiation, and growth, (McBride et al., 2006), the genetic exchange resulting in mito-nuclear discordance could affect disease progression, as well as modify the potential for transmission by sand flies. Finally, the development of hybrids and strains with mito-nuclear discordance may have biological significance for parasite evolution and adaptation.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This study was financially supported by the Ministry of Education, Culture and Sports, Science and Technology (MEXT) of Japan (Grant No. 17H01685).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Akopyants N. S., Kimblin N., Secundino N., Patrick R., Peters N., Lawyer P., et al. (2009). Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324, 265–268. 10.1126/science.1169464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A. T., Boehme L., Carbajosa G., Seitan V. C., Small K. S., Hodgkinson A. (2019). Nuclear genetic regulation of the human mitochondrial transcriptome. eLife 8, e41927. 10.7554/eLife.41927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asato Y., Oshiro M., Myint C. K., Yamamoto Y., Kato H., Marco J. D., et al. (2009). Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp. Parasitol. 121, 352–361. 10.1016/j.exppara.2008.12.013 [DOI] [PubMed] [Google Scholar]
- Bañuls A. L., Guerrini F., Le Pont F., Barrera C., Espinel I., Guderian R., et al. (1997). Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J. Eukaryot. Microbiol. 44, 408–411. 10.1111/j.1550-7408.1997.tb05716.x [DOI] [PubMed] [Google Scholar]
- Calvo-Álvarez E., Álvarez-Velilla R., Jiménez M., Molina R., Pérez-Pertejo Y., Balaña-Fouce R., et al. (2014). First evidence of intraclonal genetic exchange in trypanosomatids using two Leishmania infantum fluorescent transgenic clones. PLoS Negl. Trop. Dis. 8, e3075. 10.1371/journal.pntd.0003075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes S., Esteves C., Mauricio I., Maia C., Cristovão J. M., Miles M., et al. (2012). In vitro and in vivo behavior of sympatric Leishmania (V.) braziliensis, L. (V.) peruviana and their hybrids. Parasitology 139, 191–199. 10.1017/S0031182011001909 [DOI] [PubMed] [Google Scholar]
- Cupolillo E., Grimaldi G., Momen H. (1994). A general classification of New World Leishmania using numerical zymotaxonomy. Am. J. Trop. Med. Hyg. 50, 296–311. 10.4269/ajtmh.1994.50.296 [DOI] [PubMed] [Google Scholar]
- da Silva L. A., de Sousa C., dos S., da Grac G. C., Porrozzi R., Cupolillo E. (2010). Sequence analysis and PCR RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect. Genet. Evol. 10, 77–83. 10.1016/j.meegid.2009.11.001 [DOI] [PubMed] [Google Scholar]
- de Almeida M. E., Steurer F. J., Koru O., Herwaldt B. L., Pieniazek N. J., da Silva A. J. (2011). Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J. Clin. Microbiol. 49, 3143–3149. 10.1128/JCM.01177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado O., Cupolillo E., Bonfante-Garrido R., Silva S., Belfort E., Júnior G. G., et al. (1997). Cutaneous leishmaniasis in Venezuela caused by infection with a new hybrid between Leishmania (Viannia) braziliensis and L. (V.) guyanensis . Mem. Inst. Oswaldo Cruz 92, 581–582. 10.1590/s0074-02761997000500002 [DOI] [PubMed] [Google Scholar]
- Dujardin J. C., Bañuls A. L., Llanos-Cuentas A., Alvarez E., DeDoncker S., Jacquet D., et al. (1995). Putative Leishmania hybrids in the Eastern Andean valley of Huanuco, Peru. Acta Trop. 59, 293–307. 10.1016/0001-706x(95)00094-u [DOI] [PubMed] [Google Scholar]
- Fraga J., Veland N., Montalvo A. M., Praet N., Boggild A. K., Valencia B. M., et al. (2012). Accurate and rapid species typing from cutaneous and mucocutaneous leishmaniasis lesions of the New World. Diagn. Microbiol. Infect. Dis. 74, 142–150. 10.1016/j.diagmicrobio.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Fraga J., Montalvo A. M., Maes L., Dujardin J. C., Van der Auwera G. (2013). HindII and SduI digests of heat-shock protein 70 PCR for Leishmania typing. Diagn. Microbiol. Infect. Dis. 77, 245–247. 10.1016/j.diagmicrobio.2013.07.023 [DOI] [PubMed] [Google Scholar]
- Garcia L., Kindt A., Bermudez H., Llanos-Cuentas A., De Doncker S., Arevalo J., et al. (2004). Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J. Clin. Microbiol. 42, 2294–2297. 10.1128/jcm.42.5.2294-2297.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y., Gomez E. L., Kato H., Martini L. R., Velez L. N., Uezato H. (2016). Diffuse and disseminated cutaneous leishmaniasis: clinical cases experienced in Ecuador and a brief review. Trop. Med. Health 44, 2. 10.1186/s41182-016-0002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y., Velez L. N., Villegas N. V., Mimori T., Gomez E. A. L., Kato H. (2017). Leishmaniases in Ecuador: Comprehensive review and current status. Acta Trop. 166, 299–315. 10.1186/s41182-016-0002-0 [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y., Gomez E. A. L., Cáceres A. G., Velez L. N., Villegas N. V., Hashiguchi K., et al. (2018). Andean cutaneous leishmaniasis (Andean-CL, uta) in Peru and Ecuador: the causative Leishmania parasites and clinico-epidemiological features. Acta Trop. 177, 135–145. 10.1016/j.actatropica.2017.09.028 [DOI] [PubMed] [Google Scholar]
- Kamhawi S., Ramalho-Ortigao M., Pham V. M., Kumar S., Lawyer P. G., Turco S. J., et al. (2004). A role for insect galectins in parasite survival. Cell 119, 329–341. 10.1016/j.cell.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Kato H., Caceres A. G., Mimori T., Ishimaru Y., Sayed A. S., Fujita M., et al. (2010). Use of FTA cards for direct sampling of patient’s lesions in the ecological study of cutaneous leishmaniasis. J. Clin. Microbiol. 48, 3661–3665. 10.1128/JCM.00498-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Watanabe J., Mendoza Nieto I., Korenaga M., Hashiguchi Y. (2011). Leishmania species identification using FTA card sampling directly from patients’ cutaneous lesions in the state of Lara, Venezuela. Trans. R. Soc Trop. Med. Hyg. 105, 561–567. 10.1016/j.trstmh.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Kato H., Calvopiña M., Criollo H., Hashiguchi Y. (2013). First human cases of Leishmania (Viannia) naiffi infection in Ecuador and identification of its suspected vector species. Acta Trop. 128, 710–713. 10.1016/j.actatropica.2013.09.001 [DOI] [PubMed] [Google Scholar]
- Kato H., Gomez E. A., Martini-Robles L., Muzzio J., Velez L., Calvopiña M., et al. (2016. a). Geographic distribution of Leishmania species in Ecuador based on the cytochrome b gene sequence analysis. PLoS Negl. Trop. Dis. 10, e0004844. 10.1371/journal.pntd.0004844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Bone A. E., Mimori T., Hashiguchi K., Shiguango G. F., Gonzales S. V., et al. (2016. b). First human cases of Leishmania (Viannia) lainsoni infection and a search for the vector sand flies in Ecuador. PLoS Negl. Trop. Dis. 10, e0004728. 10.1371/journal.pntd.0004728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Cáceres A. G., Hashiguchi Y. (2016. c). First evidence of a hybrid of Leishmania (Viannia) braziliensis/L. (V.) peruviana DNA detected from the Phlebotomine sand fly Lutzomyia tejadai in Peru. PLoS Negl. Trop. Dis. 10, e0004336. 10.1371/journal.pntd.0004336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Caceres A. G., Seki C., Silupu Garcia C. R., Holguin Mauricci C., Castro Martinez S. C., et al. (2019. a). Further insight into the geographic distribution of Leishmania species in Peru by cytochrome b and mannose phosphate isomerase gene analyses. PLoS Negl. Trop. Dis. 13, e0007496. 10.1371/journal.pntd.0007496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Gomez E. A., Seki C., Furumoto H., Martini-Robles L., Muzzio J., et al. (2019. b). PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. PLoS Negl. Trop. Dis. 13, e0007403. 10.1371/journal.pntd.0007403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanra S., Datta S., Mondal D., Saha P., Bandopadhyay S. K., Roy S., et al. (2012). RFLPs of ITS, ITS1 and hsp70 amplicons and sequencing of ITS1 of recent clinical isolates of Kala-azar from India and Bangladesh confirms the association of L. tropica with the disease. Acta Trop. 124, 229–234. 10.1016/j.actatropica.2012.08.017 [DOI] [PubMed] [Google Scholar]
- Koarashi Y., Caceres A. G., Zuniga S. M. F., Palacios F. E. E., Celis T. A., Abanto A. J. L., et al. (2016). Identification of causative Leishmania species in Giemsa-stained smears prepared from patients with cutaneous leishmaniasis in Peru using PCR-RFLP. Acta Trop. 158, 83–87. 10.1016/j.actatropica.2016.02.024 [DOI] [PubMed] [Google Scholar]
- Lawton S. P., Bowen L. I., Emery A. M., Majoros G. (2017). Signatures of mito-nuclear discordance in Schistosoma turkestanicum indicate a complex evolutionary history of emergence in Europe. Parasitol 144, 1752–1762. 10.1017/S0031182017000920 [DOI] [PubMed] [Google Scholar]
- Leelayoova S., Siripattanapipong S., Hitakarun A., Kato H., Tan-ariya P., Siriyasatien P., et al. (2013). Multilocus characterization and phylogenetic analysis of Leishmania siamensis isolated from autochthonous visceral leishmaniasis cases, southern Thailand. BMC Microbiol. 13:60. 10.1186/1471-2180-13-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louradour I., Ferreira T. R., Ghosh K., Shaik J., Sacks D. (2020). In vitro generation of Leishmania hybrids. Cell Rep. 31, 107507. 10.1016/j.celrep.2020.03.071 [DOI] [PubMed] [Google Scholar]
- Luyo-Acero G. E., Uezato H., Oshiro M., Takei K., Kariya K., Katakura K., et al. (2004). Sequence variation of the cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitol 128, 483–491. 10.1017/s0031182004004792 [DOI] [PubMed] [Google Scholar]
- McBride H. M., Neuspiel M., Wasiak S. (2006). Mitochondria: more than just a powerhouse. Curr. Biol. 16, R551–R560. 10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Montalvo A. M., Fraga J., Maes I., Dujardin J. C., Van der Auwera G. (2012). Three new sensitive and specific heat shock protein 70 PCRs for global Leishmania species identification. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1453–1461. 10.1007/s10096-011-1463-z [DOI] [PubMed] [Google Scholar]
- Mouttaki T., Morales-Yuste M., Merino-Espinosa G., Chiheb S., Fellah H., Martin-Sanchez J., et al. (2014). Molecular diagnosis of cutaneous leishmaniasis and identification of the causative Leishmania species in Morocco by using three PCR-based assays. Parasitol. Vectors 7:420. 10.1186/1756-3305-7-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odiwuor S., De Doncker S., Maes I., Dujardin J. C., Van der Auwera G. (2011). Natural Leishmania donovani/Leishmania aethiopica hybrids identified from Ethiopia. Infect. Genet. Evol. 11, 2113–2118. 10.1016/j.meegid.2011.04.026 [DOI] [PubMed] [Google Scholar]
- Paranaiba L. F., Pinheiro L. J., Torrecilhas A. C., Macedo D. H., Menezes-Neto A., Tafuri W. L., et al. (2017). Leishmania enriettii (Muniz & Medin ): A highly diverse parasite is here to stay. PLoS Pathog. 13, e1006303. 10.1371/journal.ppat.1006303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel C., Cortes S., Pratlong F., Morio F., Dedet J. P., Campino L. (2006). First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major . Int. J. Parasitol. 36, 1383–1388. 10.1016/j.ijpara.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Rioux J. A., Lanotte G., Serres E., Pratlong F., Bastien P., Perieres J. (1990). Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann. Parasitol. Hum. Comp. 65, 111–125. 10.1051/parasite/1990653111 [DOI] [PubMed] [Google Scholar]
- Rogers M. B., Downing T., Smith B. A., Imamura H., Sanders M., Svobodova M., et al. (2014). Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet. 10, e1004092. 10.1371/journal.pgen.1004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotureau B., Ravel C., Couppie P., Pratlong F., Nacher M., Dedet J. P., et al. (2006). Use of PCR-restriction fragment length polymorphism analysis to identify the main new world Leishmania species and analyze their taxonomic properties and polymorphism by application of the assay to clinical samples. J. Clin. Microbiol. 44, 459–467. 10.1128/JCM.44.2.459-467.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Postigo J. A., Grouta L., Jaina S. (2020). Global leishmaniasis surveillance 2017–2018, and first report on 5 additional indicators. World Health Organ. Wkly. Epidemiological Rec. 25, 265–280. [Google Scholar]
- Sadlova J., Yeo M., Seblova V., Lewis M. D., Mauricio I., Volf P., et al. (2011). Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One 6, e19851. 10.1371/journal.pone.0019851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. O., Sato M., Yanagida T., Waikagul J., Pongvongsa T., Sako Y., et al. (2018). Taenia solium, Taenia saginata, Taenia asiatica, their hybrids and other helminthic infections occurring in a neglected tropical diseases’ highly endemic area in Lao PDR. PLoS Negl. Trop. Dis. 12, e0006260. 10.1371/journal.pntd.0006260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seblova V., Myskova J., Hlavacova J., Votypka J., Antoniou M., Volf P. (2015). Natural hybrid of Leishmania infantum/L. donovani: development in Phlebotomus tobbi, P. perniciosus and Lutzomyia longipalpis and comparison with non-hybrid strains differing in tissue tropism. Parasitol. Vectors 8, 605. 10.1186/s13071-015-1217-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L. (1986). Kinetoplast DNA in trypanosomid flagellates. Int. Rev. Cytol. 99, 119–179. 10.1016/s0074-7696(08)61426-6 [DOI] [PubMed] [Google Scholar]
- Spanakos G., Piperaki E. T., Menounos P. G., Tegos N., Flemetakis A., Vakalis N. C. (2008). Detection and species identification of Old World Leishmania in clinical samples using a PCR-based method. Trans. R. Soc Trop. Med. Hyg. 102, 46–53. 10.1016/j.trstmh.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Tabbabi A., Cáceres A. G., Bustamante Chauca T. P., Seki C., Choochartpong Y., Mizushima D., et al. (2020). Nuclear and kinetoplast DNA analyses reveal genetically complex Leishmania strains with hybrid and mito-nuclear discordance in Peru. PLoS Negl. Trop. Dis. 14, e0008797. 10.1371/journal.pntd.0008797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmi-Frank D., Nasereddin A., Schnur L. F., Schonian G., Toz S. O., Jaffe C. L., et al. (2010). Detection and identification of Old World Leishmania by high resolution melt analysis. PLoS Negl. Trop. Dis. 4, e581. 10.1371/journal.pntd.0000581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews D. P., Brelsford A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930. 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- Van den Broeck F., Savill N. J., Imamura H., Sanders M., Maes I., Cooper S., et al. (2020). Ecological divergence and hybridization of Neotropical Leishmania parasites. Proc. Natl. Acad. Sci. U. S. A. 117, 25159–25168. 10.1073/pnas.1920136117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volf P., Benkova I., Myskova J., Sadlova J., Campino L., Ravel C. (2007). Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int. J. Parasitol. 37, 589–593. 10.1016/j.ijpara.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Suzuki Y., Tachi E., Li T., Chen X., Nakao M., et al. (2012). Recent hybridization between Taenia asiatica and Taenia saginata . Parasitol. Int. 61, 351–355. 10.1016/j.parint.2012.01.005 [DOI] [PubMed] [Google Scholar]
- Yamane K., Yanagida T., Li T., Chen X., Dekumyoy P., Waikagul J., et al. (2013). Genotypic relationships between Taenia saginata, Taenia asiatica and their hybrids. Parasitology 140, 1595–1601. 10.1017/S0031182013001273 [DOI] [PubMed] [Google Scholar]
- Yanagida T., Carod J. F., Sako Y., Nakao M., Hoberg E. P., Ito A. (2014). Genetics of the pig tapeworm in Madagascar reveal a history of human dispersal and colonization. PLoS One 9, e109002. 10.1371/journal.pone.0109002 [DOI] [PMC free article] [PubMed] [Google Scholar]