Abstract
BACKGROUND AND PURPOSE:
MR imaging in neuro-oncology is challenging due to inherent ambiguities in proton signal behavior. Sodium-MR imaging may substantially contribute to the characterization of tumors because it reflects the functional status of the sodium-potassium pump and sodium channels.
MATERIALS AND METHODS:
Sodium-MR imaging data of patients with treatment-naïve glioma WHO grades I–IV (n = 34; mean age, 51.29 ± 17.77 years) were acquired by using a 7T MR system. For acquisition of sodium-MR images, we applied density-adapted 3D radial projection reconstruction pulse sequences. Proton-MR imaging data were acquired by using a 3T whole-body system.
RESULTS:
We demonstrated that the initial sodium signal of a treatment-naïve brain tumor is a significant predictor of isocitrate dehydrogenase (IDH) mutation status (P < .001). Moreover, independent of this correlation, the Cox proportional hazards model confirmed the sodium signal of treatment-naïve brain tumors as a predictor of progression (P = .003). Compared with the molecular signature of IDH mutation status, information criteria of model comparison revealed that the sodium signal is even superior to IDH in progression prediction. In addition, sodium-MR imaging provides a new approach to noninvasive tumor classification. The sodium signal of contrast-enhancing tumor portions facilitates differentiation among most glioma types (P < .001).
CONCLUSIONS:
The information of sodium-MR imaging may help to classify neoplasias at an early stage, to reduce invasive tissue characterization such as stereotactic biopsy specimens, and overall to promote improved and individualized patient management in neuro-oncology by novel imaging signatures of brain tumors.
Gliomas are the most common type of primary brain tumor.1 They are classified on the basis of MR imaging, histopathologic, and clinical criteria. However, MR imaging in neuro-oncology is challenging due to inherent ambiguities in proton (1H) signal behavior. Contrast-enhancing (CE) tumor portions in T1WI do not necessarily represent a malignant tumor.2 For example, gangliogliomas (GGs) and pilocytic astrocytomas (PAs), both low-grade gliomas, demonstrate vivid contrast enhancement.3 In turn, tissue that appears normal without contrast enhancement or physiologic T2 signal may be tumorous tissue.4,5 The evolving method of sodium (Na) MR imaging may substantially contribute to the characterization of tumors by neuro-oncologic imaging because it reflects the functional status of the sodium-potassium pump and Na channels. It allows 2 signals to be determined, which reflect the total amount of tissue Na (NaT)6,7 as well as the amount of ions with short relaxation times (NaR).7,8 The NaR signal is dependent on the microstructural environment, and, mainly, intracellular Na ions contribute to it. It correlates with the Ki-67 proliferation index of tumor cells.7 To evaluate the implications of Na-MR imaging for neuro-oncologic patient management, we analyzed Na-MR imaging data of treatment-naïve gliomas in correlation with progression-free survival (PFS) and tumor classification. We propose that the Na signal ratio NaR:NaT is a predictor for PFS and show that NaR:NaT is superior even to the prognostic information of the isocitrate dehydrogenase (IDH) mutational status. In contrast to the genetic characterization of IDH, Na-MR imaging is noninvasive and no contrast medium is needed. Moreover, Na-MR imaging yields highly relevant information that promotes tumor classification. Overall, our observations underline that Na-MR imaging is a valuable noninvasive tool for prognostic and diagnostic assessment in neuro-oncology.
Materials and Methods
Ethics Statement
The study was approved by the local medical ethics committee (Faculty of Clinical Medicine, University of Heidelberg), and all the participants gave written informed consent before enrollment. The procedures that followed were in accordance with the Declaration of Helsinki.
Patient Cohort
Inclusion criteria were 1) the suspected diagnosis of a glioma, 2) in patients >18 years, and 3) without neurologic or psychiatric illness or head trauma in their history; furthermore, 4) neoplasias had to be treatment naïve, that is, no chemotherapy, radiation or surgery was performed. We measured 34 patients whose 1H-MR imaging was suggestive of a glioma by using Na-MR imaging. Epidemiologic data and tumor characteristics are presented in On-line Table 1. Diagnoses of our patient population included PA, astrocytoma World Health Organization (WHO) grade II, anaplastic astrocytoma (AA), glioblastoma (GB), oligodendroglioma WHO grade II, anaplastic oligodendroglioma, anaplastic ependymoma, and gliomatosis cerebri. In addition, histopathologic analyses diagnosed 2 cerebral metastases (breast and prostate cancer), which mimicked GB on 1H-MR images.
Histopathologic Analysis
Tissue was evaluated according to the current WHO classification.1 Ki-67 index and IDH status were evaluated as described previously by Sahm et al.9,10
MR Imaging
Na-MR imaging was performed by using a 7T whole-body MR system (Magnetom 7T; Siemens, Erlangen, Germany) by using a double-resonant (1H/23Na) quadrature birdcage coil with an inner coil diameter of 26 cm (Rapid Biomed, Rimpar, Germany). All Na-MR images were based on a 3D attenuation-adapted projection reconstruction technique (see On-line Table 2 for details).11 1H-MR data were acquired by using a 3T whole-body system (Tim Trio 3T; Siemens) with T2-TSE, T2-FLAIR, native and contrast-enhanced T1–3D ultrafast gradient sequences.
Image Processing
Na image reconstruction was performed off-line; Na and anatomic 1H data were coregistered into the individual standard space (for details see On-line Appendix, Methods). We defined the tumor volumes of interest by their global T2 signal alterations (whole tumor VOI) and, where applicable, by their T1 signal of CE portions (CE tumor VOI) (Fig 1). In addition, we segmented healthy gray and white matter, CSF, and vessels. Segmentation was performed by using the Interactive Learning and Segmentation Toolkit (ilastik)12 (see On-line Appendix, Methods). Data were normalized to CSF as multimodally defined by the ilastik segmentation. We integrated the information from the NaR and the NaT signal into a single quantity (arbitrary units), the NaR:NaT signal ratio, which can be understood as normalizing the NaR signal with NaT. For an illustration of this quantity, we plotted the average NaR:NaT distributions of healthy gray matter, white matter, CSF, and GB (On-line Fig 1). The NaR and NaT signal behavior in both low-grade glioma and high-grade glioma is exemplarily shown in On-line Fig 2.
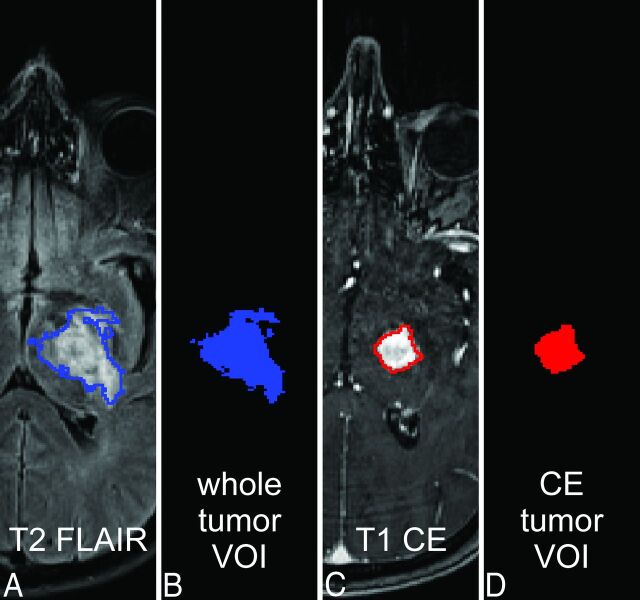
Fig 1.
Tumor masking. The T2 signal of tumor and perifocal edema is shown on an exemplary T2-FLAIR image (A) of a patient with GG (ID no. 2, On-line Table 1); it is the basis of the whole tumor VOI (A and B, blue). CE tumor portions (C) define the CE tumor VOI (C and D, red). All tumor VOIs were created by using ilastik (see Materials and Methods; Image Processing; and On-line Appendix, Methods).
Statistical Analysis
Statistical analysis was performed by using R statistical computing software (version 3.0.3; http://www.r-project.org/).13
Prediction of IDH Mutation Status from NaR:NaT
A logistic regression model was fitted to evaluate the predictive value of NaR:NaT for IDH mutation status. Based on the resulting receiver operating characteristic curve, an NaR:NaT threshold was determined, which simultaneously optimized sensitivity and specificity of the prediction (On-line Fig 3).
PFS Hazards Based on NaR:NaT
Based on the NaR:NaT threshold obtained via the IDH mutation status, we computed the Kaplan-Meier estimate of the resulting 2 groups. Further, to estimate the predictive properties of NaR:NaT for PFS, a Cox proportional hazard (PH) model was fitted to the right-censored survival data of the patient cohort (On-line Fig 4). The goodness of fit of this model was compared with a Cox PH model with IDH mutation status as a predictor (On-line Fig 5). Due to the relationship between the magnitude of NaR:NaT and IDH mutation status (as shown with the logistic regression), separate models, rather than a combined model, were fitted and compared based on the Akaike information criterion. To be able to compare both models, the models were constructed by using only data in which IDH mutation status was available.
NaR:NaT Differences among Tumor Classes
By using analysis of variance NaR:NaT from 1) whole tumor VOIs were compared among PA, astrocytoma, AA, GB, oligodendroglioma WHO grade II, anaplastic oligodendroglioma, and metastasis; and NaR:NaT from 2) CE tumor VOIs were compared among PA, AA, GB, and metastasis. Anaplastic ependymoma and gliomatosis cerebri were excluded from whole tumor VOI analysis because each group contained only a single patient. Similarly, CE tumor VOI analysis was restricted to tumors with a group size of at least 2. Post hoc analysis of significant effects was conducted with pair-wise t tests, corrected for multiple comparisons after Benjamini-Hochberg. All analyses used the mean NaR:NaT values of the VOI.
To determine the predictive discriminability of GB and metastasis from the remaining histopathologic classes (REST) based on NaR:NaT, we computed a logistic regression model for the whole tumor VOI and evaluated the receiver operating characteristic curve (On-line Fig 6). Next, to discriminate GB from metastasis and from PA plus AA, 2 logistic regressions were calculated for NaR:NaT values from CE VOI.
Results
Prediction of IDH Mutation from NaR:NaT
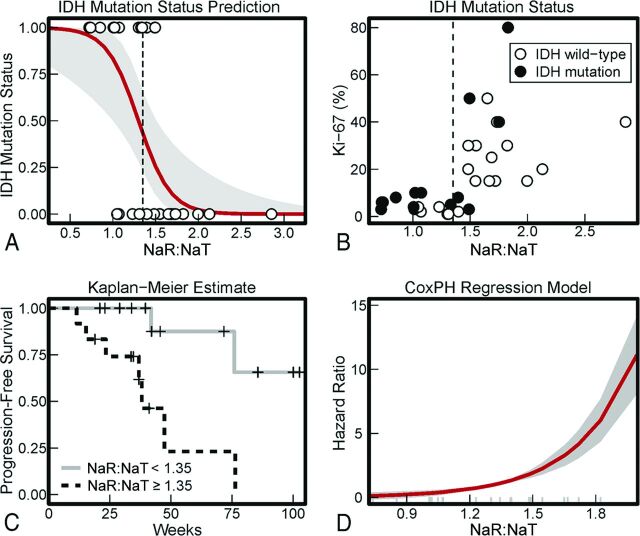
Analysis of a logistic regression model revealed that NaR:NaT of the whole tumor VOI is a significant predictor of IDH mutation status (χ2[1] = 14.47, P < .001) (Fig 2A). With an increase in NaR:NaT by 0.1, the odds ratio of an IDH mutation grows by 21.7 (β0 = 7.00, SE [β0] = 2.67; βNaR:NaT = −5.38, SE [βNaR:NaT] = 1.96). With a combined optimization of sensitivity and specificity, the model yielded an NaR:NaT threshold of 1.35 (On-line Fig 3; area under the curve = 0.87), which corresponds to an odds ratio of IDH mutation of 0.57. IDH mutations were found in 71% of tumors with an NaR:NaT below threshold and in 18% of tumors with an NaR:NaT above threshold (Fig 2B).
Fig 2.
A, Prediction of IDH mutation status from NaR:NaT. Each circular marker indicates 1 tumor's mean NaR:NaT value and its IDH mutation status. These values were entered into a logistic regression model, and the resulting probabilities of IDH wild type based on NaR:NaT are depicted in red (the gray area shows the 95% CI of the predictions). B, IDH mutation status. With a combined optimization of sensitivity and specificity, the logistic regression model yielded an NaR:NaT threshold of 1.35 (Fig 2A and On-line Fig 3; area under the curve = 0.87). IDH mutations were found in 71% of tumors with an NaR:NaT below threshold and in 18% of tumors with an NaR:NaT above threshold. C, Kaplan-Meier estimates of PFS. The estimated percentage of progression-free patients is shown in dependence on time. Patients were divided into 2 groups, having an NaR:NaT value either below or above 1.35. The threshold of 1.35 was derived based on the prediction of IDH mutation status from NaR:NaT (Fig 2A). D, The Cox PH regression model for NaT:NaR. Predicted hazard ratios in dependence on NaR:NaT mean values from the whole tumor VOI are shown. Long gray marks indicate single patient values. A positive hazard ratio indicates an increase in hazard rate that can be attributed to an increase in NaT:NaR. Practically, it describes the relative risk at the instantaneous moment, which is assumed to be constant across time.
Progression-Free Survival
Kaplan-Meier Estimates Based on NaR:NaT.
To use the prognostic information of the IDH mutation status, we used the NaR:NaT threshold of 1.35 to separate tumors into a group below threshold (mean, 1.05) and a group above threshold (mean, 1.72 [95% CI, 0.47–0.86]). Based on the NaR:NaT signal threshold of 1.35, the Kaplan-Meier estimates revealed differences in the time from MR measurement to the time of disease progression (PFS) (χ2[1] = 8.2, P = .004). Tumors below the threshold demonstrated a substantially longer PFS than those above the threshold (Fig 2C).
Cox Proportional Hazard PFS Based on NaR:NaT.
Analysis of a Cox PH model confirmed that NaR:NaT was a significant predictor of PFS (χ2[1] = 8.77, P = .003; for an analysis of the residuals see On-line Fig 4). An increase in NaR:NaT by 0.1 is associated with an increase in progression hazard rate of 340% (βNaR:NaT = 3.53, SE [βNaR:NaT] = 1.34) (Fig 2D). The 95% CI of the logarithmic hazard ratio βNaR:NaT, 2.50–271.70, indicated that, for different patient cohorts, the associated increases in hazard will, though a mild overestimation is principally possible, most probably lead to a considerable underestimation of progression rate.
In addition, the hazard ratio, that is, the change of hazard rate, of IDH mutation and wild type was estimated by using a Cox PH model (On-line Fig 5). Hazard rates were reliably predicted by IDH mutation status (χ2[1] = 7.76, P = .005). The hazard of progression is 9.9 times larger in the wild-type group than in the mutation group (βIDH = 2.30, SE [βIDH] = 0.10). A comparison of both Cox PH models revealed that NaR:NaT is a better predictor of PFS (Akaike information criterionNaR:NaT = 36.48 < Akaike information criterionIDH = 37.49) than the IDH mutational status.
Tumor Classification
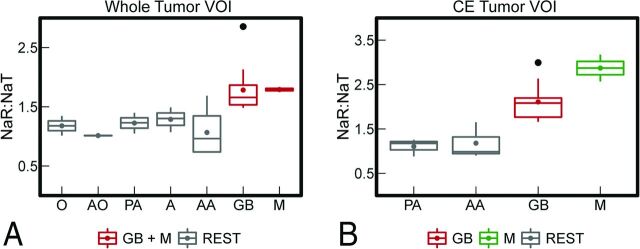
When examining NaR:NaT data of the whole tumor VOI, an ANOVA with Benjamini-Hochberg corrected P values of post hoc pair-wise t tests revealed that all brain tumors enrolled showed significantly lower NaR:NaT compared with GB and M (F[6,25] = 5.03, P = .002; no violation of homogeneity of variances assumption (Levene test F[6,25] = 0.76, P = .605); Fig 3A and On-line Table 3). An ANOVA on NaR:NaT from the CE tumor VOI (Fig 3B) revealed a significant NaR:NaT difference between GB, metastasis, and PA plus AA (F[3,14] = 11.84, P < .001; no violation of homogeneity of variances assumption [Levene test F{3,14} = 0.45, P = .718]; see On-line Table 4 for Benjamini-Hochberg corrected pair-wise t tests). A significant difference in NaR:NaT between PA and AA did not emerge. To examine the prediction quality of these significant differences in mean NaR:NaT, logistic regression models were established. The first logistic regression confirmed that NaR:NaT mean values from the whole tumor VOI are a significant predictor for the binary classification of GB plus metastasis versus REST (χ2[1] = 28.96, P < .001). With an increase in NaR:NaT of 0.001, the odds ratio of a tumor being either GB or metastasis grows by 968.77 (β0 = −20.44, SE[β0] = 8.79; βNaR:NaT = 13.78, SE[βNaR:NaT] = 5.90). At an NaR:NaT threshold of 1.50, a specificity of 94% and a sensitivity of 86% are achieved, whereas the corresponding area under the curve is 96% (On-line Fig 6). Further logistic regression models confirmed that NaR:NaT mean values from the CE tumor VOI are significant predictors for the classification of GB versus AA plus PA (χ2[1] = 4.20, P = .050) and for the classification of GB versus metastasis (χ2[1] = 4.28, P = .039).
Fig 3.
NaR:NaT values of different tumor classes. The boxplots visualize the mean NaR:NaT of the whole tumor VOI (A) and the CE tumor VOI (B) for different tumor classes. Because of the low sample size, gliomatosis cerebri (n = 1), anaplastic ependymoma (n = 1), and anaplastic oligodendroglioma (n = 1) are not depicted. For NaR:NaT of the whole tumor VOI, GB could be separated from all other gliomas (REST) but not from metastasis. NaR:NaT of the CE tumor VOI enabled all (CE) gliomas to be separated from each other except for PA versus AA (REST). Moreover, GB can be differentiated from metastasis. The box extends from the lower to upper quartile values of the data; lines represent the median; and colored filled circles depict the modal. Vertical axes indicate the range of the data; flier points are shown as black filled circles. O indicates oligodendroglioma; AO, anaplastic oligodendroglioma; AE, anaplastic ependymoma; GC, gliomatosis cerebri; A, astrocytoma; M, metastasis.
Discussion
The current study identifies NaR:NaT as a noninvasive predictor of PFS. Furthermore, we observed a strong relationship between NaR:NaT and IDH mutation status. An NaR:NaT threshold is provided for the prediction of IDH mutation status by logistic regression. Model comparison showed that NaR:NaT was even better at predicting PFS than the genetic analyses of IDH based on MR imaging data. Moreover, NaR:NaT reflects relevant tissue characteristics that provide valuable information for improved tumor classification.
Gliomas are classified on the basis of MR imaging, histopathologic, and clinical criteria. In addition, molecular abnormalities in gliomas such as IDH mutations, O6-methylguanine methyltransferase promoter methylation status,14,15 or 1p/19q co-deletions have moved into the focus of scientific interest. They are increasingly considered as supportive markers to assist diagnosis and patient management.16 The IDH mutational status of a glioma is a strong prognostic marker of outcome.17–19 Patients with WHO grades II–IV gliomas with IDH mutation have a better overall survival and PFS than those with wild-type IDH.16,20–23 Here, we found that NaR:NaT is a significant predictor of IDH mutations. Based on this prediction, the logistic regression model established a threshold that made patient stratification possible. The frequency of IDH mutations in the group above and below this threshold was in accordance with the literature data for high- and low-grade tumors,24,25 respectively, underlining that the threshold is not only sensible in theory but also when used in physiologic context. Kaplan-Meier estimates demonstrated a significantly longer PFS in patients with NaR:NaT below threshold. The established association does not necessarily imply causality. Still, Cox PH fits independently confirmed that both NaR:NaT and IDH were predictors of PFS. Moreover, model comparison based on the Akaike information criterion revealed that NaR:NaT is even superior to IDH in predicting PFS. Our observations are compatible with a genomic analysis of GB that revealed mutations in Na channel genes.26 These mutations were associated with shorter survival compared with tumors with wild-type Na channels.27 Interestingly, none of the tumors with IDH gene mutation had a Na channel gene mutation. The potential of NaR:NaT to predict IDH mutation status, as well as PFS, grants access to information that is usually restricted to genetic analyses. Hence, it extends the scope of Na-MR imaging from mere diagnosis to both diagnosis and prognosis.
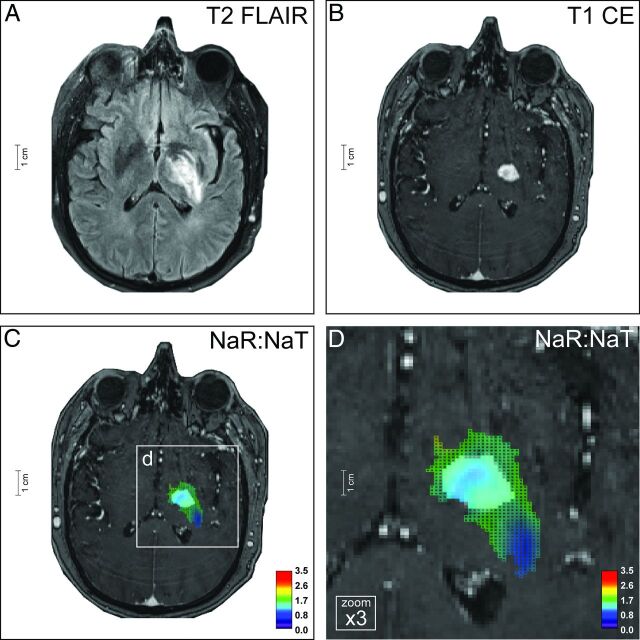
Contrast-enhanced routine 1H-MR imaging contributes to glioma classification through anatomic information and indication of areas with blood-brain barrier disruption. However, due to the limited specificity of T2- and T1-weighted imaging,3,4 a nonenhancing glioma, is not always low grade (eg, AA), and a CE glioma is not necessarily malignant (eg, PA/GG). As a consequence, often only histopathologic analysis of tumor specimens provides the diagnosis. Na-MR imaging, however, yields significant information in unraveling this diagnostic dilemma. The pathophysiologic correlate is a strong relation between NaR:NaT and the tumor proliferation index Ki-67 (On-line Fig 7) on the one hand and the significant role of Na channels in tumor cell division and migration27–31 on the other hand. NaR:NaT of the CE tumor VOI enabled differentiation of all CE tumors in our patient population except for PA versus AA. Moreover, cerebral metastases, which were initially accidentally considered to be a high-grade glioma based on 1H-MR images and thus included in our study, were correctly attributed to a separate class by NaR:NaT (Fig 3B). To translate these findings into clinical application, we present a 49-year-old patient (ID no. 2, On-line Table 1) with a supratentorial tumor of the left thalamus, internal capsule, and pallidum. The neoplasia revealed elevated T2 signals and bulky contrast enhancement with central T1 signal reduction. Based on 1H-MR imaging (Fig 4A, -B), the spectrum of differential diagnoses includes PA/GG, GB, and metastasis, and thus no reliable tumor classification could be provided. However, Na-MR imaging yielded additional information for tumor classification. NaR:NaT indicated the diagnosis of PA/GG (Fig 3 and Fig 4C, -D) and discarded the differential diagnoses of GB and M (On-line Table 1). This finding is contrasted to a case of GB in a 71-year-old patient (ID no. 21; Fig 5) which demonstrated NaR:NaT values that are indicative of GB and exclude the diagnosis of PA/GG (and metastasis) (Fig 3). Our classifications were confirmed by the histopathologic examination of the biopsy specimens (On-line Fig 8).
Fig 4.
Ganglioglioma. The neoplasia of a 49-year-old patient (ID no. 2, On-line Table 1) affects the left thalamus, pallidum, and putamen, and is characterized by a largely homogeneous elevated T2-FLAIR signal (A) and somewhat rim-like contrast enhancement (B). Based on 1H-MR imaging, differential diagnostic considerations included low-grade tumors such as GG and PA but also malignant neoplasias such as GB and cerebral metastasis. Na-MR imaging reveals a mean NaR:NaT of 1.39 (whole tumor VOI) and 1.26 (CE tumor VOI) (C and D) compatible with a low-grade tumor (Fig 3A, -B; On-line Fig 2E, -F; On-line Table 1). Thus, the differential diagnoses of GB and M could be ruled out. This result was confirmed by histopathology (On-line Fig 8). Na images are overlaid on T1-weighted postcontrast images; color mesh grid: whole tumor VOI, solid color: CE tumor VOI.
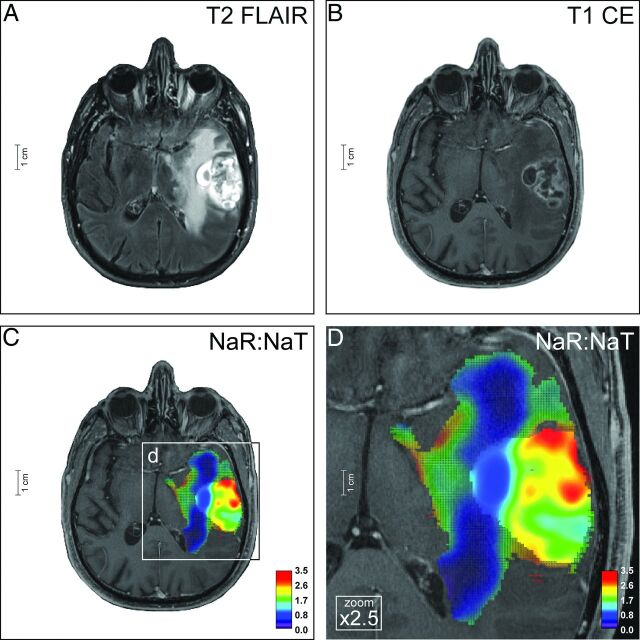
Fig 5.
Glioblastoma. The left-temporal tumor of a 71-year-old male patient (ID no. 21, On-line Table 1) shows inhomogeneous central and large homogeneous perifocal T2-FLAIR hyperintensities (A). There is a rim-like contrast enhancement of the central tumor portion, as seen on T2-FLAIR (A) and T1-weighted images (B). Na-MR imaging demonstrates a mean NaR:NaT of 1.65 (whole tumor VOI) and 2.02 (CE tumor VOI) (C and D) compatible with GB (Fig 3A, -B; On-line Fig 2G, -H; On-line Table 1). The diagnosis was histopathologically proved (On-line Fig 8). Na-MR images are overlaid onto T1-weighted postcontrast images; color mesh grid: whole tumor VOI, solid color: CE tumor VOI.
This exemplary clinical application underlines that the Na signal yielded significant prognostic and diagnostic information on PFS and tumor classification that will be able to guide clinical decision making in neuro-oncology in the future. Advanced physiologic 1H-MR imaging techniques such as perfusion-weighted imaging, diffusion-weighted imaging, and spectroscopy (1H-MR spectroscopy) provide relevant information for tumor characterization as well. For example, tumor grade is associated with relative cerebral blood volume32–34; volume transfer coefficient35–38; apparent diffusion coefficient39,40; and cerebral metabolite ratios, for example, the choline to N-acetylaspartate33 and the choline to creatine ratios.33,41 Also, relative cerebral blood volume,34,42–48 volume transfer coefficient,35,49 apparent diffusion coefficient,50–52 and metabolite ratios, for example, the choline to N-acetylaspartate ratio,53 aid prediction of PFS. Moreover, 1H-MR spectroscopy of 2-hydroxyglutarate enables the detection of IDH mutations.54 Future studies are needed to integrate the specific contribution of advanced 1H-MR imaging and X-nuclei-MR imaging techniques, respectively, to tumor grading as well as prediction of IDH mutation status and PFS.
Na-MR imaging struggles to differentiate CE PA/GG from AA. This is mainly due to the large standard deviation of NaR:NaT in AA. One cause for the large standard deviation in the Na signal ratios may be the limited spatial resolution. Larger voxel sizes intrinsically smooth local signal differences to a certain extent. Another cause of this variability in NaR:NaT may be the substantial overlap in tumor cell proliferation between astrocytoma (WHO grade II), AA (WHO grade III), and GB (WHO grade IV).1 This means that AA, which recently developed from astrocytoma, would exhibit lower NaR:NaT than AA in malignant transformation to GB. The proposed relationship between NaR:NaT and the developmental state of the tumor is supported by the strong correlation between NaR:NaT and the tumor proliferation index Ki-67 (On-line Appendix, Results; On-line Fig 7). Analysis of our data indeed indicate that NaR:NaT of AA may reflect the state of malignant transformation and that NaR:NaT as well as corresponding Ki-67 may range from values compatible with low-grade gliomas (eg, ID no. 9, On-line Table 1) to those of high-grade gliomas (eg, ID no. 10, On-line Table 1). If that is true, then future analyses of the same dataset with updated data on outcome will indicate a correlation between NaR:NaT data and the time to malignant transformation.
A future challenge in Na-MR imaging is improvement of the spatial resolution, which would, for example, enable a more-precise characterization and might allow for defining subclones of varying malignancies in high-grade tumors.55 Besides the application of high magnetic field strengths, an increased spatial resolution can be achieved by using iterative image reconstruction techniques or multichannel receiver coils. One approach incorporates a priori information from 1H-MR imaging into the image reconstruction of Na-MR data. Thereby, intensity variations in the Na image are promoted at the position of known tissue boundaries, which translate into an improved spatial resolution and enable quantification with higher accuracy.56
One limitation of this study is the low sample size, which was partly due to the inclusion criteria. Patients with a brain tumor had to be treatment naïve and be in a medical condition that permitted an additional MR imaging for scientific purposes. Clinical studies with larger patient populations are needed to prospectively test our prediction models and to analyze the interdependencies of the measures NaR:NaT and IDH as well as the effect of influencing parameters such as radiation or chemotherapy, a task that is beyond the scope of this pilot study.
Conclusions
Na-MR imaging allows for PFS prediction, which we showed is superior even to the IDH mutation status and improves the accuracy of brain tumor classification. Hence, Na-MR imaging is a promising candidate for noninvasive outcome prediction and tumor diagnosis, which may help to classify neoplasias at an early stage, to reduce invasive tissue characterization such as stereotactic biopsy specimens, and, overall, to promote improved patient management in neuro-oncology.
Supplementary Material
ABBREVIATIONS:
- AA
anaplastic astrocytoma
- CE
contrast-enhancing
- GB
glioblastoma
- GG
ganglioglioma
- IDH
isocitrate dehydrogenase
- NaR
relaxation-weighted sodium signal
- NaT
total sodium signal
- PA
pilocytic astrocytoma
- PFS
progression-free survival
- PH
proportional hazard
- WHO
World Health Organization
Footnotes
Disclosures: Armin Biller—RELATED: Grant: National Centre for Tumour Diseases.* Jan-Oliver Neumann—RELATED: Grant: National Centre of Tumour Diseases Heidelberg (NCT IFP V/3).* Wolfgang Wick—UNRELATED: Board Membership: Roche, Celldex; Grants/Grants Pending: Roche,* Boehringer Ingelheim*; Patents (planned, pending or issued): IDH antibody.* Martin Bendszus—UNRELATED: Board Membership: Data Safety Monitoring Board for Vascular Dynamics; Consultancy: Codman, Guerbet, Roche; Grants/Grants Pending: German Research Foundation,* Hopp Foundation,* Novartis,* Guerbet,* Siemens,* Codman*; Payment for Lectures (including service on Speakers Bureaus): Roche, Novartis, Guerbet, Codman. Felix Sahm—UNRELATED: Grants/Grants Pending: German Cancer Aid,* Comments: Exome-sequencing of meningioma; Patents (planned, pending or issued): Detection of antigen presentation in-situ (patent pending).* *Money paid to the institution.
A. Biller and A. Nagel were supported by an educational grant from the National Centre of Tumour Diseases Heidelberg (NCT IFP V/3). J. Kleesiek was supported by a postdoctoral fellowship from the Medical Faculty of the University of Heidelberg.
Paper previously presented at: Annual Meeting of the American Society of Neuroradiology and the Foundation of the ASNR Symposium, May 17–22, 2014; Montreal, Quebec, Canada.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97–109 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cha S. Neuroimaging in neuro-oncology. Neurotherapeutics 2009;6:465–77 10.1016/j.nurt.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. White ML, Zhang Y, Kirby P, et al. Can tumor contrast enhancement be used as a criterion for differentiating tumor grades of oligodendrogliomas? AJNR Am J Neuroradiol 2005;26:784–90 [PMC free article] [PubMed] [Google Scholar]
- 4. Scott JN, Brasher PM, Sevick RJ, et al. How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology 2002;59:947–49 10.1212/WNL.59.6.947 [DOI] [PubMed] [Google Scholar]
- 5. Price SJ, Gillard JH. Imaging biomarkers of brain tumour margin and tumour invasion. Br J Radiol 2011;84 Spec No 2:S159–67 10.1259/bjr/26838774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boada FE, LaVerde G, Jungreis C, et al. Loss of cell ion homeostasis and cell viability in the brain: what sodium MRI can tell us. Curr Topics Dev Biol 2005;70:77–101 10.1016/S0070-2153(05)70004-1 [DOI] [PubMed] [Google Scholar]
- 7. Nagel AM, Bock M, Hartmann C, et al. The potential of relaxation-weighted sodium magnetic resonance imaging as demonstrated on brain tumors. Invest Radiol 2011;46:539–47 10.1097/RLI.0b013e31821ae918 [DOI] [PubMed] [Google Scholar]
- 8. Stobbe R, Beaulieu C. In vivo sodium magnetic resonance imaging of the human brain using soft inversion recovery fluid attenuation. Magn Reson Med 2005;54:1305–10 10.1002/mrm.20696 [DOI] [PubMed] [Google Scholar]
- 9. Sahm F, Capper D, Preusser M, et al. BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood 2012;120:e28–34 10.1182/blood-2012-06-429597 [DOI] [PubMed] [Google Scholar]
- 10. Sahm F, Koelsche C, Meyer J, et al. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol 2012;123:853–60 10.1007/s00401-012-0993-5 [DOI] [PubMed] [Google Scholar]
- 11. Nagel AM, Laun FB, Weber MA, et al. Sodium MRI using a density-adapted 3D radial acquisition technique. Magn Reson Med 2009;62:1565–73 10.1002/mrm.22157 [DOI] [PubMed] [Google Scholar]
- 12. Sommer C, Straehle C, Koethe U, et al. ilastik: Interactive Learning and Segmentation Toolkit. In: Proceedings of the IEEE International Symposium on Biomedical Imaging, Chicago, Illinois. March 30–April 2, 2011:230–33 10.1109/ISBI.2011.5872394 [DOI] [Google Scholar]
- 13. R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2014 [Google Scholar]
- 14. Wick W, Weller M, van den Bent M, et al. MGMT testing: the challenges for biomarker-based glioma treatment. Nat Rev Neurol 2014;10:372–85 10.1038/nrneurol.2014.100 [DOI] [PubMed] [Google Scholar]
- 15. Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol 2014;15:e395–403 10.1016/S1470-2045(14)70011-7 [DOI] [PubMed] [Google Scholar]
- 16. Weiler M, Wick W. Molecular predictors of outcome in low-grade glioma. Curr Opin Neurol 2012;25:767–73 10.1097/WCO.0b013e32835a0217 [DOI] [PubMed] [Google Scholar]
- 17. Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 2009;27:4150–54 10.1200/JCO.2009.21.9832 [DOI] [PubMed] [Google Scholar]
- 18. van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res 2010;16:1597–604 10.1158/1078-0432.CCR-09-2902 [DOI] [PubMed] [Google Scholar]
- 19. Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol 2009;27:5743–50 10.1200/JCO.2009.23.0805 [DOI] [PubMed] [Google Scholar]
- 20. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun H, Yin L, Li S, et al. Prognostic significance of IDH mutation in adult low-grade gliomas: a meta-analysis. J Neurooncol 2013;113:277–84 10.1007/s11060-013-1107-5 [DOI] [PubMed] [Google Scholar]
- 22. Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol 2010;120:707–18 10.1007/s00401-010-0781-z [DOI] [PubMed] [Google Scholar]
- 23. Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol 2009;27:5874–80 10.1200/JCO.2009.23.6497 [DOI] [PubMed] [Google Scholar]
- 24. Kloosterhof NK, Bralten LB, Dubbink HJ, et al. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol 2011;12:83–91 10.1016/S1470-2045(10)70053-X [DOI] [PubMed] [Google Scholar]
- 25. Balss J, Meyer J, Mueller W, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 2008;116:597–602 10.1007/s00401-008-0455-2 [DOI] [PubMed] [Google Scholar]
- 26. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807–12 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Joshi AD, Parsons DW, Velculescu VE, et al. Sodium ion channel mutations in glioblastoma patients correlate with shorter survival. Mol Cancer 2011;10:17 10.1186/1476-4598-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cuddapah VA, Sontheimer H. Ion channels and transporters [corrected] in cancer. 2. Ion channels and the control of cancer cell migration. Am J Physiol Cell Physiol 2011;301:C541–49 10.1152/ajpcell.00102.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kunzelmann K. Ion channels and cancer. J Membrane Biol 2005;205:159–73 10.1007/s00232-005-0781-4 [DOI] [PubMed] [Google Scholar]
- 30. Rooj AK, McNicholas CM, Bartoszewski R, et al. Glioma-specific cation conductance regulates migration and cell cycle progression. J Biol Chem 2012;287:4053–65 10.1074/jbc.M111.311688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spector M, O'Neal S, Racker E. Phosphorylation of the beta subunit of Na+K+-ATPase in Ehrlich ascites tumor by a membrane-bound protein kinase. J Biol Chem 1980;255:8370–73 [PubMed] [Google Scholar]
- 32. Bulakbasi N, Kocaoglu M, Farzaliyev A, et al. Assessment of diagnostic accuracy of perfusion MR imaging in primary and metastatic solitary malignant brain tumors. AJNR Am J Neuroradiol 2005;26:2187–99 [PMC free article] [PubMed] [Google Scholar]
- 33. Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 2003;24:1989–98 [PMC free article] [PubMed] [Google Scholar]
- 34. Lev MH, Ozsunar Y, Henson JW, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected]. AJNR Am J Neuroradiol 2004;25:214–21 [PMC free article] [PubMed] [Google Scholar]
- 35. Mills SJ, Patankar TA, Haroon HA, et al. Do cerebral blood volume and contrast transfer coefficient predict prognosis in human glioma? AJNR Am J Neuroradiol 2006;27:853–58 [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen TB, Cron GO, Mercier JF, et al. Diagnostic accuracy of dynamic contrast-enhanced MR imaging using a phase-derived vascular input function in the preoperative grading of gliomas. AJNR Am J Neuroradiol 2012;33:1539–45 10.3174/ajnr.A3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roberts HC, Roberts TP, Brasch RC, et al. Quantitative measurement of microvascular permeability in human brain tumors achieved using dynamic contrast-enhanced MR imaging: correlation with histologic grade. AJNR Am J Neuroradiol 2000;21:891–99 [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang N, Zhang L, Qiu B, et al. Correlation of volume transfer coefficient Ktrans with histopathologic grades of gliomas. J Magn Reson Imag 2012;36:355–63 10.1002/jmri.v36.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hilario A, Sepulveda JM, Perez-Nuez A, et al. A prognostic model based on preoperative MRI predicts overall survival in patients with diffuse gliomas. AJNR Am J Neuroradiol 2014;35:1096–102 10.3174/ajnr.A3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kono K, Inoue Y, Nakayama K, et al. The role of diffusion-weighted imaging in patients with brain tumors. AJNR Am J Neuroradiol 2001;22:1081–88 [PMC free article] [PubMed] [Google Scholar]
- 41. Fountas KN, Kapsalaki EZ, Vogel RL, et al. Noninvasive histologic grading of solid astrocytomas using proton magnetic resonance spectroscopy. Stereotact Funct Neurosurg 2004;82:90–7 10.1159/000077458 [DOI] [PubMed] [Google Scholar]
- 42. Bisdas S, Kirkpatrick M, Giglio P, et al. Cerebral blood volume measurements by perfusion-weighted MR imaging in gliomas: ready for prime time in predicting short-term outcome and recurrent disease? AJNR Am J Neuroradiol 2009;30:681–88 10.3174/ajnr.A1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirai T, Murakami R, Nakamura H, et al. Prognostic value of perfusion MR imaging of high-grade astrocytomas: long-term follow-up study. AJNR Am J Neuroradiol 2008;29:1505–10 10.3174/ajnr.A1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Law M, Oh S, Babb JS, et al. Low-grade gliomas: dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging—prediction of patient clinical response. Radiology 2006;238:658–67 10.1148/radiol.2382042180 [DOI] [PubMed] [Google Scholar]
- 45. Law M, Young RJ, Babb JS, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008;247:490–98 10.1148/radiol.2472070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spampinato MV, Schiarelli C, Cianfoni A, et al. Correlation between cerebral blood volume measurements by perfusion-weighted magnetic resonance imaging and two-year progression-free survival in gliomas. Neuroradiol J 2013;26:385–95 10.1177/197140091302600404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tzika AA, Astrakas LG, Zarifi MK, et al. Spectroscopic and perfusion magnetic resonance imaging predictors of progression in pediatric brain tumors. Cancer 2004;100:1246–56 10.1002/cncr.20096 [DOI] [PubMed] [Google Scholar]
- 48. Jain R, Poisson LM, Gutman D, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 2014;272:484–93 10.1148/radiol.14131691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cao Y, Nagesh V, Hamstra D, et al. The extent and severity of vascular leakage as evidence of tumor aggressiveness in high-grade gliomas. Cancer Res 2006;66:8912–17 10.1158/0008-5472.CAN-05-4328 [DOI] [PubMed] [Google Scholar]
- 50. Ellingson BM, Malkin MG, Rand SD, et al. Volumetric analysis of functional diffusion maps is a predictive imaging biomarker for cytotoxic and anti-angiogenic treatments in malignant gliomas. J Neurooncol 2011;102:95–103 10.1007/s11060-010-0293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barajas RF Jr, Rubenstein JL, Chang JS, et al. Diffusion-weighted MR imaging derived apparent diffusion coefficient is predictive of clinical outcome in primary central nervous system lymphoma. AJNR Am J Neuroradiol 2010;31:60–66 10.3174/ajnr.A1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valles FE, Perez-Valles CL, Regalado S, et al. Combined diffusion and perfusion MR imaging as biomarkers of prognosis in immunocompetent patients with primary central nervous system lymphoma. AJNR Am J Neuroradiol 2013;34:35–40 10.3174/ajnr.A3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y, Lupo JM, Parvataneni R, et al. Survival analysis in patients with newly diagnosed glioblastoma using pre- and postradiotherapy MR spectroscopic imaging. Neuro Oncol 2013;15:607–17 10.1093/neuonc/nos334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andronesi OC, Rapalino O, Gerstner E, et al. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest 2013;123:3659–63 10.1172/JCI67229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Park CC, Hartmann C, Folkerth R, et al. Systemic metastasis in glioblastoma may represent the emergence of neoplastic subclones. J Neuropathol Exp Neurol 2000;59:1044–50 [DOI] [PubMed] [Google Scholar]
- 56. Gnahm C, Behl NGR, Biller A, et al. Anatomically Weighted 2nd Order Total Variation Reconstruction of 23Na MRI Using 1H Prior Information. In: Proceedings of the 22nd ISMRM Scientific Meeting, Milan, Italy. May 10–16, 2014 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.