Abstract
BACKGROUND AND PURPOSE:
Although perfusion and permeability MR parameters have known to have prognostic value, they have reproducibility issues. Our aim was to evaluate whether the initial area under the time-to-signal intensity curve (IAUC) derived from dynamic contrast-enhanced MR imaging can improve prognosis prediction in patients with glioblastoma with known MGMT status.
MATERIALS AND METHODS:
We retrospectively examined 88 patients with glioblastoma who underwent preoperative dynamic contrast-enhanced MR imaging. The means of IAUC values at 30 and 60 seconds (IAUC30mean and IAUC60mean) were extracted from enhancing tumors. The prognostic values of IAUC parameters for overall survival and progression-free survival were assessed with log-rank tests, according to the MGMT status. Multivariate overall survival and progression-free survival models before and after adding the IAUC parameters as covariates were explored by net reclassification improvement after receiver operating characteristic analysis for 1.5-year overall survival and 1-year progression-free survival and by random survival forest.
RESULTS:
High IAUC parameters were associated with worse overall survival and progression-free survival in the unmethylated MGMT group, but not in the methylated group. In the unmethylated MGMT group, 1.5-year overall survival and 1-year progression-free survival prediction improved significantly after adding IAUC parameters (overall survival area under the receiver operating characteristic curve, 0.86; progression-free survival area under the receiver operating characteristic curve, 0.74–0.76) to the model with other prognostic factors (overall survival area under the receiver operating characteristic curve, 0.81; progression-free survival area under the receiver operating characteristic curve, 0.69; P < .05 for all) except in the case of IAUC60mean for 1-year progression-free survival prediction (P = .059). Random survival forest models indicated that the IAUC parameters were the second or most important predictors in the unmethylated MGMT group, except in the case of the IAUC60mean for progression-free survival.
CONCLUSIONS:
IAUC can be a useful prognostic imaging biomarker in patients with glioblastoma with known MGMT status, improving prediction of glioblastoma prognosis with the unmethylated MGMT promoter status.
Glioblastoma (GBM) is the most common primary malignant tumor in the adult brain. Although its prognosis remains poor (median survival, ∼14.7 months),1 some patients with GBM show a distinct prognosis and response to chemoradiation. Previous studies have investigated the prognostic factors of GBM, including MR imaging2–4 and molecular biomarkers (O6-methylguanine-DNA methyltransferase [MGMT] promoter methylation).5
Previous studies have shown that high relative cerebral blood volume (rCBV; derived via dynamic susceptibility contrast-enhanced MR imaging) and high volume transfer constant (Ktrans; derived via dynamic contrast-enhanced [DCE] MR imaging) are associated with poor survival outcomes.6–8 However, rCBV and Ktrans measurements have reproducibility issues related to postprocessing techniques, including normalization, model-based calculation, arterial input function, and software.9–13 In contrast, the initial area under the time-to–signal intensity curve (IAUC), derived via DCE MR imaging, is a model-free parameter that does not require an arterial input function or a complicated model-based calculation and is highly reproducible.10,14 Because IAUC reflects both tumor perfusion and permeability,9,10,15 we hypothesized that IAUC might be useful for predicting the survival outcome in GBM. In this study, we used automatically calculated IAUC parameters to maximize reproducibility.
MGMT removes alkyl groups from the alkylation site of temozolomide. Methylation of the MGMT promoter inhibits MGMT activity and yields a better response to temozolomide and improved prognosis.5 Because the MGMT status is available after surgery in most patients with GBM, the predictive ability of IAUC for prognosis in patients with GBM with known MGMT status is clinically relevant. Here, we aimed to assess whether IAUC can improve prognosis prediction in patients with GBM with known MGMT status.
Materials and Methods
Our institutional review board approved this retrospective study and waived the requirement for informed consent.
Study Population
We reviewed the records of 101 consecutive patients who underwent preoperative DCE MR imaging for newly diagnosed GBM from October 2010 to July 2014. The enrollment process is shown in On-line Fig 1. The inclusion criteria were pathologically confirmed GBM, no prior biopsy or treatment, and available MGMT promoter methylation status. The exclusion criteria were previous brain biopsy or treatments (n = 6), unavailable MGMT status (n = 3), and inadequate DCE image quality, leading to processing errors (n = 4). Thus, 88 patients were enrolled.
Imaging
A 3T MR imaging unit (Achieva; Philips Healthcare, Best, the Netherlands) and an 8-channel sensitivity encoding head coil were used for preoperative MR imaging. The protocol included T1-weighted turbo spin-echo images with inversion recovery (TR, 2000 ms; TE, 10 ms; TI, 1000 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256), T2-weighted turbo spin-echo (TR, 3000 ms; TE, 80 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256), and T2-weighted fluid-attenuated inversion recovery (TR, 10,000 ms; TE, 125 ms; TI, 2500 ms; FOV, 240 mm; section thickness, 5 mm; matrix, 256 × 256) images. 3D T1-weighted turbo field echo images (TR, 9.8 ms; TE, 4.6 ms; FOV, 240 mm; section thickness, 1 mm; matrix, 224 × 224) were acquired after the injection of gadolinium-based contrast (0.1 mL/kg of gadobutrol, Gadovist; Bayer Schering Pharma, Berlin, Germany). For DCE-MR imaging, 60 dynamic phases of DCE T1-weighted images were acquired with the following parameters: TR, 6.3 ms; TE, 3.1 ms; FOV, 240 mm; matrix, 192 × 192; section thickness, 3 mm; flip angle, 15°. After we acquired the fifth phase, gadolinium-based contrast (0.1 mL/kg of gadobutrol) was injected (rate, 3 mL/s). The total acquisition time for DCE-MR imaging was 6 minutes 18 seconds and the temporal resolution was approximately 6 seconds.
Image Postprocessing and Analysis
The ROIs were drawn by the consensus of 2 neuroradiologists (Y.S.C. and S.-K.L., with 4 and 20 years of experience in neuroradiology, respectively) who were blinded to the MGMT status and other clinical information, using a semiautomatic signal-intensity-threshold method to include the entire enhanced portion of the tumors on postcontrast T1-weighted images, while avoiding necrotic or cystic portions.
IAUC values were acquired by integrating the area under the time-to-signal-intensity curve, from 0 to 30 seconds (IAUC30) and 0 to 60 seconds (IAUC60) after contrast agent arrival on the basis of previous studies.10 IAUC values were automatically calculated without any user-dependent input, to maximize interrater reliability, through automatic determination of the time of contrast agent arrival for each pixel; the time of contrast agent arrival was defined as the initial time at which a signal increase of >5% of the maximum signal increase was observed during >2 serial phases. The combination of this automatic calculation of IAUC and semiautomatic ROI segmentation of enhancing tumor yields almost perfect reproducibility.16 The IAUC values were normalized to the maximum signal increase. A detailed description of the image processing is provided in the On-line Appendix. The mean values of IAUC30 (IAUC30mean) and IAUC60 (IAUC60mean) were extracted from the enhanced tumors. Images were analyzed with Medical Image Processing, Analysis, and Visualization (Version 7.0; National Institutes of Health, Bethesda, Maryland) and R software, Version 3.0.2 (R Foundation, http://www.r-project.org/).
MGMT and Other Prognostic Parameters
MGMT promoter methylation status was evaluated with a methylation-specific polymerase chain reaction, which was retrospectively recorded from pathologic reports. Because IAUC may be related to other genetic profiles, including isocitrate dehydrogenase 1 (IDH1) mutation status, epidermal growth factor receptor (EGFR) expression, and the presence of 1p/19q codeletion, the data regarding these profiles were also recorded as described in the On-line Appendix. The presence of oligodendroglial components was also recorded because the presence of these components is related to better prognosis.17 Furthermore, we recorded the following clinical factors: age, sex, Karnofsky performance status, extent of resection, and postoperative treatment, along with conventional MR imaging findings. The extent of resection was categorized as total, subtotal (<100% but ≥75% of gross tumor removal), or partial (<75% of gross tumor removal) resection/biopsy based on the intraoperative impressions of the surgeons (J.H.C., S.-G.K., and E.H.K., with 20, 16, and 10 years of experience in neurosurgery, respectively) and postoperative MR imaging. Postoperative treatment was categorized as concurrent chemoradiation with temozolomide, radiation therapy only, temozolomide-based chemotherapy only, and no postoperative treatment.
Conventional MR imaging findings included volume of enhanced tumor, degree of edema, presence of non-contrast-enhanced tumor (nCET), presence of deep white-matter invasion, and eloquent brain involvement, which are associated with prognosis.2,3 Conventional MR imaging findings were determined on the basis of the consensus of 2 neuroradiologists (Y.S.C. and S.-K.L.), except for the enhanced tumor volume, which was automatically calculated from the ROIs on postcontrast T1-weighted images. Edema and nCET were assessed with axial T2-weighted or fluid-attenuated inversion recovery images, as described elsewhere.3,8 Edema was scored as 0 (not apparent, ≤1 cm), 1 (mild-to-moderate, >1 cm and ≤2 cm), and 2 (severe, >2 cm) on the basis of the maximum length of the edema; and nCET was classified as positive if the volume was >25% of the enhanced tumor. The presence of eloquent brain involvement and deep white-matter invasion was determined with the Visually Accessible Repository for Molecular Brain Neoplasia Data Images lexicon.2,18
Statistical Analysis
Overall survival (OS) duration was defined as the time from MR imaging–based diagnosis to death or the final follow-up in living cases. Progression-free survival (PFS) duration was defined as the time from MR imaging–based diagnosis to tumor progression, recurrence, death, or the final follow-up in cases in which no tumor progression or death was noted, according to the Response Assessment in Neuro-Oncology19 as described elsewhere.4,8
Univariate survival analyses for OS and PFS of the entire cohort were performed with Kaplan-Meier estimates and the log-rank test; the analyses included IAUC30mean, IAUC60mean, MGMT status, the presence of an oligodendroglial component, clinical factors (age, sex, Karnofsky performance status, extent of tumor resection, and postoperative treatment), and conventional MR imaging findings (enhanced tumor volume, edema, nCET, eloquent brain involvement, and deep white matter invasion). Subgroup survival analyses of the methylated and unmethylated MGMT groups were performed with the IAUC parameters. If the variables were continuous or had >3 categories, they were dichotomized by the optimal cutoff values for OS and PFS that were calculated by using the “cutp” function of the “survMisc” R package, as defined by Contal and O'Quigley.20 This method determines the optimal cutoff value yielding the most significant prognostic differences between the 2 groups based on test scores from the Cox regression model.20
The multivariate models were established with the significant factors of univariate analyses for OS and PFS in the unmethylated MGMT promoter group because the IAUC parameters were prognostic for the unmethylated MGMT group only, as shown later. In the multivariate models, all the variables that were continuous or had >3 categories were dichotomized, as in the univariate analyses. The multivariate models for OS and PFS were established from other prognostic factors only (model 0), and IAUC30mean and IAUC60mean were subsequently added as covariate parameters (IAUC30mean in model 1 and IAUC60mean in model 2). These multivariate models before (OS and PFS model 0) and after (OS and PFS models 1 and 2) adding IAUC parameters were explored and compared using 2 methods: 1) The diagnostic accuracies for 1.5-year OS and 1-year PFS prediction were assessed by receiver operative characteristic (ROC) analysis. Then, the reclassification improvement on adding the IAUC parameters was assessed using net reclassification improvement, as described by Pencina et al.21 2) The random survival forest method was used to determine the variable importance of the IAUC parameters in prognosis prediction. The forests consisted of 50,000 trees, having a minimum of 3 samples in a terminal node. The IAUC parameters and other prognostic factors were ranked by the relative importance scores, and the C-indices of the random survival forest models were calculated. Representative trees were constructed with the variables that showed the highest importance scores.22,23
We compared the IAUC parameters according to MGMT status and the other genetic profiles using the Student t test, on the basis of the results of the Shapiro-Wilk test for normality. Statistical analyses were performed with Stata (Version 12.1; StataCorp, College Station, Texas) and R for Windows, Version 3.0.2. A P value < .05 was considered significant.
Results
The clinical characteristics of the patients according to the MGMT status are summarized in On-line Table 1. None of the clinical characteristics, except for median OS and PFS, showed a significant difference between the methylated and unmethylated MGMT groups. The median OS and PFS were 442 days and 272 days, respectively, in the unmethylated MGMT group, whereas they were 790 days and 657 days, respectively, in the methylated MGMT group (log-rank test, P = .014 and .003 for OS and PFS, respectively). The other genetic characteristics of patients are summarized in On-line Table 2.
Univariate Survival Analyses
The results of the univariate analyses for OS and PFS in the entire cohort are summarized in On-line Table 3. The IAUC parameters were not significant predictors of OS and PFS in the entire cohort, though high IAUC parameters tended to be associated with poor OS (P = .090 for IAUC30mean, P = .072 for IAUC60mean). Unmethylated MGMT, 66 years of age or older, less than total resection, postoperative treatment other than concurrent chemoradiation with temozolomide, and enhanced tumor volume of ≥30.2 cm3 were associated with poor OS (P < .05 for all). Similarly, unmethylated MGMT, 70 years of age or older, less than total resection, postoperative treatment other than concurrent chemoradiation with temozolomide, and enhanced tumor volume of ≥30.2 cm3 were associated with poor PFS (P < .05 for all). Eloquent brain involvement was associated with poor OS (P = .033), but not with PFS. Sex, Karnofsky performance status, edema, nCET, and deep white-matter invasion were not associated with OS or PFS.
The results of the subgroup univariate analyses of the IAUC parameters in the methylated and unmethylated MGMT groups are summarized in Table 1 and Fig 1. High IAUC parameters were significant predictors of poor OS (P = .008 for IAUC30mean, P = .015 for IAUC60mean) and poor PFS (P = .010 for IAUC30mean, P = .015 for IAUC60mean) in the unmethylated MGMT group, but not in the methylated MGMT group. Therefore, we established multivariate models for the unmethylated MGMT group alone. Representative cases of GBMs with unmethylated MGMT promoter status are shown in Figs 2 and 3.
Table 1:
Univariate analysis of IAUC histogram parameters as predictors of OS and PFS in the unmethylated and methylated MGMT subgroups.
| Survival/Parameters | Unmethylated MGMT (n = 51) |
Methylated MGMT (n = 37) |
||||
|---|---|---|---|---|---|---|
| Cutoff Value | HR (95% CI) | P Valuea | Cutoff Value | HR (95% CI) | P Valuea | |
| OS | ||||||
| IAUC30mean | ≥10.9 | 3.04 (1.28–7.21) | .008 | ≥15.1 | 0.60 (0.17–2.14) | .429 |
| IAUC60mean | ≥28.4 | 3.48 (1.20–10.14) | .015 | ≥32.0 | 0.42 (0.16–1.14) | .079 |
| PFS | ||||||
| IAUC30mean | ≥10.9 | 2.60 (1.23–5.50) | .010 | ≥15.1 | 0.52 (0.18–1.56) | .238 |
| IAUC60mean | ≥32.6 | 2.15 (1.14–4.05) | .015 | ≥32.8 | 0.24 (0.22–1.28) | .155 |
Note:—HR indicates hazard ratio.
Calculated from the log-rank test.
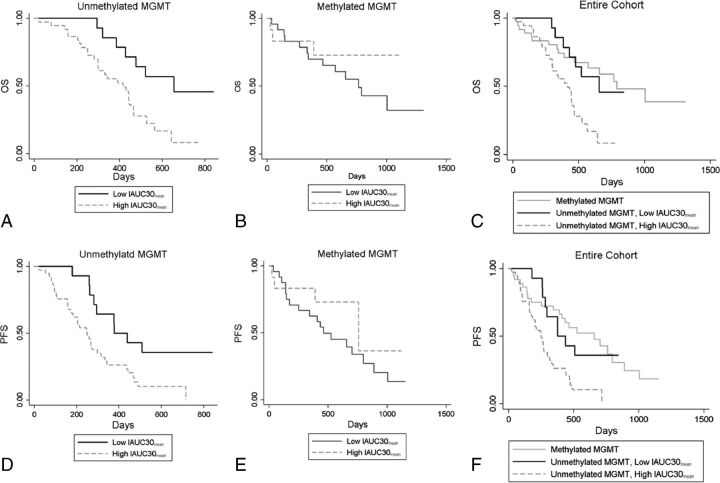
Fig 1.
Kaplan-Meier curves of overall survival (A–C) and progression-free survival (D–F) with IAUC30mean in the unmethylated MGMT (A and D, n = 51) and methylated MGMT groups (B and E, n = 31) and the entire cohort (C and F, n = 88). IAUC30mean stratified OS (log-rank test, P = .008) and PFS (log-rank test, P = .010) only in the unmethylated MGMT group, but not in the methylated MGMT group (log-rank test, P = .429 for OS, P = .238 for PFS).
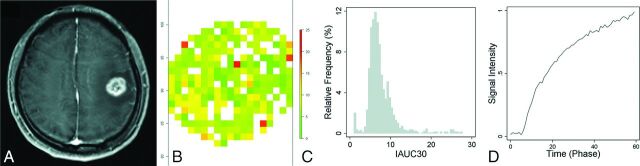
Fig 2.
A 52-year-old male patient with glioblastoma with unmethylated MGMT promoter status, whose overall survival time was 477 days. T1 weighted images (A), IAUC30 parameter map of the enhancing tumor (B), the relative frequency histogram of IAUC30 (C), and the time to the mean signal-intensity curve of the enhancing tumor (D). The volume of enhancing tumor was 8.4 cm3, and the IAUC30mean was 5.4.
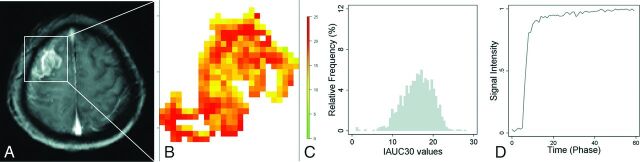
Fig 3.
A 59-year-old female patient with glioblastoma with unmethylated MGMT promoter status, whose overall survival time was 250 days. T1-weighted image (A), IAUC30 parameter map of the enhancing tumor (B), the relative frequency histogram of IAUC30 (C), and the time to the mean signal-intensity curve of the enhancing tumor (D). The volume of the enhancing tumor was 7.4 cm3, and the IAUC30mean was 16.1.
ROC Analyses and Net Reclassification Improvement for Predicting 1.5-Year OS and 1-Year PFS
The results of the ROC analyses with the multivariate models for 1.5-year OS and 1-year PFS in the unmethylated MGMT group are summarized in Table 2. Among the 51 patients in the unmethylated MGMT group, 8 were excluded from the 1.5-year OS analysis and 1 was excluded from the 1-year PFS analysis because the follow-up duration was insufficient. Both OS and PFS model 0 included age, extent of resection, postoperative treatment, and enhanced tumor volume as covariates; eloquent brain involvement was included as a covariate in the OS model 0 only, according to the results of the univariate analyses. When the IAUC30mean or IAUC60mean were added in the OS model 0 and PFS model 0 as covariates, the diagnostic accuracies improved for 1.5-year OS prediction (OS models 1 and 2 area under the receiver operating characteristic curve [AUC], 0.86) and 1-year PFS prediction (PFS model 1 AUC, 0.76; PFS model 2 AUC, 0.74), compared with model 0 (OS model 0 AUC, 0.81; PFS model 0 AUC, 0.69). Reclassification improvement by adding IAUC parameters was statistically significant, as assessed by net reclassification improvement (P = .005 and .018 for OS models 1 and 2; P = .020 with PFS model 1), except in the case of PFS model 2 with IAUC60mean, which showed borderline significance (P = .059).
Table 2:
Reclassification improvement by adding IAUC parameters for predicting 1.5-year OS (n = 43) and 1-year PFS (n = 50) in the unmethylated MGMT group
| Survival Time/Models | AUC (95% CI) | NRI (95% CI) | P Valuea | |
|---|---|---|---|---|
| 1.5-year OS (n = 43) | ||||
| Model 0 (ref) | Age, extent of resection, postoperative treatment, volume of enhancing tumors, eloquent brain involvement | 0.81 (0.65–0.96) | – | – |
| Model 1 | IAUC30mean | 0.86 (0.74–0.98) | 0.95 (0.61–1.29) | .005 |
| Model 2 | IAUC60mean | 0.86 (0.72–0.99) | 0.81 (0.47–1.15) | .018 |
| 1-year PFS (n = 50) | ||||
| Model 0 (ref) | Age, extent of resection, postoperative treatment, volume of enhancing tumors | 0.69 (0.54–0.84) | – | – |
| Model 1 | IAUC30mean | 0.76 (0.62–0.90) | 0.69 (0.39–0.98) | .020 |
| Model 2 | IAUC60mean | 0.74 (0.58–0.89) | 0.56 (0.26–0.85) | .059 |
Note:—NRI indicates net reclassification improvement; ref, reference.
Calculated from category-less NRI.
Contribution of IAUC Parameters in the Multivariate Model of Random Survival Forest
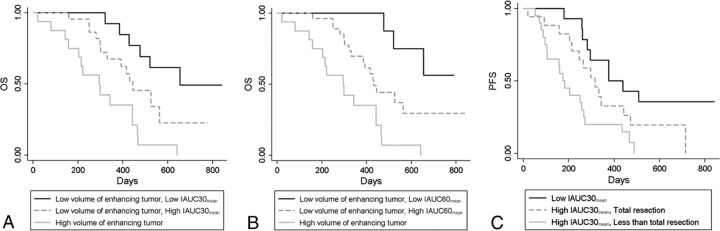
Random survival forests for OS showed that IAUC parameters were the second most important variable, following the volume of enhancing tumor as the top variable and followed by postoperative treatment as the third most important variable. The variable importance of random survival forests is shown in On-line Figs 2 and 3. The relative importance was 41% for IAUC30mean in OS model 1 and 57% for IAUC60mean in OS model 2, assuming that the relative importance of volume of enhancing tumor was 100%. Generation of a signal tree from this forest using the top 2 predictors (ie, volume of enhancing tumor and IAUC parameters) resulted in volume of enhancing tumor as the primary split and IAUC parameters as the secondary split for a group with a low volume of enhancing tumor (Fig 4). This splitting caused a significant difference in survival between the groups (P < .001 for trees with IAUC30mean and IAUC60mean, log-rank test), with a difference in mean survival of 354 days and 388 days between the high volume of enhancing tumors versus low volume of enhancing tumors with low IAUC30mean and IAUC60mean, respectively. The C-indices were higher after the IAUC parameters were added in OS models 1 and 2 (C-index = 0.67), compared with OS model 0 (C-index = 0.61).
Fig 4.
Kaplan-Meier curves depicting survival classification after random survival forest ranking of prognostic factors in glioblastomas with unmethylated MGMT promoter status (n = 51). Representative trees with IAUC30mean (A) and IAUC60mean (B) for OS caused the volume of enhancing tumor to be the primary split and then the IAUC30mean to split the subgroup with a low volume of enhancing tumor subsequently (log-rank test, P < .001). C, A representative tree with IAUC30mean for PFS caused the IAUC30mean to be the primary split and then the extent of resection to split the subgroup with high IAUC30mean subsequently (log-rank test, P < .002).
Random survival forests for PFS showed that IAUC30mean in PFS model 1 was the most important variable, followed by extent of resection and volume of enhancing tumor as the second and third most important variables, respectively, whereas in PFS model 2, IAUC60mean was the third most important variable, following the extent of resection and volume of enhancing tumor as the first and second most important variables, respectively. The relative importance was 100% for IAUC30mean and 94% for extent of resection in PFS model 1. Generation of a single tree with the top 3 predictors of PFS model 1 resulted in IAUC30mean as the primary split, and the low IAUC30mean group was subsequently split by the extent of resection; this split caused significantly different survival among the groups (Fig 4, P = .002, log-rank test). The difference in mean survival was 293 days in tumors with high IAUC30mean versus those with low IAUC30mean and less than total resection. The C-indices were higher after adding the IAUC parameters in PFS models 1 and 2 (C-indices = 0.55 and 0.54, respectively), compared with that of PFS model 0 without IAUC parameters (C-index = 0.51).
Differences in IAUC Parameters between the Unmethylated and Methylated MGMT Promoter Groups
The differences in the IAUC parameters according to MGMT status are illustrated in On-line Fig 4. The IAUC parameters were significantly higher in the methylated MGMT group than in the unmethylated group (13.5 ± 2.2 versus 12.2 ± 2.1, P = .007 for IAUC30mean; 33.7 ± 4.6 versus 31.5 ± 4.3, P = .025 for IAUC60mean). The differences in the IAUC parameters according to the other genetic profiles are described in the On-line Appendix.
Discussion
In this study, we found that higher IAUC values were predictors of poor OS and PFS in the unmethylated MGMT group and that IAUC parameters improved prognosis prediction of the unmethylated MGMT group, when added to other prognostic factors. Furthermore, IAUC values were paradoxically higher in the methylated MGMT group than in the unmethylated group.
rCBV and Ktrans values are indicators of tumor perfusion and permeability, respectively, and high values are predictors of worse prognosis.6–8,22 However, the variability of rCBV ranges from 24% to 67%, depending on the normalization method or software used for postprocessing.12 Similarly, Ktrans can vary according to the model selection, arterial input function, software, and prebolus T1 measurement.9–11 In contrast, IAUC is a model-free parameter that does not depend on model selection and arterial input function and is highly reproducible.10,14 Whether model-free parameters show high reproducibility and performance, compared with model-based parameters, was debated recently.11,14,24 While 1 study11 reported that mean Ktrans showed wider interobserver variability than model-free parameters, another study24 reported that the reproducibility between the model-free parameter and Ktrans did not differ significantly. In the latter study, the time of contrast agent arrival was manually defined to calculate the model-free parameters. In our study, the time of contrast agent arrival was automatically determined for each pixel; With this automatic determination of the time of contarst agent arrival, the IAUC has been reported to achieve almost perfect reproducibility.16
Despite the limitation of IAUC regarding the lack of a clear biologic association, IAUC values are correlated with Ktrans and rCBV.9,15 Hence, we hypothesized that IAUC would be useful for prognosis prediction because it reflects both perfusion and permeability, which are complementarily associated with prognosis. Although previous studies have reported that model-free parameters may be useful for differentiation of radiation necrosis and tumor,10,14 the prognostic value of IAUC in GBM remains unclear. Moreover, to establish the practical role of an imaging biomarker, one must elucidate whether that imaging biomarker has prognostic value after other prognostic factors are considered in the usual clinical settings. We found that IAUC parameters have prognostic value even after clinical factors and conventional MR imaging findings are considered in the unmethylated MGMT group. Therefore, IAUC represents a useful imaging biomarker that has practicality and high reproducibility. Nevertheless, considering that IAUC60mean did not achieve the same improvement in PFS prediction as IAUC30mean, the optimal time point of IAUC needs to be determined in future investigations.
The association between high IAUC values and poor prognosis in the unmethylated group is consistent with findings in previous studies, which have reported the association between high rCBV and Ktrans values and poor prognosis.6–8,22 However, in our study, the IAUC values were not prognostic when applied to the methylated MGMT group or the entire GBM cohort, and IAUC values were paradoxically higher in the methylated MGMT group than in the unmethylated group. Some studies have reported the correlation between MGMT status and rCBV and Ktrans values, with conflicting results; one study reported that rCBV values were higher in GBM cases with unmethylated MGMT than in those with methylated MGMT.25 In contrast, another study reported that Ktrans values were higher in GBMs with methylated MGMT, a finding discordant with the previously reported association between high Ktrans values and poor prognosis, and the authors hypothesized that high permeability of the methylated MGMT group allowed better penetration of temozolomide, yielding better response and prognosis.26
Although the underlying mechanism is unclear, our results may reflect IAUC values being more affected by permeability difference than perfusion difference between the methylated and unmethylated MGMT groups. Subsequently, a combination of these paradoxically high IAUC values of the methylated MGMT group and the effect of methylated MGMT as a good prognostic factor might have countervailed the association between high IAUC values and poor prognosis. This possibility is 1 explanation for our result that the IAUC values were not prognostic in the methylated MGMT group or the entire cohort. Similarly, a previous study27 reported that low Ktrans values were associated with poor prognosis in GBM, which is contradictory to the results of other studies on the association between high Ktrans values and poor prognosis.7,8 These conflicting results might be attributable to the complicated relationship among tumor permeability, MGMT status, and prognosis, as observed in our study. Future investigation with a larger cohort is necessary to analyze the relationship among tumor permeability, MGMT status, and prognosis. Given that the IAUC values have prognostic value only in the unmethylated MGMT group, the MGMT status should be known before IAUC may be considered a useful imaging biomarker. However, because the MGMT status is clinically available in most patients with GBM, in our opinion, this requirement does not mitigate the practicality of IAUC.
This study has several limitations, apart from its retrospective design. First, the IAUC parameters were not directly compared with rCBV and Ktrans. Thus, the superiority of IAUC regarding its prognostic value, reflecting both rCBV and Ktrans, was not directly proved in this study. Second, because IAUC values were prognostic in the unmethylated MGMT group only, the sample size was too small to be strictly validated to confirm the improvement in prognosis prediction by the addition of IAUC values. Third, although the IAUC is highly reproducible within the same scans from the same institutions, there may be variations among scans performed at separate times or at different institutions. Thus, this model-free parameter may have limited reproducibility.28
Conclusions
Our study demonstrated that IAUC could be a useful prognostic imaging biomarker in patients with GBM with known MGMT status, improving the prognosis prediction of GBM with unmethylated MGMT promoter status. We propose that this complementary role of IAUC and MGMT status should be considered in future investigations.
Supplementary Material
ABBREVIATIONS:
- AUC
area under the receiver operating characteristic curve
- DCE
dynamic contrast-enhanced
- EGFR
epidermal growth factor receptor
- GBM
glioblastoma
- IAUC
initial area under the time-to–signal intensity curve
- IDH1
isocitrate dehydrogenase 1
- Ktrans
volume transfer constant
- MGMT
O6-methylguanine-DNA methyltransferase
- nCET
non-contrast-enhanced tumor
- OS
overall survival
- PFS
progression-free survival
- rCBV
relative cerebral blood volume
- ROC
receiver operating characteristic
REFERENCES
- 1. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2. Wangaryattawanich P, Hatami M, Wang J, et al. Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: imaging predictors of overall and progression-free survival. Neuro Oncol 2015;17:1525–37 10.1093/neuonc/nov117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pope WB, Sayre J, Perlina A, et al. MR imaging correlates of survival in patients with high-grade gliomas. AJNR Am J Neuroradiol 2005;26:2466–74 [PMC free article] [PubMed] [Google Scholar]
- 4. Choi YS, Ahn SS, Kim DW, et al. Incremental prognostic value of ADC histogram analysis over MGMT promoter methylation status in patients with glioblastoma. Radiology 2016;281:175–84 [DOI] [PubMed] [Google Scholar]
- 5. Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 2004;10:1871–74 10.1158/1078-0432.CCR-03-0384 [DOI] [PubMed] [Google Scholar]
- 6. Law M, Young RJ, Babb JS, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008;247:490–98 10.1148/radiol.2472070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nguyen TB, Cron GO, Mercier JF, et al. Preoperative prognostic value of dynamic contrast-enhanced MRI-derived contrast transfer coefficient and plasma volume in patients with cerebral gliomas. AJNR Am J Neuroradiol 2015;36:63–69 10.3174/ajnr.A4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi YS, Kim DW, Lee SK, et al. The added prognostic value of preoperative dynamic contrast-enhanced MRI histogram analysis in patients with glioblastoma: analysis of overall and progression-free survival. AJNR Am J Neuroradiol 2015;36:2235–41 10.3174/ajnr.A4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng HL. Improved correlation to quantitative DCE-MRI pharmacokinetic parameters using a modified initial area under the uptake curve (mIAUC) approach. J Magn Reson Imaging 2009;30:864–72 10.1002/jmri.21916 [DOI] [PubMed] [Google Scholar]
- 10. Chung WJ, Kim HS, Kim N, et al. Recurrent glioblastoma: optimum area under the curve method derived from dynamic contrast-enhanced T1-weighted perfusion MR imaging. Radiology 2013;269:561–68 10.1148/radiol.13130016 [DOI] [PubMed] [Google Scholar]
- 11. Hamilton JD, Lin J, Ison C, et al. Dynamic contrast-enhanced perfusion processing for neuroradiologists: model-dependent analysis may not be necessary for determining recurrent high-grade glioma versus treatment effect. AJNR Am J Neuroradiol 2015;36:686–93 10.3174/ajnr.A4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prah MA, Stufflebeam SM, Paulson ES, et al. Repeatability of standardized and normalized relative CBV in patients with newly diagnosed glioblastoma. AJNR Am J Neuroradiol 2015;36:1654–61 10.3174/ajnr.A4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paulson ES, Schmainda KM. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology 2008;249:601–13 10.1148/radiol.2492071659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Narang J, Jain R, Arbab AS, et al. Differentiating treatment-induced necrosis from recurrent/progressive brain tumor using nonmodel-based semiquantitative indices derived from dynamic contrast-enhanced T1-weighted MR perfusion. Neuro Oncol 2011;13:1037–46 10.1093/neuonc/nor075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walker-Samuel S, Leach MO, Collins DJ. Evaluation of response to treatment using DCE-MRI: the relationship between initial area under the gadolinium curve (IAUGC) and quantitative pharmacokinetic analysis. Phys Med Biol 2006;51:3593–602 10.1088/0031-9155/51/14/021 [DOI] [PubMed] [Google Scholar]
- 16. Choi YS, Lee HJ, Ahn SS, et al. Primary central nervous system lymphoma and atypical glioblastoma: differentiation using the initial area under the curve derived from dynamic contrast-enhanced MR and the apparent diffusion coefficient. Eur Radiol 2017;27:1344–51 10.1007/s00330-016-4484-2 [DOI] [PubMed] [Google Scholar]
- 17. Appin CL, Gao J, Chisolm C, et al. Glioblastoma with oligodendroglioma component (GBM-O): molecular genetic and clinical characteristics. Brain Pathol 2013;23:454–61 10.1111/bpa.12018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vasari MRI Visual Feature Guide. The Cancer Imaging Archive (TCIA) Web site. https://wiki.cancerimagingarchive.net/dis-play/Public/VASARI+Research+Project. . Published March 25, 2015Accessed December 12, 2015.
- 19. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology working group. J Clin Oncol 2010;28:1963–72 10.1200/JCO.2009.26.3541 [DOI] [PubMed] [Google Scholar]
- 20. Contal C, O'Quigley J. An application of changepoint methods in studying the effect of age on survival in breast cancer. Computational Statistics & Data Analysis 1999;30:253–70 [Google Scholar]
- 21. Pencina MJ, D' Agostino RB, D' Agostino RB, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157–72 [DOI] [PubMed] [Google Scholar]
- 22. Jain R, Poisson LM, Gutman D, et al. Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 2014;272:484–93 10.1148/radiol.14131691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishwaran H, Kogalur UB, Blackstone EH, et al. Random survival forests. The Annals of Applied Statistics 2008;2:841–60 10.1214/08-AOAS169 [DOI] [Google Scholar]
- 24. Roberts C, Issa B, Stone A, et al. Comparative study into the robustness of compartmental modeling and model-free analysis in DCE-MRI studies. J Magn Reson Imaging 2006;23:554–63 10.1002/jmri.20529 [DOI] [PubMed] [Google Scholar]
- 25. Ryoo I, Choi SH, Kim JH, et al. Cerebral blood volume calculated by dynamic susceptibility contrast-enhanced perfusion MR imaging: preliminary correlation study with glioblastoma genetic profiles. PLoS One 2013;8:e71704 10.1371/journal.pone.0071704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahn SS, Shin NY, Chang JH, et al. Prediction of methylguanine methyltransferase promoter methylation in glioblastoma using dynamic contrast-enhanced magnetic resonance and diffusion tensor imaging. J Neurosurg 2014;121:367–73 10.3171/2014.5.JNS132279 [DOI] [PubMed] [Google Scholar]
- 27. Mills SJ, Patankar TA, Haroon HA, et al. Do cerebral blood volume and contrast transfer coefficient predict prognosis in human glioma? AJNR Am J Neuroradiol 2006;27:853–58 [PMC free article] [PubMed] [Google Scholar]
- 28. Bammer R. MR and CT Perfusion and Pharmacokinetic Imaging: Clinical Applications and Theoretical Principles: Lippincott Williams and Wilkins; 2016 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.