Abstract
Lipopeptides (LPs) are secondary metabolites produced by a diversity of bacteria and fungi. Their unique chemical structure comprises both a peptide and a lipid moiety. LPs are of major biotechnological interest owing to their emulsification, antitumor, immunomodulatory, and antimicrobial activities. To date, these versatile compounds have been applied across multiple industries, from pharmaceuticals through to food processing, cosmetics, agriculture, heavy metal, and hydrocarbon bioremediation. The variety of LP structures and the diversity of the environments from which LP-producing microorganisms have been isolated suggest important functions in their natural environment. However, our understanding of the ecological role of LPs is limited. In this review, the mode of action and the role of LPs in motility, antimicrobial activity, heavy metals removal and biofilm formation are addressed. We include discussion on the need to characterise LPs from a diversity of microorganisms, with a focus on taxa inhabiting ‘extreme’ environments. We introduce the use of computational target fishing and molecular dynamics simulations as powerful tools to investigate the process of interaction between LPs and cell membranes. Together, these advances will provide new understanding of the mechanism of action of novel LPs, providing greater insights into the roles of LPs in the natural environment.
Keyword: Microbial ecology, Lipopeptides, Bioactivity, Biosurfactant, Molecular dynamics simulation
1. Introduction
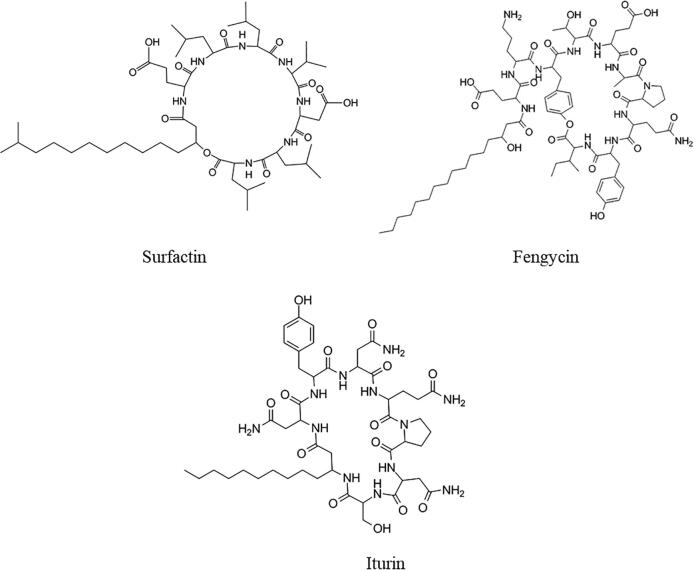
Microbial lipopeptides (LPs) are amphiphilic molecules containing both a polar and an apolar moiety in their structure. The polar moiety is a cyclic peptide, while the apolar moiety is a linear or branched fatty acid with different lengths and degrees of oxidation [1]. LPs have been isolated from a number of Gram-positive (i.e., Bacillus or Streptomyces), Gram-negative (i.e., Pseudomonas) and Gram-variable (i.e., Paenibacillus) genera of bacteria, as well as a diversity of fungal genera (i.e., Phoma and Emericella) [2], [3], [4], [5]. The amphiphilic structure of LPs imparts emulsifying properties that are of widespread use to the biotechnological industry [6]. The most extensively studied LPs are from the surfactin, fengycin and iturin families, all of which have been primarily isolated from Bacillus subtilis, a Gram-positive soil bacterium that belongs to the Firmicutes phylum (Fig. 1). Surfactins (~1.36 kDa) are composed of seven amino acids including Glu-Leu-Leu-Val-Asp-Leu-Leu and 3-hydroxy fatty acids such as tridecanoate, tetradecanoate, pentadecanoate and hexadecanoate [7], [8]. Fengycins (~1.5 kDa) contain a peptide chain of ten amino acids including Ile-Tyr-Gln-Pro-Ala or Val-Glu-Thr-Tyr-Glu-Orn linked to a β-fatty acid chain whose length can vary from C-14 to C-17 carbon atoms [9], while iturins (~1.1 kDa) are comprised of a peptide of seven amino acids including Asn-Tyr-Asn-Gln-Pro-Asn-Ser linked to a C-11 or C-12 β-fatty acid chain [10].
Fig. 1.
Structures of the LPs surfactin, fengycin and iturin. These LPs are organized into cyclopeptides linked to a β-fatty acid chain. They are produced by bacteria of the genus Bacillus through the NRPS pathway.
LPs can be found in linear or cyclic form. To date, cyclic LPs represent the most biologically active group, while research on linear LPs has been limited [11], [12]. Macrocyclization is a common characteristic of diverse natural products [13]; in LPs, this process occurs during the last stage of synthesis and it is catalysed by a C-terminal thioesterase (Te) domain [14], [15]. The macrocyclization of LPs is a strategy that ensures their structural stability and bioactivity [16]. Compared to the linear counterpart, cyclic LPs have greater physicochemical stability [13], and the number of potential structural conformations that a molecule can adopt are reduced [17]. Macrocyclization confers protection against degradation by exo and endoproteases [18], and allows the LPs to have a proper orientation for its interaction with its target [13].
LPs are biosynthesized through secondary metabolite pathways. Enzymes responsible for their synthesis are encoded by biosynthetic gene clusters (BGCs); organized arrangements of two or more genes, including regulatory elements [11], [19], [20]. For LPs, synthesis is primarily performed by non-ribosomal peptide synthetases (NRPS), and hybrid NRPS-polyketide synthases (PKS), both large multi-modular, multi-domain proteins [11], [12]. Each NRPS module has a minimum of three catalytic domains: an adenylation (A) domain, a peptidyl carrier protein (PCP) domain and a condensation (C) domain [21]. A diverse range of NRPSs have been discovered, predominantly in the bacterial phyla Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria [22]. However, many lineages have been found to harbour the genetic machinery to synthesise NRPs, including Deinococcus-Thermus, Chlorobi, Verrucomicrobia and Gemmatimonadetes [23].
LPs have important applications across multiple industries, including pharmaceuticals [24], [25], food processing [10], cosmetics [26], agriculture [27], heavy metal and hydrocarbon bioremediation [28]. Bioactivities reported for characterised LPs are diverse, and include antimicrobial [29], [30], immunomodulator [31], [32], antitumor [33] and surfactant properties [34]. Microbially synthesised LP biosurfactants have several advantages over chemically synthesised surfactants: they are biodegradable, exhibit effective bioactivities at low concentrations, have minimal toxicity, and when used in cosmetic products show a reduced risk of allergies and irritation [24], [35], [36], [37]. Additionally, LP biosurfactants maintain their bioactivity across a wide range of temperatures, pH and salinity [38].
It is proposed that the ecological roles of LP biosurfactants is to enhance interactions between the LP-producing microorganism and its environment, primarily providing strategies to survive under unfavourable conditions; yet studies on their ecological role remain scarce [6]. In this context, we review current knowledge on the ecological functions of lipopeptides in their natural environments, based on their structural properties and known modes of action.
2. Lipopeptides and their roles in bacterial motility
Bacteria utilise motility in growth and reproduction, as a survival strategy for protection against desiccation and antibiotics, to defend against competitors, and to colonize new environments [39], [40], [41], [42]. In many natural habitats, the availability of nutrients is limited, and nutrient concentrations are not homogeneous [43], [44]. Therefore, motility is a mechanism used by bacteria to not only search for more favourable, nutrient-rich environments but also to facilitate the colonisation of new habitats [44]. LPs play an essential role in active and passive bacterial motility across biotic and abiotic surfaces [6], [45]. In ground-breaking studies involving the genera Bacillus and Pseudomonas, NRPS genes encoding the enzymes responsible for the synthesis of LPs were silenced, leading to an observed reduction or total loss of motility in the bacteria. Motility was consequently restored after the addition of purified LPs or structurally similar compounds [6], [21], [45], [46], [47]. For example, Bruijn et al. (2007) used site-directed mutagenesis of the viscosin-related NRPS in Pseudomonas fluorescens SBW25 to silence the expression of viscosin, with the resulting visc- mutant no longer able to swarm [48]. While Kinsinger et al. (2003), silenced the sfp gene involved in the synthesis of surfactin in the strain JH642 to produce a non-motile mutant derived from Bacillus subtilis 168. In this experiment, motility was again restored after the addition of surfactin to B. subtilis in situ [49].
In active motility, also known as flagellum-dependent motility, LPs have a wetting agent role, reducing surface tension and allowing for bacterial colony expansion [50], [51]. Flagellated bacteria use this intrinsic form of motor-driven movement to swim and swarm in both liquid and semi-liquid media [21], [47]. This phenomenon has been observed in Serratia marcescens, Pseudomonas putida, and Bacillus subtilis species, which employ the LPs serrawetting, putisolvin, and surfactin, respectively [49], [52], [53]. In passive motility, extracellular or cell-attached LPs decrease hydrophilic interactions between biotic or abiotic surfaces and bacteria [6], [21]. Passive motility is a form of bacterial surface translocation powered by external forces to reduce adherence to surfaces and promote displacement [54], [55]. Sliding is a form of passive motility promoted by cell division that allows the bacterial colony to spread out of the point of origin and expand over surfaces [21], [45]. In sliding, bacterial secreted biosurfactants, mainly LPs, are produced to reduce the friction of hydrophilic forces between cells and the substrate [1], [56]. This enables nutrient-deprived bacteria to colonize surfaces [46].
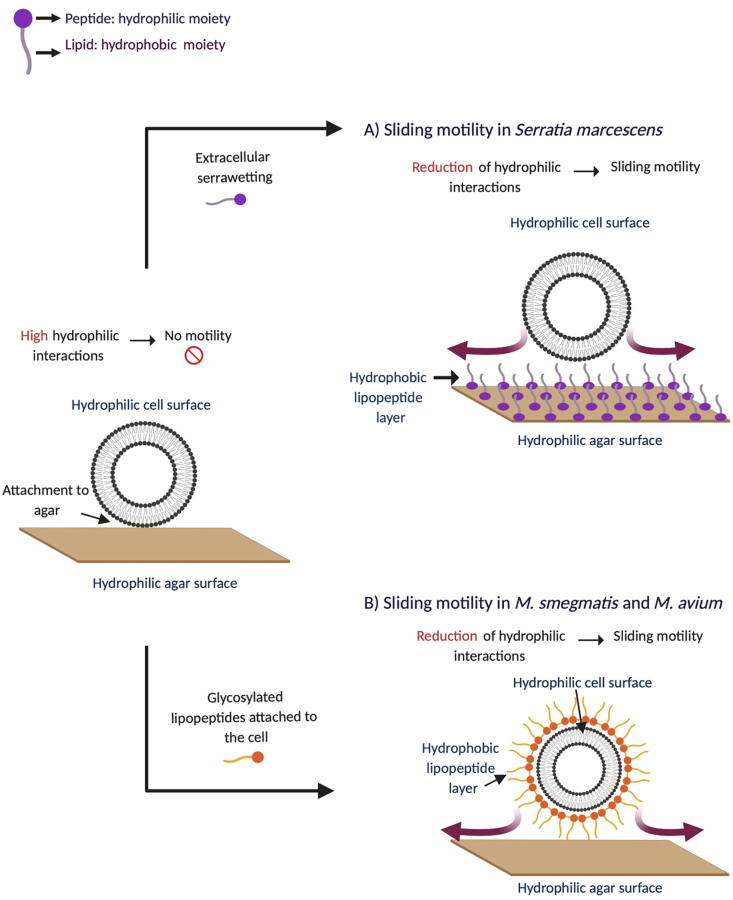
Two modes of action have been observed to explain the role of LPs in bacterial sliding on nutrient rich agar. The first mechanism, exemplified by serrawetting, which is produced by Serratia marcescens (Fig. 2A), involves secretion of LPs into the extracellular medium. The peptide (hydrophilic) moiety is adsorbed to the surface of the agar, and the aliphatic chains (hydrophobic moiety) are exposed to the medium forming a hydrophobic layer that can interact with hydrophilic cells. Interactions at the interface are thus reduced, and bacterial expansion is promoted [57]. In a second mechanism, described in Mycobacterium smegmatis and Mycobacterium avium, glycosylated LPs remain attached to the bacterial cell surface. The peptide moiety interacts with the cell by adhesion and the aliphatic tails are exposed to the medium, increasing the hydrophobicity of the cell so that it interacts with the hydrophilic surface of the cell agar promoting colony spreading (Fig. 2B) [56].
Fig. 2.
Mode of action of LPs in sliding motility. A) Serratia marcescens produces the LP serrawetting that forms a hydrophobic layer on the agar surface decreasing the hydrophilic interactions between the cell and the agar. B) Mycobacterium smegmatis and Mycobacterium avium produce glycosylated lipopeptides forming a hydrophobic layer around the cell to facilitate sliding on the hydrophilic agar surface.
Certain LPs that are known to have a role in motility also display antimicrobial activity. For instance, surfactin, a LP produced by Bacillus subtilis strains, also exhibits antimicrobial [29], antiviral [58], and antifungal activity [59]. Surfactin has also been shown to be required for swarming [60] and sliding [61]. Therefore, it has been proposed that LPs have a dual role in both promoting motility and preventing the growth and colonization of competing microorganisms [21].
3. Antimicrobial role of lipopeptides
Clinically, the application of antibiotics is to inhibit microbial growth. Yet it is unknown whether this bioactivity is the same in the ecological niche from which the microorganism producing the LP inhabits [62], [63]. In a natural environment, antibiotic concentrations are usually lower than the minimum inhibitory concentrations (MIC) employed in a therapeutic setting [64]. Additionally, antibiotics are produced in the stationary growth phase, not during the exponential phase where they could play a role in competition [65]. Therefore, the ecological role of antimicrobials has been proposed to include a dose-dependent effect, also known as hormesis [65], [66]. This principle suggests that the natural function of antibiotics at high concentrations in an ecosystem is to inhibit microbial growth by competitors, and at low concentrations, they can produce a beneficial effect on the microbial population [67].
Alternative roles of antimicrobials at subinhibitory concentrations include effects on bacteria at the cellular and microbial community level [68], [69]. At the cellular level, antibiotic molecules interact with the transcriptome modifying the phenotype [65], [70]. They also have effects on evolution, since they increase the rate of mutagenesis through the mechanism of reactive oxygen species [71], [72].
Antibiotic compounds have also been shown to promote the expression of silent biosynthetic gene clusters, with established antibiotics stimulating the synthesis of new antibiotics in an environment [73]. At the microbial community level, antibiotics inhibit or promote biofilm formation [65], [74], serve as signalling molecules [68], [74], [75], and stimulate horizontal gene transfer through conjugation within and between species [67], [76]. Thus, antibiotics can control the diversity of microbial communities in a given ecological niche, under conditions of stress cells enter dormancy, while they proliferate when conditions are more favourable [66], [77]. It is likely this situation co-occurs for different species, and it may culminate in major microbial community shifts in both the diversity of species and how they are structured [66].
The chemical structure of antibiotic LPs determines their unique mode of action. While the mechanism of action of LPs is not fully understood, there is evidence that they destabilize the bacterial membrane through their action as a detergent or in pore formation [78], [79]. All LPs studied so far interact with membranes via their lipid tails [80]. This mode of action allows LPs to act as broad-spectrum antibiotics, with a reduced potential to develop resistance. For example, mutations that lead to a change in the composition and organization of membrane lipids would require a high energetic cost for the defending microorganism, with the variety in peptide sequences reducing cleavage by proteases [80]. Characterised LPs with antimicrobial activity include crystallomycin [81], aspartocin [82], laspartomycin [83], friulimicin [84], daptomycin [30] and surfactin [29]. However, the specific mechanisms behind these antimicrobial bioactivities remain largely unknown.
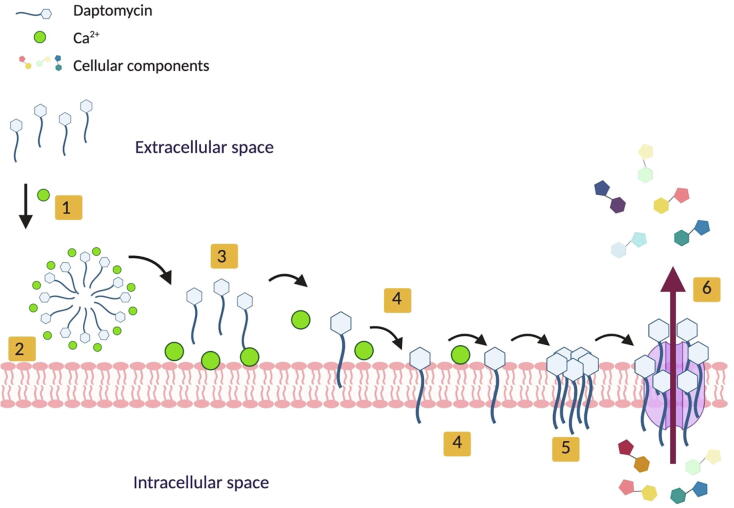
Despite extensive studies on daptomycin, several models that suggest its mode of action can be found in the literature [30], [85], [86]. According to experimental evidence, the most viable mechanism occurs when daptomycin oligomerizes and forms micelles in the presence of calcium (Fig. 3) [87]. When the micelle approaches the cell membrane, it dissociates and daptomycin molecules are assisted by calcium (Ca2+) to insert into the membrane. Daptomycin undergoes a conformational change that allows it to oligomerize and form pores in the bacterial cell membrane. Finally, the pores produce a depolarization effect that results in bacterial cell death. Furthermore, it has been proposed that daptomycin interferes with processes associated with the synthesis of cell wall components, energy and / or cell division [80].
Fig. 3.
Schematic of the mode of action of the lipopeptide antibiotic daptomycin. 1) Oligomerization of 14–16 units of daptomycin in the presence of calcium (Ca2+). 2) Interaction of daptomycin micelle with the cell membrane. 3) Dissociation of the daptomycin. 4) Insertion of daptomycin in the membrane via lipid tail. 5) Oligomerization of the daptomycin in the membrane and 6) Pore formation, depolarization, and cell death.
It has been reported that the amino acid composition in LPs affects antimicrobial activity [88]. Changes in the peptide moiety, such as substitution of amino acids or chemical modifications have produced alterations in their bioactivity [88], [89]. Residues involved in hydrophobicity have been widely studied [88], [90], [91], [92], due to their role in membrane binding, insertion and destabilization processes [93], [94]. For example, iturin A is a LP with antifungal activity [89], with a methylation at the D-Tyr residue of iturin A resulting in increased bioactivity. Methylation is a chemical alteration that increases local hydrophobicity; this affects the process that LPs use to modify the permeability of the membrane and its interaction with lipids [88]. In some cases, an increase in the LP hydrophobicity decreases its bioactivity. For example, methylation in the amino acid Asp of bacillomycin L reduced the antifungal activity; this effect was more significant when the Tyr residue was methylated [91]. It is thought that hydrophobicity affects the interaction of the LP with membrane sterols [89], [91]. Hydrophobicity is necessary for antimicrobial activity of some LPs. However, an excessive degree of this property could damage the host cell and affect the specificity on its target [95].
4. Lipopeptide relationships with heavy metals
In soil, heavy metals are defined as metals of a natural or anthropogenic origin with a density greater than 5.0 g/cm3 [96]. The most hazardous heavy metals released into the environment are arsenic (As), cadmium (Cd), chromium (Cr), mercury (Hg), zinc (Zn), nickel (Ni), lead (Pb) and copper (Cu) [97]. These are classified into two groups: those that are hazardous for humans, plants or animals, and those that a low concentrations have beneficial effects [97], [98]. In bacterial cells, high concentrations of heavy metals disrupt the function of nucleic acids and proteins. They also deactivate enzymes and electron donors, and form precipitates with essential metabolites [99]. In soil, heavy metals disturb endemic microbial communities, leading to major decreases in soil microbial community diversity and population structures [100], [101], [102].
The ecological role of LPs in heavy metal chelation is not clear [103]. It remains unknown whether microorganisms produce LPs to interact with heavy metals for their benefit or as part of an alternate process for soil detoxification [104]. Bacteria do use LPs for the chelation of heavy metals, in a similar way to siderophores, and incorporate them into their metabolism as micronutrients [6]. In the soil, some bacteria, mainly anaerobic taxa, use heavy metals as electron donors or acceptors during redox and osmoregulation reactions [105], [106]. Additionally, heavy metals participate as enzymatic cofactors [107]. During this process, heavy metals are incorporated into microbial metabolism and their solubility is modified, therefore, when they are returned to the environment, heavy metals, are bioavailable to other organisms [108].
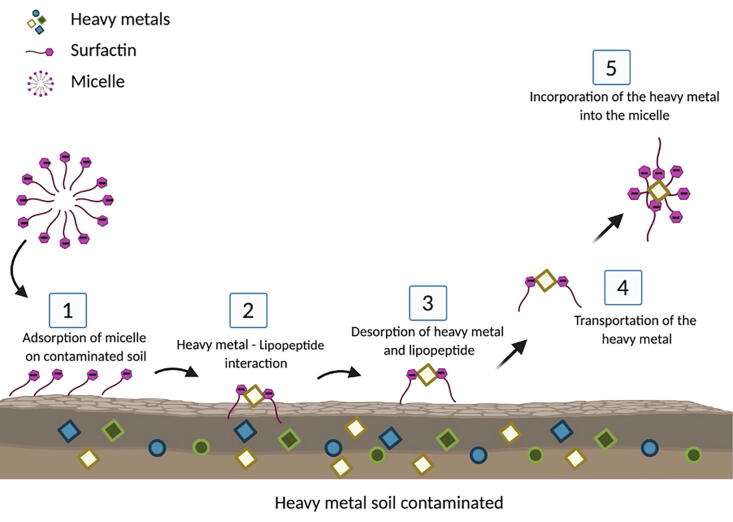
The role of LPs in the removal of heavy metals from the environment has been widely reported [109], [110], [111], [112]. Surfactin, synthesized by bacteria of the Bacillus genus, is capable of removing zinc and copper [103], [113]; Colistin, produced by Paenibacillus polymyxa, binds efficiently to copper [114]; pseudofactin II, produced by Pseudomonas fluorescens BD5, also interacts with zinc and copper [109]; while the MSI54 LP, produced by a Bacillus bacterium isolated from a marine sponge has an affinity for lead, mercury, manganese, and cadmium [115]. The ionic charge of LPs is known to play an important role in their interaction with heavy metals. While both anionic and nonionic LPs are able to adhere to soil, only those with anionic charge form complexes with metals [103]. To explain the interaction of surfactin with heavy metals, Mulligan et al. (1999) suggested a mechanism whereby surfactin micelles are adsorbed to the soil and dissociate into monomers; the negative charge of the glutamic acid and aspartate in the surfactin monomers interact with metal cations; the surfactin-metal complex is desorbed from the soil; the metal is transported and then incorporated into reformed surfactin micelles; finally the complex is stabilized with the interaction of the aliphatic chains of surfactin (Fig. 4) [103].
Fig. 4.
Schematic representation showing the mode of action of the surfactin when interacting with heavy metals. 1) Surfactin micelles are adsorbed to the soil. 2) The negative charges of the glutamic acid and aspartate of the surfactin monomers interact with the metal. 3) Desorption of the surfactin monomers and metal from soil. 4) The metal is transported away from the surface and 5) The metal is incorporated into the micelles, and the complex is stabilized with the interaction of the aliphatic chains of surfactin.
5. Lipopeptides and their role in biofilm formation
Biofilms are complex communities of microorganisms with one or more species attached to biotic or abiotic surfaces [116], [117]. The ecological functions of biofilm formation include the promotion of cell division, proliferation and resistance to environmental and chemical stressors [6], [118]. Within the biofilm, active processes of organic matter decomposition occur that increase the availability of nutrients and metabolic cooperation [118]. Another benefit for biofilm-forming bacteria is the acquisition of new genetic traits through horizontal gene transfer [6], [116].
LPs can promote or inhibit biofilm formation, depending upon the structure of the LP and the polarity of the cells and substrate [6]. The role of LPs to promote biofilm formation has been observed in Mycobacterium avium [119], a pathogenic bacterium that has a lipopeptide anchored to the cell wall. Additionally, cyclic LPs have been reported to promote biofilm formation in Pseudomonas fluorescens SBW25 [48]. Gaps exists in our knowledge of the mode of action of LPs in promoting biofilm formation. While it has been proposed that LPs have a role in adhesion through orientation of the molecule to facilitate the hydrophobic-hydrophobic or hydrophilic-hydrophilic interactions between the cell and surface, this requires validation [6].
The inhibition of biofilms by LPs isolated from bacteria of the Bacillus genus has been more widely studied. For example, bacillomycin D, a LP produced by Bacillus amyloliquefciens inhibits biofilm formation of Candida albicans [120]. While, a LP isolated from Bacillus cereus NK1 inhibited biofilm formation of the pathogenic strains P. aeruginosa and S. epidermidis [121], and putisolvin I and II LPs, isolated from P. putida, were capable of both inhibition of biofilm formation and disruption of established biofilms [53]. Furthermore, paenibacterin, a LP isolated from Paenibacillus thiaminolyticus, displays antimicrobial activity in addition to its ability to inhibit the biofilm formation by Bacillus subtilis, Micrococcus luteus, Pseudomonas aeruginosa, Staphylococcus aureus and Streptococcus bovis [122], [123].
LPs inhibit biofilm formation through changes in the hydrophobic interactions between the microorganism and a surface [124]. LPs act as pre-conditioning agents, they bind to the surface by adsorption, modifying their physicochemical properties [125]. This change prevents the adhesion of the microorganism to the surface [126]. For example, Vieira de Araujo et al. 2011 observed that addition of a 0.1% solution of surfactin to a polystyrene surface decreased the adhesion rate of 80% and 84% of Listeria monocytogenes ATCC 7644 and ATCC 19112, respectively. The authors evaluated the hydrophobicity of the polystyrene, they observed that the untreated polystyrene had a contact angle of 84.4° and it decreased to 70.3° when the 0.1% surfactin solution was added. They suggested that changes in surface properties can affect the microbial adhesion process due to the reduction of surface-microorganism interactions [17].
6. Discovery of new LPs: Where are we going?
The discovery of novel LPs occurs through both top-down and bottom-up approaches. Top-down approaches are based on culture-dependent techniques and screening for bioactivity [127]. Common assays for surfactant activity include the emulsification index [128], oil spreading and drop collapse methods [129]. Bottom-up approaches employ advance metagenomics and genomic sequencing combined with new bioinformatics tools for the discovery of genes and enzymes related to LP biosynthesis, such as NRPS and hybrid NRPS-PKS [127], [130], [131]. Omics approaches and the advent of high-throughput sequencing tools and powerful bioinformatics pipelines have allowed us to identify the presence and arrangement of biosynthetic gene clusters (BGCs) [132], [133].
High-throughput sequencing combined with bioinformatic technologies allow for the study of LPs at three levels: identification of the genes involved in the synthesis of an LP, prediction of the synthesised LP structure from genomic information, and the proposed activity and ecological role of the LP product. The antibiotics and secondary metabolite analysis shell (antiSMASH) is one such bioinformatics tool, designed for the identification, functional annotation, and analysis of BGCs [134], [135]. Its algorithm allows alignment of microbial genome sequences with previously known BGCs sequences compiled in its database. The results of analysis include an interactive map that allows a visual representation of the annotated BGCs (Fig. 5) [134], [135]. AntiSMASH has been widely used as a screening method for the identification of diverse BGCs, including those of NRPS and hybrid NRPS-PKS responsible for the synthesis of lipopeptides [2], [136], [137], [138]. Other bioinformatic tools such as the Natural Product Domain seeker (NaPDos), Prediction Informatics for Secondary Metabolomes (PRISM) and the NRPS/PKS substrate predictor also facilitate the rapid characterization of BGCs involved in the synthesis of LPs [138], [139]. Additionally, tools such as PRISM have allowed for the prediction of LPs structure [140], through identification of open reading frames in a genome sequence, with results aligned using hidden Markov models of secondary metabolites [141]. Based on the arrangement of genes and the action of the enzymes, combinatorial libraries of structures are generated. These tools can be used in conjunction with metabolite extraction and a combination of analytical chemistry techniques such as thin layer chromatography (TLC), fourier-transform infrared spectroscopy (FT-IR), nuclear magnetic resonance (NMR), matrix-assisted laser desorption/ionization time-of-flight (MALD-TOF) and mass spectrometry (MS) to inform on structure and confirmation of the final product [142], [143], [144].
Fig. 5.
Interactive map of the antiSMASH results for Bacillus velezensis FZB42. Thirteen BGCs were identified in B. velezensis FZB42. The BGCs exhibited 82–100% similarity with known BGCs. Three BGCs corresponded to NRPS, and two of those were predicted to encode the biosurfactant LPs surfactin (Region 2) and fengycin (Region 5).
To showcase capacities of antiSMASH and PRISM to identify NRPS-related BCGs and to elucidate LP structures we performed a comparative analysis using a representative LP-producing microorganism. We evaluated whether each tool was able to identify fengycin BGCs in a microorganism that has been previously reported to produce this LP [145]. We selected Bacillus velezensis FZB42 as a model bacteria as its genome has been sequenced, characterized and extensively studied [146]. Bacillus velezensis FZB42 is a Gram-positive bacterium known for its phytostimulatory and biocontrol properties. This strain produces fengycin, a LP with antifungal activity against Fusarium graminearum, a pathogenic fungi known to infect cereal crops [145].
Bioinformatic analysis revealed that both antiSMASH and PRISM identified thirteen BGCs in Bacillus velezensis FZB42 (SI Table 2 and SI Table 3, respectively). The fengycin LP cluster was identified as region 1.5 (Fig. 5) and as cluster 7 using antiSMASH and PRISM, respectively. The BGCs exhibited 82–100% similarity with known BGCs, predominantly from bacteria of the genus Bacillus, as well as three with closest similarity to BGCs from Streptomyces spp. (SI Table 1). Three BGCs corresponded to NRPS, and two of those were predicted to encode the biosurfactant LPs surfactin and fengycin. Additionally, PRISM detected two polyketide-related BGCs that contain fragments of NRPS sequences (SI Table 3). The predicted architecture of fengycin BGC included five domains involved with the synthesis and addition of the amino acids Ile, Pro, Gln, Tyr, Glu, Val, Tyr, Thr, Glu and Orn (SI Fig. 1A, B and SI Fig. 2A, B). Both bioinformatic tools allowed for a rough prediction of core scaffold at different molecular levels, antiSMASH predicted a polymer from the addition of monomers (SI Fig. 1C), while PRISM predicted structures at the level of individual atoms or bonds (SI Fig. 2C) [134], [135], [141], [147].
We found that both antiSMASH and PRISM identified similar BGCs and Fengycin domains (SI Fig. 1B and SI Fig. 2B), while antiSMASH also had the capacity to compare the BGCs of Bacillus velezensis FZB42 with the known closest match providing percentage of similarity for that LP (SI Table 2). AntiSMASH and PRISM predict compound structures, while antiSMASH provided a rough prediction of core scaffolds based on the addition of monomers, PRISM uses a bonds and individual atoms approach. We suggest that both tools can be used in a complementary way for a more comprehensive analysis of genomic data for potential LPs.
Unravelling the ecological roles of LPs represents a greater challenge. Here, the interaction between proteins and their targets can be modelled using bioinformatics-informed molecular-docking [148]. This analysis is focused on binding sites and consists of two main steps; identifying the position and orientation of protein–ligand, and evaluating the degree of affinity between them [149]. Due to their antimicrobial properties, LPs have been studied through molecular docking to identify potential therapeutic targets as treatment for infectious diseases [150], [151], [152]. In this context, molecular docking could be a promising tool for the preliminary identification of targets that interact with LPs in the natural environment, and thus inform on their ecological role.
The wide bioactivities and potential applications of LPs have increased interest in the search for new NRPS-producing microorganisms. Secondary metabolites produced by enzyme complexes occurs at a high energetic cost to that organism. Therefore, it has been proposed that microorganisms use these compounds as a protective or adaptative mechanism in specific niches [132]. While 70% of the NRPSs discovered to date have been obtained from marine organisms [153], there are few studies that have focused on terrestrial habitats. This gap in knowledge is present despite soils containing a wealth of microbial diversity; this lack of understanding is even greater when we focus on ‘extreme’ natural environments [134], [135].
In a survey by Charlop-Powers et al. (2014), 96 diverse soil microbiomes were analysed for secondary metabolite genes, and a correlation was found between biosynthetic genetic diversity and soil type. Specifically, arid soils showed a higher richness of NRP and PK synthesis related genes [154]. Similarly, Benaud et al. (2019) further showed that NRPS-encoding genes are widespread across polar desert soils spanning Antarctica and the High-Arctic [155]. In this case, the microbial inhabitants are exposed to cold temperatures, low availability of nutrients and severe moisture limitation [156], with both NRPS and PKS presence and diversity correlating with low soil moisture and carbon content [155]. In Antarctic bacteria, the majority of NRPS and PKS gene fragments have been reported to exhibit closest similarity, albeit at low levels, to known BGCs (av. 31%) present in Actinobacteria, Proteobacteria, Firmicutes and Cyanobacteria, with a high proportion likely to be LPs [135]. For example, following phylogenetic analysis of domain sequences, several Antarctic bacterial NRPS domain sequences showed similarity to those encoding for LPs, such as syringomycin and iturin, with emulsification and antimicrobial activity assays supporting their genome mining-driven discovery [140].
Moreover, a Rhodococcus strain, ADL36, also isolated from Antarctic soil has been shown to display biosurfactant activity; with chemical characterization of the compound and the genome sequencing suggesting the production of a LP with potential applications in the bioremediation of hydrocarbons [144]. In another survey, this time utilising metagenomics data, Chen et al. (2020) studied microbial mats from Shark Bay, Australia. In the ‘microbial mat’ environment microorganisms are exposed to high temperatures, UV radiation and salinity [157], with a high diversity of NRPS and hybrid NRPS-PKS genes in bacteria of the phyla Planctomycetes, Chloroflexi, Calditrichaeota and for the first time Lokiarchaeota was reported. Lokiarchaeota is an archaeal phyla not previously known to have BGCs, expanding the domain of life to which the future of natural products discovery can take place [158].
Microorganisms thrive in a myriad of ‘extreme’ environments, from hot springs and geysers, through to deep sea, hypersaline environments, evaporites, deserts, cold environments (ice, permafrost, and snow) and the Earth’s atmosphere. Given the diversity of genetic machinery recently discovered in microorganisms from Antarctic deserts and microbial matts [140], [158], we believe future investigations in a range of ecosystems at the boundaries of life will provide new insights into the ecological roles of LPs [24]. We know that microorganisms exposed to harsh conditions develop biological adaptations, metabolic processes, physiological capabilities and novel strategies to survive, and remain metabolically active in their unique environments [24]. We propose that LPs as secondary metabolites produced by such microorganisms must be assisting in strategies for survival. Therefore, bacteria and other domains of life surviving under some of the most harsh conditions on the planet must be attractive candidates in the search for novel LPs [24].
There are challenges associated with the LPs discovery. Required is the development of novel cultivation and screening tools for rare, barely studied taxa [159]. On the other hand, bioinformatics tools are limited as they can only detect BGCs based on known sequences, leading to potential mischaracterisation or failure to detect new families [160], [161]. However, with the advent of new molecular and computational technologies, the future of LPs discovery will undoubtedly expand, providing unsurpassed knowledge on the ecological roles of these important secondary metabolites [138].
7. Computational target fishing for the study of the LP-membrane interactions
One of the challenges in the study of lipopeptides is to understand the process of interaction between the LP and its target [6]. Lipopeptide-membrane interactions have traditionally been studied using experimental strategies such as spectroscopic, diffraction and microscopic methods [162]. However, limitations arise due to the complexity of membranes. For instance, membranes are dynamic environments in constant change, lipid rearrangement occurs in milliseconds [163], [164] and due to membranes being very thin, high-resolution techniques such as Nuclear Magnetic Resonance (NMR), Electron Paramagnetic Resonance (EPR) and fluorescence quenching are required [165].
Computational target fishing is a powerful tool for investigating mechanisms of interaction between bioactive molecules and their targets, at an atomic level [166], [167]. The atomic-level structure provides information about the conformational dynamics of a molecule, and it enables the prediction of intermolecular interactions to know their function [168], [169]. Molecular dynamics (MD) simulations are computational tools designed to study the structure–function relationship [168], [170]. Based on Newton's laws of motion, MD simulations allow prediction of the position in space of each atom of a molecule as a function of time. MD simulations model the force exerted on each atom and how their movement speed will be affected, thereby providing a three-dimensional model in a specific time-lapse [169]. MD simulations provide information difficult to obtain by experimental procedures. For example, MD simulations can determine the position of each atom in a specific time, predict trends of interaction or mechanisms of action, control the simulation conditions and evaluate the effect of molecular variables (such as mutations or post-translational modifications) or environmental conditions, such as temperature, pH or voltage on the membrane [169], [171].
There are two models of MD simulations, all-atoms (AA) and coarse grained (CG). While AA-MD simulations allow analysis at the atomic level of all the elements involved in a biological system [171], CG-MD simulations are a reduced version of AA-MD that focuses on a small group of atoms and their collective motions, while averaging with local movements of adjacent atoms [172], [173]. CG models have three main advantages: they are cheaper than AA models [173], simulations can be performed for longer time intervals and they decrease the degrees of freedom, such as self-assembly of biomolecules and large patches of the bilayer [172]. MD simulations have been useful for the study of membrane biophysics [172], and their application has been widely exploited to understand the mechanism of action and interaction of antimicrobial lipopeptides on the cell membrane [174], [175], [176], [177].
Polymyxin B1 (PMB1) is a lipopeptide produced by Bacilus Polymyxa [178]. It consists of a heptapeptide attached to a tripeptide side-chain acylated by a branched lipid chain (Fig. 6A). PMB1 has two irregular amino acids: D-Phenylalanine (DPhe) and α, γ-Diamino Butyric acid (DAB) [179]. Polymyxin B1 has potent antimicrobial activity mainly against Gram-negative bacteria [180]. PMB1 penetrates the bacterial outer membrane (OM) and it produces rupture of the bacterial inner membrane (IM) [174]. It is known that the peptide ring increases the permeability of the bacterial membrane. However, the mechanism triggered after the anchoring of the peptide to the cell membrane is unknown [180]. Through MD simulations in E. coli membranes, it was observed that the DAB residues of PMB1 were responsible for the binding process to the cell membrane and antimicrobial action. Consequently, the DAB residues were found to be interacting with the phosphate groups of the OM producing displacements and allowing the insertion of the lipid tail of PMB1 into the membrane. In the IM, it was observed that the hydrophobic interactions were promoted by the lipid chain of PMB1 and by the D-Phe side chain. The peptide ring remains attached to the polar heads of the phospholipids, while the lipid tail (hydrophobic part) adopts an embedded position. This study suggests that the antimicrobial action mechanism of PMB1 is a self-regulated process that produces pore formation through adsorption between the DAB residue of the lipopeptide and the insertion of the lipid tail in the cell membrane, supported by the hydrophobic residue DPhe [174].
Fig. 6.
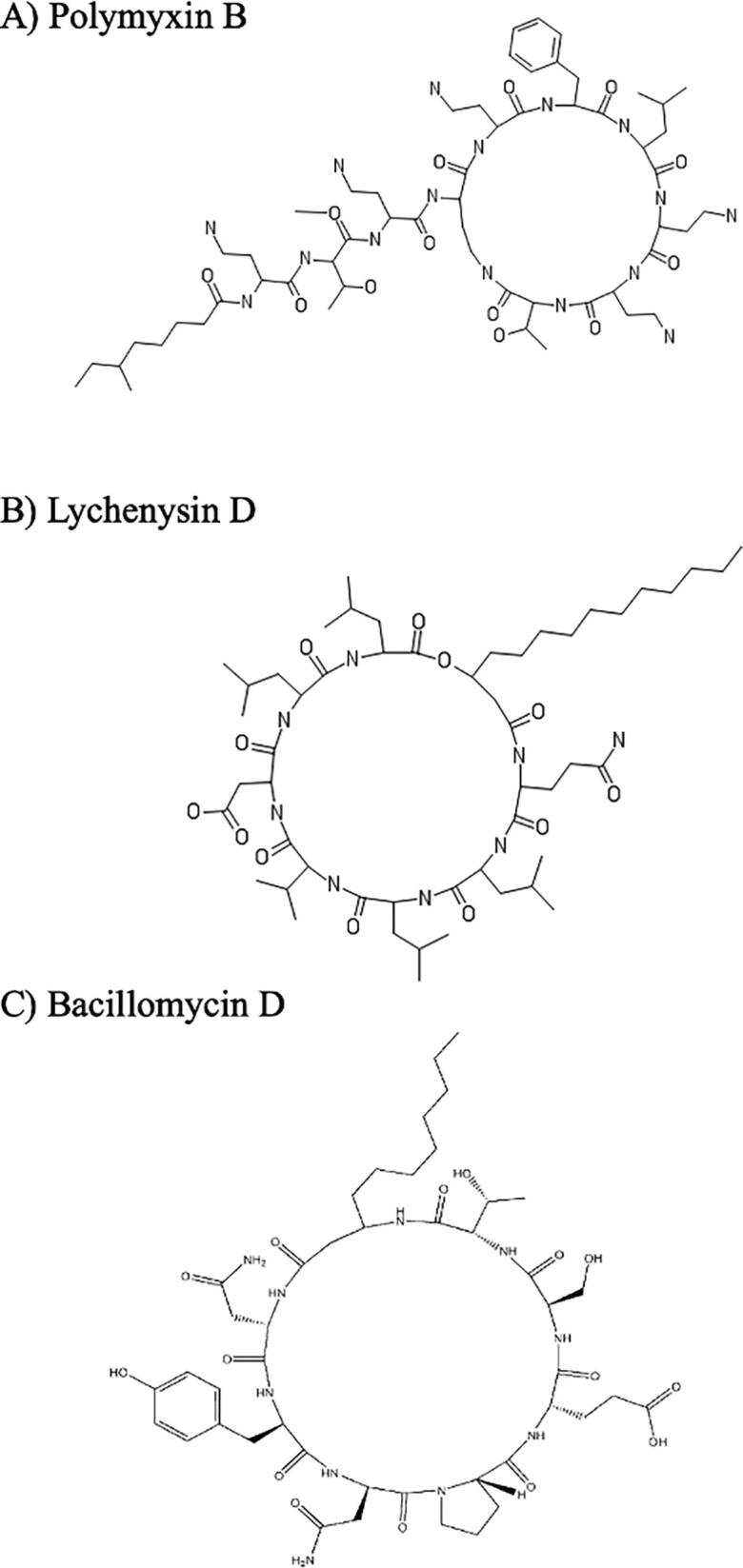
Structures of the LPs polymyxin B, lichenysin D and bacillomycin D. These LPs have been isolated from Bacilus Polymyxa, Bacillus licheniformis and Bacillus amyloliquefaciens, respectively. Polymyxin B, lichenysin D and bacillomycin D have antimicrobial activity and their mechanism of action has been studied through MD simulations.
Lichenysin is an anionic lipopeptide produced by Bacillus licheniformis [175]. It consists of a peptide ring of seven amino acids attached to a branched lipid chain (Fig. 6B) [181], and it has antimicrobial [182] and hemolytic [183] activity. Using MD simulations with a Dipalmitoylphosphatidylcholine (DPPC) membrane and supported by experimental techniques such as calorimetry, X-ray diffraction and Fourier transform infrared spectroscopy (FT-IR), Coronel et al. (2017) were able to determine the effects of lichenysin on the lipid bilayer. They found that lichenysin penetrates the DPPC bilayer via interaction with membrane phosphatidylcholines. Lichenysin induces changes in the membrane such as, dehydration of the phospholipid/water interface and increase of bilayer thickness and movement of the phospholipid acyl chains. Lichenysin increases the permeability of the membrane without its solubilization; and theses finding support the hypothesis of the pore forming mechanism [175].
Bacillomycin D is a lipopeptide of the iturin family and it has been isolated from Bacillus amyloliquefaciens [184]. It consists of a β-amino fatty acid linked to seven amino acid residues via an amide bond (Figure 6C). Bacillomycin D has antifungal, antibacterial, and hemolytic activity [185], [186], [187]. It is known that bacillomycin D produces membrane disruption of target cells; however, the exact interaction process and mechanism of action remains unclear. Through the use of a CG-MD simulations, Sun et al. (2018) investigated the insertion process of bacillomycin D into three cell membranes with differing biophysical properties: DOPC, DOPC/DPPA and DOPC/DOTAP [176]. In this case, negatively charged amino acids, Asp1 and Glu5 of bacillomycin D were reported to exhibit electrostatic interactions with cell membranes. The Pro4 residue showed similar electrostatic properties that were attributed to its hydrophobicity. Furthermore, it was observed that bacillomycin D caused changes in the curvature of the membrane which eventually damaged its integrity [176]. Accordingly, bacillomycin D exhibits higher antimicrobial activity on positively charged membranes, in this case DOPC/DOTAP. These results suggest that bacillomycin D has selective potency on specific microorganisms according to their membrane composition [176].
8. Concluding remarks
LPs are a class of compounds with broad structural diversity which results in a variety of bioactivities and ecological roles. The primary functions attributed to LPs include motility, antimicrobial, biofilm formation and association with heavy metals [6]. LPs are synthesized by NRPSs and hybrid NRPS-PKSs; these enzymes are involved in the synthesis of secondary metabolites associated with mechanisms of adaptation and/or protection that microorganisms use to survive in adverse conditions [188], [189]. Recent studies of bacteria living in challenging ecosystems, from marine [153] through to microbial mats and Antarctic desert soils [140], [190] demonstrate a wide diversity of NRPS-producing bacteria. Therefore, ‘extreme’ microorganisms are attractive sources for future mining of novel LP structures. As new technologies, from metagenomics through to genomics, bioinformatic tools and MD simulations, are coupled with rapid bioactivity screening methods and analytical chemistry techniques, the discovery of diverse LPs will no doubt lead to new insights into the ecological roles of LPs, particularly those produced under a myriad of environmental stressors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by a Hermon-Slade Foundation Grant HSF15/13 and an Australian Research Council Future Fellowship (FT170100341), both awarded to B.C.F. C.G-C. was supported by a Scientia PhD scholarship from UNSW Sydney.
Author contributions
C.G-C. performed the research. B.C.F. supervised the study. C.G-C., N.B. and B.C.F. wrote the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.02.017.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ahimou F, Jacques P, Deleu M. Surfactin and iturin A effects on Bacillus subtilis surface hydrophobicity. Enzyme Microb. Technol., vol. 27, Elsevier Science Inc; 2000, p. 749–54. https://doi.org/10.1016/S0141-0229(00)00295-7. [DOI] [PubMed]
- 2.Bekiesch P., Zehl M., Domingo-Contreras E., Martín J., Pérez-Victoria I., Reyes F. Viennamycins: lipopeptides produced by a Streptomyces sp. J Nat Prod. 2020 doi: 10.1021/acs.jnatprod.0c00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao P., Xue Y., Li X., Li J., Zhao Z., Quan C. Fungi-derived lipopeptide antibiotics developed since 2000. Peptides. 2019;113:52–65. doi: 10.1016/j.peptides.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Herath K., Harris G., Jayasuriya H., Zink D., Smith S., Vicente F. Isolation, structure and biological activity of phomafungin, a cyclic lipodepsipeptide from a widespread tropical Phoma sp. Bioorg Med Chem. 2009;17:1361–1369. doi: 10.1016/j.bmc.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Oh DC, Kauffman CA, Jensen PR, Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod 2007;70:515–20. https://doi.org/10.1021/np060381f. [DOI] [PubMed]
- 6.Raaijmakers J.M., de Bruijn I., Nybroe O., Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 7.Cooper D.G., Macdonald C.R., Duff S.J.B., Kosaric N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl Environ Microbiol. 1981;42:408–412. doi: 10.1128/aem.42.3.408-412.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Youssef N.H., Duncan K.E., McInerney M.J. Importance of 3-hydroxy fatty acid composition of lipopeptides for biosurfactant activity. Appl Environ Microbiol. 2005;71:7690–7695. doi: 10.1128/AEM.71.12.7690-7695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Fengycins Sun C. cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl Environ Microbiol. 2018:84. doi: 10.1128/AEM.00445-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meena KR, Kanwar SS. Lipopeptides as the antifungal and antibacterial agents: Applications in food safety and therapeutics. Biomed Res Int 2015;2015. https://doi.org/10.1155/2015/473050. [DOI] [PMC free article] [PubMed]
- 11.Tran P.N., Yen M.R., Chiang C.Y., Lin H.C., Chen P.Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl Microbiol Biotechnol. 2019;103:3277–3287. doi: 10.1007/s00253-019-09708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel S., Ahmed S., Eswari J.S. Therapeutic cyclic lipopeptides mining from microbes: latest strides and hurdles. World J Microbiol Biotechnol. 2015;31:1177–1193. doi: 10.1007/s11274-015-1880-8. [DOI] [PubMed] [Google Scholar]
- 13.Kopp F., Marahiel M.A. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat Prod Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- 14.Roongsawang N., Washio K., Morikawa M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int J Mol Sci. 2010;12:141–172. doi: 10.3390/ijms12010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roongsawang N., Washio K., Morikawa M. In vivo characterization of tandem C-terminal thioesterase domains in arthrofactin synthetase. ChemBioChem. 2007;8:501–512. doi: 10.1002/cbic.200600465. [DOI] [PubMed] [Google Scholar]
- 16.Hamley I.W. Lipopeptides: from self-assembly to bioactivity. Chem Commun. 2015;51:8574–8583. doi: 10.1039/c5cc01535a. [DOI] [PubMed] [Google Scholar]
- 17.de Araujo LV, Abreu F, Lins U, Anna LM de MS, Nitschke M, Freire DMG. Rhamnolipid and surfactin inhibit Listeria monocytogenes adhesion. Food Res Int 2011;44:481–8. https://doi.org/10.1016/j.foodres.2010.09.002
- 18.White C.J., Yudin A.K. Contemporary strategies for peptide macrocyclization. Nat Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 19.Martinet L, Naômé A, Deflandre B, Maciejewska M, Tellatin D, Tenconi E, et al. A single biosynthetic gene cluster is responsible for the production of bagremycin antibiotics and ferroverdin iron chelators. MBio 2019;10. https://doi.org/10.1128/mBio.01230-19. [DOI] [PMC free article] [PubMed]
- 20.Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearns D.B. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller C.A., Oberauner-Wappis L., Peyman A., Amos G.C.A., Wellington E.M.H., Berg G. Mining for nonribosomal peptide synthetase and polyketide synthase genes revealed a high level of diversity in the Sphagnum bog metagenome. Appl Environ Microbiol. 2015;81:5064–5072. doi: 10.1128/AEM.00631-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H., Fewer D.P., Holm L., Rouhiainen L., Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U S A. 2014;111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz J., Rosado A.S. Extreme environments: a source of biosurfactants for biotechnological applications. Extremophiles. 2020;24:189–206. doi: 10.1007/s00792-019-01151-2. [DOI] [PubMed] [Google Scholar]
- 25.Khan A., Butt A. Biosurfactants and their potential applications for microbes and mankind: an overview. Middle East J Bus. 2016;11:9–18. doi: 10.5742/mejb.2015.92757. [DOI] [Google Scholar]
- 26.Hamley I.W. Lipopeptides: from self-assembly to bioactivity. Chem Commun. 2015;51:8574–8583. doi: 10.1039/c5cc01535a. [DOI] [PubMed] [Google Scholar]
- 27.Penha R.O., Vandenberghe L.P.S., Faulds C., Soccol V.T., Soccol C.R. Bacillus lipopeptides as powerful pest control agents for a more sustainable and healthy agriculture: recent studies and innovations. Planta. 2020;251:1–15. doi: 10.1007/s00425-020-03357-7. [DOI] [PubMed] [Google Scholar]
- 28.Bezza F.A., Chirwa E.M.N. Production and applications of lipopeptide biosurfactant for bioremediation and oil recovery by Bacillus subtilis CN2. Biochem Eng J. 2015;101:168–178. doi: 10.1016/j.bej.2015.05.007. [DOI] [Google Scholar]
- 29.Heerklotz H., Seelig J. Leakage and lysis of lipid membranes induced by the lipopeptide surfactin. Eur Biophys J. 2007;36:305–314. doi: 10.1007/s00249-006-0091-5. [DOI] [PubMed] [Google Scholar]
- 30.Steenbergen J.N., Alder J., Thorne G.M., Tally F.P. Daptomycin: A lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother. 2005;55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 31.Kelesidis T. The interplay between daptomycin and the immune system. Front Immunol 2014;5. https://doi.org/10.3389/fimmu.2014.00052. [DOI] [PMC free article] [PubMed]
- 32.Bessler WG, Heinevetter L, Wiesmüller KH, Jung G, Baier W, Huber M, et al. Bacterial cell wall components as immunomodulators - I. Lipopeptides as adjuvants for parenteral and oral immunization. Int. J. Immunopharmacol., vol. 19, Pergamon; 1997, p. 547–50. https://doi.org/10.1016/S0192-0561(97)00054-4. [DOI] [PubMed]
- 33.Wu YS, Ngai SC, Goh BH, Chan KG, Lee LH, Chuah LH. Anticancer activities of surfactin potential application of nanotechnology assisted surfactin delivery. Front Pharmacol 2017;8. https://doi.org/10.3389/fphar.2017.00761. [DOI] [PMC free article] [PubMed]
- 34.Desai J.D., Banat I.M. Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev. 1997;61:47–64. doi: 10.1128/.61.1.47-64.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A. A review on the biosurfactants: properties, types and its applications. J Fundam Renew Energy Appl 2018;08. https://doi.org/10.4172/2090-4541.1000248.
- 36.Singh P., Patil Y., Rale V. Biosurfactant production: emerging trends and promising strategies. J Appl Microbiol. 2019;126:2–13. doi: 10.1111/jam.14057. [DOI] [PubMed] [Google Scholar]
- 37.Akbari S., Abdurahman N.H., Yunus R.M., Fayaz F., Alara O.R. Biosurfactants—a new frontier for social and environmental safety: a mini review. Biotechnol Res Innov. 2018;2:81–90. doi: 10.1016/j.biori.2018.09.001. [DOI] [Google Scholar]
- 38.Geissler M, Heravi KM, Henkel M, Hausmann R. Lipopeptide Biosurfactants From Bacillus Species. Biobased Surfactants, Elsevier; 2019, p. 205–40. https://doi.org/10.1016/b978-0-12-812705-6.00006-x.
- 39.Tamar E, Koler M, Vaknin A. The role of motility and chemotaxis in the bacterial colonization of protected surfaces. Sci Rep 2016;6. https://doi.org/10.1038/srep19616. [DOI] [PMC free article] [PubMed]
- 40.Easom C.A., Clarke D.J. Motility is required for the competitive fitness of entomopathogenic Photorhabdus luminescens during insect infection. BMC Microbiol. 2008;8:168. doi: 10.1186/1471-2180-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soutourina O.A., Semenova E.A., Parfenova V.V., Danchin A., Bertin P. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from Lake Baikal. Appl Environ Microbiol. 2001;67:3852–3859. doi: 10.1128/AEM.67.9.3852-3859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler M.T., Wang Q., Harshey R.M. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A. 2010;107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell J.G., Kogure K. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol Ecol. 2006;55:3–16. doi: 10.1111/j.1574-6941.2005.00003.x. [DOI] [PubMed] [Google Scholar]
- 44.Harshey R.M. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 45.Hölscher T., Kovács Á.T. Sliding on the surface: bacterial spreading without an active motor. Environ Microbiol. 2017;19:2537–2545. doi: 10.1111/1462-2920.13741. [DOI] [PubMed] [Google Scholar]
- 46.Fall R., Kearns D.B., Nguyen T. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol. 2006;6:31. doi: 10.1186/1471-2180-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke W.-J., Hsueh Y.-H., Cheng Y.-C., Wu C.-C., Liu S.-T. Water surface tension modulates the swarming mechanics of Bacillus subtilis. Front Microbiol. 2015;6:1017. doi: 10.3389/fmicb.2015.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Bruijn I., De Kock M.J.D., Yang M., De Waard P., Van Beek T.A., Raaijmakers J.M. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol Microbiol. 2007;63:417–428. doi: 10.1111/j.1365-2958.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- 49.Kinsinger R.F., Shirk M.C., Fall R. Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol. 2003;185:5627–5631. doi: 10.1128/JB.185.18.5627-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura S., Minamino T. Flagella-driven motility of bacteria. Biomolecules. 2019;9 doi: 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ongena M., Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 52.O’Rear J., Alberti L., Harshey R.M. Mutations that impair swarming motility in Serratia marcescens 274 include but are not limited to those affecting chemotaxis or flagellar function. J Bacteriol. 1992;174:6125–6137. doi: 10.1128/jb.174.19.6125-6137.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuiper I., Lagendijk E.L., Pickford R., Derrick J.P., Lamers G.E.M., Thomas-Oates J.E. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol Microbiol. 2004;51:97–113. doi: 10.1046/j.1365-2958.2003.03751.x. [DOI] [PubMed] [Google Scholar]
- 54.Pollitt E.J.G., Crusz S.A., Diggle S.P. Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci Rep. 2015;5:17698. doi: 10.1038/srep17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/mmbr.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martínez A., Torello S., Kolter R. Sliding motility in mycobacteria. J Bacteriol. 1999;181:7331–7338. doi: 10.1128/jb.181.23.7331-7338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuyama T., Bhasin A., Harshey R.M. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol. 1995;177:987–991. doi: 10.1128/jb.177.4.987-991.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan L., Zhang S., Wang Y., Li Y., Wang X., Yang Q. Surfactin inhibits membrane fusion during invasion of epithelial cells by enveloped viruses. J Virol. 2018:92. doi: 10.1128/jvi.00809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desmyttere H., Deweer C., Muchembled J., Sahmer K., Jacquin J., Coutte F. Antifungal activities of Bacillus subtilis lipopeptides to two venturia inaequalis strains possessing different tebuconazole sensitivity. Front Microbiol. 2019;10:2327. doi: 10.3389/fmicb.2019.02327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh A.K., Dhanjal S., Cameotra S.S. Surfactin restores and enhances swarming motility under heavy metal stress. Colloids Surf B Biointerfaces. 2014;116:26–31. doi: 10.1016/j.colsurfb.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 61.Ghelardi E., Salvetti S., Ceragioli M., Gueye S.A., Celandroni F., Senesi S. Contribution of surfactin and SwrA to flagellin expression, swimming, and surface motility in bacillus subtilis. Appl Environ Microbiol. 2012;78:6540–6544. doi: 10.1128/AEM.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kraemer SA, Ramachandran A, Perron GG. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019;7. https://doi.org/10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed]
- 63.Aminov R.I. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol. 2009;11:2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- 64.Li J., Xie S., Ahmed S., Wang F., Gu Y., Zhang C. Antimicrobial activity and resistance: Influencing factors. Front Pharmacol. 2017;8:364. doi: 10.3389/fphar.2017.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fajardo A., Linares J.F., Martínez J.L. Towards an ecological approach to antibiotics and antibiotic resistance genes. Clin Microbiol Infect. 2009;15:14–16. doi: 10.1111/j.1469-0691.2008.02688.x. [DOI] [PubMed] [Google Scholar]
- 66.Mullis M.M., Rambo I.M., Baker B.J., Reese B.K. Diversity, ecology, and prevalence of antimicrobials in nature. Front Microbiol. 2019;10:2518. doi: 10.3389/fmicb.2019.02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pishchany G., Kolter R. On the possible ecological roles of antimicrobials. Mol Microbiol. 2020;113:580–587. doi: 10.1111/mmi.14471. [DOI] [PubMed] [Google Scholar]
- 68.Tyc O., Song C., Dickschat J.S., Vos M., Garbeva P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017;25:280–292. doi: 10.1016/j.tim.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 69.Bernier S.P., Surette M.G. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol. 2013;4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen P.A., Zhu Z., van Opijnen T. Antibiotics disrupt coordination between transcriptional and phenotypic stress responses in pathogenic bacteria. Cell Rep. 2017;20:1705–1716. doi: 10.1016/j.celrep.2017.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kohanski M.A., DePristo M.A., Collins J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutierrez A., Laureti L., Crussard S., Abida H., Rodríguez-Rojas A., Blázquez J. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun. 2013;4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okada B.K., Seyedsayamdost M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev. 2017;41:19–33. doi: 10.1093/femsre/fuw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu W, Hallinen KM, Wood KB. Interplay between antibiotic efficacy and drug-induced lysis underlies enhanced biofilm formation at subinhibitory drug concentrations. Antimicrob Agents Chemother 2018;62. https://doi.org/10.1128/AAC.01603-17. [DOI] [PMC free article] [PubMed]
- 75.Zhu Y.G., Zhao Y., Zhu D., Gillings M., Penuelas J., Ok Y.S. Soil biota, antimicrobial resistance and planetary health. Environ Int. 2019;131 doi: 10.1016/j.envint.2019.105059. [DOI] [PubMed] [Google Scholar]
- 76.Lopatkin A.J., Sysoeva T.A., You L. Dissecting the effects of antibiotics on horizontal gene transfer: Analysis suggests a critical role of selection dynamics. BioEssays. 2016;38:1283–1292. doi: 10.1002/bies.201600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orman M.A., Brynildsen M.P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother. 2013;57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greber K.E., Ciura K., Belka M., Kawczak P., Nowakowska J., Bączek T. Characterization of antimicrobial and hemolytic properties of short synthetic cationic lipopeptides based on QSAR/QSTR approach. Amino Acids. 2018;50:479–485. doi: 10.1007/s00726-017-2530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buchoux S., Lai-Kee-Him J., Garnier M., Tsan P., Besson F., Brisson A. urfactin-triggered small vesicle formation of negatively charged membranes: A novel membrane-lysis mechanism. Biophys J. 2008;95:3840–3849. doi: 10.1529/biophysj.107.128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Straus S.K., Hancock R.E.W. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta - Biomembr. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Tyurin A.P., Alferova V.A., Paramonov A.S., Shuvalov M.V., Malanicheva I.A., Grammatikova N.E. Crystallomycin revisited after 60 years: Aspartocins B and C. Medchemcomm. 2018;9:667–675. doi: 10.1039/c8md00002f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siegel M.M., Kong F., Carter G.T. Aspartocin cyclic lipopeptide antibiotics: mass spectral structural confirmations and the diagnostic role played by the α, β-diaminobutyric acid residue. J Mass Spectrom. 2010;45:820–823. doi: 10.1002/jms.1755. [DOI] [PubMed] [Google Scholar]
- 83.Borders D.B., Leese R.A., Jarolmen H., Francis N.D., Fantini A.A., Falla T. Laspartomycin, an acidic lipopeptide antibiotic with a unique peptide core. J Nat Prod. 2007;70:443–446. doi: 10.1021/np068056f. [DOI] [PubMed] [Google Scholar]
- 84.Schneider T., Gries K., Josten M., Wiedemann I., Pelzer S., Labischinski H. The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob Agents Chemother. 2009;53:1610–1618. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Müller A., Wenzel M., Strahl H., Grein F., Saaki T.N.V., Kohl B. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci U S A. 2016;113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mensa B., Howell G.L., Scott R., DeGrado W.F. Comparative mechanistic studies of brilacidin, daptomycin, and the antimicrobial peptide LL16. Antimicrob Agents Chemother. 2014;58:5136–5145. doi: 10.1128/AAC.02955-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scott WRP, Baek S Bin, Jung D, Hancock REW, Straus SK. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim Biophys Acta - Biomembr 2007;1768:3116–26. https://doi.org/10.1016/j.bbamem.2007.08.034. [DOI] [PubMed]
- 88.Maget-Dana R., Ptak M., Peypoux F., Michel G. Effect of the O-methylation of tyrosine on the pore-forming properties of iturins. BBA - Biomembr. 1987;898:1–5. doi: 10.1016/0005-2736(87)90104-0. [DOI] [PubMed] [Google Scholar]
- 89.Bonmatin J-M, Laprevote O, Peypoux F. Diversity among microbial cyclic lipopeptides: iturins and surfactins. activity-structure relationships to design new bioactive agents. Comb Chem High Throughput Screen 2012;6:541–56. https://doi.org/10.2174/138620703106298716. [DOI] [PubMed]
- 90.Volpon L., Besson F., Lancelin J.M. NMR structure of active and inactive forms of the sterol-dependent antifungal antibiotic bacillomycin L. Eur J Biochem. 1999;264:200–210. doi: 10.1046/j.1432-1327.1999.00605.x. [DOI] [PubMed] [Google Scholar]
- 91.Besson F., Peypoux F., Michel G., Delcambe L. Antifungal activity upon saccharomyces cerevisiae of iturin a, mycosubtilin, bacillomycin L and of their derivatives; inhibition of this antifungal activity by lipid antagonists. J Antibiot (Tokyo) 1979;32:828–833. doi: 10.7164/antibiotics.32.828. [DOI] [PubMed] [Google Scholar]
- 92.Topman-Rakover S., Malach E., Burdman S., Hayouka Z. Antibacterial lipo-random peptide mixtures exhibit high selectivity and synergistic interactions. Chem Commun. 2020;56:12053–12056. doi: 10.1039/d0cc04493h. [DOI] [PubMed] [Google Scholar]
- 93.Makovitzki A., Avrahami D., Shai Y. Ultrashort antibacterial and antifungal lipopeptides. Proc Natl Acad Sci U S A. 2006;103:15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saint Jean K.D., Henderson K.D., Chrom C.L., Abiuso L.E., Renn L.M., Caputo G.A. Effects of hydrophobic amino acid substitutions on antimicrobial peptide behavior. Probiotics Antimicrob Proteins. 2018;10:408–419. doi: 10.1007/s12602-017-9345-z. [DOI] [PubMed] [Google Scholar]
- 95.Ebenhan T, Gheysens O, Kruger HG, Zeevaart JR, Sathekge MM. Antimicrobial peptides: their role as infection-selective tracers for molecular imaging. Biomed Res Int 2014;2014. https://doi.org/10.1155/2014/867381. [DOI] [PMC free article] [PubMed]
- 96.Srivastava V., Sarkar A., Singh S., Singh P., de Araujo A.S.F., Singh R.P. Agroecological responses of heavy metal pollution with special emphasis on soil health and plant performances. Front Environ Sci. 2017;5:64. doi: 10.3389/fenvs.2017.00064. [DOI] [Google Scholar]
- 97.Ali H, Khan E, Ilahi I. Environmental chemistry and ecotoxicology of hazardous heavy metals: environmental persistence, toxicity, and bioaccumulation. J Chem 2019;2019. https://doi.org/10.1155/2019/6730305.
- 98.Davis H.T., Marjorie Aelion C., McDermott S., Lawson A.B. Identifying natural and anthropogenic sources of metals in urban and rural soils using GIS-based data, PCA, and spatial interpolation. Environ Pollut. 2009;157:2378–2385. doi: 10.1016/j.envpol.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sobolev D, Begonia MFT. Effects of heavy metal contamination upon soil microbes: Lead-induced changes in general and denitrifying microbial communities as evidenced by molecular markers. Int. J. Environ. Res. Public Health, vol. 5, Multidisciplinary Digital Publishing Institute (MDPI); 2008, p. 450–6. https://doi.org/10.3390/ijerph5050450. [DOI] [PMC free article] [PubMed]
- 100.Chu D. Effects of heavy metals on soil microbial community. IOP Conf. Ser. Earth Environ. Sci., vol. 113, Institute of Physics Publishing; 2018, p. 12009. https://doi.org/10.1088/1755-1315/113/1/012009.
- 101.Allison S.D., Martiny J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Dorst J., Siciliano S.D., Winsley T., Snape I., Ferrari B.C. Bacterial targets as potential indicators of diesel fuel toxicity in subantarctic soils. Appl Environ Microbiol. 2014;80:4021–4033. doi: 10.1128/AEM.03939-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mulligan C.N., Yong R.N., Gibbs B.F., James S., Bennett H.P.J. Metal removal from contaminated soil and sediments by the biosurfactant surfactin. Environ Sci Technol. 1999;33:3812–3820. doi: 10.1021/es9813055. [DOI] [Google Scholar]
- 104.Abdu N., Abdullahi A.A., Abdulkadir A. Heavy metals and soil microbes. Environ Chem Lett. 2017;15:65–84. doi: 10.1007/s10311-016-0587-x. [DOI] [Google Scholar]
- 105.Ayangbenro AS, Babalola OO. A new strategy for heavy metal polluted environments: A review of microbial biosorbents. Int J Environ Res Public Health 2017;14. https://doi.org/10.3390/ijerph14010094. [DOI] [PMC free article] [PubMed]
- 106.Bruins M.R., Kapil S., Oehme F.W. Microbial resistance to metals in the environment. Ecotoxicol Environ Saf. 2000;45:198–207. doi: 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 107.De Lillo A., Cardi M., Landi S., Esposito S. Mechanism(s) of action of heavy metals to investigate the regulation of plastidic glucose-6-phosphate dehydrogenase. Sci Rep. 2018;8:13481. doi: 10.1038/s41598-018-31348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ojuederie OB, Babalola OO. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int J Environ Res Public Health 2017;14. https://doi.org/10.3390/ijerph14121504. [DOI] [PMC free article] [PubMed]
- 109.Janek T., Rodrigues L.R., Gudiña E.J., Czyżnikowska Ż. Structure and mode of action of cyclic lipopeptide pseudofactin II with divalent metal ions. Colloids Surf B Biointerfaces. 2016;146:498–506. doi: 10.1016/j.colsurfb.2016.06.055. [DOI] [PubMed] [Google Scholar]
- 110.Zhao P., Xu X., Zhao X., Ai C., Xu K., Li M. Capability of Bacillus Subtilis to remove Pb2+ via producing lipopeptides. Sci Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138941. [DOI] [PubMed] [Google Scholar]
- 111.Zhu Z., Gao C., Wu Y., Sun L., Huang X., Ran W. Removal of heavy metals from aqueous solution by lipopeptides and lipopeptides modified Na-montmorillonite. Bioresour Technol. 2013;147:378–386. doi: 10.1016/j.biortech.2013.08.049. [DOI] [PubMed] [Google Scholar]
- 112.Singh A.K., Cameotra S.S. Efficiency of lipopeptide biosurfactants in removal of petroleum hydrocarbons and heavy metals from contaminated soil. Environ Sci Pollut Res. 2013;20:7367–7376. doi: 10.1007/s11356-013-1752-4. [DOI] [PubMed] [Google Scholar]
- 113.Mulligan C.N., Yong R.N., Gibbs B.F. Heavy metal removal from sediments by biosurfactants. J Hazard Mater. 2001;85:111–125. doi: 10.1016/S0304-3894(01)00224-2. [DOI] [PubMed] [Google Scholar]
- 114.Stokowa-Sołtys K., Kasprowicz A., Wrzesiński J., Ciesiołka J., Gaggelli N., Gaggelli E. Impact of Cu2 + ions on the structure of colistin and cell-free system nucleic acid degradation. J Inorg Biochem. 2015;151:67–74. doi: 10.1016/j.jinorgbio.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 115.Ravindran A., Sajayan A., Priyadharshini G.B., Selvin J., Kiran G.S. Revealing the efficacy of thermostable biosurfactant in heavy metal bioremediation and surface treatment in vegetables. Front Microbiol. 2020:11. doi: 10.3389/fmicb.2020.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davey M.E., O’toole G.A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Besemer K. Biodiversity, community structure and function of biofilms in stream ecosystems. Res Microbiol. 2015;166:774–781. doi: 10.1016/j.resmic.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Balcázar J.L., Subirats J., Borrego C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front Microbiol. 2015;6:1216. doi: 10.3389/fmicb.2015.01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu C wei, Schmoller SK, Bannantine JP, Eckstein TM, Inamine JM, Livesey M, et al. A novel cell wall lipopeptide is important for biofilm formation and pathogenicity of Mycobacterium avium subspecies paratuberculosis. Microb Pathog 2009;46:222–30. https://doi.org/10.1016/j.micpath.2009.01.010. [DOI] [PMC free article] [PubMed]
- 120.Tabbene O., Azaiez S., Di Grazia A., Karkouch I., Ben Slimene I., Elkahoui S. Bacillomycin D and its combination with amphotericin B: Promising antifungal compounds with powerful antibiofilm activity and wound-healing potency. J Appl Microbiol. 2016;120:289–300. doi: 10.1111/jam.13030. [DOI] [PubMed] [Google Scholar]
- 121.Sriram M.I., Kalishwaralal K., Deepak V., Gracerosepat R., Srisakthi K., Gurunathan S. Biofilm inhibition and antimicrobial action of lipopeptide biosurfactant produced by heavy metal tolerant strain Bacillus cereus NK1. Colloids Surf B Biointerfaces. 2011;85:174–181. doi: 10.1016/j.colsurfb.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 122.Li R., Du W., Yang J., Liu Z., Yousef A.E. Control of Listeria monocytogenes biofilm by paenibacterin, a natural antimicrobial lipopeptide. Food Control. 2018;84:529–535. doi: 10.1016/j.foodcont.2017.08.031. [DOI] [Google Scholar]
- 123.Quinn G.A., Maloy A.P., McClean S., Carney B., Slater J.W. Lipopeptide biosurfactants from Paenibacillus polymyxa inhibit single and mixed species biofilms. Biofouling. 2012;28:1151–1166. doi: 10.1080/08927014.2012.738292. [DOI] [PubMed] [Google Scholar]
- 124.Sommer P., Martin-Rouas C., Mettler E. Influence of the adherent population level on biofilm population, structure and resistance to chlorination. Food Microbiol. 1999;16:503–515. doi: 10.1006/fmic.1999.0267. [DOI] [Google Scholar]
- 125.Zezzi do Valle Gomes M, Nitschke M. Evaluation of rhamnolipid and surfactin to reduce the adhesion and remove biofilms of individual and mixed cultures of food pathogenic bacteria. Food Control 2012;25:441–7. https://doi.org/10.1016/j.foodcont.2011.11.025.
- 126.Meylheuc T., Van Oss C.J., Bellon-Fontaine M.N. Adsorption of biosurfactant on solid surfaces and consequences regarding the bioadhesion of Listeria monocytogenes LO28. J Appl Microbiol. 2001;91:822–832. doi: 10.1046/j.1365-2672.2001.01455.x. [DOI] [PubMed] [Google Scholar]
- 127.Lee N., Hwang S., Kim J., Cho S., Palsson B., Cho B.K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput Struct Biotechnol J. 2020;18:1548–1556. doi: 10.1016/j.csbj.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang J., Xue Q., Gao H., Lai H., Wang P. Production of lipopeptide biosurfactants by Bacillus atrophaeus 5–2a and their potential use in microbial enhanced oil recovery. Microb Cell Fact. 2016;15:168. doi: 10.1186/s12934-016-0574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ghasemi A., Moosavi-Nasab M., Setoodeh P., Mesbahi G., Yousefi G. Biosurfactant production by lactic acid bacterium Pediococcus dextrinicus SHU1593 grown on different carbon sources: strain screening followed by product characterization. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-41589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Julkowska D, Obuchowski M, Holland IB, Séror SJ. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: Critical effects of surfactin and the composition of the medium. J. Bacteriol., vol. 187, American Society for Microbiology; 2005, p. 65–76. https://doi.org/10.1128/JB.187.1.65-76.2005. [DOI] [PMC free article] [PubMed]
- 131.Oueis E., Klefisch T., Zaburannyi N., Garcia R., Plaza A., Müller R. Two biosynthetic pathways in Jahnella thaxteri for thaxteramides. Distinct Types of Lipopeptides. Org Lett. 2019;21:5407–5412. doi: 10.1021/acs.orglett.9b01524. [DOI] [PubMed] [Google Scholar]
- 132.Amoutzias GD, Chaliotis A, Mossialos D. Discovery Strategies of bioactive compounds synthesized by nonribosomal peptide synthetases and type-I polyketide synthases derived from marine microbiomes. Mar Drugs 2016;14. https://doi.org/10.3390/md14040080. [DOI] [PMC free article] [PubMed]
- 133.Chen R., Wong H., Burns B. New approaches to detect biosynthetic gene clusters in the environment. Medicines. 2019;6:32. doi: 10.3390/medicines6010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 2019;47:W81–7. https://doi.org/10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed]
- 135.Medema M.H., Blin K., Cimermancic P., De Jager V., Zakrzewski P., Fischbach M.A. AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao H, Liu Y-P, Zhang L-Q. In silico and genetic analyses of cyclic lipopeptide synthetic gene clusters in Pseudomonas sp. 11K1. Front Microbiol 2019;10:544. https://doi.org/10.3389/fmicb.2019.00544. [DOI] [PMC free article] [PubMed]
- 137.Ceniceros A., Dijkhuizen L., Petrusma M., Medema M.H. Genome-based exploration of the specialized metabolic capacities of the genus Rhodococcus. BMC Genomics. 2017;18:593. doi: 10.1186/s12864-017-3966-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aleti G., Sessitsch A., Brader G. Genome mining: Prediction of lipopeptides and polyketides from Bacillus and related Firmicutes. Comput Struct Biotechnol J. 2015;13:192–203. doi: 10.1016/j.csbj.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jahanshah G., Yan Q., Gerhardt H., Pataj Z., Lämmerhofer M., Pianet I. Discovery of the cyclic lipopeptide gacamide a by genome mining and repair of the defective gaca regulator in pseudomonas fluorescens p f0–1. J Nat Prod. 2019;82:301–308. doi: 10.1021/acs.jnatprod.8b00747. [DOI] [PubMed] [Google Scholar]
- 140.Benaud N., Edwards R.J., Amos T., Gutierrez-Chavez C., Montgomery K., Nicetic I. Antarctic deser soil bacteria exhibit high novel natural product potential, evaluated through long-read genome sequencing and comparative genomics. Environ Microbiol. 2020 doi: 10.1111/1462-2920.15300. [DOI] [PubMed] [Google Scholar]
- 141.Skinnider M.A., Merwin N.J., Johnston C.W., Magarvey N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017;45:W49–W54. doi: 10.1093/nar/gkx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen Y., Liu S.A., Mou H., Ma Y., Li M., Hu X. Characterization of lipopeptide biosurfactants produced by Bacillus licheniformis MB01 from marine sediments. Front Microbiol. 2017;8:871. doi: 10.3389/fmicb.2017.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang H., Li X., Li X., Yu H., Shen Z. Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin, fengycin, and surfactin by RP-HPLC. Anal Bioanal Chem. 2015;407:2529–2542. doi: 10.1007/s00216-015-8486-8. [DOI] [PubMed] [Google Scholar]
- 144.Habib S., Ahmad S.A., Wan Johari W.L., Abd Shukor M.Y., Alias S.A., Smykla J. Production of lipopeptide biosurfactant by a hydrocarbon-degrading Antarctic Rhodococcus. Int J Mol Sci. 2020;21:6138. doi: 10.3390/ijms21176138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hanif A, Zhang F, Li P, Li C, Xu Y, Zubair M, et al. Fengycin produced by Bacillus amyloliquefaciens FZB42 inhibits Fusarium graminearum growth and mycotoxins biosynthesis. Toxins (Basel) 2019;11. https://doi.org/10.3390/toxins11050295. [DOI] [PMC free article] [PubMed]
- 146.Fan B., Wang C., Song X., Ding X., Wu L., Wu H., Bacillus velezensis FZB42 in, The gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol. 2018;2018:9. doi: 10.3389/fmicb.2018.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baspinar A., Cukuroglu E., Nussinov R., Keskin O., Gursoy A. PRISM: A web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Morris G.M., Lim-Wilby M. Molecular docking. Methods Mol Biol. 2008;443:365–382. doi: 10.1007/978-1-59745-177-2_19. [DOI] [PubMed] [Google Scholar]
- 149.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front Pharmacol. 2018;9:923. doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Eswari JS, Dhagat S, Kaser S. Homology modeling and molecular docking studies of Bacillomycin and Iturin synthetases with novel ligands for the production of therapeutic lipopeptides. Curr Drug Discov Technol 2017;14. https://doi.org/10.2174/1570163814666170816112536. [DOI] [PubMed]
- 151.Jujjavarapu S.E., Dhagat S., Kurrey V. Identification of novel ligands for therapeutic lipopeptides: daptomycin, surfactin and polymyxin. Curr Drug Targets. 2018;19:1589–1598. doi: 10.2174/1389450119666171129164932. [DOI] [PubMed] [Google Scholar]
- 152.Jujjavarapu SE, Dhagat S. Lipopeptides as Therapeutics: Molecular Docking and Drug Design. Biotechnol. Appl. Hum. Heal., Springer Singapore; 2020, p. 61–7. https://doi.org/10.1007/978-981-15-3453-9_7.
- 153.Agrawal S., Acharya D., Adholeya A., Barrow C.J., Deshmukh S.K. Nonribosomal peptides from marine microbes and their antimicrobial and anticancer potential. Front Pharmacol. 2017;8:828. doi: 10.3389/fphar.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Charlop-Powers Z., Owen J.G., Reddy B.V.B., Ternei M.A., Brady S.F. Chemical-biogeographic survey of secondary metabolism in soil. Proc Natl Acad Sci U S A. 2014;111:3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Benaud N, Zhang E, Van Dorst J, Brown M V., Kalaitzis JA, Neilan BA, et al. Harnessing long-read amplicon sequencing to uncover NRPS and Type i PKS gene sequence diversity in polar desert soils. FEMS Microbiol Ecol 2019;95. https://doi.org/10.1093/femsec/fiz031. [DOI] [PubMed]
- 156.Zhang E., Thibaut L.M., Terauds A., Raven M., Tanaka M.M., Van Dorst J. Lifting the veil on arid-to-hyperarid Antarctic soil microbiomes: A tale of two oases. Microbiome. 2020;8:1–12. doi: 10.1186/s40168-020-00809-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wong H.L., MacLeod F.I., White R.A., Visscher P.T., Burns B.P. Microbial dark matter filling the niche in hypersaline microbial mats. Microbiome. 2020;8:1–14. doi: 10.1186/s40168-020-00910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen R., Wong H.L., Kindler G.S., MacLeod F.I., Benaud N., Ferrari B.C. Discovery of an abundance of biosynthetic gene clusters in shark bay microbial mats. Front Microbiol. 2020;11:1950. doi: 10.3389/fmicb.2020.01950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Luo Y., Cobb R.E., Zhao H. Recent advances in natural product discovery. Curr Opin Biotechnol. 2014;30:230–237. doi: 10.1016/j.copbio.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Albarano L., Esposito R., Ruocco N., Costantini M. Genome mining as new challenge in natural products discovery. Mar Drugs. 2020;18:199. doi: 10.3390/md18040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kalkreuter E., Pan G., Cepeda A.J., Shen B. Targeting bacterial genomes for natural product discovery. Trends Pharmacol Sci. 2020;41:13–26. doi: 10.1016/j.tips.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Munusamy S., Conde R., Bertrand B., Munoz-Garay C. Biophysical approaches for exploring lipopeptide-lipid interactions. Biochimie. 2020;170:173–202. doi: 10.1016/j.biochi.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tarun O.B., Hannesschläger C., Pohl P., Roke S. Label-free and charge-sensitive dynamic imaging of lipid membrane hydration on millisecond time scales. Proc Natl Acad Sci U S A. 2018;115:4081–4086. doi: 10.1073/pnas.1719347115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hong C., Tieleman D.P., Wang Y. Microsecond molecular dynamics simulations of lipid mixing. Langmuir. 2014;30:11993–12001. doi: 10.1021/la502363b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mayne C.G., Arcario M.J., Mahinthichaichan P., Baylon J.L., Vermaas J.V., Navidpour L. The cellular membrane as a mediator for small molecule interaction with membrane proteins. Biochim Biophys Acta - Biomembr. 2016;1858:2290–2304. doi: 10.1016/j.bbamem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang L., Xie X.Q. Computational target fishing: What should chemogenomics researchers expect for the future of in silico drug design and discovery? Future Med Chem. 2014;6:247–249. doi: 10.4155/fmc.14.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meslamani J., Bhajun R., Martz F., Rognan D. Computational profiling of bioactive compounds using a target-dependent composite workflow. J Chem Inf Model. 2013;53:2322–2333. doi: 10.1021/ci400303n. [DOI] [PubMed] [Google Scholar]
- 168.Hospital A., Goñi J.R., Orozco M., Gelpí J.L. Molecular dynamics simulations: Advances and applications. Adv Appl Bioinforma Chem. 2015;8:37–47. doi: 10.2147/AABC.S70333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Karplus M., Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci U S A. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.[171] Monje-Galvan V, Warburton L, Klauda JB. Setting up all-atom molecular dynamics simulations to study the interactions of peripheral membrane proteins with model lipid bilayers. Methods Mol. Biol., vol. 1949, Humana Press Inc.; 2019, p. 325–39. https://doi.org/10.1007/978-1-4939-9136-5_22. [DOI] [PubMed]
- 172.Sun L., Mao J., Zhao Y., Quan C., Zhong M., Fan S. Coarse-grained molecular dynamics simulation of interactions between cyclic lipopeptide Bacillomycin D and cell membranes. Mol Simul. 2018;44:364–376. doi: 10.1080/08927022.2017.1384632. [DOI] [Google Scholar]
- 173.Wang W., Gómez-Bombarelli R. Coarse-graining auto-encoders for molecular dynamics. Npj Comput Mater. 2019;5:1–9. doi: 10.1038/s41524-019-0261-5. [DOI] [Google Scholar]
- 174.Berglund NA, Piggot TJ, Jefferies D, Sessions RB, Bond PJ, Khalid S. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: a molecular dynamics study. PLOS Comput Biol 2015;11:e1004180. https://doi.org/10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed]
- 175.Coronel J.R., Marqués A., Manresa Á., Aranda F.J., Teruel J.A., Ortiz A. Interaction of the lipopeptide biosurfactant lichenysin with phosphatidylcholine model membranes. Langmuir. 2017;33:9997–10005. doi: 10.1021/acs.langmuir.7b01827. [DOI] [PubMed] [Google Scholar]
- 176.Sun L., Mao J., Zhao Y., Quan C., Zhong M., Fan S. Coarse-grained molecular dynamics simulation of interactions between cyclic lipopeptide Bacillomycin D and cell membranes. Mol Simul. 2018;44:364–376. doi: 10.1080/08927022.2017.1384632. [DOI] [Google Scholar]
- 177.Deleu M., Bouffioux O., Razafindralambo H., Paquot M., Hbid C., Thonart P. Interaction of surfactin with membranes: A computational approach. Langmuir. 2003;19:3377–3385. doi: 10.1021/la026543z. [DOI] [Google Scholar]
- 178.Shaheen M., Li J., Ross A.C., Vederas J.C., Jensen S.E. Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem Biol. 2011;18:1640–1648. doi: 10.1016/j.chembiol.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 179.Yu Z, Qin W, Lin J, Fang S, Qiu J. Antibacterial mechanisms of polymyxin and bacterial resistance. Biomed Res Int 2015;2015. https://doi.org/10.1155/2015/679109. [DOI] [PMC free article] [PubMed]
- 180.Berglund NA, Piggot TJ, Khalid S. Interaction of the antimicrobial polymyxin B1 with the inner and outer membranes of E. coli: insights into the mechanisms of membrane disruption. Biophys J 2014;106:97a. https://doi.org/10.1016/j.bpj.2013.11.605.
- 181.Grangemard I., Wallach J., Maget-Dana R., Peypoux F. Lichenysin: A more efficient cation chelator than surfactin. Appl Biochem Biotechnol - Part A Enzym Eng Biotechnol. 2001;90:199–210. doi: 10.1385/ABAB:90:3:199. [DOI] [PubMed] [Google Scholar]
- 182.Yakimov M.M., Timmis K.N., Wray V., Fredrickson H.L. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl Environ Microbiol. 1995;61 doi: 10.1128/aem.61.5.1706-1713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Grangemard I, Wallach J, Maget-Dana R, Peypoux F. Lichenysin: A more efficient cation chelator than surfactin. Appl Biochem Biotechnol - Part A Enzym Eng Biotechnol 2001;90:199–210. https://doi.org/10.1385/ABAB:90:3:199. [DOI] [PubMed]
- 184.Gu Q., Yang Y., Yuan Q., Shi G., Wu L., Lou Z. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plantpathogenic fungus Fusarium graminearum. Appl Environ Microbiol. 2017;83:1075–1092. doi: 10.1128/AEM.01075-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Moyne A.L., Shelby R., Cleveland T.E., Tuzun S. Bacillomycin D: An iturin with antifungal activity against Aspergillus flavus. J Appl Microbiol. 2001;90:622–629. doi: 10.1046/j.1365-2672.2001.01290.x. [DOI] [PubMed] [Google Scholar]
- 186.Peypoux F., Besson F., Michel G., Delcambe L. Structure of bacillomycin D, a new antibiotic of the Iturin group. Eur J Biochem. 1981;118:323–327. doi: 10.1111/j.1432-1033.1981.tb06405.x. [DOI] [PubMed] [Google Scholar]
- 187.Tabbene O., Kalai L., Ben Slimene I., Karkouch I., Elkahoui S., Gharbi A. Anti-Candida effect of bacillomycin D-like lipopeptides from Bacillus subtilis B38. FEMS Microbiol Lett. 2011;316:108–114. doi: 10.1111/j.1574-6968.2010.02199.x. [DOI] [PubMed] [Google Scholar]
- 188.Behie S.W., Bonet B., Zacharia V.M., McClung D.J., Traxler M.F. Molecules to ecosystems: Actinomycete natural products in situ. Front Microbiol. 2017;7:2149. doi: 10.3389/fmicb.2016.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pham J.V., Yilma M.A., Feliz A., Majid M.T., Maffetone N., Walker J.R. A review of the microbial production of bioactive natural products and biologics. Front Microbiol. 2019;10:1404. doi: 10.3389/fmicb.2019.01404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hodges T.W., Slattery M., Olson J.B. Unique actinomycetes from marine caves and coral reef sediments provide novel PKS and NRPS biosynthetic gene clusters. Mar Biotechnol. 2012;14:270–280. doi: 10.1007/s10126-011-9410-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.