Abstract
Cell-free secretomes represent a promising new therapeutic avenue in regenerative medicine, and γ-irradiation of human peripheral blood mononuclear cells (PBMCs) has been shown to promote the release of paracrine factors with high regenerative potential. Recently, the use of alternative irradiation sources, such as artificially generated β- or electron-irradiation, is encouraged by authorities. Since the effect of the less hazardous electron-radiation on the production and functions of paracrine factors has not been tested so far, we compared the effects of γ- and electron-irradiation on PBMCs and determined the efficacy of both radiation sources for producing regenerative secretomes. Exposure to 60 Gy γ-rays from a radioactive nuclide and 60 Gy electron-irradiation provided by a linear accelerator comparably induced cell death and DNA damage. The transcriptional landscapes of PBMCs exposed to either radiation source shared a high degree of similarity. Secretion patterns of proteins, lipids, and extracellular vesicles displayed similar profiles after γ- and electron-irradiation. Lastly, we detected comparable biological activities in functional assays reflecting the regenerative potential of the secretomes. Taken together, we were able to demonstrate that electron-irradiation is an effective, alternative radiation source for producing therapeutic, cell-free secretomes. Our study paves the way for future clinical trials employing secretomes generated with electron-irradiation in tissue-regenerative medicine.
Keywords: regenerative medicine, cell-free secretomes, paracrine factors, biological medicinal products, γ-irradiation, electron-irradiation, γ-irradiation replacement
Graphical Abstract
γ-irradiated human peripheral blood mononuclear cells (PBMCs) are known to secrete paracrine factors with valuable regenerative potential. Since γ-irradiation represents a considerable biohazard, alternative, less hazardous stressors are urgently needed. Here, Laggner et al. demonstrate that the much safer electron-irradiation is equally effective to generate therapeutic, regenerative secretomes.
Introduction
Over the past few years, application of cell-derived, yet cell-free, secretomes, instead of transplanting cells, is being increasingly recognized as an attractive therapeutic intervention for tissue regeneration. Initial studies predominantly employed stem-cell (SC)-derived secreted factors,1, 2, 3, 4 although SCs have major drawbacks, such as a requirement of invasive procedures for isolation and limited availability. By comparison, peripheral blood mononuclear cells (PBMCs) represent a waste product of routine blood donation and are easily obtainable in larger numbers. Furthermore, secretomes of SCs and stressed PBMCs were reported to exhibit comparable regenerative effects.5 Pioneer SC transplantation studies suggested that cell death was crucial for promotion of tissue regeneration,6 and Ankersmit et al.7 were the first to demonstrate that infusing PBMCs stressed with 60 Gy γ-irradiation was superior to applying non-irradiated cell suspensions for tissue regeneration in a rodent model of acute myocardial infarction (AMI). Later studies by the same group showed that the secretome of γ-irradiated PBMCs (PBMCsec) improved cardiac outcome in rodent and porcine AMI models.8 During the last decade, PBMCsec has been successfully applied in various indications of tissue damage, including a latent porcine model of chronic post-myocardial infarction,9 and rodent models of cerebral ischemia, acute spinal cord injury, skin wounds, and diabetic wounds.10, 11, 12 Most recently, Winkler et al.13 reported that preconditioning of cardiosphere-derived cells with PBMCsec resulted in improved preservation of viability in a porcine model of reperfused myocardial infarction. Furthermore, PBMCsec improved flap surgery outcome by promoting wound healing and reducing necrosis rate.14 Based on these findings, a broad action spectrum has been attributed to the secretome, including immunomodulation, cytoprotection, vasodilation, as well as pro-angiogenic and anti-microbial effects.8,11,12,15, 16, 17, 18, 19, 20, 21, 22, 23, 24 These tissue-regenerative activities have been shown to be triggered by the interplay of many biologically active agents secreted by irradiated PBMCs, such as proteins, lipids, and extracellular vesicles (EVs).8,12,15 From a regulatory point of view, cell-free secretomes are classified as biological medicinal products. As it is impossible to analyze every single constituent, competent authorities recommend the selection of crucial identity parameters and functionalities for product characterization. These specifications then serve as a basis for the final approval of secretomes for clinical use. We successfully established a panel to demonstrate the reproducibility of PBMCsec generated under good manufacturing practice (GMP),25 and the combination of these specifications is therefore a legitimate approach to characterize pleiotropic biological medicinal products, such as secretomes, in a comprehensive manner.
The necessity of dying cells for tissue regeneration has already been recognized,6 and in contrast to other culture protocols employing hypoxia or stimulatory agents, ionizing radiation is a well-known, potent inducer of cell death. In transfusion medicine, exposure with γ-rays emitted from a radioactive nuclide is the bona fide treatment to halt donor T cell proliferation and prevent transfusion-associated graft-versus-host disease (TA-GvHD). Although highly effective in preventing TA-GvHD, γ-irradiators represent a considerable biohazard, and radioactive waste is an evident environmental problem. Additionally, increased security measures are required to reduce the risk of unauthorized use of radioactive materials. Therefore, the Cesium Irradiator Replacement Project was launched by the National Nuclear Security Administration (NNSA; Washington, DC, USA), which aims at replacing all γ-irradiators with less hazardous irradiation sources, such as β-irradiators or photon and electron beams provided by accelerators, by the end of 2027. Whereas the applicability of high-energy photon beams in transfusion medicine has already been reported,26,27 the feasibility of replacing γ-irradiation sources with β-irradiators or electron beams for the production of a regenerative secretome has not been investigated to date. Electron beams and beta-minus (β−) rays are identical from a physics point of view but have different origin.28 β−-rays are associated with radioactivity, and thus specific radiation protection aspects in terms of source handling and source depletion still apply, just as for γ-rays, whereas electron beams from an accelerator are easier to handle in this respect. So far, only few investigations comparing the biological activities of γ- and electron or β-irradiation have been conducted. Nonetheless, reported data are inconclusive due to incomparability of experimental settings, including radiation source,29 radiation dose,30 cell type investigated,30, 31, 32 and different readouts.31,33 A comprehensive and in-depth comparison of γ- and electron-irradiation has not been performed to date.
In the current study, we aimed to determine the cytotoxic and biological effects of γ- and electron-irradiation on PBMCs and comparably analyzed the composition and activity of the secretomes. First, we investigated whether γ- and electron-irradiation was equally effective in inducing PBMC death. As PBMCsec contains a range of active ingredients, the major biomolecular substance classes were analyzed, and functional assays reflecting key regenerative actions were performed to compare the biological activity of PBMCsec and secretomes obtained from electron-irradiated PBMCs.
Results
γ- and electron-irradiation equally induce cell death and DNA damage of PBMCs
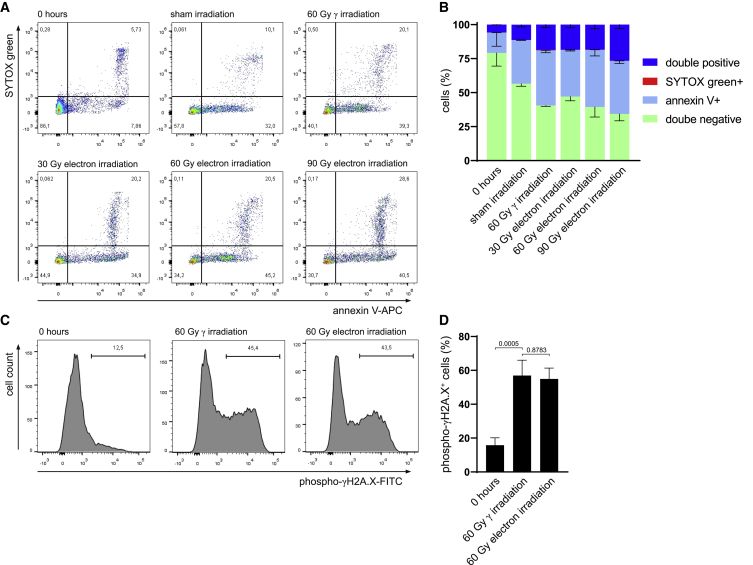
Ionizing radiation is a well-known cellular stressor, and cell death induced by 60 Gy γ-radiation was identified as a crucial trigger for secretion of regenerative factors.21 Therefore, we compared the cytotoxic effects of γ- and electron-irradiation on human PBMCs by flow cytometric assessment of Annexin V-SYTOX Green dual staining. Cells were analyzed directly after isolation (0 h) and 24 h post irradiation or sham treatment. The viability of freshly isolated PBMCs was high (79.2% ± 9.8% double-negative cells; Figures 1A and 1B), whereas culturing cells for 24 h caused a reduction of cell viability to 56.6% ± 1.8%. To determine the electron-irradiation dose equivalent to 60 Gy γ-irradiation, we employed different dosages ranging from 30 to 90 Gy electron. 24 h post irradiation, we found that the amount of viable cells was highly comparable between 60 Gy γ and 60 Gy electron (40.6% ± 0.6% and 39.4% ± 7.4% double-negative cells, respectively), whereas cell viability was significantly higher after irradiation with 30 Gy electron beams (47.1% ± 3.1%) and lower after 90 Gy electron beams (34.3% ± 5.1%). In addition, no significant differences in the amounts of early and late apoptotic and necroptotic cells were detected between 60 Gy γ-irradiation and 60 Gy electron (40.6% ± 1.8% versus 42.1% ± 4.5% Annexin V+ cells and 18.6% ± 2.1% versus 18.4% ± 2.9% double-positive cells after 60 Gy γ- and electron-irradiation, respectively). Therefore, we chose the minimum effective dose of 60 Gy electrons for our further studies.
Figure 1.
γ- and electron-irradiation exhibit comparable cytotoxic effects
(A) Representative dot plots of Annexin V-SYTOX Green-stained cells after isolation, after 24 h of culture, as well as 24 h after 60 Gy γ- and 30 Gy, 60 Gy, or 90 Gy electron-irradiation. Numbers in quadrants indicate relative number of cells. (B) Bar diagram showing average percentages of stained cells and standard deviation. Data of n = 4 donors are shown. (C) Representative histograms of phosphorylated (phospho) γH2A.X staining after isolation (0 h), 60 Gy γ-irradiation, and 60 Gy electron-irradiation. Numbers indicate relative numbers of cells. (D) Statistical analysis of phosphorylated γH2A.X-positive cells. Data are displayed as mean ± standard deviation and were analyzed by one-way ANOVA with Dunnett’s multiple comparisons. Data of n = 5 donors are shown.
As γ-irradiation is known to induce DNA double-strand breaks in lymphocytes,34 we next aimed to determine whether electron and γ-irradiation equally affected DNA damage. Although freshly isolated PBMCs displayed low levels of phosphorylated (phospho) γH2A.X (15.9% ± 9.6%; Figures 1C and 1D), cells irradiated with 60 Gy γ-irradiation and 60 Gy electron-irradiation induced comparable levels of DNA double-strand breaks (56.9% ± 20.3% and 54.9% ± 14.2% positive cells, respectively). Taken together, these data show that 60 Gy γ-irradiation and 60 Gy electron-irradiation are equally effective in inducing cell death and DNA damage of human PBMCs.
Transcriptional profiles of γ- and electron-irradiated PBMCs show comparable gene regulation
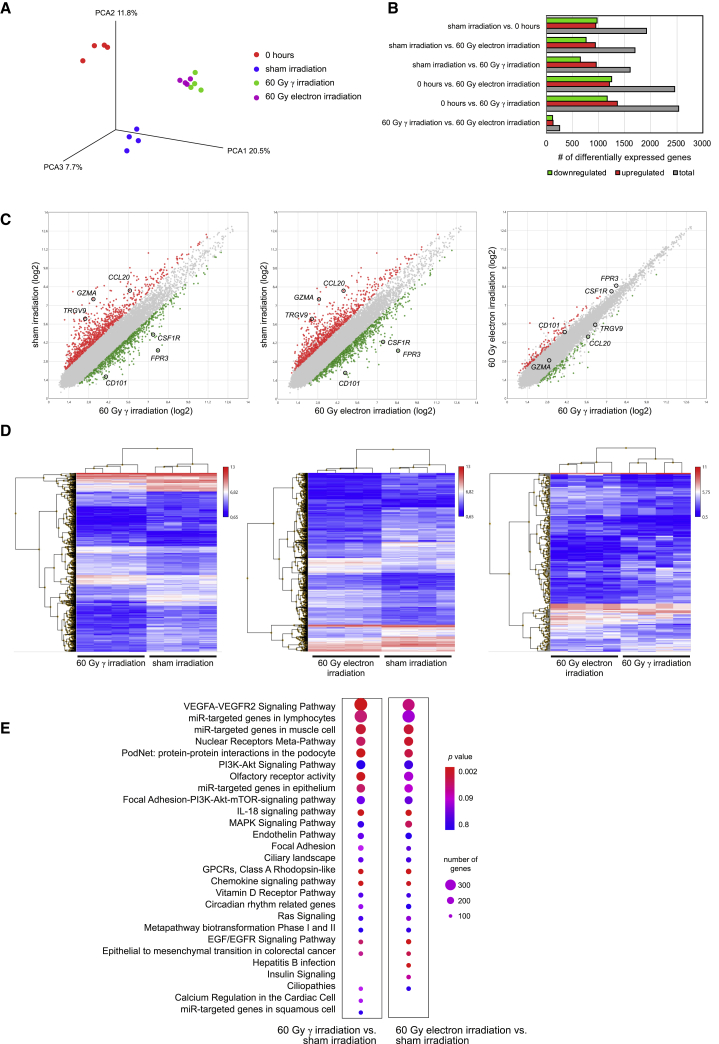
We furthermore sought to determine whether 60 Gy γ- and 60 Gy electron-irradiation might differentially affect gene expression patterns of PBMCs. To this end, we compared transcriptional profiles of freshly isolated PBMCs, sham-irradiated PBMCs after 24 h of culture, and PBMCs 24 h after 60 Gy γ- and 60 Gy electron-irradiation. Principal-component analysis (PCA) revealed that gene signatures of freshly isolated PBMCs, cultured cells without irradiation, and irradiated cells were distinct. However, γ- and electron-irradiated cells showed a very high transcriptional similarity (Figure 2A). We determined the numbers of differentially expressed genes (DEGs) with more than 2- and below 0.5-fold regulation and found that the average amount of DEGs ranged from more than 1,500 up to 2,500 when comparing 0 h and sham irradiation with 60 Gy γ- and 60 Gy electron-irradiation (Figure 2B). In contrast, roughly 250 genes were found differentially regulated when comparing γ- with electron-irradiated PBMCs. These results were corroborated by pairwise comparisons, where several up- and downregulated genes were detected between irradiated and non-irradiated samples (Figure 2C). As an example, we randomly selected three up- and downregulated genes among the top DEGs between irradiated and non-irradiated samples, i.e., CC-chemokine ligand 20 (CCL20), cluster of differentiation 101 (CD101), colony-stimulating factor 1 receptor (CSF1R), formyl peptide receptor 3 (FPR3), granzyme A (GZMA), and T cell receptor gamma variable 9 (TRGV9) (Figure 2C). Of note, these genes were equally regulated by different irradiation treatments. However, these genes were not differentially regulated when comparing γ- and electron-irradiation (Figure 2C). Gene enrichment analysis of the ~250 differentially regulated genes between γ- and electron-irradiation revealed that genes expressed higher after γ-irradiation were associated with monocyte chemotaxis, tumor necrosis factor (TNF) signaling pathway, Toll-like receptor signaling pathway, and cellular response to interleukin-1 (IL-1). Genes expressed higher after electron-irradiation compared to γ-irradiation were involved in positive regulation of T cell proliferation and glycosyl catabolic process (Figure S1). Heatmaps of global gene expression revealed that irradiated samples showed marked differences compared to non-irradiated controls, whereas similar transcriptional landscapes were observed when comparing γ- and electron-irradiated PBMCs (Figure 2D). DEGs between irradiated and non-irradiated samples were used to identify regulated cellular signaling pathways, and we found that the majority of the associated pathways were identical (Figure 2E). To corroborate our findings obtained by phosphorylated γH2A.X staining, we furthermore assessed expression values of genes associated with DNA damage responses (Figure S2). In line with our previous results, we found no differences in the expression of genes implicated in base excision repair, mismatch repair, nucleotide excision repair, homology-directed repair, and non-homologous end joining when comparing γ- and electron-irradiated PBMCs. Together, these data show that the effects of 60 Gy γ- and 60 Gy electron-irradiation on gene regulation of PBMCs are highly comparable.
Figure 2.
Effects of γ- and electron-irradiation on transcriptional regulation of PBMCs
(A) PCA of freshly isolated PBMCs, PBMCs after 24 h of culture, and PBMCs 24 h after 60 Gy γ- and 60 Gy electron-irradiation. Each dot represents one donor; color codes indicate treatments. (B) Number of differentially expressed genes among groups. (C) Scatterplots showing up- and downregulated genes (red and green, respectively) between non-irradiated and irradiated PBMCs. Each dot represents one gene. Gray dots indicate non-differentially expressed genes (fold-change cut-offs 2 and 0.5, respectively). CCL20, CC-chemokine ligand 20; CD101, cluster of differentiation 101; CSF1R, colony-stimulating factor 1 receptor; FPR3, formyl peptide receptor 3; GZMA, granzyme A; TRGV9, T cell receptor gamma variable 9. (D) Heatmaps of global gene expressions of irradiated and non-irradiated samples. Each row represents one gene; each column represents one donor with the respective treatment. Color codes indicate expression levels. (E) Differentially expressed genes between irradiated and non-irradiated cells were used to calculate associated signaling pathways. Circle sizes reflect total numbers of regulated genes per pathway, and p values are indicated by color code. Data of n = 4 donors are shown.
The secretion of proteins, lipids, and EVs by γ- and electron-irradiated PBMCs is comparable
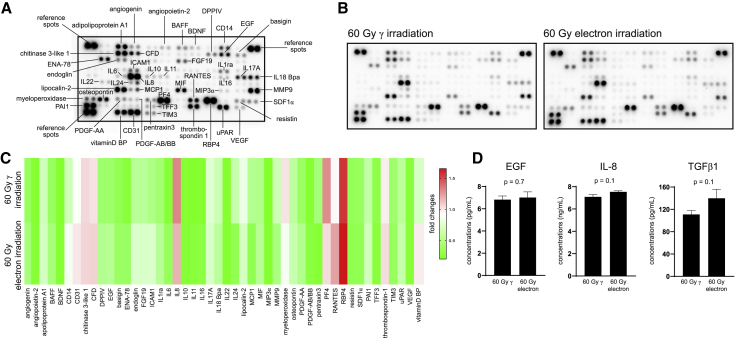
As protein release by PBMCs is promoted by γ-irradiation,8 and proteins are crucial for mediating regenerative and wound-healing promoting actions of cellular secretomes, we sought to determine whether the quality and quantity of released proteins were also comparable between γ- and electron-irradiated PBMCs. To address this question, we semiquantitatively determined cytokines using the Proteome Profiler Human XL Cytokine Array and found that the identities of released cytokines were highly comparable between γ- and electron-irradiated PBMCs (Figures 3A−3C; Figure S3). Only signals clearly discernable from background signals were included in our analysis. We furthermore selected cytokines implicated in wound healing for quantification by enzyme-linked immunosorbent assay (ELISA) and found no significant differences in the amount of secreted IL-8, transforming growth factor beta 1 (TGF-β1), and epidermal growth factor (EGF) (Figure 3D).
Figure 3.
Cytokine secretion of γ- and electron-irradiated PBMCs
(A) Immunodetection array membrane with annotations of the most abundant analytes detected in the secretomes of PBMCs. Each spot represents one analyte, and each analyte is spotted in duplicates. (B) Cytokine secretion profiles of 60 Gy γ- and 60 Gy electron-irradiated PBMCs. Pooled secretomes of n = 4 donors per condition were analyzed. (C) Analysis of integrated densities of chemiluminescent signals. Each column represents one analyte. Colors indicate fold changes of integrated intensities. (D) Pooled secretomes of n = 4 donors were measured by ELISA in technical triplicates. Bar diagrams show mean and standard deviation. For statistical analysis, two-tailed Mann-Whitney was performed. BAFF, B cell activating factor; BDNF, brain-derived neurotrophic factor; CFD, complement factor D; DPPIV, dipeptidyl-peptidase IV; EGF, epidermal growth factor; ENA-78, epithelial-derived neutrophil-activating protein 78; FGF19, fibroblast growth factor 19; ICAM1, intercellular adhesion molecule 1; IL1ra, interleukin 1 receptor antagonist; IL6, interleukin 6; IL8, interleukin 8; IL10, interleukin 10; IL11, interleukin 11; IL16, interleukin 16; IL17A, interleukin 17A; IL18BPa, interleukin 18-binding protein isoform a; IL22, interleukin 22; IL24, interleukin 24; MCP1, monocyte chemotactic protein 1; MIF, macrophage migration inhibitory factor; MIP3α, macrophage inflammatory protein 3 alpha; MMP9, matrix metallopeptidase 9; PDGF-AA, platelet-derived growth factor subunit A; PDGF-AB/BB, platelet-derived growth factor subunits A and B; PF4, platelet factor 4; RANTES, regulated and normal T cell expressed and secreted; RBP, retinol-binding protein; SDF1α, stromal cell-derived factor 1 alpha; PAI-1, plasminogen activator inhibitor-1; TFF3, trefoil factor 3; TIM3, T cell immunoglobulin and mucin-domain containing-3; uPAR, urokinase-type plasminogen activator receptor; VEGF, vascular endothelial growth factor; vitamin D BP, vitamin D-binding protein; TGFβ1, transforming growth factor beta 1.
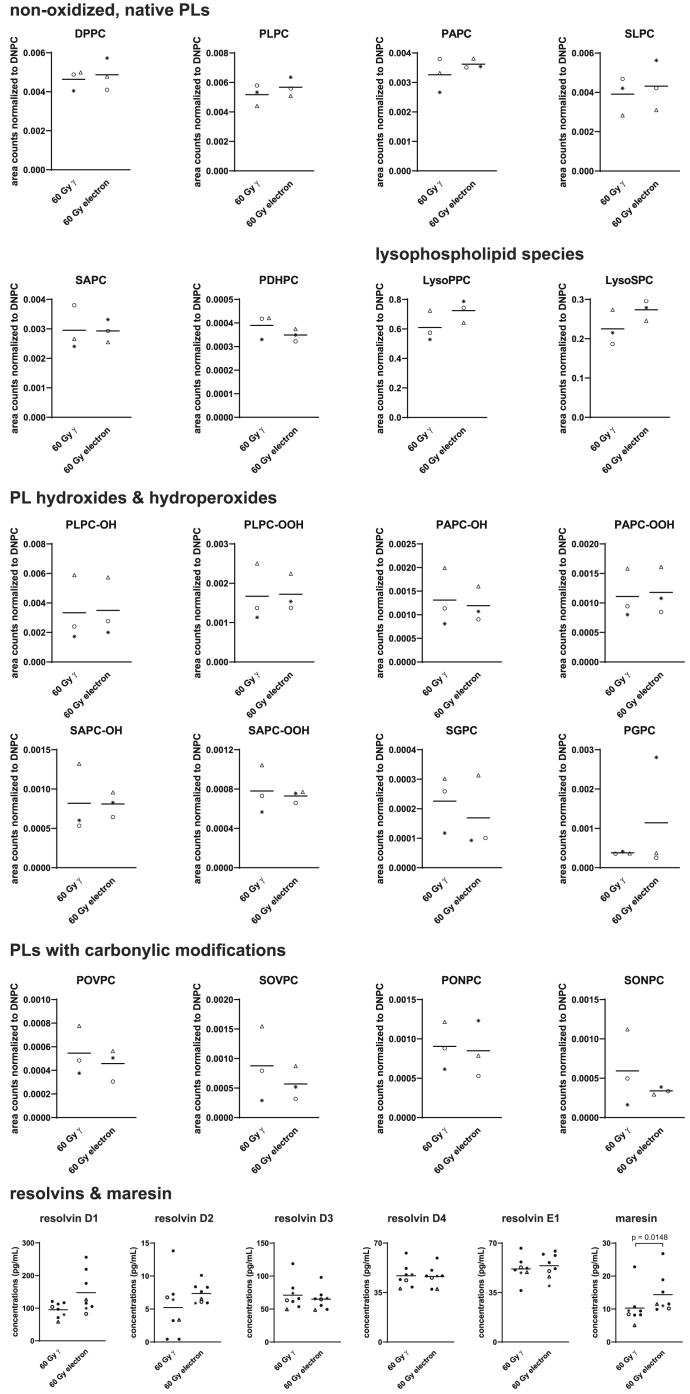
Recently, potent anti-inflammatory actions were attributed to lipids present in the PBMCsec,20,35 and γ-irradiation of PBMCs was necessary for inducing various oxidized phospholipid (PL) species15 implicated in immunomodulation.36 Therefore, we determined the presence of lipid species in PBMCsec and the secretome of electron-irradiated PBMCs by tandem mass spectrometry. When analyzing various species belonging to the groups of native PLs, lysoPLs, oxidized PLs, and PLs with carbonylic modifications, we observed no major differences among the different radiation sources (Figure 4). We furthermore chose to quantify several resolvin species and maresin 1 by ELISA, which have already been detected in PBMCsec20 and exert immunomodulatory action.37, 38, 39, 40, 41 We found no differences in resolvin concentrations between γ- and electron-irradiation (Figure 4). Only secretion of maresin was significantly increased by electron-irradiation compared to γ-irradiation.
Figure 4.
Quantification of lipids secreted by γ- and electron-irradiated PBMCs
Different classes of lipid species were analyzed by mass spectrometry, including native PLs, lysoPLs, oxidized PLs, and PLs with carbonylic modifications, resolvins, and maresin secreted by γ- and electron-irradiated PBMCs. Each dot represents one donor. For HPLC-MS/MS, data of n = 3 donors are shown, and peak areas were normalized to spiked 1,2-dinonanoyl-sn-glycero-3-phosphocholine (DNPC) controls. Horizontal bars indicate arithmetic mean. For lipidomics data, two-tailed paired t test was calculated. For statistical analysis of resolvins and maresin, two-tailed Mann-Whitney was performed. Triangles, open circles, and asterisks refer to donors 1, 2, and 3, respectively.
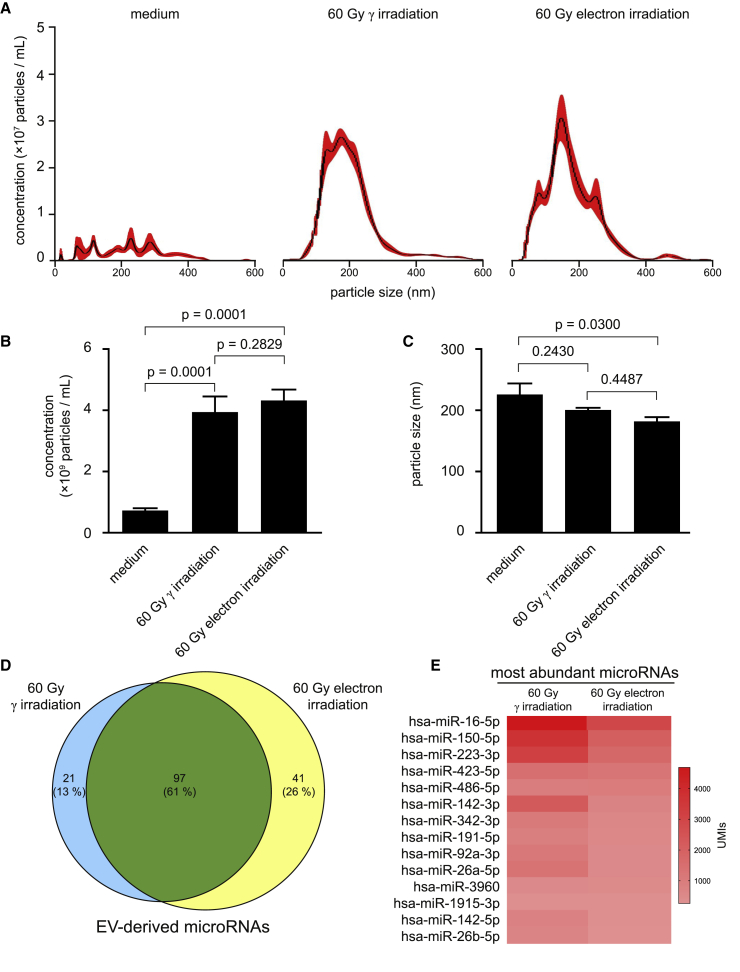
Since various regenerative effects have been attributed to EVs,42 and γ-irradiation remarkably promoted secretion of EVs,12 we compared nanoparticles secreted by irradiated PBMCs. Interestingly, we were not able to detect differences in particle concentrations and sizes between γ- and electron-irradiated PBMCs (Figures 5A−5C). As the molecular content, especially microRNAs (miRNAs), dictates the paracrine actions of EVs derived from irradiated PBMCs,12 we further analyzed the miRNA species present in EVs by RNA sequencing. We were able to detect a total of 118 different miRNAs in the EVs of γ-irradiated PBMCs and 138 miRNAs after electron-irradiation (Figure 5D; Table S1). When comparing the two forms of irradiation, the prevailing majority (61%) of the detected miRNA species were present in EVs derived from PBMCs exposed to both radiation sources. Moreover, the 14 most abundant miRNAs were identical in both groups (Figure 5E). With the use of an online database for profiling miRNAs in EVs,43 we found that the most abundant miRNA hsa-miRNA (miR)-16-5p is strongly expressed in blood exosomes and is involved in chemokine signaling, nuclear factor (NF)-kappa B signaling pathway, and p53 signaling. Functional annotations of the 14 most abundant miRNAs revealed possible involvements in several biological processes, such as cytokine signaling, immune response, cell death and proliferation, and DNA damage response (Table S2). These data show that the miRNAs present in the EVs derived from γ- and electron-irradiated PBMCs are predominantly the same and presumably exert similar biological functions.
Figure 5.
Nanoparticle measurements of EVs secreted by γ- and electron-irradiated PBMCs
(A) Averaged concentrations per sizes of detected nanoparticles in medium and in secretomes of irradiated PBMCs. Pooled secretomes of n = 4 donors were analyzed. Black lines indicate mean of 6 measurements. Red areas indicate standard errors of the mean. (B and C) Statistical analyses of particle (B) concentrations and (C) sizes. Bar diagrams show mean and standard deviation. For statistical analysis, one-way ANOVA with Tukey’s multiple comparisons was performed. (D) Venn diagram showing the amount of identical and unique microRNAs detected in the EVs of γ- and electron-irradiated PBMCs. Absolute numbers indicate the number of different microRNAs, and percentages indicate the relative amounts of identical and unique microRNAs. (E) Heatmap of the most abundant microRNAs in the EVs of γ- and electron-irradiated PBMCs. Color code indicates unique molecular identifiers (UMIs).
Taken together, these findings indicate that γ-irradiation and electron-irradiation are equally effective in inducing the release of various biologically active components by human PBMCs.
PBMCsec and secretomes obtained from electron-irradiated PBMCs display similar biological activity
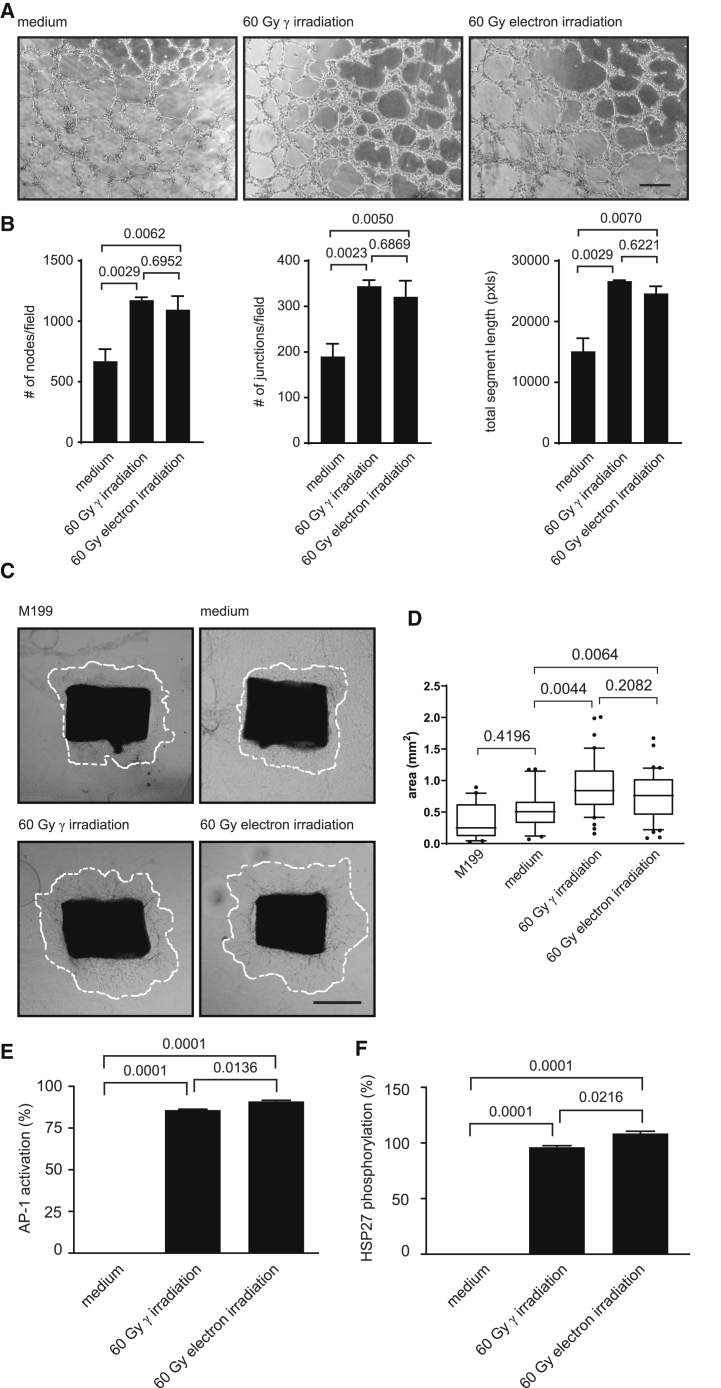
In addition to a biomolecular characterization, we aimed to determine whether the PBMCsec and secretomes obtained from electron-irradiated PBMCs share similar biological activity. As secretomes have already been successfully employed to promote wound healing,17,44 we tested the secretomes of irradiated PBMCs in several functional assays, reflecting key stages of secretome-promoted wound healing. Tube formation assay, using human umbilical vein endothelial cells (HUVECs),45 revealed that the number of nodes and junctions, as well as the total segment lengths, was comparably high when adding PBMCsec and secretomes from electron-irradiated PBMCs (Figures 6A and 6B). In contrast, medium used to culture PBMCs displayed little tube formation potency.
Figure 6.
PBMCsec and secretomes of electron-irradiated PBMCs display comparable biological activity
(A) Representative micrographs of HUVECs incubated with medium or secretomes of irradiated PBMCs. Pooled secretomes of n = 4 donors were added. Scale bar, 250 μm. (B) Statistical analyses of nodes per field, junctions per field, and total segment lengths per field. Bar diagrams show arithmetic mean and standard deviation. Groups were compared using one-way ANOVA and Tukey’s multiple comparisons. (C) Representative images of murine, thoracic,aortic rings incubated with medium or secretomes of irradiated PBMCs. Pooled secretomes of n = 4 donors were added. Scale bar, 500 μm. (D) Statistical analysis of outgrowth areas. Boxplot diagrams were generated using first and third quartiles as boxes and median as bars. Whiskers indicate 10−90 percentiles. Groups were compared using one-way ANOVA and Sidak’s multiple comparisons. (E) AP-1 activation and (F) HSP27 phosphorylation induced by the secretomes obtained from irradiated PBMCs. Values were normalized to internally established reference standards. Bar diagrams show arithmetic mean and standard deviation. Groups were compared using one-way ANOVA and Tukey’s multiple comparisons.
As formation of new blood vessels represents a crucial step of wound healing, endothelial cell migration and proliferation with secretomes of irradiated PBMCs were performed using an ex vivo murine thoracic aortic ring assay. On day 3 of explant culture, outgrowth areas of endothelial cells were low with M199 and with medium vehicle used to culture PBMCs (Figures 6C and 6D). Compared to medium alone, the secretomes of irradiated PBMCs effectively promoted cellular proliferation, and no difference in the pro-angiogenic properties of PBMCsec and secretomes obtained from electron-irradiated PBMCs was observed.
Activation of activator protein 1 (AP-1) promoter and phosphorylation of heat shock protein 27 (HSP27) have been implicated in wound healing,46,47 and biological activity of secretomes was compared in validated, cell-based assays. Although no potency was detected when medium used to culture PBMCs was added, PBMCsec and secretomes of electron-irradiated PBMCs effectively and comparably induced AP-1 activation and HSP27 phosphorylation (Figures 6E and 6F).
Discussion
Induction of cell death is a crucial step for the generation of a potent cell secretome with strong tissue-regenerative properties, and so far, γ-irradiation served as an effective inducer of cell death.7,21 Nonetheless, use of radioactive substances harbors critical safety concerns for the operator, is subject to strict legal and safety requirements, requires high economic efforts in disposal, and represents a long-lasting environmental burden. Therefore, the development and use of alternative, safer methods are highly appreciated by regulatory authorities. Electron-irradiation represents an attractive, non-radioactive substitute with lower radiation burden and fewer health concerns. In the current study, we investigated molecular composition and tissue-regenerative activity of a secretome derived from electron-irradiated PBMCs and compared it to the well-characterized PBMCsec. Our study provides evidence for comparable composition and functional characteristics of both PBMCsec and secretomes obtained from electron-irradiated PBMCs. Based on these encouraging findings, we are convinced that electron-irradiation represents a suitable, non-radioactive, and safe alternative to γ-irradiation for producing therapeutic secretomes from PBMCs.
Programmed cell death is an indispensable mechanism for embryonal development, tissue homeostasis, and non-inflammatory removal of deleterious cells.48 The idea that dying cells can promote tissue regeneration is seemingly counterintuitive. However, this concept has already been proven by Ankersmit et al.,7 and a variety of underlying mechanisms have already been elucidated.24 Here, we showed that electron-irradiation successfully induced cell death of PBMCs, which was comparable to the cytotoxicity of γ-irradiation. As irradiation is known to induce DNA damage,34 we also analyzed phosphorylated γH2A.X, a marker for DNA double-strand breaks.49 Importantly, we were able to demonstrate that irradiation of PBMCs with electron and γ-irradiation led to the formation of comparable levels of DNA double-strand breaks, suggesting similar biological functions induced by DNA damage. Our transcriptome analysis revealed only minor differences in mRNA expression after γ- and electron-irradiation of PBMCs. In addition, the molecular processes associated with DEGs were similar when γ- and electron-irradiated PBMCs were compared to non-irradiated cells. Similar to results obtained on an mRNA level, qualitative and quantitative comparisons of released proteins showed no significant differences between γ- and electron-irradiated PBMCs. Recently, we were able to demonstrate an important immunomodulatory activity of lipids present in the secretome of PBMCs exposed to γ-radiation.20,35 We thus performed lipidomics analysis and ELISA-assisted quantifications to compare the lipid composition of PBMCsec and secretomes of electron-irradiated PBMCs. We detected comparable amounts of non-oxidized and oxidized PLs and resolvins. Similar lipid species have previously been identified when non-irradiated and PBMCsec were analyzed15 and when the lipid contents of EVs were studied.12 Furthermore, the presence of immunomodulatory resolvins and maresin in the PBMCsec has already been reported.20 We therefore conclude that the secreted lipid species, both non-oxidized and oxidized, generated by γ- and electron-irradiation of PBMCs are similar. Interestingly, the lipid composition of secretomes seems to be cell type dependent. Li et al.50 showed that caspases activated during apoptotic cell death induced the secretion of proliferation-promoting factors. They also showed beneficial effects of ionizing radiation by demonstrating that proliferation of stem and progenitor cells was increased in the presence of irradiated cells. In their study, phospholipase A2, an enzyme important for synthesis and secretion of prostaglandin E2 (PGE2), was shown to be responsible for the observed effect on cell proliferation. Interestingly and in contrast to their findings, PGE2 was not detectable in our lipidomics analysis, suggesting that the regenerative and immunomodulatory effects of PBMCsec occur in a PGE2-independent manner. Intriguingly, we found increased secretion of maresin 1 after electron-irradiation compared to γ-irradiation. PBMCsec attenuated secondary tissue damage in a spinal cord injury rat model,11 and maresin 1 was reported to induce resolution of inflammation in a spinal cord injury mouse model.38 Whether the secretome derived from electron-irradiated PBMCs containing higher maresin 1 levels is equally or even more effective than that of PBMCsec in preventing neural tissue damage remains to be determined. Further elucidating the exact role of maresins in our model using electron-irradiation merits future investigations.
Previous reports have demonstrated that γ-irradiation promotes secretion of nanoparticles,12 and we were able to show a comparable particle secretion pattern by γ- and electron-irradiated PBMCs. The average concentration and average size of particles by our electron-irradiated PBMCs are in accordance with PBMCsec produced under GMP,25 indicating that γ- and electron-irradiation equally induced EV secretion by PBMCs. In addition, we performed an in-depth analysis of the EV content by sequencing miRNAs. It has been shown that irradiation of PBMCs affects the miRNA profile of EVs.12 The process of sequestering miRNAs in EVs occurs in a random and non-selective manner.51 Hence, the most abundant cytoplasmic miRNAs are also found in EVs. We found that the prevailing majority of miRNA species in EVs is the same when PBMCs were irradiated with γ- or electron-irradiation, whereas differences were mainly due to miRNAs in low abundance. Further functional studies are required to confirm whether EVs released by γ- and electron-irradiated PBMCs elicit comparable biological responses in target cells.
In addition to analytical tests, we used a panel of functional assays to determine the biological activity of PBMCsec and secretomes from electron-irradiated PBMCs. The results obtained from electron-irradiated PBMC secretomes were comparable to previous findings, where PBMCsec were tested in tube formation assays,21,25 sprouting assays,12,21,25 and potency assays.12,21,25
In spite of all efforts, our study is subject to some limitations. Pooling the secretomes of up to 120 donors is necessary to obtain large-scale batches for clinical application, and combining secretomes of several donors is crucial to diminish donor-to-donor variability.25 Since only few donors were analyzed in the current study, comparing the pooled secretomes of numerous donors exposed to γ- or electron-irradiation merits future investigations. In addition, our analyses primarily focused on obtaining an overall picture of the major constituents and key functions of the secretomes, whereas further, more sophisticated in vitro and in vivo approaches are necessary to obtain more detailed insights into the therapeutic potential of the secretome derived from electron-irradiated PBMCs. Furthermore, our semiquantitative approach of cytokine analysis by a proteome profiler was deemed a convenient way to study immunologically relevant proteins present in secretomes. However, future proteomics analyses of secretomes will delineate the overall protein signature in more detail.
In conclusion, we provide several lines of evidence that demonstrate that γ- and electron-irradiation exert similar effects on PBMCs and that electron-radiation is a suitable, less hazardous alternative to radiation emitted by radioactive substances for the generation of therapeutic secretomes. To conduct clinical trials, enormous efforts are necessary to fulfill a variety of criteria required by the regulatory authorities. These preclinical tests, including in vitro analyses of the modes of action and in vivo tests to investigate function, efficacy, and toxicology, are time consuming and expensive. We have previously conducted toxicology studies where intravenous and subcutaneous application of the PBMCsec was investigated.23 These studies allowed a phase I clinical trial with autologous PBMCsec, where no therapy-related, serious adverse events were found (ClinicalTrials.gov: NCT02284360).22 Additionally, we have recently initiated a first-in-men study with topical application of the PBMCsec in diabetic foot ulcer (EudraCT bo. 2018-001653-27; ClinicalTrials.gov: NCT04277598).52 Since it is foreseeable that the GMP production process of the secretome will use less hazardous β-radiation or even electron beams in the future, we believe that the present study justifies the extrapolation of results from toxicology and phase I clinical trials obtained from PBMCsec to electron-irradiated cells. As the Cesium Irradiator Replacement Project initiated by the NNSA intends to replace all γ-irradiator sources, many production lines worldwide currently using γ-radiators will face this problem. This study could therefore serve as a test case to avoid unnecessary, expensive, and time-consuming experiments.
Materials and methods
Ethics statements
This study was conducted in accordance with the Declaration of Helsinki and applicable local regulations. Blood was obtained from volunteers, and written, informed consent was obtained from all donors. Use of primary HUVECs and PBMCs was approved by the Institutional Review Board of the Medical University of Vienna (Ethics Committee votes 1,280/2,015 and 1,539/2,017).
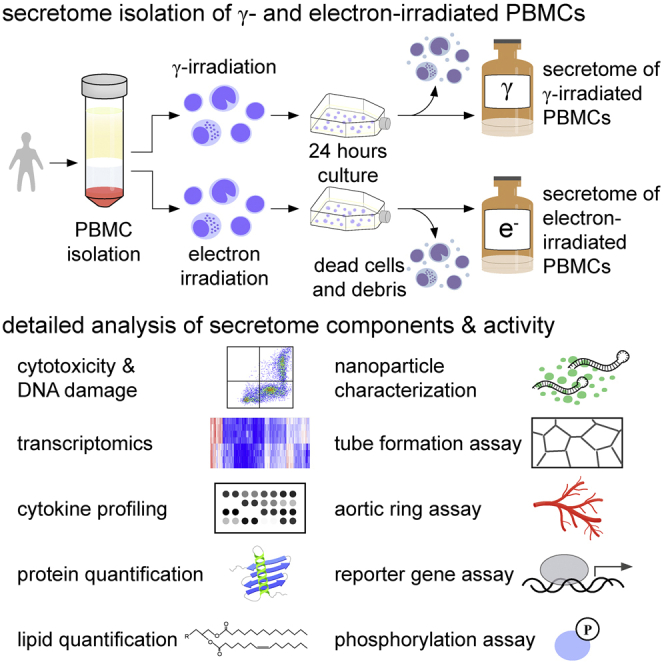
PBMC irradiation and secretome isolation
PBMCs of 9 voluntary donors were procured by the Austrian Red Cross Blood Transfusion Service for Upper Austria (Linz, Austria). PBMCs were enriched by Ficoll-Paque PLUS (GE Healthcare, Chicago, IL, USA)-assisted density centrifugation and adjusted to a concentration of 2.5 × 107 cells/mL in CellGenix granulocyte-monocyte progenitor dendritic cell medium (CellGenix, Freiburg, Germany). Cells were exposed to 60 Gy cesium 137 γ-irradiation (IBL 437C; Isotopen Diagnostik CIS, Dreieich, Germany) or 6 MeV electron-irradiation (30−90 Gy), provided by a medical linear accelerator (Synergy; Elekta, Stockholm, Sweden). Calibration data of both devices are provided in Figure S4 and Tables S3 and S4. The radiation period for 60 Gy γ and 60 Gy electron was 11 min, 12 s, and 13 min and 20 s (454 monitor units [MU]/min), respectively; i.e., very similar dose rates were applied. After 24 ± 2 h of culture, cells and cellular debris were removed by centrifugation, and conditioned medium containing the secretome was passed through a 0.22-μm filter. Secretomes were cryopreserved at −80°C until use.
Cell viability
Cell viability was determined using the Dead Cell Apoptosis Kit with Annexin V allophycocyanin (APC) and SYTOX Green (Thermo Fisher Scientific, Waltham, MA, USA), as recommended by the manufacturer. Fluorescent signals were recorded by BD FACSCanto II (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed using FlowJo software (v10.5.3; FlowJo, Ashland, OR, USA).
Assessment of DNA damage
DNA double-strand breaks were detected using fluorescein isothiocyanate (FITC)-conjugated mouse anti-human phosphorylated (Ser139) γH2A.X antibodies (BioLegend, San Diego, CA, USA) and analyzed by flow cytometry, as described above.
Transcriptomics and bioinformatics analyses
For transcriptomics analyses, total RNA was isolated using the RNeasy system (QIAGEN, Hilden, Germany). Total RNA concentrations were determined by the NanoDrop-1000 spectrophotometer (PEQLAB, Erlangen, Germany), and RNA quality was assessed by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) (Figure S5). Transcriptome profiling was carried out by the Genomics Core Facility at the Medical University of Vienna (Vienna, Austria) using the Human Gene 2.1 ST Array (Thermo Fisher Scientific). Transcriptome Analysis Console software (v.4.0; Thermo Fisher Scientific) was used for data analysis, for PCA, to determine DEGs, for hierarchical clustering, for scatterplots, and to identify pathways associated with DEGs. Gene lists of DEGs (more than 2-fold change of log2-transormed expression values, p values below 0.05) were analyzed by Cytoscape (v.3.8.5)53 using the ClueGO (v.2.5.7) plug-in.54 Biological process, immune system process, molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) were selected to identify pathways and ontologies.
Cytokine profiling
For cytokine profiling, secretomes of four donors were pooled. Cytokines were assessed by the Proteome Profiler Assay Human XL Cytokine Array Kit (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions. For quantification of chemiluminescent signals, images were converted into gray-scale images, and integrated densities of duplicate cytokine dots were calculated using the ImageJ measure tool (v.1.52p; Jana 1.8.0_172; National Institutes of Health, Bethesda, MD, USA).55 For visualization, the heatmap option of GraphPad Prism was used (v.5.01; GraphPad Software, LA Jolla, CA, USA).
Protein quantification
Concentrations of IL-8, TGF-β1, and EGF were determined with ELISAs (Human EGF Coated ELISA Kit and Human IL-8 Coated ELISA Kit, both Invitrogen, Carlsbad, CA, USA; Human TGF-β1 Quantikine ELISA, R&D Systems). All tests were performed according to standard procedures following good laboratory practice (GLP).
Lipidomics
Lipid isolation was performed as established previously.56 Reversed-phase high-performance liquid chromatography (HPLC)-electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) was carried out by the Forensic-Toxicological Laboratory (Vienna, Austria), as described elsewhere.56
Lipid quantification
The anti-inflammatory lipids resolvins and maresin were quantified by ELISAs, according to the manufacturers’ instructions (Human Resolvin D1 ELISA Kit, Human Resolvin D2 ELISA Kit, and Human Maresin ELISA Kit, all Cayman Chemical, Ann Arbor, MI, USA; Human Resolvin E1 ELISA Kit, abbkine, Wuhan, China; Human Resolvin D3 ELISA Kit and Human Resolvin D4 ELISA Kit, both MyBioSource, San Diego, CA, USA). Colorimetric measurements were performed by a microplate reader (infinite F50; TECAN, Durham, NC, USA) using Magellan F50 software (v.7.2; TECAN). Lipid concentrations were calculated using external standard curves.
Analysis of secreted EVs
EVs were isolated by ultracentrifugation, as described previously.12 Qualitative and quantitative assessments of nanoparticles were performed using the NanoSight NS500 instrument (Malvern Instruments, Malvern, UK). For each measurement, 500 μL of undiluted samples was loaded by the automatic pump control into the NanoSight system. The repetition and duration of captures were manually set to 6 captures with 30 s each. The instrument was calibrated using 100 nm particle reference controls provided by the instrument manufacturer.
EV isolation and total RNA isolation
EVs were isolated from secretomes by ultracentrifugation at 110,000 × g for 2 h at 4°C, as described previously.12 To obtain sufficient EVs for sequencing, the secretomes of 6 donors were pooled. Total RNA was isolated using peqGOLD TriFast (PEQLAB Biotechnologie, VWR International, Radnor, PA, USA), as recommended by the manufacturer, and was quality checked on a Bioanalyzer 2100 (Agilent) using a smallRNA Kit (Figure S6).
Small RNA sequencing
Sequencing libraries were prepared at the Core Facility Genomics, Medical University of Vienna, using the QIAgen smallRNA Library Prep Kit with unique molecular identifiers (UMIs), according to the manufacturer’s protocols. Libraries were quality-control (QC) checked on a Bioanalyzer 2100 (Agilent) using a High Sensitivity (HS) DNA Kit for correct insert size and quantified using Qubit double-stranded DNA (dsDNA) HS Assay (Invitrogen). Pooled libraries were sequenced on a NextSeq500 instrument (Illumina, San Diego, CA, USA) in 1 × 75 bp single-end sequencing mode. Approximately 8 million reads were generated per sample. Reads in fastq format were aligned to a database of human miRNAs and quantified considering the UMIs using the QIAseq miRNA Library Kit-Primary Quantification analysis tool from QIAgen GeneGlobe (https://geneglobe.qiagen.com/at/analyze; accessed on January 19, 2021). For functional annotations, the TAM 2.0 webtool was used.57,58
Tube formation assay
To compare pro-angiogenic properties of PBMCsec, a tube formation assay was performed with HUVECs (passage 4) as described previously.21,25 Cells were isolated as described59 and routinely cultured in endothelial cell growth basal medium-2 (EBM-2; Lonza Group AG, Basel, Switzerland), supplemented with endothelial cell growth medium-2 (EGM-2; BulletKit; Lonza). Prior to the tube formation assay, cells were maintained in EBM-2 containing 2% (vol/vol) heat-inactivated fetal bovine serum (Lonza) overnight and starved in basal EBM-2 for 4 h. Cells were seeded on growth factor-reduced Matrigel Matrix (Corning Life Sciences, Tewksbury, MA, USA) in μ-slides angiogenesis (ibidi, Graefelfing, Germany) at a density of 104 cells/cm2 and stimulated with the supernatant obtained from 4 × 106 PBMCs for 3 h. Micrographs were acquired by an inverted-phase contrast microscope (CKX41; Olympus, Tokyo, Japan) equipped with a 10× objective (CAch N, 10×/0.25 PhP; Olympus) using a SC30 camera (Olympus) and cellSens Entry software (v.1.8; Olympus). Tubule formation was quantified by the Angiogenesis Analyzer plug-in of ImageJ using default settings.60
Aortic ring sprouting assay
In addition to tube formation, the pro-angiogenic potential of PBMCsec was functionally tested by a sprouting assay using murine thoracic aortas.12,21,25 Briefly, aortic rings were sandwiched in fibrin matrices and cultured with secretome-conditioned medium equivalent to the supernatant of 4 × 106 PBMCs. Brightfield micrographs were acquired using an inverted Olympus IX83 scanning microscope (Olympus) with cellSens Imaging Software (Olympus) after culturing explants for 3 days. Outgrowth areas were determined by the ImageJ measure tool.55
Potency assays
The potencies of electron- and γ-irradiation-induced secretomes were compared by validated, GLP-compliant potency assays performed by SYNLAB Analytics and Services Switzerland AG (Birsfelden, Switzerland). AP-1 promoter activity and HSP27 phosphorylation were determined by reporter gene assay and ELISA, respectively, as described elsewhere.12,21,25 Final data were normalized to an internally established PBMCsec reference standard and are given as relative potency.
Statistical analyses
Data were collected at prospectively defined endpoints, and no outliers were excluded. Experiments were repeated using different donors and animals, respectively, in at least quadruplicates. For data analysis of tube formation assay and aortic ring sprouting assay, investigators were blinded to treatments. Data were statistically evaluated using GraphPad Prism software. Two groups were compared by Student’s t test or Mann-Whitney. Ordinary one-way ANOVA and multiple comparison post hoc tests with Dunnett’s, Sidak’s, or Tukey’s correction were calculated. Bar diagrams show arithmetic mean and standard deviation. Boxplot diagrams were generated using first and third quartiles as boxes and medians as bars. Whiskers indicate 10−90 percentiles.
Acknowledgments
We thank Dr. H.P.H. for his belief in this private-public partnership to augment patient health. We thank Prof. Dr. Ulrike Baranyi and Prof. Dr. Barbara Messner for their kind support in the cell culture experiments. The authors acknowledge the Core Facilities of the Medical University of Vienna, a member of VLSI. The datasets analyzed in the current study are available from the corresponding authors upon request. This research project was supported by the Vienna Business Agency (Vienna, Austria; grant “APOSEC to clinic” 2343727). The financial support of the Federal Ministry for Education, Science and Research (BMWFW) of Austria and the National Foundation for Research, Technology and Development, Austria to the Christian Doppler Laboratory for Biotechnology of Skin Aging is gratefully acknowledged.
Author contributions
M.L., A.G., M.S., D.G., M.M., and H.J.A. conceived and planned the experiments. M.L., A.G., D.C., M.J., M.S., A.P., C.K., S.M., M.E., and W.L. performed the experiments. M.L., A.G., D.C., M.S., C.K., M.F.-N., F.G., S.M., M.E., W.L., D.G., M.M., and H.J.A. contributed to analyses and interpretations of the results. M.L., A.G., M.M., and H.J.A. wrote the original manuscript. All authors reviewed and approved the final manuscript.
Declaration of interests
The Medical University of Vienna has claimed financial interest. H.J.A. holds patents related to this work (WO 2010/079086 A1, WO 2010/070105 A1, EP 3502692, European Patent Office application #19165340.1). All other authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2021.02.016.
Contributor Information
Michael Mildner, Email: michael.mildner@meduniwien.ac.at.
Hendrik Jan Ankersmit, Email: hendrik.ankersmit@meduniwien.ac.at.
Supplemental information
References
- 1.Luo J., Zhao S., Wang J., Luo L., Li E., Zhu Z., Liu Y., Kang R., Zhao Z. Bone marrow mesenchymal stem cells reduce ureteral stricture formation in a rat model via the paracrine effect of extracellular vesicles. J. Cell. Mol. Med. 2018;22:4449–4459. doi: 10.1111/jcmm.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Natallya F.R., Herwanto N., Prakoeswa C., Indramaya D.M., Rantam F.A. Effective healing of leprosy chronic plantar ulcers by application of human amniotic membrane stem cell secretome gel. Indian J. Dermatol. 2019;64:250. doi: 10.4103/ijd.IJD_6_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karpov A.A., Puzanov M.V., Ivkin D.Y., Krasnova M.V., Anikin N.A., Docshin P.M., Moiseeva O.M., Galagudza M.M. Non-inferiority of microencapsulated mesenchymal stem cells to free cells in cardiac repair after myocardial infarction: A rationale for using paracrine factor(s) instead of cells. Int. J. Exp. Pathol. 2019;100:102–113. doi: 10.1111/iep.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandes-Cunha G.M., Na K.S., Putra I., Lee H.J., Hull S., Cheng Y.C., Blanco I.J., Eslani M., Djalilian A.R., Myung D. Corneal wound healing effects of mesenchymal stem cell secretome delivered within a viscoelastic gel carrier. Stem Cells Transl. Med. 2019;8:478–489. doi: 10.1002/sctm.18-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korf-Klingebiel M., Kempf T., Sauer T., Brinkmann E., Fischer P., Meyer G.P., Ganser A., Drexler H., Wollert K.C. Bone marrow cells are a rich source of growth factors and cytokines: implications for cell therapy trials after myocardial infarction. Eur. Heart J. 2008;29:2851–2858. doi: 10.1093/eurheartj/ehn456. [DOI] [PubMed] [Google Scholar]
- 6.Thum T., Bauersachs J., Poole-Wilson P.A., Volk H.D., Anker S.D. The dying stem cell hypothesis: immune modulation as a novel mechanism for progenitor cell therapy in cardiac muscle. J. Am. Coll. Cardiol. 2005;46:1799–1802. doi: 10.1016/j.jacc.2005.07.053. [DOI] [PubMed] [Google Scholar]
- 7.Ankersmit H.J., Hoetzenecker K., Dietl W., Soleiman A., Horvat R., Wolfsberger M., Gerner C., Hacker S., Mildner M., Moser B. Irradiated cultured apoptotic peripheral blood mononuclear cells regenerate infarcted myocardium. Eur. J. Clin. Invest. 2009;39:445–456. doi: 10.1111/j.1365-2362.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- 8.Lichtenauer M., Mildner M., Hoetzenecker K., Zimmermann M., Podesser B.K., Sipos W., Berényi E., Dworschak M., Tschachler E., Gyöngyösi M., Ankersmit H.J. Secretome of apoptotic peripheral blood cells (APOSEC) confers cytoprotection to cardiomyocytes and inhibits tissue remodelling after acute myocardial infarction: a preclinical study. Basic Res. Cardiol. 2011;106:1283–1297. doi: 10.1007/s00395-011-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavo N., Zimmermann M., Pils D., Mildner M., Petrási Z., Petneházy Ö., Fuzik J., Jakab A., Gabriel C., Sipos W. Long-acting beneficial effect of percutaneously intramyocardially delivered secretome of apoptotic peripheral blood cells on porcine chronic ischemic left ventricular dysfunction. Biomaterials. 2014;35:3541–3550. doi: 10.1016/j.biomaterials.2013.12.071. [DOI] [PubMed] [Google Scholar]
- 10.Altmann P., Mildner M., Haider T., Traxler D., Beer L., Ristl R., Golabi B., Gabriel C., Leutmezer F., Ankersmit H.J. Secretomes of apoptotic mononuclear cells ameliorate neurological damage in rats with focal ischemia. F1000Res. 2014;3:131. doi: 10.12688/f1000research.4219.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haider T., Hoftberger R., Rüger B., Mildner M., Blumer R., Mitterbauer A., Buchacher T., Sherif C., Altmann P., Redl H. The secretome of apoptotic human peripheral blood mononuclear cells attenuates secondary damage following spinal cord injury in rats. Exp. Neurol. 2015;267:230–242. doi: 10.1016/j.expneurol.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Wagner T., Traxler D., Simader E., Beer L., Narzt M.S., Gruber F., Madlener S., Laggner M., Erb M., Vorstandlechner V. Different pro-angiogenic potential of γ-irradiated PBMC-derived secretome and its subfractions. Sci. Rep. 2018;8:18016. doi: 10.1038/s41598-018-36928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler J., Lukovic D., Mester-Tonczar J., Zlabinger K., Gugerell A., Pavo N., Jakab A., Szankai Z., Traxler D., Müller C. Quantitative hybrid cardiac [(18)F]FDG-PET-MRI images for assessment of cardiac repair by preconditioned cardiosphere-derived cells. Mol. Ther. Methods Clin. Dev. 2020;18:354–366. doi: 10.1016/j.omtm.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker S., Mittermayr R., Traxler D., Keibl C., Resch A., Salminger S., Leiss H., Hacker P., Gabriel C., Golabi B. The secretome of stressed peripheral blood mononuclear cells increases tissue survival in a rodent epigastric flap model. Bioeng. Transl. Med. 2020;6:e10186. doi: 10.1002/btm2.10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beer L., Zimmermann M., Mitterbauer A., Ellinger A., Gruber F., Narzt M.S., Zellner M., Gyöngyösi M., Madlener S., Simader E. Analysis of the secretome of apoptotic peripheral blood mononuclear cells: Impact of released proteins and exosomes for tissue regeneration. Sci. Rep. 2015;5:16662. doi: 10.1038/srep16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gugerell A., Sorgenfrey D., Laggner M., Raimann J., Peterbauer A., Bormann D., Suessner S., Gabriel C., Moser B., Ostler T. Viral safety of aposec: a novel peripheral blood mononuclear cell derived-biological for regenerative medicine. Blood Transfus. 2020;18:30–39. doi: 10.2450/2019.0249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker S., Mittermayr R., Nickl S., Haider T., Lebherz-Eichinger D., Beer L., Mitterbauer A., Leiss H., Zimmermann M., Schweiger T. Paracrine factors from irradiated peripheral blood mononuclear cells improve skin regeneration and angiogenesis in a porcine burn model. Sci. Rep. 2016;6:25168. doi: 10.1038/srep25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoetzenecker K., Assinger A., Lichtenauer M., Mildner M., Schweiger T., Starlinger P., Jakab A., Berényi E., Pavo N., Zimmermann M. Secretome of apoptotic peripheral blood cells (APOSEC) attenuates microvascular obstruction in a porcine closed chest reperfused acute myocardial infarction model: role of platelet aggregation and vasodilation. Basic Res. Cardiol. 2012;107:292. doi: 10.1007/s00395-012-0292-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasiri M.M., Beer L., Nemec L., Gruber F., Pietkiewicz S., Haider T., Simader E.M., Traxler D., Schweiger T., Janik S. Dying blood mononuclear cell secretome exerts antimicrobial activity. Eur. J. Clin. Invest. 2016;46:853–863. doi: 10.1111/eci.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laggner M., Copic D., Nemec L., Vorstandlechner V., Gugerell A., Gruber F., Peterbauer A., Ankersmit H.J., Mildner M. Therapeutic potential of lipids obtained from gamma-irradiated PBMCs in dendritic cell-mediated skin inflammation. EBioMedicine. 2020;55:102774. doi: 10.1016/j.ebiom.2020.102774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simader E., Beer L., Laggner M., Vorstandlechner V., Gugerell A., Erb M., Kalinina P., Copic D., Moser D., Spittler A. Tissue-regenerative potential of the secretome of γ-irradiated peripheral blood mononuclear cells is mediated via TNFRSF1B-induced necroptosis. Cell Death Dis. 2019;10:729. doi: 10.1038/s41419-019-1974-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simader E., Traxler D., Kasiri M.M., Hofbauer H., Wolzt M., Glogner C., Storka A., Mildner M., Gouya G., Geusau A. Safety and tolerability of topically administered autologous, apoptotic PBMC secretome (APOSEC) in dermal wounds: a randomized Phase 1 trial (MARSYAS I) Sci. Rep. 2017;7:6216. doi: 10.1038/s41598-017-06223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuschko S., Gugerell A., Chabicovsky M., Hofbauer H., Laggner M., Erb M., Ostler T., Peterbauer A., Suessner S., Demyanets S. Toxicological testing of allogeneic secretome derived from peripheral mononuclear cells (APOSEC): a novel cell-free therapeutic agent in skin disease. Sci. Rep. 2019;9:5598. doi: 10.1038/s41598-019-42057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beer L., Mildner M., Gyöngyösi M., Ankersmit H.J. Peripheral blood mononuclear cell secretome for tissue repair. Apoptosis. 2016;21:1336–1353. doi: 10.1007/s10495-016-1292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laggner M., Gugerell A., Bachmann C., Hofbauer H., Vorstandlechner V., Seibold M., Gouya Lechner G., Peterbauer A., Madlener S., Demyanets S. Reproducibility of GMP-compliant production of therapeutic stressed peripheral blood mononuclear cell-derived secretomes, a novel class of biological medicinal products. Stem Cell Res. Ther. 2020;11:9. doi: 10.1186/s13287-019-1524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janatpour K., Denning L., Nelson K., Betlach B., Mackenzie M., Holland P. Comparison of X-ray vs. gamma irradiation of CPDA-1 red cells. Vox Sang. 2005;89:215–219. doi: 10.1111/j.1423-0410.2005.00699.x. [DOI] [PubMed] [Google Scholar]
- 27.Frentzel K., Badakhshi H. Irradiation with x-rays of the energy 18 MV induces radioactivity in transfusion blood: Proposal of a safe method using 6 MV. Med. Phys. 2016;43:6517. doi: 10.1118/1.4967482. [DOI] [PubMed] [Google Scholar]
- 28.Gibbons J.P. 6th Edition. Wolters Kluwer Health; 2019. Khan’s The Physics of Radiation Therapy. [Google Scholar]
- 29.Andreassi M.G., Borghini A., Pulignani S., Baffigi F., Fulgentini L., Koester P., Cresci M., Vecoli C., Lamia D., Russo G. Radiobiological effectiveness of ultrashort laser-driven electron bunches: Micronucleus frequency, telomere shortening and cell viability. Radiat. Res. 2016;186:245–253. doi: 10.1667/RR14266.1. [DOI] [PubMed] [Google Scholar]
- 30.Babayan N., Grigoryan B., Khondkaryan L., Tadevosyan G., Sarkisyan N., Grigoryan R., Apresyan L., Aroutiounian R., Vorobyeva N., Pustovalova M. Laser-driven ultrashort pulsed electron beam radiation at doses of 0.5 and 1.0 gy induces apoptosis in human fibroblasts. Int. J. Mol. Sci. 2019;20:5140. doi: 10.3390/ijms20205140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks A.L. Chromosome damage in liver cells from low dose rate alpha, beta, and gamma irradiation: derivation of RBE. Science. 1975;190:1090–1092. doi: 10.1126/science.1188384. [DOI] [PubMed] [Google Scholar]
- 32.Elgqvist J., Timmermand O.V., Larsson E., Strand S.E. Radiosensitivity of Prostate Cancer Cell Lines for Irradiation from Beta Particle-emitting Radionuclide 177Lu Compared to Alpha Particles and Gamma Rays. Anticancer Res. 2016;36:103–109. [PubMed] [Google Scholar]
- 33.Park J.G., Yoon Y., Park J.N., Han I.J., Song B.S., Kim J.H., Kim W.G., Hwang H.J., Han S.B., Lee J.W. Effects of gamma irradiation and electron beam irradiation on quality, sensory, and bacterial populations in beef sausage patties. Meat Sci. 2010;85:368–372. doi: 10.1016/j.meatsci.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Turner H.C., Shuryak I., Weber W., Doyle-Eisele M., Melo D., Guilmette R., Amundson S.A., Brenner D.J. γ-H2AX Kinetic Profile in Mouse Lymphocytes Exposed to the Internal Emitters Cesium-137 and Strontium-90. PLoS ONE. 2015;10:e0143815. doi: 10.1371/journal.pone.0143815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panahipour L., Kochergina E., Laggner M., Zimmermann M., Mildner M., Ankersmit H.J., Gruber R. Role for lipids secreted by irradiated peripheral blood mononuclear cells in inflammatory resolution in vitro. Int. J. Mol. Sci. 2020;21:4694. doi: 10.3390/ijms21134694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blüml S., Kirchberger S., Bochkov V.N., Krönke G., Stuhlmeier K., Majdic O., Zlabinger G.J., Knapp W., Binder B.R., Stöckl J., Leitinger N. Oxidized phospholipids negatively regulate dendritic cell maturation induced by TLRs and CD40. J. Immunol. 2005;175:501–508. doi: 10.4049/jimmunol.175.1.501. [DOI] [PubMed] [Google Scholar]
- 37.Chiurchiù V., Leuti A., Dalli J., Jacobsson A., Battistini L., Maccarrone M., Serhan C.N. Proresolving lipid mediators resolvin D1, resolvin D2, and maresin 1 are critical in modulating T cell responses. Sci. Transl. Med. 2016;8:353ra111. doi: 10.1126/scitranslmed.aaf7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francos-Quijorna I., Santos-Nogueira E., Gronert K., Sullivan A.B., Kopp M.A., Brommer B., David S., Schwab J.M., López-Vales R. Maresin 1 promotes inflammatory resolution, neuroprotection, and functional neurological recovery after spinal cord injury. J. Neurosci. 2017;37:11731–11743. doi: 10.1523/JNEUROSCI.1395-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy B.D. Resolvin D1 and resolvin E1 promote the resolution of allergic airway inflammation via shared and distinct molecular counter-regulatory pathways. Front Immunol. 2012;3:390. doi: 10.3389/fimmu.2012.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalli J., Winkler J.W., Colas R.A., Arnardottir H., Cheng C.Y., Chiang N., Petasis N.A., Serhan C.N. Resolvin D3 and aspirin-triggered resolvin D3 are potent immunoresolvents. Chem. Biol. 2013;20:188–201. doi: 10.1016/j.chembiol.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherpokova D., Jouvene C.C., Libreros S., DeRoo E.P., Chu L., de la Rosa X., Norris P.C., Wagner D.D., Serhan C.N. Resolvin D4 attenuates the severity of pathological thrombosis in mice. Blood. 2019;134:1458–1468. doi: 10.1182/blood.2018886317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Jong O.G., Van Balkom B.W., Schiffelers R.M., Bouten C.V., Verhaar M.C. Extracellular vesicles: Potential roles in regenerative medicine. Front Immunol. 2014;5:608. doi: 10.3389/fimmu.2014.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T., Zhang Q., Zhang J., Li C., Miao Y.R., Lei Q., Li Q., Guo A.Y. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D89–D93. doi: 10.1093/nar/gky985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mildner M., Hacker S., Haider T., Gschwandtner M., Werba G., Barresi C., Zimmermann M., Golabi B., Tschachler E., Ankersmit H.J. Secretome of peripheral blood mononuclear cells enhances wound healing. PLoS ONE. 2013;8:e60103. doi: 10.1371/journal.pone.0060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeCicco-Skinner K.L., Henry G.H., Cataisson C., Tabib T., Gwilliam J.C., Watson N.J., Bullwinkle E.M., Falkenburg L., O’Neill R.C., Morin A. Endothelial cell tube formation assay for the in vitro study of angiogenesis. J. Vis. Exp. 2014;1:e51312. doi: 10.3791/51312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia J., Ye T., Cui P., Hua Q., Zeng H., Zhao D. Ap-1 transcription factor mediates VEGF-induced endothelial cell migration and proliferation. Microvasc. Res. 2016;105:103–108. doi: 10.1016/j.mvr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirano S., Rees R.S., Gilmont R.R. MAP kinase pathways involving hsp27 regulate fibroblast-mediated wound contraction. J. Surg. Res. 2002;102:77–84. doi: 10.1006/jsre.2001.6315. [DOI] [PubMed] [Google Scholar]
- 48.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A., Singh K., Almasan A. Histone h2ax phosphorylation: A marker for DNA damage. Methods Mol. Biol. 2012;920:613–626. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 50.Li F., Huang Q., Chen J., Peng Y., Roop D.R., Bedford J.S., Li C.Y. Apoptotic cells activate the “phoenix rising” pathway to promote wound healing and tissue regeneration. Sci. Signal. 2010;3:ra13. doi: 10.1126/scisignal.2000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abels E.R., Breakefield X.O. Introduction to extracellular vesicles: Biogenesis, rna cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gugerell A., Gouya-Lechner G., Hofbauer H., Laggner M., Trautinger F., Almer G., Peterbauer-Scherb A., Seibold M., Hoetzenecker W., Dreschl C. Safety and clinical efficacy of the secretome of stressed peripheral blood mononuclear cells in patients with diabetic foot ulcer-study protocol of the randomized, placebo-controlled, double-blind, multicenter, international phase II clinical trial MARSYAS II. Trials. 2021;22:10. doi: 10.1186/s13063-020-04948-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruber F., Bicker W., Oskolkova O.V., Tschachler E., Bochkov V.N. A simplified procedure for semi-targeted lipidomic analysis of oxidized phosphatidylcholines induced by UVA irradiation. J. Lipid Res. 2012;53:1232–1242. doi: 10.1194/jlr.D025270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Han X., Wan Y., Zhang S., Zhao Y., Fan R., Cui Q., Zhou Y. TAM 2.0: tool for MicroRNA set analysis. Nucleic Acids Res. 2018;46(W1):W180–W185. doi: 10.1093/nar/gky509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu M., Shi B., Wang J., Cao Q., Cui Q. TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreutmayer S.B., Messner B., Knoflach M., Henderson B., Niederegger H., Böck G., Van der Zee R., Wick G., Bernhard D. Dynamics of heat shock protein 60 in endothelial cells exposed to cigarette smoke extract. J. Mol. Cell. Cardiol. 2011;51:777–780. doi: 10.1016/j.yjmcc.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carpentier G. ImageJ contribution: angiogenesis analyzer. ImageJ News. 2012 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.