Abstract
Long non-coding RNAs (lncRNAs) have been shown to impact important biological functions such as proliferation, survival, and genomic stability. To analyze radiation-induced lncRNA expression in human tumors, we irradiated prostate cancer cells with a single dose of 10 Gy or a multifractionated radiotherapeutic regimen of 10 fractions with a dose of 1 Gy (10 × 1 Gy) during 5 days. We found a stable upregulation of the lncRNA LINC00261 and LINC00665 at 2 months after radiotherapy that was more pronounced after single-dose irradiation. Analysis of the The Cancer Genome Atlas (TCGA) and The Atlas of Non-coding RNAs in Cancer (TANRIC) databases showed that high expression of these two lncRNAs may be a potential negative prognostic marker for overall survival of prostate cancer patients. Knockdown of LINC00261 and LINC00665 in long-term survivors decreased survival after re-irradiation and affected DNA double-strand break repair. Mechanistically, both lncRNAs showed an interdependent expression and regulated expression of the DNA repair proteins CtIP (RBBP8) and XPC as well as the microRNA miR-329. Identifying long-term tumor adaptation mechanisms can lead to the discovery of new molecular targets, in effect opening up new research directions and improving multimodal radiation oncologic treatment.
Keywords: long non-coding RNA, targeted therapy, 3D cell culture, fractionated radiation, radiation-induced targets, radiation stress response, micro RNA, tumor adaptation, DNA repair, prostate cancer
Graphical Abstract
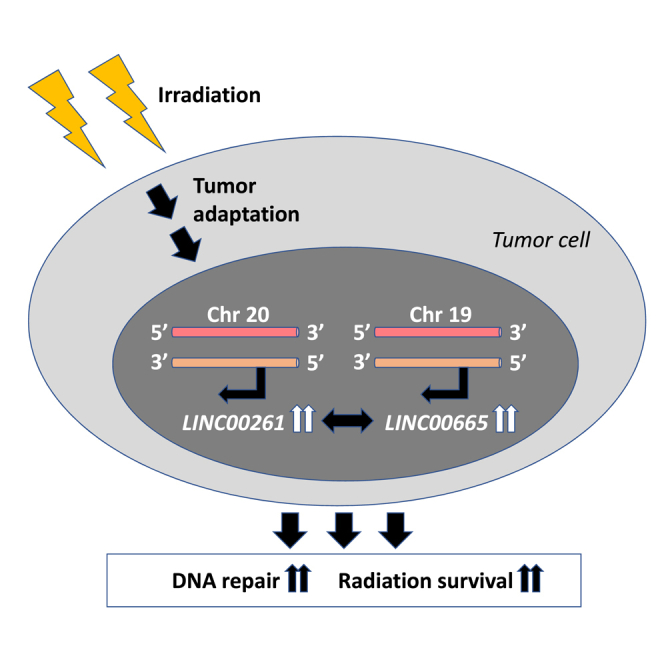
Irradiation can induce long-term effects on the expression of long non-coding RNAs in cancer cells, resulting in tumor adaptation, enhanced tumor cell survival, and radiation resistance, which can be targeted to increase radiosensitivity and treatment efficacy.
Introduction
Cancer cells can respond to stress such as radiotherapy or chemotherapy by activating survival-promoting pathways, a process called tumor adaptation. We previously reported that radiation-induced adaptation can occur through molecular changes in gene expression, protein phosphorylation, and DNA repair mechanisms, and may differ after single-dose (SD) versus clinically relevant multifractionated (MF) radiotherapy.1, 2, 3, 4, 5 In addition, some of the observed effects are stable and can be detected 2 months after irradiation or even longer.2 Targeting these adaptive responses in tumor cells is a promising strategy to decrease survival of cancer cells.1,2
During the last decade, extensive research and technical advances have uncovered that a large part of the genome is transcribed but only approximately 2% encodes for proteins.6,7 The non-coding RNA fraction is comprised of different subtypes, including microRNA (miRNA), small nucleolar RNA (snoRNA), circular RNA (circRNA), and long non-coding RNA (lncRNA). lncRNAs are defined as non-coding RNAs greater than 200 nt in length. They have been shown to impact several cellular functions by regulating gene expression not only of neighboring genes, but also of spatially distant genes by a variety of mechanisms, including epigenetic modifications,8 modulation of transcription,9 and interaction with miRNA.10,11
The potential clinical relevance of lncRNAs in human disease has recently emerged, but to date only limited knowledge about the exact mechanisms and the tissue specificity of these effects exists.12, 13, 14, 15 For example, the lncRNA HOTAIR has been found to promote breast cancer metastasis,8 while SChLAP1 (LINC00913) has an important role in tumor cell invasion of prostate cancer.16 Upregulated expression of LINC00665 was found to be a predictor of poor prognosis in lung adenocarcinoma.11 Additionally, LINC00665 impacts tumor proliferation by activating extracellular signal-regulated kinase (ERK) signaling and induces resistance to the tyrosine kinase inhibitor gefitinib in lung cancer.11,17
LINC00261 and its neighboring gene, the forkhead box transcription factor-2 (FOXA2), were described as putative tumor suppressors in lung cancer and pancreatic cancer;18, 19, 20 however, in colon cancer and esophageal cancer, FOXA2 expression correlates with advanced stage and metastatic burden, indicating that the role of molecules can differ between tissues.21, 22, 23
Survival of cancer cells after radiotherapy is highly dependent on DNA damage repair. Non-homologous end joining (NHEJ), which involves 53BP1-mediated inhibition of end resection, is the main repair pathway when cells are in the G1 phase of the cell cycle or when MF irradiation is applied.5,24 In the S/G2 phase, BRCA1 and CtIP promote end resection and facilitate error-free homologous recombination (HR).24 Loss of function in either pathway results in increased cellular radiosensitivity and decreased radiation survival. Therefore, targeting DNA repair processes is a promising approach in cancer therapy with multiple inhibitors already in clinical trials.25
The xeroderma pigmentosa-associated XPC gene is part of the nucleotide excision repair (NER) pathway which is activated by bulky DNA lesions that destabilize the DNA duplex such as cyclobutane pyrimidine dimers.26 Therefore, NER is key to repairing UV-induced DNA damage. XPC serves as damage recognition factor in this process.27 Additionally, XPC has been found to modulate NHEJ in cancer cells and thereby also affect the sensitivity to ionizing radiation.28
In this study, we show that the lncRNAs LINC00261 and LINC0065 were stably upregulated in prostate cancer cells 2 months after radiotherapy and affected radiation survival by regulating the expression of the DNA repair proteins CtIP and XPC. Furthermore, high expression of either lncRNA in malignant tissue was correlated with decreased overall survival in prostate cancer patients, indicating their potential as molecular therapeutic targets in radiation oncology.
Results
Radiation stably alters lncRNA expression in a fractionation-dependent manner
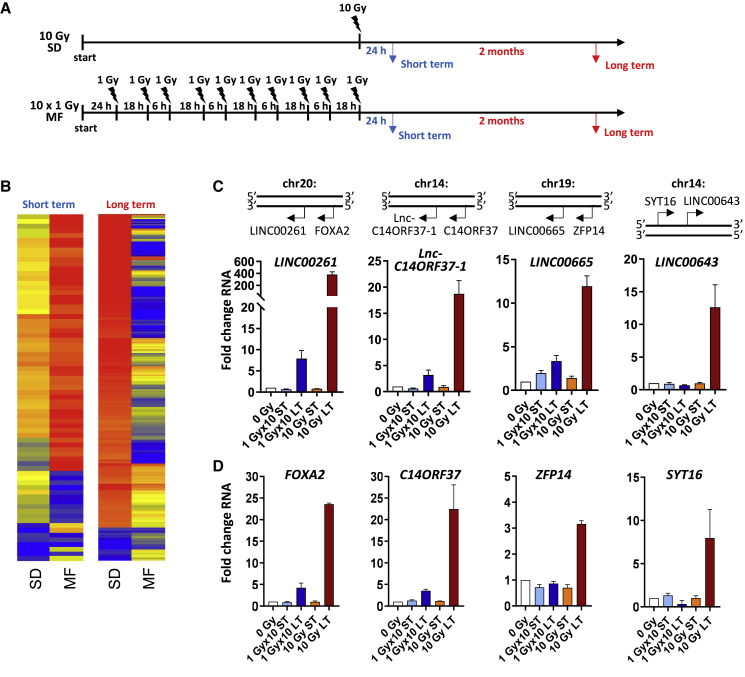
It is widely known that ionizing radiation strongly impacts gene expression. Previously, we have found that the fractionation of radiotherapy and the time point when gene expression was assessed affect the adaptive effects. Therefore, to analyze radiation-induced lncRNA expression, we irradiated prostate cancer cells with either an SD of 10 Gy of X-ray or an MF radiotherapy consisting of 10 fractions of 1 Gy delivered during 5 days (Figure 1A). RNA expression was evaluated at 24 h (short term) or at 2 months (long term) after the final radiation dose (Figure 1A). As shown in Figures 1B and 1C, lncRNA expression varied widely depending on the fractionation regimen, with a significant upregulation of multiple lncRNAs in the long-term cultures after a 10-Gy SD irradiation (Figures 1B and 1C; Figure S1). In parallel, the expression of the corresponding neighboring genes was also increased (Figure 1D; Figure S1). Similar to PC3 cells, we found a significant but less pronounced upregulation of LINC00261 and LINC00665 in LNCaP prostate cancer cells at 2 months after an SD of 10 Gy but not after MF irradiation (Figure S2).
Figure 1.
Radiotherapy leads to stable upregulation of long non-coding RNA in prostate cancer cells
(A) Schematic of treatment for single-dose (SD) and multifractionated (MF) irradiation. Short-term (ST) expression analysis was performed at 24 h after the final fraction of radiotherapy. For long-term analysis, cells were cultured for 2 months before RNA extraction was performed. (B) Heatmap of long non-coding RNA (lncRNA) expression in PC3 prostate cancer cells at 24 h (short-term) or at 2 months (long-term) after an SD irradiation of 10 Gy or MF radiotherapy of 10 times 1 Gy. (C and D) Genetic location and fold change in RNA expression of indicated lncRNA (C) and neighboring genes (D). Unirradiated cell cultures were used as control. Results show mean ± SD (n = 3). See also Figures S1 and S2.
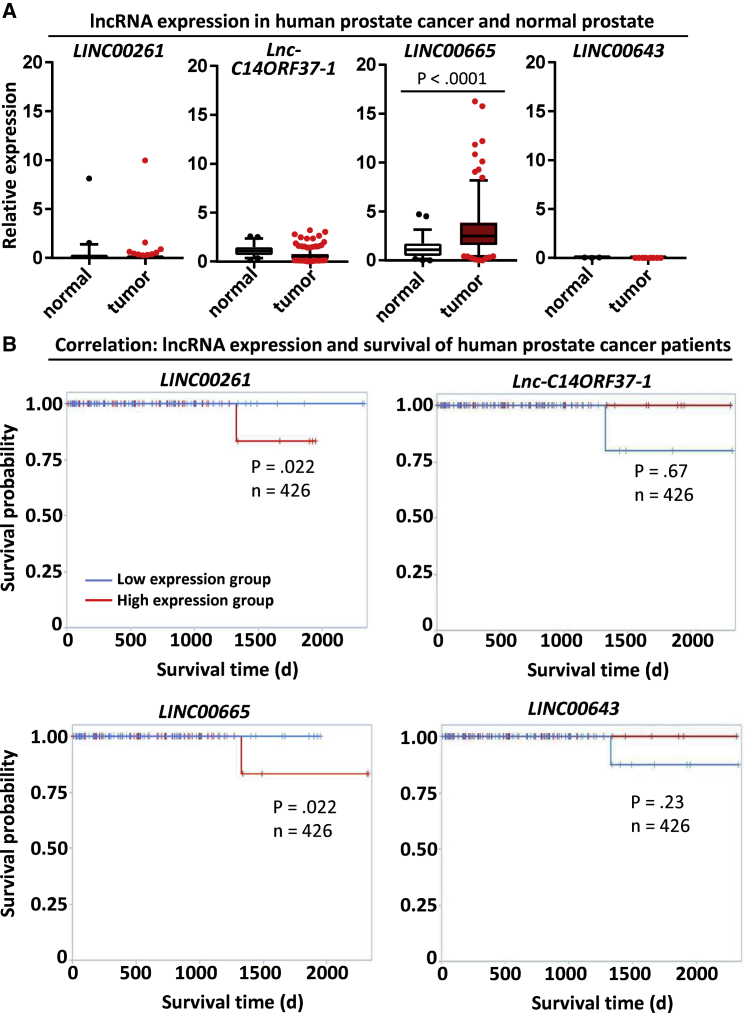
Prostate cancer patients with a high tumor expression of LINC00261 and LINC00665 tend to show reduced overall survival
Next, we analyzed the impact of the expression of upregulated lncRNA in patient samples using The Cancer Genome Atlas (TCGA) and The Atlas of Non-coding RNAs in Cancer (TANRIC) databases. Overall, LINC00261, LINC00643, and lnc-C14ORF37-1 levels were very low in both normal and malignant prostate tissue (Figure 2A). In contrast, LINC00665 was significantly overexpressed in prostate cancer (Figure 2A). Interestingly, there was a strong variation in lncRNA expression between different tissues, with LINC00261 levels being highest in normal lung and stomach tissue while LINC00665 was most strongly expressed in prostate and ovarian cancer (Figure S3). Furthermore, high expression of LINC00261 or LINC00665 was a negative prognostic marker for prostate cancer patients, with overall survival being statistically significantly reduced in comparison to the low expression group (Figure 2B). LINC00261 expression was also associated with decreased survival of patients with head and neck tumors and renal carcinoma, but it did not affect the outcome of patients with invasive breast cancer, squamous cell carcinoma of the lung, stomach cancer, or hepatocellular carcinoma (Figure S4). These data show that the role of LINC00261 seems to be tissue-specific.
Figure 2.
LINC00261 and LINC00665 expression levels negatively correlate with overall survival of prostate cancer patients
(A) lncRNA expression in normal human prostate and human prostate cancer tissue. Data were obtained from The Atlas of Non-coding RNAs in Cancer (TANRIC) database. (B) Kaplan-Meier curve for overall survival of patients with high and low expression of LINC00261, LNC-C14ORF37-1, LINC00665, and LINC00643. Data were obtained through the TANRIC database. Statistical significance was calculated using the univariate Cox proportional hazards model. See also Figures S3 and S4.
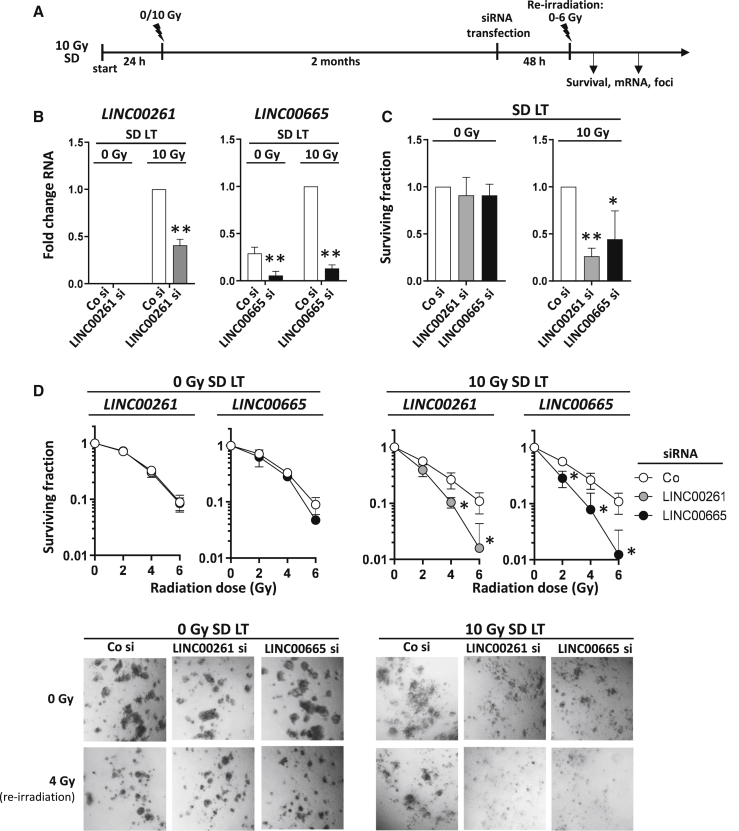
Depletion of LINC00261 or LINC00665 increases radiosensitivity of prostate cancer cells
To evaluate whether the upregulation of LINC00261 or LINC00665 in the long-term SD cells impacted survival, we performed small interfering RNA (siRNA) knockdown and re-irradiated the cells with different doses (Figure 3A). We used a 3D cell culture model for the experiments, since we found that this method more reflects the physiological conditions in vivo.29,30 Although the siRNA-mediated knockdown efficiently worked for both lncRNAs, LINC00261 expression was very low in the unirradiated (0 Gy) control cells and therefore no significant downregulation could be detected (Figure 3B). Depletion of LINC00261 or LINC00665 resulted in reduced survival of long-term 10-Gy SD cells but had no effect on 0-Gy cell cultures (Figure 3C). Additionally, radiosensitivity was significantly increased in the re-irradiated 10-Gy SD cells when LINC00261 or LINC00665 was downregulated (Figure 3D). Interestingly, the expression of FOXA2, the neighboring gene of LINC00261, was not altered by LINC00261 knockdown (Figure S5A). Similarly, modulation of FOXA2 expression had no effect on LINC00261 levels or survival of both 0-Gy and 10-Gy SD cultures (Figures S5A–S5C).
Figure 3.
Depletion of upregulated LINC00261 and LINC00665 reduces radiation survival of prostate cancer cells
(A) Schematic of the treatment schedule for establishing long-term cultures, transfecting with siRNA, and subsequent re-irradiation. (B) lncRNA expression in PC3 cells at 48 h after siRNA transfection measured with qPCR. Samples were normalized to the control siRNA (Co si) of the PC3 10-Gy SD long-term (LT) cells (because LINC00261 expression in PC3 0-Gy SD LT cells was too low for normalization). (C and D) Basal clonogenic survival (C) and radiation survival (D) of PC3 0-Gy SD. LT and PC3 10-Gy SD. LT cells after siRNA-mediated knockdown of LINC00261 and LINC00665. Co si was used as control. Results show mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01, Student’s t test. See also Figures S5A–S5C.
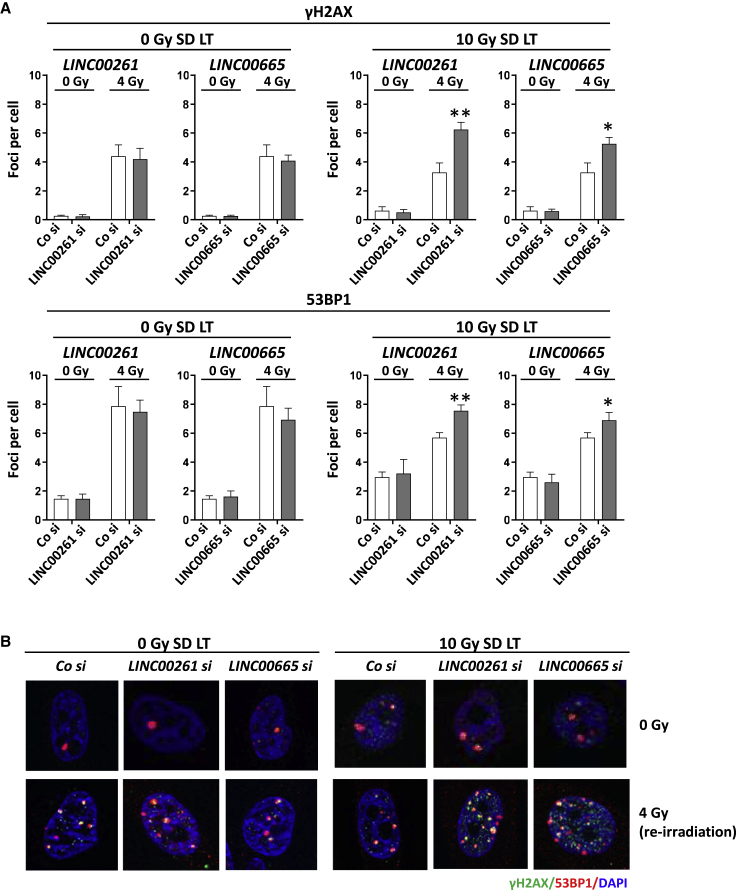
LINC00261 and LINC00665 affect radiation-induced residual DNA damage
DNA double-strand breaks (DSBs) are one of the most severe forms of DNA damage. To evaluate whether the observed effect of LINC00261 or LINC00665 knockdown on radiation survival was due to impaired DNA repair, we analyzed the number of residual DNA DSBs in the absence of LINC00261 or LINC00665. While the number of γH2AX and 53BP1 foci in PC3 0-Gy long-term cultures were unaffected by the loss of LINC00261 or LINC00665, we observed a significant increase in residual foci in the irradiated 10-Gy SD cells, indicating that LINC00261 and LINC00665 modulate DNA damage repair mechanisms (Figures 4A and 4B). Analysis of γH2AX and 53BP1 foci revealed that the slow phase of DNA repair is more compromised by LINC00261 and LINC00665 depletion than the rapid repair phase that takes place in the first 6 h after irradiation (Figure S5D).
Figure 4.
LINC00261 and LINC00665 knockdown increases the number of radiation-induced residual double-strand breaks
(A and B) Number of γH2AX and 53BP1 foci (A) and representative images (B). At 2 months after an SD of 10 Gy, cells were transfected with LINC00261 and LINC00665 siRNA. Co si and unirradiated cells (0 Gy) were used as controls. At 48 h after transfection, cells were re-irradiated with 0 and 4 Gy. Samples were fixed and stained at 24 h after irradiation. Results show mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01, Student’s t test. See also Figure S5D.
Downregulation of LINC00261 or LINC00665 affects HR and NER
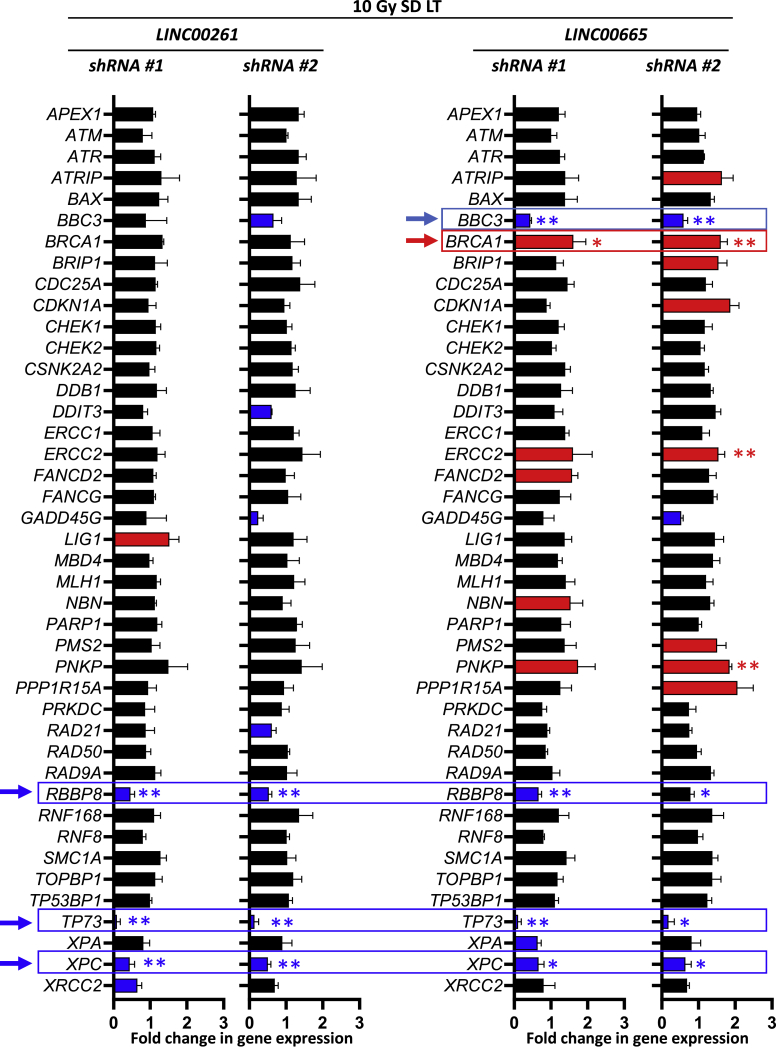
To look more in depth about how LINC00261 and LINC00665 affect the repair of DNA DSBs, we examined the gene expression of a panel of DNA damage repair genes after lncRNA depletion. To get a stable knockdown, short hairpin RNA (shRNA) vectors with two different target sequences were used. A non-targeting sequence served as the control. As shown in Figure 5, depletion of LINC00261 or LINC00665 led to a significant reduction of RBBP8, XPC, and TP73 genes. Additionally, LINC00665 modulated BBC3 and BRCA1 gene expression. BRCA1 and RBBP8, which encodes for the DNA repair protein CtIP, are part of the HR pathway, while XPC is involved in the NER. TP73 and BBC3 are associated with cell apoptosis. Interestingly, RBBP8 was upregulated at 2 months after SD irradiation compared to the unirradiated control (Figure S6A). Furthermore, silencing of LINC00261 or LINC00665 decreased and delayed radiation-induced RAD51 formation, indicating an important role for HR of these lncRNAs (Figure S6B). These results demonstrate that the lncRNAs LINC00261 and LINC00665 modulate DNA repair pathways.
Figure 5.
LINC00261 and LINC00665 depletion alters the expression of DNA damage repair genes
At 2 months (LT) after an SD of 10 Gy, PC3 cells were infected with shRNA-containing virus directed against LINC00261 or LINC00665. To exclude off-target effects, for every lncRNA, two different shRNA sequences were used. A non-targeting sequence was used as control for normalization. Gene expression of DNA damage repair-related molecules was analyzed with the RT2 Profiler PCR array human DNA repair pathway-focused array. Results show mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01, Student’s t test. See also Figure S6.
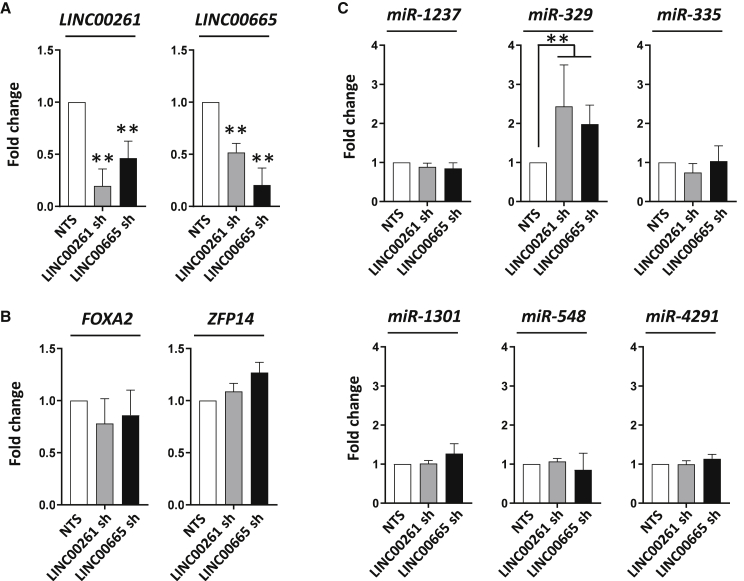
Loss of LINC00261 or LINC00665 increases the expression of miR-329
Next, we analyzed which molecules were involved in mediating the observed lncRNA-associated RBBP8 and XPC gene regulation. Interestingly, we saw an interdependent regulation of LINC00261 and LINC00665, while the expression of the neighboring genes FOXA2 and ZFP14 was not changed after lncRNA knockdown (Figures 6A and 6B). As lncRNA has been reported to impact gene expression by modulating miRNA, we evaluated the expression of different miRNAs that are predicted to interact with RBBP8 or XPC genes as well as with LINC00261 (Figure 6C). We found that the expression of miR-329 was significantly increased after depletion of LINC00261 and LINC00665, indicating a potential mechanism to regulate XPC gene expression. Furthermore, miR-329 was downregulated at 2 months after 10-Gy SD irradiation (Figure S7).
Figure 6.
Loss of LINC00261 and LINC00665 results in upregulation of miR-329
(A and B) LINC00261 and LINC00665 (A) as well as FOXA2 and ZFP14 (B) expression after an shRNA (sh-)mediated knockdown of LINC00261 or LINC00665. A vector with a non-targeting sequence (NTS) was used as control. (C) Expression changes in microRNA predicted to regulate RBBP8 and XPC genes after depletion of LINC00261 or LINC00665. Results show mean ± SD (n = 3). ∗∗p < 0.01, Student’s t test. See also Figure S7.
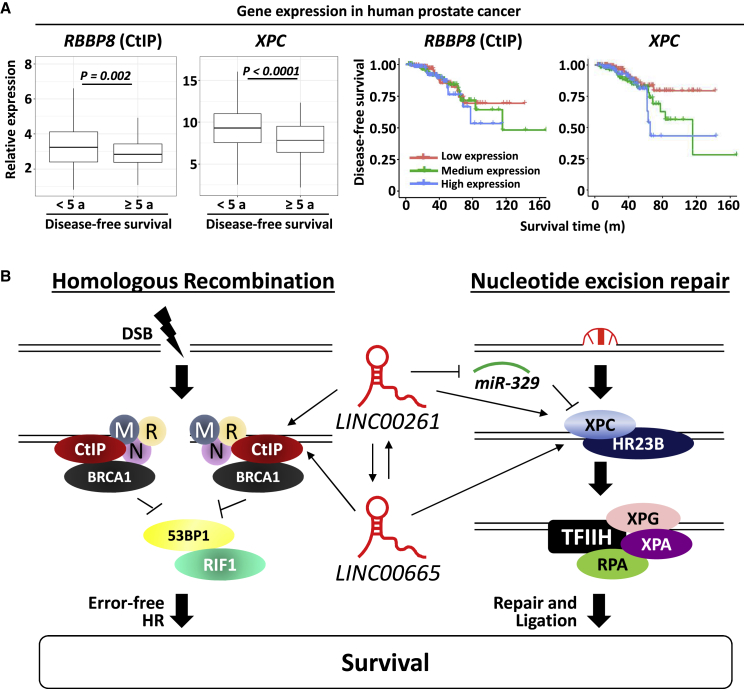
Low expression of RBBP8 or XPC is a positive prognostic marker for prostate cancer patients
To evaluate whether the tumor expression of RBBP8 or XPC correlates with patient survival, we analyzed TCGA data of prostate cancer patients. As shown in Figure 7A, low expression of both RBBP8 and XPC were associated with longer disease-free survival. Patients with disease-free survival shorter than 5 years had a significant higher tumor expression of RBBP8 or XPC than did patients with disease-free survival of more than 5 years (Figure 7A). Accordingly, patients with low tumor expression of RBBP8 or XPC had an increased disease-free survival (Figure 7A). Overall, these data indicate that inhibition of the radiation-induced upregulation of LINC00261 and LINC00665 and therefore downregulation of RBBP8 and XPC genes may be a promising strategy for treatment of prostate cancer (Figure 7B).
Figure 7.
RBBP8 and XPC are associated with decreased disease-free survival in human prostate cancer
(A) Gene expression of RBBP8 and XPC in 337 prostate cancer samples. Gene expression in patients with disease-free survival longer than 5 years and shorter than 5 years is shown as boxplots. Kaplan-Meier curves for disease-free survival of the low expression group (lower third), medium expression group (middle third), and high expression group (upper third). Expression data were obtained from TCGA database and KM-express analysis tool. For statistical analysis, the Student’s t test was used. (B) Schematic of how LINC00261 and LINC0065 regulate DNA repair and survival.
Discussion
Tumor adaptation is a phenomenon that can hamper tumor eradication and successful treatment. By inducing pro-survival processes during or after cancer therapy, tumor cells may be able to escape death, resulting in incomplete remission and disease relapse. Analyzing these adaptive changes and understanding the molecular mechanisms leading to tumor cell survival may aid in finding molecular-targeted treatment strategies to both prevent and address tumor recurrence. We previously found great differences in tumor adaptation such as gene expression and protein phosphorylation depending on the fractionation regimen of irradiation and on the time point after therapy.1,2 In this study, we show that this is also the case for lncRNA expression, with the strongest upregulation of multiple lncRNAs including LINC00261 and LINC00665 at 2 months after an SD of irradiation. Induction of lncRNA after DNA damage has been described before. Sharma et al.31 observed an increase of the lncRNA DDSR1 after treatment with DNA-damaging agents such as camptothecin or etoposide of skin fibroblasts and tumor cells as part of the cellular DNA damage response. Similarly, lncRNA ANRIL and GAS5 were upregulated at 24 h after gamma radiation in HeLa and MCF7 cells.32 However, most studies focused on the immediate changes after genotoxic stress and did not examine long-term effects of DNA damage on lncRNA expression.
Interestingly, there was a trend that high expression of both LINC00261 and LINC00665 correlated with lower survival of prostate cancer patients. However, since prostate cancer has a relatively good prognosis and high 5-year survival rates, longer observation periods are necessary to determine whether increased expression of these lncRNAs is strongly associated with poor survival. While LINC00665 is already known to be a negative prognostic marker for multiple tumor types,17,33,34 the function of LINC00261 seems to be highly tissue-dependent. In lung adenocarcinoma, LINC00261 is reported to be a tumor suppressor. LINC00261 is significantly lower expressed in non-small cell lung cancer (NSCLC) than in normal lung tissue.20,35 Upregulation of LINC00261 expression in NSCLC cells diminishes cell migration and induces cell cycle and growth arrest.20 In contrast, LINC00261 overexpression in cholangiocarcinoma (CCA) is associated with poor prognosis and tumor progression.36 Furthermore, knockdown of LINC00261 in CCA cells attenuates proliferation, migration, and tumor invasion by promoting epithelial-mesenchymal transition (EMT).36 These data show that lncRNA can have either a tumor-suppressing or a tumor-promoting effect depending on the cancer site.
Besides the previously mentioned functions, lncRNA can modulate miRNA expression by acting as a sponge to decrease miRNA levels and function.37 In line with this, we saw in parallel to increased LINC00261 and LINC00665 levels also a downregulation of miR-329 at 2 months after irradiation. Silencing of LINC00261 and LINC00665 resulted in increased miR-329 expression, indicating an inhibitory interaction. Similar effects on upregulation of miR-329 expression have been reported after knockdown of other lncRNAs such as tumor protein p73 antisense RNA 1 (TP73-AS1) or snoRNA host gene 7 (SNHG7).38,39 miR-329 is described as a tumor suppressor in several cancer types, including glioblastoma and lung cancer.39, 40, 41, 42 In cervical cancer cells, an lncRNA-mediated increase of miR-329 levels attenuates tumor proliferation and migration.43 Additionally, miR-329 overexpression correlates with sensitivity to chemotherapy in neuroblastoma and colorectal cancer, and therefore targeting lncRNA that inhibit miR-329 to increase miR-329 expression may be a potential therapeutic approach for resistant tumors.39,40
Recently, the crucial role of lncRNA in the cellular DNA damage response has emerged. Besides serving as interaction platforms for DNA repair factors at the DNA damage site and facilitating DNA focus formation,31,44 lncRNAs can also regulate radiation survival and radiosensitivity by affecting gene expression of DNA repair factors similar to the effects we have seen for LINC00261 and LINC00665. CtIP (RBBP8) is a key molecule of the HR repair pathway, while XPC is involved in the NER pathway and in NHEJ repair.24,28 We found a correlation between high expression of both molecules with reduced disease-free survival for patients suffering from prostate cancer. Although CtIP and XPC are part of the DNA repair machinery and therefore involved in preserving genome integrity by preventing DNA mutations and tumorigenesis,45,46 overexpression in cancer has been shown to be associated with poor outcome and therapy resistance in some tumor types. A high level of CtIP is a negative prognostic marker for plasma cell myeloma and gastric cancer.47,48 Furthermore, targeting of XPC in colorectal cancer increases the sensitivity to cisplatin, whereas upregulation leads to cisplatin resistance.49 Similar results have been obtained in lung carcinoma cells.50 In addition to decreased CtIP expression, RAD51 foci formation, a key step in the homologous repair pathway, was also reduced in LINC00261- and LINC00665-depleted cells, further corroborating the role of these lncRNAs for DNA repair.51
In this study we show that the radiation-induced adaptive response of tumors is not limited to changes in gene expression and protein phosphorylation but also includes modulation of non-coding RNAs. The molecular mechanisms that lead to cancer cell survival after radiation or chemotherapy are complex and still widely unknown since they are dependent on multiple factors such as tissue type, treatment regimen, and time after therapy. Targeting adaptive tumor changes is a unique and novel approach to improve cancer treatment efficacy demonstrated here for lncRNAs as an example for prostate cancer patients.
Materials and methods
Cell culture and radiation exposure
PC3 cells were obtained from the NCI tumor bank in 2015 and used up to a passage number of 15. For the experiments, asynchronously and exponentially growing cells were cultured in RPMI 1640 containing GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen). Testing for mycoplasma was performed on a monthly base. Cells were incubated at 37°C and 5% CO2.
Irradiation was performed at room temperature using SDs or multiple fractions of 320-kV X-rays with a dose rate of 2.3 Gy/min (Precision X-Ray). MF radiation was carried out as described before with two times 1 Gy per day (with a 6-h time interval between both radiations).1 For long-term PC3 cultures, irradiated and unirradiated cells were passaged twice a week and cultured for at least 8 weeks after radiation before the cells were plated for experiments.
3D colony formation assay
3D cell culture models have been shown to mimic the conditions in vivo and better predict the effects of molecular targeting on radiation survival than conventional 2D cell culture models.29,30 Therefore, we used 3D colony formation assays to assess the effect of lncRNA depletion on cell survival. 3D colony formation assays were performed as previously described.52,53 To prevent adhesion of cells to the bottom of the plates, 96-well plates were coated with 1% agarose (Sigma). Single cells were embedded in Matrigel (Corning Life Sciences) with a concentration of 0.5 mg/mL. After 8 days, images of colonies were obtained using an EVOS microscope with a ×2.5 objective. An ImageJ cell counter was used to count cell clusters with more than 50 cells. Surviving fractions were calculated as follows: (treated colony number/untreated colony number).52
Whole-genome gene expression analysis
Prostate cancer cells were irradiated with a single radiation dose of 10 Gy or MF radiation with 10 fractions of 1 Gy (two fractions per day) with a cumulative dose of 10 Gy as published. Total RNA was extracted at 24 h and 2 months after irradiation from three replicates using a QIAshredder spin column (catalog no. 79654, QIAGEN) as previously published.2 Unirradiated cells were used as controls. All extracted RNAs were purified with an RNeasy mini kit (QIAGEN). The microarray analysis was done using CodeLink Whole Genome Bioarrays representing 55,000 probes. CodeLink Expression Analysis software (GE Healthcare) was used to process the scanned images from arrays (gridding and feature intensity), and the data generated for each feature on the array were analyzed with GeneSpring software (Agilent Technologies). Raw intensity data for each gene on every array were normalized to the median intensity of the raw values from that array.
Stable shRNA-mediated knockdown of lncRNA
Stable knockdown of lncRNA was performed as previously published.1 293T cells were transfected with shRNA-containing plasmids (Dharmacon), and the psPAX2 and pMD2.VSVG packaging constructs were transfected using Lipofectamine 2000 (Invitrogen). A non-targeting sequence vector was used as the control. The virus-containing media were harvested and filtered through a 0.45-μm polyvinylidene fluoride (PVDF) syringe filter (Millipore). PC3 cells were transduced with the virus-containing media and 8 μg/mL Polybrene (Sigma). The selection was performed with puromycin (InvivoGen). Knockdown of lncRNA expression was confirmed by qPCR. For each lncRNA, two different shRNA sequences were used.
siRNA transfection
At 24 h after cell plating, cells were incubated with a pool of four siRNA duplexes (SMARTpool) targeting LINC00261 or LINC00665 RNA expression and DharmaFECT 4 transfection reagent (Dharmacon) according to the manufacturer’s protocol and as previously described.54 A nontargeted siRNA pool (Dharmacon) was used as the control. At 24 h after transfection, cell cultures were plated for colony formation assays, foci analysis, or quantitative real-time PCR. Irradiation or RNA extraction was carried out at 48 h after transfection (time point of maximal knockdown).
Real-time PCR
For quantitative real-time PCR, 1 μg of total RNA was reverse transcribed using RT2 first-strand synthesis kits (QIAGEN, 330401) as previously published.2 RT2 SYBR Green ROX qPCR master mix (QIAGEN, 330520) and RT2 qPCR primer assays (QIAGEN, 330001) were used for expression analysis of LINC00261, LINC00665, FOXA2, and ZFP14. Expression values were normalized to the housekeeping gene GAPDH. For quantitative real-time PCR reactions, an Applied Biosystems thermal cycler (QuantStudio 3) was used. After a holding stage at 95°C for 15 min, 40 cycles of alternate denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min were performed. A melt curve analysis was performed to ensure the specificity of the corresponding real-time PCR reactions. Fold change was calculated as 2−ΔΔCt, where ΔΔCt = ΔCt (test) − ΔCt (control), ΔCt = Ct (gene) – Ct (GAPDH), and Ct is the threshold cycle number. All assays were performed in triplicates. Statistical significance was calculated using an unpaired Student’s t test.
Focus array
Extracted RNA was subjected to complementary cDNA synthesis using RT2 first-strand cDNA synthesis (QIAGEN, 330404) according to the manufacturer’s instructions and as previously published.1 Quantitative real-time PCR was performed using RT2 SYBR Green qPCR master mix. The reaction (25 μL) along with cDNA was aliquoted into the wells of an RT2 Profiler PCR array (human DNA damage signaling pathway; QIAGEN, 330231, PAHS-029ZA), which contains 84 genes involved in DNA damage signaling pathway pre-dispensed, laboratory-verified, specific primer pairs. This enabled us to analyze expression of a focused panel of genes related to the ATR/ATM signaling network and transcriptional targets of DNA damage response. Data were analyzed using the RT2 Profiler array analysis software version 3.5 (SABiosciences). Relative gene expression was calculated by normalization to the arithmetic mean of five housekeeping genes (ACTB, B2M, GAPDH, HPRT1, RPLP0).
miRNA expression analysis
To identify potential miRNA that could function as a link between the lncRNAs LINC00261 and LINC00665 and the genes RBBP8 and XPC, we used DIANA Tools v2,55 the miRWalk database (http://mirwalk.umm.uni-heidelberg.de/),56 and the miRDB database (http://www.mirdb.org/).57 200 ng of total RNA was reverse transcribed into cDNA with miScript II RT kit (QIAGEN, 218160) to preferentially convert mature miRNAs into cDNA according to the manufacturer’s protocol. miScript primer assays (QIAGEN, 218300) were subsequently used to detect differential expression in irradiated versus control samples. Quantitative real-time PCR reactions were performed in an Applied Biosystems thermal cycler (ABI 7500). PCR steps included the holding stage at 95°C for 10 min followed by 40 cycles of alternate denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 70°C for 35 s. A melt curve analysis was performed to ensure the specificity of the corresponding real-time PCR reactions. snoRNA U6 was used as the normalizing control. All assays were performed in triplicate. Statistical significance was calculated using a Student’s unpaired t test.
Foci analysis
For analysis of residual DNA DSBs, immunofluorescence staining of γH2AX and 53BP1 was performed.5 At 24 h after plating, cells were re-irradiated with an SD of 4 Gy (X-ray) and incubated for 24 h at 37°C, 5% CO2. Fixation was accomplished using 3% formaldehyde/PBS for 15 min. After permeabilization with 0.25% Triton X-100/PBS for 10 min, samples were blocked with 3% BSA/PBS for 30 min. Staining of γH2AX and 53BP1 was carried out with specific antibodies overnight at 4°C and with secondary antibodies for 1 h at room temperature. Cell nuclei were stained with Vectashield/DAPI mounting medium (Alexis). Images were acquired using an AxioImager.Z1/ApoTome microscope (Zeiss).
lncRNA/gene expression analysis in patient samples
For lncRNA expression analysis, the interactive platforms TANRIC (https://www.tanric.org) and MiTranscriptome (http://mitranscriptome.org) were used.58,59 lncRNA expression data were analyzed in prostate cancer and normal prostate samples from patients included in TCGA database. For survival analysis, patient survival was analyzed in patients with a low and high expression of the indicated lncRNA. For gene expression analysis of RBBP8 and XPC, TCGA data were assessed using the online analysis tool KM-express.60 Normalized TCGA mRNA expression was evaluated for prostate cancer patients with a disease-free survival of exactly or more than 5 years and compared to patients with a disease-free survival of less than 5 years. Additionally, patients were grouped in a high expression group (upper third), a medium expression group (middle third), and a low expression group (lower third), and disease-free survival of these groups was depicted with a Kaplan-Meier curve. Significant differences in gene expression were calculated with a two-sided Student’s t test.
Data analysis
Analysis of data was performed with Microsoft Excel 2013 or R software as published.1 Fold change was calculated by normalizing the measured values to the corresponding control. Statistical significance was tested with an unpaired, two-sided Student’s t test. Results were considered statistically significant when a p value of less than 0.05 was reached.
Acknowledgments
This work was supported by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (grant ZIA BC 010670 to C.N.C.) and by the NIH Ruth L. Kirschstein National Research Service Award (NRSA) program (grant T32CA121940 to I.E.). The authors thank Jared M. May (NCI/NIH) for excellent technical assistance.
Author contributions
Conceptualization, I.E., M.J.A., and C.N.C.; methodology, I.E., M.J.A., and V.S.; investigation, I.E., M.J.A., M.A.B., S.M., and V.S.; writing – original draft, I.E.; writing – review & editing, I.E., M.J.A., M.A.B., S.M., V.S., S.C., E.E.G., and C.N.C.; funding acquisition, E.E.G. and C.N.C.; supervision, I.E., M.J.A., and C.N.C.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.02.024.
Supplemental information
References
- 1.Eke I., Makinde A.Y., Aryankalayil M.J., Sandfort V., Palayoor S.T., Rath B.H., Liotta L., Pierobon M., Petricoin E.F., Brown M.F. Exploiting radiation-induced signaling to increase the susceptibility of resistant cancer cells to targeted drugs: AKT and mTOR inhibitors as an example. Mol. Cancer Ther. 2018;17:355–367. doi: 10.1158/1535-7163.MCT-17-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eke I., Makinde A.Y., Aryankalayil M.J., Reedy J.L., Citrin D.E., Chopra S., Ahmed M.M., Coleman C.N. Long-term tumor adaptation after radiotherapy: therapeutic implications for targeting integrins in prostate cancer. Mol. Cancer Res. 2018;16:1855–1864. doi: 10.1158/1541-7786.MCR-18-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.John-Aryankalayil M., Palayoor S.T., Cerna D., Simone C.B., 2nd, Falduto M.T., Magnuson S.R., Coleman C.N. Fractionated radiation therapy can induce a molecular profile for therapeutic targeting. Radiat. Res. 2010;174:446–458. doi: 10.1667/RR2105.1. [DOI] [PubMed] [Google Scholar]
- 4.Palayoor S.T., John-Aryankalayil M., Makinde A.Y., Falduto M.T., Magnuson S.R., Coleman C.N. Differential expression of stress and immune response pathway transcripts and miRNAs in normal human endothelial cells subjected to fractionated or single-dose radiation. Mol. Cancer Res. 2014;12:1002–1015. doi: 10.1158/1541-7786.MCR-13-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eke I., Zong D., Aryankalayil M.J., Sandfort V., Bylicky M.A., Rath B.H., Graves E.E., Nussenzweig A., Coleman C.N. 53BP1/RIF1 signaling promotes cell survival after multifractionated radiotherapy. Nucleic Acids Res. 2020;48:1314–1326. doi: 10.1093/nar/gkz1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander R.P., Fang G., Rozowsky J., Snyder M., Gerstein M.B. Annotating non-coding regions of the genome. Nat. Rev. Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 7.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallen A.N., Zhou X.-B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong Z., Diao Y., Xu Y., Li X., Jiang Z., Shao C., Ji S., Shen Y., De W., Qiang Y. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 2019;10:84. doi: 10.1038/s41419-019-1361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 14.Chi Y., Wang D., Wang J., Yu W., Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8:E1015. doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossi E., Raimondi I., Goñi E., González J., Marchese F.P., Chapaprieta V., Martín-Subero J.I., Guo S., Huarte M. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 2020;11:936. doi: 10.1038/s41467-020-14623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Lu X., Zhen F., Jin S., Yu T., Zhu Q., Wang W., Xu K., Yao J., Guo R. LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol. Ther. Nucleic Acids. 2019;16:155–161. doi: 10.1016/j.omtn.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y., Shu G., Yuan X., Jing N., Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21:316–326. doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorvis C., Hatziapostolou M., Mahurkar-Joshi S., Koutsioumpa M., Williams J., Donahue T.R., Poultsides G.A., Eibl G., Iliopoulos D. Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G1124–G1137. doi: 10.1152/ajpgi.00035.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahabi S., Kumaran V., Castillo J., Cong Z., Nandagopal G., Mullen D.J., Alvarado A., Correa M.R., Saizan A., Goel R. LINC00261 is an epigenetically regulated tumor suppressor essential for activation of the DNA damage response. Cancer Res. 2019;79:3050–3062. doi: 10.1158/0008-5472.CAN-18-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B., Liu G., Ding L., Zhao J., Lu Y. FOXA2 promotes the proliferation, migration and invasion, and epithelial mesenchymal transition in colon cancer. Exp. Ther. Med. 2018;16:133–140. doi: 10.3892/etm.2018.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D.H., Tiwari A., Kim M.E., Clemons N.J., Regmi N.L., Hodges W.A., Berman D.M., Montgomery E.A., Watkins D.N., Zhang X. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett’s metaplasia. J. Clin. Invest. 2014;124:3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B., Yu J., Lu L., Dong F., Zhou F., Tao X., Sun E. Upregulated forkhead-box A3 elevates the expression of forkhead-box A1 and forkhead-box A2 to promote metastasis in esophageal cancer. Oncol. Lett. 2019;17:4351–4360. doi: 10.3892/ol.2019.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daley J.M., Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 2014;34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown J.S., O’Carrigan B., Jackson S.P., Yap T.A. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017;7:20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 27.Sugasawa K., Ng J.M., Masutani C., Iwai S., van der Spek P.J., Eker A.P., Hanaoka F., Bootsma D., Hoeijmakers J.H. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 28.Despras E., Pfeiffer P., Salles B., Calsou P., Kuhfittig-Kulle S., Angulo J.F., Biard D.S.F. Long-term XPC silencing reduces DNA double-strand break repair. Cancer Res. 2007;67:2526–2534. doi: 10.1158/0008-5472.CAN-06-3371. [DOI] [PubMed] [Google Scholar]
- 29.Eke I., Schneider L., Förster C., Zips D., Kunz-Schughart L.A., Cordes N. EGFR/JIP-4/JNK2 signaling attenuates cetuximab-mediated radiosensitization of squamous cell carcinoma cells. Cancer Res. 2013;73:297–306. doi: 10.1158/0008-5472.CAN-12-2021. [DOI] [PubMed] [Google Scholar]
- 30.Eke I., Cordes N. Radiobiology goes 3D: how ECM and cell morphology impact on cell survival after irradiation. Radiother. Oncol. 2011;99:271–278. doi: 10.1016/j.radonc.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Sharma V., Khurana S., Kubben N., Abdelmohsen K., Oberdoerffer P., Gorospe M., Misteli T. A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 2015;16:1520–1534. doi: 10.15252/embr.201540437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Özgür E., Mert U., Isin M., Okutan M., Dalay N., Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin. Exp. Med. 2013;13:119–126. doi: 10.1007/s10238-012-0181-x. [DOI] [PubMed] [Google Scholar]
- 33.Chen W., Yu Z., Huang W., Yang Y., Wang F., Huang H. LncRNA LINC00665 promotes prostate cancer progression via miR-1224-5p/SND1 axis. OncoTargets Ther. 2020;13:2527–2535. doi: 10.2147/OTT.S241578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J.-L., Zou L., Zhu T. Long non-coding RNA LINC00665 promotes metastasis of breast cancer cells by triggering EMT. Eur. Rev. Med. Pharmacol. Sci. 2020;24:3097–3104. doi: 10.26355/eurrev_202003_20674. [DOI] [PubMed] [Google Scholar]
- 35.Liao J., Dong L.-P. linc00261 suppresses growth and metastasis of non-small cell lung cancer via repressing epithelial-mesenchymal transition. Eur. Rev. Med. Pharmacol. Sci. 2019;23:3829–3837. doi: 10.26355/eurrev_201905_17810. [DOI] [PubMed] [Google Scholar]
- 36.Gao J., Qin W., Kang P., Xu Y., Leng K., Li Z., Huang L., Cui Y., Zhong X. Up-regulated LINC00261 predicts a poor prognosis and promotes a metastasis by EMT process in cholangiocarcinoma. Pathol. Res. Pract. 2020;216:152733. doi: 10.1016/j.prp.2019.152733. [DOI] [PubMed] [Google Scholar]
- 37.Militello G., Weirick T., John D., Döring C., Dimmeler S., Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief. Bioinform. 2017;18:780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y., Xu M., Che M., Von Hofe E., Abbas A., Kallinteris N.L., Lu X., Liss Z.J., Forman J.D., Hillman G.G. Curative antitumor immune response is optimal with tumor irradiation followed by genetic induction of major histocompatibility complex class I and class II molecules and suppression of Ii protein. Hum. Gene Ther. 2005;16:187–199. doi: 10.1089/hum.2005.16.187. [DOI] [PubMed] [Google Scholar]
- 39.Wang S.-Y., Wang X., Zhang C.-Y. lncRNA SNHG7 enhances chemoresistance in neuroblastoma through cisplatin-induced autophagy by regulating miR-329-3p/MYO10 axis. Eur. Rev. Med. Pharmacol. Sci. 2020;24:3805–3817. doi: 10.26355/eurrev_202004_20847. [DOI] [PubMed] [Google Scholar]
- 40.Yin J., Shen X., Li M., Ni F., Xu L., Lu H. miR-329 regulates the sensitivity of 5-FU in chemotherapy of colorectal cancer by targeting E2F1. Oncol. Lett. 2018;16:3587–3592. doi: 10.3892/ol.2018.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun C.-C., Li S.-J., Zhang F., Pan J.-Y., Wang L., Yang C.-L., Xi Y.-Y., Li J. hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. Oncotarget. 2016;7:21510–21526. doi: 10.18632/oncotarget.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao B., Tan L., He B., Liu Z., Xu R. miRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J. Transl. Med. 2013;11:172. doi: 10.1186/1479-5876-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J., Zhang J. lncRNA TP73-AS1 is a novel regulator in cervical cancer via miR-329-3p/ARF1 axis. J. Cell. Biochem. 2020;121:344–352. doi: 10.1002/jcb.29181. [DOI] [PubMed] [Google Scholar]
- 44.Michelini F., Pitchiaya S., Vitelli V., Sharma S., Gioia U., Pessina F., Cabrini M., Wang Y., Capozzo I., Iannelli F. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moynahan M.E., Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uehara Y., Ikehata H., Furuya M., Kobayashi S., He D., Chen Y., Komura J., Ohtani H., Shimokawa I., Ono T. XPC is involved in genome maintenance through multiple pathways in different tissues. Mutat. Res. 2009;670:24–31. doi: 10.1016/j.mrfmmm.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W., Song Y., He X., Liu X., Zhang Y., Yang Z., Yang P., Wang J., Hu K., Liu W. Prognosis value of RBBP8 expression in plasma cell myeloma. Cancer Gene Ther. 2020;27:22–29. doi: 10.1038/s41417-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y., Chen L., Zhao G., Li H., Guo Q., Zhu S., Li P., Min L., Zhang S. RBBP8/CtIP suppresses P21 expression by interacting with CtBP and BRCA1 in gastric cancer. Oncogene. 2020;39:1273–1289. doi: 10.1038/s41388-019-1060-7. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Cao J., Meng Y., Qu C., Shen F., Xu L. Overexpression of xeroderma pigmentosum group C decreases the chemotherapeutic sensitivity of colorectal carcinoma cells to cisplatin. Oncol. Lett. 2018;15:6336–6344. doi: 10.3892/ol.2018.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teng X., Fan X.F., Li Q., Liu S., Wu D.Y., Wang S.Y., Shi Y., Dong M. XPC inhibition rescues cisplatin resistance via the Akt/mTOR signaling pathway in A549/DDP lung adenocarcinoma cells. Oncol. Rep. 2019;41:1875–1882. doi: 10.3892/or.2019.6959. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Ding Q., Fujimori H., Motegi A., Miki Y., Masutani M. Loss of CtIP disturbs homologous recombination repair and sensitizes breast cancer cells to PARP inhibitors. Oncotarget. 2016;7:7701–7714. doi: 10.18632/oncotarget.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eke I., Hehlgans S., Sandfort V., Cordes N. 3D matrix-based cell cultures: Automated analysis of tumor cell survival and proliferation. Int. J. Oncol. 2016;48:313–321. doi: 10.3892/ijo.2015.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eke I., Deuse Y., Hehlgans S., Gurtner K., Krause M., Baumann M., Shevchenko A., Sandfort V., Cordes N. β1 Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J. Clin. Invest. 2012;122:1529–1540. doi: 10.1172/JCI61350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eke I., Zscheppang K., Dickreuter E., Hickmann L., Mazzeo E., Unger K., Krause M., Cordes N. Simultaneous β1 integrin-EGFR targeting and radiosensitization of human head and neck cancer. J. Natl. Cancer Inst. 2015;107:dju419. doi: 10.1093/jnci/dju419. [DOI] [PubMed] [Google Scholar]
- 55.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Reczko M., Maragkakis M., Dalamagas T.M., Hatzigeorgiou A.G. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sticht C., De La Torre C., Parveen A., Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J., Han L., Roebuck P., Diao L., Liu L., Yuan Y., Weinstein J.N., Liang H. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X., Miao Z., Divate M., Zhao Z., Cheung E. KM-express: an integrated online patient survival and gene expression analysis tool for the identification and functional characterization of prognostic markers in breast and prostate cancers. Database (Oxford) 2018;2018:bay069. doi: 10.1093/database/bay069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.