Abstract
The ability to direct cell behavior has been central to the success of numerous therapeutics to regenerate tissue or facilitate device integration. Biomaterial scientists are challenged to understand and modulate the interactions of biomaterials with biological systems in order to achieve effective tissue repair. One key area of research investigates the use of extracellular matrix-derived ligands to target specific integrin interactions and induce cellular responses such as increased cell migration, proliferation, and differentiation of mesenchymal stem cells (MSCs). These integrin-targeting proteins and peptides have been implemented in a variety of different polymeric scaffolds and devices to enhance tissue regeneration and integration. This review will first present an overview of integrin-mediated cellular processes that have been identified in angiogenesis, wound healing, and bone regeneration. We will then highlight research utilizing biomaterials with integrin-targeting motifs as a means to direct these cellular processes to enhance tissue regeneration. In addition to providing improved materials for tissue repair and device integration, these innovative biomaterials provide new tools to probe the complex processes of tissue remodeling in order to enhance the rational design of biomaterial scaffolds and guide tissue regeneration strategies.
Keywords: biomaterials, integrins, wound healing, angiogenesis, bone regeneration
Graphical Abstract
Integrin-targeting biomaterials can be used to guide cell behavior for improved tissue regeneration. This review summarizes integrin-mediated cellular responses involved in bone regeneration, angiogenesis, and wound healing processes. Design considerations in the development of integrin-targeting biomaterials to harness these interactions to improve regeneration are described including a perspective analysis of future research.
1. Introduction
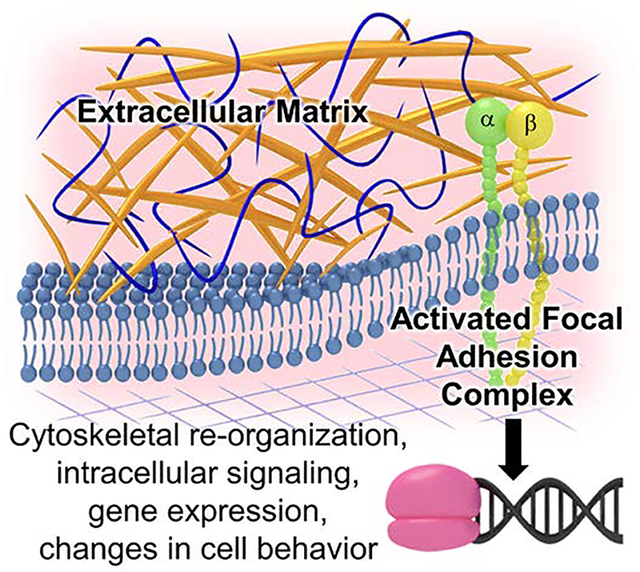
In order to address limitations of current treatment options and produce replacement tissue grafts, there is a need to develop biomaterials that can recapitulate the regenerative capacity of autografts while retaining the availability of alloplasts. Several strategies have focused on incorporation of insoluble biological cues from the extracellular matrix (ECM) into biomaterials to promote desired cell behaviors and tissue regeneration. In their native environment, cells primarily use integrins as the main receptor proteins to interact with the surrounding ECM. [1] Each integrin is composed of two noncovalently-associated transmembrane glycoprotein subunits, 18 unique α and β subunits, that combine to form 24 distinct dimers that bind to specific motifs in ECM proteins.[2] Common integrins targeted in regenerative medicine and their respective ligands in native extracellular matrix proteins are listed in Table 1. Binding of integrins to their respective ligands is dependent on divalent cations with the type of cation influencing both the affinity and the specificity of the integrin binding to the ligand.[3, 4] Integrins play a crucial role in anchoring cells to the ECM and provide the requisite link to the cytoskeleton that enables stable cell adhesion, cell spreading, mechanotransduction, and migration. In addition, binding of integrins to ECM ligands can induce integrin clustering and conformational changes that can transmit outside-in signals across the plasma membrane.[5] There are no known catalytic activities present in the cytoplasmic tails of integrins, rather downstream signaling is mediated by the activated focal adhesion complex that assembles upon integrin clustering and conformational changes. Focal adhesion complexes recruit intracellular proteins such as cytosolic kinases (e.g., focal adhesion kinase), phosphatases, and adaptor proteins that initiate cascades of signaling events that alter gene expression and cellular behavior.[4] As a result, integrin binding to the ECM is central to the regulation of cell migration, cell survival, and growth, Figure 1.[6, 7]
Table 1:
Summary of integrins and extracellular matrix proteins with complementary ligands.
| Integrin | Extracellular matrix protein ligands |
|---|---|
| α1β1 | Collagen I, Collagen IV, Laminin[8–11] |
| α2β1 | Collagen I, Collagen IV, Laminin[10–12] |
| α3β1 | Laminin, Entactin, Collagen I, Fibronectin [10, 13–17] |
| α4β1 | Fibronectin, ELIMIN1, VCAM [15,18–21] |
| α5β1 | Gelatin, Fibronectin, Fibrin, [10, 22–25] |
| α6β1 | Laminin, CCN2 [26–28] |
| α7β1 | Laminin[29] |
| α9β1 | Fibronectin, Tenascin-C, ELIMIN1, Osteopontin, ADAM8, VEGF [20, 30–34] |
| α11β1 | Collagen I, Osteolectin [11, 35] |
| αvβ1 | Fibronectin, Osteopontin [36] |
| αvβ3 | Gelatin, Fibrinogen, Vitronectin, Fibronectin, Bone Sialoprotein, CCN1 [10, 22–25, 37–41] |
| αvβ5 | Vitronectin, Fibronectin, CCN1[10, 15,26,42,43] |
| α6β4 | Laminin[26] |
Figure 1:
Integrin binding to ECM ligands induces integrin clustering and conformational changes that can transmit outside-in signals across the plasma membrane. Activated focal adhesion complexes that assemble upon integrin clustering recruit intracellular proteins that initiate cascades of signaling events that alter gene expression and cellular behavior.
Given the pivotal role of integrin binding in mediating cell behavior, there has been substantial research investigating the role of integrin-mediated signaling in tissue regeneration processes. This review will first highlight research that elucidates key integrin-mediated regenerative processes with a focus on bone regeneration, angiogenesis, and wound healing. We will then summarize design considerations in the development of integrin-targeting materials that can be used to advance tissue regeneration strategies. Finally, a critical analysis of the key challenges and future directions in material development and testing will be discussed.
2. Integrin-mediated Bone Regeneration
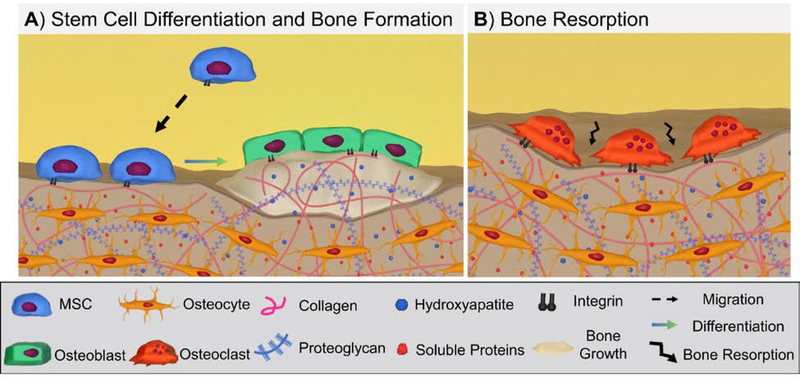
Bone healing is a complex process during which various cell types coordinate the formation and remodeling of new tissue. In the early stages after injury, the inflammatory response regulates and produces cytokines and growth factors that help recruit mesenchymal stem cells (MSCs). MSCs differentiate into osteoblasts that secrete a collagen-proteoglycan matrix that calcifies to form a bone matrix.[44–47] A secondary group of bone-specific cells called osteoclasts remodel the newly formed bone by resorption, Figure 2. The balance between new bone formation and resorption is tightly regulated and maintained to ensure sufficient bone mass or mechanical strength.[48] Although there are numerous factors involved in promoting bone regeneration, integrin-mediated interactions with the ECM play a critical role.[1] A number of studies have demonstrated the importance of integrin signaling by correlating integrin knockdown studies with detrimental effects on bone healing.[25, 49–51] Integrin-ligand interactions influence numerous cell processes, including osteogenic differentiation, bone formation, and bone remodeling.[33, 52, 53] Overall, eight integrins have been implicated in bone formation and remodeling, Figure 3 and Table S1.[54–57] This section will discuss the role that key integrins play in MSCs, osteoblasts, and osteoclasts behavior in the different stages of bone healing.
Figure 2:
Overview of key cell processes in bone formation and remodeling: (A) Recruited and resident mesenchymal stem cells (MSCs) differentiate to osteoblasts that produce new bone; (B) Osteoclast resorption of bone.
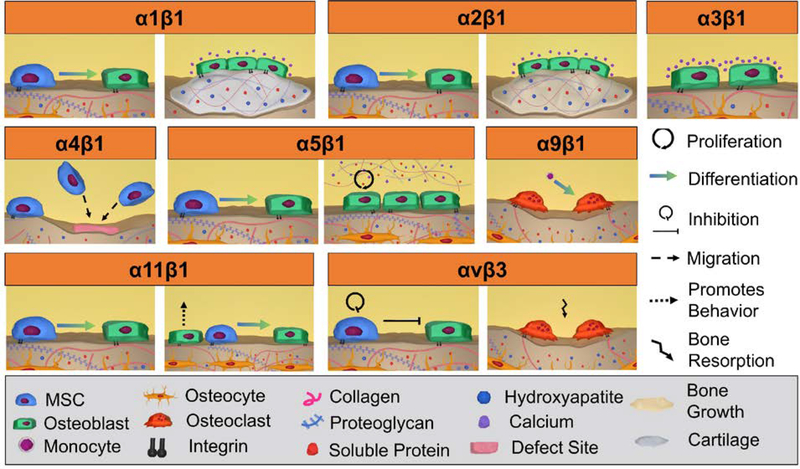
Figure 3:
Graphic illustration of integrin-mediated bone formation and remodeling. Integrins play a critical role in each of these processes: α1β1 promotes the differentiation of MSC to osteoblasts and promotes osteoblast cartilage formation and mineralization; α2β1 promotes osteogenic differentiation in MSCs and increases osteoblastic bone formation and mineralization; α3β1 increases calcium deposition of MSCs; α4β1 promotes recruitment of MSCs to the defect site; α5β1 promotes osteogenic differentiation in MSCs and the proliferation of osteoblasts and bone matrix formation; α9β1 promotes osteoclastogenesis; α11β1 directly and indirectly promotes MSC osteogenic differentiation via MSC and osteoblast integrin-binding; αvβ3 inhibits MSC proliferation and osteogenic differentiation and promotes bone resorption.
2.1. Recruitment, Attachment, and Migration
After injury, MSCs are recruited from surrounding soft tissues, bone marrow, and peripheral blood during the initial inflammatory response.[58] MSCs begin to proliferate in the defect site and later differentiate into osteoblasts and chondrocytes to promote bone formation.[58, 59] The key integrins that facilitate these processes in MSCs are α4β1, α5β1, α2β1, α11β1, and αvβ3. The α4β1 integrin plays a key role in homing these MSCs to the site of injury. Specifically, α4β1 binds to fibronectin and vascular cell adhesion protein 1 (VCAM-1), which are ECM proteins present in the defect site. [60] Kumar et al. demonstrated that ectopic expression of α4 integrin on MSCs greatly increases homing in an immunocompetent mouse model. [61] Bone marrow-derived murine MCS expressing α4 demonstrated homing and rolling to bone marrow, which can be exposed in a defect site. [63] In addition, the role of α5β1 and α2β1 in the migration of MSCs has also been studied. Veevers-Lowe et al. demonstrated that activation of α5β1 in human MSCs (hMSC) promotes the migration of the cells to fibronectin. [62] Similarly, Kolambkar et al. demonstrated that α2β1 activation by GFOGER, a peptide sequence in a triple helix structure, enhanced MSC migration in vitro. [64] Once MSCs localized in the defect site, cell survival and proliferation are central to their impact on bone regeneration.
Integrins α2β1, α11β1, and αvβ3 play a role in the survival and proliferation of MSCs. The α2β1 and α11β1 integrins bind to collagen, which is the predominant protein in the bone ECM. [65] Popov et al. found that disrupting α2β1 and α11β1 function with shRNA promoted apoptosis in hMSCs in in vitro studies. [11] Another study showed that α2β1 activation of hMSCs by a GFOGER peptide-laden hydrogel promoted adhesion and survival in the bone defect in an immunocompromised mouse model.[66] Furthermore, α2β1 has been demonstrated to promote MSC proliferation in vitro. [64] In contrast, αvβ3 has been implicated in inhibiting MSC growth. Matrino et al. showed antibody blocking of αvβ3 enhanced hMSC proliferation in vitro. [25] Additional investigation is needed to clarify the role of αvβ3 in proliferation. However, together, these integrins are shown to have a clear role in promoting the survival and proliferation of MSCs in the defect site allowing for subsequent differentiation into bone forming osteoblasts.
2.2. Osteogenic Differentiation
Once MSCs have been recruited to the defect site, they must undergo osteogenic differentiation to promote bone regeneration. Several integrins contribute to committing MSCs toward the osteogenic lineage, including α2β1, α5β1, αvβ3, α1β1, and α11β1. Each has been shown to play a role in osteogenic differentiation. The α2β1 integrin is the primary MSC adhesion receptor to collagen, the main organic component of bone tissue. [54] It has been indicated that this interaction is critical for osteoblastic differentiation and plays a significant role in mineralization during osteogenesis.[67] Specifically, studies found that α2β1-mediated adhesion activates transcription factor RunX2/Cbfa1, an important hallmark for osteoblastic differentiation. [68, 69] Further, it has been confirmed that if α2β1 is perturbed or expression knocked down, downstream phosphorylation of signaling targets decreases and the osteogenic phenotype is not observed. [49, 70, 71] Several studies demonstrated that α2β1 also activates focal adhesion kinase, increases osteoblast-specific promoters, and induces ALP and mineralization confirming its role in osteogenic differentiation of hMSCs and murine pre-osteoblasts in vitro. [69, 71–74] Furthermore, Kolambkar et al. demonstrated that α2β1 activated by GFOGER in a collagen-like tertiary structure enhanced differentiation down the osteoblastic lineage in vitro. [64] Together, these studies demonstrate the critical role α2β1 plays in MSC osteogenic differentiation.
The α5β1 integrin has also been implicated in transducing cell responses for osteoblast differentiation. Blockade studies of α5β1 have demonstrated gene expression related to bone cells is inhibited. [16] Furthermore, overexpression or priming of this integrin increased osteogenic capacity, confirming its crucial role in the process of osteogenesis. [25, 50, 51] Several studies have demonstrated that this integrin promotes osteogenic differentiation of hMSCs and murine MSCs in vitro. [25, 51, 75] Additionally, a recent study investigated the effect of fibronectin fibers stretching on hMSC differentiation to elucidate the mechanosensitivity of αvβ3 and α5β1 integrin signaling. It was observed that adjustments in fibronectin fiber strain resulted in differential integrin α5β1 and αvβ3 binding and that preferential signaling via αvβ3 over α5β1 on relaxed fibronectin fibers resulted in decreased hMSC osteogenesis.[76, 77] These studies demonstrate that this integrin plays a pivotal role in several functions in MSC osteogenic differentiation.
The function and role of αvβ3 in bone formation are debated. Its main interactions are with vitronectin and fibronectin. It should be noted that αvβ3 is only weakly expressed in hMSC populations via flow cytometry. [25] There have been conflicting reports about the role of αvβ3 in osteogenic differentiation. Salaszynyk et al. found that even in the absence of osteogenic stimulants, hMSCs cultured on vitronectin produced matrix mineralization, calcification, and alkaline phosphatase activity, which was primarily attributed to interactions with αvβ3.[78] Whereas, other studies have reported that αvβ3 had an inhibitory role in osteogenic differentiation.[25, 79] Martino et al. observed that antibody blocking of αvβ3 resulted in an enhancement of proliferation and ALP expression in hMSCs attached to fibronectin.25 Similarly, Cheng et al. demonstrated that the overexpression of αvβ3 in mouse-derived pre-osteoblasts decreased proliferation, inhibited matrix mineralization, and decreased expression of ALP, collagen type I, and bone sialoprotein.79 Due to these conflicting reports, there is a need for more research in elucidating the exact involvement of this integrin in bone formation.
Some studies suggest that both α1β1 and α11β1 also play roles in osteogenic differentiation, although possibly with less involvement. One group demonstrated that a designer protein, SclGFPGEN, targeting α1β1 in 3D hydrogel systems increased hMSC osteogenic differentiation in osteogenic medium.[74] In addition, inhibiting binding of α11β1 inhibited the osteogenic marker expression of human and mouse osteoprogenitor cells in vitro. [35] Future studies could further elucidate α1β1 and α11β1 functions in osteogenic differentiation.
As discussed, several integrins play clear roles in facilitating osteogenic differentiation of MSCs. However, this cell differentiation only lays the foundation for bone and later stages of bone healing. Mature osteoblasts and osteoclasts, in conjunction with other cells, coordinate to complete the process. Various integrins on the bone specific cells have been investigated to contribute to bone formation and remodeling.
2.3. Bone Formation and Remodeling
Bone formation occurs via two types of ossification: intramembranous and endochondral. Intramembranous ossification results in formation of compact and spongy bone directly from osteoblasts and blood cells.[80] Whereas, endochondral ossification first produces a network of cartilage that is then modified by osteoblasts and overturned to become bone. [58, 80] Both processes are critical during embryonic development, with intramembranous ossification producing the clavicle and both facial and cranial bones, while the rest of the skeleton is formed by endochondral ossification. It is also understood that post injury bone healing is a combination of both these two processes. [58] In order to develop biomaterials for bone injuries, it is important to understand how to target the key biological facilitators that enable proper bone formation. Although there are a host of factors that orchestrate osteogenesis, the cells involved, specifically the osteoblasts, are of critical importance.
Just as integrins play a vital role in promoting osteogenic differentiation of MSCs to osteoblasts, these surface receptors are also important in bone formation and tissue maintenance. Through various function ablating studies in vitro and in vivo, it has been identified that there are several key integrins implicated in these processes. An in vitro study demonstrated that the β1 subunit impairment resulted in defective proliferation, differentiation, and cell function in osteoblasts. The specific integrins which have been identified to be involved in osteogenesis include α3β1, α5β1, and α11β1. [14, 71, 81] Among these three, α5β1 is largely responsible for osteoblast survival, maintenance of the differentiated phenotype, and mineralization of bone. First, osteoblast survival is highly dependent on α5β1 interaction with a non-collagenous protein in bone ECM called fibronectin.[25] Both in vitro and in vivo studies confirmed that disrupting the receptor-ligand interactions resulted in significant levels of osteoblast apoptosis. [14, 81] To confirm α5β1 role in osteoblast functionality, similar integrin blocking studies were conducted. The results demonstrated that interference suppressed mineralized bone nodule formation.[71] Second, α5β1 is also responsible for load sensing and mechanotransduction in osteoblasts. It was observed that in rats with skeletal unloading, expression of the α5β1 was downregulated, which resulted in low osteoblast survival, altered bone matrix, and bone loss. Contrastingly, overexpression of the integrin using transforming growth factors β1increased osteoblast survival. [82]
The other two integrins, α11β1 and α3β1, maintain similar function to α5β1 integrin, although their role may be considered secondary. α11β1 is responsible for binding the growth factor osteolectin and the resulting integrin-ligand complex is hypothesized to enable proper maintenance of bone mass. [11, 35] In a study conducted by Shen et al., knocking down α11β1 expression impaired osteogenic differentiation of MSCs as well as their ability to bind osteolectin. Through this knockdown, adult mice maintained low levels of osteogenesis and had accelerated bone loss. [35] Lastly, studies also investigated how other integrins, including α3β1 and αvβ5 are relevant in osteogenesis. Isolated studies show that these integrins are actively expressed on the surface of bone cells throughout differentiation, bone production, and maintenance life cycle, but their exact function is yet to be identified. [55]
In the bone growth cycle tissue remodeling, the process of old bone matrix resorption, and new matrix deposition, is vital to ensure that bone tissue remains healthy and functional within the body. [83] The main cells that are involved in the process of bone resorption are called osteoclasts. These cells are not derived from MSCs but differentiate from monocyte-macrophage cell lineages. [84] While bone formation and resorption are explained as two distinct processes, it is important to understand that they happen in conjunction during the lifetime of bone tissue as well as in the event of injury.
Bone resorption by osteoclasts also involves integrin-based signaling, similar to osteogenesis, to ensure proper function. The main integrins involved in bone resorption include α2β1, α9β1, and αvβ3. The α9β1 integrins expressed on the surface of osteoclasts are essential for osteoclast formation, recruitment, and mobility of the cells to the bone tissue matrix. Rao et al. demonstrated that antibody blocking of α9β1 in osteoclast precursors significantly decreased the formation of mature osteoclasts. [33] This was further confirmed when preosteoclast cells were cultured from mice with double knockdown for the α9 subunit alongside wildtype cells. The study results demonstrated that mutant preosteoclasts did not mature, and the cells formed less resorption pits compared to the wild type cells. Further, the double knockdown mice also maintained an increased bone volume compared to control, demonstrating the effect on the function of mature osteoclasts in resorption. [33]
The αvβ3 integrin has also been identified to be critical in osteoclast function and bone resorption. [85] Furthermore, αvβ3 was identified as an important adhesive integrin for osteoclasts as, without proper integrin functionality, osteoclasts maintained poor adhesion to the substrate, resulting in high level of apoptosis. [38] β3 null mice also developed osteopetrosis (increased bone volume compared to normal) due to dysfunctional osteoclasts. [86] A study performed by a separate group of researchers demonstrated that by designing an antagonist highly specific to αvβ3 integrin, the function ablation inhibited bone resorption in women and increased bone mineral density, thus confirming the previously understood role of this subunit pair. [87, 88] Lastly, although α2β1 is primarily involved in adhesion and osteogenic differentiation of hMSCs, several studies have shown its involvement in maintaining resorptive capabilities in osteoclasts. Helfrich et al. reported that α2β1 continued remodeling of bone tissue with an αvβ3 knockdown.[89] This remodeling ability of α2β1 is noteworthy because of the difference in functionality based on the cell type that expresses it, MSC vs. osteoclast.
In summary, integrin interactions have been implicated in each phase of bone regeneration from MSC recruitment, survival, and differentiation to neotissue formation and resorption. Much of the work described here is the result of rigorous fundamental studies using knockdown or knockout models. This elucidation of individual integrin roles coupled with improved integrin-targeting biomaterials has led to the advent of osteoinductive matrices to improve bone regeneration using ECM-ligands.
3. Integrin-mediated Wound Healing
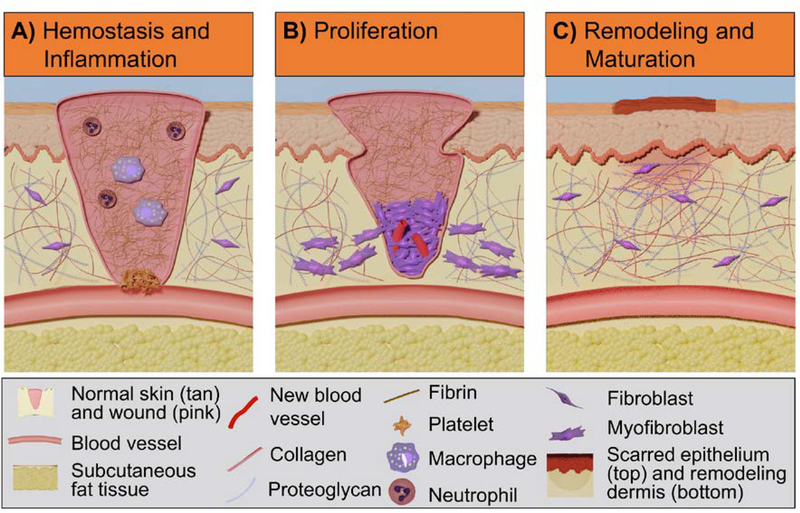
Wound healing is comprised of three overlapping phases: hemostasis and inflammation, proliferation, and maturation and remodeling, Figure 4.[90, 91] In these phases, interactions between cells (fibroblasts, keratinocytes, and endothelial cells) and the ECM play a significant role in regulating regeneration.[92, 93] To better understand the integrin-ECM interactions, researchers have utilized both knockdown/knockout or overexpression animal models and in vitro cell studies to investigate the effect of integrin signaling on wound healing outcomes. These studies have demonstrated that loss or induction of specific integrins can result in impaired re-epithelialization, delayed granulation tissue formation, or other healing-related processes.[30, 94] A summary of the integrins and respective ECM proteins involved in the phases of wound healing is provided in Table S2. Figure 5 presents the integrins that are involved in wound healing in addition to their specific activities. In this section, we will discuss the relevant integrin-mediated cellular processes in re-epithelialization and granulation tissue formation. Understanding the role of integrin interactions in these processes and the resulting effect on wound healing outcomes can provide design strategies to improve chronic wound healing.[95–97]
Figure 4:
Overview of wound healing phases and key cellular processes. (A) Hemostasis and inflammation: blood clot is formed after formation of a platelet plug and fibrin matrix when macrophages and neutrophils are recruited to induce innate immune responses against invading microbes and foreign substances; (B) Proliferation: re-epithelialization and granulation tissue formation occur with migration, proliferation, and differentiation of fibroblasts and keratinocytes; (C) Remodeling and maturation: Matrix remodeling and wound closure lead to the restoration of skin barrier and reconstruction of connective tissue.
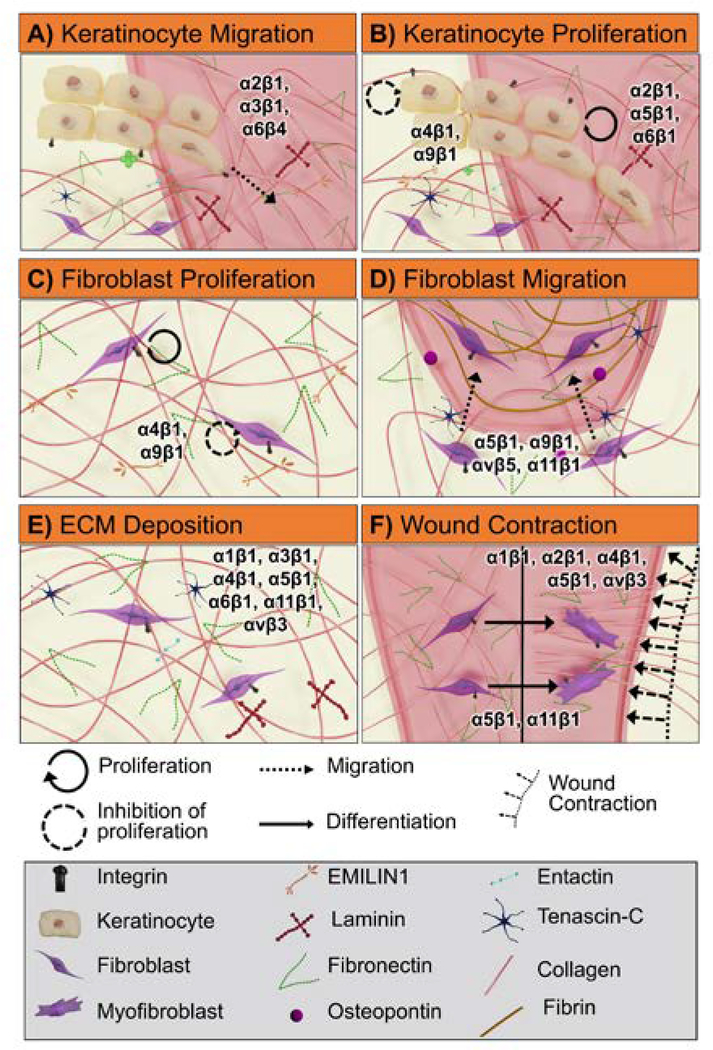
Figure 5:
Graphic illustration of integrin-mediated cellular processes in wound healing that begins with basal keratinocyte migration into the wound bed (A) and proliferation above the basal keratinocyte layer (B) followed by recruitment of activated fibroblasts and proliferation (C). Subsequent fibroblasts migration into the fibrin-fibronectin provisional matrix (D) and production and deposition of essential ECM components (collagen, fibronectin, and laminin) occurs after fibroblasts infiltrate into the blood clot (E). Finally, fibroblasts differentiation into myofibroblasts induces wound contraction by interacting with reconstructed ECM matrix (F). In each of these processes, integrins play a critical role: (A) keratinocyte migration is regulated by integrins α2β1, α3β1, and α6β4; (B) keratinocyte proliferation is regulated by α2β1, α4β1, α5β1, α6β1, and α9β1; (C) fibroblast proliferation is regulated by α4β1 and α9β1; (D) fibroblast migration is regulated by α5β1, α9β1, α11β1, and αvβ5; (E) ECM deposition (collagen, fibronectin, and laminin) is regulated by α1β1, α3β1, α4β1, α5β1, α6β1, α11β1, and αvβ3; (F) myofibroblast differentiation is regulated by α5β1 and α11β1 and wound contraction regulated by α1β1, α2β1, α4β1, α5β1, and αvβ3.
3.1. Re-epithelialization
Re-epithelialization restores the epidermis barrier and is dependent on the migration and proliferation of epithelial keratinocytes. [98] In new wounds, keratinocytes are exposed to a new pericellular environment, which facilitates the release of sequestered growth factors and cytokines from the disrupted ECM. The released factors include epidermal growth factors (EGFs), transforming growth factor-β (TGF-β), and fibroblast growth factors (FGFs). These activating signals trigger the transition of keratinocytes to a pro-migratory phenotype (epithelial-mesenchymal transition). [99–102] The reconstruction of cell-cell adhesion and cellular attachment to the basement membrane relies on keratinocyte migration and proliferation that are mediated by several different integrins including α2β1, α3β1, α4β1, α5β1, α9β1, and α6β4.
Directional migration of keratinocytes into the provisional matrix initiates re-epithelialization and is mediated by the interactions of integrins α2β1, α6β4, and α3β1 with collagen and laminin. [103, 104] Pilcher et al. demonstrated that keratinocytes showed upregulation of matrix metalloproteinase-1 (MMP-1) after binding to wound-edge collagen via integrin α2β1. [105] Also, primary keratinocytes migration on collagen gel was inhibited by collagenase antibodies. Therefore, the α2β1-collagen interaction can induce the production of MMP-1 that degrades collagen fibrils and detaches the keratinocytes from the matrix, initiating the cell migration. [106] In contrast, α3β1 and α6β4 bind to laminin-332 and mediate the keratinocyte migration. Multiple studies highlighted that the interaction between laminin-332 and α3β1 could activate intracellular polarization, therefore promoting the keratinocyte migration. [17, 107] Keratinocyte polarization is essential to determine the migration direction and relies on Rac-1 activation by α3β1-mediated FAK/Src signaling pathway. [108] Integrin α6β4 can also regulate keratinocyte migration by colocalizing with Rac1 to form a Rac1/α6β4 complex. [109] β4 integrin-deficient keratinocytes demonstrate a circular migration track, indicating the crucial role of this complex. It was hypothesized that the Rac1/α6β4 complex could remodel laminin-332 fibrils into linear tracks and facilitate the linear migration of the cells. [109] Studies have confirmed that keratinocytes are anchored onto the basement membrane through the attachment of integrin α6β4 to hemidesmosome component, plectin. [110, 111] This finding is essential for understanding the stabilized cell attachment to the newly formed basement membrane after wound closure. Furthermore, Spinardi et al. demonstrated that keratinocyte migration could be enhanced with connective tissue growth factor (CCN2) via α5β1-fibronectin interaction and activation of FAK-MAPK signaling. [112]
Basal keratinocytes and epidermal stem cells start proliferating 48 to 72 hours after the onset of migration. [113, 114] Recent studies uncovered the influence of β1 integrins on keratinocyte proliferation. Keratinocyte proliferation is promoted by α5β1 binding fibronectin and when α6β1 recognizes a laminin-based peptide sequence YIGSR, leading to enhanced epidermal development. [115, 116] In addition, an α2β1 integrin-expressing mice model showed a substantial keratinocyte hyperproliferation and offered an insight of α2β1 integrin participation in re-epithelialization. [117] Although the α9β1-mediated intracellular signaling pathway is not entirely understood, it can result in up or down regulation of keratinocyte proliferation. Specifically, an induced α9β1 integrin-deficient mice model demonstrated a poor re-epithelialization and wound healing outcome, but keratinocytes proliferation is inhibited when α9β1 binds to elastic microfibril interface-located protein 1 (EMILIN1). [20, 118, 119] EMILIN1 can also inhibit keratinocyte proliferation via α4β1-mediated signaling. [20]
3.2. Granulation Tissue Formation and Wound Contraction
In parallel to re-epithelialization, disrupted dermal connective tissue is repaired with granulation tissue formation followed by wound remodeling. Granulation tissue formation aids in skin tissue regeneration by restoring the blood supply and the mechanical and functional integrity of the connective tissue. In association with peripheral immune cells, pericytes, and keratinocytes, fibroblasts play a central role in this process by regulating matrix formation and inducing wound contraction. Upon wounding, factors secreted by macrophages and keratinocytes (TGF-β, PDGF, FGF, CTGF) can activate fibroblasts. [120–123] Also, the change in mechanical properties and oxygen tension of the local tissue can also induce the fibroblast activation. [124–126] Then, activated fibroblast cells undergo proliferation and migrate into the blood clot, where the deposition of ECM molecules occurs sequentially to restore the connective tissue strength. The following event is the differentiation of fibroblasts into myofibroblasts that contributes to wound contraction and closure. Eventually, the granulation tissue ECM initiates remodeling after wound contraction, resulting in either normal or scar tissue. These cellular activities are temporally overlapped and depend on the reciprocal interactions between cells and ECM molecules. Fibroblast proliferation, migration, and differentiation are regulated by different ECM components, while ECM deposition and remodeling are modulated by cell protein production. Therefore, ECM-cell interactions through integrins are essential in directing granulation tissue formation and wound contraction.
The β1 integrins, including α1β1, α2β1, α4β1, and α9β1, regulate fibroblast proliferation. An α1-deficient mice model displayed a reduction in fibroblasts and a hypocellular dermis, revealing the need for α1β1 in regulating fibroblast proliferation. [9] Binding to collagen triggers a unique regulatory growth pathway of α1β1 with the adaptor protein Shc. Integrin linking to Shc can respond to mitogenic growth factors and promote cell survival-related gene expression through the G1 phase of the cell cycle, which enhances cell proliferation. [127] In contrast, both α4β1 and α9β1 can inhibit fibroblast proliferation when interacting with ELIMIN1 when the loss of ELIMIN1 results in dermal hyperproliferation and accelerated wound closure. [20] As elucidated in the study, ELIMIN1 binds to α4/α9β1 and activates Phosphatase and tensin homolog (PTEN), and PTEN inhibits PI3k/Akt pathway and Erk1/2 phosphorylation that are pro-proliferation signals. Furthermore, α2β1 also regulates fibroblast proliferation as decreased α2β1 expression accounts for enhanced fibroblast proliferation in idiopathic pulmonary fibrosis. [128] A synthetic laminin sequence, EF1, has been shown to enhance fibroblast proliferation via interaction with α2β1, as well as a recombinant collagen-mimetic protein, SclGFPGER. [129, 130] Although the mechanism of integrin-mediated fibroblast proliferation in the early stage of granulation tissue formation has not been fully understood, β1 integrins modulate fibroblast mitosis and can be employed to improve skin tissue regeneration.
Following proliferation, fibroblasts migrate and infiltrate the blood clot and the provisional matrix that consists of fibrin and fibronectin fibrils. [131] Blood clot ECM remodeling and fibroblast motility rely on multiple integrins-ECM interactions in different stages, including α5β1, α9β1, α11β1, and αvβ5. Both crosslinking of fibronectins to fibrin matrix and fibronectin fibril formation are required for cell adhesion and migration to the provisional matrix. Researchers have revealed that dermatopontin, a protein abundant in the provisional matrix and wound fluid, can induce fibronectin fibril formation and enhance fibroblast adhesion via integrin α5β1. [132] Dermatopontin-knock out mice demonstrate an abnormal ECM architecture and decreased skin tissue flexibility. Another study found that integrin α9β1 was required for dermal fibroblast migration as blocking α9 integrin with antibody impaired the interaction between fibroblasts and tenascin-C, reducing cell adhesion and migration. [31] However, it was found that the α9β1 blockade does not affect myofibroblast differentiation and wound contraction in the later stage of granulation tissue formation. Fibroblast migration was also impaired in an α11β1 knock-out mice model, showing that α11β1 was essential for fibroblast migration. [133] Additionally, CCN1/CYR61 can function as an extracellular matrix signaling molecule and mediate fibroblast migration via direct interaction with integrins αvβ5 in granulation tissue. [40]
After fibroblast infiltration into the blood clot, ECM deposition is initiated by fibroblasts and other cells. The significant components synthesized during this period include collagen, laminin, and fibronectin. Collagen synthesis can be modulated by integrins α1β1 and α6β1 and accounts for later collagen fibril formation and wound contraction. α1β1 has been found to participate in the downregulation of collagen synthesis by fibroblasts as the dermis of α1-null mice shows higher production levels of both collagen and collagenase. [134] Additionally, dermal fibroblasts derived from α1-null mice demonstrate reduced sensitivity to collagen gel, indicating that the fibroblast adhesion to collagen and adhesion-dependent cellular signaling are also regulated by α1β1 integrin. In a word, the interaction between α1β1 and collagen allows a feedback regulation of collagen production. Lastly, α6β1 has also been shown to stimulate collagen deposition by binding to CCN2/CTGF. [27] Also, Aumailley et al. showed that α6β1 played a significant role in initiating basement membrane formation and mediating the deposition of laminin 1 as α6β1 knock-out mice demonstrated a shutdown of laminin α1 chain synthesis. [135]
In addition to collagen and laminin, fibronectin is also crucial in wound contraction, indicating its importance in skin tissue regeneration. Integrins α3β1, α4β1, and α5β1 participate in the regulation of fibronectin deposition in different manners. α3β1 can adhere to and interact with entactin, enhancing the deposition of entactin and fibronectin. [13] This interaction also promotes cell adhesion to fibronectin via α3β1. Integrin α5β1 regulates fibronectin deposition and blockade of α5β1 by antibody results in a reduction of fibronectin accumulation. [136, 137] Binding to EDA domain of fibronectin, integrin α4β1 mediates fibronectin synthesis and fiber assembly by fibroblast. [138] In addition, αvβ3 has been shown to modulate the deposition of tenascin-C with the activation of the Src/MAPK/MMP signal pathway. [139]
Towards the end of granulation tissue formation, myofibroblast differentiation and wound contraction take place to promote wound closure. Integrins α5β1 and α11β1 have been shown to regulate myofibroblast differentiation by interacting with fibronectin or collagen. [133, 140] Thannickal et al. demonstrated that TGF-β1-mediated myofibroblast differentiation required cell adhesion via integrin α5β1. As the most significant step of wound closure, wound contraction relies on the deposition of ECM components and the maturation of myofibroblasts. There are two ways that myofibroblasts can be anchored to collagen fibrils to induce the collagen matrix contraction. First, myofibroblasts can indirectly anchor to the synthesized collagen fibrils by attaching to the fibronectin matrix. By binding to the fibronectin matrix, integrin α5β1 can promote focal adhesion and mediate matrix contraction via RhoA-GTP and FAK signal pathways. [141, 142] Fragmented fibronectin (the V region) also enhances the fibronectin-matrix contraction when interacting with α4β1. [143] Collagen gel contraction by fibroblasts can be inhibited by αvβ3 antibody, indicating that αvβ3 mediates collagen contraction via interaction with fibronectin.[37] Second, fibroblasts can bind to collagen directly via integrins α1β1 and α2β1. [144, 145] For instance, blocking of integrin α1β1 results in a reduction of collagen lattice remodeling and gel contraction by fibroblasts, indicating that collagen contraction in wound healing requires integrin α1β1. [146, 147] Also, α2β1 regulates the reorganization and contraction of a collagen matrix, presumably with the participation of PI3K activation. [37, 148]
Wound angiogenesis is another crucial process in wound healing through the entire process of granulation tissue formation. New blood vessels are responsible for revascularizing the regenerated connective tissue and supplying the tissue with oxygen and nutrients for growth and contraction. From capillary sprouting to blood vessel remodeling, ECM guides the cell migration by providing scaffold support and regulating the cell behaviors with integrin-mediated signaling. [149] Several essential integrins have been identified in wound angiogenesis, including αvβ3, α2β1, and α3β1. Particularly in the provisional matrix of a healing wound, αvβ3 is expressed on the tip of the capillary sprouts and has key interactions with fibrin and fibronectin.[150] Blockade of αvβ3 with an antibody or an RGD-containing peptide also results in delayed angiogenesis in a murine wound model. [151] Zweers et al. showed that the ablation of the α2β1 gene resulted in enhanced neovascularization via a shift of collagen-integrin binding signaling. [152] Additionally, Mitchell et al. demonstrated the significance of α3β1 in the crosstalk between keratinocytes and endothelial cells that promoted endothelial cell migration.[153] More relevant integrin-ECM interactions will be detailed in the next section to provides a comprehensive review of integrins in angiogenesis.
In summary, there are dynamic changes to the ECM through each phase of wound healing with corollary integrin-ECM interactions that guide re-epithelialization, granulation tissue formation, and wound contraction. Research has highlighted the myriad roles of integrin interactions in the restoration of skin tissue structure and functions. As a corollary, the investigation into the integrin-mediated cellular mechanisms of delayed wound healing, scar tissue formation, and other cutaneous pathologies can be used to identify therapeutic targets for improved wound dressings and treatments.
4. Integrin-mediated Angiogenesis
Angiogenesis, the growth of new blood vessels from pre-existing blood vessels, occurs throughout development, regeneration, and disease. It is a vital process in supplying tissue with necessary oxygen and nutrients for survival.[154, 155] Angiogenesis involves four phases: stimulation and basement membrane breakdown, sprouting, tube formation, and maturation, Figure 6.[156] Each of these phases is associated with changes to the extracellular matrix and a corresponding integrin-mediated cellular response, Figure 7. In their quiescent state, endothelial cells adhere to a basement membrane consisting of laminins and collagen type IV through integrins α1β1, α2β1, α6β1 and α6β4.[10, 157, 158] Integrins α1β1, α2β1, αvβ3, α9β1 have been implicated in various cellular responses to proangiogenic factors, thereby stimulating basement membrane breakdown and other key angiogenic processes.[34, 159, 160] Basement membrane breakdown reveals collagen and laminin cryptic sites supporting initial migration.[10] In addition, the breakdown of the basement membrane exposes an interstitial/provisional ECM consisting of collagen type I, fibronectin, fibrinogen/fibrin, and vitronectin that promote sprouting and subsequent tube formation.[155, 157, 161, 162] The interstitial/provisional ECM composition presents new ligands for the involvement of integrins α1β1, α2β1, α3β1, α5β1, αvβ3, and αvβ5.[156, 163–165] Interstitial collagen, fibrin, and laminin are shown to support tube formation implicating α2β1, α5β1, α6β1, and αvβ3.[156, 166–169] During maturation, the ECM basement membrane begins to recover its composition through collagen type IV assembly and laminin deposition along the sprout with pericyte reassociation.[170, 171] As such, a new set of ligands are again presented with respective integrins α1β1, α2β1, α4β1, α6β1, and α6β4.[10, 21, 42, 157, 158, 170] A full list of these integrins with their respective protein ligands is provided in Table S3. Overall, these changes in the ECM through the phases of angiogenesis highlight that integrin signaling is dynamic through this process and can be targeted to mitigate cellular responses. This section provides an overview of the identified roles of integrins in each phase of angiogenesis and highlights the potential for integrin targeting strategies to promote vascularization of biomaterials.
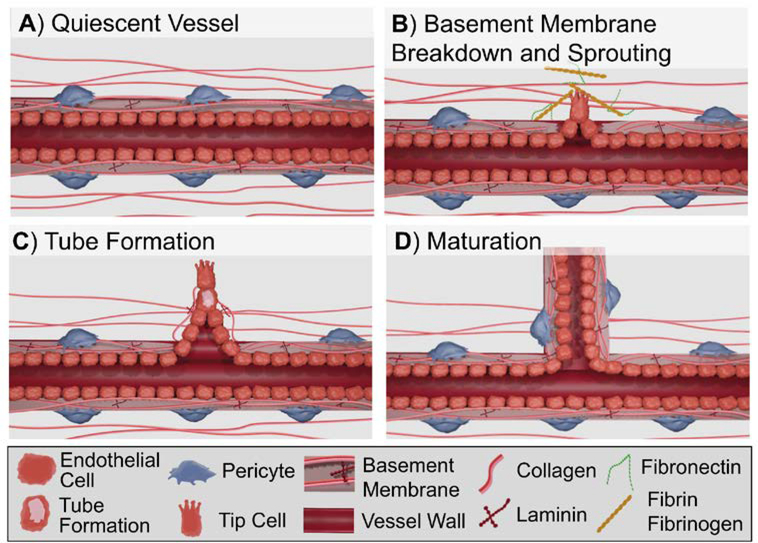
Figure 6:
Overview of key cell processes in angiogenesis: (A) Vessel in non-proliferative quiescent state; (B) Stimulation by proangiogenic factors results in basement membrane breakdown and sprouting; (C) Vascular tube formation via vacuole formation; (D) Pericyte reassociation and reformation of the basement membrane stabilizes the newly formed vessel.
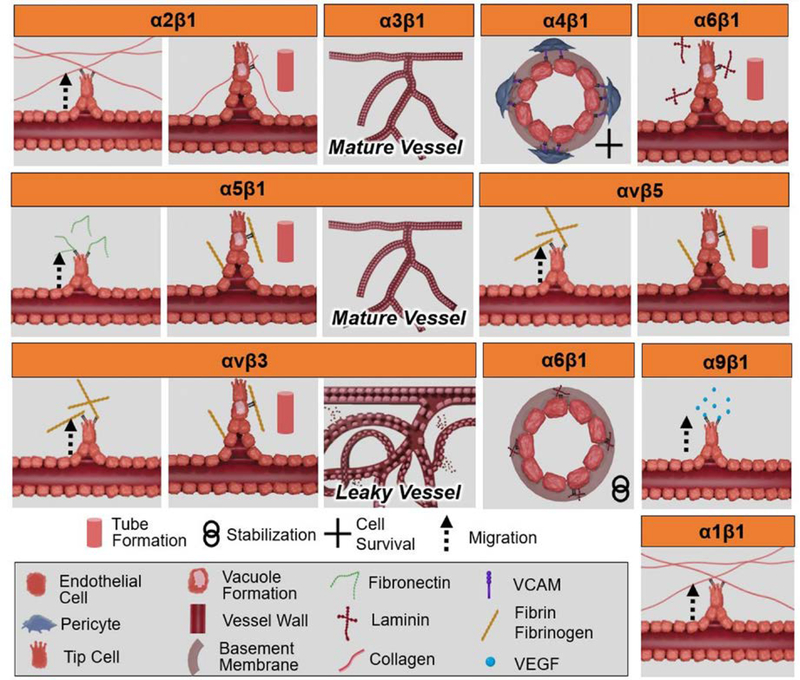
Figure 7:
Graphic illustration of integrin-mediated cellular processes in angiogenesis. Integrins play a critical role in each phase of angiogenesis: α2β1 promotes endothelial migration and tube formation; α3β1 represses pathogenic angiogenesis and forms mature vessels; α4β1 supports pericyte-endothelial interaction during angiogenesis supporting survival; α6β1 promotes tube formation; α5β1 promotes endothelial migration, tube formation, and the promotion of mature vasculature; αvβ5 promotes endothelial migration and tube formation; αvβ3 supports endothelial migration and tube formation, but has been associated in the development of an immature and leaky vascular network; α6β1 supports vessel stabilization; α9β1 promotes endothelial cell migration; and α1β1 promotes endothelial migration.
4.1. Stimulation and Basement Membrane Breakdown
Angiogenesis is initiated and sustained through proangiogenic signals, such as vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF). [161, 172] Additional proangiogenic factors include tumor necrosis factor-α (TNF α), transforming growth factor-β (TGF β), nitric oxide, hypoxia-inducible factor, angiopoietin-1 (ang-1), angiopoietin-2 (ang-2), and PDGF.[161] Proangiogenic factors interact with a variety of cellular receptors to activate endothelial cells from their quiescent state resulting in the production of matrix metalloproteinases (MMPs), migration, and proliferation with each functioning toward the formation of a new vascular network.[172, 173] Recently, integrins, such as αvβ3 and α9β1, have been shown to aid in this process either directly or in cooperation with growth factor receptors, thereby supporting the induced angiogenic response.[34, 159, 174] α9β1 has been shown to directly bind VEGF-A. Antibody blocking of the integrin resulted in the inhibition of VEGF-A binding and subsequent VEGF induced angiogenesis.[34] Additionally, VEGFR2 phosphorylation is improved when endothelial cells are plated on αvβ3 ligands, such as vitronectin and fibrinogen, with colocalization of the receptor and integrin suggesting a collaboration between the receptor and integrin.[159, 174–176] Additional studies collaborated the cooperation showing reduced responses to VEGF via antibody blocking of αvβ3.[177] Stimulation by proangiogenic factors results in alternations in the expressions of integrins, such as α1β1, α2β1, αvβ3, and α5β1 and α6β4, and the expression of MMPs thereby facilitating membrane breakdown and subsequent angiogenic processes. [178–181]
The breakdown of the basement membrane has been noted as a necessary step in angiogenesis, allowing for subsequent endothelial migration.[182, 183] MMPs degrade the basement membrane exposing cryptic sites, reveal an interstitial matrix that directs migration, participate in ECM remodeling, and release sequestered proangiogenic factors.[182, 184, 185] Additionally, MMPs help to detach pericytes from the basement membrane and cleave endothelial cell-cell adhesions.[161] The make-up of the basement membrane being primarily laminin and collagen type IV implicate MT1-MMP, MMP-2, MMP-9, for its breakdown, although others are likely to play a role.[161, 182, 186, 187] MMP-2 and MMP-9 are known to be able to break down collagen type IV and laminins, whereas MT1-MMP breaks down laminins and participates in MMP-2 activation.[182] Various integrins have been implicated for several roles with MMPs. Integrins α2β1 and αvβ3 have been connected in MMP-2 activation, synthesis, and directed function.[184] Shed vesicles containing MMPs may bind the basement membrane/ECM via β1, potentially facilitating localized degradation.[186] Additionally, MMP-2 is found to localize to the endothelial surface via αvβ3 in a functionally active form allowing for directed degradation. [188] The importance of this cooperation has been detailed through blockage studies of MMP-2 binding to αvβ3, resulting in inhibition of cell mediated collagen type IV degradation and invasion. [189] Basement membrane breakdown by MMPs and endothelial activation results in hyperpermeability of the vessel allowing for leakage of the blood plasma proteins fibrinogen, vitronectin, and fibronectin. [190] These proteins contribute to the provisional/interstitial matrix in which endothelial infiltration occurs.[191]
4.2. Sprouting and Organization
The breakdown of the basement membrane and activation of endothelial cells are critical to the subsequent sprouting phase of angiogenesis. Degradation of the basement membranes results in the loss of collagen binding sites recognized by various integrins such as α2β1 and exposure of αVβ3 cryptic sites, thereby supporting endothelial migration.[10, 41, 192] Basement membrane breakdown facilitates exposure to an interstitial ECM and deposition of blood borne proteins that constitute the interstitial/provisional ECM. The interstitial/provisional ECM is composed of collagen type I, fibronectin, fibrinogen/fibrin, and vitronectin, resulting in integrin transduction promoting subsequent cell infiltration/migration and tube formation. [155, 157, 161, 162] As noted previously, the activation of endothelial cells and exposure to new ligands results in the upregulation of integrins α1β1, α2β1, αvβ3, and α5β1 and down regulation of others such as α6β4 that allow for endothelial migration. [178–181] Continued production of MMPs supports endothelial migration into the ECM. [193]
The loss of collagen type IV binding sites and exposure to interstitial collagen results in collagen-based haptotaxis into the interstitial matrix facilitated by α1β1 and α2β1.[181, 182, 194] Senger et al. demonstrated the haptotactic roles of α1β1 and α2β1 on collagen type I gradients and antibody blocking of one or both integrins that resulted in an individual migration reduction of ~40% and a combined migration reduction of nearly 90%.[160] α1β1 and α2β1 have also been shown to support VEGF stimulated chemotaxis with blockage of the integrins significantly reducing angiogenesis.[160, 181] In addition to the roles of the collagen integrins in endothelial migration, the α1 subunit may also contribute to sustaining endothelial proliferation during angiogenesis. The α1 subunit has been demonstrated to regulate the production of MMP-9, thereby mediating MMP derived angiostatin, an inhibitor of endothelial proliferation. [195, 196]
Although the impact of α9β1 in angiogenesis has not been as extensively depicted in literature, researchers have demonstrated its potential role in chemotaxis. Vlahakis et al. illustrated that α9β1 can directly bind VEGF-A and that antibody blocking of the integrin considerably reduced induction and migration to specific isoforms of VEGF-A. [34] This result details a potential role of α9β1 in mediating migration of endothelial cells via VEGF-A induced chemotaxis. Studies have also implicated the roles of α4β1 in migration and the stimulation of new vessels. Utilizing alginate with REDV modifications, Wang et al. demonstrated the angiogenic potential of the peptide sequence that is a ligand for α4β1. [197] Others have detailed similar results utilizing the synthetic peptide sequence. [198]
The α5β1 integrin is upregulated during angiogenesis in response to bFGF and serves numerous functions, including migration, vessel structuring, and endothelial cell survival. [179, 199] Studies have shown that exposure to the PHSRN sequence, present in fibronectin, upregulates α5β1 promoting invasion of the endothelial cells into the provisional matrix and upregulation of MMP-1. [200, 201] The expression of MMPs in the provisional/interstitial matrix aid in guiding the migrating and proliferating endothelial cells.[200] In addition to migration, α5β1 has demonstrated roles in the vascular structure. Utilizing α5 null mice, Francis et al. detailed distended vasculature and a loss of pattern complexity. [202] Similarly, utilizing hydrogels with recombinant fibronectin fragments that preferentially bound α3β1/α5β1, Li et al. was able to produce non-tortuous, organized and, non-leaky vessels compared to hydrogels that preferentially bound αvβ3. [203]
Compared to precise roles of other integrins, α3β1 has a much more ubiquitous impact on vascular development, function, and pathogenic angiogenesis. Utilizing conditional knockout α3 mice that were endothelial cell specific, da Silva et al. demonstrated enhanced tumor growth and tumor angiogenesis. They demonstrated this effect in an in-vivo tumor transplant and hypoxia-induced retinal assay suggesting the role of α3β1 in repressing pathological angiogenesis.[204] As noted prior, Li et al. illustrated a similar role of α3β1 in producing non-tortuous, organized and, non-leaky vessels.[203] These studies suggest α3β1 represses pathogenic angiogenesis and produces vasculature with proper structure and function.
Researchers disagree on the roles of αvβ3 and αvβ5 with reports of necessitated function while others report adverse vessel organization. αvβ3 and αvβ5 have been described as necessary to angiogenesis with antagonists resulting in the inhibition of cell migration and new vessel growth. [173, 178, 199, 205, 206] αvβ3 is found to localize MMP-2 on the surface of endothelial cells and support infiltration into the provisional matrix, thereby allowing for migration.[207] Researchers have demonstrated that endothelial migration on vitronectin, a provisional ECM component, is mediated by the integrin. [208] Additionally, it has been detailed that αvβ3 promotes proliferation and prevents apoptosis of endothelial cells.[209, 210] Conversely to supportive roles in infiltration and migration, αvβ3 has been implicated in adverse vessel patterning and function. For example, αvβ3 specific fibronectin materials result in networks that are tortuous, disorganized, and leaky. [203, 211]
Migration and proliferation are critical components to the growing vascular sprout and subsequent network. Integrin binding mediates these processes by binding to the matrix proteins presented. As shown, the composition of the matrix is vital in producing a normal and functional network, although further research in this area is needed. As angiogenesis progresses, two components of the sprout are present, the tip cell and the stalk. Endothelial proliferation and subsequent tube formation are noted to occur within the stalk. Tube formation is mediated by ECM composition and integrin binding.
4.3. Tube Formation
Endothelial tube formation occurs within the stalk of the endothelial sprout.[212] Two mechanisms in which lumen formation occurs have been described. The first occurs via pinocytosis, vacuole development, and vacuole merging thereby creating the vascular tube. [213] The second method proposes that the vascular lumen is shaped by hemodynamic forces.[212] A variety of integrins have been shown to mediate the process, including α2β1, αvβ3, α5β1, and α6β1 which suggests the roles of laminin, collagen, and fibrin.
Various models have depicted the role of α6β1 in tube formation. In vitro studies using antibodies against α6β1 have blocked differentiation into capillary tubes. [214] For example, Lee et al. showed that antibody blocking of α6β1 in Matrigel inhibited capillary morphogenesis that was otherwise shown to occur spontaneously without the addition of growth factors.[168] Similarly, Kubota et al. demonstrated that tubule formation proceeded with the addition of laminin to collagen type I gels, but was inhibited through the addition of antibodies to laminin.[215] Other studies have utilized the IKVAV laminin sequence to bind α6β1 and support tube formation.[216, 217] Tube formation by α6β1 is consistent with reports on the role of laminin in capillary formation.[15, 169, 218, 219]
In addition to laminin, collagen type I has been shown to mediate tube formation and is driven by the collagen receptor α2β1.[220] In contrast to studies showing that laminin is needed, Liu and Senger demonstrated endothelial capillary morphogenesis occurred on collagen type I substrates, but not on laminin I substrates. They detailed that Src and Rho activation initiates capillary morphogenesis by collagen type I that is dependent on β1.[221] Singh et al. similarly utilized a PEG-collagen type I hydrogel and was able to demonstrate tube formation of endothelial cells within the 3D matrix. [222] Sweeney et al. determined that tube morphogenesis required ligation of α2β1 to the collagen sequence GFP*GER, where P* is hydroxyproline. [163] Furthermore, binding of α2β1 to a collagen matrix was shown to mediate pinocytosis that lead to intracellular vacuoles. The vacuoles subsequently combine to form a lumen thereby driving tube formation. [166]
The roles of αvβ3, αvβ5, and α5β1 have been investigated due to their roles in binding the provisional matrix. Studies have primarily utilized fibrin matrices for their investigations with blocking of the integrins to determine their roles in lumen formation. However, they appear to have overlapping roles in the process. Lauren et al. showed that only by simultaneously blocking both integrins is there a substantial reduction in tube formation on fibrin matrices. [167] The RGD sequence of fibrin has been shown to drive lumen formation and is dependent on both αvβ3 and α5β1.[223] Other studies have also detailed the roles of αvβ3, αvβ5, and α5β1 in tube formation, although α5β1 potentially plays a larger role. [173, 223, 224] For example, studies using α5 null mice have depicted the role of the α5β1 integrin in regulating tube diameter. [202]
The tube formation process may be mediated by multiple integrins. However, the above studies may also indicate that the regulation of morphogenesis through integrins is dependent on the matrix utilized. Depending on the integrins that are targeted by the matrix, the resulting vasculature can present with a normal and functional network, whereas others may promote an altered network in both appearance and functionality. Thus, further investigation is warranted to delineate integrin-mediate tube formation in biomaterials.
4.4. Maturation and Stability
Following tube network formation, the maturation process proceeds with pericyte reassociation and basement membrane deposition, which facilitates vessel stabilization. Pericyte recruitment along the abluminal wall is primarily cytokine driven, such as endothelial produced PDGF-B.[225, 226] The pericyte-endothelial cell interaction mediated by α4β1 is critical during angiogenesis and supports survival, whereas antagonists of α4β1 result in apoptosis of both cell types.[21] The association also drives the assembly of the basement membrane by endothelial cells that may be facilitated through CCN2 binding mediated by α5β1 and α6β1. [42, 227] Integrins α5β1, α3β1, and α6β1 that recognize the remodeled matrix are upregulated further supporting maturity of the vessels.[42] Additionally, as α6β4 is present on quiescent endothelial cells, it is logical that the assembly of a new basement membrane rich in laminin and maturing of the vessel would result in the re-expression of α6β4.[180] The combined results of pericyte reassociation, basement membrane deposition, and cell-ECM interactions provide stability to the vessel.[42, 228]
In summary, research is still unraveling the interplay between integrins and ECM ligands in angiogenesis. It has been demonstrated that integrins are heavily involved in the four stages of angiogenesis that include basement membrane breakdown, sprouting, tube formation, and maturation. As such, these integrin-mediated processes are design targets to promote neovascularization in tissue engineering constructs, a grand challenge in the field and critical to the success in developing large tissue grafts. Notably, research in this area can also be used to advance research to inhibit angiogenesis as a means to limit tumor growth or other disease states.
5. Integrin-targeting Biomaterials
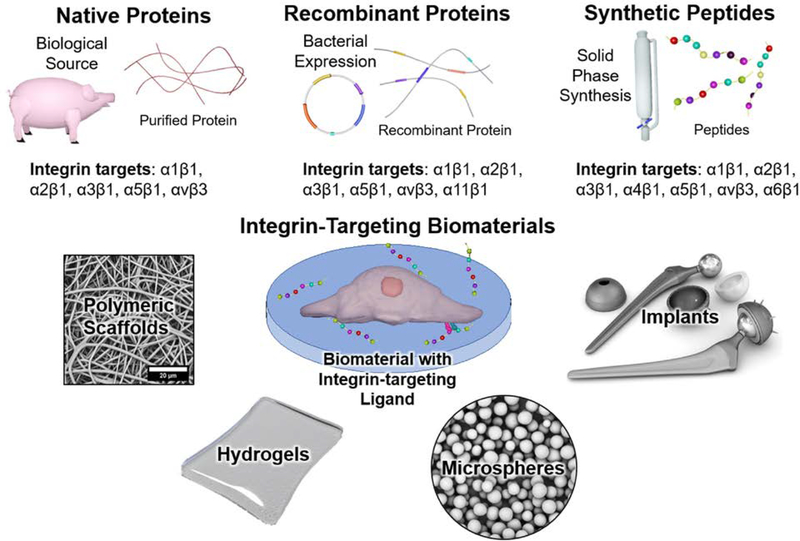
Integrin-targeting biomaterials have recently garnered attention as a method to enhance the regenerative capabilities of materials by taking advantage of the integrin-mediated processes described above. Researchers have shown the ability to direct the integrin-mediated processes through ligand presentation in a variety of biomaterials, Figure 8. The first generation of biomaterials utilized native proteins obtained from natural sources, such as bovine or porcine collagen. However, the lack of specificity and batch variability led to later generations of materials with recombinant proteins or peptide sequences identified from binding motifs. Each of these ligand presentation methods has distinct advantages and disadvantages that stem from their production process, material incorporation, and specificity. The following section will provide a comparative analysis of ligand selection in biomaterial design and example applications to illustrate the potential of integrin-targeting biomaterials.
Figure 8:
A schematic of different integrin-targeting ligand sources and biomaterial matrices used for ligand presentation.
5.1. Native Proteins
Native ECM proteins have been used to modify synthetic biomaterials or directly serve as scaffolds to promote targeted integrin-mediated cell responses that enhance tissue regeneration. By breaking down dissected bovine, porcine, marine, or murine tissue, ECM proteins are extracted with enzymes or solvents. [229–232]The proteins are then precipitated or reconstituted with solvents, such as trichloroacetic acid and acetone. [233] Consequently, these tissue-derived products maintain the full length and most functionalities of the native protein. In our previous sections, we identified several ECM molecules that interact with the integrins involved in regenerative processes, including collagen, fibronectin, laminin, and elastin. The key integrins that bind to collagen include α1β1 and α2β1, while fibronectin regulates the cell behaviors by mainly interacting with α5β1 and αvβ[234]3 and elastin contains ligands for αvβ3.[8, 11, 25, 97, 115, 137] Additionally, gelatin is obtained as a hydrolysis product of collagen and able to trigger integrins α5β1 and αvβ3-mediated signaling via RGD-containing motifs. [235, 236]
Synthetic biomaterials, including polymers, metals, or ceramics, can be modified or engineered with tissue-derived proteins by physical absorption, encapsulation, or surface coating by covalent conjugation.[237–243] ECM proteins can also be fabricated into hydrogels, meshes, and matrices via self-assembly or electrospinning, independently or with other synthetic polymers. [244, 245]For instance, collagen hydrogels are prepared by neutralizing collagen solutions as collagen fibrils self-assemble into bundled fibers at neutral pH.[246–252] Here, applications of extracted ECM proteins and decellularized scaffolds will be discussed in the scope of tissue regeneration, focusing on the integrin-protein interactions and regenerative outcomes.
Collagen-based matrices and hydrogels have been utilized to promote different regenerative processes. For wound healing, a collagen-chitosan scaffold loaded with adipose-derived mesenchymal stem cells (aMSCs) promoted aMSC differentiation into keratinocytes, resulting in the reconstruction of dermis and epidermis. [253] Other types of protein-based biomaterials have enhanced fibroblast and keratinocyte proliferation or migration, including fibroin/elastin matrix, electrospun fibronectin meshes, gelatin/PCL-coated polyurethane dressing, and alginate-gelatin crosslinked hydrogel. [22, 23, 254, 255] These biomaterials showed potential in accelerating re-epithelialization and wound closure via interactions with different integrins.
Collagen-based matrices also improved capillary and endothelial cell morphogenesis via interactions with α1β1 and α2β1, stimulating the formation of new blood vessels. [220, 256] Laminin is a main component of the basement membrane and plays a central role in angiogenesis and re-epithelialization, as mentioned before. It was observed that in a collagen-based matrix, laminin improved angiogenesis by promoting the expression of integrin subunit α6, which led to a significant increase in VEGF uptake by ECs, resulting in stimulated EC network aggregation. [169] Besides, fibrin is essential to understand the mechanism of blood coagulation, serving as a critical component of the blood clot. [257] Therefore, researchers have focused on improving angiogenesis with fibrin. A fibrin-based matrix developed by Hall et al. demonstrated that the fibrillar structure and RGD-containing binding sites facilitated the angiogenetic activities of endothelial cells, such as adhesion and spreading on the substrate. [258]
In bone regeneration, collagen remains a common design choice as the major insoluble fibrous protein in bone ECM. Salaszynk et al. demonstrated that stem cells cultured on well plates coated with collagen, vitronectin, and fibronectin had increased calcium deposition compared to the tissue culture polystyrene control, as well as increased protein levels including ALP, osteocalcin, and osteopontin. [78] This result suggested these whole proteins can individually or synergistically promote osteogenesis, presumably via regulation of integrins α1β1, α2β1, α5β1, and αvβ3. Tissue-derived proteins remain one of the most commonly used means of introducing bioactivity to synthetic materials.
5.2. Recombinant Proteins
The first recombinant protein approved for commercial use was human insulin in 1982. [259] Since then, recombinant proteins have been used for various biomedical applications such as drug delivery, therapeutics, and, more recently, tissue engineering. Advances in recombinant DNA technology has led to the production of recombinant fibrous proteins such as collagen-like proteins and fibronectin fragments in prokaryotic systems.[25, 260] To produce the recombinant proteins, the desired gene is amplified by PCR and then inserted into a cloning vector for expression in prokaryotes. For example, plasmids, circular, double-stranded DNA molecules, are most commonly used in the Escherichia coli (E. coli) expression system. After expression, the proteins are isolated and the structure verified using Western blot or similar assays.[261] Unfortunately, it remains challenging to generate a recombinant collagen due to the expensive post-translational modification needed to achieve its native triple helical structure. The (Gly-Xaa-Yaa) repeating sequence is stabilized by post-translational modification with hydroxyproline (Hyp) and this triple helical structure is required to maintain integrin binding affinity.[262–264] Although there is superior design control with recombinant proteins, there are few available options due to the costs and challenges associated with protein engineering and expression. Streptococcal collagen-like protein (Scl-2) and recombinant fibronectin (rFN) fragments are recombinant proteins expressed in E. coli systems for investigation as integrin-targeting proteins.[25, 265] Scl-2 variants are used to target α1β1 and α2β1 in tissue engineering applications, while other variants of Scl-2 have been developed to target other integrins such as α11β1. [74, 266] Several variations of rFN were developed to target α3β1, α5β1, and αvβ1. [203, 267] The integrin specificity of these recombinant proteins have been investigated in various tissue regeneration applications.
The recombinant Scl-2 protein serves as an alternative for collagen with improved integrin specificity. Despite the absence of Hyp and a shorter triple helix domain than human type I collagen, Scl-2 maintains the triple helix structure at physiological temperatures like human collagen.[262] Scl-2 is a “biological blank slate” with no known native binding sites.[265] This allows Scl-2 bioactivity to be readily customizable by site-directed mutagenesis to introduce different peptide sequences for specific integrin binding. [268] Scl2 proteins have been modified to contain the peptide sequences GFPGER based on the GF/LOGER sequence (O; hydroxyproline) identified in collagen that interacts with α1β1 and α2β1. Molecular modeling was used to identify a novel and selective integrin-binding sequence, GFPGEN, with specificity for the α1 I-domain over the α2 I-domain. Several studies have demonstrated that Scl2 proteins modified to contain these integrin-binding sequences support cell-specific binding, spreading and migration. [129, 265, 269]
Unlike native fibrillar collagens, these Scl proteins do not form stable networks and must be conjugated or adsorbed to synthetic matrices for use in tissue engineering. Scl-2-GFPGER functionalized hydrogels were used as a tool to promote luminal endothelial cell adhesion in vascular grafts. [265] Cereceres et al. demonstrated that Scl-2-GFPGER enhanced fibroblast adhesion and cell proliferation in a wound dressing application.[129] Two strains of Scl-2 were evaluated for the potential to promote osteogenic differentiation. Hydrogel-encapsulating hMSCs with Scl-2-GFPGER and Scl-2-GFPGEN demonstrated an increase in expression of an osteogenic marker in vivo. [74]
rFN fragments are composed of discrete fragments of whole fibronectin. Whole fibronectin contains three types of modules. Specifically, type III contains several binding domains that have different functionalities. For instance, III10 contains the RGD sequence and PHSRN sequence located within III9. [270, 271] III12 thru III14 has been shown to bind to several growth factors, including VEGF and BMP. [267] rFN can be customized to target specific integrins by selecting which domains are included in the recombinant protein fragment. For example, rFN III9–10/12–14 contains both III9–10 and III12–14 to promote cell adhesion and growth factor binding. [267] The flexibility of these recombinant proteins allows for diverse applications in tissue engineering.
rFN has also been shown to be a driver for integrin-mediated tissue regeneration. rFN III9–10 hydrogels with recombinant fibronectin targeting α3/α5β1 with VEGF promoted non-tortuous blood vessel formation and non-leaky blood vessels in vivo. Hydrogels with either rFN III9*−10 (structurally stabilized with (Leu1408 to Pro)) or rFN III9–10 with a flexible linker between the domains of recombinant fibronectin targeting α3β1/a5b1 promoted non-tortuous blood vessel formation and non-leaky blood vessels in vivo. [203] Hydrogels with rFN III9–10/12–14 and growth factors increased bone volume and skin wound healing compared to growth factors alone or rFN III9–10/12–14 alone in mouse models in vivo. [267] rFN III9*−10 increased ALP expression in a fibrin matrix compared to fibrin alone with hMSCs. [25] Furthermore, Agarwal et al. found that rFN7–10 coated screws promoted bone fixation and bone implant ingrowth compared to uncoated screws in vivo. [268] These studies demonstrate the potential of recombinant proteins to improve the healing potential of biomaterials.
5.3. Synthetic Peptides
Peptides must be carefully designed to promote ligand-integrin interactions as observed in native cell-ECM protein interactions. These short fragments are heteropolymers of amino acid residues linked together with peptide bonds. There is no distinct definition of chain length, but peptides typically range in the number of residues from 2–50 amino acids. [272] Synthetic peptide fabrication and use was first developed in the 1960s.[273] There are several methods to produce peptides including chemical synthesis, enzyme technologies, extraction from natural sources, and recombinant DNA technology.[274] The following discussion will focus on the solid-phase peptide synthesis where the peptide chain is anchored to a matrix and elongated via addition of amino acids linked by peptide bonds between the carboxyl and amino groups. [275] The general procedure for this synthesis includes anchoring to the matrix, coupling of amino acids, and cleavage of the fragment for release.[272] In addition to the specific integrin-binding motif, the peptide sequence can be designed to present requisite tertiary and quaternary structures (e.g. triple helix), include conjugation sites, and optimize spacing for binding affinity. Biomaterials research with synthetic peptides demonstrates the ability to target a range of different integrins including a1β1, α2β1, α5β1, αvβ3, α6β1, α4β1, α3β1, and α6β1that diversify the biological responses that can be achieved.
Physical or chemical incorporation of these peptides has been used to confer bioactivity to a range of substrates including hydrogels, microspheres, meshes, and metal implants. Briefly, physical incorporation includes methodologies that cause peptides to be either adsorbed or precipitated on to the substrate. These techniques include coatings, precipitates, or self-assembly of peptides. [272, 276, 277] Chemical conjugation of the peptide to the matrix involves a direct reaction with the peptide such as acrylate conjugation, click chemistries, or other coupling mechanisms. [276] [278] Given the diversity in techniques, significant considerations must be given to how efficiently the peptide is immobilized to the substrate. There are several factors to consider when deciding between the two techniques including adsorption efficiency, surface area of biomaterial to retention efficiency, as well as efficiency of the modification reaction, volume of inclusion, accessibility of the peptides, and conformation. [278]
Peptide design for biomaterial modification should be based on the desired cellular response, the integrin(s) implicated in the response, and the ECM protein/peptide motif that both targets the integrin and elicits the response. For example, laminins are a protein component of the ECM that facilitate numerous integrin-mediated cellular responses in angiogenesis and wound healing. In angiogenesis, IKVAV from the laminin α1 chain has been demonstrated to promote various functions within angiogenesis and has been utilized in a myriad of constructs. For instance, Chen et al. conjugated a SIKVAV sequence to a chitosan hydrogel and demonstrated enhanced angiogenesis. [279] Similarly, Nakumara et al. immobilized the IKVAV sequence in a collagen membrane and demonstrated endothelial tube formation. [216] This sequence has been shown to promote angiogenic cellular responses via interactions with the integrins αvβ3, α3β1, and α6β1. [12, 217, 280] However, the interplay of the integrins with the IKVAV sequences and the corresponding responses require further elucidation.
The laminin sequences SIKVAV, YIGSR, and A5G81 have shown analogous capacities for wound healing. The SIKVAV sequence covalently bonded to an alginate dressing demonstrated enhanced re-epithelialization and regeneration of tissue compared to controls. (Hashimoto, 2004). As previously mentioned, the integrins αvβ3, α3β1, and α6β1 have been shown to bind to the IKVAV sequence; however, non-integrin binding receptors for the sequence may exist and therefore further research is needed. [12, 217, 280, 281] Zhu et al. demonstrated the hydrogel immobilized A5G81 laminin-derived dodecapeptide sequence accelerated both dermal and epidermal cell proliferation and faster tissue regeneration mediated by the α3β1 and α6β1 integrins. [282] Salber et al. demonstrated that the YIGSR sequence from residues 929–933 of the laminin β1 chain enhances keratinocyte and fibroblast proliferation and the upregulation of α6 integrin in a type I collagen construct. [28] The studies demonstrate the promise of using these peptides in promoting wound healing.
In addition to laminin sequences, collagen motifs have also been shown to direct numerous functions and as such been applied in peptide engineering. The first class of these peptides include collagen-mimetic peptides which target the α2β1 integrin. The GFOGER hexapeptide sequence is found on residues 502–507 α1(I) chain of type I collagen. This sequence is often included in a longer synthetic triple-helical peptide, mimicking the triple helical structure of type I collagen, to permit integrin binding.[283] In vitro studies with passive adsorption and surface modification of meshes with GFOGER enhanced cell migration, proliferation, and differentiation down the osteoblastic lineages. [64] In vivo, the use of this peptide has produced promising results for bone regeneration as well. Incorporating the peptide inside a hydrogel system improved encapsulated cell engraftment and increased bone volume in radial segmental defects of immunodeficient mice. [66] In separate studies, passive adsorption on PCL discs and titanium implants promoted bone formation in critical size defects solidifying the strong potential of GFOGER for bone healing applications. [73, 284] Other collagen mimetic peptides include DGEA and P15 – both target the same integrins that natural collagen acts as a ligand for, but P15 lacks the RGD domain, and is dependent on the GIAG residue for successful binding to cells. [283] All three of these peptides have demonstrated promotion of cell adhesion on scaffolds, cell spreading, osteogenic differentiation, and in the case of P15 peptide, good osseointegration in animal models. [285, 286] On the wound healing side, a unique collagen-mimetic peptide (GEFYFDLRLKGDK) was functionalized on PDMS constructs resulting in enhanced keratinocytes and fibroblasts migration and proliferation. [28] Despite the success of collagen-mimetic peptides in promoting osteogenesis and wound healing, the use of these peptides in improving angiogenic outcomes is limited and needs further investigation.
A separate class of popular peptides includes the RGD-based sequence. This sequence is found in several different ECM molecules including fibronectin, vitronectin, and bone sialoprotein. [56, 287] RGD-based peptide studies have shown promising results in multiple areas including osteogenesis, wound healing, and angiogenesis. Choi et al. demonstrated that coating an elastin-like polypeptide with RGD accelerated wound closure, re-epithelization, and upregulated expression of dermal tissue. Similarly, Yu et al. demonstrated incorporating RGD into an alginate-based hydrogel enhanced angiogenesis. [288] Lastly, to improve osteogenesis, several different authors were able to show that modifying PLGA films or microspheres with RGD peptides improved MSC attachment, proliferation, and osteoblastic differentiation. [289–291] One note to address here is the observation that the RGD sequence is promiscuous - multiple integrins can bind this motif sequence including α5β1, αvβ1, αvβ3, and αvβ8. In order to target a specific integrin pair for example α5β1, secondary peptides with the sequence PHSRN must be incorporated along with RGD, as this synergy helps with targeted attachment of α5β1. [292]
The last class of peptides have no biological similarity to ECM proteins and include peptides that have been generated with artificial sequences of amino acids that can promote integrin priming using phage display. This technique is a highly efficient selection methodology in which a library of peptide sequences or variants is expressed on a bacteriophage coat protein. [293] Each variant is then evaluated for binding specific integrin targets. One example of a peptide sequence isolated using this technique is RRETAWA. This sequence binds the α5β1 integrin with very high affinity but bears no homology or similarity to any amino acid sequence found in ECM proteins. [294, 295] Within this peptide, the β1subunit has been shown to have strong interactions with the Arg-Arg-Glu motif while α5 interacts with the hydrophobic Trp residue. In addition to the original study, other studies have also confirmed RRETAWA’s ability to improve osteogenic phenotypes after incorporation in a biomaterial system. Gandavarapu et al. incorporated cyclic RRETAWA in hydrogels and the results demonstrated a highly specific interaction with the α5β1 integrin based on attachment studies. Further, MSCs successfully differentiated and maintained key markers of osteogenic differentiation. [294]
5.4. Design Considerations of Integrin-targeting Biomaterials
The different types of ligands available presents biomaterials researchers with a large selection of options for designing integrin-targeting materials. Each ligand has a different source and production method that affect integrin-targeting biomaterial design due to feasibility, scaling, and expense of technique. Considering the growing understanding of the effect of biomaterial physical cues on cell behavior, it is critical to delineate the individual and synergistic effects of integrin interactions and material properties on the desired outcome. As such, researchers must consider not just what ligand is presented but also how the full biomaterial landscape affects cell behavior. In the following section, we will discuss key design features of materials from ligand production to material physical cues that should be considered to effectively harness the integrin-mediated cell processes to enhance regeneration.
After identifying the target integrin and corresponding ligand, researchers first select the method of production that is dependent on both the selected ligand and target application. There is a long history of isolating proteins from mammalian or marine animal sources and the production and purification methods have been optimized to allow for large-scale production of these native proteins. Unfortunately, extraction and purification procedures, such as the use of solvents, can result in the altered conformation of the proteins, diminishing bioactivity, and shortening shelf-life. Further, due to biological variability in the source and isolation methods, there can be batch-to-batch variability. Finally, tissue-derived proteins and decellularized tissue commonly maintain the full tertiary and quaternary structures post isolation. With this full structure, an important point to consider is that whole proteins can have off-target biological responses as they usually contain multiple integrin-binding sites. These native proteins also offer little opportunity to optimize ligand specificity/affinity or conjugation strategies. Recombinant protein expression and solid-phase peptide synthesis for ligand production can address some of these concerns. As mentioned above, recombinant protein production is carefully engineered and heavily relies on vector expression and purification from bacterial systems.[265, 267] This method also uses well-established molecular cloning techniques and knowledge of bacterial reproduction that can facilitate iterative design and scale-up. As compared to isolation from tissues, protein purification of recombinant proteins is relatively mild with fewer resulting effects on protein conformation and improved batch consistency once optimized. A unique feature of recombinant expression is the ability to optimize ligand presentation and properties through protein engineering strategies such as site directed mutagenesis. [296] These proteins, including Scl-2 and rFN, allow for designated insertion of integrin binding sites so they can be customized to target specific integrins, allowing improved control over cell behavior and thus tissue healing outcomes.[265] However, there are few recombinant expression systems that have been developed for integrin-targeting proteins so the number of available engineered proteins is limited. Similar to the design control of recombinant expression, the direct control of amino acid sequence in solid-phase synthesis of peptides also enables systematic refinement of their structure and precise manipulation.[278] In addition, this specificity can also contribute to understanding and tuning the magnitude of the cellular response observed. The peptide sequences provide more consistent bioactivity owing to the sequences being rather short and thus having limited folding capability that may impact bioactivity. Although this attribute can be considered an advantage, it may also limit the method as certain sequences, such as GFOGER, need to be presented in a specific conformation. [73] Thus, the lack of conformational ability of peptides, primarily due to their short sequences, can be a significant downside. However, significant advancements have been made in increasing synthesized peptide length, such as backbone protection and heating that are pushing the limits above 100 residues. [297] Further, the motif utilized needs to be carefully considered as binding motifs may be a ligand for multiple integrins, such as the previously mentioned sequence of RGD. Regardless, even if the motif is ubiquitous and promotes binding of multiple integrins, the presentation of a single defined ligand limits the potential of non-specific integrin binding. On the contrary, this specificity can also be a hindrance to peptides as the ability to target multiple integrins may be desired. Compared to whole proteins and some recombinant proteins that may be able to interact with a wide variety of integrins, multiple peptide sequences would need to be employed in order to derive the same activity synergistically. This may dramatically increase the cost as multiple peptide sequences would need to be synthesized. Synthesis and purification protocols for peptides are well established and typically do not result in adverse effects on the bioactivity of isolated peptides. Despite these advantages, peptide synthesis remains costly making scalability difficult with cost increasing with peptide length.[278]
Lastly, potential adverse host responses or immunological reactions to these biomaterials should be considered. Although there is strong conservation of structural proteins across species, there is a growing concern of potential sources of immunogenicity of proteins isolated from non-human sources. Tissue-derived proteins and recombinant proteins isolated from biological systems may have immunogenic epitopes and potential lipopolysaccharide contamination, respectively.[298, 299] In contrast, synthetic peptides likely have little immunogenicity concerns due to the fabrication methodology and their synthetic nature. Thus, there are critical tradeoffs to consider when selecting a ligand production route and the need for specificity, scalability, availability, and cost must be weighed for each application.
In addition to the ligand source and specificity, there is a growing understanding of the effect of substrate properties on cellular behavior including seminal work showing that substrate modulus[300], surface morphology[301], and 2D versus 3D structure[302] modulate cellular responses in tissue regeneration. Given that integrins provide the physical link between the biomaterial and the cytoskeleton, it follows that integrins in these activated focal adhesion complexes are integral to the observed effects on mechanotransduction and mediate cytoskeleton tension.[300, 303] For example, MSCs cultured on matrices of high stiffness (e.g. metal implants, thermoplastics) maintain a large spreading area and enhance focal adhesions with a high level of osteogenic differentiation. Conversely, MSCs grown on low stiffness matrices (e.g. soft hydrogels) maintain relatively poorly defined actin cytoskeletons, leading to different phenotypes.[300] The modulus of substrates has also shown to impact the growth of fibroblasts and endothelial cells. Fibroblasts demonstrated preferential growth on a higher modulus hydrogel (13.7kPa), whereas endothelial cells had preferential growth on a low modulus hydrogel (0.3kPa).[304] Researchers have attributed the modulation of cell behavior via mechanical stiffness to integrin-mediated transduction pathways.[300, 305, 306] Additionally, Murikipudi et al. determined that substrate modulus can affect the expression of integrins and changes in downstream signaling.[307]
In addition to modulus, studies have shown that controlling surface topography and architecture can direct cellular responses.[301, 308–310] For example, Guvendiren et al. demonstrated how the introduction of lamellar surface wrinkling of hydrogels promoted stem cell differentiation down the osteogenic lineage. Changing the surface morphology of the hydrogels to hexagonal patterns caused the cells to remain rounded and differentiate into adipogenic lineage.[311] Topographical effects also extend to angiogenesis and wound healing. Bauer et al. demonstrated that aligned scaffolds could impact sprout extension speeds with alignment parallel to a VEGF gradient resulting in a greater speed compared to alignment that is perpendicular to the gradient. [301] Similarly, in dermal wound healing models, it was observed that differences in the topography such as widths in between nanogrooves or mesh size affected cell migration speed, cell phenotype, and ECM production. [309, 310] In addition to topography, the 3D structure of the matrix has been shown to influence stem cell differentiation and cell responses.[311] Additionally, architectural features have also been shown to impact integrin expression with 3D substrates markedly increasing integrin expression over 2D substrates.[302] It is hypothesized that 3D matrices improve cell attachment through increased surface area and provide a dynamic interface that occurs naturally in their native niche.[312] Researchers have also found that integrin clusters are larger with longer lifetimes in traditional two-dimensional assays as compared to more relevant three-dimensional environments.[313] This requires additional investigation in translating much of the current work completed in 2D to more relevant 3D systems.
Researchers have several tools to modulate stiffness, surface topography, and 3D architecture in integrin-targeting biomaterials. Protein-based biomaterials (e.g. collagen and gelatin) are typically chemically crosslinked to achieve desired structural and degradation profiles.[314–316] Modulating the degree of crosslinking is the most common method of achieving variations in substrate modulus. Given that crosslinking typically occurs through lysine residues, there is poor control of which crosslinking sites are reacted and this can interfere with integrin-binding. For example, Kishan et al. reported that increased crosslinking of gelatin matrices increased stiffness and degradation time but reduced cell adhesion.[314] Thus, it is often challenging to optimize both physical properties and integrin-binding in protein scaffolds. Synthetic materials offer a broader range of moduli from soft hydrogels (100s Pa to 100s kPa) to synthetic polymers (100s kPa to 10 GPa) and metals (10s to 100s GPa). There are a variety of chemical and physical methodologies for ligand incorporation including surface adsorption, chemical conjugation, and blending. Surface coating can be implemented with physical adsorption or covalent bioconjugation. Physical adsorption leads to short retention times of proteins on the material surface; whereas, covalent conjugation via a functionalized group may interfere with the protein conformation or block the integrin-binding sites.[269, 317, 318] Recombinant proteins can be designed to mitigate this effect by removing the conjugation residue (e.g. lysine) from proximity of the integrin-binding motif or adding in additional conjugation residues at terminal ends (e.g. cysteine).[129] However, this requires advanced protein engineering methodology that ensures both retention of desired protein structures and expression viability. Solid-phase synthesis of peptides offers more facile means to optimize binding affinity and conjugation strategies through direct design of the peptide sequence with rapid iterations.[278] Biomaterial fabrication method provides researchers with control of the topography and geometry.[319, 320] Hydrogels derived from synthetic and natural polymers have been studied in a variety of forms including cell-encapsulating hydrogels,[321] coatings[322], and porous formulations.[323] A variety of other processing methods can be used to generate a wide-range of geometries including non-woven meshes with electrospinning,[314] porous foams with freeze drying,[324] salt-leaching,[325] and emulsion templating,[326] and more complex architectures with 3D printing.[327] There are several excellent reviews that cover the relevant structures available and the corollary effects on cell behavior.[314, 328–330]
In summary, the design of integrin-targeting materials revolves around balancing the advantages and limitations of the selected ligand source and material physical properties. The availability and known responses of native proteins such as collagen is mitigated by its batch-to-batch variability, lack of design flexibility, and off-target interactions. New generations of materials include recombinant proteins and peptide sequences that provide improved integrin specificity and targeted cellular responses as compared to native proteins. Incorporation of ligands into synthetic materials provides researchers with greater versatility over the material physical properties with corollary synergistic effects on cell behavior.
6. Critical Analysis and Future Directions
Although several studies have already demonstrated the potential impact of integrin-targeting biomaterials in regenerative medicine, current research suffers from limitations that can affect outcomes and mitigate the potential impact of this technology. We previously discussed the design considerations and need for improved biomaterials to advance integrin targeting. In this section, we will discuss the fundamental studies needed to continue to unravel the complexity of integrin-ligand interactions as well as the influence of biological variables that can confound cell behavior testing and downstream outcomes.
6.1. Ligand presentation and integrin clustering
Beyond individual integrin-ligand binding, it is the clustering of integrins that strengthens and stabilizes cellular binding to the extracellular matrix and results in activated focal adhesions complexes and downstream signaling. As such, ligand presentation (density, spatial organization) can play an important role in integrin clustering and the resulting cellular response from these integrin-ligand interactions.[331] Traditional methods of functionalizing a biomaterial non-specifically decorate the surface with cell adhesive ligands or blend ligands into the bulk of the material. Although the global density of the ligands can effectively modulate the bioactivity of the surface, the random distribution of ligands only promotes occupancy of cell integrins rather than effectively promoting integrin clustering. Researchers are now investigating the use of multivalent ligands, where the ligands are grouped together in islands, to better promote integrin clustering for an amplified response. Karimi et al. has done an extensive review of the different strategies being studied to engineer biomaterials that present multivalent ligands including polymer bioconjugates, nanolithography patterning, and protein engineering approaches.[332] Nanoscale clusters on the surface can be generated by first synthesizing a multivalent bioconjugate with a star or comb polymer base and then using these to modify synthetic materials using standard processes.[333, 334] One of the disadvantages of this approach is the lack of spatial control when presenting the multivalent ligands. The ambiguity in spacing between clusters or number of ligands per cluster has motivated the development of nanopatterning for better control of the material surface chemistry. The primary methods used for nanopatterning include nanoimprint lithography and block copolymer micelle nanolithography.[332, 335] These techniques have enabled fabrication of surfaces with highly engineered spatial geometries and great versatility in the designs with promising results.[336, 337] Additionally, researchers have also utilized recombinant protein expression to generate engineered sequences that control the number of integrin binding sites within linear polypeptides or recombinant proteins.[74, 331] The ability to control the polypeptide length and binding site patterns enables similar control over spatial patterning to promote integrin clustering. In parallel to performing cell culture studies for these newer generation materials, biomaterial scientists are using computer modeling to understand effects of local versus global ligand densities and identify potential cell binding thresholds using parameters such as ligand dissociation coefficients.[331] Lastly, typical in vitro studies evaluate the effects of ligand concentration on a cellular length scale often with single cell densities. Another factor to consider is how that dynamic might change in vivo when the biomaterial interfaces with different cell types or processes such as protein adsorption that may hide the ligands, thereby changing integrin binding, clustering, and downstream signaling. As the science is elucidated, this understanding should be used to thoroughly evaluate the robustness of biomaterials to confirm their true potential for triggering desired cell responses. Some of the current studies in recent papers have been careful to address this point performing ligand vs. focal adhesion studies, but broader adoption of this practice is needed to meticulously evaluate the capabilities of biomaterials to elicit targeted cell responses.
Synergy of integrin-targeting with other biological compounds
One of the limitations of much of the in vitro integrin-targeting investigation to date is that it is done in isolation of other biological cues that are present during regeneration. It has been established that there is cross-talk between integrins and other biological compounds (e.g. growth factors). [338] After integrin-ligand binding, activated integrins cluster together and can recruit growth factors to amplify the signaling effects.[140] By potentially combining integrin-targeting biomaterials with growth factor delivery, these new biomaterials may be able to deliver smaller doses of growth factors along with integrin-ligand priming to push activation of cell responses to larger magnitudes.[339] This synergistic approach may provide ways to produce biomaterials with enhanced regenerative capacity. Growth factor therapy remains a promising approach for regenerative purposes but delivery and dosing remain a challenge. More often than not, supraphysiological dosing is required and this results in high risk as well as expensive treatment options. Therefore, a combination of integrin-ligand signaling with the use of growth factors may provide an improved regenerative strategy. Although this strategy has shown promise, there is limited understanding of how integrin-mediated signaling and growth factor signaling converge to affect the downstream cell behavior. Researchers have identified key pathways that are involved in integrin-mediated signaling including MAPK/ERK, Wnt, β-catenin, and RTK. [340, 341] Understandably, this biological phenomenon gets more complicated when considering the involvement of growth factors and certain pieces of the biology behind synergistic signaling still remain unclear. The potential strategy of using integrin targeting with growth factor release in biomaterial design requires more efforts in the area of fundamental biology to elucidate these synergistic mechanisms. Biomaterial scientists may not have the tools to parse out these biological contingencies and it is likely that progress in this area is dependent on continued efforts in the field of molecular biology. Regardless, the observed benefit from empirical studies highlight the promise of this synergistic approach.
6.3. Translation of In Vitro Findings to In Vivo Outcomes
Integrin-targeting materials must undergo in vitro and in vivo studies to evaluate their regenerative potential for future clinical use. Both methods are essential in understanding the bioactivity of integrin-targeting materials, but there are many considerations to be made for in vitro and in vivo models. Although in vitro studies are relatively cheaper and more cost-effective, the role of other cells, extracellular cytokines, and mechanical signals that could affect integrin binding and signaling are challenging to replicate. Another consideration is the effect of a controlled cell culture environment in vitro compared to the dynamic environment of in vivo models. It has been demonstrated that the pattern of integrin expression on cultured cells does not always mimic that of cells in tissues, which highlights the limitation of in vitro studies.[342, 343] One way that researchers have verified the integrins involved in cellular processes is by using antibody-blocking studies.[61, 71] Although this can be used to confirm initial cell adhesion, antibody-blocking studies cannot be used in long term experiments as their efficacy rapidly diminishes with time. Therefore, this method cannot be used as negative controls for experiments that evaluate angiogenesis, proliferation, and differentiation. A method to circumvent the limitations of antibody blocking is by developing synthetic peptides and recombinant proteins that lack integrin-targeting sequences as controls to investigate the role of integrin activations in longer-term studies. [73, 74] Despite these limitations, in vitro models have been successful in elucidating specific integrin activities in tissue regeneration and determining which biomaterials should be explored further in vivo.
In vivo studies address many of the problems found with in vitro studies but are faced with other challenges. Transgenic mice have been a useful tool in evaluating the role of integrins in biological processes in vivo through the induced ablation or deletion of the specific integrin genes which help elucidate the roles specific integrins play in tissue regeneration.[11, 33, 195] However, transgenic animal models can result in unknown downstream consequences and thus could give an inaccurate picture for the processes in an unmodified animal model.[344] Furthermore, the participation of native factors or cells is disadvantageous for in vivo studies as the specific integrin-mediated interaction may be countered or compensated. For example, it is difficult to determine whether experimental outcomes are caused by activating one type of cell or if the effects are being compounded or diminished due to integrin activation by multiple types of cells. Additionally, in vivo models have difficulties in tracing cellular activities, mapping of the cell phenotypes, and detailing the downstream signal cascades so direct cause and effect of the integrin-targeting materials can be challenging to determine. Although using these in vivo and in vitro models elucidates the interactions of integrin-ligand binding, their limitations should be considered when evaluating experimental outcomes.
6.4. Cell-based Biological Variability
Innate biological variability is another factor that must be considered when evaluating the ability of biomaterials to direct cell responses. If not considered, these biological variables may confound the results of experiments utilized to delineate the role of integrins and evaluate integrin-targeting materials. First, non-human cell sources can result in different outcomes due to variable phenotypes. Several researchers have shown that cell surface marker expression, including integrins, varies among species.[345–348] These differences should be taken into consideration as it could affect the magnitude of the cell response to ligand-presenting materials. High variability in trends for integrin-mediated processes may be observed when comparing data across species simply due to differences in expression levels of integrins. Similarly, intraspecies donor variability in human cell lines should be considered. Studies have shown how cell behaviors such as migration, proliferation, and differentiation can differ due to factors such as donor age or sex. For example, Phinney et al. and Zhukareva et al. demonstrated how physiological states and age of donors can affect in vitro stem cell proliferation as well as osteogenic differentiation capabilities. [349] This observation may also extend to MSCs involved in regenerative capacities for other processes like wound healing and cardiovascular applications. [350, 351] To the best of our knowledge, researchers have yet to publish studies evaluating effects of donor variability on migration or proliferative capabilities for other cell types like fibroblasts or endothelial cells. Regardless, this should be accounted for when evaluating the bioinstructive capabilities of integrin-targeted materials by observing cell-material interactions from cultures isolated from different donors to confirm trends with added rigor.
One last consideration is the homogeneity of cell cultures that are expanded and culture conditions used for in vitro studies. This specific inspection may be more significant for certain cell lines like MSC that can be a heterogeneous mixture of cells, resulting in variable osteogenic potentials. It has been noted that this difference can cause variable expression levels and patterns of specific osteogenic markers such as RUNX2 expression, alkaline phosphatase activity, and bone ECM proteins.[349] Differing osteogenic potentials can be further confounded due to different culture conditions (media, cell densities, time frames) that are used across different research groups, further impacting the levels of differentiation observed in studies.[343] All of these factors can culminate to affect how MSCs behave when they interact with ligand-presenting materials. To alleviate some of these concerns, verification procedures can be performed to limit the effect of these biological variables - new technologies now allow for isolation and expansion of more homogenous MSC cultures. Additionally, standard practices such as verifying cell populations using flow cytometry before use in studies are being implemented. It is important to note that the whole field should become more standardized in these practices, including other specific like culture conditions as well. Understanding these biological nuances can provide means to develop new tools for evaluating these biomaterials and improve their capabilities in the in vivo setting.
6.5. Concluding Remarks and Outlook
Tissue regeneration is a complex process that is partially orchestrated through interactions with the intricate and multifaceted extracellular matrix. This review highlights the role of integrin binding in mediating cellular responses at each stage of osteogenesis, angiogenesis, and wound healing. These three areas were selected as illustrative examples only. There is a growing body of research elucidating integrin-mediated cellular processes across numerous regenerative medicine applications. In addition, there is a growing understanding of the role of integrin interactions in medical device integration and host response. For example, we recently reviewed the roles of integrin and syndecan-binding in endothelial cell phenotype and hemostatic regulation. Identification of endothelium-substrate interactions that limit platelet aggregation and thrombosis is central to the design of improved biomaterials with long-term thromboresistance.[352] The design of integrin-targeting biomaterials is the key research hurdle to taking advantage of these key mediators of cell behavior in order to improve regeneration strategies. Research spans the use of native proteins, recombinant proteins, and engineered peptides with integration in a diverse set of substrates from conjugation to hydrogel matrices to adsorption coatings of titanium implants. Although current studies have already demonstrated the benefit of these strategies, critical consideration should be given to how variables in the ligand, substrate, and cell sourcing affect downstream outcomes. Additional research is needed to advance both our understanding of these factors and generate improved materials with optimized integrin-targeting and clustering. Overall, the introduction of integrin-targeting to material design provides numerous opportunities to enhance tissue regeneration and device integration while also providing new tools to probe the complex processes of tissue remodeling.
Supplementary Material
Acknowledgements
Funding was provided by the National Science Foundation, grant number 1822196, and National Institutes of Health, grant number R21AR076107.
Biography

Elizabeth Cosgriff-Hernandez is a Professor of Biomedical Engineering at The University of Texas at Austin and holder of the L.B. (Preach) Meaders Professorship in Engineering. She received a B.S. in Biomedical Engineering and Ph.D. in Macromolecular Science and Engineering from Case Western Reserve University. Her laboratory specializes in the synthesis of hybrid biomaterials with targeted integrin interactions and scaffold fabrication strategies including injectable foams, 3D printing emulsion inks, reactive and in-line blending electrospinning.
Footnotes
Conflict of Interest
Elizabeth Cosgriff-Hernandez reports a stakeholder interest in ECM Biosurgery which seeks to commercialize Designer Collagens based on the Scl2 protein.
Supporting Information
Supporting information is available from the WileyOnline Library or from the author.
References
- [1].Danen EH, 2013. [Google Scholar]
- [2].Plow EF, Haas TA, Zhang L, Loftus J, Smith JW, Journal of Biological Chemistry. 2000, 275, 21785. [DOI] [PubMed] [Google Scholar]
- [3].Stupack DG, Oncology (Williston Park, NY) 2007, 21, 6. [PubMed] [Google Scholar]
- [4].Kim S-H, Turnbull J, Guimond S, Journal of Endocrinology. 2011, 209, 139. [DOI] [PubMed] [Google Scholar]
- [5].Kim C, Ye F, Ginsberg MH, Annual review of cell and developmental biology. 2011, 27, 321. [DOI] [PubMed] [Google Scholar]
- [6].Bourdoulous S, Orend G, MacKenna DA, Pasqualini R, Ruoslahti E, The Journal of cell biology. 1998, 143, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ruoslahti E, Annual review of cell and developmental biology. 1996, 12, 697. [DOI] [PubMed] [Google Scholar]
- [8].Eble J, Golbik R, Mann K, Kühn K, EMBO J. 1993, 12, 4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pozzi A, Wary KK, Giancotti FG, Gardner HA, J. Cell Biol. 1998, 142, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stupack DG, Cheresh DA, J. Cell Sci. 2002, 115, 3729. [DOI] [PubMed] [Google Scholar]
- [11].Popov C, Radic T, Haasters F, Prall W, Aszodi A, Gullberg D, Schieker M, Docheva D, Cell Death Dis. 2011, 2, e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kikkawa Y, Hozumi K, Katagiri F, Nomizu M, Kleinman HK, Koblinski JE, Cell adhesion & migration. 2013, 7, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu C, Chung AE and McDonald JA, Journal of Cell Science. 1995, 108, 2511. [DOI] [PubMed] [Google Scholar]
- [14].Moursi AM, Globus RK, Damsky CH, Journal of cell science. 1997, 110, 2187. [DOI] [PubMed] [Google Scholar]
- [15].Strömblad S, Cheresh DA, Chemistry & biology. 1996, 3, 881. [DOI] [PubMed] [Google Scholar]
- [16].Moursi AM, Damsky CH, Lull J, Zimmerman D, Doty SB, Aota S.-i., Globus RK, Journal of cell science. 1996, 109, 1369. [DOI] [PubMed] [Google Scholar]
- [17].Choma DP, Pumiglia K and DiPersio CM, Journal of Cell Science. 2004, 117, 3947. [DOI] [PubMed] [Google Scholar]
- [18].Kumar G, Narayan B, in Classic Papers in Orthopaedics, (Eds: Banaszkiewicz PA, Kader DF), Springer London, London: 2014, 503. [Google Scholar]
- [19].Guan J-L, Hynes RO, Cell. 1990, 60, 53. [DOI] [PubMed] [Google Scholar]
- [20].Danussi C, Petrucco A, Wassermann B, Pivetta E, Modica TM, Del Bel Belluz L, Colombatti A, Spessotto P, J Cell Biol. 2011, 195, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Garmy-Susini B, Jin H, Zhu Y, Sung R-J, Hwang R, Varner J, The Journal of clinical investigation. 2005, 115, 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kim SE, Heo DN, Lee JB, Kim JR, Park SH, Jeon SH and Kwon IK, Biomedical Materials. 2009, 4, p.004106. [DOI] [PubMed] [Google Scholar]
- [23].Chong EJ, Phan TT, Lim IJ, Zhang YZ, Bay BH, Ramakrishna S and Lim CT, Acta biomaterialia. 2007, 3, 321. [DOI] [PubMed] [Google Scholar]
- [24].Sarker B, Singh R, Silva R, Roether JA, Kaschta J, Detsch R, Schubert DW, Cicha I and Boccaccini AR, PLOS one. 2014, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Martino MM, Mochizuki M, Rothenfluh DA, Rempel SA, Hubbell JA, Barker TH, Biomaterials. 2009, 30, 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Avraamides CJ, Garmy-Susini B, Varner JA, Nature Reviews Cancer. 2008, 8, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Heng EC, Huang Y, Black SA Jr and Trackman PC, J. Cell. Biochem. 2006, 98, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Salber J, Gräter S, Harwardt M, Hofmann M, Klee D, Dujic J, Jinghuan H, Ding J, Kippenberger S, Bernd A and Groll J, Small. 2007, 3, 1023. [DOI] [PubMed] [Google Scholar]
- [29].Belkin AM, Stepp MA, Microsc. Res. Tech. 2000, 51, 280. [DOI] [PubMed] [Google Scholar]
- [30].Singh P, Chen C, Pal-Ghosh S, Stepp MA, Sheppard D, Van De Water L, J Invest Dermatol. 2009, 129, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nakayama Y, Kon S, Kurotaki D, Morimoto J, Matsui Y and Uede T, Laboratory investigation. 2010, 90, 881. [DOI] [PubMed] [Google Scholar]
- [32].Egusa H, Kaneda Y, Akashi Y, Hamada Y, Matsumoto T, Saeki M, Thakor DK, Tabata Y, Matsuura N, Yatani H, Biomaterials. 2009, 30, 4676. [DOI] [PubMed] [Google Scholar]
- [33].Rao H, Lu G, Kajiya H, Garcia-Palacios V, Kurihara N, Anderson J, Patrene K, Sheppard D, Blair HC, Windle JJ, Choi SJ, Roodman GD, Journal of Bone and Mineral Research. 2006, 21, 1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vlahakis NE, Young BA, Atakilit A, Hawkridge AE, Issaka RB, Boudreau N, Sheppard D, Journal of Biological Chemistry. 2007, 282, 15187. [DOI] [PubMed] [Google Scholar]
- [35].Shen B, Vardy K, Hughes P, Tasdogan A, Zhao Z, Yue R, Crane GM, Morrison SJ, Elife. 2019, 8, e42274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Humphries JD, Byron A, Humphries MJ, J. Cell Sci. 2006, 119, 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cooke ME, Sakai T and Mosher DF, Journal of cell science. 2000, 113, 2375. [DOI] [PubMed] [Google Scholar]
- [38].Horton MA, The international journal of biochemistry & cell biology. 1997, 29, 721. [DOI] [PubMed] [Google Scholar]
- [39].Rezania A, Thomas CH, Branger AB, Waters CM, Healy KE, Journal of Biomedical Materials Research: An Official Journal of The Society for Biomaterials and The Japanese Society for Biomaterials. 1997, 37, 9. [DOI] [PubMed] [Google Scholar]
- [40].Grzeszkiewicz TM, Kirschling DJ, Chen N and Lau LF, Journal of Biological Chemistry. 2001, 276, pp.21943. [DOI] [PubMed] [Google Scholar]
- [41].Davis GE, Biochemical and biophysical research communications. 1992, 182, 1025. [DOI] [PubMed] [Google Scholar]
- [42].Stratman AN, Malotte KM, Mahan RD, Davis MJ, Davis GE, Blood, The Journal of the American Society of Hematology. 2009, 114, 5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qin Z, Fisher GJ and Quan T, Journal of Biological Chemistry. 2013, 288, 12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Aubin JE, Triffitt JT, Principles of Bone Biology. 2002, 1, 59. [Google Scholar]
- [45].Branemark P-I, The Journal of prosthetic dentistry. 1983, 50, 399. [DOI] [PubMed] [Google Scholar]
- [46].Rutkovskiy A, Stensløkken K-O, Vaage IJ, Medical science monitor basic research. 2016, 22, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Franceschi RT, Ge C, Xiao G, Roca H, Jiang D, Annals of the New York Academy of Sciences. 2007, 1116, 196. [DOI] [PubMed] [Google Scholar]
- [48].Florencio-Silva R, Sasso G. R. d. S., Sasso-Cerri E, Simões MJ, Cerri PS, BioMed research international. 2015, 2015. [Google Scholar]
- [49].Tamura Y, Takeuchi Y, Suzawa M, Fukumoto S, Kato M, Miyazono K, Fujita T, Journal of bone and mineral research. 2001, 16, 1772. [DOI] [PubMed] [Google Scholar]
- [50].Keselowsky BG, Collard DM, García AJ, Proceedings of the National Academy of Sciences. 2005, 102, 5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hamidouche Z, Fromigué O, Ringe J, Häupl T, Vaudin P, Pagès J-C, Srouji S, Livne E, Marie PJ, Proceedings of the National Academy of Sciences. 2009, 106, 18587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Castoldi M, Pistone M, Caruso C, Puddu A, Filanti C, Piccini D, Tacchetti C, Manduca P, Cell biology international. 1997, 21, 7. [DOI] [PubMed] [Google Scholar]
- [53].Bruder SP, Jaiswal N, Ricalton NS, Mosca JD, Kraus KH, Kadiyala S, Clinical Orthopaedics and Related Research®. 1998, 355, S247. [DOI] [PubMed] [Google Scholar]
- [54].Gronthos S, Stewart K, Graves SE, Hay S, Simmons PJ, Journal of Bone and Mineral Research. 1997, 12, 1189. [DOI] [PubMed] [Google Scholar]
- [55].Bennett J, Carter D, Alavi A, Beresford J, Walsh S, Archives of Oral Biology. 2001, 46, 229. [DOI] [PubMed] [Google Scholar]
- [56].Grzesik WJ, Robey PG, Journal of Bone and Mineral Research. 1994, 9, 487. [DOI] [PubMed] [Google Scholar]
- [57].Hughes D, Salter D, Dedhar S, Simpson R, Journal of bone and mineral research. 1993, 8, 527. [DOI] [PubMed] [Google Scholar]
- [58].Marsell R, Einhorn TA, Injury. 2011, 42, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gómez-Barrena E, Rosset P, Lozano D, Stanovici J, Ermthaller C, Gerbhard F, Bone. 2015, 70, 93. [DOI] [PubMed] [Google Scholar]
- [60].Hartz B, Volkmann T, Irle S, Loechelt C, Neubauer A, Brendel C, Blood, The Journal of the American Society of Hematology. 2011, 118, 2362. [DOI] [PubMed] [Google Scholar]
- [61].Kumar S, Ponnazhagan S, The FASEB Journal. 2007, 21, 3917. [DOI] [PubMed] [Google Scholar]
- [62].Veevers-Lowe J, Ball SG, Shuttleworth A, Kielty CM, Journal of cell science. 2011, 124, 1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rüster B, Göttig S, Ludwig RJ, Bistrian R, Müller S, Seifried E, Gille J, Henschler R, Blood. 2006, 108, 3938. [DOI] [PubMed] [Google Scholar]
- [64].Kolambkar YM, Bajin M, Wojtowicz A, Hutmacher DW, Garcia AJ, Guldberg RE, Tissue Engineering Part A 2013, 20, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Oryan A, Monazzah S, Bigham-Sadegh A, Biomedical and environmental sciences. 2015, 28, 57. [DOI] [PubMed] [Google Scholar]
- [66].Amy AJG, Clark Y, Nature Communications. 2020. [Google Scholar]
- [67].Mizuno M, Kuboki Y, The Journal of Biochemistry. 2001, 129, 133. [DOI] [PubMed] [Google Scholar]
- [68].Suzawa M, Tamura Y, Fukumoto S, Miyazono K, Fujita T, Kato S, Takeuchi Y, Journal of Bone and Mineral Research. 2002, 17, 240. [DOI] [PubMed] [Google Scholar]
- [69].Takeuchi Y, Suzawa M, Kikuchi T, Nishida E, Fujita T, Matsumoto T, Journal of Biological Chemistry. 1997, 272, 29309. [DOI] [PubMed] [Google Scholar]
- [70].Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, Franceschi RT, Journal of Biological Chemistry. 2000, 275, 4453. [DOI] [PubMed] [Google Scholar]
- [71].Schneider GB, Zaharias R, Stanford C, J. Dent. Res. 2001, 80, 1540. [DOI] [PubMed] [Google Scholar]
- [72].Xiao G, Wang D, Benson MD, Karsenty G, Franceschi RT, J. Biol. Chem. 1998, 273, 32988. [DOI] [PubMed] [Google Scholar]
- [73].Reyes CD, García AJ, Journal of Biomedical Materials Research Part A: An Official Journal of The Society for Biomaterials, The Japanese Society for Biomaterials, and The Australian Society for Biomaterials and the Korean Society for Biomaterials. 2004, 69, 591. [Google Scholar]
- [74].Becerra‐Bayona SM, Guiza‐Arguello VR, Russell B, Höök M, Hahn MS, J. Biomed. Mater. Res., Part A. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Oh S-H, Kim J-W, Kim Y, Lee MN, Kook M-S, Choi EY, Im S-Y, Koh J-T, PloS one. 2017, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Li B, Moshfegh C, Lin Z, Albuschies J, Vogel V, Scientific reports. 2013, 3, 2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lee J, Abdeen AA, Tang X, Saif TA, Kilian KA, Biomaterials. 2015, 69, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Salasznyk RM, Williams WA, Boskey A, Batorsky A, Plopper GE, BioMed Res. Int. 2004, 2004, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cheng SL, Lai CF, Blystone SD, Avioli LV, Journal of Bone and Mineral Research. 2001, 16, 277. [DOI] [PubMed] [Google Scholar]
- [80].Setiawati R, Rahardjo P, in Osteogenesis and bone regeneration, IntechOpen, 2018. [Google Scholar]
- [81].Keselowsky B, Wang L, Schwartz Z, Garcia A, Boyan B, Journal of Biomedical Materials Research Part A. 2007, 80, 700. [DOI] [PubMed] [Google Scholar]
- [82].Dufour C, Holy X, Marie PJ, American Journal of Physiology-Endocrinology and Metabolism. 2008, 294, E794. [DOI] [PubMed] [Google Scholar]
- [83].Horton MA, Helfrich MH, in Madame Curie Bioscience Database [Internet], Landes Bioscience, 2013. [Google Scholar]
- [84].Boyle WJ, Simonet WS, Lacey DL, Nature. 2003, 423, 337. [DOI] [PubMed] [Google Scholar]
- [85].Nakamura I, Pilkington MF, Lakkakorpi PT, Lipfert L, Sims SM, Dixon SJ, Rodan GA, Duong LT, Journal of cell science. 1999, 112, 3985. [DOI] [PubMed] [Google Scholar]
- [86].McHugh KP, Hodivala-Dilke K, Zheng M-H, Namba N, Lam J, Novack D, Feng X, Ross FP, Hynes RO, Teitelbaum SL, The Journal of clinical investigation. 2000, 105, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Horton MA, Proceedings of the Nutrition Society. 2001, 60, 275. [DOI] [PubMed] [Google Scholar]
- [88].Cacciari B, Spalluto G, Current medicinal chemistry. 2005, 12, 51. [DOI] [PubMed] [Google Scholar]
- [89].Helfrich M, Nesbitt S, Lakkakorpi P, Barnes M, Bodary S, Shankar G, Mason W, Mendrick D, Väänänen H, Horton M, Bone. 1996, 19, 317. [DOI] [PubMed] [Google Scholar]
- [90].Wild T, Rahbarnia A, Kellner M, Sobotka L, Eberlein T, Nutrition. 2010, 26, 862. [DOI] [PubMed] [Google Scholar]
- [91].Li J, Chen J, Kirsner R, Clin Dermatol. 2007, 25, 9. [DOI] [PubMed] [Google Scholar]
- [92].Tracy LE, Minasian RA, Caterson EJ, Adv Wound Care (New Rochelle). 2016, 5, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Xue M, Jackson CJ, Adv Wound Care (New Rochelle). 2015, 4, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sisco M, Chao JD, Kim I, Mogford JE, Mayadas TN, Mustoe TA, Wound Repair and Regeneration. 2007, 15, 566. [DOI] [PubMed] [Google Scholar]
- [95].Goodman S. L. a. P., M., Trends in pharmacological sciences. 2012, 33, 405. [DOI] [PubMed] [Google Scholar]
- [96].Hosoyama K, Lazurko C, Munoz M, McTiernan CD, Alarcon EI, Front Bioeng Biotechnol. 2019, 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Almine JF, Wise SG and Weiss AS, Birth Defects Research Part C: Embryo Today: Reviews. 2012, 96, 248. [DOI] [PubMed] [Google Scholar]
- [98].Raja, Sivamani K, Garcia MS, Isseroff RR, Front Biosci. 2007, 12, 2849. [DOI] [PubMed] [Google Scholar]
- [99].Räsänen K. a. V., A., Journal of dermatological science. 2010, 58, 97. [DOI] [PubMed] [Google Scholar]
- [100].Al Moustafa AE, Achkhar A and Yasmeen A, Front Biosci (Schol Ed). 2012, 4, 671. [DOI] [PubMed] [Google Scholar]
- [101].Billottet C, Tuefferd M, Gentien D, Rapinat A, Thiery JP, Broët P and Jouanneau J, Journal of cellular biochemistry. 2008, 104, 826. [DOI] [PubMed] [Google Scholar]
- [102].Masola V, Onisto M, Zaza G, Lupo A and Gambaro G, Journal of translational medicine. 2012, 10, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Brakebusch C, Cell Migration in Development and Disease. 2005, 275, 298. [Google Scholar]
- [104].Parks WC, The Journal of investigative dermatology. 2007, 127, 264. [DOI] [PubMed] [Google Scholar]
- [105].Pilcher BK, Dumin JA, Sudbeck BD, Krane SM, Welgus HG and Parks WC, The Journal of cell biology. 1997, 137, 1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Grenache DG, Zhang Z, Wells LE, Santoro SA, Davidson JM and Zutter MM, Journal of Investigative Dermatology. 2007, 127, 455. [DOI] [PubMed] [Google Scholar]
- [107].Frank DE, Carter WG, J Cell Sci. 2004, 117, 1351. [DOI] [PubMed] [Google Scholar]
- [108].Choma DP, Milano V, Pumiglia KM, DiPersio CM, J Invest Dermatol. 2007, 127, 31. [DOI] [PubMed] [Google Scholar]
- [109].Sehgal BU, DeBiase PJ, Matzno S, Chew TL, Claiborne JN, Hopkinson SB, Russell A, Marinkovich MP, Jones JC, J Biol Chem. 2006, 281, 35487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Spinardi L, Einheber S, Cullen T, Milner TA and Giancotti FG, The Journal of cell biology. 1995, 129, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].De Pereda JM, Lillo MP and Sonnenberg A, The EMBO journal. 2009, 28, 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kiwanuka E, Andersson L, Caterson EJ, Junker JP, Gerdin B, Eriksson E, Exp Cell Res. 2013, 319, 2938. [DOI] [PubMed] [Google Scholar]
- [113].Patel GK, Wilson CH, Harding KG, Finlay AY, Bowden PE, J Invest Dermatol. 2006, 126, 497. [DOI] [PubMed] [Google Scholar]
- [114].Morasso MI, Tomic-Canic M, Biol Cell. 2005, 97, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bata-Csorgo Z, Cooper KD, Ting KM, Voorhees JJ, Hammerberg C, J Clin Invest. 1998, 101, 1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kim YY, Li H, Song YS, Jeong HS, Yun HY, Baek KJ, Kwon NS, Shin YK, Park KC and Kim DS, Journal of tissue viability. 2018, 27, 117. [DOI] [PubMed] [Google Scholar]
- [117].Teige I, Backlund A, Svensson L, Kvist PH, Petersen TK, Kemp K, Int Immunopharmacol. 2010, 10, 107. [DOI] [PubMed] [Google Scholar]
- [118].Singh P, Reimer CL, Peters JH, Stepp MA, Hynes RO, Van De Water L, J Invest Dermatol. 2004, 123, 1176. [DOI] [PubMed] [Google Scholar]
- [119].Roy S, Bingle L, Marshall JF, Bass R, Ellis V, Speight PM, Whawell SA, J Oral Pathol Med. 2011, 40, 755. [DOI] [PubMed] [Google Scholar]
- [120].Grotendorst GR, Martin GR, Pencev D, Sodek J and Harvey AK, The Journal of clinical investigation. 1985, 76, 2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Greenhalgh DG, Sprugel KH, Murray MJ and Ross R, The American journal of pathology. 1990, 136, 1235. [PMC free article] [PubMed] [Google Scholar]
- [122].Frazier K, Williams S, Kothapalli D, Klapper H and Grotendorst GR, Journal of Investigative Dermatology. 1996, 107, 401. [DOI] [PubMed] [Google Scholar]
- [123].Schmid P, Itin P, Cherry G, Bi C and Cox DA, The American journal of pathology. 1998, 152, 485. [PMC free article] [PubMed] [Google Scholar]
- [124].Wong VW, Akaishi S, Longaker MT and Gurtner GC, 131. 2011. [DOI] [PubMed] [Google Scholar]
- [125].Lokmic Z, Musyoka J, Hewitson TD and Darby IA, International review of cell and molecular biology. 2012, 296, 139. [DOI] [PubMed] [Google Scholar]
- [126].Tandara A. A. a. M., T.A., World journal of surgery. 2004, 28, 294.14961188 [Google Scholar]
- [127].Wary KK, Mainiero F, Isakoff SJ, Marcantonio EE and Giancotti FG, Cell. 1996, 87, 733. [DOI] [PubMed] [Google Scholar]
- [128].Xia H, Seeman J, Hong J, Hergert P, Bodem V, Jessurun J, Smith K, Nho R, Kahm J, Gaillard P and Henke C, The American journal of pathology. 2012, 181, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Cereceres S, Touchet T, Browning MB, Smith C, Rivera J, Hoeoek M, Whitfield-Cargile C, Russell B and Cosgriff-Hernandez E, Advances in wound care. 2015, 4, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Suzuki N, Nakatsuka H, Mochizuki M, Nishi N, Kadoya Y, Utani A, Oishi S, Fujii N, Kleinman HK and Nomizu M, Journal of Biological Chemistry. 2003, 278, 45697. [DOI] [PubMed] [Google Scholar]
- [131].Corbett SA, Lee L, Wilson CL and Schwarzbauer JE, Journal of Biological Chemistry. 1997, 272, 24999. [DOI] [PubMed] [Google Scholar]
- [132].Kato A, Okamoto O, Ishikawa K, Sumiyoshi H, Matsuo N, Yoshioka H, Nomizu M, Shimada T and Fujiwara S, Journal of Biological Chemistry. 2011, 286, 14861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Schulz JN, Zeltz C, Sørensen IW, Barczyk M, Carracedo S, Hallinger R, Niehoff A, Eckes B and Gullberg D, Journal of Investigative Dermatology. 2015, 135, 1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Gardner H, Broberg A, Pozzi A, Laato M, Heino J, Journal of Cell Science. 1999, 112, 263. [DOI] [PubMed] [Google Scholar]
- [135].Aumailley M, Pesch M, Tunggal L, Gaill F and Fassler R, Journal of cell science. 2000, 113, 259. [DOI] [PubMed] [Google Scholar]
- [136].Wang Z, Collighan RJ, Gross SR, Danen EH, Orend G, Telci D and Griffin M, Journal of Biological Chemistry. 2010, 285, 40212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Moir LM, Burgess JK and Black JL, Journal of Allergy and Clinical Immunology. 2008, 121, 1034. [DOI] [PubMed] [Google Scholar]
- [138].Shinde AV, Kelsh R, Peters JH, Sekiguchi K, Van De Water L and McKeown-Longo PJ, Matrix biology. 2015, 41, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Yang Y, Dang D, Mogi S and Ramos DM, Biochemical and biophysical research communications. 2004, 322, 935. [DOI] [PubMed] [Google Scholar]
- [140].Thannickal VJ, Lee DY, White ES, Cui Z, Larios JM, Chacon R, Horowitz JC, Day RM and Thomas PE, J. Biol. Chem. 2003, 278, 12384. [DOI] [PubMed] [Google Scholar]
- [141].Midwood KS, Valenick LV, Hsia HC and Schwarzbauer JE, Molecular biology of the cell. 2004, 15, 5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Tomasek J. J. a. A., S.K., The Anatomical Record. 1992, 234, 153. [DOI] [PubMed] [Google Scholar]
- [143].Valenick LV, Hsia HC and Schwarzbauer JE, Experimental cell research. 2005, 309, 48. [DOI] [PubMed] [Google Scholar]
- [144].Gullberg D, Tingström A, Thuresson AC, Olsson L, Terracio L, Borg TK and Rubin K, Experimental cell research. 1990, 186, 264. [DOI] [PubMed] [Google Scholar]
- [145].Gutierrez J, Droppelmann CA, Contreras O, Takahashi C and Brandan E, PloS one. 2015, 10, p.e0135005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Carver W, Molano I, Reaves TA, Borg TK and Terracio L, Journal of cellular physiology. 1995, 165, 425. [DOI] [PubMed] [Google Scholar]
- [147].Racine-Samson L, Rockey DC and Bissell DM, Journal of Biological Chemistry. 1997, 272, 30911. [DOI] [PubMed] [Google Scholar]
- [148].Schiro JA, Chan BM, Roswit WT, Kassner PD, Pentland AP, Hemler ME, Eisen AZ and Kupper TS, Cell. 1991, 67, 403. [DOI] [PubMed] [Google Scholar]
- [149].Li J, Zhang YP and Kirsner RS, Microscopy research and technique. 2003, 60, 107. [DOI] [PubMed] [Google Scholar]
- [150].Tonnesen MG, Feng X and Clark RA, Journal of Investigative Dermatology Symposium Proceedings. 2000, 5, 40. [DOI] [PubMed] [Google Scholar]
- [151].Jang YC, Arumugam S, Gibran NS and Isik FF, Wound Repair and Regeneration. 1999, 7, 375. [DOI] [PubMed] [Google Scholar]
- [152].Zweers MC, Davidson JM, Pozzi A, Hallinger R, Janz K, Quondamatteo F, Leutgeb B, Krieg T and Eckes B, Journal of Investigative Dermatology. 2007, 127, 467. [DOI] [PubMed] [Google Scholar]
- [153].Mitchell K, Szekeres C, Milano V, Svenson KB, Nilsen-Hamilton M, Kreidberg JA and DiPersio CM, Journal of cell science. 2009, 122, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Carmeliet P, Nature medicine. 2000, 6, 389. [DOI] [PubMed] [Google Scholar]
- [155].Briquez PS, Clegg LE, Martino MM, Gabhann FM, Hubbell JA, Nature Reviews Materials. 2016, 1. [Google Scholar]
- [156].Senger DR, Davis GE, Cold Spring Harbor perspectives in biology. 2011, 3, a005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Mongiat M, Andreuzzi E, Tarticchio G, Paulitti A, Int J Mol Sci. 2016, 17, 1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Desai D, Singh P, Van De Water L, LaFlamme SE, Advances in wound care. 2013, 2, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Soldi R, Mitola S, Strasly M, Defilippi P, Tarone G, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Senger DR, Perruzzi CA, Streit M, Koteliansky VE, de Fougerolles AR, Detmar M, The American journal of pathology. 2002, 160, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Rundhaug JE, Journal of cellular and molecular medicine. 2005, 9, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Davis GE, Stratman AN, Sacharidou A, Koh W, Molecular Basis for Endothelial Lumen Formation and Tubulogenesis During Vasculogenesis and Angiogenic Sprouting, Vol. 288, Elsevier Science & Technology, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Sweeney SM, DiLullo G, Slater SJ, Martinez J, Iozzo RV, Lauer-Fields JL, Fields GB, San Antonio JD, Journal of Biological Chemistry. 2003, 278, 30516. [DOI] [PubMed] [Google Scholar]
- [164].Hynes R, Journal of Thrombosis and Haemostasis. 2007, 5, 32. [DOI] [PubMed] [Google Scholar]
- [165].Silva R, D’Amico G, Hodivala-Dilke KM, Reynolds LE, Arteriosclerosis, thrombosis, and vascular biology. 2008, 28, 1703. [DOI] [PubMed] [Google Scholar]
- [166].Davis GE, Camarillo CW, Experimental cell research. 1996, 224, 39. [DOI] [PubMed] [Google Scholar]
- [167].Laurens N, Engelse MA, Jungerius C, Löwik CW, van Hinsbergh VW, Koolwijk P, Angiogenesis. 2009, 12, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Lee T-H, Seng S, Li H, Kennel SJ, Avraham HK, Avraham S, Journal of Biological Chemistry. 2006, 281, 40450. [DOI] [PubMed] [Google Scholar]
- [169].Stamati K, Priestley JV, Mudera V and Cheema U, Exp. Cell Res. 2014, 327, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Bignon M, Pichol-Thievend C, Hardouin J, Malbouyres M, Bréchot N, Nasciutti L, Barret A, Teillon J, Guillon E, Etienne E, Blood, The Journal of the American Society of Hematology. 2011, 118, 3979. [DOI] [PubMed] [Google Scholar]
- [171].Ucuzian AA, Gassman AA, East AT, Greisler HP, Journal of Burn Care & Research. 2010, 31, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Takahashi T, Ueno H, Shibuya M, Oncogene. 1999, 18, 2221. [DOI] [PubMed] [Google Scholar]
- [173].Nisato RE, Tille J-C, Jonczyk A, Goodman SL, Pepper MS, Angiogenesis. 2003, 6, 105. [DOI] [PubMed] [Google Scholar]
- [174].Somanath PR, Malinin NL, Byzova TV, Angiogenesis. 2009, 12, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Bussolino F, Serini G, Mitola S, Bazzoni G, Dejana E, EMBO reports. 2001, 2, 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Zanella S, Mingozzi M, Dal Corso A, Fanelli R, Arosio D, Cosentino M, Schembri L, Marino F, De Zotti M, Formaggio F, ChemistryOpen. 2015, 4, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Masson-Gadais B, Houle F, Laferrière J, Huot J, Cell stress & chaperones. 2003, 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Brooks PC, Clark RA, Cheresh DA, Science. 1994, 264, 569. [DOI] [PubMed] [Google Scholar]
- [179].Kim S, Bell K, Mousa SA, Varner JA, The American journal of pathology. 2000, 156, 1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Hiran TS, Mazurkiewicz JE, Kreienberg P, Rice FL, LaFlamme SE, Journal of cell science. 2003, 116, 3771. [DOI] [PubMed] [Google Scholar]
- [181].Senger DR, Claffey KP, Benes JE, Perruzzi CA, Sergiou AP, Detmar M, Proceedings of the National Academy of Sciences. 1997, 94, 13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Sang QXA, Cell research. 1998, 8, 171. [DOI] [PubMed] [Google Scholar]
- [183].Haas T, Madri JA, Trends in cardiovascular medicine. 1999, 9, 70. [DOI] [PubMed] [Google Scholar]
- [184].Stetler-Stevenson WG, The Journal of clinical investigation. 1999, 103, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Lafleur MA, Handsley MM, Edwards DR, Expert Reviews in Molecular Medicine. 2003, 5, 1. [DOI] [PubMed] [Google Scholar]
- [186].Taraboletti G, D’Ascenzo S, Borsotti P, Giavazzi R, Pavan A, Dolo V, The American journal of pathology. 2002, 160, 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].McCawley LJ, Matrisian LM, Current opinion in cell biology. 2001, 13, 534. [DOI] [PubMed] [Google Scholar]
- [188].Silletti S, Kessler T, Goldberg J, Boger DL, Cheresh DA, Proceedings of the National Academy of Sciences. 2001, 98, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Brooks PC, Silletti S, von Schalscha TL, Friedlander M, Cheresh DA, Cell. 1998, 92, 391. [DOI] [PubMed] [Google Scholar]
- [190].Senger DR, Am. J. Pathol. 1996, 149, 1. [PMC free article] [PubMed] [Google Scholar]
- [191].Senger DR, The American journal of pathology. 1996, 149, 1. [PMC free article] [PubMed] [Google Scholar]
- [192].Eliceiri BP, Cheresh DA, Molecular medicine. 1998, 4, 741. [PMC free article] [PubMed] [Google Scholar]
- [193].Stratman AN, Saunders WB, Sacharidou A, Koh W, Fisher KE, Zawieja DC, Davis MJ, Davis GE, Blood, The Journal of the American Society of Hematology. 2009, 114, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Gardner H, in I Domain Integrins, Springer, 2014, 21. [Google Scholar]
- [195].Pozzi A, Moberg PE, Miles LA, Wagner S, Soloway P, Gardner HA, Proceedings of the National Academy of Sciences. 2000, 97, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Hodivala-Dilke KM, Reynolds AR, Reynolds LE, Cell and tissue research. 2003, 314, 131. [DOI] [PubMed] [Google Scholar]
- [197].Wang W, Guo L, Yu Y, Chen Z, Zhou R, Yuan Z, Journal of Biomedical Materials Research Part A. 2015, 103, 1703. [DOI] [PubMed] [Google Scholar]
- [198].Massia SP, Hubbell JA, Journal of Biological Chemistry. 1992, 267, 14019. [PubMed] [Google Scholar]
- [199].Kim S, Bakre M, Yin H, Varner JA, The Journal of clinical investigation. 2002, 110, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Zeng Z-Z, Yao H, Staszewski ED, Rockwood KF, Markwart SM, Fay KS, Spalding AC, Livant DL, Translational oncology. 2009, 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Yamada KM, The Journal of clinical investigation. 2000, 105, 1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Francis SE, Goh KL, Hodivala-Dilke K, Bader BL, Stark M, Davidson D, Hynes RO, Arteriosclerosis, thrombosis, and vascular biology. 2002, 22, 927. [DOI] [PubMed] [Google Scholar]
- [203].Li S, Nih LR, Bachman H, Fei P, Li Y, Nam E, Dimatteo R, Carmichael ST, Barker TH, Segura T, Nature materials. 2017, 16, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].da Silva RG, Tavora B, Robinson SD, Reynolds LE, Szekeres C, Lamar J, Batista S, Kostourou V, Germain MA, Reynolds AR, The American journal of pathology. 2010, 177, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Brooks PC, Strömblad S, Klemke R, Visscher D, Sarkar FH, Cheresh DA, The Journal of clinical investigation. 1995, 96, 1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Friedlander M, Brooks PC, Shaffer RW, Kincaid CM, Varner JA, Cheresh DA, Science. 1995, 270, 1500. [DOI] [PubMed] [Google Scholar]
- [207].Brooks PC, Strömblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, Cheresh DA, Cell. 1996, 85, 683. [DOI] [PubMed] [Google Scholar]
- [208].Leavesley D, Schwartz M, Rosenfeld M, Cheresh D, The Journal of cell biology. 1993, 121, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Brooks PC, Montgomery AM, Rosenfeld M, Reisfeld RA, Hu T, Klier G, Cheresh DA, Cell. 1994, 79, 1157. [DOI] [PubMed] [Google Scholar]
- [210].Wang J, Milner R, Journal of Neurochemistry. 2006, 96, 148. [DOI] [PubMed] [Google Scholar]
- [211].Blatchley MR, Gerecht S, Nature materials. 2017, 16, 881. [DOI] [PubMed] [Google Scholar]
- [212].Betz C, Lenard A, Belting H-G, Affolter M, Development. 2016, 143, 2249. [DOI] [PubMed] [Google Scholar]
- [213].Pauty J, Usuba R, Cheng IG, Hespel L, Takahashi H, Kato K, Kobayashi M, Nakajima H, Lee E, Yger F, EBioMedicine. 2018, 27, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Bauer J, Margolis M, Schreiner C, Edgell CJ, Azizkhan J, Lazarowski E, Juliano R, Journal of cellular physiology. 1992, 153, 437. [DOI] [PubMed] [Google Scholar]
- [215].Kubota Y, Kleinman HK, Martin GR, Lawley TJ, The Journal of cell biology. 1988, 107, 1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Nakamura M, Yamaguchi K, Mie M, Nakamura M, Akita K, Kobatake E, Bioconjugate chemistry. 2009, 20, 1759. [DOI] [PubMed] [Google Scholar]
- [217].Taubenberger AV, Bray LJ, Haller B, Shaposhnykov A, Binner M, Freudenberg U, Guck J, Werner C, Acta biomaterialia. 2016, 36, 73. [DOI] [PubMed] [Google Scholar]
- [218].Shakado S, Sakisaka S, Noguchi K, Yoshitake M, Harada M, Mimura Y, Sata M, Tanikawa K, Hepatology. 1995, 22, 969. [PubMed] [Google Scholar]
- [219].Ingber DE, Folkman J, Vol. 58, Elsevier Inc, 1989, 803. [Google Scholar]
- [220].Whelan MC, Senger DR, J. Biol. Chem. 2003, 278, 327. [DOI] [PubMed] [Google Scholar]
- [221].LIU Y, SENGER DR, The FASEB journal. 2004, 18, 457. [DOI] [PubMed] [Google Scholar]
- [222].Singh RK, Seliktar D, Putnam AJ, Biomaterials. 2013, 34, 9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Bayless KJ, Salazar R, Davis GE, The American journal of pathology. 2000, 156, 1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Rocha LA, Learmonth DA, Sousa RA, Salgado AJ, Biotechnology advances. 2018, 36, 208. [DOI] [PubMed] [Google Scholar]
- [225].Stratman AN, Davis GE, Microscopy and Microanalysis. 2012, 18, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Adams RH, Alitalo K, Nature reviews Molecular cell biology. 2007, 8, 464. [DOI] [PubMed] [Google Scholar]
- [227].Hall-Glenn F, De Young RA, Huang B-L, van Handel B, Hofmann JJ, Chen TT, Choi A, Ong JR, Benya PD, Mikkola H, PloS one. 2012, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Armulik A, Abramsson A, Betsholtz C, Circulation research. 2005, 97, 512. [DOI] [PubMed] [Google Scholar]
- [229].Schmidt MM, Dornelles RCP, Mello RO, Kubota EH, Mazutti MA, Kempka AP and Demiate IM, International Food Research Journal. 2016, 23, 913. [Google Scholar]
- [230].Bray BA, Mandl I and Turino GM, Science. 1981, 214, 793. [DOI] [PubMed] [Google Scholar]
- [231].Humbert‐David N, Chandrasekaran S, Tanzer ML and Garrone R, Biology of the Cell. 1995, 83, 39. [Google Scholar]
- [232].Kleinman HK, McGarvey ML, Liotta LA, Robey PG, Tryggvason K and Martin GR, Biochemistry. 1982, 21, 6188. [DOI] [PubMed] [Google Scholar]
- [233].Jiang L, He L and Fountoulakis M, J. Chromatogr. A. 2004, 1023, 317. [DOI] [PubMed] [Google Scholar]
- [234].Qian Y, Zhou X, Zhang F, Diekwisch TG, Luan X and Yang J, ACS Appl. Mater. Interfaces. 2019, 11, 37381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Dormond O, Bezzi M, Mariotti A and Rüegg C, J. Biol. Chem. 2002, 277, 45838. [DOI] [PubMed] [Google Scholar]
- [236].Davidenko N, Schuster CF, Bax DV, Farndale RW, Hamaia S, Best SM and Cameron RE, Journal of Materials Science: Materials in Medicine. 2016, 27, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Joddar B, Ito Y, Title J. Mater. Chem. 2011, 21, 13737. [Google Scholar]
- [238].Browning MB, Russell B, Rivera J, Höök M. and Cosgriff-Hernandez EM, Biomacromolecules. 2013, 14, 2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Campos DM, Gritsch K, Salles V, Attik GN and Grosgogeat B, BioResearch Open Access. 2014, 3, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Francisco AT, Hwang PY, Jeong CG, Jing L, Chen J and Setton LA, Acta Biomater. 2014, 10, 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Hsu SH, Kuo WC, Chen YT, Yen CT, Chen YF, Chen KS, Huang WC and Cheng H, Acta Biomater. 2013, 9, 6606. [DOI] [PubMed] [Google Scholar]
- [242].Kijeńska E, Prabhakaran MP, Swieszkowski W, Kurzydlowski KJ and Ramakrishna S, Eur. Polym. J. 2014, 50, 30. [DOI] [PubMed] [Google Scholar]
- [243].Suri S. a. S., C.E., Tissue Eng., Part A. 2010, 16, 1703. [DOI] [PubMed] [Google Scholar]
- [244].Hinderer S, Layland SL and Schenke-Layland K, Adv. Drug Delivery Rev. 2016, 97, 260. [DOI] [PubMed] [Google Scholar]
- [245].Wade R. J. a. B, J.A., Mater. Today. 2012, 15, 454. [Google Scholar]
- [246].Antoine EE, Vlachos PP and Rylander MN, Tissue Eng., Part B. 2014, 20, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Antoine EE, Vlachos PP and Rylander MN, Tissue Engineering Part B: Reviews. 2014, 20, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Reichert JC, Heymer A, Berner A, Eulert J, Noth U, Biomed Mater. 2009, 4, 065001. [DOI] [PubMed] [Google Scholar]
- [249].Pensalfini M, Ehret AE, Stüdeli S, Marino D, Kaech A, Reichmann E and Mazza E, Journal of the mechanical behavior of biomedical materials. 2017, 69, 85. [DOI] [PubMed] [Google Scholar]
- [250].Helary C, Zarka M and Giraud‐Guille MM, Journal of tissue engineering and regenerative medicine. 2012, 6, 225. [DOI] [PubMed] [Google Scholar]
- [251].Ananta M, Brown RA and Mudera V, Tissue Eng., Part A. 2012, 18, 353. [DOI] [PubMed] [Google Scholar]
- [252].Alvarez GS, Hélary C, Mebert AM, Wang X, Coradin T and Desimone MF, Journal of Materials Chemistry B. 2014, 2, 4660. [DOI] [PubMed] [Google Scholar]
- [253].Shokrgozar MA, Fattahi M, Bonakdar S, Kashani IR, Majidi M, Haghighipour N, Bayati V, Sanati H and Saeedi SN, Iranian biomedical journal. 2012, 16, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Vasconcelos A, Gomes AC and Cavaco-Paulo A, Acta Biomater. 2012, 8, 3049. [DOI] [PubMed] [Google Scholar]
- [255].Chantre CO, Campbell PH, Golecki HM, Buganza AT, Capulli AK, Deravi LF, Dauth S, Sheehy SP, Paten JA, Gledhill K and Doucet YS, Biomaterials. 2018, 166, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Davis GE, Black SM and Bayless KJ, In Vitro Cellular & Developmental Biology-Animal. 2000, 36, 513. [DOI] [PubMed] [Google Scholar]
- [257].Weisel JW, Adv. Protein Chem. 2005, 70, 247. [DOI] [PubMed] [Google Scholar]
- [258].Hall H, Baechi T and Hubbell JA, Microvasc. Res. 2001, 62, 315. [DOI] [PubMed] [Google Scholar]
- [259].Buckel P, Trends in pharmacological sciences. 1996, 17, 450. [DOI] [PubMed] [Google Scholar]
- [260].Farajollahi MM, Hamzehlou S, Mehdipour A, Samadikuchaksaraei A, BioImpacts: BI. 2012, 2, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Staunton D, Millard CJ, Aricescu AR, Campbell ID, in Extracellular Matrix Protocols, Springer, 2009, 73. [DOI] [PubMed] [Google Scholar]
- [262].Peng YY, Yoshizumi A, Danon SJ, Glattauer V, Prokopenko O, Mirochnitchenko O, Yu Z, Inouye M, Werkmeister JA, Brodsky B, Biomaterials. 2010, 31, 2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Ivaska J, Käpylä J, Pentikäinen O, Hoffrén A-M, Hermonen J, Huttunen P, Johnson MS, Heino J, Journal of Biological Chemistry. 1999, 274, 3513. [DOI] [PubMed] [Google Scholar]
- [264].Katagiri F, Ishikawa M, Yamada Y, Hozumi K, Kikkawa Y, Nomizu M, Archives of biochemistry and biophysics. 2012, 521, 32. [DOI] [PubMed] [Google Scholar]
- [265].Cosgriff-Hernandez E, Hahn MS, Russell B, Wilems T, Munoz-Pinto D, Browning M, Rivera J, Höök M, Acta Biomater. 2010, 6, 3969. [DOI] [PubMed] [Google Scholar]
- [266].Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC, The Journal of cell biology. 2008, 183, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Martino MM, Tortelli F, Mochizuki M, Traub S, Ben-David D, Kuhn GA, Müller R, Livne E, Eming SA, Hubbell JA, Science translational medicine. 2011, 3, 100ra89. [DOI] [PubMed] [Google Scholar]
- [268].Agarwal R, González-García C, Torstrick B, Guldberg RE, Salmerón-Sánchez M, García AJ, Biomaterials. 2015, 63, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Browning MB, Guiza V, Russell B, Rivera J, Cereceres S, Höök M, Hahn MS, Cosgriff-Hernandez EM, Tissue Eng., Part A. 2014, 20, 3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Takahashi S, Leiss M, Moser M, Ohashi T, Kitao T, Heckmann D, Pfeifer A, Kessler H, Takagi J, Erickson HP, The Journal of cell biology. 2007, 178, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Brown AC, Dysart MM, Clarke KC, Stabenfeldt SE, Barker TH, Journal of Biological Chemistry. 2015, 290, 25534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Guzmán F, Barberis S, Illanes A, Electronic Journal of Biotechnology. 2007, 10, 279. [Google Scholar]
- [273].Lau JL, Dunn MK, Bioorganic & medicinal chemistry. 2018, 26, 2700. [DOI] [PubMed] [Google Scholar]
- [274].Kyle S, Aggeli A, Ingham E, McPherson MJ, Trends in biotechnology. 2009, 27, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Nilsson BL, Soellner MB, Raines RT, Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Perlin L, MacNeil S, Rimmer S, Soft matter. 2008, 4, 2331. [Google Scholar]
- [277].Stawikowski M, Fields GB, Current protocols in protein science. 2012, 69, 18.1. 1. [DOI] [PubMed] [Google Scholar]
- [278].Collier JH, Segura T, Biomaterials. 2011, 32, 4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Chen S, Zhang M, Shao X, Wang X, Zhang L, Xu P, Zhong W, Zhang L, Xing M, Zhang L, Journal of Materials Chemistry B. 2015, 3, 6798. [DOI] [PubMed] [Google Scholar]
- [280].Ali S, Saik JE, Gould DJ, Dickinson ME, West JL, BioResearch open access. 2013, 2, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Mecham RP, Annual review of cell biology. 1991, 7, 71. [DOI] [PubMed] [Google Scholar]
- [282].Zhu Y, Cankova Z, Iwanaszko M, Lichtor S, Mrksich M, Ameer GA, Proceedings of the National Academy of Sciences. 2018, 115, 6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Shekaran A, Garcia AJ, Journal of biomedical materials research Part A. 2011, 96, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Wojtowicz AM, Shekaran A, Oest ME, Dupont KM, Templeman KL, Hutmacher DW, Guldberg RE, García AJ, Biomaterials. 2010, 31, 2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Hennessy KM, Pollot BE, Clem WC, Phipps MC, Sawyer AA, Culpepper BK, Bellis SL, Biomaterials. 2009, 30, 1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Bhatnagar RS, Qian JJ, Wedrychowska A, Sadeghi M, Wu YM, Smith N, Tissue Engineering. 1999, 5, 53. [DOI] [PubMed] [Google Scholar]
- [287].Ganss B, Kim RH, Sodek J, Critical Reviews in Oral Biology & Medicine. 1999, 10, 79. [DOI] [PubMed] [Google Scholar]
- [288].Yu J, Gu Y, Du KT, Mihardja S, Sievers RE, Lee RJ, Biomaterials. 2009, 30, 751. [DOI] [PubMed] [Google Scholar]
- [289].Park JS, Yang HN, Jeon SY, Woo DG, Na K, Park K-H, Biomaterials. 2010, 31, 6239. [DOI] [PubMed] [Google Scholar]
- [290].Anderson JM, Kushwaha M, Tambralli A, Bellis SL, Camata RP, Jun H-W, Biomacromolecules. 2009, 10, 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Yang X, Roach H, Clarke N, Howdle S, Quirk R, Shakesheff K, Oreffo R, Bone. 2001, 29, 523. [DOI] [PubMed] [Google Scholar]
- [292].Redick SD, Settles DL, Briscoe G, Erickson HP, Journal of Cell Biology. 2000, 149, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Koivunen E, Gay DA, Ruoslahti E, Journal of Biological Chemistry. 1993, 268, 20205. [PubMed] [Google Scholar]
- [294].Gandavarapu NR, Alge DL, Anseth KS, Biomaterials science. 2014, 2, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Koivunen E, Wang B, Ruoslahti E, The Journal of cell biology. 1994, 124, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Seo N, Russell BH, Rivera JJ, Liang X, Xu X, Afshar-Kharghan V, Höök M, Journal of Biological Chemistry. 2010, 285, 31046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Behrendt R, White P, Offer J, Journal of Peptide Science. 2016, 22, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].De Groot AS, Martin W, Clinical Immunology. 2009, 131, 189. [DOI] [PubMed] [Google Scholar]
- [299].Schwarz H, Schmittner M, Duschl A, Horejs-Hoeck J, PloS one. 2014, 9, e113840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Lv H, Li L, Sun M, Zhang Y, Chen L, Rong Y, Li Y, Stem Cell Res. Ther. 2015, 6, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Bauer AL, Jackson TL, Jiang Y, PLoS Comput. Biol. 2009, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Hong H, Stegemann JP, J. Biomater. Sci., Polym. Ed. 2008, 19, 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Marie PJ, Haÿ E, Saidak Z, Trends in Endocrinology & Metabolism. 2014, 25, 567. [DOI] [PubMed] [Google Scholar]
- [304].Robinson KG, Nie T, Baldwin AD, Yang EC, Kiick KL, Akins RE Jr, Journal of biomedical materials research Part A. 2012, 100, 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Shih YRV, Tseng KF, Lai HY, Lin CH, Lee OK, J. Bone Miner. Res. 2011, 26, 730. [DOI] [PubMed] [Google Scholar]
- [306].Du J, Chen X, Liang X, Zhang G, Xu J, He L, Zhan Q, Feng X-Q, Chien S, Yang C, Proc. Natl. Acad. Sci. 2011, 108, 9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Murikipudi S, Methe H, Edelman ER, Biomaterials. 2013, 34, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Persson M, Lehenkari PP, Berglin L, Turunen S, Finnilä MAJ, Risteli J, Skrifvars M, Tuukkanen J, Sci. Rep. 2018, 8, 10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Parkinson LG, Rea SM, Stevenson AW, Wood FM, Fear MW, Tissue Eng., Part A. 2012, 18, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Kim HN, Hong Y, Kim MS, Kim SM, Suh K-Y, Biomaterials. 2012, 33, 8782. [DOI] [PubMed] [Google Scholar]
- [311].Persson M, Lehenkari PP, Berglin L, Turunen S, Finnilä MAJ, Risteli J, Skrifvars M, Tuukkanen J, Scientific reports. 2018, 8, 10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Meng X, Leslie P, Zhang Y, Dong J, Springerplus. 2014, 3, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Lepzelter D, Bates O, Zaman M, Langmuir. 2012, 28, 5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Kishan A, Nezarati R, Radzicki C, Renfro A, Robinson J, Whitely M, Cosgriff-Hernandez E, J. Mater. Chem. B. 2015, 3, 7930. [DOI] [PubMed] [Google Scholar]
- [315].Yang G, Xiao Z, Long H, Ma K, Zhang J, Ren X, Zhang J, Sci. Rep. 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Yahyouche A, Zhidao X, Czernuszka JT, Clover A, Acta Biomater. 2011, 7, 278. [DOI] [PubMed] [Google Scholar]
- [317].Gon S, Bendersky M, Ross JL, Santore MM, Langmuir. 2010, 26, 12147. [DOI] [PubMed] [Google Scholar]
- [318].Rabe M, Verdes D, Seeger S, Adv. Colloid Interface Sci. 2011, 162, 87. [DOI] [PubMed] [Google Scholar]
- [319].Norman JJ, Desai TA, Ann. Biomed. Eng. 2006, 34, 89. [DOI] [PubMed] [Google Scholar]
- [320].Jeon H, Simon CG Jr, Kim G, J. Biomed. Mater. Res., Part B. 2014, 102, 1580. [DOI] [PubMed] [Google Scholar]
- [321].Pupkaite J, Rosenquist J, Hilborn J. n, Samanta A, Biomacromolecules. 2019, 20, 3475. [DOI] [PubMed] [Google Scholar]
- [322].Dalton E, Chai Q, Shaw MW, McKenzie TJ, Mullins ES, Ayres N, J. Polym. Sci., Part A: Polym. Chem. 2019, 57, 1389. [Google Scholar]
- [323].Verbeke CS, Mooney DJ, Adv. Healthcare Mater. 2015, 4, 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Liu H, Nakagawa K, Chaudhary D, Asakuma Y, Tadé MO, Chem. Eng. Res. Des. 2011, 89, 2356. [Google Scholar]
- [325].Zhang D, Burkes WL, Schoener CA, Grunlan MA, Polymer. 2012, 53, 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Robinson JL, Moglia RS, Stuebben MC, McEnery MA, Cosgriff-Hernandez E, Tissue Eng., Part A. 2014, 20, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Sears NA, Seshadri DR, Dhavalikar PS, Cosgriff-Hernandez E, Tissue Eng., Part B. 2016, 22, 298. [DOI] [PubMed] [Google Scholar]
- [328].Saha K, Pollock JF, Schaffer DV, Healy KE, Curr. Opin. Chem. Biol. 2007, 11, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Ermis M, Antmen E, Hasirci V, Bioactive materials. 2018, 3, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Bettinger CJ, Langer R, Borenstein JT, Angew. Chem. Int. Ed. 2009, 48, 5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Benitez PL, Mascharak S, Proctor AC, Heilshorn SC, Integr. Biol. 2016, 8, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Karimi F, O’Connor AJ, Qiao GG, Heath DE, Advanced Healthcare Materials. 2018, 7, 1701324. [DOI] [PubMed] [Google Scholar]
- [333].Frith JE, Mills RJ, Cooper-White JJ, J. Cell Sci. 2012, 125, 317. [DOI] [PubMed] [Google Scholar]
- [334].Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG, J. Cell Sci. 2000, 113, 1677. [DOI] [PubMed] [Google Scholar]
- [335].Spatz JP, Mössmer S, Hartmann C, Möller M, Herzog T, Krieger M, Boyen H-G, Ziemann P, Kabius B, Langmuir. 2000, 16, 407. [Google Scholar]
- [336].Cavalcanti-Adam EA, Micoulet A, Blümmel J, Auernheimer J, Kessler H, Spatz JP, European journal of cell biology. 2006, 85, 219. [DOI] [PubMed] [Google Scholar]
- [337].Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP, Biophys. J. 2007, 92, 2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Gómez-Lamarca MJ, Cobreros-Reguera L, Ibáñez-Jiménez B, Palacios IM, Martín-Bermudo MD, J. Cell Sci. 2014, 127, 4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Shekaran A, García JR, Clark AY, Kavanaugh TE, Lin AS, Guldberg RE, García AJ, Biomaterials. 2014, 35, 5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Saidak Z, Le Henaff C, Azzi S, Marty C, Da Nascimento S, Sonnet P, Marie PJ, J. Biol. Chem. 2015, 290, 6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Mitra SK, Hanson DA, Schlaepfer DD, Nature reviews Molecular cell biology. 2005, 6, 56. [DOI] [PubMed] [Google Scholar]
- [342].Albelda SM, Buck CA, The FASEB journal. 1990, 4, 2868. [PubMed] [Google Scholar]
- [343].Siebers M, Ter Brugge P, Walboomers X, Jansen J, Biomaterials. 2005, 26, 137. [DOI] [PubMed] [Google Scholar]
- [344].Doyle A, McGarry MP, Lee NA, Lee JJ, Transgenic research. 2012, 21, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin M, Tacke F, Blood, The Journal of the American Society of Hematology. 2010, 115, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Wang RN, Paraskevas S, Rosenberg L, Journal of Histochemistry & Cytochemistry. 1999, 47, 499. [DOI] [PubMed] [Google Scholar]
- [347].Schittenhelm L, Hilkens CM, Morrison VL, Frontiers in immunology. 2017, 8, 1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [348].Byrareddy SN, Sidell N, Arthos J, Cicala C, Zhao C, Little DM, Dunbar P, Yang GX, Pierzchalski K, Kane MA, The Journal of Immunology. 2015, 194, 5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Zhukareva V, Obrocka M, Houle JD, Fischer I, Neuhuber B, Cytokine. 2010, 50, 317. [DOI] [PubMed] [Google Scholar]
- [350].Tamama K, Kerpedjieva SS, Advances in wound care. 2012, 1, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Sabbah N, Tamari T, Elimelech R, Doppelt O, Rudich U, Zigdon-Giladi H, Biomolecules. 2019, 9, 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Post A, Wang E, Cosgriff-Hernandez E, Ann. Biomed. Eng. 2019, 47, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Ekholm E, Hankenson KD, Uusitalo H, Hiltunen A, Gardner H, Heino J, Penttinen R, The American journal of pathology. 2002, 160, 1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [354].Liu Q, Limthongkul W, Sidhu G, Zhang J, Vaccaro A, Shenck R, Hickok N, Shapiro I, Freeman T, Journal of Orthopaedic Research. 2012, 30, 1626. [DOI] [PubMed] [Google Scholar]
- [355].Mizuno M, Fujisawa R, Kuboki Y, Journal of cellular physiology. 2000, 184, 207. [DOI] [PubMed] [Google Scholar]
- [356].Xiao G, Wang D, Benson MD, Karsenty G, Franceschi RT, Journal of Biological Chemistry. 1998, 273, 32988. [DOI] [PubMed] [Google Scholar]
- [357].García JR, Clark AY, García AJ, Journal of biomedical materials research Part A. 2016, 104, 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Clover J, Dodds R, Gowen M, Journal of cell science. 1992, 103, 267. [DOI] [PubMed] [Google Scholar]
- [359].Saidak Z, Le Henaff C, Azzi S, Marty C, Da Nascimento S, Sonnet P, Marie PJ, Journal of Biological Chemistry. 2015, 290, 6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [360].Petrie TA, Raynor JE, Reyes CD, Burns KL, Collard DM, García AJ, Biomaterials. 2008, 29, 2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].Stephansson SN, Byers BA, García AJ, Biomaterials. 2002, 23, 2527. [DOI] [PubMed] [Google Scholar]
- [362].Elmengaard B, Bechtold JE, Søballe K, Biomaterials. 2005, 26, 3521. [DOI] [PubMed] [Google Scholar]
- [363].Ferris DM, Moodie GD, Dimond PM, Giorani CWD, Ehrlich MG, Valentini RF, Biomaterials. 1999, 20, 2323. [DOI] [PubMed] [Google Scholar]
- [364].Yamazaki CM, Kadoya Y, Hozumi K, Okano-Kosugi H, Asada S, Kitagawa K, Nomizu M and Koide T, Biomaterials. 2010, 31, 1925. [DOI] [PubMed] [Google Scholar]
- [365].Liu XJ, Kong FZ, Wang YH, Zheng JH, Wan WD, Deng CL, Mao GY, Li J, Yang XM, Zhang YL and Zhang XL, Plos one. 2013, 8, p.e67124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Fleischmajer R, Utani A, MacDonald ED, Perlish JS, Pan TC, Chu ML, Nomizu M, Ninomiya Y and Yamada Y, Journal of cell science. 1998, 111, 1929. [DOI] [PubMed] [Google Scholar]
- [367].Kim JP, Zhang K, Chen JD, Wynn KC, Kramer RH and Woodley DT, Journal of cellular physiology. 1992, 151, 443. [DOI] [PubMed] [Google Scholar]
- [368].Weston BS, Wahab NA and Mason RM, Journal of the American Society of Nephrology. 2003, 14, 601. [DOI] [PubMed] [Google Scholar]
- [369].Ghatak S, Niland S, Schulz J-N, Wang F, Eble JA, Leitges M, Mauch C, Krieg T, Zigrino P, Eckes B, The American Journal of Pathology. 2016, 186. [DOI] [PubMed] [Google Scholar]
- [370].San Antonio JD, Zoeller JJ, Habursky K, Turner K, Pimtong W, Burrows M, Choi S, Basra S, Bennett JS, DeGrado WF, The American journal of pathology. 2009, 175, 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [371].Fukushi J.-i., Makagiansar IT, Stallcup WB, Molecular biology of the cell. 2004, 15, 3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Wu C-C, Wang L-C, Su Y-T, Wei W-Y, Tsai K-J, Biomaterials. 2018, 185, 142. [DOI] [PubMed] [Google Scholar]
- [373].Malinda KM, Nomizu M, Chung M, Delgado M, Kuratomi Y, Yamada Y, Kleinman HK, Ponce ML, The FASEB Journal. 1999, 13, 53. [PubMed] [Google Scholar]
- [374].Hynes RO, Nature medicine. 2002, 8, 918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.