Abstract
The use of mouse derived mammary organoids can provide a unique strategy to study mammary gland development across a normal life cycle, as well as offering insights into how malignancies form and progress. Substantial cellular and epigenomic changes are triggered in response to pregnancy hormones, a reaction that engages molecular and cellular changes that transform the mammary epithelial cells into “milk producing machines”. Such epigenomic alterations remain stable in post-involution mammary epithelial cells and control the reactivation of gene transcription in response to re-exposure to pregnancy hormones. Thus, a system that tightly controls exposure to pregnancy hormones, epigenomic alterations, and activation of transcription will allow for a better understanding of such molecular switches. Here, we describe the characterization of ex vivo cultures to mimic the response of mammary organoid cultures to pregnancy hormones and to understand gene regulation and epigenomic reprogramming on consecutive hormone exposure. Our findings suggest that this system yields similar epigenetic modifications to those reported in vivo, thus representing a suitable model to closely track epigenomic rearrangement and define unknown players of pregnancy-induced development.
Supplementary Information
The online version of this article (10.1007/s10911-020-09465-0) contains supplementary material, which is available to authorized users.
Keywords: Mammary organoids, Pregnancy-induced development, Epigenomics
Introduction
The mammary gland is one of the few organs that undergoes multi-stage development post-birth. Although significant changes mark the pre- and post-pubescence mammary developmental stages, those associated with pregnancy have the greatest effect on cellular and tissue reorganization.
The adaptation of hormonal conditions to mimic many of the cellular and molecular events brought to the mammary epithelial cells (MECs) by pregnancy may allow for better dissection of signaling and responses that would otherwise be masked during tissue processing or untimed hormone exposure. Most importantly, such strategy would support the development of systems to understand the influence of pregnancy hormones in MECs from a variety of animal species, thus providing a suitable platform to understand pregnancy induced mammary development from an evolutionary perspective.
For the past several years, optimized three dimensional (3D) organoid cultures have provided a strong and reliable platform to dissect normal and malignant mammary development. In fact, several studies have demonstrated that inclusion of several hormone cocktails to 3D cultures induces branching morphogenesis [1–5] and developmental stage transitions, such as lactation and involution [6]. In addition, 3D organoid cultures allow for the validation of mammary focused phenotypes observed in genetically engineered mouse models [7–9] and during mammary oncogenesis [10, 11]. These findings support its relevance to define nuances of mammary development in a more tightly controlled strategy.
Several studies have reported that a pregnancy cycle (gestation, lactation and involution) induces stable modifications to the epigenome of MECs, which influence normal development and breast cancer risk [12, 13]. More specifically, post-pregnancy MECs were shown to retain an epigenetic memory from previous pregnancies, marked by stable loss of DNA methylation at specific regulatory regions, which influences the expression of milk-associated proteins in consecutive exposure to pregnancy hormones [14, 15]. More recently, this rapid increase on gene expression was also observed in organoid cultures derived from post-pregnancy mammary tissue, therefore supporting a cell autonomous regulation of such epigenetic memory [1].
Here, we demonstrate that organoid cultures also represent a suitable system to understand the molecular changes brought to MECs by pregnancy hormones. Using this approach, we provide a more in-depth picture of the dynamics of gene expression and active regulatory regions of 3D organoid cultures derived from pre- and post-pregnancy mammary tissue. Our strategy demonstrates how readily the epigenomic and transcriptomic changes are in response to pregnancy hormones, and how such changes are enhanced in cells re-exposed to pregnancy hormones. Moreover, we utilized our system to validate the role of a known mammary regulatory factor, EZH2, on organoid development, a strategy that yielded hypothesis for its role in controlling gene expression in response to re-exposure to pregnancy hormones. Further utilization of such robust and highly controlled 3D culturing system will undoubtedly improve our knowledge of activation or repression of processes that influence oncogenesis in a pregnancy dependent manner.
Results
Pregnancy Hormones Induce Robust Upregulation of Casein 2 (Csn2) in Post-Pregnancy Mammary Organoid Cultures
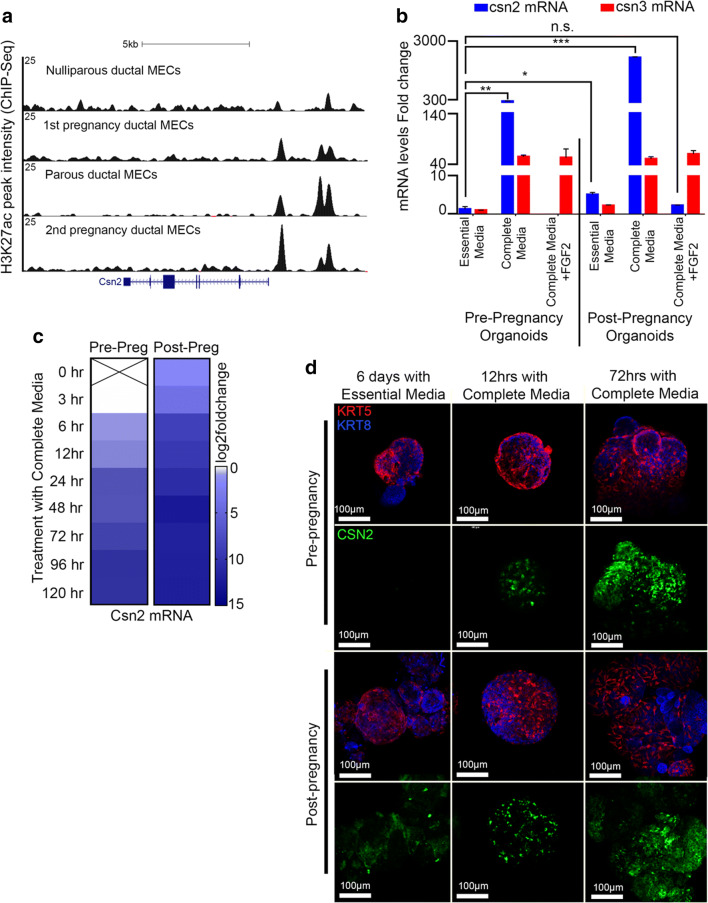
Our previous work identified pregnancy induced changes in the epigenome of MECs [14]. More recently, we utilized chromatin immune precipitation sequencing (ChIP-seq) and demonstrated a substantial expansion of the active regulatory landscape of post-pregnancy MECs [1]. Amongst the alterations brought by a full pregnancy cycle, we found two regions upstream of the Csn2 genomic locus, a milk associated protein, to stably gain H3K27ac marks after pregnancy (Fig. 1a). Such pregnancy-induced alterations were associated with a 10-fold increase in Csn2 mRNA levels and protein levels in flow cytometry isolated, post-pregnancy luminal cells and organoid cultures treated with pregnancy hormones [1]. Based on these findings, we predict that Csn2 levels can act as a reporter marker to track gene reactivation of pregnancy-induced epigenome.
Fig. 1.
Pregnancy hormones induce robust upregulation of Casein 2 (Csn2) in post-pregnancy mammary organoid cultures. (a) Genome browser tracks showing distribution of H3K27Ac peaks at the Csn2 locus in luminal ductal MECs harvested from pre- and post-pregnancy mice at distinct pregnancy cycles. (b) mRNA levels (qPCR) of casein genes, Csn2 and Csn3, in pre- and post-pregnancy mammary organoids grown with essential medium, complete medium (supplemented with pregnancy hormones) or complete medium with FGF2 n = 2 biological replicates, with 3 technical replicates each. ***p = 0.0002, **p = 0.002, *p = 0.01 (c) Csn2 mRNA levels (2^deltadeltaCT) in pre-pregnancy mammary organoids grown with complete medium. Normalized to pre- pregnancy organoids treated with complete medium for 3 h. (d) Immunofluorescence (IF) imaging of whole mounted pre- and post-pregnancy mammary organoids before or after complete medium exposure for 12 h and 72 h. KRT8 (blue), KRT5 (red) and CSN2 (green). Scale: 100 μm. For analyses, error bars indicate standard error of the mean (SEM) across samples of same experimental group. p values were defined using Welch’s t-test
In order to investigate the conditions that support increased gene expression in response to pregnancy hormones, we derived organoid cultures utilizing pre- and post-pregnancy mammary tissue and measured Csn2 and Casein 3 (Csn3), an additional milk associated protein, mRNA levels from cultures grown at several conditions (Supplementary Fig.S1). Our results confirmed that Csn2 mRNA levels were ~ 9-fold higher in post-pregnancy organoids grown with pregnancy hormones, than in pre-pregnancy organoids grown under the same conditions (Fig. 1b). Interestingly, Csn2 mRNA levels failed to increase in pre- and post-pregnancy organoids grown with pregnancy hormones and Fibroblast Growth Factor 2 (FGF2), suggesting that the proliferation and self-renewal programs known to be regulated by FGF2 [16, 17] may have interfere with the differentiation and specialization process driven by pregnancy hormones in mammary organoids. Moreover, Csn3 mRNA levels were equally induced in pre- and post-pregnancy organoids grown with complete medium (~47- and ~ 48-fold, respectively), independently of FGF2 presence, thus suggesting that its expression is regulated by pregnancy hormones independent of a previous exposure to pregnancy hormones.
We next set out to more precisely define the dynamics of Csn2 mRNA levels in pre- and post-pregnancy organoids using a time-course response to pregnancy hormones. Our results suggested that Csn2 mRNA level differences peaked after 6 hours (h) of pregnancy hormones treatment, with post-pregnancy organoids expressing ~20x fold higher mRNA levels than pre-pregnancy ones (Fig. 1c). Such differences in mRNA levels were normalized after 96 h of pregnancy hormone treatment, a time point where pre- and post-pregnancy organoids expressed similar levels of Csn2 mRNA. Accordingly, CSN2 protein levels peaked higher in post-pregnancy mammary organoids in response to pregnancy hormone treatment, a result that agrees with previous observations [1] (Fig. 1d). Collectively, these results confirm that rapid gene expression activation in response to pregnancy hormones are maintained in organoid cultures derived from post-pregnancy MECs, and suggests the utilization of Csn2 levels as a reporter marker to track pregnancy-induced epigenetic gene expression reactivation.
Pregnancy Hormones Drive Changes to the Active Regulatory Landscape of Pre-Pregnancy Mammary Organoid Cultures
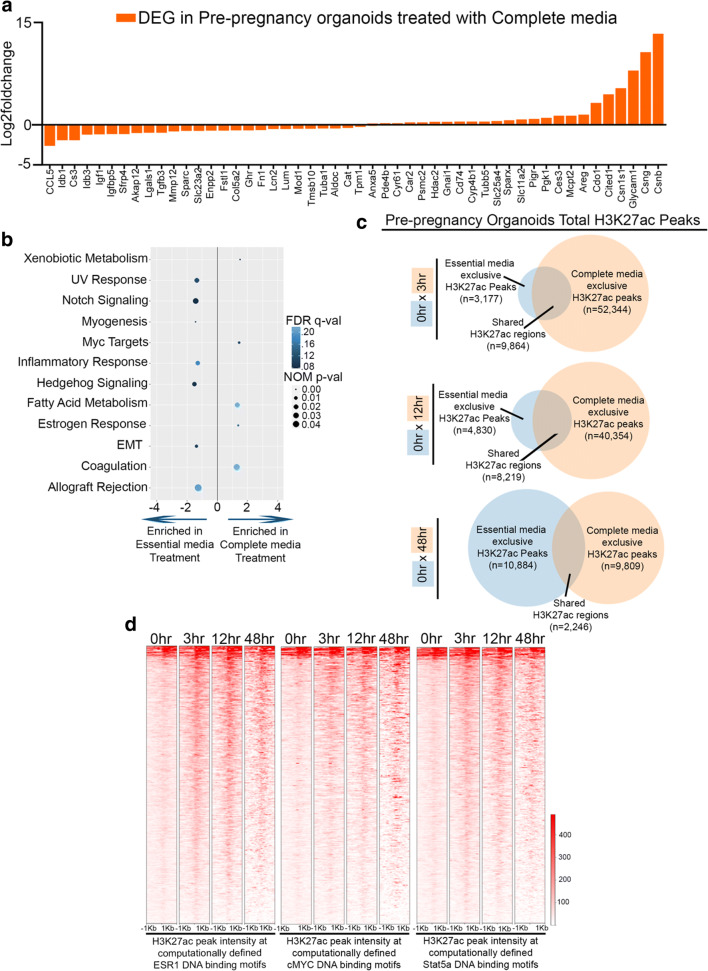
We next decided to fully characterize the alteration to the transcription output and epigenome of pre-pregnancy mammary organoids in response to pregnancy hormones. Thus, we utilized a previously published RNA-seq data set that compared the transcriptome of whole mammary tissue from nulliparous and parous rats and mice with whole mouse mammary gland [18], to define whether a signature of parity was present in pre-pregnancy organoid cultures grown with pregnancy hormones (Complete medium, 9 days of treatment). We found that 31% of such parity signature was present in pre-pregnancy organoid cultures grown with pregnancy hormones, thus suggesting that a signature of parity was established during in vitro exposure to pregnancy hormones, (Fig. 2a and Supplementary Table S1). Gene Set Enrichment Analysis (GSEA) demonstrated that signatures of active proliferation [19, 20] (Myc targets), milk synthesis [21] (fatty acid metabolism) and estrogen response [22] were enriched in pre-pregnancy organoid cultures grown with pregnancy hormones, thus supporting their pregnancy- like development (Fig. 2b). Interestingly, signatures associated with immune response and stem-like state developmental processes, known to be suppressed during pregnancy [23–25], were downregulated in pre-pregnancy organoid cultures grown with pregnancy hormones, thus supporting that our system incorporates a complete array of transcriptional modifications associated with mammary pregnancy development.
Fig. 2.
Pregnancy hormones drive changes to the active regulatory landscape of pre-pregnancy mammary organoid cultures. (a) RNAseq quantification of parity gene signature in pre-pregnancy organoids treated with complete medium for 9 days. n = 2 biological replicates. (b) GSEA of gene networks differentially expressed in pre-pregnancy organoids treated with complete medium for 9 days. (c) Venn diagrams comparing H3K27ac peaks of pre-pregnant mammary organoid cultures before complete medium treatment (0 h) and after 3 h, 12 h and 48 h treatment with complete medium. (d) Density plot showing H3K27ac levels at computationally defined DNA binding motifs recognized by the transcription factors ESRa, MYC and STAT5 in pre-pregnancy mammary organoids before complete medium treatment (0 h) and after 3 h, 12 h, 48 h treatment with complete medium
Given that treatment with pregnancy hormones induced pregnancy associated transcriptional changes to organoid cultures, we next investigated whether changes to the active regulatory landscape were also established by such treatment. In doing so, we utilized Cleavage under targets and release using nuclease (Cut&Run), a strategy that allows for immunoprecipitation of native chromatin [26] to profile the active histone mark H3K27ac in pre-pregnancy mammary organoids treated with pregnancy hormones for 3 h, 12 h and 48 h. Total H3K27ac peak analysis demonstrated a ~ 40-fold expansion of the number of H3K27ac peaks in mammary organoids treated with pregnancy hormones for the first 3 h and 12 h (Fig. 2c). Such expansion likely activated putative enhancer regions, given that analyses of total H3K27ac peaks genomic distributions indicated a ~ 5-fold and ~ 4-fold increase, respectively, in the number of peaks mapping to intergenic and genic regions, supporting the notion that pregnancy induces changes to the enhancer landscape of MECs (Supplementary Fig. S2A and S2B). Our analyses also demonstrated a 4 to 3-fold decrease on the number of H3K27ac peaks shared between organoids with no treatment (0 h) to those grown with pregnancy hormones for 48 h, in comparison to those shared between 0 h and 3 h, and 0 h and 12 h, respectively, suggesting a greater divergence of active regulatory landscape after prolonged exposure to hormones (Fig. 2c, bottom panel).
Gene Ontology (GO) analyses suggested that H3K27ac peaks exclusive to organoids after hormone treatment for 3 h were associated with genes that play a role in adherent junction organization and response to epidermal growth factor, while peaks exclusive to organoids treated with hormones for 12 h were enriched for genes associated with negative regulation of notch signaling and positive regulation of actin filament bundle assembly, all processes previously linked with pregnancy development [27–30] (Supplementary Fig. S2C). Amongst the networks predicted to be associated with H3K27ac peaks exclusive to organoids after hormone treatment for 48 h, we identified terms associated with unfolded protein response, protein catabolism and endoplasmic reticulum response, all pathways previously described to be essential for the post-pregnancy involution process of mammary glands [31–35] (Supplementary Fig. S2C, bottom panel). In fact, it was recently demonstrated that pregnancy-induced mammary involution can be recapitulated in vitro [6], thus supporting an involution-like state to organoid cultures treated for 48 h with pregnancy hormones, an observation that we are now supplementing with molecular signatures. Moreover, we also observed that a fraction of all pregnancy hormone exclusive peaks were shared across all 3 timepoints, suggesting sustainability of epigenetic changes established by pregnancy signals throughout the treatment time-course (Supplementary Fig. S2D).
We next set out to define whether specific Transcription Factor (TF) DNA motifs were enriched in regions that gained H3K27ac peaks after hormone treatment (complete medium exclusive peaks). Our analyses identified gain of H3K27ac peaks at DNA motifs predicted to be occupied by the TFs Interferon responsive factor 1 (IRF1) and Forkhead box protein C2 (FOXC2), previously described to enhance proliferation and suppress apoptosis of MECs [36, 37], in organoid cultures treated with pregnancy hormones for 3 h and 12 h, suggesting that signals of immediate response to pregnancy hormones remained active for many hours (Supplementary Fig. S2E). Conversely, our analyses identified DNA motifs for a different set of TFs in organoid cultures treated with pregnancy hormones for 48 h, including the Specificity protein 3 (SP3), which has been implicated in regulating gap junction formation during pregnancy and lactation [38]. These results further suggest that prolonged hormone exposure in vitro drives molecular programs that could be demarking a differential developmental stage, as suggested by the analyses of terms associated with H3K27ac peaks exclusive to organoid cultures treated with pregnancy hormones for 48 h (Fig. 2c, bottom panel).
Surprisingly, this unbiased TF DNA motif analyses did not indicate enrichment of classical regulators of mammary pregnancy-induced development such as Estrogen receptor α (ERα) [39, 40], cellular Myelocytomatosis (cMYC) [20] and Signal transducer and activator of transcription 5 (STAT5) [41, 42], at H3K27ac peaks exclusive to organoid treated with pregnancy hormones, suggesting that perhaps these TFs could be associating with chromatin, even in the absence of pregnancy hormones. To address this point, we analyzed H3K27ac peaks intensity of computationally defined DNA motifs recognized by ERα (EREs), cMYC (eBOX) and STAT5 (GAS), and found increased peak intensity at these DNA motifs in response to treatment with pregnancy hormones, suggesting that some of these regions may be occupied by such TFs during organoid growth and expansion, and further gain of H3K27ac levels in response to pregnancy hormones which may represent increased TF binding and enhanced gene expression activation (Fig. 2d).
Altered Epigenomic Landscape Drives Post-Pregnancy Mammary Organoids Response to Re-Exposure to Pregnancy Hormones
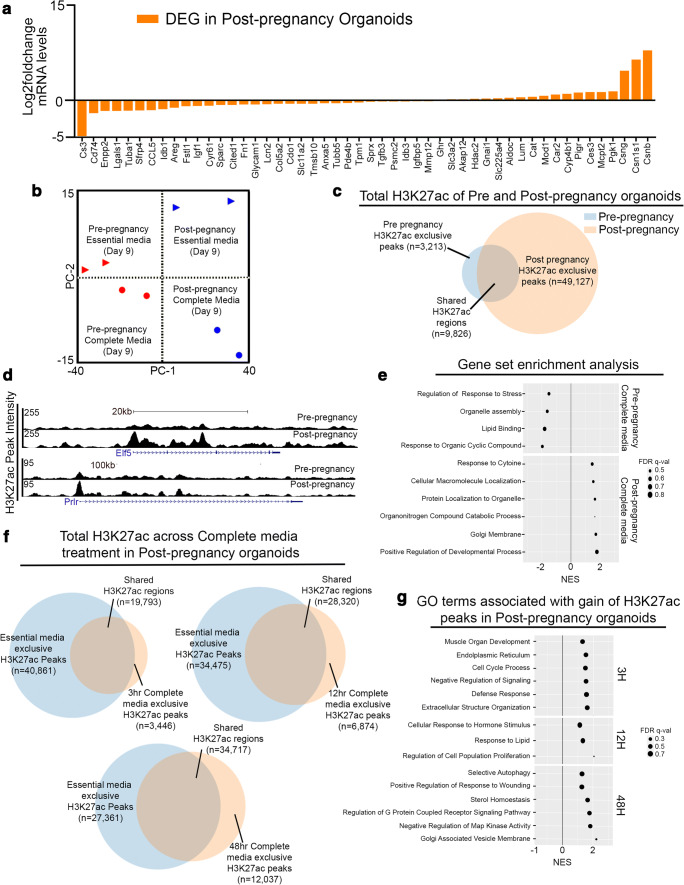
Having established the effects of pregnancy hormone treatment on gene expression and on the epigenome of pre-pregnancy mammary organoids, we next investigated the dynamics of epigenomic remodeling in post-pregnancy organoids cultures in response to pregnancy hormones. We confirmed that post-pregnancy mammary organoids grown with essential medium (no hormones) retained a parity gene signature with 36% of the genes matching the originally described gene expression analyses, suggesting that mammary organoid culturing does not erase a transcription state established by a previous pregnancy (Fig. 3a and Supplementary Table S2). Moreover, principal component analysis of RNAseq datasets derived from untreated and hormone-treated pre- and post-pregnancy organoid cultures demonstrate grouping of transcriptomes based on treatment with pregnancy hormones (PC-1) and parity (PC-2), further suggesting that the post-pregnancy organoids, independent of complete medium treatment, display a unique gene expression profile in organoid cultures. This is further supported by the clustering of pre- and post-pregnancy organoid cultures with complete medium, which shows their expression profiles are different from their untreated counterparts, but similar to each other (Fig. 3b).
Fig. 3.
Altered epigenomic landscape drives post-pregnancy mammary organoids response to re-exposure to pregnancy hormones. (a) RNAseq quantification of parity gene signature in post-pregnancy mammary organoids grown with essential media (no pregnancy hormones) for 9 days. n = 2 biological replicates. (B) Principal component analyses of gene expression datasets from pre- and post-pregnancy organoids treated with and without complete medium for 9 days. (c) Venn diagram comparing total H3K27ac peaks of untreated pre-pregnant mammary organoids and post-pregnancy mammary organoids. (d) Genome browser tracks showing distribution of H3K27ac peaks in untreated pre- and post-pregnancy mammary organoid cultures for the Elf5 and Prlr loci. (e) S-plot showing untreated post-pregnancy organoid exclusive H3K27ac peaks intersected with gene expression from untreated post-pregnancy organoids day 9. (f) Venn diagrams comparing H3K27ac peaks from post-pregnant mammary organoid cultures before complete medium treatment (0 h) and after 3 h, 12 h and 48 h of treatment with complete medium (g) GSEA of gene networks exclusive to post-pregnancy mammary organoids treated with complete medium for 3 h, 12 h and 48 h
In order to define whether the unique transcriptome of post-pregnancy mammary organoids have an epigenetic basis, we employed active histone mark H3K27ac Cut&Run. Initial comparisons of total H3K27ac peaks in untreated organoids demonstrated a ~ 15-fold expansion of the active epigenomic landscape in post-pregnancy mammary organoids, a substantial alteration to the epigenomic landscape brought by pregnancy that was previously reported [1, 14] and that was retained after organoid culturing (Fig. 3c). Such expansion to the active epigenomic landscape was mostly observed on intergenic and genic regions, given the 5-fold and 4.6-fold increase of H3K27ac peaks at these regions in post-pregnancy organoids, in comparison to pre-pregnancy organoids (Supplementary Fig.S2 A and Supplementary Fig. S3A). Furthermore, we identified 14,441 genes that were associated with H3K27ac peaks exclusive to untreated post-pregnancy organoids, which included genomic regions associated with E74-like factor 5 (Elf5) and Prolactin receptor (Prlr) genes, known players of mammary gland development and lactation [43, 44] (Fig. 3d). We utilized this list of genes to identify the dynamics of gene expression regulation using RNAseq datasets from pre- and post-pregnancy organoid treated with pregnancy hormones.
In total, we identified 350 Differentially Expressed Genes (DEGs, fold change greater than 4) across pre- and post-pregnancy mammary organoids treated with pregnancy hormones, that associated with the H3K27ac peaks exclusive to untreated post-pregnancy organoids, thus supporting the notion that many of the genes with expression influenced by pregnancy hormones are epigenetically altered in post-pregnancy MECs (Supplementary Table S3). Further gene set enrichment analyses of H3K27ac-associated DEGs demonstrated enrichment of pathways associated with Golgi and Endoplasmic reticulum processes (post-pregnancy organoids treated with pregnancy hormones), and cytoplasmic transport and lipid-binding (pre-pregnancy organoids treated with pregnancy hormones), further suggesting that an array of regulatory networks that demark milk production-like processes and those associated with involution are represented in the epigenome of post-pregnancy MECs (Fig. 3e).
We next set out to define changes to post-pregnancy epigenomic landscape in response to re-exposure to pregnancy hormones. Genomic distributions analyses of H3K27ac peaks across untreated (0 h) and hormone treated (3 h, 12 h and 48 h) post-pregnancy mammary organoids indicated a minor increase on the percentage of intergenic regions across all treated timepoints (3 h = 11.8%, 12 h = 15.0% and 48 h = 18.9%) compared to untreated (0 h = 19.3%) suggesting that pregnancy hormones do not dramatically modify the post-pregnancy landscape (Supplementary Fig. S3A). Interestingly, comparison of total H3K27ac peaks from untreated post-pregnancy mammary organoids with peaks present in pre-pregnancy organoids treated for 3 h with pregnancy hormones, demonstrated an overlap of 55% of all peaks, thus supporting that the epigenome of post-pregnancy mammary organoids were established by signals present during pregnancy, and that a substantial fraction of these regions were associated with a response to pregnancy hormones (Supplementary Fig. S3B).
Further analyses of total H3K27ac peaks demonstrated an overall 1.7-fold increase on the percentage of shared peaks and a 3.5-fold increase on the percentage of hormone-treated exclusive peaks across the pregnancy hormone treatment time points, further supporting that hormone treatment did not dramatically modify the post-pregnancy landscape (Fig. 3f). Nonetheless, regions that gained H3K27ac peaks in post-pregnancy organoids treated with hormones were associated with a series of genes involved with branching morphogenesis (3 h), cell contraction and calcium transport (12 h), and endoplasmic reticulum process (48 h). These results suggest a progression from tissue expansion (3 h), lactation-like (12 h) and involution-like (48 h) stages of development (Supplementary Fig. S3C). In fact, IF analyses of organoids grown with pregnancy hormones illustrates a differential morphology of myoepithelial cells (Cytokeratin 5, KRT5+ cells, red) specifically in post-pregnancy organoid cultures, a phenotype that may indicate contractibility of such cells [45, 46] and further support their advanced developmental stage in response to pregnancy hormones (Supplementary Fig. S3D).
Surprisingly, our analyses showed that ~50% of all H3K27ac peaks in post-pregnancy organoids detected in untreated cultures failed to gain active histone marks in cultures treated with pregnancy hormones, suggesting that a fraction of the pregnancy-induced epigenome does not get immediately altered in response to pregnancy hormones (Fig. 3f). Analyses of the genes associated with these regions indicated an enrichment for pathways controlling DNA damage, response to oxidative stress and immune communication (Supplementary Fig. S3E). These results suggest that a series of tissue homeostasis pathways are constantly activated in post-pregnancy MECs, and their suppression in response to pregnancy hormones highlight additional mechanisms that may play a role on enhancing post-pregnancy MECs development in consecutive exposure to pregnancy hormones.
With the notion that post-pregnancy mammary organoids display many of the H3K27ac peak gain present in pre-pregnancy organoids treated with hormones (Supplementary Fig. S3B), we set out to define differential H3K27ac levels across hormone treatment in pre- and post-pregnancy mammary organoids. In doing so, we identified that regions associated with genes controlling Extracellular matrix (ECM) remodeling (3 h), response to lipids (12 h), and wound healing (4 8 h) gained higher levels of H3K27ac mark in post-pregnancy mammary organoids (Fig. 3g). Given that these pathways control tissue remodeling during pregnancy-induced mammary expansion (ECM, 3 h) [47, 48], milk associated components (lipids, 12 h) [49, 50], and involution-like processes (wound healing, 48 h) [51, 52], our analyses further suggests that post-pregnancy organoids treated with pregnancy hormones may have an advanced developmental progress across all stages of pregnancy-induced development.
Utilization of Organoid Cultures to Define Players in Pregnancy-Induced Development and Post-Pregnancy Epigenome
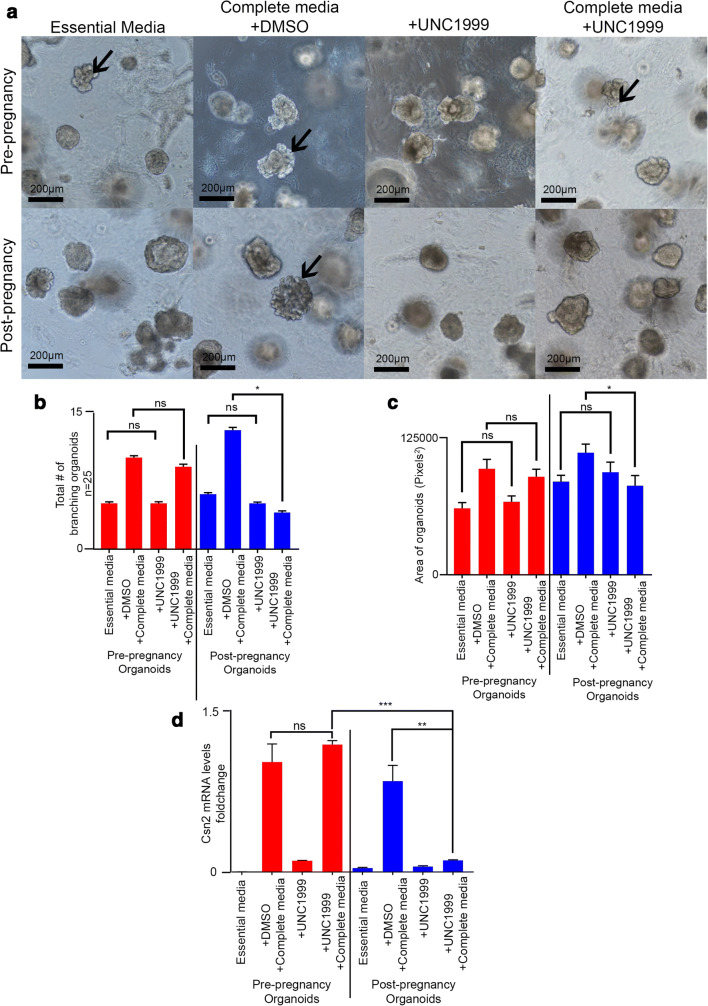
The identity of factors that establish and maintain the post-pregnancy epigenome remains unclear. In contrast, many factors have been described to play a role during overall pregnancy-induced development of the mammary gland. In fact, loss of the histone methyltransferase factor Enhancer of zest homolog 2 (EZH2), a key component of the Polycomb Repressive Complex 2 (PRC2), has been shown to impact pregnancy-induced stages of mammary gland development and duct elongation defects in vivo [9]. In order to illustrate the robustness of utilizing organoid systems to understand the pregnancy-induced enhancer landscape, we utilized an EZH2 chemical inhibitor (UNC1999) to block its activity in pregnancy hormone treated pre- and post-pregnancy organoids, and characterize its role on branching morphogenesis and Csn2 mRNA levels.
We found that EZH2 inhibition had no significant effect on branching morphogenesis of pre-pregnancy mammary organoids, independent of growth conditions (Fig. 4a-b). Conversely, we found that EZH2 inhibition blocked the effects of pregnancy hormones on post-pregnancy mammary organoids with a 3.25-fold decrease in the number of branching organoids, thus suggesting a specific role for EZH2 on controlling the branching of organoids previously exposed to pregnancy hormones (Fig. 4a-b). We also observed EZH2 inhibition had a small, but statistically significant effect on post-pregnancy organoid size, yielding an ~1.35-fold smaller organoids than those grown with complete media treatment without EZH2 inhibition, an effect not observed in pre-pregnancy organoids (Fig. 4c). Moreover, we also observed an ~11-fold downregulation of Csn2 mRNA in post-pregnancy organoids grown with pregnancy hormones and the EZH2 inhibitor, in comparison to pre-pregnancy organoids under the same conditions (Fig. 4d), further suggesting that EZH2 may work in conjunction with some of the pregnancy-induced epigenetic modifications. Overall, this proof-of-principal analyses illustrates the robustness of organoid systems to understand gene activation and epigenomic reprogramming. In conclusion, we demonstrate that organoid cultures are a suitable system to define specific drivers of gene regulation from a pregnancy-induced landscape perspective.
Fig. 4.
Utilization of organoid cultures to define players in pregnancy-induced development and post-pregnancy epigenome. (a) Representative brightfield images of mammary organoids treated with essential medium or complete medium, supplemented with either DMSO (Dimethyl sulfoxide, control), or EZH2 inhibitor UNC1999 for 48 h. Arrows indicated examples of branching organoids. Scale = 200 μm. (b) Branching quantification of pre- and post-pregnancy mammary organoid cultures treated with essential medium or complete medium, supplemented with either DMSO (control) or EZH2 inhibitor UNC1999. 13 fields of view per well/replicate. n = 25 organoids. ns = not significant; *p = 0.027 differences between post-pregnancy organoids treated with complete media and complete media with UNC1999. Error bars indicate standard error of the mean (SEM) across samples of same experimental group. p values were defined using Students t-test. (c) Size quantification of pre- and post-pregnancy mammary organoid cultures treated with essential medium or complete medium, supplemented with either DMSO (control) or EZH2 inhibitor UNC1999. n = 20 organoids per condition. ns = not significant; *p = 0.018 differences between post-pregnancy organoids treated with complete medium and DMSO and post-pregnancy organoids treated with complete medium and UNC1999. For analyses, error bars indicate standard error of the mean (SEM) across samples of same experimental group. p values were defined using Welch’s t-test. (d) Csn2 mRNA levels (qPCR) in pre- and post-pregnancy mammary organoid cultures treated with either essential medium or complete medium, with DMSO control or UNC1999. *p = 0.0364 differences between post-pregnancy organoids treated with complete medium and DMSO and post pregnancy organoids treated with complete medium and UNC1999. ***p = 0.0009 differences between pre-pregnancy organoids treated with complete medium and UNC1999, and post-pregnancy organoids treated with complete medium and UNC1999. Error bars indicate standard error of the mean (SEM) across samples of same experimental group. p values were defined using Welch’s t-test
Discussion
Our studies demonstrate that organoid cultures are a suitable approach for investigating the effects of pregnancy hormones on the epigenome and transcriptional output of MECs, as well as providing mechanistic insight into how such responses occur in cells previously exposed to a pregnancy cycle in vivo. Utilizing our 3D organoid culturing method to dissect molecular mechanisms controlled by pregnancy signals in MECs presents a series of advantages. For example, the pre-culturing timing of ~6 days, designed to allow mammary organoids to acclimate to culturing, provides a normalization period to remove non-MECs from culture, and to diminish MEC-specific signals present in mice at distinct stages of the estrous cycle. In addition, our proposed method provides a platform to dissect into cell-autonomous mechanisms that operate within a pregnancy-induced epigenome, given that changes to ECM and immune composition have been described in mammary glands after a pregnancy cycle [51, 53–55] . Nonetheless, the incorporation of different ECM substrates and immune cells into organoid cultures may highlight further signaling pathways that together with MECs, influence a myriad of mechanisms controlling pregnancy-induced development.
The present study validated observations proposed in flow cytometry isolated MECs and supports the idea that a full pregnancy cycle induces stable epigenomic changes that alter the transcriptional output of MECs [1]. By analyzing markers of active regulatory regions (H3K27ac), our current method allowed for a snapshot of epigenetic changes in a timely fashion, thus illustrating some of the immediate molecular responses to pregnancy hormones. Given that additional organoid methods can mimic phenotypic alterations observed during involution [6], a deep molecular analysis of such developmental stage would provide further insights into how the epigenome is shaped to assume a post-involution state. Moreover, incorporating additional histone marks of poised enhancers (H3K4me1) or repressed enhancers (H3K27me) will illustrate parts of the epigenome that may became less active after pregnancy. Interestingly, EZH2 has been implicated to contribute to the memory machinery of cells either functioning as or recruiting DNA methyltransferases to certain genes, which could play a part in how post-pregnancy cells turn off certain genes during pregnancy [56]. Specifically, mammary epithelial cells from pre-pregnancy mice lacking EZH2 express high levels of milk related genes Csn2 and Whey acidic protein (Wap) earlier in pregnancy than wild-type mice [57], supporting its specialized role on controlling an epigenomic landscape brought by a previous pregnancy cycle. Further investigation regarding the relationship between EZH2, epigenetic memory, and gene re-activation, in response to pregnancy hormones, is needed to precily define molecular dynamics that control mammary development and parity-induced epigenome.
The application of an organoid strategy could also help to define the effects that the age of pregnancy has on controlling molecularly relevant and evolutionary conserved epigenetic modifications, which govern post-pregnancy breast tissue homeostasis and development in consecutive pregnancies. For example, the age of first pregnancy in women has a strong influence on milk supply and breast cancer development. Even though several studies report that human females have a significantly increased milk supply during a second pregnancy [58–61], women that experienced their first pregnancy after 35 years of age are at risk to require medical intervention to improve milk production and breastfeeding [62]. Thus, employing tissue fragments collected from women and rodents spanning a variety of age of first pregnancy and those from aged subjects never exposed to pregnancy hormones will address outstanding questions about the impact of aging on evolution and molecular adaptation of breast cells to control development, gene expression and milk production in response to pregnancy signals.
In addition, a series of large-scale population studies found correlations between the age of first full-term pregnancy and breast cancer development [63, 64]. Women younger than the age of 25 have an approximate 30% decrease in the incidence of breast cancer. In contrast, pregnancy in women older than 38 years of age correlates with a 30–50% increase in developing more aggressive subtypes of breast cancer within the first ten years after giving birth [63, 65]. Notably, we recently reported that a full pregnancy cycle blocked cancer initiation and epigenetic reprograming in murine MECs after overexpressing the potent oncogene cMYC, suggesting that pregnancy-induced molecular changes may impact the transcriptional output that can drive cancer initiation [1]. Such effects were also observed in organoid cultures derived from post-pregnancy MECs, which supports the conclusion that cell autonomous signals that block cancer initiation can also be studied in 3D culturing systems.
Lastly, we provided a proof-of-principal perspective regarding the robustness of organoid systems. The incorporation of small chemical inhibitors, or more direct genetic manipulation of regulatory factors, to the culturing system presented here may also represent a cost- and time-effective screening strategy to define new players that control the mammary epithelial epigenome and block cancer initiation. Such strategies could also be employed in organoid cultures derived from healthy breast tissue of women with genetic or familial predisposition to develop breast cancer, as an additional tool to search for strategies that may prevent on further decrease the risk of cancer.
Methods
Antibodies. All antibodies were purchased from companies listed below and used without further purification. Antibodies for IF: anti-β-casein/csn2 mouse monoclonal antibody (Santa Cruz Biotechnology Inc., #sc-166,530, 200 μg/mL, 1:300 dilution, RRID: AB_2084348), anti-Cytokeratin5 rabbit monoclonal antibody (Abcam, #EP1601Y, 0.5 mg/ML 1:300 dilution RRID:AB_869890), anti-Cytokeratin-8 mouse monoclonal antibody (Abcam, #EP1628Y, 0.5 mg/mL 1:300 dilution, RRID:AB_869901). Antibody for Cut&Run: H3K27ac histone marks (Abcam, #ab4729, RRID:AB_2118291).
Isolation of Primary Mammary Epithelial Organoids
Primary mammary organoids were derived from either pre-pregnant or post-pregnant female Balb/c mice as previously described [2]. In short, pipettes and tubes were pre-coated with 5% BSA solution (in 1X PBS, Gibco #A10008–01). Female mice (~15 weeks old) were euthanized via CO2 asphyxiation, and mammary glands from 3 animals (thoracic and inguinal mammary glands pairs) were removed and collected into 10 cm culture dishes. The pooled glands were then minced under sterile conditions using scalpels, cutting around 50 times in a crisscross pattern to loosen the tissue of the glands. The minced glands were then transferred to a 50 mL falcon tube that contained 20 mL of collagenase solution which consisted of AdDf+++ (Advanced DMEM F12 (Dulbecco’s Modified Eagle Medium/F-1,5 mM GlutaMax 5 mM HEPES, 1x Penicillin/Streptomycin), FBS (1%, Corning #35–010-CV), Insulin (5μg/mL, Sigma #I9278) and Collagenase A (2 mg/mL, type IV from Clostridium histolyticum, Sigma #C5138). The tubes with the glands and collagenase solution were shaken at 200 RPM for 30–40 min until solution became cloudy, and large chunks of tissue dissipated. After digestion, 1 mL of FBS was added, and then solution was passed up and down a 5 mL pipette 10 times to ensure complete disassociation of the tissue. The solution was spun down in a centrifuge at 300 x g for 5 min at room temperature and the fat-containing supernatant was removed. The pellet was resuspended in 10 mL of AdDf+++ and then passed through a 100 μm strainer (Falcon #352360) to ensure no large tissue pieces would proceed. An additional room temperature 5-min centrifugation step at 300 x g was used to wash any remaining enzyme from the pelleted epithelium. The pellet was then resuspended in 10 mL of AdDf+++, pulse centrifugated to 500 x g, then had supernatant removed. This was repeated a total of 3 times.
Organoid Cultures
The organoid pellet was resuspended using a pre-chilled pipette tip in desired amount of Matrigel (100%, Corning #354230). In a 37 °C pre-warmed 24 well plate, three small domes were made using a total of 50 μl of Matrigel in a triangle-shaped pattern. The plates were then inverted and placed into a CO2 incubator (5% CO2, 37 °C) for 20 min to allow Matrigel to solidify. Each of the wells was filled with 0.5 mL of Essential organoid medium AdDf+++, supplemented with 1x ITS (Insulin/Transferrin/Sodium Selenite, Gibco #41400–045) and FGF-2 (Final concentration: 5 nm, PeproTech #450–33) for 6 days. Medium was changed every two days. The cultured mammary organoids were then grown in medium lacking FGF2 for 24 h and then incubated with complete medium (AdDf+++, supplemented with ITS (Final Concentration:1x, Insulin/Transferrin/Sodium Selenite, Gibco, #41400–045), 17-β-Estradiol (Final concentration: 40 ng/mL, Sigma #E2758), Progesterone (Final concentration: 120 ng/mL, Sigma #P8783), Prolactin (Final concentration: 120 ng/mL,Sigma #L4021). Organoids were isolated from Matrigel using Cell Recovery solution (0.5 mL, Corning #354253).
Organoid Branching and Size Quantification
Organoid visualization and image collection were performed on a Nikon Eclipse TI microscope utilizing NIS-Elements BR software (Nikon). For branching and size quantification, at least 13 fields of view and 20 to 25 organoids were analyzed. The area (size) of mammary organoids in each image was measured via ImageJ. A branching classification was given to organoids displaying three or more elongated buds.
RNA Isolation and RT-qPCR
Medium was removed from each of the wells and then washed with 0.5 mL 1x PBS. RNA was extracted by adding Trizol (0.5 mL, Thermo Fisher Scientific, #15596018) to each well with organoids. Reverse transcription was carried out using SuperScript III ™ kit (Thermo Fisher Scientific). RTqPCR was performed using a Quantstudio 6 with SYBR Green Master mix (Applied Biosystems #4368577). Each reaction was run in at least duplicate. Relative gene expression was calculated via the deltadeltact method in which the values for the measured genes were normalized to the house keeping gene, mouse β-actin mRNA. Primers sequences targeting mRNA of β-actin, Csn2 and Csn3 genes were designed as previously described [1].
Mammary Organoids Wholemount Immunofluorescence Staining
Each well was washed with 0.5 mL of 1X PBS, followed by the addition of Cell recovery solution (0.5 mL, Corning #354253). Culture plates were them incubated at 4 °C for 30 min or until Matrigel domes were no longer visible, followed by the addition of 1 mL of AdDf+++ and gentle pipetting all organoids were released from Matrigel. Organoids were then transferred to a 1% BSA (in 1X PBS, Gibco #A10008–01) pre-coated 15 mL conical tube, and spun at 500 x g for 5 min at room temperature (RT). Organoid pellets were gently resuspended in 1 mL of 4% PFA (in 1X PBS, Electron Microscopy Sciences, #15711) for 1 h at RT, following centrifugation at 500 x g for 5 min at RT. Fixed organoids were permeabilized with 1 mL of Permeabilization solution (0.5% Triton X-100, Sigma #93443 in 1X PBS) and incubated for 30 min at RT, followed by centrifugation at 500 x g for 5 min at RT. Permeabilized organoids were washed once with 1 mL of washing solution (0.1% Tween-20 solution, MP, #9005-64-5 in 1X PBS), followed by centrifugation at 500 x g for 5 min at RT. Fixed and permeabilized organoid pelleted were gently resuspended with 0.5 mL of blocking buffer (1X PBS; 300 mM Glycine, Fischer-Scientific, #G45–212; 10 mg/mL BSA, Sigma #A2153; 5% Goat serum, Abcam #ab7481) and incubated for 1 h at RT, followed by centrifugation at 500 x g for 5 min at RT. Blocked organoids were then resuspended in blocking buffer containing indicated concentrations of antibodies, and incubated overnight at 4 °C with constant agitation. Staining solution was removed, and organoids were washed with 1 mL of washing solution, centrifugation at 500 x g for 5 min at RT for a total of 3 times. Stained organoids were then stained with Propidium Iodide (PI, for nuclear staining, Invitrogen, 1:1000 dilution), at room temperature for 15 min. Staining solution was removed, and organoids were washed with 1 mL of washing solution and then underwent centrifugation at 500 x g for 5 min at RT for a total of 3 times. At the end of the last centrifugation, the supernatant was removed, and one drop of Prolong™ Glass Antifade Mountant (Invitrogen #P36982) was added to stained organoids, followed by mounting on a glass slide and allowed to cure overnight in the dark. All imaging was acquired on a Zeiss 780 Confocal Microscope.
RNA-Seq Library Preparation and Analyses
Mammary organoids (n = 2–3 wells per technical replicate) were collected, dissociated and resuspended in Trizol (Thermo Fisher Scientific, #10296010) for RNA extraction. Double stranded cDNA synthesis and Illumina libraries were prepared utilizing the Ovation RNA-seq system (V2) (Nugen Technologies, #7102–32). RNA-seq libraries were prepared utilizing the Ovation ultralow DR multiplex system (Nugen Technologies, #0331–32). Each library (n = 2 per experimental condition) was barcoded with Illumina True-seq adaptors to allow sample multiplexing, followed by sequencing on an Illumina NextSeq500, 76 bp single-end run. Bioinformatics analyses were performed with command-line interfaced tools such as FastQC [66] for quality control and Trimmomatic [67] for sequence trimming. We used STAR [68] for mapping reads and deepTools [69] for principal component analysis. Further, we utilized FeatureCounts [70] for assigning reads to genomic features and DESeq [71] to assess changes in expression levels. Gene Set Enrichment Analysis (GSEA) was used for global analyses of differentially expressed genes [72]. For the parity signature analysis (total of 47 genes [18]), differentially expressed genes above 0.5 log2foldchange were counted as upregulated, while genes below −0.5 log2foldchange were counted as downregulated.
Cut&Run Library Preparation and Analyses
Mammary organoids (n = 2–3 wells per technical replicate) were collected, dissociated, and permeabilized with digitonin, following overnight incubation with antibody against H3K27ac histone marks (Abcam, #ab4729) at 4°C with constant agitation. Antibody-chromatin complexes were fragmented with pA-MNAse and purified utilizing Phenol-Chloroform. Cut&Run libraries (n = 2 per experimental condition) were amplified and barcoded using Clontech DNA Smart ChIP-Seq kit (Clontech, #634866) in accordance with the manufacturer’s instructions, then pooled for sequencing on an Illumina NextSeq500, 76 bp paired-end run. Reads were mapped to the indexed mm9 genome using bowtie2 short-read aligner tool [73] using default settings. Sparse Enrichment Analysis for Cut&Run (SEACR) peak-calling program [74] was used to identify enriched genomic regions with an empirical threshold of n = 0.01, returning the top n fraction of peaks based on total signal within peaks. The stringent argument was implemented, which used the summit of each curve. Further downstream analyses were performed using various command-line interfaced programs including deepTools for investigating H3K27ac peak intensity at DNA binding motifs and bedtools [75] for defining regions that are shared between conditions as well as peaks exclusive to each condition. Open source software such as Enrichr [76, 77] for comparing peaks against publicly available data and GREAT [78] for gene ontology analyses served to support condition-exclusive analyses. Additionally, UCSC’s Genome Browser [79] was used to investigate region specific H3K27ac peak intensity. H3K27ac Cut&Run peaks were utilized as input for an unbiased transcription factor analyses using Analysis of Motif Enrichment (AME) [80] and Find Individual Motif Occurrences (FIMO) [81] was used to computationally define DNA binding motif regions. DESeq2 [82] was utilized to generate genomic regions with differential H3K27ac levels sample groups (FDR < 0.05).
Supplementary Information
(DOCX 2185 kb)
Acknowledgements
This work was performed with assistance from CSHL Animal Facility, the CSHL NextGen Sequencing Shared Resources, which are supported by the CSHL Cancer Center Support Grant 5P30CA045508. This work was financially supported by the CSHL and Northwell Health affiliation, the CSHL and Simons Foundation Award, the Gladowsky Foundation Award, the Rita Allen Scholar Award, the AACR-Breast Cancer Research Foundation Award, the Pershing Square Sohn Prize for Cancer Research, and the NIH/NCI grant R01CA248158-01 (C.O.D.S.).
Data Availability Statement
RNA-seq and Cut&Run datasets are available on NCBI database [https://www.ncbi.nlm.nih.gov/], BioProject ID: PRJNA656955.
Abbreviations
- 3D
Three-dimensional
- AdDf+++
Advanced DMEM F12 (Dulbecco’s modified eagle medium/F-1,5 mM glutaMax 5 mM HEPES, 1× penicillin/streptomycin)
- AME
Analysis of motif enrichment
- β-actin
Beta actin
- BSA
Bovine serum albumin
- Csn1s2a
Casein alpha s2-like A
- cDNA
Complementary DNA
- ChIP-seq
Chromatin immune precipitation sequencing
- Cited1
Cbp/P300 interacting transactivator with Glu/Asp Rich Carboxy-Terminal Domain 1
- cMYC
Cellular myelocytomatosis
- CO2
Carbon dioxide
- Csn2
Casein 2
- Csn3
Casein 3
- Cut&Run
Cleavage under targets and release using nuclease
- DEGs
Differentially expressed genes
- DESeq
Program for differential gene expression analysis
- DMSO
Dimethyl sulfoxide
- eBOX
cMYC motif
- ECM
Extracellular matrix
- Elf5
E74-like factor 5 gene
- ERα
Estrogen Receptor alpha
- EREs
ER alpha motif
- EZH2
Enhancer of zest homolog 2
- FastQC
Program for quality control check of high throughput sequencing dataset
- FBS
Fetal bovine serum
- FDR
False discovery rate
- FGF2
Fibroblast growth factor 2
- FIMO
Find individual motif occurrences
- FOXC2
Forkhead box protein C2
- GAS
STAT5 motif
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- GREAT
Program for annotation of sequencing dataset
- H3K27ac
Histone 3 lysine 27 acetylation
- H3K4me1
Histone 3 lysine 4 monomethylation
- H4K27me3
Histone 4 lysine 27 trimethylation
- Hrs
hours
- IF
Immunofluorescence
- IRF1
Interferon responsive factor 1
- ITS
Insulin/Transferrin/Sodium Selenite
- KRT5
Cytokeratin 5
- KRT8
Cytokeratin 8
- MECs
Mammary epithelial cells
- mRNA
messenger RNA
- NCBI
National Center for Biotechnology Information
- ns
not significant
- pA-MNAse
fusion protein produced by the pK19-pA-MN plasmid
- PBS
Phosphate buffered saline
- PC-1
Principal Component 1
- PC-2
Principal Component 2
- PFA
Paraformaldehyde
- PI
Propidium Iodide
- RNA-seq
RNA sequencing
- PRC2
Polycomb repressive complex 2
- Prlr
Prolactin receptor
- RPM
revolutions per minute
- RT
Room temperature
- RT-qPCR
Real time quantitative polymerase chain reaction
- SEACR
Sparse enrichment analysis for cut & run
- SEM
Standard error of the mean
- SP3
Specificity protein 3
- STAR
Program for mapping sequencing dataset
- STAT5
Signal transducer and activator of transcription 5
- TF
Transcription factor
- UCSC
University of California, Santa Cruz
- UNC1999
Chemical inhibitor of EZH2
- Vdr
Vitamin D receptor
- Wap
Whey acidic protein
- × g
times gravity
Author Contributions
C.O.D.S. designed and supervised the research; C.O.D.S, M.F.C., and M.C.T. wrote the manuscript, performed bioinformatics analyses, and analyzed results; M.F.C performed experiments.
Compliance with Ethical Standards
Competing Interests
The authors have no competing interests to disclose.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Michael F. Ciccone and Marygrace C. Trousdell contributed equally to this work.
References
- 1.Feigman MJ, Moss MA, Chen C, Cyrill SL, Ciccone MF, Trousdell MC, Yang ST, Frey WD, Wilkinson JE, dos Santos CO. Pregnancy reprograms the epigenome of mammary epithelial cells and blocks the development of premalignant lesions. Nat Commun. 2020;11(1):2649. doi: 10.1038/s41467-020-16479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewald AJ. Isolation of mouse mammary organoids for long-term time-lapse imaging. Cold Spring Harb Protoc. 2013;2013(2):130–133. doi: 10.1101/pdb.prot072892. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen-Ngoc KV, et al. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol Biol. 2015;1189:135–162. doi: 10.1007/978-1-4939-1164-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo AT, Mori H, Mott J, Bissell MJ. Constructing three-dimensional models to study mammary gland branching morphogenesis and functional differentiation. J Mammary Gland Biol Neoplasia. 2012;17(2):103–110. doi: 10.1007/s10911-012-9251-7. [DOI] [PubMed] [Google Scholar]
- 5.Reginato MJ, Muthuswamy SK. Illuminating the center: mechanisms regulating lumen formation and maintenance in mammary morphogenesis. J Mammary Gland Biol Neoplasia. 2006;11(3–4):205–211. doi: 10.1007/s10911-006-9030-4. [DOI] [PubMed] [Google Scholar]
- 6.Sumbal J, Chiche A, Charifou E, Koledova Z, Li H. Primary mammary Organoid model of lactation and involution. Front Cell Dev Biol. 2020;8:68. doi: 10.3389/fcell.2020.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava V, Huycke TR, Phong KT, Gartner ZJ. Organoid models for mammary gland dynamics and breast cancer. Curr Opin Cell Biol. 2020;66:51–58. doi: 10.1016/j.ceb.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruitt HC, Metge BJ, Weeks SE, Chen D, Wei S, Kesterson RA, Shevde LA, Samant RS. Conditional knockout of N-Myc and STAT interactor disrupts normal mammary development and enhances metastatic ability of mammary tumors. Oncogene. 2018;37(12):1610–1623. doi: 10.1038/s41388-017-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalak EM, Milevskiy MJG, Joyce RM, Dekkers JF, Jamieson PR, Pal B, Dawson CA, Hu Y, Orkin SH, Alexander WS, Lindeman GJ, Smyth GK, Visvader JE. Canonical PRC2 function is essential for mammary gland development and affects chromatin compaction in mammary organoids. PLoS Biol. 2018;16(8):e2004986. doi: 10.1371/journal.pbio.2004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muthuswamy SK. Organoid models of Cancer explode with possibilities. Cell Stem Cell. 2018;22(3):290–291. doi: 10.1016/j.stem.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Duarte AA, Gogola E, Sachs N, Barazas M, Annunziato S, R de Ruiter J, Velds A, Blatter S, Houthuijzen JM, van de Ven M, Clevers H, Borst P, Jonkers J, Rottenberg S. BRCA-deficient mouse mammary tumor organoids to study cancer-drug resistance. Nat Methods. 2018;15(2):134–140. doi: 10.1038/nmeth.4535. [DOI] [PubMed] [Google Scholar]
- 12.Husby A, Wohlfahrt J, Øyen N, Melbye M. Pregnancy duration and breast cancer risk. Nat Commun. 2018;9(1):4255. doi: 10.1038/s41467-018-06748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh S, Gu F, Wang CM, Lin CL, Liu J, Wang H, Ravdin P, Hu Y, Huang THM, Li R. Genome-wide DNA methylation profiling reveals parity-associated hypermethylation of FOXA1. Breast Cancer Res Treat. 2014;147(3):653–659. doi: 10.1007/s10549-014-3132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dos Santos CO, Dolzhenko E, Hodges E, Smith AD, Hannon GJ. An epigenetic memory of pregnancy in the mouse mammary gland. Cell Rep. 2015;11(7):1102–1109. doi: 10.1016/j.celrep.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh SJ, Clement K, Jee D, Merlini A, Choudhury S, Maruyama R, Yoo R, Chytil A, Boyle P, Ran FA, Moses HL, Barcellos-Hoff MH, Jackson-Grusby L, Meissner A, Polyak K. Age- and pregnancy-associated DNA methylation changes in mammary epithelial cells. Stem Cell Reports. 2015;4(2):297–311. doi: 10.1016/j.stemcr.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman-Krnacik S, Rosen JM. Differential temporal and spatial gene expression of fibroblast growth factor family members during mouse mammary gland development. Mol Endocrinol. 1994;8(2):218–229. doi: 10.1210/mend.8.2.8170478. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Martinez D, Koledova Z, Qiao G, Streuli CH, Lu P. FGF ligands of the postnatal mammary stroma regulate distinct aspects of epithelial morphogenesis. Development. 2014;141(17):3352–3362. doi: 10.1242/dev.106732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, D'Cruz CM, Chodosh LA. Hormone-induced protection against mammary tumorigenesis is conserved in multiple rat strains and identifies a core gene expression signature induced by pregnancy. Cancer Res. 2006;66(12):6421–6431. doi: 10.1158/0008-5472.CAN-05-4235. [DOI] [PubMed] [Google Scholar]
- 19.Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1(4):533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes NE, Stoelzle T. Key signalling nodes in mammary gland development and cancer: Myc. Breast Cancer Res. 2009;11(5):210. doi: 10.1186/bcr2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rijnkels M, Kabotyanski E, Montazer-Torbati MB, Beauvais CH, Vassetzky Y, Rosen JM, Devinoy E. The epigenetic landscape of mammary gland development and functional differentiation. J Mammary Gland Biol Neoplasia. 2010;15(1):85–100. doi: 10.1007/s10911-010-9170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y, Manka D, Wagner KU, Khan SA. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc Natl Acad Sci U S A. 2007;104(37):14718–14723. doi: 10.1073/pnas.0706933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiede B, Kang Y. From milk to malignancy: the role of mammary stem cells in development, pregnancy and breast cancer. Cell Res. 2011;21(2):245–257. doi: 10.1038/cr.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bansal AS, Bora SA, Saso S, Smith JR, Johnson MR, Thum MY. Mechanism of human chorionic gonadotrophin-mediated immunomodulation in pregnancy. Expert Rev Clin Immunol. 2012;8(8):747–753. doi: 10.1586/eci.12.77. [DOI] [PubMed] [Google Scholar]
- 25.Betts CB, Pennock ND, Caruso BP, Ruffell B, Borges VF, Schedin P. Mucosal immunity in the female murine mammary gland. J Immunol. 2018;201(2):734–746. doi: 10.4049/jimmunol.1800023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6. [DOI] [PMC free article] [PubMed]
- 27.Radice GL, Ferreira-Cornwell MC, Robinson SD, Rayburn H, Chodosh LA, Takeichi M, Hynes RO. Precocious mammary gland development in P-cadherin-deficient mice. J Cell Biol. 1997;139(4):1025–1032. doi: 10.1083/jcb.139.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snedeker SM, Brown CF, DiAugustine RP. Expression and functional properties of transforming growth factor alpha and epidermal growth factor during mouse mammary gland ductal morphogenesis. Proc Natl Acad Sci U S A. 1991;88(1):276–280. doi: 10.1073/pnas.88.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3(4):429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Haaksma CJ, Schwartz RJ, Tomasek JJ. Myoepithelial cell contraction and milk ejection are impaired in mammary glands of mice lacking smooth muscle alpha-actin. Biol Reprod. 2011;85(1):13–21. doi: 10.1095/biolreprod.110.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woessner JF., Jr Catabolism of collagen and non-collagen protein in the rat uterus during post-partum involution. Biochem J. 1962;83:304–314. doi: 10.1042/bj0830304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warri A, et al. Autophagy and unfolded protein response (UPR) regulate mammary gland involution by restraining apoptosis-driven irreversible changes. Cell Death Discov. 2018;4:40. doi: 10.1038/s41420-018-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, LaVail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo J, Long Y, Ren G, Zhang Y, Chen J, Huang R, Yang L. Punicalagin reversed the hepatic injury of Tetrachloromethane by Antioxidation and enhancement of autophagy. J Med Food. 2019;22(12):1271–1279. doi: 10.1089/jmf.2019.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elswood J, Pearson SJ, Payne HR, Barhoumi R, Rijnkels M, W. Porter W. Autophagy regulates functional differentiation of mammary epithelial cells. Autophagy. 2020:1–19. [DOI] [PMC free article] [PubMed]
- 36.Chapman RS, Duff EK, Lourenco PC, Tonner E, Flint DJ, Clarke AR, Watson CJ. A novel role for IRF-1 as a suppressor of apoptosis. Oncogene. 2000;19(54):6386–6391. doi: 10.1038/sj.onc.1204016. [DOI] [PubMed] [Google Scholar]
- 37.Gan L, Liu Z, Jin W, Zhou Z, Sun C. Foxc2 enhances proliferation and inhibits apoptosis through activating Akt/mTORC1 signaling pathway in mouse preadipocytes. J Lipid Res. 2015;56(8):1471–1480. doi: 10.1194/jlr.M057679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu ZJ, Kollander R, Kiang DT. Differential up-regulation of gap junction connexin 26 gene in mammary and uterine tissues: the role of Sp transcription factors. Mol Endocrinol. 1998;12(12):1931–1938. doi: 10.1210/mend.12.12.0208. [DOI] [PubMed] [Google Scholar]
- 39.Mallepell S, Krust A, Chambon P, Brisken C. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc Natl Acad Sci U S A. 2006;103(7):2196–2201. doi: 10.1073/pnas.0510974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bocchinfuso WP, Lindzey JK, Hewitt SC, Clark JA, Myers PH, Cooper R, Korach KS. Induction of mammary gland development in estrogen receptor-alpha knockout mice. Endocrinology. 2000;141(8):2982–2994. doi: 10.1210/endo.141.8.7609. [DOI] [PubMed] [Google Scholar]
- 41.Barash I. Stat5 in the mammary gland: controlling normal development and cancer. J Cell Physiol. 2006;209(2):305–313. doi: 10.1002/jcp.20771. [DOI] [PubMed] [Google Scholar]
- 42.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24(18):8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, Tsao S, Tellis I, Zavarsek S, Xu D, Lapinskas EJ, Visvader J, Lindeman GJ, Thomas R, Ormandy CJ, Hertzog PJ, Kola I, Pritchard MA. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24(3):635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HJ, Gallego-Ortega D, Ledger A, Schramek D, Joshi P, Szwarc MM, Cho C, Lydon JP, Khokha R, Penninger JM, Ormandy CJ. Progesterone drives mammary secretory differentiation via RankL-mediated induction of Elf5 in luminal progenitor cells. Development. 2013;140(7):1397–1401. doi: 10.1242/dev.088948. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson AJ, et al. Multiscale activity imaging in the mammary gland reveals how oxytocin enables lactation. bioRxiv. 2019: p. 657510.
- 46.Stewart TA, et al.. Mammary mechanobiology: PIEZO1 mechanically-activated ion channels in lactation and involution. bioRxiv. 2019: p. 649038. [DOI] [PubMed]
- 47.Kim HY, Nelson CM. Extracellular matrix and cytoskeletal dynamics during branching morphogenesis. Organogenesis. 2012;8(2):56–64. doi: 10.4161/org.19813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keely PJ, Wu JE, Santoro SA. The spatial and temporal expression of the alpha 2 beta 1 integrin and its ligands, collagen I, collagen IV, and laminin, suggest important roles in mouse mammary morphogenesis. Differentiation. 1995;59(1):1–13. doi: 10.1046/j.1432-0436.1995.5910001.x. [DOI] [PubMed] [Google Scholar]
- 49.Enami J, Nandi S. Hormonal control of milk protein synthesis in cultured mouse mammary explants. Cell Differ. 1977;6(3–4):217–227. doi: 10.1016/0045-6039(77)90017-3. [DOI] [PubMed] [Google Scholar]
- 50.Naylor MJ, Oakes SR, Gardiner-Garden M, Harris J, Blazek K, Ho TWC, Li FC, Wynick D, Walker AM, Ormandy CJ. Transcriptional changes underlying the secretory activation phase of mammary gland development. Mol Endocrinol. 2005;19(7):1868–1883. doi: 10.1210/me.2004-0254. [DOI] [PubMed] [Google Scholar]
- 51.Martinson HA, Jindal S, Durand-Rougely C, Borges VF, Schedin P. Wound healing-like immune program facilitates postpartum mammary gland involution and tumor progression. Int J Cancer. 2015;136(8):1803–1813. doi: 10.1002/ijc.29181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, Bell AK, Ferrier RK, Sandilands GP, Gusterson BA. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6(2):R75–R91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCready J, Arendt LM, Glover E, Iyer V, Briendel JL, Lyle SR, Naber SP, Jay DG, Kuperwasser C. Pregnancy-associated breast cancers are driven by differences in adipose stromal cells present during lactation. Breast Cancer Res. 2014;16(1):R2. doi: 10.1186/bcr3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schedin P, Mitrenga T, McDaniel S, Kaeck M. Mammary ECM composition and function are altered by reproductive state. Mol Carcinog. 2004;41(4):207–220. doi: 10.1002/mc.20058. [DOI] [PubMed] [Google Scholar]
- 55.Shakhar K, Valdimarsdottir HB, Bovbjerg DH. Heightened risk of breast cancer following pregnancy: could lasting systemic immune alterations contribute? Cancer Epidemiol Biomark Prev. 2007;16(6):1082–6. doi: 10.1158/1055-9965.EPI-07-0014. [DOI] [PubMed] [Google Scholar]
- 56.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 57.Yoo KH, Oh S, Kang K, Hensel T, Robinson GW, Hennighausen L. Loss of EZH2 results in precocious mammary gland development and activation of STAT5-dependent genes. Nucleic Acids Res. 2015;43(18):8774–8789. doi: 10.1093/nar/gkv776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuppa AA, Tornesello A, Papacci P, Tortorolo G, Segni G, Lafuenti G, Moneta E, Diodato A, Sorcini M, Carta S. Relationship between maternal parity, basal prolactin levels and neonatal breast milk intake. Biol Neonate. 1988;53(3):144–147. doi: 10.1159/000242775. [DOI] [PubMed] [Google Scholar]
- 59.Ingram JC, Woolridge MW, Greenwood RJ, McGrath L. Maternal predictors of early breast milk output. Acta Paediatr. 1999;88(5):493–499. doi: 10.1080/08035259950169486. [DOI] [PubMed] [Google Scholar]
- 60.De Amici D, et al. Does ethnicity predict lactation? A study of four ethnic communities. Eur J Epidemiol. 2001;17(4):357–362. doi: 10.1023/a:1012731713393. [DOI] [PubMed] [Google Scholar]
- 61.Ingram J, Woolridge M, Greenwood R. Breastfeeding: it is worth trying with the second baby. Lancet. 2001;358(9286):986–987. doi: 10.1016/S0140-6736(01)06126-8. [DOI] [PubMed] [Google Scholar]
- 62.Kitano N, Nomura K, Kido M, Murakami K, Ohkubo T, Ueno M, Sugimoto M. Combined effects of maternal age and parity on successful initiation of exclusive breastfeeding. Prev Med Rep. 2016;3:121–126. doi: 10.1016/j.pmedr.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terry MB, et al. The Influence of Number and Timing of Pregnancies on Breast Cancer Risk for Women With BRCA1 or BRCA2 Mutations. JNCI Cancer Spectr. 2018;2(4):pky078. doi: 10.1093/jncics/pky078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichols HB, et al. Breast Cancer risk after recent childbirth: a pooled analysis of 15 prospective studies. Ann Intern Med. 2018. [DOI] [PMC free article] [PubMed]
- 65.Nichols HB, Schoemaker MJ, Cai J, Xu J, Wright LB, Brook MN, Jones ME, Adami HO, Baglietto L, Bertrand KA, Blot WJ, Boutron-Ruault MC, Dorronsoro M, Dossus L, Eliassen AH, Giles GG, Gram IT, Hankinson SE, Hoffman-Bolton J, Kaaks R, Key TJ, Kitahara CM, Larsson SC, Linet M, Merritt MA, Milne RL, Pala V, Palmer JR, Peeters PH, Riboli E, Sund M, Tamimi RM, Tjønneland A, Trichopoulou A, Ursin G, Vatten L, Visvanathan K, Weiderpass E, Wolk A, Zheng W, Weinberg CR, Swerdlow AJ, Sandler DP. Breast Cancer risk after recent childbirth: a pooled analysis of 15 prospective studies. Ann Intern Med. 2019;170(1):22–30. doi: 10.7326/M18-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.FastQC: A Quality Control Tool for High Throughput Sequence Data. 2015.
- 67.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 71.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meers MP, Tenenbaum D, Henikoff S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019;12(1):42. doi: 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McLeay RC, Bailey TL. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 2185 kb)
Data Availability Statement
RNA-seq and Cut&Run datasets are available on NCBI database [https://www.ncbi.nlm.nih.gov/], BioProject ID: PRJNA656955.