Abstract
Apis mellifera is an important provider of ecosystem services, and during flight and foraging behaviour is exposed to environmental pollutants including airborne particulate matter (PM). While exposure to insecticides, antibiotics, and herbicides may compromise bee health through alterations of the gut microbial community, no data are available on the impacts of PM on the bee microbiota. Here we tested the effects of ultrapure Titanium dioxide (TiO2) submicrometric PM (i.e., PM1, less than 1 µm in diameter) on the gut microbiota of adult bees. TiO2 PM1 is widely used as a filler and whitening agent in a range of manufactured objects, and ultrapure TiO2 PM1 is also a common food additive, even if it has been classified by the International Agency for Research on Cancer (IARC) as a possible human carcinogen in Group 2B. Due to its ubiquitous use, honey bees may be severely exposed to TiO2 ingestion through contaminated honey and pollen. Here, we demonstrated that acute and chronic oral administration of ultrapure TiO2 PM1 to adult bees alters the bee microbial community; therefore, airborne PM may represent a further risk factor for the honey bee health, promoting sublethal effects against the gut microbiota.
Subject terms: Entomology, Microbial ecology, Environmental impact
Introduction
Honey bees (Apis mellifera Linnaeus) are important providers of ecosystem services, both regulating, through pollination of a wide range of crops and uncultivated plants, and provisioning, for the delivery of honey, pollen, propolis and other products to humans. Moreover, the honey bee is an important bioindicator of environmental contamination, and both the insect and its products are used for the detection of environmental pollutants1–5.
In the last decade, one of the major problems plaguing honey bees is a phenomenon named Colony Collapse Disorder (CCD) which causes loss of colonies worldwide6–8. The multifactorial origin of CCD is widely acknowledged, and environmental stressors such as pesticides, heavy metals, or airborne particulate matter (PM) pollutants may play a key role in driving the bees’ decline9.
While the effects of pesticides and heavy metals on bees are widely recognised, till now very few data are available on the effects of PM on bees’ health. These studies include lethal and sublethal effects (e.g., behaviour, gene expression, and cellular alterations) of the exposure to lead and cadmium oxides and TiO2 nanoparticles10–13.
In polluted environments, airborne PM is known to stuck to the body of the bee and can be also ingested through contaminated pollen and honey that represent the food sources of the bee colony3,5,14. If ingested, dusts can come into contact with the gut microbiome lining the intestinal epithelium posing a hazard to the bacterial community. Recent evidence suggests that environmental stressors can indirectly compromise bee health through gut microbiota disruption15. The honey bee harbors a simple and distinct gut community, thought to be the result of a long-lasting evolutionary relationship15. While no evidence has been reported until now on the impacts of PM on the bee health, alterations of the gut microbial community composition were demonstrated in bees exposed to antibiotics16, glyphosate17, insecticides18,19 and sublethal doses of cadmium and selenite20.
The gut of worker bees is dominated by nine clusters of bacterial species comprising between 95% and 99.9% of total diversity in almost all individuals, based on investigations carried out on 16S rDNA21–23 and on total DNA metagenomics of intestinal samples24. The two omnipresent Gram-negative species are Snodgrassella alvi and Gilliamella apicola, both members of Phylum Proteobacteria25. Among Gram-positive bacteria, two groups of species in the Firmicutes phylum are ubiquitous and abundant, referred to as the Lactobacillus Firm-4 and Lactobacillus Firm-5 clades26. Although often less abundant, the cluster of the species Bifidobacterium asteroides27 is also found in most adult worker bees. Less numerous and even less prevalent are the Proteobacteria species Frischella perrara28, Bartonella apis29, Parasaccharibacter apium23 and a group of Gluconobacteria species designated Alpha2.1. These species have narrow niches in the intestines of bees (e.g., F. perrara) or are generalists, i.e., they are also found in the hive environment (for example, P. apium, Lactobacillus kunkeei and the Alpha2.1 group), which may explain their relatively lower frequency in bee gut detections. While other bacteria may occasionally be present, these nine species groups represent bacterial lineages that appear to be specifically adapted to life alongside their hosts, bees. This gut microbiota organization is well described by Kwong & Moran30.
The newly hatched larvae are free of bacteria that start colonizing the gut thanks to interactions with worker bees and the hive environment31,32. During the metamorphosis of the larvae to pupae and finally, into adult bees, the lining of the intestine is renewed: the newly emerged adult bees have very few bacteria in the intestine and are readily colonized by the typical intestinal microbial community33.
Titanium dioxide (TiO2) is a naturally occurring metal oxide. As rutile, TiO2 is not rare in nature and may concentrate in the heavy fraction of sediments. Synthetic TiO2 in form of sub-micrometric PM, i.e., PM1, less than 1 µm in diameter, is widely used as a filler and whitening agent in a range of manufactured objects, such as plastics, paints, paper, printing inks, textiles, catalysts, floor and roofing materials, and vehicles components. TiO2 is also a common ingredient in cosmetics, pharmaceuticals, sunscreen and as a food additive. While TiO2 used for non-food applications has surface coatings of alumina and silica, to reduce photoactivity, and organic surface treatments, conferring hydrophobic properties, TiO2 as a food additive is ultrapure, and PM size ranges from 400 µm to 30 nm34,35.
The potential ecological and human health impact of exposure to TiO2 is of growing concern. Indeed, inhalation and intra-tracheally administration of PM of TiO2, including nano-meter sized particles, induces lung cancer in rats36. TiO2 has indeed been classified by the International Agency for Research on Cancer (IARC) as a possible human carcinogen in Group 2B (IARC, 2010). New evidence suggests that oral exposure to nanosized TiO2 promotes chronic intestinal inflammation and carcinogenesis in rats35, as well as dysbiosis of gut microbiota37.
Trace element analyses already demonstrated that Titanium can be detected in honey, pollen and bees, especially in high density urban and residential/suburban areas38–41. Titanium can be only found in combined form and the most widespread chemical form both from natural and anthropogenic sources is TiO2. Therefore, honey bees may be severely exposed to TiO2 ingestion through contaminated honey and pollen, and in severely polluted areas, PM1 of TiO2 can be detected in pollen grains and honey (Negri, personal communication).
The toxicity of TiO2 PM1at nano-scale has been already studied in honey bee with histological and immunohistochemical approaches12 shedding light on the negative effects at high doses. However, in-depth studies on the effects on the gut environment of the TiO2 ingested by bees are still lacking, and no studies on specific impacts on the bee’s gut microbiota are available. Here we wanted to fill this gap by studying the effect of TiO2 on honey bees gut microbiota in the experimental context of controlled oral administration, providing evidence that ingestion of PM1 of TiO2 can alter the bee microbial community. Sub-lethal doses were employed, while acute and chronic exposures were assessed at 24 and 96 h post-emergency for acute and chronic experiments respectively.
Results
SEM–EDX results
Scanning electron microscope (SEM) coupled with X-ray (EDX) analyses were used to assess the morphology, chemical composition and size of TiO2 particles delivered to the bees.
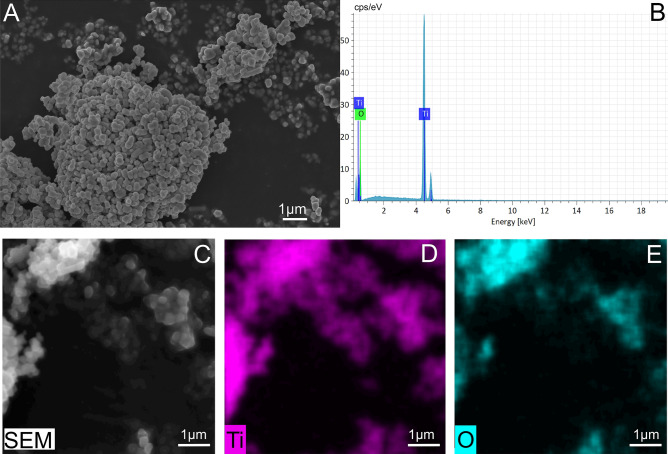
SEM analyses showed that the TiO2 stock powder had a sub-micrometre size < 1 µm (between 800 and 200 nm) and an ellipsoidal/spherical shape (Fig. 1A). Moreover, EDX analyses and compositional mapping confirm the purity of TiO2 rutile (Fig. 1B–E).
Figure 1.
Control sample of TiO2 rutile powder and elemental mapping. (A) SEM image showing the morphology and size and (B) EDX spectrum showing the purity of the TiO2 powder. (C) SEM-BSE image and the (D) titanium and (E) oxygen element maps.
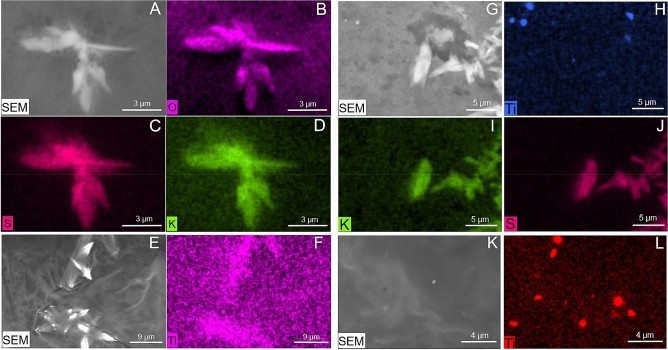
SEM–EDX analyses demonstrated the absence of TiO2 in haemolymph collected from the acute, chronic and control samples (Figure S1A,B). In the haemolymph, EDX spectra (Figure S1B) showed the presence of many elements e.g., Mg, Ca, Na, S, P, K, Cl42.
SEM observation performed on chronic and control gut highlighted the presence of many lanceolate crystals likely due to precipitation of salts. EDX analysis confirmed their chemical composition as K2SO4 (Fig. 2A–D). In the rectum of treated bees, TiO2 was found both associated (Fig. 2E,F) and not associated (Fig. 2G–L) with K2SO4 crystals.
Figure 2.
Elemental mapping of the rectum in control (A–D) and chronical samples (E–L). (A,E,G,K) SEM-BSE images; (B–D) element analysis highlighting the presence of K2SO4 crystals in the control sample; (F) titanium dioxide associated with K2SO4 crystals; (H–L) titanium dioxide not associated with K2SO4 crystals.
Bacterial diversity in the studied honey bees
The gut bacterial diversity was assessed by means of Illumina HTS of bacterial 16S amplicons covering the V3-V4 regions. A total of 537,641 sequences were produced, filtered and downscaled to 216,000 (i.e., 12,000 per each sample) after elimination of homopolymers, sequences not aligning to the target region, chimaeras, non-bacterial sequences and rarefaction to the least populated sample. After this downscaling, one out of 18 samples (a replicate of the 100X acute test) was eliminated because it had a lower number of sequences. The resulting Good’s index of coverage of the rarefied samples was 94.7 ± 1.2%, indicating that the vast majority of bees gut bacterial diversity was covered by the sequencing.
The structure of honey bee gut bacterial community was investigated at OTUs level, testing with a CCA model if the treatment (control vs chronic vs acute) and the dose had significant effects on the bacterial gut communities of the studied bees. Results (Fig. 3) indicated that the bees from the acute and the chronic experiments, being sampled at different life stages, hosted very different gut microbiota. Within each exposure time, it was found that samples were clearly grouped among doses for chronic exposure, but not for the acute. Similar outcomes were obtained by Principal Component Analyses (Figure S2).
Figure 3.
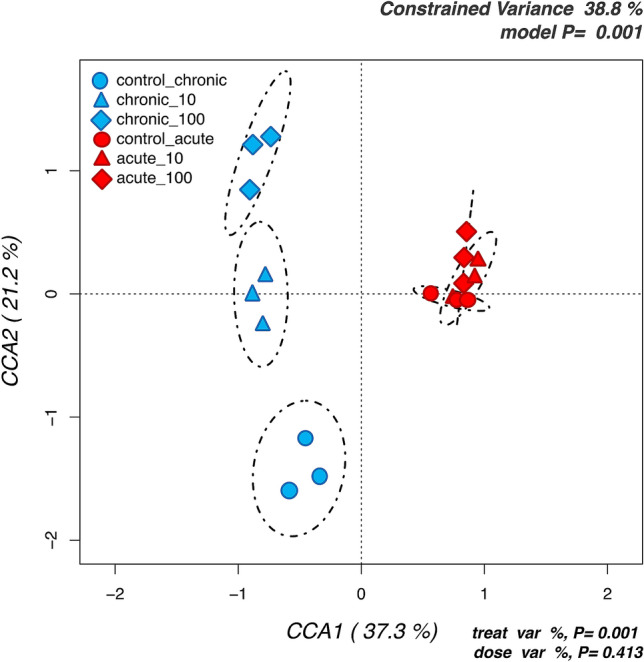
Hypothesis-driven canonical correspondence analysis (CCA) of honey bees gut microbiota testing the significance of the treatment and dose effects on the abundances of all analysed OTUs. Samples are labelled according to the 6 groups studied.
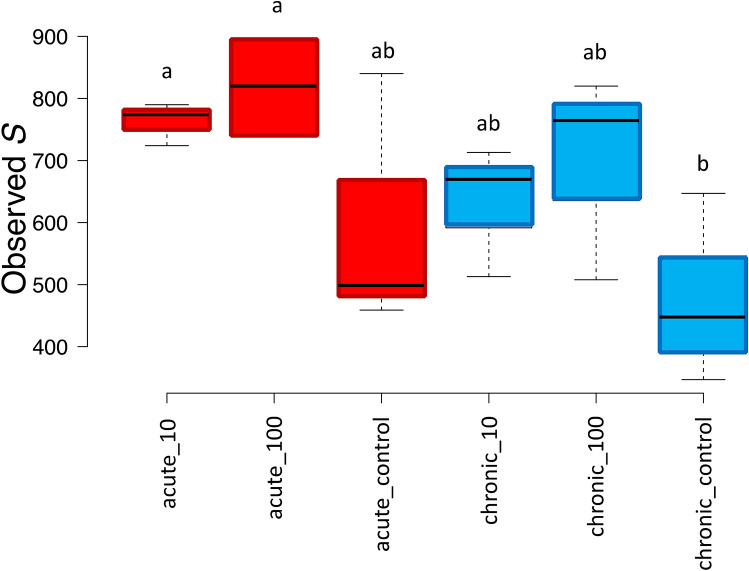
Differences among samples were also reflected by α-diversity analyses on the total number of OTUs identified in each treatment, which highlighted a clear trend: bees exposed to both chronic and acute TiO2 PM displayed a higher diversity as compared to their relative controls. The difference was significant according to LSD test between the chronic control and the two acute treatments (Fig. 4).
Figure 4.
Number of observed OTUs (S) for the chronic and acute treatments and their respective controls. Significant differences are highlighted with different letters according to Tukey’s HSD test for comparison of means.
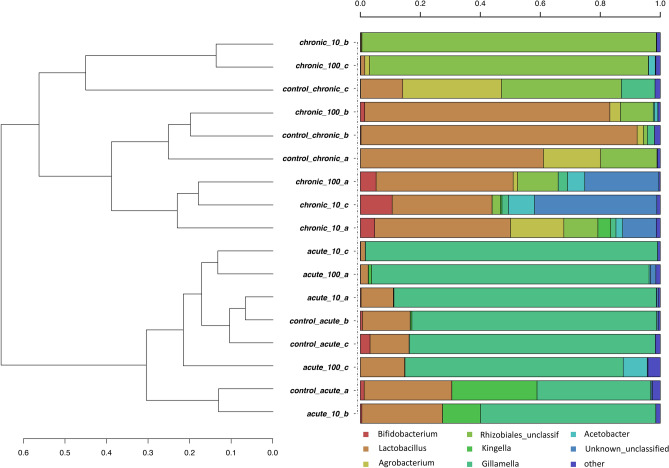
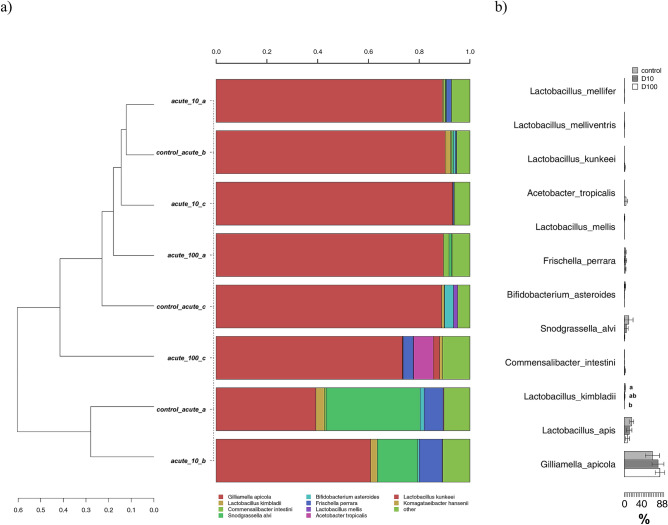
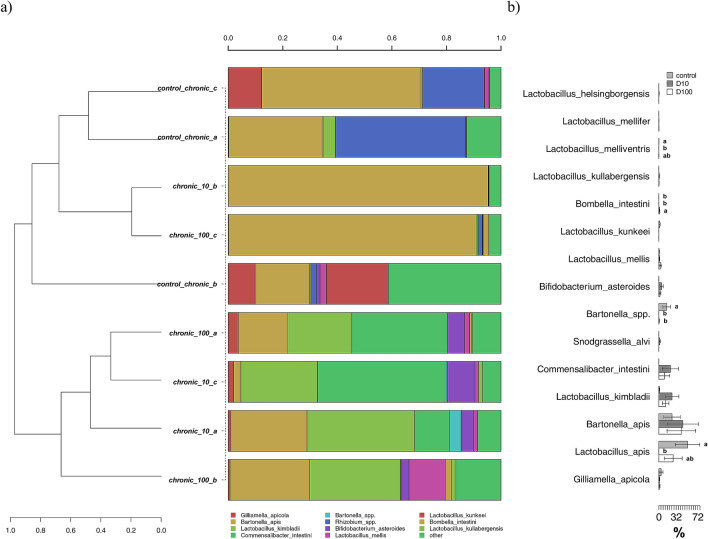
The distinct bacterial community structures induced by TiO2 PM exposure were also highlighted by hierarchical clustering of sequences classified at the genus level (Fig. 5): in agreement with the CCA results, chronic and acute experiments were grouped in two main separate clusters sharing less than 70% of similarity. The two controls for each treatment representing adults sampled at two different life stages were also grouped separately one from the other. Gilliamella was the dominant genus in the acute experiments (24 h), with relative percentages reaching more than 80% of total abundance in a number of acute treatments at both 10X and 100X, followed by Lactobacillus and Acetobacter. A very different composition was identified in 96 h bees of the chronic exposure experiment (Fig. 5): here the dominant genus was Lactobacillus, which decreased in the TiO2 exposed bees. The latter was however enriched in Bifidobacterium, reaching relative concentration between 5 and 12% in a number of chronically exposed bees. A particular feature was detected in two treated replicates at 10X and 100X, with Acetobacter covering the vast majority (i.e., > 98%) of the observed diversity at the genus level.
Figure 5.
Hierarchical clustering of sequences classified at the genus level for both chronic and acute treatments. Bars of different colours indicated the relative percentage of the genera identified in the honey bees gut. Only genera participating with > 1% in at least one sample are shown, while taxa with lower participations were added to the “other” group. Similar samples were clustered using the average linkage algorithm.
Distinctive features in the gut microbiota of honey-bees acutely exposed to TiO2
After defining that the two bee groups from the acute and the chronic exposure experiments had very different gut bacterial compositions (Figs. 3 and 5), separate analyses were carried out on the two groups, focusing on the relative presence of the most abundant OTUs classified at the species level.
In the acute exposure experiments, G.apicola was the most abundant species with an average of ca 70% of the total bacterial community, followed by Lactobacillus apis, S. alvi, Lactobacillus kimbladii and Acetobacter tropicalis (Fig. 6a). The clustering did not show clear discrimination between control and treated bees, but a Metastats model on the same OTUs revealed a number of significant differences (Fig. 6b). Specifically, a significant reduction with increasing acute doses of TiO2 was found for L. kimbladii. The same trend was detected for L. apis and S. alvi, but with no statistical significance.
Figure 6.
(a) Hierarchical clustering of sequences of OTUs participating with > 1% in at least one sample for the acute exposure experiment. Rare OTUs with lower participation were added to the “other” group; (b) metastats model showing differences among treatments for the most abundant OTUs. Significant differences are highlighted with different letters according to Tukey’s HSD test for comparison of means.
Distinctive features in the gut microbiota of honey-bees chronically exposed to TiO2
A stronger differentiation between control and TiO2 exposed bees was found in the chronic experiments. Here, the composition at the species level was very different, reflecting the age differences between this and the previous group of bees. G. apicola was still present, but with much lower relative abundances (< 2%). On the contrary, a strong increase in abundances was found for B. apis, L. apis, L. kimbladii, Commensalibacter intestini, and Bartonella spp. (Fig. 7a). Much higher was also the number of species displaying significant differences between treatments and controls (Fig. 7b), thus highlighting significant effects of chronic TiO2 exposure on the honey bees gut microbiota. Specifically, L. apis, Lactobacillus melliventris and Bartonella spp. were significantly reduced by the exposure to TiO2, in most cases with a dose dependent effect. On the contrary, Bombella intestini was found to be significantly enriched only in the X100 treatment.
Figure 7.
(a) Hierarchical clustering of sequences of OTUs participating with > 1% in at least one sample for the chronic exposure experiment. Rare OTUs with lower participation were added to the “other” group; (b) metastats model showing differences among treatments for the most abundant OTUs. Significant differences are highlighted with different letters according to Tukey’s HSD test for comparison of means.
Discussion
In the present study, we explored the potential effects of acute and chronic oral administration of pure TiO2 rutile on honey bee gut microbiota. The particles used were PM1; ranging from 800 to 200 nm as characterized by SEM–EDX analysis. In a preliminary assay, we fed newly emerged worker bees with four different concentrations of TiO2 in 1 M sucrose solution, including 100 ng/µL and 10 ng/µL as test doses, after the rejection of the two higher doses 104 ng/µL and 103 ng/µL. Newly emerged worker bees were orally exposed to the selected doses of TiO2 in an acute (single treatment, sampling at 24 h) and a chronic application (treatments repeated every 24 h, sampling at 96 h). In both assays, we obtained no mortality for the entire trials. This is partly in contrast with similar experiments on the honey bee, that however applied higher doses than the one tested here: TiO2 resulted highly toxic at 1000 ng/µL12, while in another study the LC50 at 96 h resulted in 5.9 ng/µL13. Nevertheless, TiO2 showed high mortality at a very high dose of 2400 ng/µL) in cutworm (Spodoptera litura)43.
Unfortunately, the studies by Özkan et al.13 and Ferrara et al.12 do not provide a specific characterization of the size, morphology and purity of TiO2 dust delivered to the bees. In our research, TiO2 dust were carefully characterised by SEM/EDX, demonstrating the absence of contaminants or coatings and the dimensions greater than 200 nm. Such size might also explain the absence of TiO2 PM in the haemolymph since particles might not be able to cross the barrier of the gut epithelium. However, a more in-depth study involving the analysis of the gut epithelium should be carried out to exclude the presence of TiO2 PM from the epithelial cells or cytological abnormalities.
An indirect impact of TiO2 on haemolymph has been thoroughly investigated in other organisms. In Mytilus galloprovincialis, TiO2 has been shown to affect several immune parameters in both circulating haemocytes and haemolymph serum, resulting in immunomodulation44–46. In larvae of Galleria mellonella (Lepidoptera: Pyralidae), exposure with dietary TiO2 nanoparticles (NPs) has dose-dependent toxic effects and can enhance the stress-resistant capacity with a significant increase in the total protein amount and content of malondialdehyde (MDA) and glutathione S-transferase activity at 100, 500 and 1000 ppm47. In our experiments, SEM–EDX reveals the typical haemolymph elements spectrum (Mg, Ca, Na, S, P, K, Cl) of the honey bee48.
The analyses of bacterial community composition allowed by HTS of 16S amplicons clearly show that the acute and chronic exposed bees populations had hosted very different bacterial gut populations (Figs. 5, 6 and 7). This is expected since analyses were carried out after 1 dpe (day post-emergence) for the acute experiment and at 4 dpe for the chronic experiment. The β-diversity analyses here carried out by means of unconstrained (PCA and hierarchical clustering, Fig. 5 and Figure S2) and constrained (CCA, Fig. 3) analyses showed a clear distinction in bacterial community structure between the chronic and the acute experiments, and in the chronic experiments among controls and treated bees. These results are in line with several studies on other chemical stressors16–20, thus confirming that also PM modulates the honey bees gut bacterial community.
The results obtained in the unexposed controls are quite in agreement with previous evidence and showed the typical community of adult worker bee microbiota that is dominated by Lactobacillus, Bifidobacterium, Gilliamella, Snodgrassella. The presence of Bartonella and Commensalibacter in 4 dpe bees from the chronic experiment is in agreement with the characteristic gut microbial community of the so-called “winter bees”, i.e., the last generations of workers characterised by overwintering individuals with an extended lifespan to ensure colony survival until spring49. The variability occurring in control bees is in agreement with other studies, where it has been demonstrated that, while many of the phylotypes are consistently present in adult worker bees, their relative abundance can vary across individuals49,50, and representative of these two genera can be found also in early stages of development20,51.
In the treated populations, the two tested doses of TiO2 had significant impacts on the gut bacterial communities, with more differences between exposed and control bees in the chronic as compared to the acute exposure. As far as we know, this is the first study where the effects of TiO2 particles where specifically studied on bees gut microbiota, but they can be compared with a number of studies that previously assessed the detrimental impacts of airborne PM on the bees physiology12,13. Regarding α-diversity indexes, we found a dose-dependent increase in diversity, (Fig. 3), while the opposite was found for the neonicotinoid insecticide Thiacloprid18, polystyrene particles at µm levels52, and antibiotics16. It must be highlighted that in these cited works a significant mortality was detected, whereas in our study sub-lethal doses were tested and no mortality registered. It can thus be speculated that the microbiota responded to the sub-lethal stressors by increasing the diversity, as postulated by the ecological theory of the intermediate disturbance hypothesis firstly proposed by Connell in 197853.
Concerning the changes in species abundances, we found that in the acute test only one species, L. kimbladii, was significantly reduced by the highest dose of TiO2 applied (Fig. 6). Firstly described in 2014 by Olofsson54 and colleagues, this species was more recently proposed as a possible probiotic55,56. Interestingly, L. kimbladii was instead found at higher abundances in the 96 h bees from the chronic exposure, with values higher than the control in the two treated theses (Fig. 7). This change may point to an adaptation in time of the microbial community, with a probiotic species becoming more abundant to counteract the chronic effects of a chemical stressor, as previously shown for Bifidobacterium species in bees exposed to the insecticide Nitenpyram19.
While in the acute experiment only L. kimbladii was significantly inhibited by the TiO2 particles (Fig. 6), in the chronic exposure three species were inhibited: L. apis, L. melliventris and Bartonella spp. (Fig. 7). L. apis is one of the most studied components of the bees gut microbiome, whose functions as probiotics were demonstrated by processes such as the attenuation of immune dysregulation57 and the inhibition of Paenibacillus larvae and other pathogens58; the induction of resistance towards bacterial infection was also demonstrated for L. melliventris, which was accordingly proposed as a bee probiotic59. Finally, the reduction of species belonging to Bartonella was also found in bees parasitized by Varroa,60, but this genus was also found to be increased after exposure to glyphosate17. The only taxon whose relative presence was significantly increased by chronic TiO2 exposure was B. intestinii, an acetic acid bacterium (AAB) firstly isolated in honey bees in 2017 by Yun and colleagues61. This species, together with other AAB, may play a role in the regulation of the innate immune system homeostasis62 and may thus represent an adaptation of the gut bacterial community to counteract the stressors sub-lethal effects.
Titanium oxide nanoparticles are known to exert a toxic effect on several bacteria, with proposed modes of action related to both chemical and physical interactions with the cells envelopes63, and alterations in human gut bacterial communities are often found including detrimental effects, as recently reviewed by Lams et al.,. (2020)64. Here we provide for the first time modulation of the honey bees gut microbiota induced by both acute and chronic exposure to TiO2 PM1, with stronger effects in the latter case. The effects we report are related to doses that are probably higher than the ones that can be found under field conditions and can be considered sub-lethal since no mortality was observed among all treated bees, and an increase in bacterial diversity was even found. On the other side, some negative effects related to the decrease in the relative percentages of some beneficial lactobacilli were however observed, and for this reason the role of airborne particulate as a further risk factor for Apis mellifera health in addition to other chemical stressors should be further explored.
Materials and methods
Honey bees
Experiments were conducted during October 2018 with Apis mellifera ligustica colonies maintained in the experimental apiary of the University of Napoli “Federico II”, Department of Agricultural Sciences. Brood frames with capped cells from two colonies were selected and kept in a climatic chamber at 36 °C and 60% relative humidity for approximately 16 h. Newly emerged workers were randomly selected and used for the bioassays.
Preparation of TiO2 feeding solution
Titanium dioxide rutile (TiO2) powder was acquired from 2B Minerals S.r.l. (Modena, Italy). Suspensions were prepared by dissolving 0.5 g TiO2 in 50 mL distilled H2O, vortexed for ~ 20 s and sonicated for 15 min to increase dispersion and ensure the maximum distribution of particles in water13. A 1 M sucrose solution was then prepared with TiO2 suspensions.
Preliminary palatability tests on bees were carried out with the following dilutions: 104 ng/µL, 103 ng/µL, 100 ng/µL, 10 ng/µL. The last two concentrations were chosen for the experiments following rejection of the more concentrated solutions by the bees.
Chronic and acute exposure
Two groups of 30 newly eclosed bees were randomly collected from the combs. The bees were fed ad libitum with 1 µL of 10 ng/µL and 100 ng/µL TiO2 solutions (1 M sucrose), respectively (hereinafter CH10- and CH100-bees), and then placed into plastic cages containing 1.5 mL of the same solution to which they were previously fed. Control bees consisted of 30 individuals fed ad libitum with an untreated sucrose solution (1 M).
The solution was changed every 24 h. During the experiments, all caged bees were kept in a climatic chamber at 36 °C and 60% relative humidity. After 96 h the bees were anesthetized with CO2 for ~ 30 s and the whole gut dissected as previously described65. Guts were stored in absolute ethanol and immediately refrigerated at -80 °C for the subsequent microbiological analysis. Experiments were conducted in triplicate (i.e., 3 replicates each made by three guts pooled together). Bees survival was recorded each day.
The experimental design of acute exposure was the same as in chronic exposure except for the duration of the experiment that in acute exposure lasted only 24 h instead of 96, and bees were fed only once with 10 ng/µL or 100 ng/µL TiO2 solutions (hereinafter AC10- and AC100-bees).
TiO2 detection in the gut
To assess morphology, average size and chemical purity of TiO2 powder, few µgrams were mounted onto stubs and analysed through a Scanning Electron Microscope (SEM) provided with X-ray spectroscopy (EDX) (Zeiss Gemini SEM 500—Bruker Quanta X-Flash 61|31). Secondary Electrons (SE), BackScattered Electrons (BSE) images, and EDX point analyses, were acquired as previously described3,14.
The presence of TiO2 in the haemolymph and gut of chronic bees was investigated by SEM–EDX. Haemolymph from 4 randomly selected CH10- and CH100-bees plus 4 randomly selected control bees was sampled from the dorsal aorta, following the method standardized by Garrido and colleagues66. The bee gut (rectum with excrements) from CH10- and CH100-bees plus control bees (n = 4 randomly selected, for each concentration and control) were mounted onto stub and dried in a sterilized oven at 20 °C for 40 min. Samples were carbon coated and analyzed with SEM/EDX.
Microbiota analysis
DNA extraction
DNA was extracted from gut samples collected for microbiological analyses. For each dose—acute and chronical—n. 3 groups of three guts were analysed. From these samples, the total microbial DNA was extracted using the Fast DNA SPIN Kit for Soil (MP Biomedicals, USA) with the following modifications: each sample was homogenized in the FastPrep for 40 s at speed setting of 6.5 twice, keeping it in ice between the two homogenisation steps, while the final centrifugation was carried out at 14,000×g for 15 min, and the final resuspension of the Binding Matrix was carried out in 50 µL of nuclease-free water.
The DNA extractions were checked with electrophoresis on a 1% agarose gel, and then quantified using a QuBit fluorometer (Invitrogen, United Kingdom).
DNA amplification
The V3-V4 region of the bacterial 16S rRNA gene (between 480 and 490 bp) was amplified by PCR using the universal primers 343f. (5′-TACGGRAGGCAGCAG-3′), and 802r (5′-TACNVGGGTWTCTAATCC-3′)67. A two-step PCR protocol was implemented in order to reduce the possibility of generating non-specific primer annealing, as detailed in Berry et al.68. The PCR reaction mix comprised of 20.5 µL of MegaMix (Microzone Limited, United Kingdom), 1.25 μL of each primer (10 μM), and 2 μL (1 ng/μL concentration) of DNA template. Thermal cycling conditions were as follows: Step 1: an initial denaturation at 94 °C for 5 min, followed by 25 cycles at 94 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. Step 2: initial hold at 95 °C for 5 min, followed by 10 cycles of 95 °C for 30 s, 50 °C for 30 s, and 30 °C for 30 s; then, a final extension at 72 °C for 10 min. At the second step, each sample was amplified using a dedicated forward primer with a 9- base extension at the 5′ end, which acts as a tag, in order to make simultaneous analyses of all samples in a single sequencing run possible The DNA amplifications were checked with electrophoresis on a 1% agarose gel, and then quantified using a QuBit fluorometer (Invitrogen, United Kingdom). PCR products generated from the second step were multiplexed as a single pool using equivalent molecular weights (20 ng). The pool was then purified using the solid phase reversible immobilization (SPRI) method with Agencourt AMPure XP kit (REF A63880, Beckman Coulter, Milan, Italy), then sequenced by Fasteris S.A. (Geneva, Switzerland). The TruSeq DNA sample preparation kit (REF 15026486, Illumina Inc, San Diego, CA) was used for amplicon library preparation, whereas the sequencing was carried out with the MiSeq Illumina instrument (Illumina Inc., San Diego, CA) generating 300 bp paired-end reads.
Sequence data preparation and statistical analyses
High-throughput sequencing data filtering, multiplexing and preparation for concomitant statistical analyses were carried out as previously detailed69. In summary, paired-reads were assembled to reconstruct the full V3-V4 amplicons using the “pandaseq” script70 with a maximum of 2 allowed mismatches and at least 30 bp of overlap between the read pairs. was then carried out with the Fastx-toolkit was then employed for samples demultiplexing (http://hannonlab.cshl.edu/fastx_toolkit/).
Mothur v.1.32.171 was applied in order to remove sequences with large homopolymers (≥ 10), sequences that did not align within the targeted V3-V4 region, chimeric sequences72 and sequences not classified as bacterial after alignment against the Mothur version of the RDP training data set. Mothur and R (http://www.R-proje ct.org/) were employed to analyze the resulting high-quality sequences following the operational taxonomic unit (OTU) and the taxonomy-based approach. For the OTU approach, sequences were first aligned against the SILVA reference aligned database for bacteria73 using the NAST algorithm and a kmer approach74,75, and then clustered at the 3% distance using the average linkage algorithm. OTUs having a sum of their abundances across all samples of than 0.1% of the total were grouped into a single “rare OTUs” group. For taxonomy based analyses, sequences were classified into taxa using an amended version of the Greengenes database76.
Mothur and R were also employed for statistical analyses on OTU and taxonomy matrixes using hierarchical clustering with the average linkage algorithm at different taxonomic levels, Principal component analysis (PCA) for unconstrained samples grouping, Canonical correspondence analyses (CCA) to assess the significance of different treatments on the analysed diversity. Features that were significantly different between treatments were identified with Metastats76.
Sequence data were submitted to the National Centre for Biotechnology Information Sequence Read Archive (BioProject accession number PRJNA693145).
Supplementary Information
Acknowledgements
GP was partially supported by the Doctoral School on the Agro-Food System (Agrisystem) of the Università Cattolica del Sacro Cuore (Italy). We thank Tiziano Catelani and Paolo Gentile for advices on the SEM-EDX analyses and Francesco Pennacchio for the permission to use the Entomology laboratory at the Federico II University of Naples (Italy).
Author contributions
G.P., I.N., conceived and designed the experiments; G.P., G.P.D., G.S. performed the experiments; G.P., I.N., E.P., analysed data G.P., G.D.P., I.N., E.P. wrote the article. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85153-1.
References
- 1.Devillers, J. & Pham-Delegue, M.-H. Honey Bees: Estimating the Environmental Impact of Chemicals. (Taylor & Francis, 2002).
- 2.Satta A, et al. Combination of beehive matrices analysis and ant biodiversity to study heavy metal pollution impact in a post-mining area (Sardinia, Italy) Environ. Sci. Pollut. Res. 2012;19:3977–3988. doi: 10.1007/s11356-012-0921-1. [DOI] [PubMed] [Google Scholar]
- 3.Negri, I., Mavris, C., Di Prisco, G., Caprio, E. & Pellecchia, M. Honey bees (Apis mellifera, L.) as active samplers of airborne particulate matter. PLoS ONE10, 1–22 (2015). [DOI] [PMC free article] [PubMed]
- 4.Goretti E, et al. Heavy metal bioaccumulation in honey bee matrix, an indicator to assess the contamination level in terrestrial environments. Environ. Pollut. 2020;256:113388. doi: 10.1016/j.envpol.2019.113388. [DOI] [PubMed] [Google Scholar]
- 5.Papa, G., Capitani, G., Capri, E., Pellecchia, M. & Negri, I. Vehicle-derived ultrafine particulate contaminating bees and bee products. Sci. Total Environ. 141700 (2020). doi:10.1016/j.scitotenv.2020.141700 [DOI] [PubMed]
- 6.Brodschneider R, et al. Preliminary analysis of loss rates of honey bee colonies during winter 2015/16 from the COLOSS survey. J. Apic. Res. 2016;55:375–378. doi: 10.1080/00218839.2016.1260240. [DOI] [Google Scholar]
- 7.Dainat B, VanEngelsdorp D, Neumann P. Colony collapse disorder in Europe. Environ. Microbiol. Rep. 2012;4:123–125. doi: 10.1111/j.1758-2229.2011.00312.x. [DOI] [PubMed] [Google Scholar]
- 8.vanEngelsdorp, D. & Meixner, M. D. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invert. Pathol.103, S80–S95 (2010). [DOI] [PubMed]
- 9.Feldhaar H, Otti O. Pollutants and their interaction with diseases of social hymenoptera. Insects. 2020 doi: 10.3390/insects11030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.AL Naggar, Y. et al. Sublethal effects of chronic exposure to CdO or PbO nanoparticles or their binary mixture on the honey bee (Apis millefera L.). Environmental Science and Pollution Research27, 19004–19015 (2020). [DOI] [PubMed]
- 11.Dabour, K., Al Naggar, Y., Masry, S., Naiem, E. & Giesy, J. P. Cellular alterations in midgut cells of honey bee workers (Apis millefera L.) exposed to sublethal concentrations of CdO or PbO nanoparticles or their binary mixture. Sci. Total Environ.651, 1356–1367 (2019). [DOI] [PubMed]
- 12.histological and immunohistochemical assays Ferrara, G. et al. Toxicity assessment of nano-TiO 2 in Apis mellifera L., 1758. Microsc. Res. Tech. 2019;83:332–337. doi: 10.1002/jemt.23418. [DOI] [PubMed] [Google Scholar]
- 13.Özkan Y, Irende İ, Akdeniz G, Kabakçi D, Münevver S. Evaluation of the comparative acute toxic effects of TiO2, Ag-TiO2 and ZnO-TiO2 composite nanoparticles on honey bee (Apis mellifera) J. Int. Environ. Appl. Sci. 2014;10:26–36. [Google Scholar]
- 14.Pellecchia, M. & Negri, I. Particulate matter collection by honey bees (Apis mellifera, L.) near to a cement factory in Italy. PeerJ2018, 1–21 (2018). [DOI] [PMC free article] [PubMed]
- 15.Raymann K, Moran NA. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018;26:97–104. doi: 10.1016/j.cois.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raymann K, Shaffer Z, Moran NA. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017;15:1–22. doi: 10.1371/journal.pbio.2001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motta EVS, et al. Oral or topical exposure to glyphosate in herbicide formulation impacts the gut microbiota and survival rates of honey bees. Appl. Environ. Microbiol. 2020;86:1–21. doi: 10.1128/AEM.01150-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YJ, et al. Thiacloprid exposure perturbs the gut microbiota and reduces the survival status in honeybees. J. Hazard. Mater. 2020;389:121818. doi: 10.1016/j.jhazmat.2019.121818. [DOI] [PubMed] [Google Scholar]
- 19.Zhu, L., Qi, S., Xue, X., Niu, X. & Wu, L. Nitenpyram disturbs gut microbiota and influences metabolic homeostasis and immunity in honey bee (Apis mellifera L.). Environ. Pollut.258, 113671 (2020). [DOI] [PubMed]
- 20.Rothman JA, Leger L, Kirkwood JS, McFrederick QS. Cadmium and selenate exposure affects the honey bee microbiome and metabolome, and bee-associated bacteria show potential for bioaccumulation. Appl. Environ. Microbiol. 2019;85:1–18. doi: 10.1128/AEM.01411-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS ONE. 2012;7:1–10. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabree, Z. L., Hansen, A. K. & Moran, N. A. Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS ONE7, (2012). [DOI] [PMC free article] [PubMed]
- 23.Corby-Harris, V., Maes, P. & Anderson, K. E. The bacterial communities associated with honey bee (apis mellifera) foragers. PLoS ONE9, (2014). [DOI] [PMC free article] [PubMed]
- 24.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA. 2012;109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong, W. K. & Moran, N. A. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a memb. Int. J. Syst Evol. Microbiol.63, 2008–2018 (2013). [DOI] [PubMed]
- 26.Martinson VG, et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2011;20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 27.Bottacini F, et al. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS ONE. 2012;7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel, P., Kwong, W. K. & Moran, N. A. Frischella perrara gen. nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. Int. J. Syst. Evol. Microbiol.63, 3646–3651 (2013). [DOI] [PubMed]
- 29.Kešnerová, L., Moritz, R. & Engel, P. Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol.66, 414–421 (2016). [DOI] [PubMed]
- 30.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016;14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hroncova Z, et al. Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0118707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vojvodic, S., Rehan, S. M. & Anderson, K. E. Microbial gut diversity of Africanized and European honey bee larval instars. PLoS ONE8, (2013). [DOI] [PMC free article] [PubMed]
- 33.Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014;80:7378–7387. doi: 10.1128/AEM.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Younes, M. et al. Scientific opinion on the proposed amendment of the EU specifications for titanium dioxide (E 171) with respect to the inclusion of additional parameters related to its particle size distribution. EFSA J.17, (2019). [DOI] [PMC free article] [PubMed]
- 35.Bettini S, et al. Food-grade TiO2 impairs intestinal and systemic immune homeostasis, initiates preneoplastic lesions and promotes aberrant crypt development in the rat colon. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baan RA. Carcinogenic hazards from inhaled carbon black, titanium dioxide, and talc not containing asbestos or asbestiform fibers: Recent evaluations by an IARC monographs working group. Inhal. Toxicol. 2007;19:213–228. doi: 10.1080/08958370701497903. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, Han S, Zhou D, Zhou S, Jia G. Effects of oral exposure to titanium dioxide nanoparticles on gut microbiota and gut-associated metabolism: In vivo. Nanoscale. 2019;11:22398–22412. doi: 10.1039/C9NR07580A. [DOI] [PubMed] [Google Scholar]
- 38.Smith KE, et al. Honey as a biomonitor for a changing world. Nat. Sustain. 2019;2:223–232. doi: 10.1038/s41893-019-0243-0. [DOI] [Google Scholar]
- 39.Solayman M, et al. Physicochemical properties, minerals, trace elements, and heavy metals in honey of different origins: A comprehensive review. Comprehensive Rev. Food Sci. Food Saf. 2016;15:219–233. doi: 10.1111/1541-4337.12182. [DOI] [PubMed] [Google Scholar]
- 40.Ribeiro, R. de O. R. et al. Determination of trace elements in honey from different regions in Rio de Janeiro State (Brazil) by total reflection X-ray fluorescence. J. Food Sci.79, (2014). [DOI] [PubMed]
- 41.Omode PE, Ademukola SA. Determination of trace metals in southern Nigerian honey by use of atomic absorption spectroscopy. Spectrosc. Lett. 2008;41:328–331. doi: 10.1080/00387010802371239. [DOI] [Google Scholar]
- 42.Chan QWT, Howes CG, Foster LJ. Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics. 2006;5:2252–2262. doi: 10.1074/mcp.M600197-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Chakravarthy, A. K. DNA-tagged nano gold: A new tool for the control of the armyworm, Spodoptera litura Fab. (Lepidoptera: Noctuidae). Afr. J. Biotechnol.11, (2012).
- 44.Barmo C, et al. In vivo effects of n-TiO2 on digestive gland and immune function of the marine bivalve Mytilus galloprovincialis. Aquat. Toxicol. 2013;132–133:9–18. doi: 10.1016/j.aquatox.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Auguste M, et al. In vivo immunomodulatory and antioxidant properties of nanoceria (nCeO2) in the marine mussel Mytilus galloprovincialis. Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 2019;219:95–102. doi: 10.1016/j.cbpc.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Corsi I, et al. Common strategies and technologies for the ecosafety assessment and design of nanomaterials entering the marine environment. ACS Nano. 2014;8:9694–9709. doi: 10.1021/nn504684k. [DOI] [PubMed] [Google Scholar]
- 47.Zorlu, T., Nurullahoğlu, Z. & Altuntaş, H. Influence of Dietary Titanium Dioxide Nanoparticles on the Biology and Antioxidant System of Model Insect, Galleria mellonella (L.) (Lepidoptera: Pyralidae). J. Entomol. Res. Soc.20, (2018).
- 48.Mikulecky M, Bounias M. Worker honeybee hemolymph lipid composition and synodic lunar cycle periodicities. Braz. J. Med. Biol. Res. 1997;30:275–279. doi: 10.1590/S0100-879X1997000200018. [DOI] [PubMed] [Google Scholar]
- 49.Kešnerová L, et al. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020;14:801–814. doi: 10.1038/s41396-019-0568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamei S, et al. Feeding honeybee colonies with honeybee-specific lactic acid bacteria (Hbs-LAB) does not affect colony-level Hbs-LAB composition or paenibacillus larvae spore levels, although american foulbrood affected colonies harbor a more diverse Hbs-LAB community. Microb. Ecol. 2020;79:743–755. doi: 10.1007/s00248-019-01434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong Z-X, et al. Colonization of the gut microbiota of honey bee (Apis mellifera) workers at different developmental stages. Microbiol. Res. 2019;231:126370. doi: 10.1016/j.micres.2019.126370. [DOI] [PubMed] [Google Scholar]
- 52.Wang, K. et al. Gut microbiota protects honey bees (Apis mellifera L.) against polystyrene microplastics exposure risks. J. Hazard. Mater.402, 123828 (2021). [DOI] [PubMed]
- 53.Connell JH. Diversity in tropical rain forests and coral reefs. Science. 1978;199:1302–1310. doi: 10.1126/science.199.4335.1302. [DOI] [PubMed] [Google Scholar]
- 54.Olofsson, T. C., Alsterfjord, M., Nilson, B., Butler, È. & Vásquez, A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isol. Int. J. Syst. Evol. Microbiol.64, 3109–3119 (2014). [DOI] [PMC free article] [PubMed]
- 55.Duong BTT, et al. Investigation of the gut microbiome of Apis cerana honeybees from Vietnam. Biotech. Lett. 2020;42:2309–2317. doi: 10.1007/s10529-020-02948-4. [DOI] [PubMed] [Google Scholar]
- 56.Mårtensson A, et al. Effects of a honeybee lactic acid bacterial microbiome on human nasal symptoms, commensals, and biomarkers. Int. Forum Allergy Rhinol. 2016;6:956–963. doi: 10.1002/alr.21762. [DOI] [PubMed] [Google Scholar]
- 57.Daisley BA, et al. Novel probiotic approach to counter Paenibacillus larvae infection in honey bees. ISME J. 2020;14:476–491. doi: 10.1038/s41396-019-0541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamei S, et al. The secretome of honey bee-specific lactic acid bacteria inhibits Paenibacillus larvae growth. J. Apic. Res. 2019;58:405–412. doi: 10.1080/00218839.2019.1572096. [DOI] [Google Scholar]
- 59.Hyrsl P, et al. Plant alkaloid sanguinarine and novel potential probiotic strains lactobacillus apis, lactobacillus melliventris and Gilliamella apicola promote resistance of honey bees to nematobacterial infection. Bull. Insectol. 2017;70:31–38. [Google Scholar]
- 60.Hubert J, et al. Bacteria detected in the honeybee parasitic mite Varroa destructor collected from beehive winter debris. J. Appl. Microbiol. 2015;119:640–654. doi: 10.1111/jam.12899. [DOI] [PubMed] [Google Scholar]
- 61.Yun, J.-H., Lee, J.-Y., Hyun, D.-W., Jung, M.-J. & Bae, J.-W. Bombella apis sp. nov., an acetic acid bacterium isolated from the midgut of a honey bee. International Journal of Systematic and Evolutionary Microbiology67, 2184–2188 (2017). [DOI] [PubMed]
- 62.Crotti E, et al. Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microbiol. 2010;76:6963–6970. doi: 10.1128/AEM.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baysal, A., Saygin, H. & Ustabasi, G. S. Interaction of PM2.5 airborne particulates with ZnO and TiO2 nanoparticles and their effect on bacteria. Environmental Monitoring and Assessment190, 34 (2018). [DOI] [PubMed]
- 64.Lamas B, Martins Breyner N, Houdeau E. Impacts of foodborne inorganic nanoparticles on the gut microbiota-immune axis: potential consequences for host health. Part. Fibre Toxicol. 2020;17:19. doi: 10.1186/s12989-020-00349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carreck NL, et al. Standard methods for Apis mellifera anatomy and dissection. J. Apic. Res. 2013;52:1–40. doi: 10.3896/IBRA.1.52.4.03. [DOI] [Google Scholar]
- 66.Garrido PM, Martin ML, Negri P, Eguaras MJ. A standardized method to extract and store haemolymph from Apis mellifera and the ectoparasite Varroa destructor for protein analysis. J. Apic. Res. 2013;52:67–68. doi: 10.3896/IBRA.1.52.2.13. [DOI] [Google Scholar]
- 67.Połka J, Rebecchi A, Pisacane V, Morelli L, Puglisi E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015;46:342–356. doi: 10.1016/j.fm.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 68.Berry, D., Ben Mahfoudh, K., Wagner, M. & Loy, A. barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol.77, 7846–7849 (2011). [DOI] [PMC free article] [PubMed]
- 69.Vasileiadis, S. et al. Changes in soil bacterial communities and diversity in response to long-term silver exposure. FEMS Microbiol. Ecol.91, fiv114 (2015). [DOI] [PubMed]
- 70.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeSantis TZ, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.