Abstract
Excessive production of inflammatory chemokines and reactive oxygen species (ROS) can cause a feedback cycle of inflammation response that has a negative effect on cutaneous wound healing. The use of wound-dressing materials that simultaneously absorb chemokines and scavenge ROS constitutes a novel ‘weeding and uprooting’ treatment strategy for inflammatory conditions. In the present study, a composite hydrogel comprising an amine-functionalized star-shaped polyethylene glycol (starPEG) and heparin for chemokine sequestration as well as Cu5.4O ultrasmall nanozymes for ROS scavenging (Cu5.4O@Hep-PEG) was developed. The material effectively adsorbs the inflammatory chemokines monocyte chemoattractant protein-1 and interleukin-8, decreasing the migratory activity of macrophages and neutrophils. Furthermore, it scavenges the ROS in wound fluids to mitigate oxidative stress, and the sustained release of Cu5.4O promotes angiogenesis. In acute wounds and impaired-healing wounds (diabetic wounds), Cu5.4O@Hep-PEG hydrogels outperform the standard-of-care product Promogram® in terms of inflammation reduction, increased epidermis regeneration, vascularization, and wound closure.
Keywords: Inflammatory chemokines, Reactive oxygen species, Nanozymes, Hydrogels, Wound healing
Graphical abstract
Highlights
-
•
Cu5.4O@Hep-PEG hydrogels capture pro-inflammatory factors, decreasing immune cell influx and inflammatory level in wounds.
-
•
Hydrogels can scavenge ROS, reducing secretion of pro-inflammatory factors and inflammatory cell infiltration in wounds.
-
•
This strategy disrupts the feedback relationship between ROS and inflammation and improve the wound-healing environment.
1. Introduction
Cutaneous wounds are a global health problem and cause significant economic losses to society [1,2]. Efficient reconstruction of the function and integrity of the damaged tissues is currently the central focus for their clinical treatment [3]. Multiple therapeutic options, including novel dressing products [4,5], growth factors [6,7], and bioengineered skin grafts [8] are available. However, strategies for the regenerative healing of the injured tissues, especially for chronic wounds, remain elusive. This may be partly due to the fact that modulating the microenvironment of the inflammation site by taking advantage of the intrinsic regenerative capacity of the host remains largely unexplored.
Skin injuries initiate a series of biological events, among which large amounts of reactive oxygen species (ROS) are produced in the wound microenvironment as a feature of the defense response to invading pathogens [9]. Although this process is generally beneficial to healing, unregulated overproduction of ROS can cause damage to DNA, proteins, lipids, and even cells [9,10]. Furthermore, elevated ROS production in injured tissues can trigger deleterious effects such as cellular senescence [11], fibrotic scarring [12], and uncontrolled inflammation [13,14]. Accordingly, the successful healing and regeneration of wounded tissues relies on an optimal balance between the constructive and harmful roles of ROS. Therefore, the use of biomaterials that manage ROS levels and control oxidative damage in injured tissues shows promise as a therapeutic strategy for regenerative wound healing.
Numerous studies [15,16] including our own [17] have explored the use of nanomaterials with ROS-scavenging properties for the repair of injured tissues. Although these treatments achieved beneficial therapeutic effects, their success remains limited. These strategies focused on the scavenging of ROS rather than preventing their persistent and excessive production by controlling the level of inflammation.
It is difficult to remove ROS completely. Massive ROS production initiates many pathological signaling pathways [13,14], especially the nuclear factor kappa-light-chain-enhancer (NF-κB) pathway, resulting in increased production of pro-inflammatory factors such as interleukin-8 (IL-8) and monocyte chemoattractant protein-1 (MCP-1). Furthermore, MCP-1 and IL-8 promote the invasion of inflammatory monocytes/macrophages and polymorphonuclear neutrophils (PMNs) into the inflamed tissue [18,19]. These immune cells overproduce several pro-inflammatory cytokines, such as IL-6 and tumor necrosis factor α (TNF-α) as well as numerous ROS, thus perpetuating a feedback cycle that leads to chronic non-healing wounds. To break this cycle completely, it is essential to disrupt the mutual interaction of the different phenomena involved by scavenging ROS and capturing pro-inflammatory factors, preventing damage from excessive ROS both downstream and upstream simultaneously, thus accelerating wound healing.
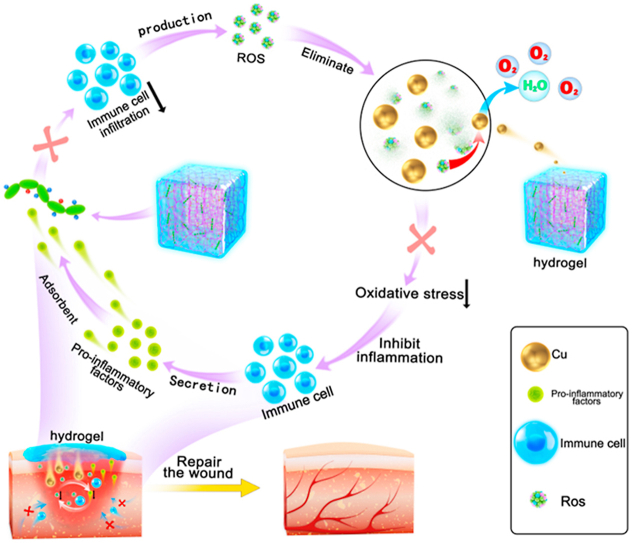
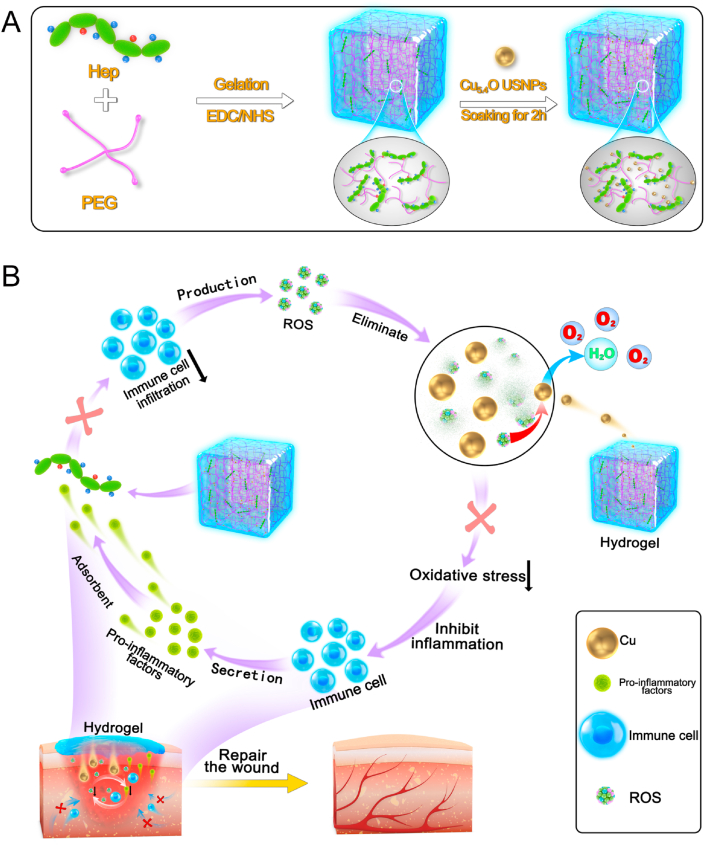
Accordingly, in the present study, Cu5.4O@Hep-PEG hydrogels comprising star-shaped polyethylene glycol (starPEG), heparin, and Cu5.4O ultrasmall nanoparticles (USNPs) were developed and optimized as a means to simultaneously suppress persistent pro-inflammatory chemokine gradients and ameliorate high oxidative stress in inflammatory diseases (Fig. 1A). The Hep-PEG hydrogels effectively scavenge the pro-inflammatory factors MCP-1 and IL-8 in wound fluids [20], thus decreasing the migratory activity of polymorphonuclear neutrophils and monocytes. In our previous work, Cu5.4O USNPs were demonstrated to exhibit superoxide dismutase (SOD), catalase, and glutathione peroxidase and protect against ROS-related damage to wound tissue at very low dosages simultaneously [17]. Hep-PEG gels can be customized to incorporate Cu5.4O USNPs through coordination between the amine groups and copper (Fig. 1A). These Cu5.4O@Hep-PEG hydrogels capture chemokines from inflamed wound tissue, inhibiting the excessive infiltration of immune cells, while simultaneously mitigating ROS generation by sustained release of Cu5.4O USNPs to the wound tissue, thus preventing further inflammation-activation signaling (Fig. 1B). Our approach is not to suppress inflammation completely but in parallel maintaining the balance for fighting pathogens and avoiding unwanted side effects. Models of acute normal wound healing and delayed diabetic wound healing were used to assess the overall pro-regenerative effect of Cu5.4O@Hep-PEG hydrogels.
Fig. 1.
Schematic of Cu5.4O@Hep-PEG hydrogels in the treatment of diabetic wounds. (A) Preparation of Cu5.4O@Hep-PEG hydrogels. I The Hep-PEG gel network is prepared by covalent cross-linking between the carboxyl groups in heparin sodium and the amino groups in 4-arm PEG-NH2. II The Cu5.4O@Hep-PEG hydrogel is prepared by soaking Hep-PEG hydrogel in a Cu5.4O USNP solution for 2 h. (B) In diabetic wounds, high levels of inflammation generate intracellular ROS, which modulates the production of chemokines associated with inflammation. Furthermore, the pro-inflammatory factors generated induce the migration and infiltration of more inflammatory cells. The infiltrated inflammatory cells produce excessive ROS to further aggravate inflammation, leading to a feedback cycle. To break this cycle completely, it is essential to disrupt their mutual interaction by simultaneously scavenging ROS and capturing pro-inflammatory factors. Cu5.4O@Hep-PEG can release Cu5.4O USNPs to scavenge ROS, capture pro-inflammatory factors, improve the environment of wound sites, and accelerate diabetic wound healing.
2. Materials and methods
2.1. Chemicals and materials
Copper chloride (CuCl2, 98%), l-ascorbic acid (AA, 99%) and N-hydroxysuccinimide (NHS, 98%) were purchased from J&K Scientific Co., Ltd. (Beijing, China). Acetate (≥99.8%), anhydrous sodium acetate (≥99.0%), iodine (≥99.8%), barium chloride dihydrate (99%), 2,2-diphenyl-1-picrylhydrazyl (DPPH), and sodium hydroxide (NaOH, 97%) were obtained from Aladdin biotechnology Co., Ltd. (Shanghai, China). Phosphate buffered saline (PBS), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDAC), recombinant murine tumor necrosis factor-alpha (TNF-α), interleukin 6 (IL-6), TNF-α ELISA kits, and IL-6 ELISA kits were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Heparin sodium (Hep, Mw 14,000 Da) and Roswell Park Memorial Institute (RPMI) medium 1640 were obtained from Solarbio Science & Technology Co., Ltd. (Beijing, China). Fetal bovine serum (FBS) was obtained from Gibco (New York, USA). 4-Arm PEG-NH2 (PEG, Mw 10,000 Da) was purchased from Xian Ruixi Biological Technology Co., Ltd. (Xian, China). Anhydrous ethanol (99.5%) and Methylene Blue (MB, ≥98%) were obtained from Macklin biochemical Co., Ltd. (Shanghai, China). Assay kits to assess scavenging capacity for hydrogen peroxide (H2O2), hydroxyl radicals (·OH), and superoxide anions (O2·-) were purchased from Nanjing Jiancheng bioengineering institute Co., Ltd. (Nanjing, China). Recombinant mouse MCP-1 and recombinant human IL-8 were provided by Bioworld Technology, Inc. (Minnesota, USA). ELISA Kits for mouse MCP-1 and human IL-8 were purchased from Cloud-clone Corp. (Wuhan, China) and ELISA Kits for mouse CXCL-1 (There is no IL-8 in the murine system, CXCL-1 in mouse has similar function as IL-8 in human) were purchased from AMEKO Life Science (Shanghai, China). Milli-Q water was used in all experiments.
2.2. Instrumentation
The morphologies of the hydrogels were defined using scanning electron microscopy (SEM, Quanta FEG 250, Thermo Fisher, USA). Energy dispersive spectroscopy (EDS) analysis was performed using a JSM-7610F scanning electron microscope (JEOL, Japan). The concentration of copper was determined using a 5110 inductively coupled plasma mass spectrometer (ICP-MS, Agilent, USA). Optical absorption spectra were obtained using a Hitachi U3010 spectrometer (Hitachi, Japan). Fourier-transform infrared (FTIR) spectra in the range of 4000 to 400 cm−1 were obtained using a Nicolet iS10 spectrometer (Thermo Fisher, USA). The storage (G′) moduli of the hydrogel networks were obtained using a HR-1 Discovery hybrid rheometer (TA, USA). The crystal structures of the Cu5.4O USNPs and hydrogels were obtained using X-Ray diffractometry (XRD, D8, Bruker Axs, Germany). X-Ray photoelectron spectroscopy (XPS) was performed using an Escalab 250 Xi (Thermo Scientific, USA). Gel cross-linking degree was measured by a VTMR20-010V-I NMR cross-link density imaging analyzer (Shanghai Newmai, China).
2.3. Preparation of Cu5.4O@Hep-PEG hydrogels
The heparin-PEG hydrogels (Hep-PEG gels) were obtained according to previous methods with slight changes [21]. The formation of Hep-PEG gels depends on the covalent cross-links between carboxyl groups in heparin sodium and the amino groups in 4-arm PEG-NH2 [22,23]. Briefly, a solution of heparin (0.025 g) was activated using NHS and EDAC (molar ratio of NHS to EDAC = 1:2) in 0.5 mL MES solution (50 mM) on ice with stirring for 15 min. Subsequently, amine end-functionalized 4-arm star PEG was dissolved in water on ice, and then added into the activated heparin solution dropwise at a series molar ratios (heparin/PEG = 1:5, 1:7.5, 1:10, 1:12.5, and 1:15). After stirring for another 15 min, the solution was spread on a clean glass culture dish. After overnight polymerization, gels were swollen in PBS for 24 h to remove unreacted molecules.
The Cu5.4O USNPs were loaded into Hep-PEG gels by simple soaking treatment to obtain Cu5.4O@Hep-PEG hydrogels [24]. In brief, the gels prepared above were immersed in PBS solutions containing Cu5.4O USNPs at different concentrations (6–960 μg mL−1) for 0–12 h at room temperature to determine the optimal soaking conditions. The excess Cu5.4O USNPs were removed by washing with PBS solution three to five times.
2.4. Characterization
Swelling behavior To investigate the swelling behavior of the gels, they were freeze-dried and immersed in PBS solution at 37 °C for 24 h [21]. When equilibrium had been reached, the samples were drained through a tissue and weighed again. Swelling ratios were calculated using S (%) = (Wt − W0)/W0 × 100, where Wt is the weight of the swollen gel at equilibrium and W0 is the weight of dry gel.
Rheological analysis The storage (G′) moduli of the swollen gels were determined by rheological analysis using a Modular Compact Rheometer MCR 302 (Anton Paar Co., Austria) with a measuring gap size of 1.5 mm. G′ was measured with the variation of the frequency increasing from 0.1 to 100 rad s−1 at 37 °C and a shear stress of 1 Pa.
Mesh size analysis The average pore size (ξ) of a purely elastic gel can be estimated using rubber-elasticity theory (RET), which extends the relationship between storage modulus (G′) and the number of cross-linkers [25,26]. RET is based on the assumptions that all covalent bound chains of gels play a role in the retraction force after an affine deformation ignoring end effects of the single chains of gels and excluding any effects of physical entanglement. The mesh sizes (ξ) of the gels were evaluated according to the following formula (the affine network model) [26]:
where G′ is the storage modulus of the swollen gel, NA is the Avogadro constant, R is the molar gas constant, and T is the temperature.
Cross-linking degree analysis The degree of cross-linking in the heparin-PEG hydrogels was estimated from the NH2-PEG and heparin contents. Assuming that the carboxylic acid groups on heparin (28 per heparin molecule) bind to three of the four amino groups on PEG, the following equation was used to calculate the cross-linking degree (C%) of a gel [21,27]:
where P is the molality of PEG in the final washed gel and H is the molality of heparin in the final washed gel. The NH2-PEG and heparin levels in the gels were obtained by dialysis and subsequent absorption spectrophotometry analysis. More specifically, the gels (20 μL, 3.2 wt%) were dialyzed against Milli-Q water (Mw cutoff: 100,000 Da) for 2 days, and the water was changed every 4 h. The heparin was quantified by MB bleaching assay with slight modifications [28]. A reaction mixture containing 1.5 μM MB, sodium acetate (15 mM)/acetic acid (28 mM) buffer (pH 4.6), and the free heparin solution obtained above was prepared, and its absorbance at 664 nm was measured after 10 min. The concentration of PEG was determined by measuring the absorbance of the reaction mixture in Milli-Q water containing barium chloride (44 mM), iodine (1.5 mM in anhydrous ethanol), and the free PEG obtained above at 456 nm after 10 min [29]. The free PEG and heparin molecules were quantified using standard absorbance curves (n = 3).
2.5. Adsorption and release behavior of Cu5.4O USNPs from gels
In order to determine the saturation adsorption time of Cu5.4O USNPs in Hep-PEG hydrogels, dry gels with a molar ratio of 1:7.5 were immersed in PBS solutions containing Cu5.4O USNPs (10 μg mL−1) for different times (5, 10, 20, 30, 60, 120, 240, 480, and 720 min). In order to study the effect of the concentration of the Cu5.4O USNPs solution on the amount of Cu5.4O USNPs adsorbed, gels (molar ratio of 1:7.5) were immersed in Cu5.4O USNP solutions with different concentrations (6, 15, 30, 60, 120, 240, 480, and 960 μg mL−1) for 2 h. To explore the Cu5.4O USNP adsorption capacity of different molar ratio gels, gels prepared with Hep/PEG molar ratios of 1:5, 1:7.5, 1:10, 1:12.5, and 1:15 were immersed in PBS containing Cu5.4O USNPs (30 μg mL−1) for 2 h. The release of Cu5.4O USNPs from gels after different durations was measured at room temperature. Briefly, 20 μL Cu5.4O@Hep-PEG gels (3.2 wt%, Cu 16 ng μL−1 in gel) were immersed in centrifuge tubes, which contained 1 mL of PBS buffer, with shaking, and the Cu5.4O@Hep-PEG gels were removed from the solutions at various time points (3, 6, 12, 24, 48, 72, 96, 120, 240, and 360 h). The excess Cu5.4O USNPs were removed by washing the gels with PBS solution three times and the concentration of Cu in the gels obtained were detected by ICP-MS using the aqua regia ablation method (n = 3).
2.6. Pro-inflammatory factors binding to gels in vitro
The capability of Hep-PEG (1:7.5) gels to sequester wound-relevant pro-inflammatory factors (MCP-1, IL-8, IL-6 and TNF-α) was quantified by ELISA. Briefly, a total volume of 1 mL RPMI medium with FBS (2%, v/v) containing a Hep-PEG gel (3.2 wt%, 20 μL) and pro-inflammatory factors (100 ng mL−1 of mouse MCP-1, 100 ng mL−1 of human IL-8, 100 ng mL−1 of mouse IL-6, 100 ng mL−1 of mouse TNF-α) were incubated together at 37 °C. The supernatant was sampled at different time points (0, 0.5, 1, 2, 4, 6, 8, 12 and 24 h) and then directly stored at −80 °C until analysis using the corresponding ELISA kit. To explore the effect of the Cu5.4O USNP content in the Cu5.4O@Hep-PEG gels on pro-inflammatory factor binding, Cu5.4O@Hep-PEG gels (3.2 wt%, 20 μL) soaked in Cu5.4O USNP solutions (0, 15, 30, and 60 μg mL−1) were incubated separately with MCP-1 and IL-8 by the above mentioned method. The concentration-dependent binding capacities of Cu5.4O@Hep-PEG gel to MCP-1 and IL-8 were assessed by quantifying the mouse MCP-1 (0.4–850 μg mL−1) or human IL-8 (0.34–700 μg mL−1) after incubation with the Cu5.4O@Hep-PEG gel for 24 h. Then, the supernatant concentrations of MCP-1 and IL-8 were determined by ELISA kits. All experiments were performed with three independent samples. In addition, the dissociation constants (Kd) of MCP-1 and IL-8 for Cu5.4O@Hep-PEG gel were evaluated according to the Scatchard formula:
where B is the molarity of heparin binding pro-inflammatory factors, F is the molarity of the free pro-inflammatory factors, Kd is the dissociation constant, and Bmax is the maximum saturation binding molarity of heparin to pro-inflammatory factors.
2.7. Free -radical-scavenging assays
To evaluate the free-radical-scavenging capacity of Cu5.4O@ Hep-PEG gels (1:7.5), 20 μL of Cu5.4O@Hep-PEG gel (3.2 wt%, Cu 16 ng μL−1 in gel) was immersed in a centrifuge tube containing 10 mL PBS solution and shaken at 50 rpm and room temperature. Then, 100 μL of the above solution was removed from the tube at various time points (3, 6, 12, 24, 48, 72, 96, 120, 240, and 600 h) and stored at 4 °C and the medium in the tube was replenished with the same volume of PBS. The radical-scavenging assay results for the Cu5.4O@Hep-PEG gels were obtained by absorption spectrophotometry analysis of these Cu5.4O USNP storage solutions.
DPPH assay The DPPH-scavenging capacity of Cu5.4O@Hep-PEG gel was evaluated by a previously published method with a slight change [30]. Briefly, a total volume of 100 μL anhydrous ethanol containing 2 ng of DPPH and solutions of Cu5.4O USNPs released from 20 μL of Cu5.4O@Hep-PEG gel (3.2 wt%, Cu 16 ng μL−1 in gel) were incubated together for 30 min in the dark. The same volume of DPPH anhydrous ethanol solution without Cu5.4O USNPs was defined as the control (n = 3). All the solutions used were freshly prepared.
H2O2 assay The H2O2-scavenging capacity of Cu5.4O@Hep-PEG gel was measured using a H2O2 assay kit. The Cu5.4O USNP storage solution from a specific time point was added to 2 mM of H2O2 solution in the dark. The absorbance at 550 nm was measured 2 h later. A mixture with water instead of Cu5.4O USNPs was used as a control (n = 3). All the solutions used were freshly prepared.
·OH assay The ·OH-scavenging capacities of Cu5.4O@Hep-PEG gels were determined using an ·OH assay kit. Briefly, a reaction mixture containing the working solution from the kit and a Cu5.4O USNPs storage solution from a particular time point was incubated in the dark for 20 min. The same volume of water instead of Cu5.4O USNP solution was used as a control (n = 3). All the solutions used were freshly prepared.
Superoxide anion radical assay The O2·--scavenging capacity of Cu5.4O@Hep-PEG gel was evaluated using O2·--scavenging capacity assay kit. A mixture comprising the working solution from the kit and a Cu5.4O USNPs storage solution from a specific time point was incubated in the dark. The same volume of water instead of Cu5.4O USNP solution was used as a control (n = 3). All the solutions used were freshly prepared.
2.8. Cell culture
The NIH-3T3 mouse embryonic fibroblast cells (3T3) and human umbilical vein endothelial cells (HUVEC) were purchased from the American Type Culture Collection (ATCC). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM), which contained 10% fetal bovine serum (FBS), 100 μg mL−1 streptomycin, and 100 U mL−1 penicillin under a humidified 5% CO2 atmosphere at 37 °C.
2.9. Evaluation of cytotoxicity in vitro and in vivo
The cytotoxicity of Cu5.4O USNPs was determined by the cell counting kit-8 assay (CCK-8, Dojindo, Japan) in vitro [31]. 3T3 cells were seeded in 96-well culture plates (1 × 104 cells per well). After 24 h, Cu5.4O USNPs at different concentrations (300 ng mL−1, 600 ng mL−1) were introduced. After co-incubation for a predetermined time (24 or 48 h), we carefully washed the cells for three times with sterile PBS, treated them with 90 μL fresh culture medium and 10 μL CCK-8 solution, and incubated the cells again at 37 °C for 2 h. Then, the absorbance at 450 nm was measured using a microplate reader (Thermo Varioskan Flash, USA) to evaluate cell viability. Cell viability was calculated using the formula below:
| Cell viability (%) = [(As-Ab)/(Ac-Ab)] × 100 |
where As is the absorbance of the sample (CCK-8 solution with cells and Cu5.4O USNPs); Ac is the absorbance of the control (CCK-8 solution with cells, without Cu5.4O USNPs); and Ab is the absorbance of the blank (CCK-8 solution without cells or Cu5.4O USNPs).
To further evaluate the biocompatibility of Cu5.4O in vivo, BALB/c mice (male, aged 8–10 weeks and weighing 20–25 g) were intravenously administrated with Cu5.4O USNPs at a dose of 16 μg kg−1. To evaluate the biocompatibility of the hydrogels in vivo, 20 μL of Hep-PEG (3.2 wt%) and Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel) were implanted subcutaneously in BALB/c mice (male, aged 8–10 weeks and weighing 20–25 g) and removed at 30 days. Untreated mice were employed as a control group. At 30 days post-treatment, blood samples were taken and subjected to complete blood panel and serum biochemistry analysis. Serum biochemistry analysis focused on the indicators of hepatic function, aspartate aminotransferase (AST) and alanine aminotransferase (ALT) and the indicators of kidney function, blood urea nitrogen (BUN) and creatinine (CRE). In addition, another five mice from each group were sacrificed on day 30, and the heart, liver, spleen, lung, and kidney tissues were subjected to hematoxylin and eosin (H&E) staining.
2.10. Pro-inflammatory factor absorption by Cu5.4O@Hep-PEG in vitro
We assessed the absorption of pro-inflammatory factors by Cu5.4O@Hep-PEG in vitro by transmigration assay [20]. Mouse peripheral blood monocytes were isolated using a mouse peripheral blood monocyte isolation kit (Solarbio, China) according to the manufacturer's instructions. Briefly, 4 mL fresh mouse blood was collected into a centrifuge tube and 3 mL reagent A and 2 mL reagent D were added to the tube in sequence. The mixture was then centrifuged at 800 g and room temperature for 30 min. The segment between the blood plasma and reagent D was carefully removed and washed with 5 mL PBS. This was then centrifuged at 250 g for 10 min. The cells were cultured in RPMI medium 1640 (Solarbio, China) containing 10% FBS (Gibco, USA), 100 μg mL−1 streptomycin, and 100 U mL−1 penicillin at 37 °C under a humidified 5% CO2 atmosphere. Cells were then supplemented with 10 ng mL−1 granulocyte-macrophage colony stimulating factor (GM-CSF, Sigma, USA) to stimulate macrophage formation.
Mouse peripheral blood neutrophils were isolated using a mouse peripheral blood neutrophil isolation kit (Solarbio, China) according to the manufacturer's instructions. Briefly, 4 mL fresh mouse blood was collected in a centrifuge tube and 4 mL reagent A and 2 mL reagent C were added to the centrifuge tube in sequence. The mixture was then centrifuged at 1000 g and room temperature for 30 min. Then, the segment between reagent A and reagent C (neutrophils) was carefully collected and washed with 5 mL PBS followed by centrifuging at 250 g centrifuge for 10 min. The cell culture medium and conditions were the same as those used to isolate the monocytes.
Freshly mice macrophages and neutrophils were used for the transmigration assays. Mice macrophages and neutrophils were suspended in culture medium and placed (2.5 × 105 cells per insert) in the upper compartment of a transwell chamber (pore size 3 μm, Corning, USA). Conditioned medium with recombinant human IL-8 (5 ng mL−1) and recombinant mouse MCP-1 (10 ng mL−1) was added to the lower well after incubation with 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel) for 2 h. Mice macrophages and neutrophils were allowed to migrate for 2 h. Then, cell migration to the lower well was quantified microscopically.
Next, we assessed pro-inflammatory secretion under high-level oxidative stress. Mice macrophages were seeded in 24-well plates (15 × 104 cells per well) with 100 μM H2O2 and treated with 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel). Cells without treatment were used as a control. After 2 h treatment, the cell medium was collected. For ELISA assay, the concentrations of MCP-1 and CXCL-1 (There is no IL-8 in the murine system, CXCL-1 in mice has similar function like IL-8 in humans) in the conditioned medium were quantified with the corresponding ELISA kits according to the protocols provided by the manufacturer. For transmigration assays, conditioned medium was added to the lower well after incubation with 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel) for 2 h. Mice monocytes and neutrophils were allowed to migrate for 2 h. Then, cell migration to the lower well was quantified microscopically.
2.11. ROS-scavenging effects of Cu5.4O@Hep-PEG hydrogels in vitro
To investigate the ROS-scavenging ability of Cu5.4O@Hep-PEG hydrogel in vitro, 3T3 fibroblasts were seeded into 24-wells plates at a density of 5 × 104 cells per well for 24 h. Then, H2O2 was supplemented to a final concentration of 250 μM. After incubation for 1 h, 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG hydrogel (3.2 wt%, Cu 16 ng μL−1 gel) was added to the well, which was incubated at 37 °C for 24 h, respectively.
Cell-protecting effect assay The cell viabilities of 3T3 fibroblasts following different treatments was detected by CCK-8 [31]. After 24 h H2O2 incubation, 3T3 fibroblasts were carefully rinsed three times with sterile PBS and treated them with 450 μL fresh culture medium and 50 μL CCK-8 solution. The fibroblasts were then incubated at 37 °C for a further 2 h. The absorbance at 450 nm was measured using a microplate reader (Thermo Varioskan Flash, USA) to quantify cell viability.
Cell apoptosis assay Cell apoptosis assays were performed using an Annexin V-propidium iodide (PI) apoptosis detection kit (C1062, Beyotime, China) according to the manufacturer's protocol. Briefly, 3T3 cells were incubated with H2O2 and hydrogels for 24 h in a 24-well plate then collected carefully, rinsed with cold PBS, and resuspended in 195 μL binding buffer. Then, we added 5 μL Annexin V-FITC and 10 μL PI in sequencewere sequentially added, and incubated the cell suspension was incubated at room temperature in the dark for 15 min. Finally, the percentage of apoptotic cells was determined by fluorescence-activated cell sorting (FACS) using an Attune Acoustic Focusing Cytometer (Life Technologies, USA). The data were analyzed using the FlowJo software package (Tree Star Incorporation, USA).
Oxygen-free-radical quantitative measurement For oxygen free radical assays, we used CellRox (CellRox Green Reagents, Invitrogen, USA), a novel fluorogenic probe for measuring oxidative stress in live cells [32]. Adherent 3T3 cells were harvested and stained with CellRox (1:200) at 37 °C in the dark for 30 min. Finally, the fluorescence intensity of cells was detected with FACS and the data were analyzed using the FlowJo software package (Tree Star Incorporation, USA).
2.12. Effects of Cu5.4O USNPs on endothelial cell proliferation in vitro
Immunofluorescence staining We used Ki67 antibody to determine the promotion effect of Cu5.4O on HUVECs [33]. HUVECs were seeded in 24-well plates (5 × 104 cells per well) and treated with 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel) for 24 h. Then, cells were permeabilized with Triton X-100 (Boster, China) and blocked with 10% goat serum (Boster, China). Cells were incubated with rabbit anti-human Ki67 (1:200, Abcam, America) at 4 °C overnight and then further incubated with goat anti-rabbit Alexa 488 (Abcam, America) diluted 1:1000 in PBS at room temperature for 1 h. Nuclei were stained with DAPI (1:1000, Thermo Fisher, America). Then, the cells were imaged using a fluorescence microscope (Leica).
Cell migration assay To investigate the effect of Cu5.4O@Hep-PEG on HUVEC transmigration, cell scratch assays were performed. HUVECs were seeded in 24-well plates (8 × 104 cells per well) until formation of a confluent monolayer. After treatment with FBS-free medium for 24 h, an incisional wound was simulated by scratching the cell monolayer in a straight line using a 200 μL pipette tip. The cells were then rinsed with PBS to remove debris and incubated with 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel) at 37 °C in medium containing 1% FBS. Cells were photographed at regular intervals and cell migration (%) was calculated using:
| Cell migration (%) = ((A0-At) / A0) × 100 |
where A0 is the area of the scratch at 0 h and At is the area of the scratch without cell migration at time t.
2.13. Evaluation of wound healing using an impaired wound model
To assess the wound-healing effect of Cu5.4O@Hep-PEG on impaired wounds, a Type I diabetic mouse model was established following the procedure according to the literature [34]. In brief, after fasting for 24 h, Balb/c mice (male, aged 8–10 weeks and weighing 20–25 g) were given intraperitoneal injections of 10 mg kg−1 streptozocin (STZ, Sigma-Aldrich, USA) (10 mg mL−1 in sodium citrate buffer, pH 4.0–4.5) for 5 days. All mice were provided with normal food and water. Blood glucose levels were monitored by taking tail-vein blood every day and using a blood glucose meter (Roche Diagnostics, Shanghai, China). STZ-treated mice with whole-blood glucose levels in excess of 16.7 mmol L−1 that also showed symptoms of weight loss, polyuria, polydipsia, and increased appetite were considered to be Type I diabetic. Before wound-creating surgery, type I diabetic mice were fed a high-fat and high-sugar diet for 2 weeks.
To build a full-thickness cutaneous wound model, diabetic mice were anesthetized with 1% pentobarbital sodium (60 mg kg−1) and fixed on a surgical corkboard. The backs of the mice were completely depilated 12 h before establishing the wound model. After anesthetization, two rounds, full-thickness wounds 6 mm in diameter were created on the back of each mouse. The mice were separated into four groups, which were treated with 3 M Tegaderm™ wound dressing (3 M Health Care, USA) (as control), 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu 16 ng μL−1 gel). The gels were fixed to the wounds using 3 M Tegaderm™ wound dressing. During the study, the hydrogels were not replaced. The wounds were photographed at different time points (0, 7, and 14 days post-surgery) and wound closure rates were determined using:
| Wound closure rate (%) = (S0-St) / S0 × 100 |
where S0 is the original wound area and St is the area of the wound at a time t.
At 3, 7, and 14 days post-surgery, the wound site tissues were equally cut into two sections. One section was homogenized for ELISA assay. The other was fixed in 4% paraformaldehyde and embedded in paraffin for macrophage staining. Furthermore, at these three time points, the hydrogel covering the wound site was collected for ELISA assay with mouse MCP-1, CXCL-1. On day 14 post-surgery, the wound site tissues were equally cut into two sections. One was frozen and embedded in optimum cutting temperature (OCT) matrix (Sakura, Leiden, The Netherlands) for cryostat sectioning at −20 °C, while the other was fixed with 4% paraformaldehyde and embedded in paraffin for H&E staining.
2.14. Quantitative evaluation of pro-inflammatory factors in wound tissues and hydrogels
The whole wound tissue was excised, rinsed with cold PBS, and homogenized at 4 °C for 5 min. The homogenates were mixed with protease inhibitor and centrifuged at 12,000 rpm at 4 °C for 10 min. The supernatants were collected and stored at −80 °C. The concentrations of MCP-1, CXCL-1, TNF-α, SOD and VEGF in the supernatant were quantified by the corresponding ELISA kits according to the protocols provided by the manufacturer.
Pro-inflammatory factors in the hydrogels were detected as previously described [20]. Briefly, hydrogels recovered from the wounds were immersed in PBS and mechanically disrupted at 30 Hz for 30 s using a bead mill (7-mm steel beads, TissueLyser LT, Qiagen). 1% Triton X-100 was added 5 min before centrifugation for mediator displacement. Concentrations of MCP-1 and CXCL-1 in the supernatant were determined by the corresponding ELISA kit.
2.15. Histological examination
The whole wound tissue with a margin of around 2 mm of ambient unwounded skin was excised, fixed with 4% paraformaldehyde for 24 h, embedded in paraffin, and sectioned into 7-μm-thickness slices.
H&E staining Sections were deparaffinized in xylene for 30 min and rehydrated using a descending-ethanol series (100%, 95%, and 80%), and distilled water for 5 min each. Then, the samples were immersed in hematoxylin for 5 min then in PBS for 3 min to prevent background staining. The sections were then immersed in eosin for 2 min, distilled water for 5 min, and 5 min each in ethanol/water mixture solutions (80%, 90%, and 100%) for dehydration, followed by 15 min in xylene. Slides were mounted using neutral resin and coverslipped. The stained sections were observed and digitally photographed using a Nikon microscope (Nikon, Tokyo, Japan). Based on these H&E staining sections, the length of the regenerated epidermis was measured using Image J software.
Masson's trichrome staining Sections were deparaffinized and rehydrated by the same method as that used for H&E staining and then stained using a Masson's trichrome staining kit (Solarbio, China) following the manufacturer's protocol. Briefly, the cell nuclei were stained with A1:A2 (1:1) for 5 min before the sections were thoroughly rinsed with water and then submerged in acid alcohol for 3 s to achieve differentiation. Ponceau acid fuchsin solution treatment for 5 min was used to stain fibrous tissue, followed by 2% acetic acid solution (1 min), phosphomolybdic acid solution (30 s) for differentiation, and aniline blue (20 s). Then, we dehydrated, mounted, and coverslipped the sections using the same method as that used for H&E staining. The stained sections were observed and digitally photographed under a Nikon microscope (Nikon, Tokyo, Japan). Collagen index measurement was performed using Image J software.
Immunofluorescence and immunohistochemistry staining Sections were stained with CD31 to measure the number of micro-vessels in the wound edge and with CD11b to evaluate the number of CD11b+ immune cells in the wound site. Briefly, after deparaffinization and rehydration, the nonspecific binding sites were blocked using 10% goat serum at 37 °C for 30 min. The sections were then incubated with rabbit anti-mouse CD31 and rabbit anti-mouse CD11b (Abcam, America), respectively, diluted 1:200 in PBS containing 1% bovine serum albumin (BSA) at 4 °C. The sections were then left overnight, washed with PBS, and incubated with secondary antibodies of goat anti-rabbit Alexa Fluor 594 (Abcam, America) diluted 1:1000 in PBS at 37 °C for 60 min before staining with DAPI (Sigma, America) and mounting using antifade mounting medium (Thermo Fisher, America). Images of the sections were obtained using a laser confocal microscope (Zeiss LSM780, Germany) and analyzed using the Image J software package.
For immunohistochemistry (IHC) staining, each section was deparaffinized with xylene, rehydrated, and the endogenous peroxidase in the sections was inactivated in 3% H2O2 for 15 min at room temperature before blocking with 10% goat serum. The sections were then incubated with rabbit anti-mouse CD31 antibody (Abcam, America) diluted 1:200 in PBS containing 1% BSA. A horseradish peroxidase (HRP) detection system (Beyotime, China) and a 3′-diaminobenzidine (DAB) reagent kit (Beyotime, China) were used for IHC staining. A Nikon microscope (Nikon, Tokyo, Japan) was used to observe and digitally photograph the sections.
Tissue ROS measurement To detect the ROS levels in dermal wounds, tissue samples were collected and frozen in liquid nitrogen. Following sucrose cryoprotection, the tissue samples were embedded in optimal cutting temperature (OCT) cryoembedding medium and subsequently sectioned. The cyro-sections were stored at −20 °C until use. After washing with PBS three times, the cyro-sections were incubated with dihydroethidium (DHE, Sigma, USA) diluted to 1:200 at 37 °C for 30 min. DHE indicates the ROS level of the wound tissue, as previous reported [35]. Images were captured using laser confocal microscopy (Zeiss LSM780, Germany) and analyzed using the Image J software package.
2.16. Tandem mass tag (TMT) technology for quantitative proteomic analysis
Tissue samples for proteomic analysis were obtained from wounded skin tissues on day 14 of treatment with Cu5.4O@Hep-PEG or 3 M Tegaderm™ wound dressing. The tissue samples were homogenized and suspended in SDT lysis buffer (1 mM DTT, 100 mM Tris-HCl, 4% SDS, pH 7.6). Samples were stored at −80 °C prior to analysis. TMT-labeling quantitative proteomic analysis was performed by Shanghai Applied Protein Technology. The protocol was followed as previously described [36]. MS/MS spectra were analyzed with Proteome Discoverer 1.4 running the MASCOT engine (version 2.2, Matrix Science, London, UK).
For bioinformatics analysis, we used the platform of Shanghai Applied Protein Technology (http://cloud.aptbiotech.com/#/main-page), and for analysis of protein-protein interactions, we used the Search Tool for the Retrieval of Interacting Proteins (STRING) algorithm (http://www.string-db.org/).
2.17. Statistical analysis
Statistical differences were established using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA) by means of one-way analysis of variance (ANOVA) followed by Tukey's test. For all results *p < 0.05; **p < 0.01; n.s. no significance.
3. Results and discussion
3.1. Characterization of Hep-PEG hydrogels
StarPEG-heparin gels have been reported to bind numerous pro-inflammatory factors, such as MCP-1 and IL-8 [20]. In the current study, we designed a Hep-PEG gel as a depot for chemokine adsorption from an inflamed wound based on the interaction between heparin and pro-inflammatory factors in the wound fluids, avoiding further accumulation of immune cells and thus modifying the inflammatory process and facilitating wound healing [37,38]. To promote local skin wound repair and remove pro-inflammatory factors as much as possible, the network structure and physical properties of Hep-PEG gels should be optimized [21]. Thus, Hep-PEG gels with different molar ratios of heparin to PEG (1:5, 1:7.5, 1:10, 1:12.5, and 1:15) were prepared through the conjugation of 4-arm PEG-NH2 and heparin using EDC and NHS as coupling agents. First, the chemical structure of a Hep-PEG gel with a molar ratio of heparin to PEG of 1:5 was investigated by FT-IR. As shown in Fig. S1A, the peaks for heparin at 3300-3600 cm−1 and 1648 cm−1 correspond to the hydroxyl and carboxyl groups of heparin, respectively [21]. The reduction in amplitude of the above peaks for the Hep-PEG gel indicates successful conjugation between the amide groups in PEG and the carbonyl group in heparin. Compared with the spectrum of 4-arm PEG-NH2 alone, the new peak at 1564 cm−1 for the Hep-PEG gel is attributed to the N-H bending vibration of the amide groups (amide II) [21], further indicating the successful cross-linking between PEG-NH2 and heparin. The degree of cross-linking in the gels was calculated based on the hypothesis that the carboxylic acid groups on heparin (28 per heparin molecule) bond to three of the four amino groups on PEG [21]. Increasing the molar ratio of heparin to PEG (ranging from 1:5 to 1:15) leads to a similar increase in the cross-linking degree of these gels (Fig. S1B), which is significantly related to the network structure and physical properties of the gels. As expected, a reduced swelling ratio (ranging from 13.2% to 10.5%) and increased storage moduli (ranging from 0.2 kPa to 23.9 kPa) are obtained by increasing the cross-linking degree of the gels (Fig. S1C-D). Assuming a Poisson ratio of 0.5, the Young's modulus of Hep-PEG gel is three-times the storage modulus [20]. The corresponding Young's moduli of the gels were calculated as Fig. S1E. An increased cross-linking degree results in a denser network structure that is more rigid (larger storage modulus) and a decrease in water absorption due to greater contractility, which is caused by the increase in the number of covalent bridges in the gels (smaller swelling) [21,26]. The mesh sizes of the gels were estimated using RET [26] to be in the nanoscale region. A diminishing mesh size (ranging from ~25.3 to ~5.6 nm) with increasing cross-linking degree is observed for the gels (Fig. S1F).
Accordingly, the Young's moduli and mesh sizes of the gels meet the requirements of local skin wound treatment and chemokine adsorption, respectively. The Hep-PEG (1:7.5) gel has a Young's modulus of 6.6 kPa, which is similar to that of human skin (4.5–8 kPa) [20], and a mesh size of ~12.3 nm, which allows the adsorption of wound-related chemokines (MCP-1 and IL-8 have gyration radii of less than 2 nm) [39,40]. Consequently, the molar ratio of heparin to PEG was fixed at 1:7.5 for subsequent studies, and the concentration was set at 3.2 wt% (heparin 1.2 wt% and PEG 2.0 wt%).
3.2. Characterizations of Cu5.4O@ Hep-PEG hydrogels
First, we synthesized highly monodisperse Cu5.4O USNPs via a green, economic, simple, and quick approach according to our previous work [17]. Then, Cu5.4O@Hep-PEG hydrogels were prepared by a simple soaking method. The pre-prepared Hep-PEG (1:7.5) gels were soaked in Cu5.4O USNPs (10 μg mL−1) solutions for different durations (5–720 min) to determine the time point of adsorption saturation. Quantitative analysis revealed that gels with a mesh size of ~12.3 nm allow rapid penetration of Cu5.4O USNPs with a diameter of ~4.5 nm in just 30 min, and the saturation time point is ~2 h (Fig. S2A). Then, the Hep-PEG gels were immersed in Cu5.4O USNP solutions with different concentrations (6–960 μg mL−1) for 2 h to obtain functional gels with the highest Cu5.4O USNPs loading amount (Fig. S2B). A linear relationship of initial concentration of Cu5.4O USNPs solution and loading amount of Cu5.4O USNPs in observed, which indicates that the Hep-PEG gel has low utilization rate for Cu5.4O USNPs. According to the results of cytotoxicity in vitro (Fig. S2C), we discovered a relatively safe concentration of Cu5.4O USNPs for subsequently biological experiments. 300 ng mL−1 and 600 ng mL−1 Cu5.4O USNPs showed low cytotoxicity. The resulting Cu5.4O@Hep-PEG gels (3.2 wt%, 20 μL) contain 320 ng Cu5.4O USNPs, which is absolutely safe for cells.
Compared to the XRD pattern of Hep-PEG gel, that of Cu5.4O@Hep-PEG gel shows a new diffraction peak that corresponds to CuxO, which provides strong evidence for the successful loading of Cu5.4O into the Hep-PEG gels (Fig. S3A). To further confirm the presence of Cu5.4O USNPs on the gels, XPS was used to characterize the surface of the prepared gels. Compared with the Hep-PEG gel, the XPS survey spectrum of the Cu5.4O@Hep-PEG gel shows an additional weak peak corresponding to Cu2p, which verifies the low content of Cu5.4O USNPs in the gel (Fig. S3B). TEM in combination with TEM-EDS elemental mapping of the Cu5.4O@Hep-PEG gel indicated that the content of Cu is much lower than those of C and O in the matrix (Fig. 2A). After soaking in Cu5.4O USNPs solution, the fitted Cu2p peak shows two main signals at binding energies of 952.4 and 932.5 eV, which correspond to the binding energies of Cu2p1/2 and Cu2p3/2 in Cu5.4O USNPs [17] (Fig. S3C), respectively. It is worth mentioning that two new satellite peaks appear at binding energies of 951.1 and 931.6 eV, which are 1.3 eV and 0.9 eV lower than those for Cu2p1/2 and Cu2p3/2 in Cu5.4O USNPs, respectively. In addition, the corresponding N1s XPS peak for Hep-PEG gel could be fitted to two peaks centered at 399.3 and 400.7 eV, which correspond to the N atoms in free amino groups and NH-acetyl groups, respectively [41,42] (Fig. S3D). From the N1s XPS spectrum for Cu5.4O@Hep-PEG gel, most of the N1s binding energies are unchanged upon soaking, while a new weak signal at 401.7 eV is observed, indicating the reduction of electron density for N atoms upon coordination with Cu [43]. Furthermore, the O1s XPS spectrum for Cu5.4O@Hep-PEG gel shows almost no difference compared with that of Hep-PEG gel, so it can be inferred that O atoms are not involved in the coordination reaction (Fig. S3E). Therefore, it may be that the Cu5.4O USNPs are bound to Hep-PEG gels via coordination and physical adsorption interactions (such as van der Waals and hydrogen-bonding interactions) [[44], [45], [46]]. The coordination process involves the lone-pair electrons of N being transferred to vacant orbitals on Cu, thus forming N-Cu coordination bonds [43]. The saturation adsorption of Cu5.4O USNPs by Hep-PEG gels with different molar ratios (1:5, 1:7.5, 1:10, 1:12.5, 1:15) was explored by soaking them in Cu5.4O USNPs (30 μg mL−1) solution for 2 h (Fig. S2D). As expected, an increase of Cu5.4O USNPs loading (8.5–30.7 ng μL−1 gel) is obtained by increasing the molar ratio of Hep-PEG gels, which may be attributed to the increase in NH2-PEG, which forms N-Cu coordination bonds, with increasing cross-linking degree.
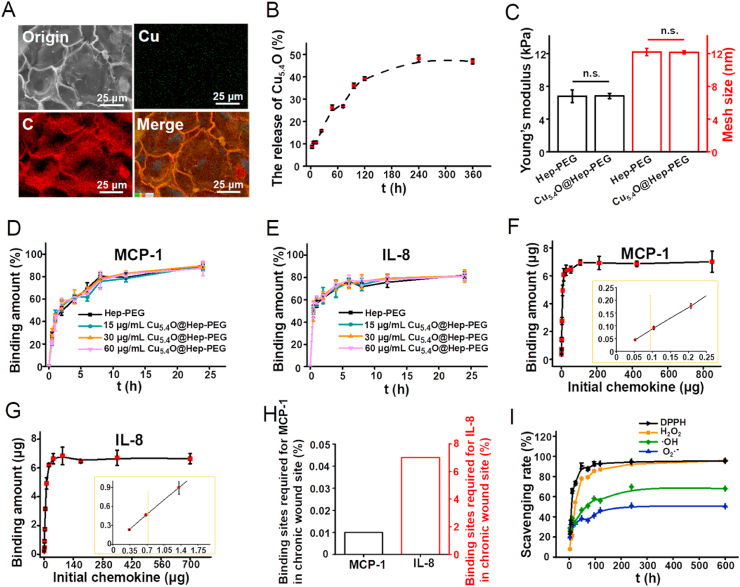
Fig. 2.
(A) SEM and elemental mapping images of Cu5.4O@Hep-PEG hydrogel. (B) In vitro cumulative release of Cu5.4O USNPs from Cu5.4O@Hep-PEG over time. (C) Young's moduli and mesh sizes of Hep-PEG and Cu5.4O@Hep-PEG hydrogels. (D) Binding kinetics of MCP-1 and (E) IL-8 for hydrogels with different Cu5.4O USNP loading amounts over 24 h of co-incubation. (F) MCP-1 and (G) IL-8 binding amounts for Cu5.4O@Hep-PEG hydrogels after co-incubation for 24 h at different chemokine concentrations. (H) Chemokine saturation of Cu5.4O@Hep-PEG and the binding sites required for chemokines in chronic wound sites. (I) In vitro DPPH, H2O2, ·OH, and O2·- scavenging capabilities of Cu5.4O@Hep-PEG hydrogels over time.
In vitro release of Cu5.4O USNPs from Cu5.4O@Hep-PEG gel was investigated by ICP-MS. A 20 μL sample of Cu5.4O@Hep-PEG gel containing 320 ng Cu5.4O USNPs was immersed in PBS buffer with slight shaking. During incubation for 24 h, ~16% of the Cu5.4O USNPs are released from the Cu5.4O@Hep-PEG gel (Fig. 2B). These Cu5.4O USNPs are held on the Hep-PEG gel by non-covalent forces, so they are easily released from the gel. However, the Cu5.4O USNPs that form N-Cu coordination bonds are more difficult to release. Thus, only ~48% of the Cu5.4O USNPs are released from the Cu5.4O@Hep-PEG gel during further incubation for 9 d, whereupon equilibrium is reached.
In addition, special attention was paid to the effect of introducing Cu5.4O USNPs to a Cu5.4O@Hep-PEG gel on its storage modulus. After immersion in Cu5.4O USNPs solution, the Hep-PEG gel exhibits the same storage modulus, which is still similar to that of human skin (Fig. 2C). Thus, the average mesh sizes were calculated for Cu5.4O USNPs-loaded gels and unloaded one according to the storage modulus of gels. The Cu5.4O@Hep-PEG gel must have an average mesh size above 10 nm to ensure that wound-relevant chemokines can enter (Fig. 2C). Furthermore, on the basis of SEM morphology observation, it is possible to state that introduction of Cu5.4O USNPs to the Hep-PEG gel does not lead to noticeable changes in the micromorphology and structure of the gel (Fig. S3F).
3.3. Pro-inflammatory factors binding of gels in vitro
We studied the binding capacities of Hep-PEG and Cu5.4O@Hep-PEG gels for pro-inflammatory factors (MCP-1, IL-8, IL-6, TNF-α), which are highly expressed in chronic wounds [39,40]. The kinetics of the binding between pro-inflammatory factors and the gels were investigated though co-incubation of one piece of gel (20 μL) and the pro-inflammatory factors in 1 mL of solution. The rapid increase in the binding amounts of MCP-1 and IL-8 at the beginning might be attributed to the early swelling of the gel. After 24 h, nearly 89% of the MCP-1 and 77% of the IL-8 are bound to the Hep-PEG gel (Fig. 2D–E). Conversely, only ~14% of the TNF-α is scavenged by the Hep-PEG gel, almost no IL-6 is bound to the Hep-PEG gel (Fig. S4A), which is consistent with previous reports [20]. The pro-inflammatory-factor-binding ability of Hep-PEG gel is determined by the intense electrostatic interaction between negatively charged sulfate groups in the extracellular matrix glycosaminoglycans (heparin, heparan sulfate) and positively charged amino acid residues in the pro-inflammatory factors [47]. Because heparin is the dominant functional part of the Hep-PEG network, it exhibits strong binding to MCP-1 and IL-8, which has positively charged amino acid residues [48]. In contrast, the ability of Hep-PEG gel to bind TNF-α, IL-6, which have poor heparin affinity, is observed [20]. The specific pro-inflammatory factors that bind to Hep-PEG gels are in line with those identified previous reports [20]. It is also worth mentioning that the loading amount of Cu5.4O USNPs has no obvious effect on adsorption capacity for MCP-1 and IL-8 (Fig. 2D–E). Thus, it may be stated that the addition of Cu5.4O USNPs to gels does not affect the interaction between heparin and pro-inflammatory factors.
To further explore the binding capacity of Cu5.4O@Hep-PEG gel for pro-inflammatory factors, incubation experiments were performed with increasing concentrations of MCP-1 (0–106 μg mL−1) and IL-8 (0–88 μg mL−1) for 24 h (Fig. 2F–G). A positive correlation between the initial and bound amounts of pro-inflammatory factors is observed discovered for Cu5.4O@Hep-PEG gel, and the bound amount of chemokines reaches saturation when the initial pro-inflammatory factors exceed a certain concentration. Despite slight differences in the saturation amounts of Cu5.4O@Hep-PEG gel for MCP-1 (350 ng μL−1 gel) and IL-8 (332 ng μL−1 gel), the saturation amounts are far higher than chronic-wound-site concentrations (0.9 ng mL−1 for MCP-1, 702 ng mL−1 for IL-8) [20]. At their wound-site concentrations, most of the MCP-1 (~89%) and IL-8 (~66%) are bound after 24 h (Fig. 2F–G), and fewer than 0.01% and 7.0% of the usable binding sites on the Cu5.4O@Hep-PEG gel would be occupied (Fig. 2H). The dissociation constants (Kd) of MCP-1 and IL-8 for Cu5.4O@Hep-PEG gel were calculated based on the Scatchard formula (Fig. S4B-C), and the lower Kd values (75 nM for MCP-1 and 334 nM for IL-8) may provide information on the intense electrostatic interaction between the pro-inflammatory factors and the gel. In addition, the saturation degree of heparin binding sites was calculated to be 79% for MCP-1 and 73% for IL-8. Thus, the pure Hep-PEG gel network acts as an efficient pro-inflammatory-factor trap, reducing their concentration and ultimately eliminating inflammation.
3.4. ROS-scavenging ability of Cu5.4O@Hep-PEG gel
Cu5.4O USNPs exhibit a broad ability to scavenge ROS, including H2O2, O2·-, ·OH, and other free radicals [17]. The ROS-scavenging ability of the Cu5.4O@Hep-PEG gel for a period of 25 d in vitro was measured. As shown in Fig. 2I, the Cu5.4O@Hep-PEG gel exhibits efficient ROS scavenging in a time-dependent pattern. First, the DPPH free-radical-scavenging ability of Cu5.4O@Hep-PEG gel was evaluated [49]. It was found that the DPPH-scavenging capacity is proportional to the amount of Cu5.4O USNPs released (Fig. 2I). The cumulative release amount of Cu5.4O USNPs from gels is ~51 ng mL−1 after 24 h, leading to ~73.4% elimination of DPPH radicals. The maximum scavenging capability of Cu5.4O@Hep-PEG gel for DPPH exceeds 90% (Fig. S4D), corresponding to a cumulative release of ~154 ng mL−1 Cu5.4O USNPs. Then, the scavenging abilities of Cu5.4O@Hep-PEG gels for H2O2, O2·-, and ·OH were evaluated using commercially available kits. As expected, time-dependent scavenging profiles were also observed for these radicals (Fig. 2I). After release of Cu5.4O USNPs for 24 h, the H2O2, O2·-, and ·OH elimination rates are around 54.3%, 33.6%, and 33.9%, respectively. As shown in Fig. S4D, Cu5.4O@Hep-PEG gels can scavenge up to approximately 95.9% of H2O2, 68% of ·OH, and 50% of the O2·- with the corresponding Cu5.4O USNP concentration of ~154 ng mL−1. Thus, Cu5.4O@Hep-PEG gel is a broad-spectrum ROS-scavenging agent that can be obtained by simple soaking.
3.5. Cellular ROS scavenging and pro-inflammatory-factor capture in vitro
The effect of scavenging ROS of Cu5.4O@Hep-PEG was examined by quantitatively analyzing intracellular ROS levels via flow cytometry. As shown in Fig. S5A, C, the ratio of fluorescence-positive cells after H2O2 treatment (with higher intracellular ROS levels) significantly decreases from 58.4% to 17.8% and 18.0% when the cells are treated with Cu5.4O (p < 0.01) and Cu5.4O@Hep-PEG (p < 0.01), respectively. This low ROS ratio is comparable to that of normal cells (14.7%). However, for the Hep-PEG group, the ROS ratio remains at a high level (57.3%). These results demonstrate that the strong ROS-scavenging ability of Cu5.4O@Hep-PEG hydrogel is due to the Cu5.4O nanozyme. We also examined the effects of Cu5.4O@Hep-PEG against H2O2-induced cell apoptosis by flow cytometry. In Fig. S5B, D, the Annexin V-FITC+ PI− and Annexin V-FITC+ PI+ quadrants indicate apoptotic cells. Cu5.4O (p < 0.01) and Cu5.4O@Hep-PEG (p < 0.01) significantly decrease the occurrence of cell apoptosis and necrosis induced by H2O2 treatment, which further demonstrate the ROS-scavenging and cytoprotective abilities of Cu5.4O@Hep-PEG at the cellular level. CCK-8 analysis (Fig. S5E) show that Cu5.4O and Cu5.4O@Hep-PEG significantly increase cell viability under high levels of oxidative stress compared to Hep-PEG (p < 0.01) and a non-treated control (p < 0.01), demonstrating that Cu5.4O USNPs have a cytoprotective effect against ROS-induced damage. The intracellular ROS levels clearly decrease when the cells are pre-treated with Cu5.4O and Cu5.4O@Hep-PEG.
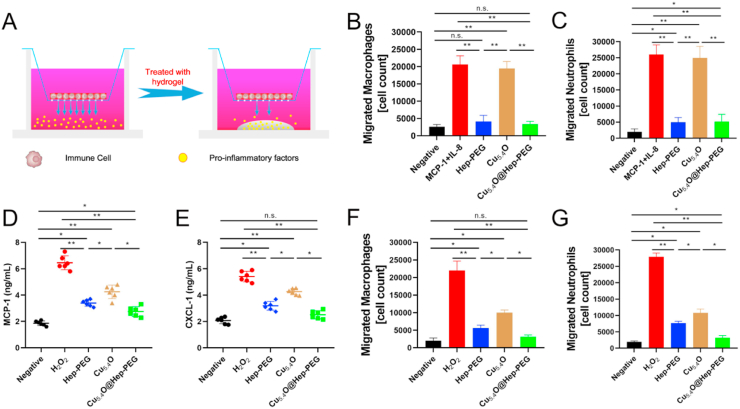
Moreover, the capture of pro-inflammatory factors by the hydrogels was investigated by means of transmigration assays using primary mouse macrophages and neutrophils (Fig. 3A), which are predominantly attracted by MCP-1 and IL-8, respectively, and found abundantly in inflammatory tissue [50]. Medium conditioned with recombinant mouse MCP-1 (10 ng mL−1) and human IL-8 (5 ng mL−1) was employed as a chemoattractant stimulus to illicit the migration of macrophages and neutrophils. Transmigration of both macrophages and neutrophils are significantly reduced upon incubation with both Hep-PEG (macrophages: p < 0.01; neutrophils: p < 0.01) and Cu5.4O@Hep-PEG (macrophages: p < 0.01; neutrophils: p < 0.01) compared to those for Cu5.4O alone and a non-treated control (Fig. 3B–C). Cu5.4O exhibits no effect on the chemoattractant activity of the conditioned medium compared to the non-treated control (p > 0.05). Notably, we found that human IL-8 could also promote mouse neutrophils transmigration. These results may also contribute to sufficient homology between human and murine [51,52]. Human IL-8 is highly similar to mouse CXCL-1. For example, human IL-8 could activate neutrophils migration through CXCR1 which was involved in mouse neutrophils migration [53]. These results demonstrate that Hep-PEG captures chemokines to reduce the transmigration of inflammatory cells. Moreover, high levels of oxidative stress stimulate the secretion of pro-inflammatory factors in macrophages, which can result in massive inflammatory-cell infiltration. To further examine the scavenging of ROS to inhibit inflammation levels, macrophages were first incubated with 100 μM H2O2 under different conditions for 2 h, and the cell medium was collected for ELISA and transmigration analysis. As shown in Fig. 3D–E, after H2O2 treatment, the concentrations of MCP-1 and CXCL-1 for the H2O2 group are 6.2 and 5.5 ng mL−1, respectively. Compared to the H2O2 group, the levels of MCP-1 and CXCL-1 in the Cu5.4O (MCP-1: p < 0.05; CXCL-1: p < 0.05), Hep-PEG (MCP-1: p < 0.01; CXCL-1: p < 0.01), and Cu5.4O@Hep-PEG (MCP-1: p < 0.01; CXCL-1: p < 0.01) groups are significantly decreased. Moreover, Hep-PEG (p < 0.01), Cu5.4O (p < 0.01), and Cu5.4O@Hep-PEG (p < 0.01) significantly reduce the transmigration of macrophages (Fig. 3F) and neutrophils (Fig. 3G), while the H2O2-treated group exhibits a high level of transmigration. Notably, we found that Cu5.4O also decreases cell transmigration, demonstrating that scavenging ROS can inhibit the secretion of MCP-1 and CXCL-1, thus decreasing the consequent macrophage and neutrophil migration. Compared to Cu5.4O (MCP-1: p < 0.05; CXCL-1: p < 0.05) and Hep-PEG (MCP-1: p < 0.05; CXCL-1: p < 0.05), Cu5.4O@Hep-PEG shows more significant inhibition of transmigration, indicating that scavenging ROS and capturing pro-inflammatory factors simultaneously with Cu5.4O@Hep-PEG significantly reduces macrophage and neutrophil infiltration and decreases inflammation levels.
Fig. 3.
Cellular pro-inflammatory-factor capture with Cu5.4O@Hep-PEG. (A) Schematic of the transmigration assay procedure used to evaluate pro-inflammatory-factor capture by Cu5.4O@Hep-PEG. A transwell is inserted into the well of cell culture plate. The small brown circles represent immune cells, the small yellow dots represent pro-inflammatory factors, and the grey semi-ellipse at the bottom represents the hydrogel. (B) The number of macrophages and (C) neutrophils in the lower chamber of the transwell incubated with human IL-8 (5 ng mL−1) and mouse MCP-1 (10 ng mL−1) under different treatment conditions. (D) Concentrations of MCP-1 and (E) CXCL-1 incubated with 100 μM H2O2 under different treatment conditions. (F) The number of macrophages and (G) neutrophils in the lower chamber of a transwell incubated with conditioned medium collected after incubation with 100 μM H2O2 under different treatment conditions. The data in (B)–(G) are means ± s.d. from six independent replicates. (*p < 0.05; **p < 0.01; n.s.: no significance, One-way ANOVA).
3.6. Effects of Cu5.4O@Hep-PEG on HUVEC proliferation in vitro
Enhanced proliferation and migration of vascular endothelial cells are beneficial for angiogenesis. The angiogenesis effect of Cu has been reported to stimulate the expression of platelet-derived growth factor (PDGF), basic fibroblast growth factor (bFGF), and vascular endothelial growth factor (VEGF) [54]. Accordingly, we investigated whether Cu5.4O USNPs promote HUVEC proliferation and migration in vitro. We stained HUVECs with Ki67, which reflects cells proliferation, after treatment with 20 μL of Hep-PEG (3.2 wt%), Cu5.4O (16 ng μL−1), or Cu5.4O@Hep-PEG (3.2 wt%, Cu: 16 ng μL−1 gel) for 24 h. The expression of Ki67 increases significantly for the Cu5.4O and Cu5.4O@Hep-PEG treated groups compared with that for the non-treated control (p < 0.05, p < 0.05) and Hep-PEG-treated groups (p < 0.05, p < 0.05), respectively (Fig. S6A-B). However, there is no obvious difference between the Cu5.4O and Cu5.4O@Hep-PEG groups (p > 0.05). These results indicate that Cu5.4O@Hep-PEG promotes HUVEC proliferation, which is mainly attributed to the action of Cu5.4O USNPs.
Additionally, the ability of Cu to stimulate cell migration was investigated using a scratch assay. The effect of Cu5.4O@Hep-PEG on cell migration was evaluated (Fig. S6C). The migration speed of cells exposed to Cu5.4O and Cu5.4O@Hep-PEG increase significantly when compared to those for the non-treated control (p < 0.01, p < 0.01) and Hep-PEG groups (p < 0.01, p < 0.01) (Fig. S6D). However, there is no obvious difference between the Cu5.4O and Cu5.4O@Hep-PEG groups (p > 0.05), demonstrating that the migration promotion is due to the Cu5.4O USNPs. The higher cell migration speeds of cells treated with Cu5.4O and Cu5.4O@Hep-PEG are attributed to the presence of Cu at concentrations that are sufficiently low to avoid cytotoxicity but physiologically relevant. Thus, Cu5.4O@Hep-PEG increases cell migration speed and the expression of Ki67 protein relative to those for the non-treated control and Hep-PEG groups, which may be attributed to the release of Cu5.4O and its angiogenic effects.
3.7. Cu5.4O@Hep-PEG hydrogels enhance acute normal wound healing
The murine skin wound model was utilized to assess the effect of Cu5.4O@Hep-PEG on wound healing. As shown in Fig. S7A, B, after 7 days of treatment, the wound for the Cu5.4O@Hep-PEG group appears to be healed, while the wounds for the wound-dressing, Hep-PEG, and Cu5.4O groups persist. We further observed that wounds treated with Hep-PEG, Cu5.4O, and Cu5.4O@Hep-PEG are absolutely healed at 12.4 ± 1.0, 14.2 ± 1.2, and 10.2 ± 0.7 days after surgery, respectively, while the wound treated with the standard wound dressing is healed 16.6 ± 1.3 days after surgery, demonstrating the excellent therapeutic efficacy of the Cu5.4O@Hep-PEG hydrogel for acute wounds (Fig. S7C).
We also assessed the inflammation levels of wound tissues after treatment. First, the ROS level in the wound tissue of normal mice was detected using DHE staining. As shown in (Fig. S8A-B), the fluorescence intensities for the wound-dressing (p < 0.01), Hep-PEG (p < 0.05), and Cu5.4O (p < 0.05) groups are significantly higher than that of the Cu5.4O@Hep-PEG group. Interestingly, the fluorescence intensity for the Cu5.4O@Hep-PEG group is much lower than that for the Cu5.4O group (p < 0.05). Similarly, although Hep-PEG and Cu5.4O both increase SOD activity compared to the standard wound dressing, the SOD activity for the Cu5.4O@Hep-PEG group is higher than those for the Hep-PEG (p < 0.05) and Cu5.4O groups (p < 0.05) (Fig. S8C). The overall results indicate that Cu5.4O@Hep-PEG significantly decreases ROS levels in wound tissues by simultaneously scavenging ROS with Cu5.4O directly and sequestering pro-inflammatory factors with Hep-PEG to prevent further recruitment of immune cells and subsequent ROS generation.
Furthermore, the pro-inflammatory factors MCP-1 (Fig. S8D), CXCL-1 (Fig. S8E), and TNF-α (Fig. S8F) are significantly decreased in the Hep-PEG (MCP-1: p < 0.01; CXCL-1: p < 0.05; TNF-α: p < 0.05), Cu5.4O (MCP-1: p < 0.01; CXCL-1: p < 0.05; TNF-α: p < 0.01), and Cu5.4O@Hep-PEG groups (MCP-1: p < 0.01; CXCL-1: p < 0.01; TNF-α: p < 0.01) compared with the wound-dressing group, indicating that either scavenging ROS by Cu5.4O or capturing pro-inflammatory factors by Hep-PEG could decrease the concentration of pro-inflammatory factors in wounds. Notably, for the wound tissues from the Cu5.4O@Hep-PEG group, the concentration of pro-inflammatory factors is significantly lower than those in the Hep-PEG (p < 0.05) and Cu5.4O (p < 0.05) groups. We also measured the concentrations of MCP-1 and CXCL-1 in the hydrogel. As shown in Fig. S8G-H, the amounts of adsorbed MCP-1 and CXCL-1 for Cu5.4O@Hep-PEG are much lower compared to those for the Hep-PEG group (MCP-1: p < 0.01; CXCL-1: p < 0.01). These results indicate that Cu5.4O@Hep-PEG has a more sustainable effect on inflammation inhibition by simultaneously scavenging ROS through Cu5.4O and capturing pro-inflammatory factors through Hep-PEG.
To determine whether scavenging ROS using Cu5.4O@Hep-PEG aggravates wound infection, a mouse full-thickness infected wound model was established. As shown in Fig. S9A, on day 2 post-infection, a yellow puss was observed, indicating that wound infection was achieved. On days 3, 5, and 7 post-infection, there was no significant difference between the bacterial loads of the wounds for the control and Cu5.4O@Hep-PEG groups (Fig. S9B-C). Therefore, scavenging ROS with Cu5.4O@Hep-PEG does not make the wound more prone to bacterial infection.
3.8. Cu5.4O@Hep-PEG hydrogels enhance impaired diabetic wound healing
A type I diabetes mice model was employed to investigate the effect of Cu5.4O@Hep-PEG on impaired wound healing. In our study, wound dressing, Hep-PEG, Cu5.4O, and Cu5.4O@Hep-PEG were applied immediately after wounding. The hydrogels were not replaced during the experiments so as to prevent discomfort to the mouse caused by frequent dressing changes and anesthesia.
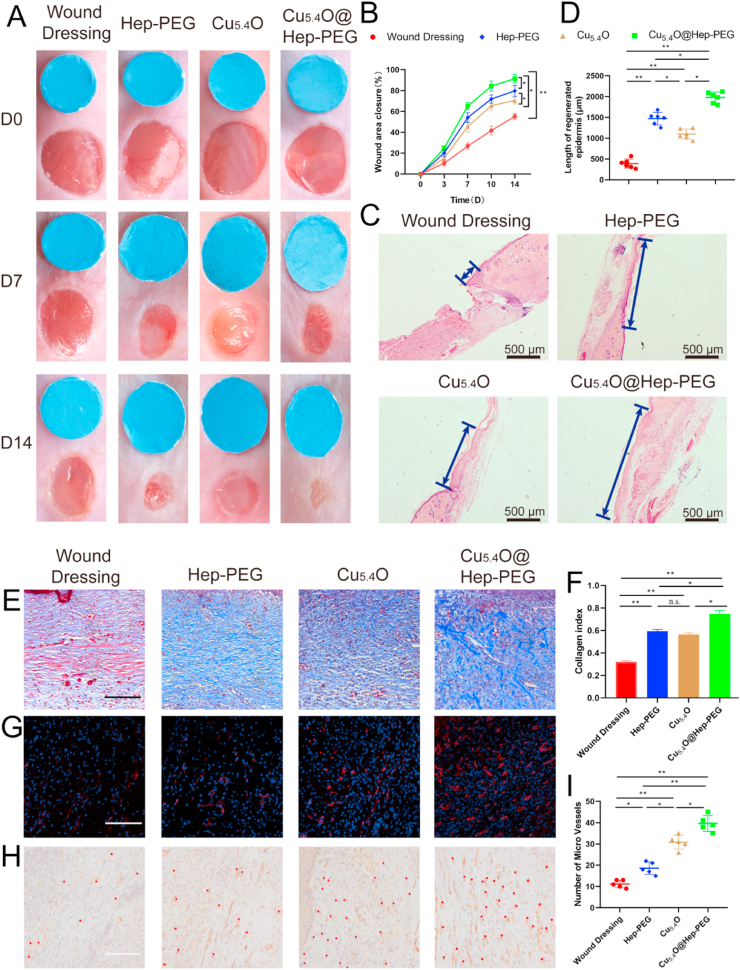
As shown in Fig. 4A, compared to the wound dressing, Hep-PEG, Cu5.4O, and Cu5.4O@Hep-PEG accelerate wound healing at different levels. The percentage of closed-wound area for the Cu5.4O@Hep-PEG group is higher than 90% on day 14 post-surgery, while for the wound-dressing, Hep-PEG, and Cu5.4O groups, the percentages of closed wound area are 55.3%, 79.8%, and 70.5%, respectively (Fig. 4B). This demonstrates that Cu5.4O@Hep-PEG promotes diabetic cutaneous wound healing.
Fig. 4.
Efficiency of Cu5.4O@Hep-PEG for diabetic wound healing. (A) Representative images of diabetic wounds at different time points (blue 6-mm-diameter disc provided for scale reference). (B) Area percentages of closed wounds. (C) Representative histological images and (D) lengths of regenerated epidermis 14 days after surgery (blue arrows indicate regenerated epidermis) (Scale bar: 500 μm). (E) Masson's trichrome staining results for diabetic wounds under different treatment regimens (Scale bar: 100 μm). (F) Collagen index measurements performed with Image J software. (G) CD31 (red) and DAPI (blue) staining results for diabetic wound tissues from each group (Scale bar: 100 μm). (H) CD31 IHC staining of diabetic wound tissues from each group. The red dots indicate blood vessels (Scale bar: 100 μm). (I) Statistical analysis of the number of blood vessels per wound field. The data in (B), (C), and (E) are mean ± s.d. from six independent replicates. (*p < 0.05; **p < 0.01; n.s.: no significance, One-way ANOVA).
We also measured the lengths of regenerated epidermis through histological analysis, as it is an essential parameter used to evaluate wound healing. As shown in Fig. 4C–D, the newly regenerated epidermis length for the Cu5.4O@Hep-PEG group are significantly greater than those for the wound dressing (p < 0.01), Hep-PEG (p < 0.05), and Cu5.4O groups (p < 0.05), further demonstrating that Cu5.4O@Hep-PEG promotes diabetic wound healing by scavenging ROS and capturing pro-inflammatory factors more efficiently than Hep-PEG (by only capturing pro-inflammatory factors) and Cu5.4O (by only scavenging ROS).
In addition, we evaluated the collagen contents of diabetic wound sites by Masson trichrome staining. The collagen fibers are labeled blue and the staining strength indicates the collagen content. As shown in Fig. 4E–F, wounds treated with Hep-PEG, Cu5.4O, and Cu5.4O@Hep-PEG show higher collagen contents than that for the wound-dressing group. Furthermore, the Cu5.4O@Hep-PEG group shows more collagen deposition than the Hep-PEG and Cu5.4O groups. Since we have demonstrated that Cu5.4O promotes the proliferation and migration of vascular endothelial cells, which are beneficial for wound healing, it is intriguing to evaluate the in vivo angiogenic effects of Cu5.4O@Hep-PEG. Blood vessels carry nutrients to the wound site and promote wound healing. As shown in Fig. 4G–I, CD31 immunofluorescence and IHC staining demonstrated that the number of blood vessels for the Cu5.4O (p < 0.01) and Cu5.4O@Hep-PEG treated wounds (p < 0.01) are significantly higher than that for the wound-dressing group, demonstrating that Cu5.4O and Cu5.4O@Hep-PEG have angiogenic effects, as indicated by the in vitro experiments. We also performed ELISA analysis to detect the concentrations of VEGF in diabetic wound sites (Fig. S10). The wound treated with Cu5.4O@Hep-PEG shows a higher concentration of VEGF than those treated with the standard wound dressing (p < 0.01), Hep-PEG (p < 0.01), and Cu5.4O (p < 0.05). Cu5.4O@Hep-PEG releases Cu5.4O to promote angiogenesis directly and also protects vascular endothelial cells from high levels of inflammation and oxidative stress. Notably, the Cu5.4O group shows a higher concentration of VEGF in the wound tissue than the wound-dressing (p < 0.01) and Hep-PEG (p < 0.01) groups. This trend is similar to that observed for the results of CD31 staining; further confirming that Cu5.4O@Hep-PEG has an angiogenic effect.
3.9. Mechanisms of diabetic wound healing by Cu5.4O@Hep-PEG
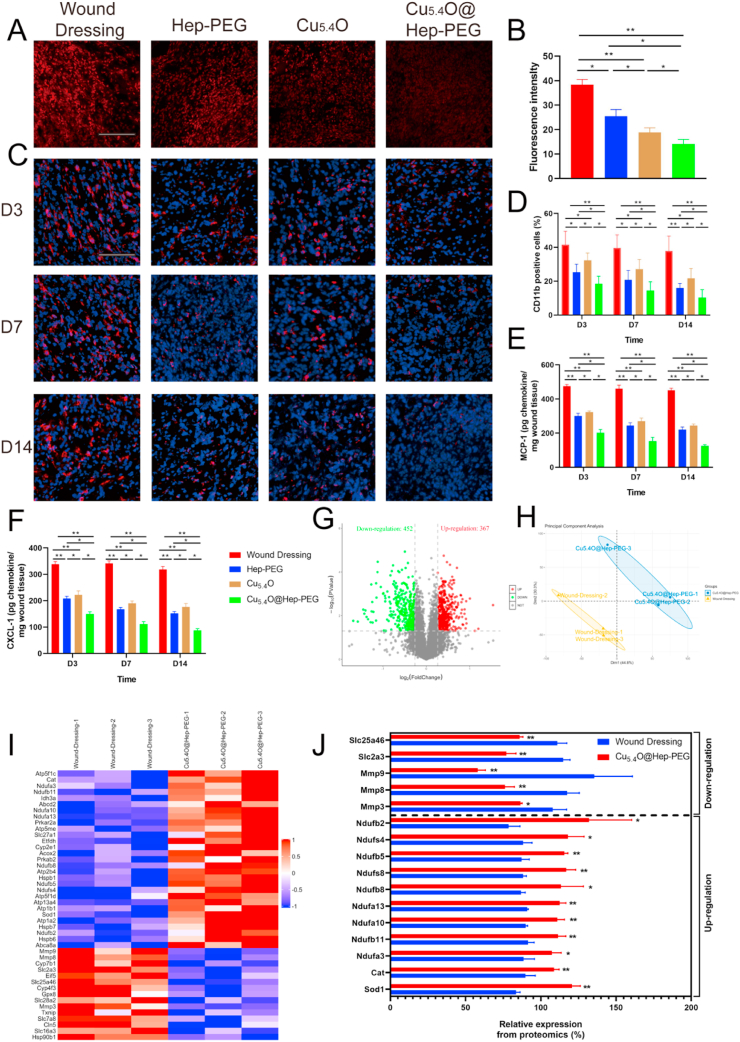
Next, we investigated the mechanism of this effect. First, to evaluate the ROS-scavenging activities of Cu5.4O@Hep-PEG in diabetic wounds, the wound tissue was harvested and frozen quickly in liquid nitrogen to produce frozen sections, followed by staining with the fluorescent dye DHE. As shown in Fig. 5A–B, the fluorescence intensities for the Hep-PEG (p < 0.05), Cu5.4O (p < 0.01), and Cu5.4O@Hep-PEG groups (p < 0.01) are significantly lower than that for the wound-dressing group. The fluorescence intensity for Cu5.4O@Hep-PEG is lower than those for Hep-PEG (p < 0.05) and Cu5.4O (p < 0.05) owing to its ROS-scavenging and pro-inflammatory-factor-capturing effects. SOD is a natural enzyme that scavenges ROS to keep the natural redox balance in the biological system, but this enzyme is prone to inactivation in high-ROS environments. Antioxidants that scavenge ROS can maintain SOD activity, as previously reported [55]. The results shown in Fig. S11A demonstrate that the SOD activity for the Cu5.4O@Hep-PEG group remains at a high level, whereas that for the wound dressing group is significantly lower (p < 0.01). Both Hep-PEG and Cu5.4O increase SOD activity to some extent, but significantly less than Cu5.4O@Hep-PEG (Hep-PEG: p < 0.01; Cu5.4O: p < 0.05) due to the synergistic effect of ROS scavenging and pro-inflammatory-factor capture. The results demonstrate that Cu5.4O@Hep-PEG protects cutaneous cells by scavenging ROS and maintain SOD activity in vivo.
Fig. 5.
Anti-inflammatory and anti-oxidative-stress effects of Cu5.4O@Hep-PEG in diabetic wounds. (A) Representative confocal images and (B) statistical analysis of the fluorescence intensity of DHE (red) in diabetic wounds from each group. (Scale bar: 100 μm) (C) Representative images and (D) statistical analysis of the CD11b staining (red) and DAPI staining (blue) of diabetic wounds from each group at different time points. (Scale bar: 100 μm) (E) Concentrations of MCP-1 and (F) CXCL-1 in wounds at different times. (G) Volcano plots showing the proteins upregulated and downregulated by Cu5.4O@Hep-PEG. (H) PCA results for differentially expressed proteins in the wound tissues in the wound-dressing and Cu5.4O@Hep-PEG groups. Each data point corresponds to PCA analysis result for a single sample. (I) Heat maps showing the proteins involved in inflammation and oxidative stress that are significantly upregulated and downregulated upon Cu5.4O@Hep-PEG treatment, and (J) relative expression of these proteins. The data in (B) and (D–F) are means ± s.d. from six independent replicates (One-way ANOVA). The data in (J) are means ± s.d. from three independent replicates (t-test). (*p < 0.05; **p < 0.01).
Moreover, CD11b staining was performed to quantify the CD11b+ immune cells in wound sites, which reflects the level of inflammation. We chose 3, 7, and 14 days post-surgery as observation time points representing the early, middle, and late periods of wound healing. As shown in Fig. 5C–D, the number of CD11b+ immune cells in the wound tissue is significantly lower for the Cu5.4O (p < 0.05), Hep-PEG (p < 0.05), and Cu5.4O@Hep-PEG (p < 0.01) groups compared to that for the wound-dressing group due to the inflammation inhibiting effects of Cu5.4O, Hep-PEG, and Cu5.4O@Hep-PEG. Furthermore, at 3, 7, and 14 days post-surgery, the number of CD11b+ immune cells for Hep-PEG is lower than that for Cu5.4O (p < 0.05) because of its pro-inflammatory-factor capture. However, the number of CD11b+ immune cells for Hep-PEG (p < 0.05) and Cu5.4O (p < 0.05) are much more than that for Cu5.4O@Hep-PEG because scavenging pro-inflammatory factors or ROS alone cannot inhibit inflammation sustainably. Importantly, the Cu5.4O@Hep-PEG group maintains the lowest CD11b+ immune cell level (p < 0.05) because Cu5.4O@Hep-PEG scavenges ROS, captures pro-inflammatory factors, and inhibits inflammatory cell migration synergistically.
Additionally, we collected wound tissues at 3, 7, and 14 days post-surgery as the early, middle, and late periods of wound healing to detect the concentration of pro-inflammatory factors. The results of ELISA assays (Fig. 5E–F) show that the concentrations of MCP-1 and CXCL-1 for the Hep-PEG (p < 0.01), Cu5.4O (p < 0.01), and Cu5.4O@Hep-PEG (p < 0.01) groups are lower than those for the wound-dressing group. This demonstrates that both scavenging ROS with Cu5.4O and capturing pro-inflammatory factors with Hep-PEG decrease the concentrations of pro-inflammatory factors in wounds. Interestingly, at 3, 7, and 14 days post-surgery, the concentrations of MCP-1 and CXCL-1 for the Hep-PEG group are lower than those for the Cu5.4O group (MCP-1: p < 0.05; CXCL-1: p < 0.05) owing to the chemokine-capturing effect of Hep-PEG. Notably, for wound tissues treated with Cu5.4O@Hep-PEG, the concentrations of pro-inflammatory factors (MCP-1 and CXCL-1) are significantly lower than those for the wound dressing (p < 0.01), Hep-PEG (p < 0.05), and Cu5.4O (p < 0.05) groups at 3, 7, and 14 days post-surgery. At these three time points, the concentrations of MCP-1 and CXCL-1 continuously decrease for the Hep-PEG, Cu5.4O, and Cu5.4O@Hep-PEG groups. This is a similar trend to that observed for CD11b+ immune cell infiltration (Fig. 5D). Thus, Cu5.4O@Hep-PEG exerts a sustainable and synergistic inflammation-inhibition effect by slowly releasing Cu5.4O and simultaneously scavenging ROS and capturing pro-inflammatory factors with Hep-PEG.
We also measured the concentrations of MCP-1 and CXCL-1 in the gels to further investigate their pro-inflammatory-factor capturing effect. As shown in Fig. S11B-C, on day 3 (MCP-1: p < 0.01; CXCL-1: p < 0.01), day 7 (MCP-1: p < 0.01; CXCL-1: p < 0.01), and day 14 post-surgery (MCP-1: p < 0.01; CXCL-1: p < 0.01), the MCP-1 and CXCL-1 levels for Hep-PEG are higher than those for Cu5.4O@Hep-PEG. Furthermore, on day 14, the Cu5.4O@Hep-PEG gel is still not saturated, while the Hep-PEG gel is saturated with MCP-1 and CXCL-1 only 7 days after surgery. This proves that scavenging ROS and capturing pro-inflammatory factors decrease the secretion of MCP-1 and CXCL-1.
The pro-inflammatory factors in inflamed wound sites promote the recruitment of immune cells into the wound to produce excessive ROS and pro-inflammatory factors. In turn, the ROS further promote the production of pro-inflammatory factors and aggravate inflammation. Thus, simultaneous pro-inflammatory-factors capture by Hep-PEG and ROS scavenging by Cu5.4O not only reduces the levels of pro-inflammatory factors and ROS in wound tissues, they also synergistically disrupt the mutual interaction between ROS and inflammation, thereby achieving a better therapeutic effect.
Macrophages play an important role in wound repair. After monocytes are recruited to an inflamed wound site, macrophages are polarized (M1 pro-inflammatory or M2 anti-inflammatory) and influence the wound healing process [56]. The polarization of macropahges is affected by the wound microenvironment. Uncontrolled and excessive inflammation obstructs polarization from M1 to M2, leading to massive accumulation of the M1 form. Moreover, ROS are important mediators in pro-inflammatory signaling pathways, which lead to the induction of pro-inflammatory (M1) macrophages [57]. A previous study demonstrated that scavenging excess ROS promotes macrophage polarization to M2 [58]. In our strategy, we used Cu5.4O@Hep-PEG hydrogels that scavenge ROS, capture MCP-1 and IL-8 as well as preventing the infiltration of CD11b+ immune cells, thus inhibiting excessive inflammation and ROS production. After Cu5.4O@Hep-PEG treatment, MCP-1 (Fig. 5E), CXCL-1 (Fig. 5F), and TNF-α (Fig. S11D), which are mainly secreted by M1 macrophages [59], are significantly decreased, while VEGF (Fig. S10), which is secreted by M2 macrophages [59], is increased, indicating that the polarization from M1 to M2 macrophages is promoted by Cu5.4O@Hep-PEG. Therefore, inhibiting uncontrolled inflammation by simultaneously adsorbing chemokines and scavenging ROS may promote macrophage polarization from M1 to M2. In our future studies, we will investigate macrophage polarization in more detail.
TMT technology was used to perform quantitative proteomic analysis as a means to explore the protein changes in wounds treated with Cu5.4O@Hep-PEG and further investigate its effect on wound healing. Volcano plots of the results reveal 1533 differentially expressed proteins (p < 0.05) of which 452 are downregulated (based on a cutoff value of 0.83-fold) and 367 are upregulated (based on a cutoff value of 1.2-fold) after Cu5.4O@Hep-PEG treatment (Fig. 5G). Furthermore, Cu5.4O@Hep-PEG-treated and wound-dressing-treated diabetic mouse cutaneous wounds show substantially different proteomics profiles (Fig. 5H), according to unguided principal component analysis (PCA) of the data.
We also investigated the impact of Cu5.4O@Hep-PEG on the proteins related to oxidative stress and inflammation. Compared with the wound-dressing group, several important antioxidant genes in the Cu5.4O@Hep-PEG treated group, including SOD1, CAT, Acox2, Ndufa3, Ndufa10 Ndufa13, Ndufb2, Ndufb5, Ndufb8, and Ndufb11 are significantly upregulated upon Cu5.4O@Hep-PEG treatment, while MMP3, MMP8, and MMP9 are downregulated (Fig. 5I–J). High levels of inflammation increase the expression of matrix metalloproteinases (MMPs) [60]. MMPs induce an increase in cellular ROS, which causes oxidative damage to DNA and thus genomic instability [61]. The trend in the expression of SOD genes observed here is consistent with the results for SOD protein levels (Fig. S11A) in mouse wound tissue, further confirming that Cu5.4O@Hep-PEG maintains the expression of antioxidant genes by scavenging ROS. Furthermore, the TNF-α level in wound tissue treated with Cu5.4O@Hep-PEG is significantly lower than those for tissues treated with the standard wound dressing (p < 0.01), Hep-PEG (p < 0.05), and Cu5.4O (p < 0.05) (Fig. S11D), demonstrating that the level of inflammation in the wound site is substantially decreased by Cu5.4O@Hep-PEG treatment and that it provides an improved environment for wound healing.
Moreover, we also found that several proteins related to collagen synthesis are upregulated by Cu5.4O@Hep-PEG treatment. As shown in Fig. S12A-C, Col4a1, Col4a3, Col6a1, Col17a1, Col24a1, Col1a2, Col3a1, Col5a1, Col5a2, Col1a1, Col2a1, Col16a1, Col11a2, and Col25a1 are significantly upregulated. These 14 proteins are crucial for many biological processes, such as cell adhesion, migration, and differentiation as well as skin development and wound healing [36]. Collagen is the most abundant extracellular protein and is an essential component of connective tissues. It forms cross-striated fibrils that provide support for cell growth and mechanical resilience to connective tissue [62] and is also fundamental to cell-cell interactions and cell attachment, which are vital for tissue repair [63]. Under high levels of inflammation and oxidative stress, collagen synthesis can be obstructed by excessive pro-inflammatory factors and DNA damage caused by ROS. The upregulation of proteins related to collagen synthesis further demonstrates that Cu5.4O@Hep-PEG decreases inflammation and oxidative stress, accelerating wound healing.
In diabetic wounds, uncontrolled inflammation causes inflammatory cell infiltration and further induces ROS production [64]. Furthermore, hyperglycemia-induced ROS overproduction is also caused by the interaction of advanced glycation end-products (AGEs) with their receptors (RAGEs) [65]. This interaction generates intracellular ROS, which modulates the expression of many genes associated with inflammation and vascular remodeling, including IL-8, TNF-α, and MCP-1 [66,67]. Meanwhile, the chemokines generated induce the migration and infiltration of more inflammatory cells. The infiltrated inflammatory cells then produce excessive ROS to further aggravate inflammation, leading to a feedback cycle. To break this cycle completely, it is essential to disrupt this interaction by simultaneously scavenging ROS and decreasing pro-inflammatory factors.
3.10. Biocompatibility of Cu5.4O@Hep-PEG in vivo
We evaluated the biocompatibility of Cu5.4O@Hep-PEG with reference to the blood chemistry and organ histopathology of healthy mice. As shown in Fig. S13A, at 30 days after treatment with Hep-PEG, Cu5.4O, and Cu5.4O@Hep-PEG, no necrosis, congestion, and hemorrhaging are observed in the heart, liver, spleen, and lung tissues. Serum biochemistry analysis (Fig. S13B-D) revealed that the serum concentrations of the liver-function indicators AST and ALT and the kidney-function indicators BUN and CRE for the treatment groups are similar to those for the control group (p > 0.05), demonstrating the biocompatibility of the gels. Moreover, the complete blood panel analysis (Fig. S13E-I) revealed no observable difference in the hematology of the treatment groups as compared to that of the control group (p > 0.05), further demonstrating the excellent biocompatibility of Cu5.4O@Hep-PEG.
4. Conclusions
We have developed a Cu5.4O@Hep-PEG hydrogel as a means to control the inflammation of wound sites and thus promote wound healing. Cu5.4O@Hep-PEG captures several inflammatory chemokines, decreasing immune cell influx and inflammatory signaling in wound sites. Moreover, Cu5.4O@Hep-PEG gel has the ability to scavenge ROS from wound sites, resulting in the decreased secretion of pro-inflammatory factors and inflammatory cell infiltration. Furthermore, this treatment strategy will disrupt the feedback relationship between ROS and inflammation and improve the wound-healing environment. Thus, our findings provide a novel therapeutic strategy for the treatment of conditions caused by uncontrolled inflammation.
Data availability
The raw/processed data will be available due to reasonable request.
Author contribution
Yuan Peng: Conceptualization, Methodology, Writing - original draft. Danfeng He: Conceptualization, Methodology, Writing - original draft. Xin Ge: Data curation, Formal analysis. Yifei Lu: Methodology, Formal analysis. Yuanhao Chai: Methodology, Formal analysis. Yixin Zhang: Software, Formal analysis. Zhengwei Mao: Writing - review & editing, Validation. Gaoxing Luo: Funding acquisition, Formal analysis. Jun Deng: Funding acquisition, Writing - review & editing, supervision. Yan Zhang: Funding acquisition, Supervision.
Declaration of competing interest
The authors have no competing financial interests or personal relationships that could influence the work published in this paper.
Acknowledgement
This work was financially supported by National Natural Science Foundation of China (Grant No. 51703243, 81630055, 81920108022), Project of Science and Technology Commission of Shanghai municipality (Grant No. 18441904500, 19441912300). Y.P., D.H., Z.M, G.L., J.D. and Y.Z. conceived and designed the research. Y.P. and D.H. conducted the experiment and analyzed the data with assistance from X.G., Y.L., Y.C and Y.X.Z. Y.P., D.H., Z.M, G.L., J.D. and Y.Z. co-wrote the paper.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.02.006.
Contributor Information
Zhengwei Mao, Email: zwmao@zju.edu.cn.
Gaoxing Luo, Email: logxw@hotmail.com.
Jun Deng, Email: djun.123@163.com.
Yan Zhang, Email: zhangy1330@sh9hospital.org.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Singer A.J., Clark R.A. Cutaneous wound healing. N. Engl. J. Med. 1999;1:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong D.G., Boulton A.J.M., Bus S.A. Diabetic foot ulcers and their recurrence. N. Engl. J. Med. 2017;376(24):2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 3.Kurita M., Araoka T., Hishida T., O'Keefe D.D., Takahashi Y., Sakamoto A., Sakurai M., Suzuki K., Wu J., Yamamoto M., Hernandez-Benitez R., Ocampo A., Reddy P., Shokhirev M.N., Magistretti P., Delicado E.N., Eto H., Harii K., Belmonte J.C.I. In vivo reprogramming of wound-resident cells generates skin epithelial tissue. Nature. 2018;561(7722):243–247. doi: 10.1038/s41586-018-0477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiekh P.A., Singh A., Kumar A. Exosome laden oxygen releasing antioxidant and antibacterial cryogel wound dressing OxOBand alleviate diabetic and infectious wound healing. Biomaterials. 2020;249:120020. doi: 10.1016/j.biomaterials.2020.120020. [DOI] [PubMed] [Google Scholar]
- 5.Tang Q., Lim T., Wei X.J., Wang Q.Y., Xu J.C., Shen L.Y., Zhu Z.Z., Zhang C.Q. A free-standing multilayer film as a novel delivery carrier of platelet lysates for potential wound-dressing applications. Biomaterials. 2020;255:120138. doi: 10.1016/j.biomaterials.2020.120138. [DOI] [PubMed] [Google Scholar]
- 6.Desmet C.M., Préat V., Gallez B. Nanomedicines and gene therapy for the delivery of growth factors to improve perfusion and oxygenation in wound healing. Adv. Drug Deliv. Rev. 2018;129:262–284. doi: 10.1016/j.addr.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Dunba C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M. Gene therapy comes of age. Science. 2018;359(6372) doi: 10.1126/science.aan4672. eaan4672. [DOI] [PubMed] [Google Scholar]
- 8.Su T., Zhang M., Zeng Q., Pan W., Huang Y., Qian Y., Dong W., Qi X., Shen J. Mussel-inspired agarose hydrogel scaffolds for skin tissue engineering. Bioact. Mater. 2020;6(3):579–588. doi: 10.1016/j.bioactmat.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunnill C., Patton T., Brennan J., Barrett J., Dryden M., Cooke J., Leaper D., Georgopoulos N.T. Reactive oxygen species (ROS) and wound healing: the functional role of ROS and emerging ROS-modulating technologies for augmentation of the healing process. Int. Wound J. 2017;14(1):89–96. doi: 10.1111/iwj.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eming S.A., Martin P., Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med. 2014;6(265):265sr6. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ungvari Z., Tarantini S., Kiss T., Wren J.D., Giles C.B., Griffin C.T., Murfee W.L., Pacher P., Csiszar A. Endothelial dysfunction and angiogenesis impairment in the ageing vasculature. Nat. Rev. Cardiol. 2018;15(9):555–565. doi: 10.1038/s41569-018-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney B.C., Chen J.H., Kent R.A., Rummani M., Alkhalil A., Moffatt L.T., Rosenthal D.S., Shupp J.W. Reactive oxygen species scavenging potential contributes to hypertrophic scar formation. J. Surg. Res. 2019;244:312–323. doi: 10.1016/j.jss.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Lopresti S.T., Popovic B., Kulkarni M., Skillen C.D., Brown B.N. Free radical-decellularized tissue promotes enhanced antioxidant and anti-inflammatory macrophage response. Biomaterials. 2019;222:119376. doi: 10.1016/j.biomaterials.2019.119376. [DOI] [PubMed] [Google Scholar]
- 14.Di A., Gao X.P., Qian F., Kawamura T., Han J., Hecquet C., Ye R.D., Vogel S.M., Malik A.B. The redox-sensitive cation channel TRPM2 modulates phagocyte ROS production and inflammation. Nat. Immunol. 2011;13(1):29–34. doi: 10.1038/ni.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni D., Wei H., Chen W., Bao Q., Rosenkrans Z.T., Barnhart T.E., Ferreira C.A., Wang Y., Yao H., Sun T., Jiang D., Li S., Cao T., Liu Z., Engle J.W., Hu P., Lan X., Cai W. Ceria Nanoparticles meet hepatic ischemia-reperfusion injury: the perfect imperfection. Adv. Mater. 2019;31(40) doi: 10.1002/adma.201902956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni D., Jiang D., Kutyreff C.J., Lai J., Yan Y., Barnhart T.E., Yu B., Im H.J., Kang L., Cho S.Y., Liu Z., Huang P., Engle J.W., Cai W. Molybdenum-based nanoclusters act as antioxidants and ameliorate acute kidney injury in mice. Nat. Commun. 2018;9(1):5421. doi: 10.1038/s41467-018-07890-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T., Xiao B., Xiang F., Tan J., Chen Z., Zhang X., Wu C., Mao Z., Luo G., Chen X., Deng J. Ultrasmall copper-based nanoparticles for reactive oxygen species scavenging and alleviation of inflammation related diseases. Nat. Commun. 2020;11(1):2788. doi: 10.1038/s41467-020-16544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerard C., Rollins B.J. Chemokines and disease. Nat. Immunol. 2001;2(2):108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 19.Martins-Green M., Petreaca M., Wang L. Chemokines and their receptors are key players in the orchestra that regulates wound healing. Adv. Wound Care. 2013;2(7):327–347. doi: 10.1089/wound.2012.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohmann N., Schirmer L., Atallah P., Wandel E., Ferrer R.A., Werner C., Simon J.C., Franz S., Freudenberg U. Glycosaminoglycan-based hydrogels capture inflammatory chemokines and rescue defective wound healing in mice. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai9044. [DOI] [PubMed] [Google Scholar]
- 21.Freudenberg U., Hermann A., Welzel P.B., Stirl K., Schwarz S.C., Grimmer M., Zieris A., Panyanuwat W., Zschoche S., Meinhold D. A star-PEG-heparin hydrogel platform to aid cell replacement therapies for neurodegenerative diseases. Biomaterials. 2009;30(28):5049–5060. doi: 10.1016/j.biomaterials.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi N., Zhang L., Chae B., Palla C.S., Furst E.M., Kiick K.L. Growth factor mediated assembly of cell receptor-responsive hydrogels. J. Am. Chem. Soc. 2007;129(11):3040–3041. doi: 10.1021/ja0680358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benoit D.S.W., Anseth K.S. Heparin functionalized PEG gels that modulate protein adsorption for hMSC adhesion and differentiation. Acta Biomater. 2005;1(4):461–470. doi: 10.1016/j.actbio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Thoniyot P., Tan M.J., Karim A.A., Young D.J., Loh X.J. Nanoparticle-hydrogel composites: concept, design, and applications of these promising, multi-functional materials. Adv. Sci. 2015;2:1400010. doi: 10.1002/advs.201400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peppas N.A., Hilt J.Z., Khademhosseini A., Langer R. Hydrogels in biology and medicine: from molecular principles to bionanotechnology. Adv. Mater. 2006;18(11):1345–1360. [Google Scholar]
- 26.Podgornik R. Book review: polymer physics. M. Rubinshtein and R. H. Colby, oxford university press. J. Stat. Phys. 2003;115:1757–1761. 2004. [Google Scholar]
- 27.Capila I., Linhardt R.J. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 2002;41(3):391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Jeevarathinam A.S., Humphries K., Jhunjhunwala A., Chen F., Hariri A., Miller B.R., Jokerst J.V. A mechanistic investigation of methylene blue and heparin interactions and their photoacoustic enhancement. Bioconjugate Chem. 2018;29(11):3768–3775. doi: 10.1021/acs.bioconjchem.8b00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong G.W., Wei D.Z., He M.L., Xiong Y.C. Discarded free PEG-based assay for obtaining the modification extent of pegylated proteins. Talanta. 2007;71(1):381–384. doi: 10.1016/j.talanta.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Zhong G., Yang X., Jiang X., Kumar A., Long H., Xie J., Zheng L., Zhao J. Dopamine-melanin nanoparticles scavenge reactive oxygen and nitrogen species and activate autophagy for osteoarthritis therapy. Nanoscale. 2019;11(24):11605–11616. doi: 10.1039/c9nr03060c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frith J.E., Menzies D.J., Cameron A.R., Ghosh P. Effects of bound versus soluble pentosan polysulphate in PEG/HA-based hydrogels tailored for intervertebral disc regeneration. Biomaterials. 2014;35(4):1150–1162. doi: 10.1016/j.biomaterials.2013.10.056. [DOI] [PubMed] [Google Scholar]
- 32.Davison C.A., Durbin S.M., Thau M.R., Zellmer V.R., Chapman S., Diener J.M., Wathen C.A., Leevy W.M., Schafer Z.T. Antioxidant enzymes mediate survival of breast cancer cells deprived of extracellular matrix. Canc. Res. 2013;73(12):3704–3715. doi: 10.1158/0008-5472.CAN-12-2482. [DOI] [PubMed] [Google Scholar]
- 33.Wang L., Smith B.A., Balanis N.G., Tsai B., Nguyen K., Cheng M.W., Obusan M.B., Esedebe F.N., Patel S.J., Zhang H. A genetically defined disease model reveals that urothelial cells can initiate divergent bladder cancer phenotypes. Proc. Natl. Acad. Sci. U.S.A. 2020;117(1):563–572. doi: 10.1073/pnas.1915770117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Bai Y., Li Y., Liang G., Jiang Y., Liu Z., Liu M., Hao J., Zhang X., Hu X. IL-15 enhances activation and IGF-1 production of dendritic epidermal T cells to promote wound healing in diabetic mice. Front. Immunol. 2017;8:1557. doi: 10.3389/fimmu.2017.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M., Topalovski M., Toombs J.E., Wright C., Moore Z.R., Boothman D.A., Yanagisawa H., Wang H., Witkiewicz A.K., Castrillon D.H. Fibulin-5 blocks microenvironmental ROS in pancreatic cancer. Canc. Res. 2015;75(23):5058–5069. doi: 10.1158/0008-5472.CAN-15-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Liu X., Zhou Z., Tan L., Wang X., Zheng Y., Han Y., Chen D.F., Yeung K.W.K., Cui Z., Yang X., Liang Y., Li Z., Zhu S., Wu S. Lysozyme-assisted photothermal eradication of methicillin-resistant staphylococcus aureus infection and accelerated tissue repair with natural melanosome nanostructures. ACS Nano. 2019;13(10):11153–11167. doi: 10.1021/acsnano.9b03982. [DOI] [PubMed] [Google Scholar]
- 37.Witt D.P., Lander A.D. Differential binding of chemokines to glycosaminoglycan subpopulations. Curr. Biol. 1994;4(5):394–400. doi: 10.1016/s0960-9822(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 38.Tsurkan M.V., Chwalek K., Prokoph S., Zieris A., Levental K.R., Freudenberg U., Werner C. Defined polymer-peptide conjugates to form cell-instructive starPEG-heparin matrices in situ. Adv. Mater. 2013;25(18):2606–2610. doi: 10.1002/adma.201300691. [DOI] [PubMed] [Google Scholar]
- 39.Gillitzer R., Goebeler M. Chemokines in cutaneous wound healing. J. Leukoc. Biol. 2001;69(4):513–521. [PubMed] [Google Scholar]
- 40.Fivenson D.P., Faria D.T., Nickoloff B.J., Poverini P.J., Kunkel S.L., Burdick M.D., Strieter R.M. Chemokine and inflammatory cytokine changes during chronic wound healing. Wound Repair Regen. 1997;5(4):310–322. doi: 10.1046/j.1524-475X.1997.50405.x. [DOI] [PubMed] [Google Scholar]
- 41.Xie X., Liu J., Li T., Song Y., Wang F. Post-formation copper-nitrogen species on carbon black: their chemical structures and active sites for oxygen reduction reaction. Chemistry. 2018;24(39):9968–9975. doi: 10.1002/chem.201801764. [DOI] [PubMed] [Google Scholar]
- 42.Sepahvand S., Jonoobi M., Ashori A., Gauvin F., Brouwers H., Oksman K., Yu Q. A promising process to modify cellulose nanofibers for carbon dioxide (CO) adsorption. Carbohydr. Polym. 2020;230:115571. doi: 10.1016/j.carbpol.2019.115571. [DOI] [PubMed] [Google Scholar]
- 43.Bratskaya S., Voit A., Privar Y., Ziatdinov A., Ustinov A., Marinin D., Pestov A. Metal ion binding by pyridylethyl-containing polymers: experimental and theoretical study. Dalton Trans. 2016;45(31):12372–12383. doi: 10.1039/c6dt01435f. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X., Xue N., Li C., Li N., Wang H., Kocic N., Beniwal S., Palotas K., Li R., Xue Q. Coordination-controlled C-C coupling products via ortho-site C-H activation. ACS Nano. 2019;13(2):1385–1393. doi: 10.1021/acsnano.8b06885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent B. The van der Waals attraction between colloid particles having adsorbed layers. II. Calculation of interaction curves. J. Colloid Interface Sci. 1973;42:270–285. [Google Scholar]
- 46.Sato N., Aoyama Y., Yamanaka J., Toyotama A., Okuzono T. Particle adsorption on hydrogel surfaces in aqueous media due to van der Waals attraction. Sci. Rep. 2017;7(1):6099. doi: 10.1038/s41598-017-06257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Handel T.M., Johnson Z., Crown S.E., Lau E.K., Proudfoot A.E. Regulation of protein function by glycosaminoglycans as exemplified by chemokines. Annu. Rev. Biochem. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 48.Johnson Z., Proudfoot A.E., Handel T.M. Interaction of chemokines and glycosaminoglycans: a new twist in the regulation of chemokine function with opportunities for therapeutic intervention. Cytokine Growth Factor Rev. 2005;16(6):625–636. doi: 10.1016/j.cytogfr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 49.Li L., Guo J., Wang Y., Xiong X., Tao H., Li J., Jia Y., Hu H., Zhang J. A broad-spectrum ROS-eliminating material for prevention of inflammation and drug-induced organ toxicity. Adv. Sci. 2018;5(10):1800781. doi: 10.1002/advs.201800781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerszten R.E., Garciazepeda E.A., Lim Y., Yoshida M., Ding H.A., Gimbrone M.A., Luster A.D., Luscinskas F.W., Rosenzweig A. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature. 1999;398(6729):718–723. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- 51.Goodwin R.G., Friend D., Ziegler S.F., Jerzy R., Falk B.A., Gimpel S., Cosman D., Dower S.K., March C.J., Namen A.E. Cloning of the human and murine interleukin-7 receptors: demonstration of a soluble form and homology to a new receptor superfamily. Cell. 1990;60(6):941–951. doi: 10.1016/0092-8674(90)90342-c. [DOI] [PubMed] [Google Scholar]
- 52.Xue Z., Huang K., Cai C., Cai L., Jiang C., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y.E., Liu J., Horvath S., Fan G. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan X., Patera A.C., Kennedy A.P., Deno G., Gonsiorek W., Manfra D.J., Vassileva G., Zeng M., Jackson C., Sullivan L., Rodriguez W.S., Opdenakker G., Damme J.V., Hedrick J.A., Lundell D., Lira S.A., Hipkin R.W. Murine CXCR1 is a functional receptor for GCP-2/CXCL6 and interleukin-8/CXCL8. J. Biol. Chem. 2007;282(16):11658–11666. doi: 10.1074/jbc.M607705200. [DOI] [PubMed] [Google Scholar]
- 54.Fujiwara T., Duscher D., Rustad K.C., Kosaraju R., Rodrigues M., Whittam A.J., Januszyk M., Maan Z.N., Gurtner G.C. Extracellular superoxide dismutase deficiency impairs wound healing in advanced age by reducing neovascularization and fibroblast function. Exp. Dermatol. 2016;25(3):206–211. doi: 10.1111/exd.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L., Sun S., Tan L., Wang Y., Wang L., Zhang Z., Zhang L. Polystyrene nanoparticles reduced ROS and inhibited ferroptosis by triggering lysosome stress and TFEB nucleus translocation in a size-dependent manner. Nano Lett. 2019;19(11):7781–7792. doi: 10.1021/acs.nanolett.9b02795. [DOI] [PubMed] [Google Scholar]
- 56.Diegelmann R.F., Evans M.C. Wound healing: an overview of acute, fibrotic and delayed healing. Front. Biosci. 2004;9:283–289. doi: 10.2741/1184. [DOI] [PubMed] [Google Scholar]
- 57.Rendra E., Riabov V., Mossel D.M., Sevastyanova T., Harmsen M.C., Kzhyshkowska J. Reactive oxygen species (ROS) in macrophage activation and function in diabetes. Immunobiology. 2019;224(2):242–253. doi: 10.1016/j.imbio.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Padgett L.E., Burg A.R., Lei W., Tse H.M. Loss of NADPH oxidase-derived superoxide skews macrophage phenotypes to delay type 1 diabetes. Diabetes. 2015;64(3):937–946. doi: 10.2337/db14-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bashir S., Sharma Y., Elahi A., Khan F. Macrophage polarization: the link between inflammation and related diseases. Inflamm. Res. 2016;65(1):1–11. doi: 10.1007/s00011-015-0874-1. [DOI] [PubMed] [Google Scholar]
- 60.Croft A.P., Campos J., Jansen K., Turner J.D., Marshall J.L., Attar M., Savary L., Wehmeyer C., Naylor A., Kemble S. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570(7760):246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radisky D.C., Levy D., Littlepage L.E., Liu H., Nelson C.M., Fata J.E., Leake D., Godden E.L., Albertson D.G., Nieto M.A. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436(7047):123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bainbridge P. Wound healing and the role of fibroblasts. J. Wound Care. 2013;22(8):407–412. doi: 10.12968/jowc.2013.22.8.407. [DOI] [PubMed] [Google Scholar]
- 63.Malhotra V., Erlmann P. The pathway of collagen secretion. Annu. Rev. Cell Dev. Biol. 2015;31:109–124. doi: 10.1146/annurev-cellbio-100913-013002. [DOI] [PubMed] [Google Scholar]
- 64.Wu H., Li F., Wang S., Lu J., Li J., Du Y., Sun X., Chen X., Gao J., Ling D. Ceria nanocrystals decorated mesoporous silica nanoparticle based ROS-scavenging tissue adhesive for highly efficient regenerative wound healing. Biomaterials. 2018;151:66–77. doi: 10.1016/j.biomaterials.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 65.Maamoun H., Benameur T., Pintus G., Munusamy S., Agouni A. Crosstalk between oxidative stress and endoplasmic reticulum (ER) stress in endothelial dysfunction and aberrant angiogenesis associated with diabetes: a focus on the protective roles of heme oxygenase (HO)-1. Front. Physiol. 2019;10:70. doi: 10.3389/fphys.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 67.Hotamisligil G.S. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542(7640):177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data will be available due to reasonable request.