Abstract
Biotic stresses represent a serious threat to rice production to meet global food demand and thus pose a major challenge for scientists, who need to understand the intricate defense mechanisms. Proteomics and metabolomics studies have found global changes in proteins and metabolites during defense responses of rice exposed to biotic stressors, and also reported the production of specific secondary metabolites (SMs) in some cultivars that may vary depending on the type of biotic stress and the time at which the stress is imposed. The most common changes were seen in photosynthesis which is modified differently by rice plants to conserve energy, disrupt food supply for biotic stress agent, and initiate defense mechanisms or by biotic stressors to facilitate invasion and acquire nutrients, depending on their feeding style. Studies also provide evidence for the correlation between reactive oxygen species (ROS) and photorespiration and photosynthesis which can broaden our understanding on the balance of ROS production and scavenging in rice-pathogen interaction. Variation in the generation of phytohormones is also a key response exploited by rice and pathogens for their own benefit. Proteomics and metabolomics studies in resistant and susceptible rice cultivars upon pathogen attack have helped to identify the proteins and metabolites related to specific defense mechanisms, where choosing of an appropriate method to identify characterized or novel proteins and metabolites is essential, considering the outcomes of host-pathogen interactions. Despites the limitation in identifying the whole repertoire of responsive metabolites, some studies have shed light on functions of resistant-specific SMs. Lastly, we illustrate the potent metabolites responsible for resistance to different biotic stressors to provide valuable targets for further investigation and application.
Keywords: Proteomics, Metabolomics, Biotic stress, Magnaporthe oryzae, Brown Plant hopper, Xanthomonas oryzae, Rice stripe virus, Nematode, Rice
Background
Due to their sessile nature, plants are exposed to various biotic and abiotic stresses. Biotic stress is induced by pathogenic bacteria, fungi, viruses, and nematodes; pest attacks; and invasion of parasitic plants. Abiotic stress is caused by adverse environmental conditions such as drought, excess salt, flood, extreme heat and cold, heavy metals, and radiation (McDowell and Dangl 2000; Sarwat et al. 2013). Pathogens and pests, which induce biotic stress, are responsible for significant yield losses in rice, of around 30.0% globally in 2019 (Savary et al. 2019); therefore, these stresses present a threat to food supply. The main goal of pathogens and pests is to obtain nutrients from plants; however, to achieve this, they cause disease and weaken the plant to enable easy access to obtain nutrients. Pathogens can be biotrophs, necrotrophs, or hemibiotrophs, based on the method of nutrient acquisition (Freeman and Beattie 2008). Plant parasitic nematodes are migratory or sedentary biotrophic obligate parasites, which feed on plant tissues by initiating special feeding structures or incorporating cell wall-degrading enzymes, virulence proteins (Ali et al. 2017; Sato et al. 2019). Insects, particularly herbivorous species, can be divided into chewing and piercing-sucking insects. Chewing insects break and chew plant tissues, resulting in mechanical damage to the plants. Conversely, piercing-sucking insects penetrate plant cells and obtain nutrients from vascular tissues (Fujita et al. 2013). Furthermore, invading nematodes, insects, and tools used for agricultural practices act as vectors and transmit viruses into host plants (Alexander and Cilia 2016).
In response to biotic stressors, plants have developed an array of dynamic constitutive and inducible defense mechanisms to protect themselves against the damage caused by invading pathogens. Cell walls, waxy epidermal cuticles, and barks serve as constitutive defense mechanisms, which, along with structural firmness, act as a first line of defense. Pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) are inducible defense mechanisms (Bigeard et al. 2015), which have been explained using a “zigzag” model (Jones and Dangl 2006). Importantly, host plants activate intricate networks of signaling cascades associated with the generation of reactive oxygen species (ROS) and the activation of hormones. Additionally, these cascades regulate kinase signaling to induce defense-related genes via the activation of transcription factors (TFs). Consequently, various secondary metabolites (SMs) and antimicrobial compounds such as phytoalexins and phenolics, are synthesized (Jain et al. 2019).
Proteins and metabolites, the final genome products, are involved in fundamental life processes. To overcome biotic stress, plants utilize multiple classes of proteins, including: (1) catalytic enzymes involved in cell wall modifications, phytohormones, ROS, and pathogenesis-related (PR) proteins; (2) TFs and post-translational factors; and (3) receptors and receptor-like kinases (Wu et al. 2016; Wu et al. 2017; Meng et al. 2019). Meanwhile, plant metabolites have distinct functions. More than 200,000 plant metabolites (Kang et al. 2019) have been classified into three dominant groups: primary metabolites, secondary (or specialized) metabolites, and hormones, which have overlapping functions (Erb and Kliebenstein 2020). Furthermore the contribution of primary metabolites to cellular energy supply and structure, phytohormones, and SMs is also important (Jwa et al. 2006). Four well-characterized hormones, abscisic acid (ABA), salicylic acid (SA), jasmonates (JA), and ethylene, play a critical role in modulating cellular mechanisms and activating plant immunity (Verma et al. 2016). SMs include phenolics produced via the shikimic and malonic acid pathways, terpenes via the mevalonic acid (MVA) and 2-C-methylerythritol 4-phosphate (MEP) pathways, and nitrogen-containing compounds from nitrogen-containing amino acids (Cheah et al. 2020; Khare et al. 2020). SMs function as antimicrobial compounds, damage–associated molecular patterns (DAMPs), and virulence factors for pathogens and are involved in callose deposition and the regulation of programmed cell death (Piasecka et al. 2015; Zaynab et al. 2018). In rice, metabolites involved in defense against biotic stress include volatile indole, glucosinolates, benzoxazinoids, phenylpropanoid phytoalexins, diterpenoid phytoalexins, and phenylamides (Erb and Kliebenstein 2020). Accordingly, studying proteins and metabolites is critical to understand the sophisticated plants’ responses to different biotic stressors under the view of proteome and metabolome. Proteomics approach is used to detect and analyze proteins. This tool can identify wide array of proteins including observation of any change in protein level during specific developmental stage of plants or plants under stresses (Tan et al. 2017; Liu et al. 2019). Moreover, proteomics can reflect the metabolic processes and their possibilities to interact with important regulatory pathways. Metabolomics approach is used to detect metabolites which are the end products of different regulatory processes and elucidate the molecular mechanisms behind any kind of variation in plants more efficiently compared to the levels of transcripts and proteins (Arbona et al. 2013). This approach differs from other omics approaches as metabolites are highly complex and requires more than one analytical platform to analyze them efficiently (Salem et al. 2020).
Since 2010, studies have investigated global changes in the composition of proteins and metabolites in plants exposed to biotic stress (for review, see Draper et al. 2011; Kushalappa and Gunnaiah 2013; Sarwat et al. 2013; Feussner and Polle 2015; Alexander and Cilia 2016; Hong et al. 2016; Meena et al. 2017; Peyraud et al. 2017; Chen et al. 2019). These studies have expanded our understanding of the regulatory mechanisms underlying plant responses and the invasive success of pests (Parker et al. 2009; Okazaki and Saito 2016). More recently, studies conducted in rice, one of the most important cereal crops, have revealed responses common to other plants as well as those specific to rice. In this review, we summarize recent findings from the proteomics and metabolomics studies in rice upon attack of various biotic stress agents, including rice mutants with altered disease resistance (Table 1 and Table 2). Finally, we suggest a framework for the improvement of rice performance.
Table 1.
Global proteomics studies investigating biotic stress responses in rice
| Method | Bacteria | Cultivars | Key finding | Reference |
| 2-DE, MALDI-TOF MS | Xoo races T7174 (IC) and Xo7435 (C) | Java 14 | Thaumatin-like protein (PR5) and probenazole (PBZ) were triggered by JA | Mahmood et al. 2006 |
| 2-DE, MALDI-TOF/TOF MS | Xoo race PXO99A (IC) and DY89031 (C) | Xa21-transgenic suspension cells | Nine putative PM-associated proteins with potential functions in rice defense were identified | Chen et al. 2007 |
| 2-DE, MALDI-TOF-TOF MS | Xoo strain Zhe173 (IC) | somatic hybrid line SH76 (R) | Majority of DEPs were involved in photosynthesis | Yu et al. 2008 |
| 2-DE, NanoLC MS/MS | P. fluorescens strain KH-1 | Co43 | P. fluorescens modulated rice metabolic pathways including energy metabolism and defense | Kandasamy et al. 2009 |
| 2-DE, MALDI-TOF MS | Xoo race Xo7435 | Thaumatin-like protein gene transgenic-OX line | Variation in oxidative stress and energy metabolism associated proteins was observed in disease resistance | Mahmood et al. 2009a |
| 2-DE, MALDI-TOF MS | Xoo races T7174 (IC) and Xo7435 (C) | Java 14 treated with probenazole | PR5 was highly induced in PBZ pretreated plants during their interaction with Xoo | Mahmood et al. 2009b |
| 2-DE, MALDI-TOF MS | Sinorhizobium meliloti 1021 | NPB | Defense related proteins were highly induced in root, whereas photosynthesis related proteins induced in leaf | Chi et al. 2010 |
| LC-MALDI-MS/MS | Xoo XKK.12 | Baldo | Virulence- associated factors were identified | González et al. 2012 |
| 2-DE, MALDI-TOF-MS | Xoo strain 89,773–1-1 | 9311 | Disease resistance signal transduction, pathogenesis, and regulation of cell metabolism were activated | Li et al. 2012a |
| 2-DE, MALDI-TOF MS and nESI-LC-MS/MS | Xoo strain K3 | Dongjin | DUF26, β-1,3-glucanase, and basic secretory protein family proteins as the main host defense related proteins | Wang et al. 2013 |
| LC-MS/MS | Xoo strain Zhe173 | IRBB5 (R) | Several epigenetic factors regulated plant disease resistance pathway by alternating phosphorylation and dephosphorylation | Hou et al. 2015 |
| 2-DE and MALDI-TOF MS | Xoo isolate DX133 | PB1 (S) and O. longistaminata (R) | Proteins related to defense response were mainly found expressed in the resistant plants | Kumar et al. 2015 |
| 2D-DIGE, MALDI-TOF-MS | Xoo strain PXO124 | O. meyriana | Peroxidase was critical in the early response of O. meyriana | Chen et al. 2016 |
| Method | Fungi | Cultivars | Key finding | Reference |
| 2-DE, MALDI-TOF-MS | M. oryzae race KJ401 (IC) and KJ101 (C) | Jinheung | Receptor-like protein kinases, pathogenesis-related proteins, and JA were induced in incompatible interaction | Sun et al. 2004 |
| 2-DE, ESI Q-TOF MS | Rhizoctonia solani strain LR 172 | Labelle (S) and LSBR-5 (R) | 3- β-hydroxysteroid dehydrogenase/isomerase was first identified in resistant rice | Lee et al. 2006 |
| 2DE, MALDI-TOF MS | M. oryzae isolate Hoku1 | ZTS (S) and ZTR (R, carryig Pi-zt) | Whole plant-specific resistance was associated with thaumatin-like protein | Koga et al. 2012 |
| 2-DE, MALDI-TOF/TOF MS | M. oryzae race ZC13 isolate 97-151a | CO39 (S) and C101LAC (R) | Resistant cultivar was more sensitive to SA signaling system | Li et al. 2012b |
| 2-DE, MALDI-TOF-MS or nESI-LC-MS/MS | M. oryzae race KJ401 and KJ301 | Jinheung | Different defense responses of rice and pathogenicity of M. oryzae | Kim et al. 2013 |
| 2-DE, MALDI-TOF/TOF-MS and nESI-LC-MS/MS | Cochliobolus miyabeanus strain SHS-2 | Dongjin | Enzymes involved in the Calvin cycle and glycolysis were decreased; but the TCA cycle, amino acids, and ethylene biosynthesis were increased | Kim et al. 2014a |
| 2-DE, MALDI-TOF/TOF and nanoLC-MS/MS | M. oryzae race ZC13 | CO39 (S) and C101LAC (R) | Resistant rice had more and rapid signal transduction cascades | Li et al. 2015 |
| iTRAQ, LC-MS/MS | M. oryzae ZHONG-10-8-14 | NPB | Activation of ABA signaling in the early stage of infection, but CK signaling in later stages of infection | Cao et al. 2016 |
| 2-DE, MALDI-ToF | R. solani (WGL-12-1) of AG-1 IA | Four susceptible and two tolerant cultivars | Novel factors associated with susceptibility and resistance | Prathi et al. 2018 |
| iTRAQ, HPLC-MS/MS | M. oryzae isolates KJ201 and RB22 | NPB (WT) and NPB-Piz-t | Seven common proteins induced by Piz-t compatible and incompatible interactions | Tian et al. 2018 |
| 2-DE, MS/MS | M. oryzae | 14 rice varieties | Functional correlation between nuclear reprogramming and immune response during blast disease | Narula et al. 2019 |
| iTRAQ, LC-MS/MS | R. solani isolate AG1 IA | Lemont (S) and Teqing (R) | A network of SA, JA, ROS, and the TCA cycle related proteins conferred resistance against R. solani | Ma et al. 2020a |
| iTRAQ, HPLC-MS/MS | M. oryzae isolates Guy-11 and YN716 | VP-1636 (WT) and GN-5 (pi21-mutant) | JA, SA, and ethylene metabolisms were upregulated in mutant line | Nawaz et al. 2020 |
| 2-DE, LC-MS/MS | M. oryzae (race 007.0), and its elicitor chitin | NPB and suspension cell derived from NPB calli | Rapid alkalinization factors, phytosulfokines, and novel immune response peptide were identified | Wang et al. 2020 |
| Method | Virus | Cultivars | Key finding | Reference |
| 2-DE, MALDI-TOF/TOF-MS | Rice black-streaked dwarf virus | Huai 5 (S) | Overaccumulation of H2O2 disrupted photosynthesis and metabolism and caused oxidative stress and abnormal plant growth | Xu et al. 2013 |
| 2-DE, MALDI-TOF MS | Rice stripe virus (RSV) | Wuyujing 3 (S) and Xudao 3 (R) | Downregulation of heat shock protein, protein disulfide isomerase, glyoxalase in Wuyuing 3 | Yang et al. 2013 |
| iTRAQ and RP-HPLC, LC-MS/MS | RSV | Wuyujing 3 | Changes of chlorosis, cell death and plant defense by RSV | Wang et al. 2015 |
| 1-DE, LC-MS/MS | Southern rice black-streaked dwarf virus | NPB treated with cytosinpeptidemycin | PR and HSP were triggered by cytosinpeptidemycin | Yu et al. 2018 |
| Method | Insect | Cultivars | Key finding | Reference |
| iTRAQ, nano-LC ESI QqTOF MS | BPH, Nilaparvata lugens Stål) | TN1 (S) and TN1 carrying Bph15 (R) | Glycine cleavage system protein was upregulated in the resistant lines. | Wei et al. 2009 |
| 2D-DIGE; and 2-DE, MALDI TOF/TOF-MS | SBPH, Laodelphax striatellus Fallén) | Rice lines 02428 (S) and Pf9279–4 (R) | ROS scavenging and SA-mediated SAR were more active in resistant rice | Dong et al. 2017 |
| iTRAQ and LC-MS/MS | BPH | PS (Indica, S), PR (O. officinalis, R), and their hybrid line HR | SMs, carbon metabolism, and glyoxylate and dicarboxylate metabolism were key markers of resistance | Zhang et al. 2019 |
| nano-LC-MS/MS | Cnaphalocrocis medinalis | TN1 (S) and Qingliu (R) | Phenylalanine ammonia lyase and chalcone synthase were higher in resistant rice | Cheah et al. 2020 |
| iTRAQ and nano-LC-MS/MS | BPH Biotype I and Biotype Y | TN1 (S) and YHY15 (moderately-R) | Post-translational modifications, protein turnover, and chaperones presented a significant difference between two BPH biotypes | Zha and You 2020 |
| Method | Nematode | Cultivars | Key finding | Reference |
| HPLC-MS/MS | Meloidogyne graminicola | NPB and Khao Pahk Maw | α-linolenic acid, glutathione, and phenylpropanoid biosynthesis were involved in resistance | Xiang et al. 2020 |
| Method | Others | Cultivars | Key finding | Reference |
| 2-DE, MS/MS | NA | Kinmaze (WT) and LMM cdr2 | Active metabolic changes were associated with programmed cell death | Tsunezuka et al. 2005 |
| 2-DE, MALDI-TOF-MS | NA | LMMs spl1 | PBZ1 served as a cell death marker and defense protein | Kim et al. 2008 |
| 2-DE, MALDI-TOF/TOF | NA | Zhefu802 (WT) and LMM spl5 | Defense response was induced, and amino acid metabolism and photosynthesis were reduced in spl5 | Chen et al. 2013 |
| 2-DE, MALDI-TOF/TOF MS | NA | CO39 (S) and C101LAC (R) treated with Me-JA | MeJA induced higher ROS in resistant rice | Li et al. 2014 |
| LC-MS/MS | NA | ZH11 (WT) and LMM oscul3a | Differentially expressed proteins were chloroplast and cytoplasm proteins | Gao et al. 2019 |
| 2-DE, MALDI-TOF/TOF MS and NanoLC-MS/MS | NA | CO39 (S) and C101LAC (R, carrying Pi-1 gene) treated with SA | Phosphorylation regulation by SA contributed differently in resistant and susceptible rice | Sun et al. 2019 |
Abbreviations: Xoo Xanthomonas oryzae pv. oryzae, IC Incompatible, C Compatible, PM Plasma membrane, S Susceptible, R Resistant, DEP Differentially expressed proteins, P. fluorescens Pseudomonas fluorescens, OX Over-expression, NPB Nipponbare, WT Wild type, PB1 Pusa Basmati1, M. oryzae Magnaporthe oryzae, ABA Abscisic acid, CK Cytokinin, SA Salicylic acid, JA Jasmonic acid, ROS Reactive oxygen species, HSP Heat shock protein, TCA Tricarboxylic acid, BPH Brown plant hopper, TN1 Taichung Native-1, SM Secondary metabolite, SBPH Small brown planthopper, SAR Systemic acquired resistance, LMM Lesion mimic mutant, 1-DE One-dimensional gel electrophoresis, 2-DE Two-dimensional gel electrophoresis, 2D-DIGE Two-dimensional difference gel electrophoresis, MS Mass spectrometry, MALDI-TOF Matrix-assisted laser desorption ionization time of flight, TOF Time-of-Flight, Q-TOF Quadrupole time-of-flight, QqTOF Quadrupole-quadrupole-time-of-flight, iTRAQ Isobaric tags relative and absolute quantification, LC-MS Liquid chromatography mass spectrometry, HPLC High-performance liquid chromatography, RP-HPLC Reverse phase-high performance liquid chromatography, UHPLC Ultra high performance liquid chromatography, GC-MS Gas chromatography mass spectrometry, CE-MS Capillary electrophoresis mass spectrometry, nESI Nano-electrospray ionization source, NA Not applicable
Table 2.
Global metabolomics studies investigating biotic stress responses in rice
| Method | Bacteria | Cultivars | Key finding | Reference |
| LC-TOF-MS and GC-TOF-MS | Xoo PXO99 and ∆PXO994rax- ST | TP309 (S) and TP309_Xa21 | Alkaloid biosynthesis was increased specifically in TP309_Xa21 to PX099 but not ∆PXO994rax- ST | Sana et al. 2010a |
| RP-HPLC-MS | Azospirillum strains 4B and B510 | Cigalon and NPB | Phenolic compounds were mainly affected | Chamam et al. 2013 |
| LC-MS | Burkholderia glumae AU6208 and Escherichia coli B6 | Cigalon and NPB treated with Azospirillum lipoferum 4B | Flavonoid compounds and hydroxycinnamic acid (HCA) derivatives changed differently upon each bacterium | Chamam et al. 2015 |
| LC-MS/MS | Xoo | Basmati 385 treated with Pseudomonas aeruginosa BRp3 | Rice defense-related enzymes were activated by P. aeruginosa | Yasmin et al. 2017 |
| HPLC | Pseudomonas putida RRF3 | TKM 9 | P. putida stimulated plant defense responses and altered rhizosphere chemical constituents | Kandaswamy et al. 2019 |
| UHPLC-QE Orbitrap/MS | Bacillus pumilus LZP02 | Longgeng 46 | Bacillus pumilus enhanced carbohydrate metabolism and phenylpropanoid biosynthesis | Liu et al. 2020 |
| UHPLC-DAD/ESI-QTOF | 10 PGPR strains and B. glumae AU6208 | NPB | Common metabolomics signature of nine compounds as rice response to different PGPR | Valette et al. 2020 |
| Method | Fungi | Cultivars | Key finding | Reference |
| FIE-MS, GC-TOF-MS | M. oryzae strain Guy11 | B. distachyon ABR1, H. vulgare Golden Promise and CO39 | Common metabolic re-programming strategy was deployed by M. oryzae in different hosts | Parker et al. 2009 |
| HPLC-MS/MS | Fusarium fujikuroi strain VE13 | Dorella (S) and Selenio (R) | Sakuranetin accumulated in resistant cultivar | Siciliano et al. 2015a |
| GC-MS | Harpophora oryzae strain R5–6-1 and M. oryzae strain Guy11 | CO39 | Different induction patterns of metabolites of the shikimate and lignin against pathogenic and mutualistic fungi | Xu et al. 2015 |
| GC-MS | R. solani | Narayan with and without Bacillus amyloliquefaciens (SN13) treatment | Identified novel aspect of rare sugar induced by Bacillus amyloliquefaciens | Srivastava et al. 2016 |
| CE/TOF-MS | R. solani AG-1 isolate C-154 | 29S (S) and 32R (R) | Canavanine was significantly higher in resistant rice | Suharti et al. 2016aa |
| CE/TOF-MS | R. solani AG-1 isolate C-154 | 29S (S) and 32R (R) | Chlorogenic acid specifically induced in resistant rice | Suharti et al. 2016ba |
| CE/TOF-MS | R. solani AG-1 isolate C-154 | 29S (S) and 32R (R) | Distinct responses of susceptible and resistant rice | Suharti et al. 2016ca |
| GC-MS | R. solani AG1-IA isolate BRS1 | PB1 (S) | Altered carbon metabolism and perturbed hormonal signaling | Ghosh et al. 2017 |
| 2-DE, MALDI-TOF MS/MS & GC-MS | R. solani isolate AGI-IA | IR-64 (WT) and AtNPR1-OX line | Novel immunity-related prognostic proteins induced by AtNPR1 | Karmakar et al. 2019a,b |
| QTOF-UPHPLC MS | M. oryzae strain Guy11 | CO39, NPB, and LTH (S); Pi-gm, Pi-4B, and Pi-B (R) | Bayogenin 3-O-Cellobioside, a saponin compound, was first identified in rice for the first time | Norvienyeku et al. 2020a |
| Method | Insect | Cultivars | Key finding | Reference |
| 1H NMR | BPH | TN1 (S) and B5 (R) | Activation of GABA shunt and shikimate metabolisms was vital for BPH resistance | Liu et al. 2010a |
| GC-MS | Rice gall midge biotype 1 (GMB1) | TN1, Kavya, and RP2068 | Potential biomarkers of rice-gall midge interaction were identified | Agarrwal et al. 2014a |
| GC-MS | GMB1 | RP2068-18-3-5 (R) | During HR, upregulation of LPO and LPO marker metabolite azelaic acid; and higher accumulation of GABA at the feeding site | Agarrwal et al. 2016a |
| UHPLC-MS and GC-MS | Rice stem borer (Chilo suppressali) | Minghui 63 | Activation of phytohormones and shikimate-mediated and terpenoid-related secondary metabolism | Liu et al. 2016 |
| GC-MS | BPH | TN1 (S) and YHY15 (R) | Resistance to BPH was mediated by SM synthesis through the shikimate pathway | Peng et al. 2016a |
| 1H NMR and GC-FID/MS | BPH | TN1 (S) and NIL-Bph15 (R) | BPH adapts and recovers at different stage in susceptible and resistant plants | Liu et al. 2017 |
| UPLC-Q-TOF MS | BPH | Dongjin treated with B. velezensis YC7010 | B. velezensis induced SA, JA, and secondary metabolites to enhance resistance | Harun-Or-Rashid et al. 2018 |
| GC-MS | BPH | NPB (S) and Bph6-transgenic line R6 (R) | Bph6 resistance gene affected lipid levels in leaf sheath only | Zhang et al. 2018a |
| GC-MS & LC-MS | BPH | TN1 (S), IR36 and IR56 (R) | Defense-related metabolites, cyanoamino acids, and lipid metabolism were increased by BPH and were more stable in resistant cultivars | Kang et al. 2019a |
| UPLC-QToF-MS | BPH | KDML105 (S) and IL308 (R) | Susceptible and resistant rice induced common SMs at different levels | Uawisetwathana et al. 2019a |
| LC-ESI-MS/MS | Cnaphalocrocis medinalis | Minghui 63 | JA-dependent signaling pathway was found vital in response to leaf folder | Wang et al. 2020 |
| Method | Nematode | Cultivars | Key finding | Reference |
| HPLC | Ditylenchus angustus | Two susceptible and five resistant cultivars | Induction and accumulation of phenolic compounds in the resistant varieties | Gill et al. 1996a |
| Method | Others | Cultivars | Key finding | Reference |
| LC-MS and Q-TOF MS/MS | NA | ZH17 (WT), wrky62, wrky76 and dsOW62/76 | SA, JA, and phenolamides were increased and free pools of flavonoids were decreased in the double mutant | Liang et al. 2017 |
Abbreviations: single asterisk “a” indicates comparative metabolomics studies of resistant and susceptible plants; “b” indicates the study applied both proteomics and metabolomics approaches; Xoo Xanthomonas oryzae pv. oryzae, S Susceptible, R Resistant, WT Wild type, OX Over-expression, NPB Nipponbare, PB1 Pusa Basmati1, PGPR Plant growth promoting rhizobacteria, B. glumae Burkholderia glumae, M. oryzae Magnaporthe oryzae, B. distachyon Brachypodium distachyon, H. vulgare Hordeum vulgare, SA Salicylic acid, JA Jasmonic acid, BPH Brown plant hopper, TN1 Taichung Native-1, HR Hypersensitive response, LPO Lipid peroxidation, SM Secondary metabolite NIL Near-isogenic line, B. velezensis Bacillus velezensis, LMM Lesion mimic mutant, dsOW62/76 Mutant containing RNA interfering constructs of OsWRKY62 and OsWRKY76, 2-DE Two-dimensional gel electrophoresis, MS Mass spectrometry, MALDI-TOF Matrix-assisted laser desorption Ionization time of flight, TOF Time-of-Flight, Q-TOF Quadrupole time-of-flight, LC-MS Liquid chromatography mass spectrometry, HPLC High-performance liquid chromatography, RP-HPLC Reverse phase-high performance liquid chromatography, UHPLC Ultra high performance liquid chromatography, GC-MS Gas chromatography mass spectrometry, CE-MS Capillary electrophoresis mass spectrometry, 1H NMR Proton nuclear magnetic resonance, FID Flame-ionization detection, DAD Diode array detector, NA Not applicable
Methods Used to Study Plant Proteomics and Metabolomics
Proteomics and metabolomics analyses involve a variety of techniques. For example, proteomics studies can involve gel-based or gel-free techniques. Gel-based methods are the most commonly used for global protein analyses, and include two-dimensional gel electrophoresis (2-DE) and difference gel electrophoresis (DIGE). In combination with advanced mass spectrometry (MS) techniques, hundreds of proteins can be detected in a single polyacrylamide gel, allowing analyses of their mass-to-charge ratio and post-translational modifications (Vanderschuren et al. 2013; Tan et al. 2017). Gel-free techniques were developed to address the limitations associated with gel-based techniques, such as reproducibility, bias, the need for technical expertise, and difficulties in detecting proteins present at low abundance or those that are highly acidic or basic (Tan et al. 2017). This approach utilizes three labeling methods: tag-based labeling such as isotope-coded affinity tag (ICAT), isobaric tags relative and absolute quantification (iTRAQ), tandem mass tag (TMT), and dimethyl and 18O labeling; metabolic labeling such as stable isotope labeling by amino acids in cell culture (SILAC) and 15N labeling; and label-free techniques, which use multi-dimensional capillary liquid chromatography (LC) coupled to nano-electrospray ionization-tandem mass spectrometry (NS-ESI-MS/MS) methods, such as sequential window acquisition of all theoretical mass spectra (SWATH-MS) (Tan et al. 2017; Ludwig et al. 2018).
Progression in analytical chemistry has led to the development of a range of metabolomics techniques, including gas chromatography (GC), liquid chromatography (LC), and capillary electrophoresis (CE) in combination with mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy (Fukusaki and Kobayashi 2005; Piasecka et al. 2019). GC-MS is a popular method capable of quantifying the levels of volatile and semi-volatile organic compounds from diverse samples. Conversely, LC-MS is a more comprehensive method, and crude extracts can be used to quantify a wide variety of metabolites. Over time, LC-MS has been optimized, allowing the collection of more effective metabolomics data by introducing ultra-performance liquid chromatography coupled with high-resolution mass analysis methods, such as time-of-flight, Fourier transform, and Orbitrap-based MS (Salem et al. 2020). Another powerful technique is CE-MS; however, this is rarely used to analyze plant metabolites because of the time-consuming and diverse extraction requirements. However, this highly sensitive technique can classify metabolites into classes that other techniques cannot, particularly highly charged metabolites (Fukusaki and Kobayashi 2005; Salem et al. 2020). NMR spectroscopy is considered to be less biased due to its independence of ionization. In addition, this method is highly reproducible, requires minimum sample preparation, and can identify novel compounds (Valentino et al. 2020). NMR is rarely used to study rice metabolomics in response to biotic stress (Table 2). Proteomics and metabolomics require the use of multiple analysis tools and databases to efficiently detect and classify proteins and metabolites based on their specific functions and related pathways. Different analysis tools, as well as their specificities, have been well documented (Piasecka et al. 2019; Sarim et al. 2020). Commonly used databases include Gene Ontology knowledgebase (http://geneontology.org/), protein database of the National Center for Biotechnology Information, RiceCyc (http://pathway.gramene.org/gramene/ricecyc.shtml), OryzaCyc in the Plant Metabolic Network database (www.plantcyc.org), and Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/).
Response of Rice to Various Biotic Stresses Considering Proteomics and Metabolomics
Transcriptomics studies of rice have provided a wealth of information and a global view of mixed gene regulation in response to various biotic stressors (Anderson and Mitchel-Olds 2011). With the emergence of advanced methods for the validation of proteins and metabolites, progress has been made in elucidating the subsequent systematic changes that follow transcription in rice (Table 1 and Table 2).
Response of Rice to Bacteria
Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight, which is the most important disease of rice caused by bacterial pathogens. Studies investigating global changes in rice proteins in response to Xoo infection have been performed. Central carbon catabolism is reduced, whereas signal transduction associated with disease resistance, pathogenesis, and the regulation of cell metabolism are upregulated, including several putative resistance (R) genes, putative receptor-like kinases, and PR (Mahmood et al. 2006; Yu et al. 2008; Mahmood et al. 2009b; Sana et al. 2010; Li et al. 2012a). Particularly, thaumatin-like protein (PR5), probenazole (PBZ), Domain of Unknown Function 26 (DUF26), and β-1,3-glucanase were reported as the key findings in the early studies (Mahmood et al. 2006; Wang et al. 2013). A secretome analysis against Xoo identified virulence-associated factors and plant-specific proteins such as proteases or peptidases and proteins involved in host defense, the transport system, and maintaining redox balance (González et al. 2012; Wang et al. 2013; Kim et al. 2014b). A similar analysis was performed using a suspension of Oryza meyeriana, a wild species that is strongly resistant to Xoo. Upregulation of the signal transduction protein, LysM receptor-like kinase and defense protein, and downregulation of a ROS enzyme (peroxidase) and cell wall modifications (via expansins and pectin acetylesterase) were reported (Chen et al. 2016). Besides, phosphosites identification suggested that phosphorylation of TFs, kinases, epigenetic controlling factors, and disease resistant proteins may be functionally relevant to Xoo resistance in IRBB5 (Hou et al. 2015). Furthermore, a metabolomics study revealed differences between resistant and susceptible phenotypes against Xoo in the accumulation of metabolites before and after infection (Sana et al. 2010). Particularly, XA21-expressing plants differed from the wild-type (WT) plants, with higher levels of sugar alcohols, tricarboxylic acid cycle (TCA) intermediates, and miscellaneous compounds in the absence of treatment. After treatment, XA21 plants contained more responsive metabolites, including rutin, pigments, fatty acids and lipids, and arginine, which are likely required for polyamine biosynthesis and alkaloid metabolism. Notably, the virulence signal acetophenone was depressed in XA21.
Plant growth-promoting rhizobacteria (PGPR), whose growth is stimulated by root exudates, assist plants via nutrient uptake and phytohormone production. In proteomics analysis, photosynthesis and defense related proteins were found to accumulate by Pseudomonas fluorescens and Sinorhizobium meliloti (Kandasamy et al. 2009; Chi et al. 2010). Metabolite profiling was first performed in 2013, when two rice cultivars were infected with rice-associated Azospirillum species (Azospirillum lipoferum 4B and Azospirillum sp. B510). In that study, changes in phenolic compounds, such as flavonoids and hydroxycinnamic derivatives, were found to differ depending on the cultivar-PGPR strain interaction (Chamam et al. 2013). Additionally, Nipponbare inoculated with 10 PGPR strains presented common metabolomics signatures, including reduced alkylresorcinol [5-tridecyl resorcinol, 5-pentadecyl resorcinol, 5 (12-heptadecyl) resorcinol] levels and the differential induction of two antimicrobial compounds, N-p-coumaroylputrescine and N-feruloylputrescine, but in different manners (Valette et al. 2020). Pseudomonas is a PGPR used as a biocontrol agent against rice disease, due to its antagonism towards other bacteria and fungi. Analysis of roots and root exudates of rice infected with Pseudomonas putida by High-performance liquid chromatography (HPLC) revealed the induction of SA (Kandaswamy et al. 2019). In another study, Pseudomonas aeruginosa was found to produce compounds associated with systemic acquired resistance (SAR), including siderophores (1-hydroxy-phenazine, pyocyanin, and pyochellin), and antibacterial compounds, including 4-hydroxy-2-alkylquinolines and rhamnolipids (Yasmin et al. 2017). This newly isolated strain can also induce defense-related enzymes in rice plants, suggesting that it may have potential for improving rice plant performance against pathogens.
Response of Rice to Fungi
Magnaporthe oryzae, the causal agent of rice blast disease, has caused huge losses in rice (Dean et al. 2012). Recently, proteomics and metabolomics studies investigating rice response to M. oryzae infection were reviewed (Meng et al. 2019; Azizi et al. 2019). In addition to the influence of M. oryzae on the basic biological processes of the host, such as photosynthesis and primary metabolism, global studies over the past decade have elucidated the detailed interaction between rice and M. oryzae considering whole proteins and metabolites (for review, see Azizi et al. 2019; Meng et al. 2019). Accordingly, metabolomics studies emphasize the difference between biotrophic and necrotrophic stages, such as the accumulation of metabolic photosynthetic sinks at biotrophic stage or phenolic compounds at necrotrophic stage (for review, see Azizi et al. 2019). In addition, a class of SMs also help to prevent fungal invasion via the antimicrobial activity of phytoalexins, including N-benzoyl tryptamine, N-cinnamoyl tryptamine, sakuranetin, and phenylamides, or the ROS-scavenging activity of serotonin. The induction or suppression of these compounds is tightly related to phytohormones such as ABA, JA, SA, and auxin. Furthermore, proteomics studies have revealed the roles of DUF26, nucleotide binding-leucine rich repeat, PRs, ROS production-scavenging enzymes, heat shock proteins (HSPs), nuclear reorganization-related proteins, TFs, and phytohormone signaling in rice resistance, whereby proteins related to pathogen perception and signal transduction are important during the early stage of infection (for review, see Meng et al. 2019). A recent study using iTRAQ found that probenazole-inducible protein 1 (PBZ1) and phenylpropanoid accumulated in both resistant and susceptible cultivars, which was in contrast to reports from previous studies utilizing the 2DE approach (Ma et al. 2020b). Interestingly, a metabolomic assay using HPLC identified a rice saponin, Bayogenin 3-O-cellobioside, which is the first saponin found in rice (Norvienyeku et al. 2020). Accordingly, the accumulation of Bayogenin 3-O-cellobioside is well corelated with blast resistance. Thus, improvements in these methods provide a more precise view of the interaction between rice and rice blast fungus on a case-by-case basis.
Rhizoctonia solani is a necrotrophic fungus that causes sheath blight in rice. In contrast to M. oryzae, this fungus causes cell death from the early stage of infection; thus, studies of R. solani infection have identified many distinct changes. The most notable changes have been observed in photosynthesis and sugar metabolism. The reduction of Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo) large subunits was observed in an early study (Lee et al. 2006). Two metabolomics studies reported the induction of glycolysis and TCA cycle intermediates (succinate, pyruvate, and aconitate) and decreased levels of sugar metabolites (sucrose, glucose, fructose, glucosone, galactose, hexopyranose, turanose, maltose, and glucopyranose), suggesting that respiration in the infected tissue was enhanced for energy production. ROS, SA, JA, aromatic aliphatic amino acids, and phenylpropanoid intermediates also accumulated, accompanied by the suppression of myo-inositol, indicating the loss of antioxidant activity, which is consistent with the formation of lesions by necrotrophic pathogens (Suharti et al. 2016b; Ghosh et al. 2017). The increase of SA, known as an inducer of defense responses against biotrophic pathogens, is consistent with the following research which found the hemobiotrophic nature of R. solani (Kouzai et al. 2018). Global proteome and metabolome studies also suggested the distinct responses of resistant and susceptible phenotypes. Resistant cultivars were found to produce higher levels of antifungal proteins (β-1–3 glucanase and chitinase), ROS scavenging machinery, and 3β-hydroxysteroid dehydrogenase (3β-HSD) for the synthesis or regulation of plant steroids. Additionally, 14–3-3 protein, which is involved in protein interactions, and the chaperonin 60 β precursor were reduced (Lee et al. 2006). Glycolysis or gluconeogenesis and fatty acid β-oxidation for adenosine triphosphate (ATP) production were upregulated, whereas energy consumption was reduced in resistant plants via the reassimilation of photorespiratory ammonia or the regulation of energy metabolism. A high abundance of proteins related to glycolysis, α-amino acid biosynthesis, and stress response have also been observed in R. solani resistance through the analysis of transgenic rice expressing AtNPR1, a key regulator of SAR (Karmakar et al. 2019). Consistent with JA, lignification and signaling were found to be stable in resistant plants (Suharti et al. 2016c). Additionally, differences in ROS regulation between resistant and susceptible plants were noted (Ma et al. 2020a). Consistent with this, pipecolic acid, which regulates SAR and induces necrotic symptoms, was upregulated in susceptible plants (Suharti et al. 2016b). However, some studies reported unexpected findings, including the downregulation of Casparian strip membrane domain-like protein 2B1, which is passively required for lignin deposition in resistant cultivars, and the upregulation of spermidine hydroxycinnamoyl transferase 1, which is involved in the biosynthesis or modification of alkaloids, terpenoids, and phenolics in susceptible plants (Prathi et al. 2018). A study investigating protein changes after infection with another necrotrophic fungus, Cochliobolus miyabeanus, which causes brown spot disease in rice, suggested a pattern similar to that seen in response to R. solani infection (Kim et al. 2014a). Proteins involved in the Calvin cycle (fructose bisphosphate aldolase, sedoheptulose-1, 7-bisphosphatase, and RuBisCO) were reduced; however, oxaloacetate aspartate and aminotransferase, which are required for amino acid biosynthesis, and enzymes involved in redox homeostasis (peroxiredoxins, glutathione reductase, and NADP-dependent isocitrate dehydrogenase) were found to be accumulated.
Those studies in necrotrophic pathogens suggested a distinct response compared with hemibiotrophic pathogens at an early stage, which was exemplified by the reduction in photosynthesis, increase in energy production, and accumulation of ROS. However, current data from a limited number of proteomics and metabolomics studies indicate that different cultivars may use different mechanisms to respond to fungal pathogens.
Response of Rice to Virus
Viruses transmitted to rice by either plants or insects induce physiological changes such as suppressed photosynthesis and chlorosis (for review, see Alexander and Cilia 2016). The key changes introduced by viral infection through insects as the vector are those involving carbon metabolism. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a glycolytic enzyme, has emerged as a multifunctional protein in several non-metabolic processes and is increased in several plant species following viral infection (for review, see Alexander and Cilia 2016; Chen et al. 2013). Additionally, fluctuations in amino acids, which provide materials for viral replication, plant defense, ROS accumulation for chlorotic damage, and respiration in terms of energy supply, are also important responses to viral infection. These findings have been confirmed and clarified by proteome and metabolome studies. For example, chloroplast proteins are degraded, chlorophyll a and chlorophyll b synthesis is inhibited, and 26S proteasome is enhanced during rice stripe virus (RSV, Tenuivirus genus) infection (Wang et al. 2015). Furthermore, enhanced H2O2 production by rice black-streaked dwarf virus (RBSDV, Reoviridae family, Fijivirus genus) may diminish light absorption, resulting in reduced photosynthesis (Xu et al. 2013). Studies on uninfected and virus-infected rice, or on resistant and susceptible cultivars, have also identified a class of proteins and metabolites that underly viral resistance. Accordingly, RBSDV induces the production of ABA and cytokinins and reduces the production of indole-3-acetic acid, gibberellins, JA, and SA by suppressing expression of the related genes (Huang et al. 2018). In that study, gibberellic acid (GA) was able to rescue the typical dwarfing symptom, and pre-application of SA was able to reduce the severity of the disease. ROS, which has a dual function in chlorosis and in viral defense mechanisms, was found to accumulate at different levels in susceptible and resistant cultivars (Xu et al. 2013). In line with this, treatment with the antiviral bioactive SM, cytosinpeptidemycin, was shown to upregulate peroxidase, superoxide dismutase, and catalase in response to southern black-streaked dwarf virus infection (Yu et al. 2018). Additionally, the increased production of PR proteins, including PR5, PR10, and Bet v1 allergen or HSPs, has also been associated with resistance to virus in rice (Yang et al. 2013; Wang et al. 2015; Yu et al. 2018).
Response of Rice to Insects
Interactions between plants and insects include dynamic defense mechanisms in plants and weapons in insects to enable successful invasion. Plants avoid being eaten in two ways, both involving a dominant role of SMs: repelling ovipositing herbivores along with attracting enemies and causing herbivore mortality. Insects may overcome this by secreting effectors in salivary proteins or capitalizing SMs (Lu et al. 2018). Although the strategies used by plants to defend against each kind of insect may vary (Harun-Or-Rashid et al. 2018), common mechanisms involve JA signaling, detoxification, cell wall modifications, photosynthesis, phytohormones, and defensive SMs (for review, see Ling et al. 2019; Zogli et al. 2020).
The brown planthopper (BPH, Nilaparvata lugens Stål, Hemiptera: Delphacidae) is a typical monophagous vascular feeder. Proteomics and metabolomics studies on rice response to BPH infection have revealed the occurrence of dynamic changes. Lipid transport and metabolism, SM biosynthesis, amino acid transport and metabolism, and phytohormone signaling are commonly induced by BPH in both susceptible and resistant cultivars (Wei et al. 2009; Dong et al. 2017; Zhang et al. 2018; Zhang et al. 2019; Zha and You 2020). Notably, studies that have utilized resistant and susceptible cultivars to observe changes on protein and metabolite levels have identified markers and features associated with resistance to BPH infection. This has also been demonstrated in time-course studies during each stage of BPH infection. A metabolomics study in leaf sheath and honeydew revealed enhanced fatty acid oxidation, glyoxylate cycle, gluconeogenesis, and γ-aminobutyric acid (GABA) shunt in susceptible cultivars, whereas glycolysis was upregulated in resistant cultivars, resulting in the production of substrates for SM synthesis via the shikimate pathway (Liu et al. 2010; Peng et al. 2016). Lower levels of amino acids in nymphs at the early, but not the late, stage of infection were reported in resistant cultivars compared with susceptible cultivars, indicating the rapid adaptation of BPH (Liu et al. 2017). In another study, there were clear differences in the response of resistant and susceptible cultivars to small BPH (Laodelphax striatellus Fallén, Homoptera: Delphacidae) during the early stage, when the resistant cultivar displayed less tissue damage due to the upregulation of ROS removal machinery and SA (Dong et al. 2017). A lipidomics study reported that the resistance conveyed by Bph6 gene involves wax biosynthesis (for example fatty acid methyl esters) (Zhang et al. 2018). Recently, Kang et al. (2019) reported that primary metabolism was inhibited in all cultivars at the early stage (24 h) but only recovered in the late stage (96 h) in resistant cultivars. In that study, amino acids, organic acids, and fatty acids were also found to be stable in resistant cultivars. Higher levels of flavonoid glycosides (schaftoside, iso-schaftoside, rhoifolin, and apigenin 6-C-α-l-arabinoside-8-C-β-l-arabinoside) were induced in resistant rice compared with susceptible rice (Uawisetwathana et al. 2019).
Phytohormones play an important role in the interaction between rice and hopper. The function of SA in this interaction appears ambiguous, as it has been reported to be upregulated in both susceptible (Peng et al. 2016) and resistant (Dong et al. 2017) cultivars. Enhanced JA metabolism is the main difference in the resistance of wild rice Oryza officinalis compared to BPH-susceptible O. sativa (Zhang et al. 2019). Interestingly, both SA and JA signaling was enhanced in the endophytic strain Bacillus velezensis YC7010, which induces resistance to BPH infection in rice (Harun-Or-Rashid et al. 2018).
Studies on the response of rice to other insects have also revealed specific responses. The response of a resistant rice line (cultivar Qingliu) to rice leafroller (Cnaphalocrocis medinalis) involved the activation of the Calvin cycle and the light reaction of photosynthesis, followed by the biosynthesis of amino acids and other metabolites (Cheah et al. 2020). Furthermore, resistance was determined by flavonoid biosynthesis at a specific rate and time. Rice also defends against the rice stem borer (Chilo suppressalis) using a similar mechanism. An integrated transcriptomics and metabolomics study suggested increased photosynthesis via the accumulation of monosaccharides and not of oligosaccharides, galactinol, and various amino acids (Liu et al. 2016). Conversely, the resistance of rice to rice gall midge was found to involve differences in fatty acids before and after infection, whereas glutamine and 23-oxotetracosanoic acids were associated with susceptibility (Agarrwal et al. 2014).
Response of Rice to Nematodes
Nematodes are universally present in nature and include species that are parasitic to plants, including rice (Sato et al. 2019). Studies investigating rice-parasitic nematode interactions have generally involved mutants and transcriptome analyses, with a notable lack of proteomics and metabolomics studies. In 1996, a group of researchers used HPLC to evaluate differences in the phenolic profiles of five resistant and two susceptible deep-water rice upon Ditylenchus angustus infection. They reported changes in SMs, such as chlorogenic acids and phytoalexin sakuranetin, which were mainly identified in the resistant rice (Gill et al. 1996). A previous proteomics study on the rice and root-knot-nematode (Meloidogyne graminicola) interaction revealed new proteins as well as changes in existing proteins (Xiang et al. 2020). Importantly, proteins involved in stress, metabolic pathways, and SM biosynthesis were found to accumulate at the early stage of infection, and this continued to the later stage of infection. Additionally, an integration of transcript analyses revealed that four specific proteins related to α-linolenic acid metabolism, phenylpropanoid biosynthesis, glutathione metabolism, and plant–pathogen interaction pathways were downregulated in the susceptible cultivar but upregulated in the resistant cultivar (Xiang et al. 2020).
Proteomics and Metabolomics Studies in Mutants with Altered Defense Response
Lesion mimic mutants have been used to study resistance for over two decades. Therefore, proteomics studies on lesion mimic mutants of rice have widened our understanding of the fundamentals of hypersensitive response (HR)-like symptom. A 2DE study on the lesion-mimic mutant Cell death and resistance 2 (cdr2) and Rice spotted leaf 5 (spl5) reported the accumulation of defense-related proteins, including probenazole-induced (PBZ1) protein (Tsunezuka et al. 2005; Chen et al. 2013). PBZ1 protein is also highly inducible in the Squamosa promoter-binding-like protein 1 (spl1) mutant (Kim et al. 2008). Regarding central metabolism, photosynthesis is inhibited whereas respiration is enhanced via the down and upregulation of the associated proteins, respectively. Interestingly, the overproduction of ROS in lesion mimic mutants induces ROS-scavenging enzymes, such as L-ascorbate peroxidase 7, but suppresses superoxide dismutase in cdr2 mutant, confirming that the tight regulation of ROS is correlated with the formation time and density of lesions. In addition, pathogen-infected mimic responses, such as enhanced lipid metabolism, were found to suppress carbon and nitrogen metabolism and the accumulation of SA and SMs in oscul3a mutants (Gao et al. 2019).
Proteomics and metabolomics have been studied to understand PTI, also known as basal resistance. Loss-of-function of Pi21, a quantitative resistance gene encoding a proline-rich protein that includes a putative heavy metal-binding domain and putative protein-protein interaction motifs, results in non-race specific and durable blast resistance (Fukuoka et al. 2009). Protein profiling of a Pi21-knockout mutant in the absence of pathogen infection revealed the accumulation of photosynthates, carbohydrate metabolites, and small molecule metabolites, compared with the WT plants (Nawaz et al. 2020). Additionally, metabolomics studies have shown that the enhanced basal resistance associated with loss-of-function of WRKY62 and WRKY76 (dsOW62/76) is associated with the accumulation of amino acids, constituents of TCA, phenolic acids derived from the phenylpropanoid pathway, upregulated SA and JA, and antimicrobial phytoalexins, such as sakuranetin and phenolamides (Liang et al. 2017).
Proteomics has also been studied to understand ETI in Pizt-expressing plants in response to avirulent and virulent isolates, which suggested that various specific responses are induced by Pizt (Tian et al. 2018). Accordingly, fluctuations in 56 proteins were common between Pizt and WT plants after infection and included PR proteins, proteins related to hormonal regulation and defense and stress response, receptor-like kinases, and cytochrome P450. Interestingly, the incompatible interaction differed significantly from the compatible interaction in only a few proteins, including alcohol dehydrogenase I, receptor-like protein kinase, endochitinase, similar-to-rubisco large subunit, NADP-dependent malic enzyme, and two hypothetical proteins. This finding raises the question of whether variation in only those compounds could lead to different ETI outcomes.
Conclusions and Prospects
Common Metabolites and Specific Metabolites upon each Biotic Stress
Studies investigating the response to biotic stresses have commonly reported changes in photosynthesis, possibly due to the abundant related proteins and metabolites. Photosynthesis is upregulated or downregulated in susceptible or resistant phenotypes in response to different pathogens. In response to insects, two theories have been proposed to explain this phenomenon. The first notes that the intrinsic activation of photosynthesis provides organic compounds for the synthesis of defense-related metabolites as a result of pathogen manipulation for food resources (Cheah et al. 2020). An opposing theory states that plants suppress photosynthesis to conserve energy and reduce food supply to pathogens. Photosynthesis is enhanced by Xoo (Sana et al. 2010; Li et al. 2012a), insect (Liu et al. 2016; Cheah et al. 2020), and M. oryzae infection in the biotrophic stage (Azizi et al. 2019) but suppressed by the necrotrophic fungus R. solani (Lee et al. 2006; Karmakar et al. 2019) and viral infection (Xu et al. 2013). Therefore, photosynthetic activity varies depending on the feeding style of the pathogen, which is consistent with the response in other species (for review Chen et al. 2019). In addition, the appearance of cell death lesions during the necrotrophic infection stage is likely to underly the significantly lower level of photosynthesis-related enzymes and metabolites.
ROS exert a positive effect on defense to various pathogens by acting as signaling molecules or inhibiting pathogens by inducing local cell death. However, enhanced accumulation of ROS might result in cell death, thus facilitating the virus (Xu et al. 2013) and necrotrophic pathogen (Suharti et al. 2016b), or it might suppress normal plant metabolism due to the occurrence of oxidative stress. Therefore, the balance of ROS production and scavenging must be tightly regulated. The global profiling studies reviewed herein support the correlation between ROS and photorespiration and photosynthesis. Particularly, enhanced photorespiration is important for the induction of ROS (Kangasjärvi et al. 2012), whereas ROS, such as H2O2, suppress photosynthesis and plant processes in response to stress (Xu et al. 2013). ROS are also induced by SA (Li et al. 2012b) or the inactivation of GABA shunt (Suharti et al. 2016b). Although ROS accumulation has been reported in response to most biotic stresses, the timing and intensity of ROS vary significantly depending on the pathogen and cultivar, which present different levels of susceptibility.
Phytohormones are also a key response exploited by rice and pathogens. In order to induce a defense response, rice plants upregulate SA signaling when exposed to M. oryzae (Meng et al. 2019; Azizi et al. 2019), JA when exposed to insects (Zhang et al. 2019; Wang et al. 2020), and both SA and JA when exposed to R. solani (Ma et al. 2020a). PGPR also support rice defense via the induction of SA, as reported for Pseudomonas (Kandaswamy et al. 2019), or the induction of ABA signaling and suppression of SA signaling via the bioactive SM cytosinpeptidemycin in response to Streptomyces (Yu et al. 2018). Conversely, ABA and cytokinins are activated during viral infection or during the early stage of M. oryzae invasion (Cao et al. 2016) in order to facilitate infection. Viral infection downregulates IAA, GA, JA, and SA (Huang et al. 2018). SA activates ROS faster in resistant cultivars and alleviates the decrease in plant photosynthesis (Li et al. 2012b), whereas JA suppresses photosynthesis (Rakwal and Komatsu 2000). Moreover, ABA signaling influences the calcium (Ca2+) signaling that is considered as a front line of signaling events and is involved in resistance response of rice against M. oryzae (Wang et al. 2019). In a proteomics study of resistant Gangyuan8 (GY8) and susceptible Lijiangxintuanheigu (LTH) cultivars infected by M. oryzae, Ca2+ − dependent protein kinase, Ca2+ sensor calmodulin, and calmodulin-like protein were particularly found upregulated in LTH but remained the same in GY8, suggesting that the proteins possibly regulate blast resistance negatively in those cultivars (Ma et al. 2020b). Another proteomics study found several Ca2+-binding proteins in saliva of phloem-feeding insects and honeydews of BPH and green rice leafhopper, supporting the function of Ca2+-binding proteins in counteracting the sieve-tube occlusion defenses in host plants (Will et al. 2013; Zhu et al. 2020). These findings suggest that phytohormone regulation, as part of plant defense mechanisms, via different target pathways is complicated, and that cooperation occurs between the pathways.
Signaling components and SMs are highly diversified molecules, dependent on the type of biotic stress. For example, different interactions between rice and PGPR result in different metabolic changes (Chamam et al. 2013). This is explained by the diverse types and functions of these molecules, especially SMs (Erb and Kliebenstein 2020). SMs are less well-conserved, multifunctional metabolites, which guarantee the response to various biotic factors, but resist manipulation and save costs associated with biosynthesis. Therefore, differences in SMs are associated with the resistance of different cultivars to pathogens at different stages of infection. Moreover, each cultivar has a set of differentially expressed proteins (Prathi et al. 2018), diversifying the response.
Limitations in Global Proteomics and Metabolomics Studies in Rice
Studying proteins and metabolites is more difficult than studying transcriptomics for several reasons: (1) the complexity of proteins and metabolites with different properties makes them difficult to identify using the same method. In addition, proteins undergo various post-translational modifications, resulting in the generation of different isoforms (for review, see Wu et al. 2016; Tan et al. 2017). (2) Technical obstacles, including an appropriate extraction method, the sensitivity of detection, and the detection of post-translational modifications, can limit the detection of proteins or metabolites present at a low abundance (for review, see Castro-moretti et al. 2020). Due to the cost of producing SMs, rice plants are required to maintain a high level of regulation, processing, and storage, to ensure that some SMs are produced at trace amounts (for review, see Erb and Kliebenstein 2020). To detect those SMs, an appropriate experimental design, pipelines, and standard methods are critical (for review, see Alexander and Cilia 2016; Chen et al. 2019). (3) Lack of an information library or database to identify new molecules results in a comparatively large amount of unknown proteins or metabolites in each study (for review, see Chen et al. 2019; Castro-moretti et al. 2020). For example, 15 out of 21 general differentially expressed proteins were unknown in the study of Zhang (Zhang et al. 2019). Thirty-three identified metabolites were undefined in the study of Madhavan (Madhavan et al. 2019). Additionally, 7% and 9% of metabolites extracted from R. solani infection in susceptible and resistant cultivars, respectively, were undefined (Suharti et al. 2016b). Rice plants possess specific metabolites (for review, see Okazaki and Saito 2016), which cannot be identified based on the libraries of other species. However, with recently developed methods, we have made progress and expanded our knowledge in this area, exemplified by the identification of new biomarkers (Agarrwal et al. 2014; Duan et al. 2020).
Consistency in Global Studies of Transcripts and Proteins
Global studies of proteins and metabolites are usually combined with transcription profiling (Table 1 and Table 2). In general, the fluctuation of major molecules is well corelated with gene expression (Sana et al. 2010; Zha and You 2020); however, inconsistencies have also been observed. Peroxidase expression is not associated with the activity between Nipponbare and O. meyeriana (Chen et al. 2016). In the study of Zhang, the expression patterns of four out of eight genes were in contrast to the expression pattens of the proteins (Zhang et al. 2019). A low correlation between mRNA and protein levels has also been observed in half of all genes examined in secreted proteins from rice suspensions (Dong et al. 2017). This may be due to post-transcriptional regulation, for example by RNA binding proteins (Xu et al. 2013), or post-translational modifications, which were recently shown to be a significant response to insect invasion (Zha and You 2020). Furthermore, the challenges of methods used in proteomics have limited the identification of all possible isoforms, consequently influencing correlation studies.
Use of Proteomics and Metabolomics to Improve Rice Performance
Time-series studies have complemented our understanding on the conflict between pathogens and rice at each stage of infection in susceptible and resistant cultivars. Accordingly, the outcome of this conflict is determined by the up or downregulation of certain molecules as well as the intensity of these molecules. For example, defense-related metabolites, cyanoamino acids, and lipid metabolism were increased in both susceptible and resistant cultivars but were more stable in rice resistant to BPH (Kang et al. 2019). Resistant rice infected with M. oryzae displayed a higher sensitivity to SA (Li et al. 2012b), and resistance to R. solani is dependent on the stability of JA and lignification (Suharti et al. 2016c). These studies confirmed a potential metabolic target but emphasized the limitations associated with studying and utilizing metabolites, especially phytohormones, whose balance is critical for plant growth and development (Peleg and Blumwald 2011). One necessary approach for more effective outcome from rice-pathogen interaction studies would be integration of omics approaches, such as combining transcriptomics with proteomics or metabolomics (Prathi et al. 2018; Wang et al. 2020) or combining proteomics with metabolomics (Karmakar et al. 2019). Moreover, combining separate studies on specific stressor may serve as excellent approach, even though this may bring out some inconsistency. Resultant proteins and metabolites in response to a common biotic stressor may belong to similar pathways, which will eventually increase the efficacy of outcome to get more detailed insight into the intricate cellular activities during rice responses to that stressor. For instance, two separate proteomics and metabolomics studies on rice response to leafroller insect (Cnaphalocrocis medinalis) suggest that the JA biosynthesis pathway related proteins and metabolites are critical for resistance (Cheah et al. 2020; Wang et al. 2020).
The limited identification of molecules in proteomics and metabolomics studies as compared to transcriptomics studies has implied the simpler outcome at the final products than the gene regulation. In this context, metabolomics studies on resistance genes have recalled a long-standing question: whether different genes associated with resistance result in different outcomes via different metabolic pathways. Studies on Pizt- and Pi21-mediated resistance and basal resistance in dsOW62/76 have suggested a different outcome. If this is the case, SMs represent a tool that is guaranteed to perform well. However, the greatest obstacle is the cost of SM biosynthesis. In most studies, the number of upregulated SMs is greater than the number of downregulated SMs, affirming the tight regulation of SMs due to their cost. Therefore, more studies on resistance genes and how to deploy SMs in plant resistance are needed.
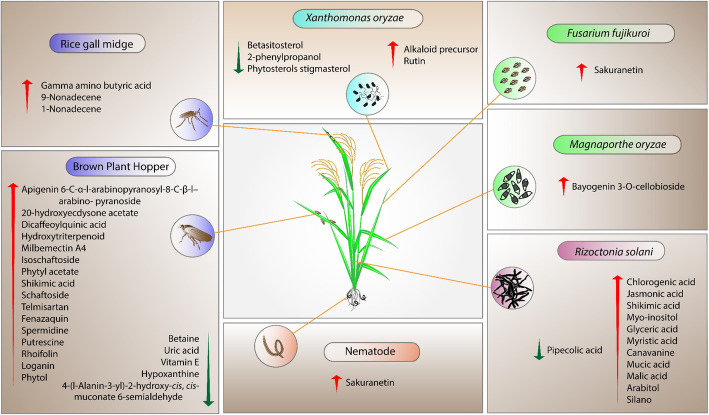
The application of metabolomics to improve plant performance to stresses has been previously suggested (Hong et al. 2016). An example was proposed by Kushalappa and Gunnaiah (2013), who suggested 10 heuristic steps to streamline metabolomics-proteomics studies to identify resistance genes. The main difference in the metabolic profiles of resistant and susceptible cultivars provided us with scaffolds to produce stable resistant rice, which is sometimes confined by a resistance gene approach. Thus, we collected the potent metabolites responsible for resistance to different stressors, as illustrated in Fig. 1. These metabolites are specifically induced or reduced in resistant rice cultivars (Table 2). Due to the multiple functions of primary metabolites in rice, we limited our analysis to SMs or primary metabolites that function in resistance via a non-primary pathway. These SMs provide targets for further investigation and use.
Fig. 1.
Potent metabolites linked to stress resistance in rice. Those metabolites were specifically increased (red arrow) or decreased (green arrow) in resistant plants as compared to susceptible plants in indicated studies (Table 2, marked by a). SA: salicylic acid, JA: jasmonic acid
Acknowledgements
Not applicable.
Abbreviations
- 2-DE
Two-dimensional gel electrophoresis
- ABA
Abscisic acid
- ETI
Effector-triggered immunity
- GC
Gas chromatography
- iTRAQ
Isobaric tags relative and absolute quantification
- JA
Jasmonic acid
- LC
Liquid chromatography
- MS
Mass spectrometry
- PAMP
Pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- SA
Salicylic acid
- SM
Secondary metabolite
- TF
transcription factor
Authors’ Contributions
J.J. designed and directed the project. K.T.X.V., M.R., M.R., K.T.T.T., S.T.K., and J.J. contributed to the writing of the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Basic Research Lab program (Project No. 2018R1A4A1025158) and the Mid-Career Researcher program (Project No. 2020R1A2C2012976), National Research Foundation of Korea.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
The manuscript has been approved by all authors.
Competing Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kieu Thi Xuan Vo and Md Mizanor Rahman contributed equally to this work.
Contributor Information
Kieu Thi Xuan Vo, Email: vtxkieu@khu.ac.kr.
Md Mizanor Rahman, Email: mizanor@khu.ac.kr.
Md Mustafizur Rahman, Email: mr10bau2@khu.ac.kr.
Kieu Thi Thuy Trinh, Email: tttkieu.biotech@khu.ac.kr.
Sun Tae Kim, Email: stkim5505@gmail.com.
Jong-Seong Jeon, Email: jjeon@khu.ac.kr.
References
- Agarrwal R, Bentur JS, Nair S. Gas chromatography mass spectrometry based metabolic profiling reveals biomarkers involved in rice-gall midge interactions. J Integr Plant Biol. 2014;56:837–848. doi: 10.1111/jipb.12244. [DOI] [PubMed] [Google Scholar]
- Agarrwal R, Padmakumari AP, Bentur JS, et al. Metabolic and transcriptomic changes induced in host during hypersensitive response mediated resistance in rice against the Asian rice gall midge. Rice. 2016;9:1–15. doi: 10.1186/s12284-016-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MM, Cilia M. A molecular tug-of-war: global plant proteome changes during viral infection. Curr Plant Biol. 2016;5:13–24. doi: 10.1016/j.cpb.2015.10.003. [DOI] [Google Scholar]
- Ali MA, Azeem F, Li H, et al. Smart parasitic nematodes use multifaceted strategies to parasitize plants. Front Plant Sci. 2017;8:1–21. doi: 10.3389/fpls.2017.01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Mitchel-Olds T. Ecological genetics and genomics of plant defenses: evidence and approaches. Funct Ecol. 2011;25:312–324. doi: 10.1111/j.1365-2435.2010.01785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbona V, Manzi M, Ollas CD, et al. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int J Mol Sci. 2013;14:4885–4911. doi: 10.3390/ijms14034885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi P, Osman M, Hanafi MM, et al. Adaptation of the metabolomics profile of rice after Pyricularia oryzae infection. Plant Physiol Biochem. 2019;144:466–479. doi: 10.1016/j.plaphy.2019.10.014. [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI) Mol Plant. 2015;8:521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Cao J, Yang C, Li L, et al. Rice plasma membrane proteomics reveals Magnaporthe oryzae promotes susceptibility by sequential activation of host hormone signaling pathways. Mol Plant Microbe Interact. 2016;29:902–913. doi: 10.1094/MPMI-08-16-0165-R. [DOI] [PubMed] [Google Scholar]
- Castro-Moretti FR, Gentzel IN, Mackey D, et al. Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites. 2020;10:1–23. doi: 10.3390/metabo10020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamam A, Sanguin H, Bellvert F, et al. Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry. 2013;87:65–77. doi: 10.1016/j.phytochem.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Chamam A, Wisniewski-Dyé F, Comte G, et al. Differential responses of Oryza sativa secondary metabolism to biotic interactions with cooperative, commensal and phytopathogenic bacteria. Planta. 2015;242:1439–1452. doi: 10.1007/s00425-015-2382-5. [DOI] [PubMed] [Google Scholar]
- Cheah BH, Lin HH, Chien HJ, et al. SWAtH-MS-based quantitative proteomics reveals a uniquely intricate defense response in Cnaphalocrocis medinalis-resistant rice. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-63470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Ma R, Chen XL. Advances of metabolomics in fungal pathogen–plant interactions. Metabolites. 2019;9:169. doi: 10.3390/metabo9080169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Yuan Y, Li Q, et al. Proteomic analysis of rice plasma membrane reveals proteins involved in early defense response to bacterial blight. Proteomics. 2007;7:1529–1539. doi: 10.1002/pmic.200500765. [DOI] [PubMed] [Google Scholar]
- Chen X, Dong Y, Yu C et al. (2016) Analysis of the proteins secreted from the oryza meyeriana suspension-cultured cells induced by Xanthomonas oryzae pv. oryzae. PLoS One 11:1-16. 10.1371/journal.pone.0154793. [DOI] [PMC free article] [PubMed]
- Chen X, Fu S, Zhang P, et al. Proteomic analysis of a disease-resistance-enhanced lesion mimic mutant spotted leaf 5 in rice. Rice. 2013;6:1–10. doi: 10.1186/1939-8433-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi F, Yang P, Han F, et al. Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics. 2010;10:1861–1874. doi: 10.1002/pmic.200900694. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, et al. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Fang X, Yang Y, et al. Comparative proteomic analysis of susceptible and resistant rice plants during early infestation by small brown planthopper. Front Plant Sci. 2017;8:1–14. doi: 10.3389/fpls.2017.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J, Rasmussen S, Zubair H. Biology of plant metabolomics. 2011. Metabolite analysis and metabolomics in the study of biotrophic interactions between plants and microbes annual plant reviews volume 43. [Google Scholar]
- Duan G, Li C, Liu Y et al. (2020) Magnaporthe oryzae Systemic Defense Trigger 1 (MoSDT1)-Mediated Metabolites Regulate Defense Response in Rice. BMC Plant Biol 21, 40. 10.1186/s12870-020-02821-6. [DOI] [PMC free article] [PubMed]
- Erb M, Kliebenstein D. Plant secondary metabolites as defenses, regulators and primary metabolites- the blurred functional trichotomy. Plant Physiol. 2020;184:00433.2020. doi: 10.1104/pp.20.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Polle A. What the transcriptome does not tell - proteomics and metabolomics are closer to the plants’ patho-phenotype. Curr Opin Plant Biol. 2015;26:26–31. doi: 10.1016/j.pbi.2015.05.023. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Beattie GA (2008) An overview of plant defenses against pathogens and herbivores. Plant Heal Instr. 10.1094/phi-i-2008-0226-01
- Fujita D, Kohli A, Horgan FG. Rice resistance to Planthoppers and leafhoppers. Crit Rev Plant Sci. 2013;32:162–191. doi: 10.1080/07352689.2012.735986. [DOI] [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- Fukusaki E, Kobayashi A. Plant metabolomics: potential for practical operation. J Biosci Bioeng. 2005;100:347–354. doi: 10.1263/jbb.100.347. [DOI] [PubMed] [Google Scholar]
- Gao Z, Liu Q, Zhang Y, et al. A proteomic approach identifies novel proteins and metabolites for lesion mimic formation and disease resistance enhancement in rice. Plant Sci. 2019;287:110182. doi: 10.1016/j.plantsci.2019.110182. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Kanwar P, Jha G. Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of Rhizoctonia solani. Sci Rep. 2017;7:1–12. doi: 10.1038/srep41610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill JR, Harbornez JB, Plowright RA, et al. The induction of phenolic compounds in rice after infection by the stem nematode Ditylenchus angustus. Nematologica. 1996;42:564–578. doi: 10.1163/004625996X00063. [DOI] [Google Scholar]
- González JF, Degrassi G, Devescovi G, et al. A proteomic study of Xanthomonas oryzae pv. Oryzae in rice xylem sap. J Proteomics. 2012;75:5911–5919. doi: 10.1016/j.jprot.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Harun-Or-Rashid M, Kim HJ, Yeom SI, et al. Bacillus velezensis YC7010 enhances plant defenses against brown planthopper through transcriptomic and metabolic changes in rice. Front Plant Sci. 2018;9:1–15. doi: 10.3389/fpls.2018.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Yang L, Zhang D, et al. Plant metabolomics: an indispensable system biology tool for plant science. Int J Mol Sci. 2016;17:767. doi: 10.3390/ijms17060767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Qiu J, Tong X, et al. A comprehensive quantitative phosphoproteome analysis of rice in response to bacterial blight. BMC Plant Biol. 2015;15:1–15. doi: 10.1186/s12870-015-0541-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Li Y, Tang G, et al. Dynamic phytohormone profiling of rice upon rice black-streaked dwarf virus invasion. J Plant Physiol. 2018;228:92–100. doi: 10.1016/j.jplph.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Jain P, Dubey H, Singh PK, et al. Deciphering signalling network in broad spectrum near isogenic lines of rice resistant to Magnaporthe oryzae. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-50990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jwa NS, Agrawal GK, Tamogami S, et al. Role of defense/stress-related marker genes, proteins and secondary metabolites in defining rice self-defense mechanisms. Plant Physiol Biochem. 2006;44:261–273. doi: 10.1016/j.plaphy.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Kandasamy S, Loganathan K, Muthuraj R, et al. Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci. 2009;7:1–8. doi: 10.1186/1477-5956-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy R, Ramasamy MK, Palanivel R, et al. Impact of Pseudomonas putida RRF3 on the root transcriptome of rice plants: insights into defense response, secondary metabolism and root exudation. J Biosci. 2019;44:1–13. doi: 10.1007/s12038-019-9922-2. [DOI] [PubMed] [Google Scholar]
- Kang K, Yue L, Xia X, et al. Comparative metabolomics analysis of different resistant rice varieties in response to the brown planthopper Nilaparvata lugens Hemiptera: Delphacidae. Metabolomics. 2019;15:1–13. doi: 10.1007/s11306-019-1523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjärvi S, Neukermans J, Li S, et al. Photosynthesis, photorespiration, and light signalling in defence responses. J Exp Bot. 2012;63:1619–1636. doi: 10.1093/jxb/err402. [DOI] [PubMed] [Google Scholar]
- Karmakar S, Datta K, Molla KA, et al. Proteo-metabolomic investigation of transgenic rice unravels metabolic alterations and accumulation of novel proteins potentially involved in defence against Rhizoctonia solani. Sci Rep. 2019;9:1–16. doi: 10.1038/s41598-019-46885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Singh NB, Singh A, et al. Plant secondary metabolites synthesis and their regulations under biotic and abiotic constraints. J Plant Biol. 2020;63:203–216. doi: 10.1007/s12374-020-09245-7. [DOI] [Google Scholar]
- Kim JY, Wu J, Kwon SJ, et al. Proteomics of rice and Cochliobolus miyabeanus fungal interaction: insight into proteins at intracellular and extracellular spaces. Proteomics. 2014;14:2307–2318. doi: 10.1002/pmic.201400066. [DOI] [PubMed] [Google Scholar]
- Kim SG, Wang Y, Lee KH, et al. In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J Proteomics. 2013;78:58–71. doi: 10.1016/j.jprot.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Kim ST, Kim SG, Agrawal GK, et al. Rice proteomics: a model system for crop improvement and food security. Proteomics. 2014;14:593–610. doi: 10.1002/pmic.201300388. [DOI] [PubMed] [Google Scholar]
- Kim ST, Kim SG, Kang YH, et al. Proteomics analysis of rice lesion mimic mutant (sp/1) reveals tightly localized probenazole-induced protein (PBZ1) in cells undergoing programmed cell death. J Proteome Res. 2008;7:1750–1760. doi: 10.1021/pr700878t. [DOI] [PubMed] [Google Scholar]
- Koga H, Dohi K, Nishiuchi T, et al. Proteomic analysis of susceptible rice plants expressing the whole plant-specific resistance against Magnaporthe oryzae: involvement of a thaumatin-like protein. Physiol Mol Plant Pathol. 2012;77:60–66. doi: 10.1016/j.pmpp.2011.12.001. [DOI] [Google Scholar]
- Kouzai Y, Kimura M, Watanabe M, et al. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 2018;217:771–783. doi: 10.1111/nph.14849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bimolata W, Kannan M, et al. Comparative proteomics reveals differential induction of both biotic and abiotic stress response associated proteins in rice during Xanthomonas oryzae pv. oryzae infection. Funct Integr Genomics. 2015;15:425–437. doi: 10.1007/s10142-014-0431-y. [DOI] [PubMed] [Google Scholar]
- Kushalappa AC, Gunnaiah R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 2013;18:522–531. doi: 10.1016/j.tplants.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Lee J, Bricker TM, Lefevre M, et al. Proteomic and genetic approaches to identifying defence-related proteins in rice challenged with the fungal pathogen Rhizoctonia solani. Mol Plant Pathol. 2006;7:405–416. doi: 10.1111/j.1364-3703.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Teng S, et al. Proteomics analysis of rice proteins up-regulated in response to bacterial leaf streak disease. J Plant Biol. 2012;55:316–324. doi: 10.1007/s12374-011-0346-2. [DOI] [Google Scholar]
- Li Y, Nie Y, Zhang Z, et al. Comparative proteomic analysis of methyl jasmonate-induced defense responses in different rice cultivars. Proteomics. 2014;14:1088–1101. doi: 10.1002/pmic.201300104. [DOI] [PubMed] [Google Scholar]
- Li Y, Ye Z, Nie Y, et al. Comparative phosphoproteome analysis of Magnaporthe oryzae-responsive proteins in susceptible and resistant rice cultivars. J Proteomics. 2015;115:66–80. doi: 10.1016/j.jprot.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Nie Y, et al. Proteomic analysis of salicylic acid-induced resistance to Magnaporthe oryzae in susceptible and resistant rice. Proteomics. 2012;12:2340–2354. doi: 10.1002/pmic.201200054. [DOI] [PubMed] [Google Scholar]
- Liang X, Chen X, Li C, et al. Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci Rep. 2017;7:1–15. doi: 10.1038/s41598-017-02643-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Ang L, Weilin Z. Current understanding of the molecular players involved in resistance to rice planthoppers. Pest Manag Sci. 2019;75:2566–2574. doi: 10.1002/ps.5487. [DOI] [PubMed] [Google Scholar]
- Liu C, Du B, Hao F, et al. Dynamic metabolic responses of brown planthoppers towards susceptible and resistant rice plants. Plant Biotechnol J. 2017;15:1346–1357. doi: 10.1111/pbi.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Hao F, Hu J, et al. Revealing different systems responses to brown planthopper infestation for pest susceptible and resistant rice plants with the combined metabonomic and gene-expression analysis. J Proteome Res. 2010;9:6774–6785. doi: 10.1021/pr100970q. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang Z, Xu W, et al. Bacillus pumilus LZP02 promotes rice root growth by improving carbohydrate metabolism and phenylpropanoid biosynthesis. Mol Plant Microbe Interact. 2020;33:1222–1231. doi: 10.1094/MPMI-04-20-0106-R. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang X, Tzin V, et al. Combined transcriptome and metabolome analyses to understand the dynamic responses of rice plants to attack by the rice stem borer Chilo suppressalis (Lepidoptera: Crambidae) BMC Plant Biol. 2016;16:1–17. doi: 10.1186/s12870-016-0946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lu S, Liu K, et al. Proteomics: a powerful tool to study plant responses to biotic stress. Plant Methods. 2019;15:1–20. doi: 10.1186/s13007-019-0515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HP, Luo T, Fu HW, et al. Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat Plants. 2018;4:338–344. doi: 10.1038/s41477-018-0152-7. [DOI] [PubMed] [Google Scholar]
- Ludwig C, Gillet L, Rosenberger G, et al. Data-independent acquisition-based SWATH - MS for quantitative proteomics: a tutorial. Mol Syst Biol. 2018;14:1–23. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Sheng C, Qiao L, et al. A comparative proteomic approach to identify defence-related proteins between resistant and susceptible rice cultivars challenged with the fungal pathogen Rhizoctonia solani. Plant Growth Regul. 2020;90:73–88. doi: 10.1007/s10725-019-00551-w. [DOI] [Google Scholar]
- Ma Z, Wang L, Zhao M, et al. ITRAQ proteomics reveals the regulatory response to Magnaporthe oryzae in durable resistant vs. susceptible rice genotypes. PLoS One. 2020;15:1–20. doi: 10.1371/JOURNAL.PONE.0227470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Paranidharan V, Erban A, et al. The metabolic response of suspension-cultured cells from blast-resistant and -susceptible rice (Oryza sativa L.) genotypes to a Pyricularia oryzae elicitor. Indian Phytopathol. 2019;72:195–202. doi: 10.1007/s42360-019-00131-y. [DOI] [Google Scholar]
- Mahmood T, Jan A, Kakishima M, et al. Proteomic analysis of bacterial-blight defense-responsive proteins in rice leaf blades. Proteomics. 2006;6:6053–6065. doi: 10.1002/pmic.200600470. [DOI] [PubMed] [Google Scholar]
- Mahmood T, Jan A, Komatsu S. Proteomic analysis of bacterial blight defence signalling pathway using transgenic rice overexpressing thaumatin-like protein. Biol Plant. 2009;53:285–293. doi: 10.1007/s10535-009-0052-9. [DOI] [Google Scholar]
- Mahmood T, Kakishima M, Komatsu S. Proteome analysis of Probenazole-effect in Rice-bacterial blight interactions. Protein Pept Lett. 2009;16:1041–1052. doi: 10.2174/092986609789055331. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dangl JL. Signal transduction in the plant immune response. Trends Biochem Sci. 2000;25:79–82. doi: 10.1016/S0968-0004(99)01532-7. [DOI] [PubMed] [Google Scholar]
- Meena KK, Sorty AM, Bitla UM, et al. Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci. 2017;8:1–25. doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Gupta R, Min CW, et al. Proteomics of Rice—Magnaporthe oryzae interaction: what have we learned so far? Front Plant Sci. 2019;10:1–14. doi: 10.3389/fpls.2019.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula K, Choudhary P, Ghosh S, et al. Comparative nuclear proteomics analysis provides insight into the mechanism of signaling and immune response to blast disease caused by Magnaporthe oryzae in Rice. Proteomics. 2019;19:1800188. doi: 10.1002/pmic.201800188. [DOI] [PubMed] [Google Scholar]
- Nawaz G, Usman B, Peng H, et al. Knockout of pi21 by crispr/cas9 and itraq-based proteomic analysis of mutants revealed new insights into M. oryzae resistance in elite rice line. Genes. 2020;11:1–24. doi: 10.3390/genes11070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norvienyeku J, Lin L, Waheed A et al (2020) Bayogenin 3-O-cellobioside confers non cultivar-specific defense against the rice blast fungus Pyricularia oryzae. Plant Biotechnol J. 10.1111/pbi.13488 [DOI] [PMC free article] [PubMed]
- Okazaki Y, Saito K. Integrated metabolomics and phytochemical genomics approaches for studies on rice. Gigascience. 2016;5:1–7. doi: 10.1186/s13742-016-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Beckmann M, Zubair H, et al. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009;59:723–737. doi: 10.1111/j.1365-313X.2009.03912.x. [DOI] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14:290–295. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Peng L, Zhao Y, Wang H, et al. Comparative metabolomics of the interaction between rice and the brown planthopper. Metabolomics. 2016;12:132. doi: 10.1007/s11306-016-1077-7. [DOI] [Google Scholar]
- Peyraud R, Dubiella U, Barbacci A, et al. Advances on plant–pathogen interactions from molecular toward systems biology perspectives. Plant J. 2017;90:720–737. doi: 10.1111/tpj.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka A, Jedrzejczak-Rey N, Bednarek P. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 2015;206:948–964. doi: 10.1111/nph.13325. [DOI] [PubMed] [Google Scholar]
- Piasecka A, Kachlicki P, Stobiecki M. Analytical methods for detection of plant metabolomes changes in response to biotic and abiotic stresses. Int J Mol Sci. 2019;20:379. doi: 10.3390/ijms20020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathi NB, Palit P, Madhu P, et al. Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani. Plant Physiol Biochem. 2018;130:258–266. doi: 10.1016/j.plaphy.2018.07.012. [DOI] [PubMed] [Google Scholar]
- Rakwal R, Komatsu S. Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis. 2000;21:2492–2500. doi: 10.1002/1522-2683(20000701)21:12<2492::AID-ELPS2492>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Salem MA, De Souza LP, Serag A, et al. Metabolomics in the context of plant natural products research: from sample preparation to metabolite analysis. Metabolites. 2020;10:1–30. doi: 10.3390/metabo10010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TR, Fischer S, Wohlgemuth G, et al. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics. 2010;6:451–465. doi: 10.1007/s11306-010-0218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarim KM, Srivastava R, Ramteke PW (2020) Next-generation Omics Technologies for Exploring Complex Metabolic Regulation during plant-microbe interaction microbial services in restoration ecology. Elsevier Inc. 10.1016/b978-0-12-819978-7.00009-9
- Sarwat M, Ahmad A, Abdin MZ. Stress signaling in plants: genomics and proteomics perspective, volume 1. 2013. [Google Scholar]
- Sato K, Kadota Y, Shirasu K. Plant immune responses to parasitic nematodes. Front Plant Sci. 2019;10:1–14. doi: 10.3389/fpls.2019.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary S, Willocquet L, Pethybridge SJ, et al. The global burden of pathogens and pests on major food crops. Nat Ecol Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- Siciliano I, Amaral Carneiro G, Spadaro D, et al. Jasmonic acid, Abscisic acid, and salicylic acid are involved in the Phytoalexin responses of Rice to Fusarium fujikuroi, a high gibberellin producer pathogen. J Agric Food Chem. 2015;63:8134–8142. doi: 10.1021/acs.jafc.5b03018. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Bist V, Srivastava S, et al. Unraveling aspects of Bacillus amyloliquefaciens mediated enhanced production of rice under biotic stress of Rhizoctonia solani. Front Plant Sci. 2016;7:1–16. doi: 10.3389/fpls.2016.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharti WS, Nose A, Zheng SH. Metabolite profiling of sheath blight disease resistance in rice: in the case of positive ion mode analysis by CE/TOF-MS. Plant Prod Sci. 2016;19:279–290. doi: 10.1080/1343943X.2016.1140006. [DOI] [Google Scholar]
- Suharti WS, Nose A, Zheng SH. Metabolomic study of two rice lines infected by Rhizoctonia solani in negative ion mode by CE/TOF-MS. J Plant Physiol. 2016;206:13–24. doi: 10.1016/j.jplph.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Suharti WS, Nose A, Zheng S-H. Canavanine involvement in the interaction of rice lines and Rhizoctonia solani. Acta Physiol Plant. 2016;39:37. doi: 10.1007/s11738-016-2331-3. [DOI] [Google Scholar]
- Sun R, Qin S, Zhang T, et al. Comparative phosphoproteomic analysis of blast resistant and susceptible rice cultivars in response to salicylic acid. BMC Plant Biol. 2019;19:1–15. doi: 10.1186/s12870-019-2075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TK, Sang GK, Du HH, et al. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics. 2004;4:3569–3578. doi: 10.1002/pmic.200400999. [DOI] [PubMed] [Google Scholar]
- Tan BC, Lim YS, Lau SE. Proteomics in commercial crops: an overview. J Proteomics. 2017;169:176–188. doi: 10.1016/j.jprot.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Tian D, Yang L, Chen Z, et al. Proteomic analysis of the defense response to Magnaporthe oryzae in rice harboring the blast resistance gene Piz-t. Rice. 2018;11:47. doi: 10.1186/s12284-018-0240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunezuka H, Fujiwara M, Kawasaki T, et al. Proteome analysis of programmed cell death and defense signaling using the rice lesion mimic mutant cdr2. Mol Plant Microbe Interact. 2005;18:52–59. doi: 10.1094/MPMI-18-0052. [DOI] [PubMed] [Google Scholar]
- Uawisetwathana U, Chevallier OP, Xu Y, et al. Global metabolite profiles of rice brown planthopper-resistant traits reveal potential secondary metabolites for both constitutive and inducible defenses. Metabolomics. 2019;15:151. doi: 10.1007/s11306-019-1616-0. [DOI] [PubMed] [Google Scholar]
- Valentino G, Graziani V, D’Abrosca B, et al. NMR-based plant metabolomics in nutraceutical research: an overview. Molecules. 2020;25:1444. doi: 10.3390/molecules25061444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valette M, Rey M, Gerin F, et al. A common metabolomic signature is observed upon inoculation of rice roots with various rhizobacteria. J Integr Plant Biol. 2020;62:228–246. doi: 10.1111/jipb.12810. [DOI] [PubMed] [Google Scholar]
- Vanderschuren H, Lentz E, Zainuddin I, et al. Proteomics of model and crop plant species: status, current limitations and strategic advances for crop improvement. J Proteomics. 2013;93:5–19. doi: 10.1016/j.jprot.2013.05.036. [DOI] [PubMed] [Google Scholar]
- Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:1–10. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hajano JUD, Ren Y, et al. iTRAQ-based quantitative proteomics analysis of rice leaves infected by Rice stripe virus reveals several proteins involved in symptom formation. Virol J. 2015;12:1–21. doi: 10.1186/s12985-015-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Zhang A, et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019;29:820–831. doi: 10.1038/s41422-019-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim SG, Wu J, et al. Secretome analysis of the rice bacterium Xanthomonas oryzae (Xoo) using in vitro and in planta systems. Proteomics. 2013;13:1901–1912. doi: 10.1002/pmic.201200454. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu Q, Du L et al (2020) Transcriptomic and Metabolomic responses of Rice plants to Cnaphalocrocis medinalis Caterpillar infestation. Insects 11. 10.3390/insects11100705 [DOI] [PMC free article] [PubMed]
- Wei Z, Hu W, Lin Q, et al. Understanding rice plant resistance to the Brown Planthopper (Nilaparvata lugens): a proteomic approach. Proteomics. 2009;9:2798–2808. doi: 10.1002/pmic.200800840. [DOI] [PubMed] [Google Scholar]
- Will T, Furch AC, Zimmermann MR. How phloem-feeding insects face the challenge of phloem-located defenses. Front Plant Sci. 2013;4:1–12. doi: 10.3389/fpls.2013.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Gong F, Cao D, et al. Advances in crop proteomics: PTMs of proteins under abiotic stress. Proteomics. 2016;16:847–865. doi: 10.1002/pmic.201500301. [DOI] [PubMed] [Google Scholar]
- Wu Y, Mirzaei M, Haynes PA (2017) Proteomics of Rice-our Most valuable food crop proteomics. In: Food science: from farm to fork. Elsevier Inc. 10.1016/B978-0-12-804007-2.00002-3
- Xiang C, Yang X, Peng D, et al. Proteome-wide analyses provide new insights into the compatible interaction of Rice with the root-knot nematode Meloidogyne graminicola. Int J Mol Sci. 2020;21:5640. doi: 10.3390/ijms21165640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Ni H, Chen Q, et al. Comparative proteomic analysis reveals the cross-talk between the responses induced by H2O2and by long-term rice black-streaked dwarf virus infection in rice. PLoS One. 2013;8:1–14. doi: 10.1371/journal.pone.0081640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XH, Wang C, Li SX, et al. Friend or foe: differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci Rep. 2015;5:1–14. doi: 10.1038/srep13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Dai L, Xia H, et al. Comparative proteomic analysis of rice stripe virus (RSV)-resistant and-susceptible rice cultivars. Aust J Crop Sci. 2013;7:588–593. [Google Scholar]
- Yasmin S, Hafeez FY, Mirza MS et al (2017) Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front Microbiol 8. 10.3389/fmicb.2017.01895 [DOI] [PMC free article] [PubMed]
- Yu CL, Yan SP, Wang CC, et al. Pathogenesis-related proteins in somatic hybrid rice induced by bacterial blight. Phytochemistry. 2008;69:1989–1996. doi: 10.1016/j.phytochem.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Yu L, Wang W, Zeng S, et al. Label-free quantitative proteomics analysis of Cytosinpeptidemycin responses in southern rice black-streaked dwarf virus-infected rice. Pestic Biochem Physiol. 2018;147:20–26. doi: 10.1016/j.pestbp.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Zaynab M, Fatima M, Abbas S, et al. Microbial pathogenesis role of secondary metabolites in plant defense against pathogens. Microb Pathog. 2018;124:198–202. doi: 10.1016/j.micpath.2018.08.034. [DOI] [PubMed] [Google Scholar]
- Zha W, You A. Comparative iTRAQ proteomic profiling of proteins associated with the adaptation of brown planthopper to moderately resistant vs. susceptible rice varieties. PLoS One. 2020;15:1–13. doi: 10.1371/journal.pone.0238549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li Y, Guo J, et al. Lipid profiles reveal different responses to brown planthopper infestation for pest susceptible and resistant rice plants. Metabolomics. 2018;14:0. doi: 10.1007/s11306-018-1422-0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yin F, Xiao S, et al. Proteomic analysis of the rice (Oryza officinalis) provides clues on molecular tagging of proteins for brown planthopper resistance. BMC Plant Biol. 2019;19:1–11. doi: 10.1186/s12870-018-1622-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhu K, Li L et al (2020) Proteomics of the honeydew from the Brown Planthopper and green Rice leafhopper reveal they are rich in proteins from insects, Rice Plant and bacteria. Insects 11. 10.3390/insects11090582 [DOI] [PMC free article] [PubMed]
- Zogli P, Pingault L, Grover S, et al. Ento(o)mics: the intersection of ‘omic’ approaches to decipher plant defense against sap-sucking insect pests. Curr Opin Plant Biol. 2020;56:153–161. doi: 10.1016/j.pbi.2020.06.002. [DOI] [PubMed] [Google Scholar]