Abstract
Background/Aims
This study aimed to characterize the changes in the gut microbiota of irritable bowel syndrome (IBS) patients and to investigate the consequent alterations in bacterial functions.
Methods
We performed 16S rRNA metagenomic sequencing and a phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analyses using fecal samples from control (n=12) and diarrhea-dominant IBS patients (n=7).
Results
The samples were clustered by the principal coordinates analysis depending on the presence of IBS (p=0.003). In the IBS patients, the abundances of Acidaminococcaceae, Sutterellaceae, and Desulfovibrionaceae were significantly increased, while those of Enterococcaceae, Leuconostocaceae, Clostridiaceae, Peptostreptococcaceae, and Lachnospiraceae were significantly decreased. The PICRUSt results indicated that two orthologues involved in secondary bile acid biosynthesis were significantly decreased in IBS patients. Modules involved in multidrug resistance, lipopolysaccharide biosynthesis, the reductive citrate cycle, and the citrate cycle were significantly increased in the IBS patients. In contrast, modules involved in cationic antimicrobial peptide resistance, and some transport systems were more abundant in controls than in IBS patients.
Conclusions
Changes in the gut microbiota composition in IBS patients lead to alterations in bacterial functions, such as bile acid transformation and the induction of inflammation, which is a known pathophysiological mechanism of IBS.
Keywords: Irritable bowel syndrome, Gastrointestinal microbiome
INTRODUCTION
Irritable bowel syndrome (IBS) is a functional bowel disorder that is characterized by recurrent abdominal pain associated with a change in bowel habits or defecation.1 Multiple pathophysiological mechanisms have been suggested as inducing factors of IBS, including gut-brain dysregulation, changes in bile acid metabolism, altered gut permeability, immune dysfunction, and alterations in gut microbiota.2,3
The IBS symptom is associated with distinct compositions of fecal microbiota.4-7 Several groups of Firmicutes and Proteobacteria have been reported to have correlations with IBS symptom scores in patients.7 In addition, the severity of IBS symptoms may be associated with microbial richness, presence of methanogens, and enrichment of Clostridiales or Prevotella species.6 However, previous studies have limitations in the functional analysis.
The gut microbiota is associated with the bile acid modification, which is involved in the occurrence of IBS. The gut microbiota affects the metabolism of bile acids and increases their excretion.8 The modification of bile acid profiles has been suggested as one of the mechanisms for symptom generation in patients with IBS and is thus a potential therapeutic target for IBS. Therefore, it is necessary to determine whether the compositional changes of microbiota in IBS are associated with the bile acid profiles. Regarding this issue, a previous study proposed that the alterations of bile acid profiles in IBS may be secondary to dysbiosis.9 However, the study investigated the fecal microbiota composition by real-time polymerase chain reaction (PCR), not by metagenomic sequencing, which may limit the analysis of the entire microbial community.
Therefore, we hypothesized that the changes in the gut microbiota and the consequent alterations in the bacterial functions may contribute to the occurrence and progression of IBS. Thus, the aim of this study was to characterize the changes in the gut microbiota in IBS and to investigate the consequent alterations in bacterial functions.
MATERIALS AND METHODS
1. Subjects
Subjects were prospectively enrolled at the Department of Gastroenterology of Seoul National University Bundang Hospital between September 2013 and September 2017. All subjects were from Korea and received colonoscopies and questionnaires about IBS symptoms under the supervision of a well-trained interviewer. We investigated the patients’ symptoms by a validated questionnaire, and classified the subjects into IBS or control groups according to the Rome III criteria. The control group was defined as subjects with no gastrointestinal symptoms and no colonoscopic lesions. The control group consisted of subjects who underwent colonoscopy for the purpose of health checkup. We excluded post-infectious IBS patients, and those who took antibiotics or probiotics within three months. Subjects were excluded if there was a history of inflammatory bowel disease, abdominal operation, severe systemic disease, and malignancy. Age and sex matching could not be performed due to the small number of subjects. The IBS patients were treated with concomitant medications for IBS: antispasmotic drug such as pinaverium bromide, tiropramide, trimebutine maleate, or loperamide and ramosetron.
2. Consent and institutional review board
All patients provided written informed consent to participate in this study. The study was performed in accordance with the 53rd World Medical Association Declaration of Helsinki. This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (IRB number: B-1305/202-003). This study protocol has been registered at ClinicalTrials.gov (NCT03675100).
Some of the control samples (FMT04, 07, 09, 11, 14, 17, 20, 23, 26, and 29) were obtained from the cohort collected by Prof. H.Y. under the topic of fecal microbiota transplantation for Clostridium difficile infection, which was approved by the IRB of Seoul National University Bundang Hospital (IRB number: B-1507-305-006).
3. Symptom assessment questionnaire
All subjects completed a validated Rome III criteria-based bowel disease questionnaire, which was translated from the original Bowel Disease Questionnaire into Korean (Korean Bowel Disease Questionnaire).10-12 According to the validated questionnaires, patients were asked to rate the frequency and severity of symptoms experienced over the last 3 months prior to the study. Patients who met IBS on the Rome III criteria were consecutively enrolled in the patient group;13 IBS was defined as recurrent abdominal pain or discomfort for at least 3 days per month in the past 3 months with at least 2 of the following; improvement with defecation, onset associated with a change in frequency of stool, and onset associated with a change in form of stool. The IBS patients were categorized as having IBS with diarrhea (IBS-D) on the basis of the Rome III criteria.
4. Microbiota analysis in fecal samples
Fecal samples (≥5 g per subject) were collected by patients at home on the day before the hospital visit, immediately frozen at –20°C, brought frozen to the laboratory, and stored frozen at –80°C until analysis.
Total DNA was extracted from the fecal samples (n=19) using QIAamp® DNA stool mini kits (Qiagen, Venlo, Netherlands) following the manufacturer’s recommendations, as described previously.14 The V3-V4 region of the 16S rRNA gene was amplified by PCR using the primers 341F (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3’) and 805R (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’). The PCR products were confirmed by electrophoresis and were purified using a QIAquick PCR purification kit (Qiagen). The purified PCR products were tagged with Illumina indices and adapters from a Nextera® XT Index Kit (Illumina, San Diego, CA, USA). Short DNA fragments were eliminated using a FavorPrepTM DNA purification kit (Favorgen, Ping-Tung, Taiwan). The PCR amplicons were quantified using a Quant-iTTM PicoGreenTM dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). After pooling 300 ng of DNA per sample, the PCR products were purified with a FavorPrepTM DNA gel extraction kit (Favorgen). The DNA extraction and PCR processes for the preparation of the control samples were conducted by ChunLab. Inc. (Seoul, Korea), with an identical protocol.
The quality assessment for confirmation of the DNA integrity and product size was conducted on a Bioanalyzer 2100 instrument (Agilent, Santa Clara, CA, USA) using a DNA 7500 chip at ChunLab, Inc. Metagenomic sequencing was performed using the Illumina MiSeq platform at ChunLab, Inc.
The raw reads were processed starting with the quality check and filtering of low quality (<Q25) reads using Trimmomatic 0.32.15 The paired-end sequence data were merged using PANDAseq.16 The primer sequences were then trimmed with an in-house program of ChunLab, Inc. at a similarity cutoff of 0.8. Nonspecific amplicons that did not encode 16S rRNA were identified by using the HMMER program hmmsearch with 16S rRNA profiles.17 Sequences were denoised using DUDE-Seq,18 and nonredundant reads were extracted through UCLUST clustering.19 The EzBioCloud database was utilized for taxonomic assignment using USEARCH (8.1.1861_i86linux32)19 followed by more precise pairwise alignment.20 UCHIME21 and the nonchimeric 16S rRNA database from EzBioCloud were used to detect chimeras for reads that contained a less than 97% best hit similarity rate. The sequence data were then clustered using CD-HIT22 and UCLUST.19
To visualize the sample differences, the principal coordinates analysis was performed with unweighted UniFrac.23 Principal coordinates analysis plots were generated using the ade4 package and s.class function in R. Microbial diversity (observed OTU count, Chao1, Shannon indices, and phylogenetic diversity) was examined using BIOiPLUG (Chunlab, Inc.).
5. Phylogenetic investigation of communities by reconstruction of unobserved states
To predict the microbial community’s functional capabilities, we performed a phylogenetic investigation of the communities by reconstruction of the unobserved states (phylogenetic investigation of communities by reconstruction of unobserved states, PICRUSt).24 This analysis enabled us to infer alterations in functional markers of microbiota based on the Kyoto Encyclopedia of Genes and Genomes database.
6. Linear discriminant analysis effect size analysis
A linear discriminant analysis effect size (LEfSe) analysis (http://huttenhower.sph.harvard.edu/galaxy/) was conducted to determine the taxonomic compositions that were significantly different between the control and IBS groups.25 The conditions for the LEfSe analysis were as follows: (1) an α value of less than 0.05 for the factorial Kruskal-Wallis test between the control and IBS groups; (2) an α value of less than 0.05 for the pairwise Wilcoxon test among the taxonomic compositions; (3) a threshold of the logarithmic linear discriminant analysis score for discriminative features of less than 2.0; and (4) a multiclass analysis set as all-against-all.
7. Statistical analysis
Statistical analyses were performed using PASW Statistics version 18.0 (2019; SPSS Inc., Chicago, IL, USA). Data were expressed as mean±standard error of the mean. We performed the Wilcoxon rank sum test to compare continuous variables between two groups, control and IBS. p-values <0.05 were considered statistically significant.
RESULTS
1. Control and IBS patient samples and sequencing results
We sequenced 19 fecal samples from 19 individuals, including subjects with IBS-D (n=7) and controls (n=12) (Table 1). Thirteen men and six women were included. The mean age was 48.2 years, and showed no significant difference according to sex: 46.1 years in men and 52.7 years in women (p=0.455) (Supplementary Table 1).
Table 1.
Baseline Characteristics of Subjects and Samples
| Group | Study ID | Sex | Age, yr |
Disease | Total valid reads |
OTUs count (normalization at 25,592 reads) |
|---|---|---|---|---|---|---|
| Control | CNC03 | M | 54 | Normal | 27,394 | 646 |
| CNC11 | F | 53 | Normal | 48,617 | 584 | |
| FMT04 | M | 20 | Normal | 107,670 | 833 | |
| FMT07 | M | 57 | Normal | 75,414 | 457 | |
| FMT09 | M | 46 | Normal | 76,712 | 271 | |
| FMT11 | M | 45 | Normal | 66,871 | 756 | |
| FMT14 | M | 43 | Normal | 49,952 | 272 | |
| FMT17 | F | 57 | Normal | 53,404 | 393 | |
| FMT20 | F | 40 | Normal | 50,668 | 126 | |
| FMT23 | F | 38 | Normal | 82,479 | 333 | |
| FMT26 | M | 40 | Normal | 57,878 | 215 | |
| FMT29 | M | 47 | Normal | 85,164 | 947 | |
| IBS | IBS25 | M | 40 | IBS-D | 29,733 | 568 |
| IBS90 | M | 51 | IBS-D | 140,628 | 610 | |
| IBS100 | F | 55 | IBS-D | 59,812 | 438 | |
| IBS101 | F | 73 | IBS-D | 48,611 | 567 | |
| IBS102 | M | 60 | IBS-D | 85,835 | 510 | |
| IBS103 | M | 36 | IBS-D | 71,490 | 564 | |
| IBS105 | M | 60 | IBS-D | 25,592 | 454 |
OTU, operational taxonomic unit; M, male; F, female; IBS, irritable bowel syndrome; IBS-D, IBS with diarrhea.
After data processing, on average, each sample had 65,470±6,481 valid reads and 502±48 operational taxonomic units (OTUs) (when normalized at 25,592 reads) (Table 1). The rarefaction curve analysis showed that the sequencing depth was sufficient to recover most of the microbial community diversity (Supplementary Fig. 1).
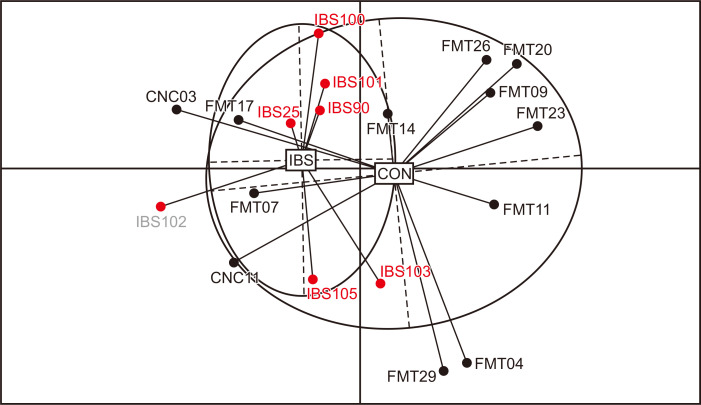
According to the principal coordinates analysis based on OTU counts, samples were significantly separated depending on the presence of IBS (UniFrac, p=0.003) (Fig. 1).
Fig. 1.
Clustering of samples according to irritable bowel syndrome (IBS) status. Principal coordinates analysis (PCoA) of 19 samples. Control (CON) samples are indicated with black dots, and IBS samples are indicated with red dots.
2. Diversity of the fecal microbiota
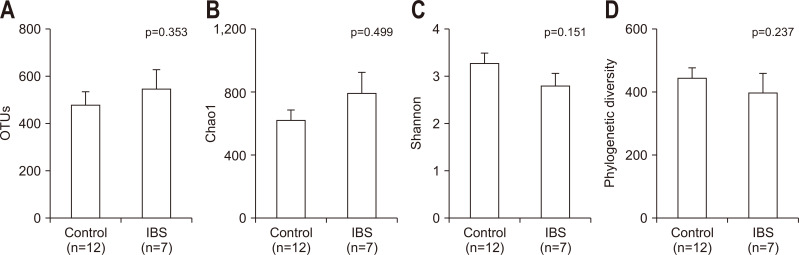
The number of observed OTUs, the Chao1 index, the Shannon index, and the phylogenetic diversity index were used to evaluate the bacterial diversity in different disease statuses. However, the difference was not significant between IBS patients and normal controls (Fig. 2).
Fig. 2.
Diversity indices of the gut microbiota. The indices indicating the species richness, i.e., the (A) observed operational taxonomic unit (OTU)count and (B) Chao1 indices, and those indicating the species diversity, i.e., the (C) Shannon and (D) phylogenetic diversity indices, did not differ significantly between the normal control and irritable bowel syndrome (IBS) groups. p-values were evaluated with the Wilcoxon rank sum test.
3. Composition of the fecal microbiota
The fecal microbiota of the normal control and IBS samples were primarily composed of Firmicutes (control, 72.796%±4.936%; IBS, 56.643%±10.453%) and Bacteroidetes (control, 20.446%±4.866%; IBS, 36.329%±11.369%) (Supplementary Fig. 2A). Other OTUs included Verrucomicrobia (control, 1.805%±1.713%; IBS, 0.764%±0.579%), Proteobacteria (control, 0.507%±0.133%; IBS, 3.139%±1.969%), Fusobacteria (control, 0.364%±0.184%; IBS, 2.273%±2.270%), and Actinobacteria (control, 4.074%±1.471%; IBS, 0.818%± 0.307%). The Firmicutes/Bacteroidetes ratio (control, 43.781±33.594; IBS, 15.568±10.511), and the abundance of Firmicutes, Verrucomicrobia, and Actinobacteria were reduced in IBS samples on average in each group (Supplementary Fig. 2B, C, E, H). In contrast, Bacteroidetes, Proteobacteria, and Fusobacteria were relatively enriched in IBS samples compared to normal controls (Supplementary Fig. 2D, F, G). However, the difference was not significant. The microbiota compositions at each taxonomic level (phylum, class, order, family, genus, and species) and the p-values for the comparisons of the proportion of bacterial compositions are shown in Supplementary Dataset 1.
4. Alterations of fecal microbiota in IBS
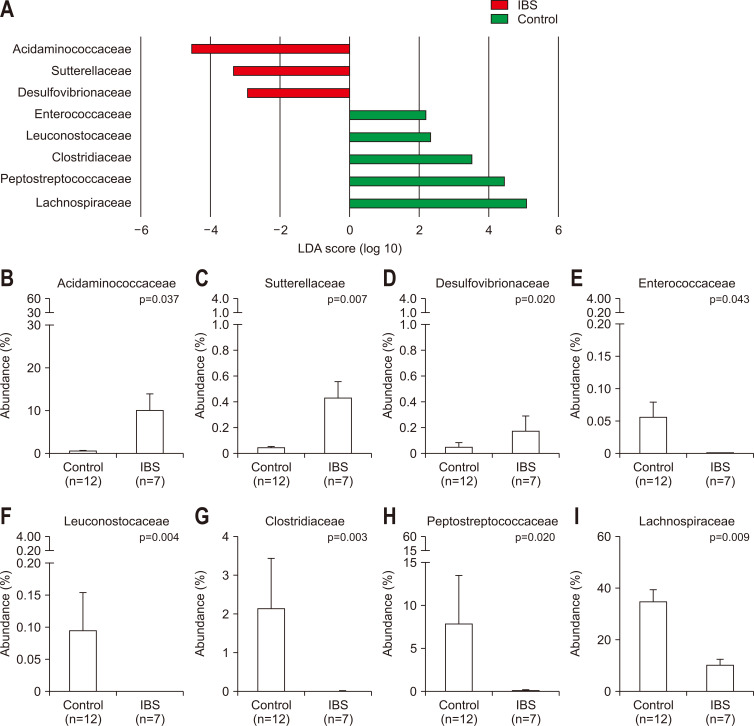
Although the comparison of phyla compositions did not show significant differences, the analysis at the family level identified significant changes in the microbial compositions in IBS patients. According to the LEfSe analysis results, the relative abundance of Desulfovibrionaceae, Sutterellaceae, and Acidaminococcaceae significantly increased in the IBS samples (Fig. 3A). In contrast, the relative abundance of Lachnospiraceae, Peptostreptococcaceae, Erysipelotrichaceae, Clostridiaceae, Leuconostocaceae, Enterococcaceae, and Actinomycetaceae significantly decreased in IBS samples (Fig. 3A). Acidaminococcaceae was significantly enriched in IBS samples (9.886%±4.001%) compared to normal controls (0.387%±0.211%, p=0.037) (Fig. 3B). In addition, Sutterellaceae and Desulfovibrionaceae, which were present in low abundance (<1% on average), were significantly more abundant in IBS (Sutterellaceae, 0.040%±0.015% in normal controls and 0.429%±0.128% in IBS patients, p=0.007; Desulfovibrionaceae, 0.052%±0.035% in normal controls and 0.174%±0.116% in IBS patients, p=0.020) (Fig. 3C and D). In contrast, Enterococcaceae and Leuconostocaceae with low abundance proportions (<1%) were significantly decreased in IBS patients (Enterococcaceae, 0.054%±0.026% in normal controls and 0.001%±0.001% in IBS patients, p=0.043; Leuconostocaceae, 0.093%±0.060% in normal controls and 0.000%±0.000% in IBS patients, p=0.004) (Fig. 3E and F). Clostridiaceae decreased from 2.134%±1.311% in the normal controls to 0.013%±0.007% in IBS patients (p=0.003) (Fig. 3G). Similarly, Peptostreptococcaceae was depleted in IBS patients (7.782%±5.631% in normal controls and 0.056%±0.024% in IBS patients, p=0.020) (Fig. 3H). In addition, Lachnospiraceae was significantly reduced in IBS patients (34.397%±4.899% in normal controls and 9.798%±2.935% in IBS patients, p=0.009) (Fig. 3I).
Fig. 3.
Differences in the microbiota composition between irritable bowel syndrome (IBS) samples and normal control samples. (A) Linear discriminant analysis (LDA) effect size (LEfSe) of the gut microbiota at the family level. Abundance ratios of the families that differed between the IBS and normal control groups according to LEfSe: (B) Acidaminococcaceae, (C) Sutterellaceae, (D) Desulfovibrionaceae, (E) Enterococcaceae, (F) Leuconostocaceae, (G) Clostridiaceae, (H) Peptostreptococcaceae, and (I) Lachnospiraceae. p-values were assessed with the Wilcoxon rank sum test.
5. Changes of functional capabilities associated with bile acids
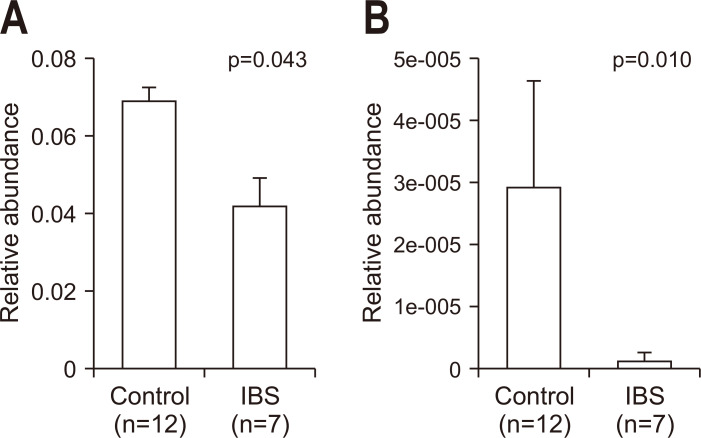
Two orthologues associated with secondary bile acid biosynthesis (pathway ko00121) were significantly decreased in IBS patients. K07007, the 3-dehydro-bile acid delta 4,6-reductase (baiN) was significantly decreased in IBS patients (p=0.043) (Fig. 4). In addition, K15874, bile acid 7 beta-dehydratase (baiI), was significantly reduced in IBS patients (p=0.010).
Fig. 4.
Changes in orthologues involved in secondary bile acid biosynthesis (pathway ko00121). Abundance of (A) K07007 (baiN, 3-dehydro-bile acid delta 4,6-reductase) and (B) K15874 (baiI, bile acid 7 beta-dehydratase) in normal controls (n=12) and IBS patients (n=7). p-values were evaluated with the Wilcoxon rank sum test.
IBS, irritable bowel syndrome.
6. Change of modules
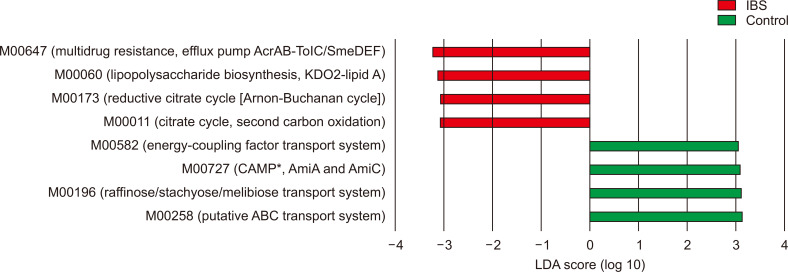
According to the LEfSe analysis of the PICRUSt results (the relative abundance of modules), the modules involved in multidrug resistance, lipopolysaccharide biosynthesis, reductive citrate cycle and citrate cycle were more abundant in IBS patients than controls (Fig. 5). In contrast, the modules contributing to the energy-coupling factor transport system, cationic antimicrobial peptide resistance, the raffinose/stachyose/melibiose transport system, and the putative ATP-binding cassette transport system were more abundant in normal controls than IBS patients.
Fig. 5.
Linear discriminant analysis (LDA) effect size of modules according to the occurrence of irritable bowel syndrome (IBS).
ABC, ATP-binding cassette. *CAMP, cationic antimicrobial peptide; Ami, N-acetylmuramoyl-L-alanine amidase.
DISCUSSION
In this study, we characterized the microbial changes in IBS and inferred their roles in the development and progression of IBS based on 16S rRNA metagenomic sequencing and PICRUSt, which predicts the changes in the functional markers of the microbiota. Our results suggest that the compositional alterations of the gut microbiota in IBS patients may result in changes in the bacterial functions, such as bile acid transformation and the induction of inflammation.
In the present study, an increase in the pathogenic bacteria, Desulfovibrionaceae, and a decrease in the beneficial bacteria, Lachnospiraceae, was observed in IBS patients (Fig. 3D and I). Members of Desulfovibrionaceae are sulfate-reducing bacteria, which produce H2S that can contribute to the progression of cancer in the colonic epithelium.26 The increase of Desulfovibrionaceae has not been identified in previous IBS studies. The present results showing an increased abundance of Desulfovibrionaceae are inconsistent with the decrease in Desulfovibrio spp. in IBS-D patients.27 However, this study27 was limited by analyzing the abundance using quantitative real-time PCR assays estimating approximately 300 species, which could not cover the same breadth of the bacterial spectrum as metagenomic sequencing covers (e.g., 1,301 bacterial species in this study). On the other hand, another study suggested an increased abundance of Desulfovibrio spp., which would be more plausible regarding the pathogenicity of the taxa.28 In contrast, our results showed a decrease in Lachnospiraceae, the butyrate-producing bacteria, in IBS. This result is consistent with the decreased abundance of Lachnospiraceae in cirrhosis exhibiting reduced levels of fecal bile salts, which reflects a drop in the transformation from primary to secondary bile acids.29 The Firmicutes fraction has been shown to be unstable in IBS patients.30 However, the abundance of Firmicutes decreased in patients with IBS in this study, which is inconsistent with the previous results.
Other taxa that differed between IBS and controls in the present study seem to have some association with IBS. As we investigated some evidence for the association with IBS, we found that Acidaminococcaceae, which was more abundant in IBS patients than controls in our study, was also found to be enriched in the fecal microbiota of patients with active major depressive disorder.31 This similar trend is plausible since IBS patients commonly suffer from psychiatric disorders such as anxiety and depression.32 Abundance of an unclassified member of the Peptostreptococcaceae correlates with the volume of a part of the brain (the right inferior segment of the circular sulcus) in IBS patients.33 Associations of other minor taxa (Sutterellaceae, Enterococcaceae, Leuconostocaceae, and Clostridiaceae) with IBS have yet to be identified.
To infer the alterations in the functional capabilities of the gut microbiota, we first investigated the microbial changes in patients with IBS in association with bile acid transformation. The bile acids and the gut microbiota interact with each other therefore influencing the composition of each. Primary bile acids synthesized in the liver are converted to secondary bile acids by bacterial activity in the intestine. Therefore, excessive bacterial activity can increase the proportion of secondary bile acids in the bile acid pool.34 In addition, not all gut bacteria have the ability to transform bile acids and survive the bactericidal effect of bile acids; therefore, alterations in gut microbiota composition can result in changes in bile acid composition. As shown in the present study, the gut microbiota of IBS patients had fewer orthologues involved in secondary bile acid synthesis. The decrease in orthologues in secondary bile acid synthesis in our study is consistent with a previous study that measured sera and fecal bile acid profiles and observed an increased proportion of primary bile acids in the fecal bile acids of IBS patients compared to healthy controls.9 Therefore, we presume that the dysbiosis in IBS may affect the bile acid profiles to induce or aggravate IBS.
Second, in IBS patients, there were higher levels of microbial orthologues involved in lipopolysaccharide (LPS) biosynthesis compared to normal controls. The increase in LPS biosynthesis indicates an increase in gram-negative bacteria. The increase of gram-negative bacteria and LPS induces intestinal inflammatory responses and increases intestinal tight junction permeability through the upregulation of Toll-like receptor-4-dependent expression of a membrane-associated protein, CD14.35 Consistently, IBS-D patients have higher serum levels of LPS than controls.36 Our results support the previous hypothesis that immune reactivity to bacterial-derived LPS may be involved in the development of IBS-D.36 Furthermore, the enhanced release of LPS-induced tumor necrosis factor-α in IBS patients has been linked to psychiatric comorbidities, specifically anxiety.37 Therefore, the increase of LPS biosynthesis modules in IBS may suggest a role for the enriched gram-negative bacteria in the development, symptoms, and psychiatric comorbidities of IBS.
The gut microbiota in IBS had an increased abundance of M00011, the module constituting the second carbon oxidation part of the citrate cycle. The citrate cycle is a metabolic pathway used by aerobic organisms to obtain energy from carbohydrates, fats, and proteins. Anaerobic bacteria are known to have an incomplete citrate cycle. However, there was no significant change in the abundance of the pathway indicating the entire citrate cycle (map00020 in the Kyoto Encyclopedia of Genes and Genomes). Changes in the transport system modules show alterations in the systems by which the bacteria import nutrients and export toxic molecules,38,39 reflecting the compositional changes of the microbiota. However, the roles of the modules were broad and general, inferring less association with IBS.
This study has a limitation in that the bacterial functional genes were not directly measured. The abundance of bacterial functional genes was presumed through the abundance of the bacteria that possess the functional genes. Despite this limitation, the present results suggest what the compositional changes of microbiota actually mean biologically and how they may influence the symptoms of IBS. In addition, the number of control and IBS patients is small. The sample size was limited since most of the patients felt discomfort in bringing the samples to the hospital although we had cordially requested. The sample size of this study (IBS-D, n=7; controls, n=12) is modest according to a systematic review which suggested 20 per group as the appropriate sample size for statistical difference in a microbiota analysis.40 However, despite the small sample size, our results are clear regarding the changes in the composition of the gut microbiota, which led to alterations in bacterial functions such as bile acid transformation and induction of inflammation, which is a well-known pathophysiological mechanism of IBS. Although the previous studies have assessed fecal microbial composition between healthy controls and IBS patients,5,27,40 this study has novelty in that we conducted the PICRUSt to predict the changes in the microbial community’s functional capabilities. This study also has a limitation that we could not include the IBS patients with constipation. Although this study was aimed to conduct in both of IBS patients with constipation and IBS-D patients, we could not persuade IBS patients with constipation to bring stool samples for this research. Lastly, the fecal samples may be less representative of gut microbial ecology as compared with mucosal samples. However, samples may have advantages on representing the luminal microbiome of the whole gastrointestinal tract.
In conclusion, changes in the gut microbiota composition in IBS patient may lead to alterations in bacterial functions, such as bile acid transformation and the induction of inflammation.
Supplemental Materials
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the government of the Republic of Korea (2016R1A2B4013133). In addition, this study was supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (MIST) of the Republic of Korea (project number: NRF-2016M3A9F3947027).
The raw datasets generated during the current study are available in the NCBI Sequence Read Archive (SRA accession number SRR9017435 ~ SRR9017453), https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=search_obj. The processed data generated and analyzed during this study are included in this published article as a Supplementary Dataset file.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
AUTHOR CONTRIBUTIONS
Guarantor of the article: N.K. Data analysis, statistical analysis, data interpretation and manuscript drafting: S.M.L. Study design, enrollment of the subjects, data interpretation, and critical revision: N.K. Enrollment of subjects: H.Y. Supervision of the manuscript: Y.S.K. Conduct of the experiments: S.I.C., J.H.P. Study supervision and funding acquisition: D.H.L. Approval of final manuscript: all authors.
REFERENCES
- 1.Mearin F, Lacy BE, Chang L, et al. Bowel disorders. Gastroenterology. 2016;150:1393–1407. doi: 10.1053/j.gastro.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 2.Camilleri M, Ford AC. Irritable bowel syndrome: pathophysiology and current therapeutic approaches. Handb Exp Pharmacol. 2017;239:75–113. doi: 10.1007/164_2016_102. [DOI] [PubMed] [Google Scholar]
- 3.Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointest Liver Physiol. 2017;312:G52–G62. doi: 10.1152/ajpgi.00338.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffery IB, O'Toole PW, Öhman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 6.Tap J, Derrien M, Törnblom H, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152:111–123. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 7.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 8.Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Dior M, Delagrèverie H, Duboc H, et al. Interplay between bile acid metabolism and microbiota in irritable bowel syndrome. Neurogastroenterol Motil. 2016;28:1330–1340. doi: 10.1111/nmo.12829. [DOI] [PubMed] [Google Scholar]
- 10.Choi YJ, Hwang SW, Kim N, Park JH, Oh JC, Lee DH. Association between SLC6A4 serotonin transporter gene lainked polymorphic region and ADRA2A -1291C>G and irritable bowel syndrome in Korea. J Neurogastroenterol Motil. 2014;20:388–399. doi: 10.5056/jnm14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YJ, Kim N, Yoon H, et al. Overlap between irritable bowel syndrome and functional dyspepsia including subtype analyses. J Gastroenterol Hepatol. 2017;32:1553–1561. doi: 10.1111/jgh.13756. [DOI] [PubMed] [Google Scholar]
- 12.Lee JY, Kim N, Park JH, et al. Expression of neurotrophic factors, tight junction proteins, and cytokines according to the irritable bowel syndrome subtype and sex. J Neurogastroenterol Motil. 2020;26:106–116. doi: 10.5056/jnm19099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology. 2006;130:1377–1390. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Lee SM, Kim N, Yoon H, Nam RH, Lee DH. Microbial changes and host response in F344 rat colon depending on sex and age following a high-fat diet. Front Microbiol. 2018;9:2236. doi: 10.3389/fmicb.2018.02236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee B, Moon T, Yoon S, Weissman T. DUDE-Seq: fast, flexible, and robust denoising for targeted amplicon sequencing. PLoS One. 2017;12:e0181463. doi: 10.1371/journal.pone.0181463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 20.Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 21.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbonero F, Gaskins HR. Sulfate-reducing bacteria in the human gut microbiome. In: Highlander SK, Rodriguez-Valera F, White BA, editors. Encyclopedia of metagenomics. Springer; Boston: 2015. pp. 617–619. [DOI] [Google Scholar]
- 27.Malinen E, Rinttilä T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 28.Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. 2015;9:318–331. doi: 10.5009/gnl14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajilić-Stojanović M, Jonkers DM, Salonen A, et al. Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am J Gastroenterol. 2015;110:278–287. doi: 10.1038/ajg.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang H, Ling Z, Zhang Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Lee C, Doo E, Choi JM, et al. The increased level of depression and anxiety in irritable bowel syndrome patients compared with healthy controls: systematic review and meta-analysis. J Neurogastroenterol Motil. 2017;23:349–362. doi: 10.5056/jnm16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labus JS, Hollister EB, Jacobs J, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017;5:49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith ME, Morton DG. Digestion and absorption. In: Smith ME, Morton DG, editors. The digestive system. 2nd ed. Churchill Livingstone; London: 2010. pp. 129–152. [DOI] [Google Scholar]
- 35.Guo S, Al-Sadi R, Said HM, Ma TY. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am J Pathol. 2013;182:375–387. doi: 10.1016/j.ajpath.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dlugosz A, Nowak P, D'Amato M, et al. Increased serum levels of lipopolysaccharide and antiflagellin antibodies in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2015;27:1747–1754. doi: 10.1111/nmo.12670. [DOI] [PubMed] [Google Scholar]
- 37.Liebregts T, Adam B, Bredack C, et al. Immune activation in patients with irritable bowel syndrome. Gastroenterology. 2007;132:913–920. doi: 10.1053/j.gastro.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 38.Zhang P. Structure and mechanism of energy-coupling factor transporters. Trends Microbiol. 2013;21:652–659. doi: 10.1016/j.tim.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pittayanon R, Lau JT, Yuan Y, et al. Gut microbiota in patients with irritable bowel syndrome: a systematic review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.