Abstract
Cadmium (Cd) contamination of rice is a serious food safety issue that has recently been gaining significant public attention. Therefore, reduction of Cd accumulation in rice grains is an important objective of rice breeding. The use of favourable alleles of Cd accumulating genes using marker-assisted selection (MAS) is theoretically feasible. In this study, we validated a segment covering OsHMA3-OsNramp5-OsNramp1 on chromosome 7 of japonica for establishing low-cadmium accumulating indica rice variety. The OsHMA3-OsNramp5-OsNramp1jap haplotype significantly decreased grain Cd concentration in middle-season indica genetic background. The improved 9311 carrying the OsHMA3-OsNramp5-OsNramp1jap haplotype with recurrent parent genome recovery of up to 91.6% resulted in approximately 31.8% decrease in Cd accumulation in the grain and with no penalty on yield. There is a genetic linkage-drag between OsHMA3-OsNramp5-OsNramp1 jap and the gene conditioning heading to days (HTD) in the early-season indica genetic background. Because the OsHMA3-OsNramp5-OsNramp1-Ghd7jap haplotype significantly increases grain Cd concentration and prolongs growth duration, the linkage-drag between OsHMA3-OsNramp5-OsNramp1 and Ghd7 should be broken down by large segregating populations or gene editing. A novel allele of OsHMA3 was identified from a wide-compatibility japonica cultivar, the expression differences of OsNramp1 and OsNramp5 in roots might contribute the Cd accumulating variation between japonica and indica variety.
Subject terms: Genetics, Plant sciences
Introduction
Contamination of agricultural soils with heavy metals is a serious threat to crop production. Cadmium (Cd), one of the most dangerous heavy metals, has long been recognised as a major threat to human health through contamination of the food chain1. Rice (Oryza sativa L.) is the staple food for more than half of the world’s population, as well as a crop with high Cd accumulating abiltiy2. To suppress Cd pollution of food, rice has a strict upper level of Cd content. According to the Codex Alimentarius Commission of the FAO/WHO, the maximum Cd concentration in polished rice should not exceed 0.4 mg/kg, whereas stricter limits were implemented in China, with a maximum level for Cd of 0.2 mg/kg3,4. Rice-derived products are subject to the risk of Cd contamination, and consequent threats to consumer health if rice is grown in Cd-contaminated agricultural soil5. In some areas of China, Japan, Thailand, Bangladesh, Indonesia and Korea, the production of rice grains with high Cd levels, and subsequent transfer of the contaminant to the human food chain, is a serious environmental and food safety issue6,7. Fortunately, rice germplasm carries a large number of genetic variations related to grain Cd concentration. Japonica cultivars generally accumulate lower concentrations of Cd than indica and aus cultivars2,8–10, suggesting that Cd accumulation is genetically controlled in rice. It should, therefore, be possible to breed rice cultivars with a low Cd accumulation based on genetic improvement. Cd, from soil to grains in rice, has been a subject of considerable research and several relevant genes have been identified in recent years, including OsIRT1, OsHMA3, OsNramp1, OsNramp5, OsHMA2, OsLCD and OsLCT111–19. Although genotypic differences in Cd concentration and several genes have been studied, only limited efforts have been made to breed rice with reduced Cd content, especially in indica cultivars.
Of the Cd transport-related genes of rice, OsHMA3, which is a major quantitative trait locus (QTL) mapped and cloned from low Cd-accumulating japonica cultivars (Akita 63 and Nipponbare), plays a transporter in Cd compartmentation into root cell vacuoles. Non-functional allele of OsHMA3 results in high efficiency of root-to-shoot Cd translocation12,13. OsNramp1 and OsNramp5 are metal transporter genes that have been suggested to mediate the adsorption of Cd from soil to root14–16. The expression of OsNRAMP1 in roots was found to be higher in high Cd-accumulating indica cultivars (Habataki, Anjana Dhan, Jarjan) than in low Cd-accumulating japonica varieties (Sasanishiki, Nipponbare)14. The expression of OsHMA3 and OsNRAMP5 in roots was lower in high Cd-accumulating indica cultivars (9311) than in low Cd-accumulating varieties, including PA64S, which was derived from an indica/japonica cross20–22. Variations in the promoter sequences was found between indica and japonica cultivars, which might lead to differences between in the expression levels of OsHMA3, OsNRAMP1 and OsNRAMP5, as well as in Cd accumulation in the grains14,20–22. In addition, mutant osnramp5 produced by carbonion-beam irradiation greatly decreases Cd uptake by roots, resulting in decreased Cd in grain and exhibit no agriculturally adverse traits15. The combined action of OsHMA3, OsNRAMP1 and OsNRAMP5 might contribute to the difference in grain Cd accumulation between indica and japonica varieties. Coincidentally, these three genes—OsHMA3, OsNramp5 and OsNramp1—are all located on 7.4 Mb to 9.0 Mb of chromosome 7 within a region of about 1565 kb, according to the Rice Genome Annotation Project Database23. The segment covering OsHMA3-OsNramp5-OsNramp1 from a low Cd-accumulating japonica cultivar may be valuable for improving indica varieties to produce low Cd accumulation.
The severity of rice grain Cd pollution in some areas of China led to the initiation of a national key breeding programme to reduce the Cd content of indica rice in 2014. This study was conducted with the objectives of (1) validating the effect of the OsHMA3-OsNramp5-OsNramp1 region on chromosome 7 for grain Cd accumulation; (2) evaluating the use of OsHMA3-OsNramp5-OsNramp1jap for indica improvement with low Cd accumulation in grains and (3) improving the elite indica cultivar with low Cd grain accumulation by MAS.
Results
Phenotypic variation and the effects of OsHMA3-OsNramp5-OsNramp1 in recombinant inbred lines derived from an indica–japonica cross
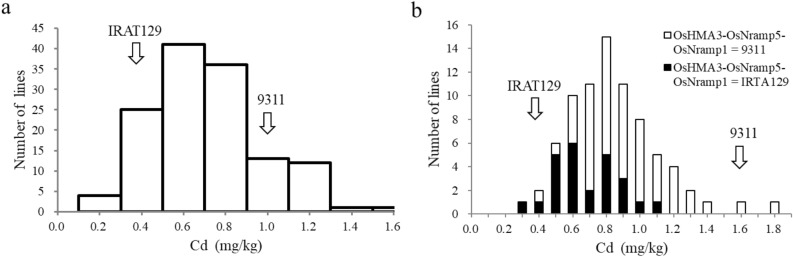
A recombinant inbred line (RIL) population was bred from crosses between indica variety 9311 and a japonica landrace IRAT129. The 9311 variety is an elite two-line middle-season indica hybrid restorer in China that accumulates Cd at high levels, and the IRAT129 variety is a high-compatibility japonica landrace that has a low ability to accumulate Cd. IRAT129, 9311, and RILs with 133 lines were grown in a Cd-polluted paddy field at Ningxiang in the 2016 middle-season rice growing season (Table 1). The Cd concentrations of IRAT129 and 9311 brown rice were 0.493 and 1.020 mg/kg, respectively. The segregation of Cd concentration in brown rice RILs was continuously distributed with Cd concentrations ranging from 0.133 to 1.463 mg/kg, averaging to 0.609 mg/kg (Fig. 1a). The kurtosis (0.174) and skewness (0.649) of the distribution of Cd concentrations were < 1, indicating a normal distribution. Transgressive segregation in both directions was observed, indicating predominance of additive gene action contributed by both parental alleles.
Table 1.
Populations used to validate the effect of OsHMA3-OsNramp5-OsNramp1 from IRAT129. The IRAT129, 9311, H611 and H819 presented in the column of samples indicate the haplotype of OsHMA3-OsNramp5-OsNramp1.
| Population | Generation | Segregating target genes | Samples | Growing season | Trait measured |
|---|---|---|---|---|---|
| RILs | F6 | 133 lines, containing 25 lines of IRAT129, 53 lines of 9311 | Middle-season rice cropping season in 2016 | Cd | |
| F7 | OsHMA3-OsNramp5-OsNramp1 | 25 lines of IRAT129, 53 lines of 9311 | Middle-season rice cropping season in 2017 | Cd | |
| NIL611 | BC3F2:3 | 30 lines of IRAT129, 30 lines of H611 and 60 lines of heterozygote | Early-season rice cropping season in 2017 | HTD, Cd | |
| NIL819 | BC3F2:3 | 30 lines of IRAT129, 28 lines of H819 and 60 lines of heterozygote | |||
| NIL9311 | BC3F2:3 | 23 lines of IRAT129, 30 lines of 9311 and 59 lines of heterozygote | Middle-season rice cropping season in 2017 |
Figure 1.
Phenotypic distributions of brown rice Cd concentration in RILs. (a) Distributions of brown rice Cd concentration of RILs in the 2016 middle-season rice growing season. (b) Distribution of brown rice Cd concentration and its association with the two genotypic groups of 78 lines from RILs across 2016 and 2017 middle-season rice growing season. OsHMA3-OsNramp5-OsNramp = 9311, carrying OsHMA3-OsNramp5-OsNramp from 9311; OsHMA3-OsNramp5-OsNramp = IRAT129, carrying OsHMA3-OsNramp5-OsNramp from IRAT129.
A total of 133 RILs were genotyped using three pairs of intragenic molecular markers for OsHMA3, OsNramp5 and OsNramp1 genes (Table 2). Overall, 78 F7 lines with a homozygous genomic region covering OsHMA3, OsNramp5 and OsNramp1 genes (25 OsHMA3-OsNramp5-OsNramp1jap homozygotes and 53 OsHMA3-OsNramp5-OsNramp1ind homozygotes) were selected and their phenotypes were investigated in 2017. The frequency distribution of Cd concentration was not distinguishable among different genotypes, but OsHMA3-OsNramp5-OsNramp1jap homozygous lines tended to have lower values and OsHMA3-OsNramp5-OsNramp1ind homozygous lines had higher values (Fig. 1b). Results of a two-way ANOVA on the phenotypic differences between OsHMA3-OsNramp5-OsNramp1jap and OsHMA3-OsNramp5-OsNramp1ind homozygous lines in 2016 and 2017 are presented in Table 3. Significant variation was apparent between the two homozygous genotypic groups over the 2 years. Genotypes of OsHMA3-OsNramp5-OsNramp1 explained 10.9% of the phenotypic variance in Cd concentration in 2016 and 8.6% of the variance in 2017. The OsHMA3-OsNramp5-OsNramp1 haplotype of IRAT129 appeared to be associated with decreased Cd concentration in brown rice.
Table 2.
Three developed intra-genic markers used to amplify Cd accumulating genes.
| Gene | Marker | Primer sequence | Expected size (bp) of IRAT129 and 9311 (IRAT129/9311) | Restriction enzyme | |
|---|---|---|---|---|---|
| Forward (5′-3′) | Reverse (5′-3′) | ||||
| OsHMA3 | HMA3 | AATCCATCCAACCAAACCCGGAA | ATGCCCAGCGATCCAAGCTC | 280 bp | BsaHI |
| OsNramp5 | Nramp5 | TTGTCATGCCGTACGCTT | CACCAAGGCAGAATGCAA | 140/144 bp | |
| OsNramp1 | Nramp1 | GGCTGTAGATACCACATCACC | GCTGCCTCAAGAATTATACCTG | 761/355 bp | |
Table 3.
Brown rice Cd concentration and HTD of different genotypes differing in the OsHMA3-OsNramp5-OsNramp1 region.
| Population | Year | Trait | Phenotype of different genotypes (Mean ± SD) | ANOVA | A | D | D/[A] | R2 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| japonica | indica | H | F | P | |||||||
| RIL | 2016 | Cd | 0.486 ± 0.186 | 0.694 ± 0.274 | n.a | 12.05 | < .0001 | 0.10 | 10.9 | ||
| 2017 | Cd | 0.769 ± 0.260 | 1.033 ± 0.372 | n.a | 10.07 | < .0001 | 0.13 | 8.6 | |||
| NIL9311 | 2017 | Cd | 1.491 ± 0.160 | 1.981 ± 0.294 | 1.736 ± 0.200 | 35.92 | < .0001 | 0.25 | 0.00 | 0.01 | 17.9 |
| HTD | 106.7 ± 2.0 | 107.5 ± 1.4 | n.a | 2.99 | 0.0212 | 0.50 | 6.8 | ||||
| NIL611 | 2017 | Cd | 0.791 ± 0.081 | 0.527 ± 0.078 | 0.800 ± 0.095 | 101.48 | < .0001 | − 0.13 | 0.14 | − 1.09 | 37.1 |
| HTD | 89.0 ± 2.7 | 75.0 ± 2.1 | n.a | 274.29 | < .0001 | − 7.0 | 84.3 | ||||
| NIL819 | 2017 | Cd | 0.820 ± 0.092 | 0.392 ± 0.118 | 0.765 ± 0.113 | 137.42 | < .0001 | − 0.21 | 0.17 | − 0.79 | 45.4 |
| HTD | 92.0 ± 1.9 | 73.0 ± 1.6 | n.a | 1574.57 | < .0001 | − 9.5 | 96.6 | ||||
| IRAT129 | 2017 | Cd | 0.465 ± 0.107 | ||||||||
| 9311 | 2017 | Cd | 1.838 ± 0.216 | ||||||||
| H611 | 2017 | Cd | 0.412 ± 0.044 | ||||||||
| H819 | 2017 | Cd | 0.436 ± 0.062 | ||||||||
HTD is the days to heading. Japonica and indica are lines carrying homozygous OsHMA3-OsNramp5-OsNramp1 haplotype from IRAT129 and indica variety (9311, H611 or H819), respectively. H, the lines carrying heterozygous OsHMA3-OsNramp5-OsNramp1 haplotype. A, additive effect of replacing OsHMA3-OsNramp5-OsNramp1ind haplotype with OsHMA3-OsNramp5-OsNramp1jap haplotype. D, degree of dominance. R2, the proportion of phenotypic variance explained by OsHMA3-OsNramp5-OsNramp1.
Validation of OsHMA3-OsNramp5-OsNramp1jap effects in the indica genetic background
Three sets of NILs of OsHMA3-OsNramp5-OsNramp1 in an indica genetic background replaced with of 9311 (an elite two-line middle-season indica hybrid restorer in China), and H611 and H811 (two elite two-line early-season indica hybrid restorers in China) were developed to validate the practicality and effectiveness of OsHMA3-OsNramp5-OsNramp1jap in molecular marker-assisted improvement of indica rice with low Cd-accumulating ability. Discrete distributions of Cd concentration were not observed in the three sets of NILs, but the frequency distributions were distinguishable among the different genotypes. OsHMA3-OsNramp5-OsNramp1jap homozygous, OsHMA3-OsNramp5-OsNramp1ind homozygous and heterozygous lines tended to have lower, higher and higher values in NIL9311, whereas it exhibited the opposite response in NIL611 and NIL819. These results indicated that different genotypic groups of NIL9311, NIL611 and NIL819 might carry different alleles for Cd, whereas OsHMA3-OsNramp5-OsNramp1jap in NIL611 and NIL819 has a different genetic effect direction from that in NIL9311. Two-way ANOVAs were used to examine differences among the three genotypic groups in each of the two NIL sets. As expected, major allele effects were detected for Cd in all the three NIL sets and explained 17.9%, 37.1% and 45.4% of the phenotypic variance in NIL9311, NIL611 and NIL819, respectively. The OsHMA3-OsNramp5-OsNramp1jap haplotype decreased Cd concentration in NIL9311 by 0.25 mg/kg (Table 3). However, in contrast to the results from RIL and NIL9311, the favourable Cd-decreasing allele was found in indica cultivars H611 and H819, and the OsHMA3-OsNramp5-OsNramp1jap haplotype increased Cd by 0.13 mg/kg in NIL611 and 0.21 mg/kg in NIL819 (Table 3).
Major effects were detected for HTD in NIL611 and NIL819. The enhancing alleles were all from IRAT129; they increased HTD by 7.0 and 9.5 days and explained 84.3% and 96.6% of the phenotypic variance in NIL611 and NIL819, respectively. A minor effect was detected for HTD in NIL9311, and the enhancing allele came from 9311, increasing HTD by 0.5 days and explaining 6.8% of the phenotypic variance. These results indicated that the segment containing OsHMA3-OsNramp5-OsNramp1 from IRAT129 has major effects on HTD in early-season indica rice and minor effects in middle-season indica rice and the directions of the genetic effects are opposite.
Heading date and yield potential gene (Ghd7) analysis of NIL population
A cloned gene Ghd7, an important regulator of heading date and yield potential in rice, which plays a crucial role in increasing productivity and adaptability of rice globally is located close to OsNramp1, within a physical distance of about 181.5 kb (Chr.7: 8,970,856–9,152,377), according to the Rice Genome Annotation Project Database23,24. To investigate the role of Ghd7 gene on the expression of OsHMA3-OsNramp5-OsNramp1jap in early-season and middle-season indica genetic backgrounds, we obtained the Ghd7 sequence of IRAT129, H611, H819 and 9311. Comparison of the predicted protein sequences identified three alleles, equivalent to Ghd7-0, Ghd7-1 and Ghd7-2 in a previous study24. IRAT129 and 9311 carried a functionally weaker allele of Ghd7-2 and a fully functional allele of Ghd7-1, respectively (Fig. 2). The Ghd7 locus of H611 and H819 was completely deleted (Ghd7-0). The genotyping of the three sets of NILs with an intragenic marker for Ghd7 showed that the segment of OsHMA3-OsNramp5-OsNramp1jap also contained Ghd7jap in three sets of NILs. The HTD effects in early-season and middle-season indica rice could be a result from the effects of different Ghd7 alleles.
Figure 2.
The predicted protein sequences of GHD7 for IRAT129, 9311, H611 and H819.
Genetic background and agronomic traits of improved 9311 with the OsHMA3-OsNramp5-OsNramp1-Ghd7jap haplotype
Eight improved homozygous plants from the NIL9311 population were genotyped using high-density SNP markers to check the genome recovery rate of the recurrent parent in the improved lines. Of the 1125 SNP markers covering 12 chromosomes, 630 SNPs showed polymorphism between the donor IRAT129 and recipient 9311. The selected improved plants showed a high genome recovery rate of the recurrent parent, at 90.4–91.6% (Table 4). A graphical genotype map of NIL9311-58 was constructed based on the SNP genotyping results (Fig. 3), assuming that the size of the target chromosome segment containing the OsHMA3-OsNramp5-OsNramp1jap was approximately 14.3 Mb. The genetic background of NIL9311-58 was also analysed with 48 SSR makers, which are used for the identification of rice varieties (NY/T 1433-2014)26. The result showed only one polymorphic SSR marker RM542 (Chr. 7: 12,712,017–12,712,178), neighbouring OsHMA3-OsNramp5-OsNramp1-Ghd7 (Chr. 7: 7,405,745–9,155,030)27, was identified between improved 9311 (NIL9311-58) and original 9311.
Table 4.
Genome recovery rate of eight improved homozygous plants from NIL9311 population.
| Line no | No. of SNP | No. of SNP allele | Genome ratio (%) | |||
|---|---|---|---|---|---|---|
| Recipient | Donor | Hetero | Recipient | Donor | ||
| NIL9311-11 | 617 | 558 | 56 | 3 | 90.7 | 9.3 |
| NIL9311-12 | 619 | 560 | 56 | 3 | 90.7 | 9.3 |
| NIL9311-21 | 620 | 561 | 56 | 3 | 90.7 | 9.3 |
| NIL9311-54 | 619 | 563 | 53 | 3 | 91.2 | 8.8 |
| NIL9311-58 | 617 | 565 | 52 | 0 | 91.6 | 8.4 |
| NIL9311-62 | 619 | 560 | 57 | 2 | 90.6 | 9.4 |
| NIL9311-61 | 619 | 562 | 54 | 3 | 91.0 | 9.0 |
| NIL9311-62 | 618 | 556 | 57 | 5 | 90.4 | 9.6 |
Figure 3.
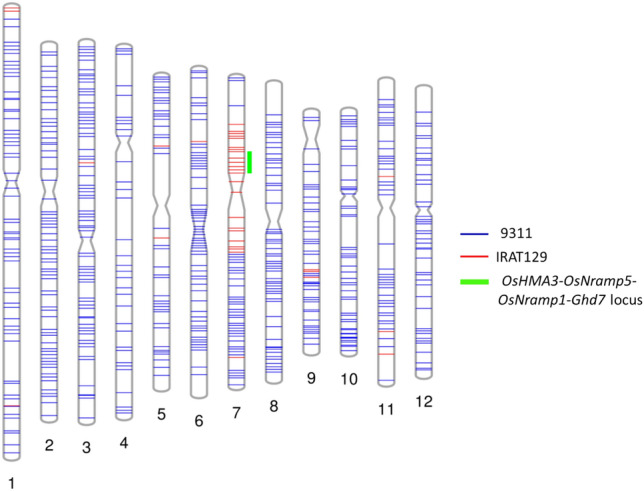
Graphical genotype maps of the improved 9311 (NIL9311-58). NIL9311-58 with the OsHMA3-OsNramp5-OsNramp1-Ghd7jap haplotype was genotyped using 1.2 K SNP markers. Six hundred and thirty polymorphic SNPs between bi-parents were used for the construction of the genotype maps.
To test whether the OsHMA3-OsNramp5-OsNramp1-Ghd7jap haplotype has a negative effect on the yield, the agronomic traits and grain metallic element of eight lines derived from NIL9311-58 (improved 9311carrying OsHMA3-OsNramp5-OsNramp1jap with favourable agronomic performance and lower Cd accumulation in the grain) and the control 9311 were evaluated in the middle rice-cropping season at Niangxiang in 2018. The improved 9311 showed a significant increase in Mn concentration, panicle numbers and yield, and a significant decrease in DTH and grain Cd accumulation compared with the control 9311 (Table 5). Overall, the improved 9311 had no negative effect on yield, whereas it decreased grain Cd accumulation by about 31.8% and increased Mn accumulation by about 21.2%.
Table 5.
Agronomic performance of improved 9311 (NIL9311-58) lines carrying OsHMA3-OsNramp5-OsNramp1jap in the genetic background of 9311.
| Genotype | NL | DTH | PH | PN | NGP | SF (%) | TGW (g) | YP (g) | Cd (mg/kg) | Fe (mg/kg) | Mn (mg/kg) | Cu (mg/kg) | Zn (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9311 | 5 | 92.0 ± 0.3 | 119.0 ± 1.1 | 8.3 ± 1.1 | 208.7 ± 7.1 | 89.1 ± 7.8 | 29.9 ± 1.0 | 36.7 ± 2.7 | 1.57 ± 0.25 | 19.97 ± 0.01 | 42.73 ± 0.75 | 3.29 ± 0.26 | 31.7 ± 0.70 |
| NIL9311-58 | 8 | 89.4 ± 1.5* | 117.9 ± 2.6 | 9.6 ± 1.2* | 213.9 ± 15.2 | 88.1 ± 1.9 | 29.1 ± 0.6 | 38.4 ± 2.1* | 1.07 ± 0.12* | 20.8 ± 0.07 | 51.77 ± 2.17* | 3.45 ± 0.20 | 31.37 ± 1.31 |
NL: number of lines; DTH: days to heading; PH: plant height; PN: number of panicles per plant; NGP: number of grains per panicle; SF: spikelet fertility; TGW: 1000-grain weight; YP: yield per plant; Cd, Fe, Mn, Cu, Zn: Cd, Fe, Mn, Cu and Zn content in brown grains.
*Significance at P < 0.01.
Haplotype and expression of OsHMA3, OsNramp1 and OsNramp5 in IRAT129, 9311, NIL9311-58, H611 and H819
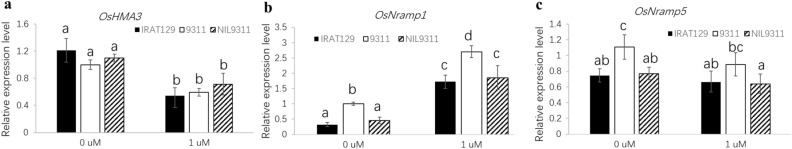
At least eight alleles (I to VIII) of OsHMA3 have identified in previous reports25. The coding regions of OsHMA3 of IRTA129, 9311, H611 and H819 were therefore sequenced. Results indicated that 9311, H611 and H819 have the same functional allele of type V, whereas IRTA129 did not belong to any of the eight haplotypes, which has a single amino acid mutation at 550th position with IIe being substituted by Val compared to type V allele (designated as type X here) (Table 6). Liu et al. reported that the expression difference of OsHMA3, which due to natural variation in the promoter of OsHMA3, contributes to differential grain cadmium accumulation between indica and japonica rice20. We therefore compared the expression level of OsHMA3 in roots of IRTA129, 9311 and NIL9311-58. In inconsistent with the results of Liu et al., there were no significant differences in OsHMA3 expression between the three cultivars tested in either the absence or the presence of Cd. The presence of Cd tended to decrease the expression of OsHMA3, and the effect was significant in all tested lines (Fig. 4a). Sequence variations in the about 2 kb OsHMA3 promoter were therefore investigated. The results showed that promoter sequences in 9311, H611 and H819 are the same as that of type 1 (indica type) in the research of Liu et al20. Thirteen nucleotide variations were identified between type 1 and IRTA129 promoter (Table S1). Liu et al. revealed that the differential transcriptional activity of OsHMA3 promoter between indica and japonica could be attributed to the seven nucleotide changes occurring in the region between − 683 and − 557 bp20. However, there was no nucleotide variation between type1indica and IRTA129 japonica promoter.
Table 6.
OsHMA3 haplotypes of 9311, H611 and IRAT129.
| Cultivar | Subspecies | Amino acid position | HMA3 haplotype* | Functional* | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 80 | 299 | 380 | 550 | 614 | 638 | 678 | 728 | 736 | 752 | 775 | 826–878 | ||||
| Nipponbarea | Japonica | R | F | S | V | S | A | C | T | G | V | E | I | Yes | |
| Akita 64b | Japonica | R | F | S | V | S | A | C | T | G | V | E | I | Yes | |
| Dulara | Aus | R | F | S | V | S | A | C | T | C | V | E | IV | Yes | |
| Fengaizhana | Indica | R | L | S | I | G | A | R | T | G | A | E | V | Yes | |
| Cho-Ko-Kokub | Indica | H | L | S | V | D | V | R | T | G | A | E | Deletion | VIII | No |
| 9311 | Indica | R | L | S | I | G | A | R | T | G | A | E | V | Yes | |
| H611 | Indica | R | L | S | I | G | A | R | T | G | A | E | V | Yes | |
| H819 | Indica | R | L | S | I | G | A | R | T | G | A | E | V | Yes | |
| IRAT129 | Japonica | R | L | S | V | G | A | R | T | G | A | E | X | ||
Figure 4.
Relative expression of OsHMA3 (a), OsNramp1 (b) and OsNramp5 (c) in rice roots of different lines in the presence or absence of 1 μM Cd. Plants of IRAT129, 9311 and NIL9311-58 were treated with and without 1 μM Cd for 24 h. Data were means ± SD (n = 3). Means with different letters are significantly different at P < 0.05 (one-way ANOVA, Tukey HSD test).
The coding regions of OsNramp1 and OsNramp5 of IRTA129, 9311, H611 and H819 were also sequenced. Results indicated that 9311, H611 and H819 have the different allele of OsNramp1 with that of IRTA129, which has three single amino acid mutation at 34th, 252th and 512th position, respectively (Table S2). Consistent with results of Takahashi et al.14 and Chang et al.21, the level of OsNramp1 expression in the roots of indica 9311 was higher than that of japonica IRAT129 and NIL9311-58 in either the absence or the presence of Cd. The presence of Cd tended to increase the expression of Nramp1, and the effect was significant in all tested lines (Fig. 4b). Sequence variations in the about 2 kb OsNramp1 promoter were therefore investigated. The results showed that promoter sequences in 9311, H611 and H819 are the same as that of type 1 (indica type) in the research of Liu et al.20. Nineteen nucleotide variations including the 406 bp InDel variation were identified between IRTA129 and 9311 promoters (Table S3). The 406 bp were missing in 9311, as well as in H611 and H819. Sequencing analysis results showed that 9311, H611, H819 and IRTA129 have the same allele of OsNramp5 with no amino acid variation. Similar results with the research of Liu et al.22, the level of OsNramp5 expression in the roots of 9311 was significantly higher than that of IRAT129 and NIL9311-58 in either the absence or the presence of Cd (Fig. 4c). Twelve nucleotide variations were found in the promoter region (Table S4). These sequence variations in the promoter region could lead to the expression level difference of OsNramp1 and OsNramp5 between 9311 and NIL9311-58.
Discussion
Accumulation of Cd in crops is a serious agricultural issue. Reducing Cd concentration in the edible parts of crops through breeding is a promising option for decreasing risks to human health without any additional cost to farmers. The incorporation of favourable alleles of major Cd accumulation QTLs/genes could be an important measure to decrease the grain Cd concentration of rice. Japonica rice generally accumulates lower amounts of Cd in the grain than do indica and aus cultivars. Three genes (OsHMA3, OsNramp5 and OsNramp1), located in a region of about 1565 kb on chromosome 7 (physical location: 7,405,745–8,970,856), have previously been identified as playing critical roles in Cd accumulation in rice. However, there is no evidence that OsHMA3, OsNramp5 or OsNramp1 is/are the casual gene(s) for general differences in Cd accumulation between indica and japonica rice, and there is no previous report on the use of OsHMA3, OsNramp5 or OsNramp1 from low Cd accumulation japonica cultivars in indica improvement for low Cd accumulation. In this study, we investigated the role of genomic segment covering OsHMA3-OsNramp5-OsNramp1, by introgressing it in an indica breeding programme aimed at decreasing potential grain Cd pollution risk in elite indica cultivars. For the evaluation of the effect on Cd accumulation, the OsHMA3-OsNramp5-OsNramp1jap haplotype was tested in an indica–japonica cross background and a different season-type indica cultivar background using RILs and NILs population over two cropping seasons.
In this study, OsHMA3-OsNramp5-OsNramp1 was found to explain 8.6–10.9% of the grain Cd concentration variance in RILs derived from indica/japonica crossing (9311 × IRAT129) in two cropping seasons, and the favourable allele came from japonica, as expected. For the further evaluation of the OsHMA3-OsNramp5-OsNramp1jap haplotype in the indica breeding programme, we removed the japonica donor genome from the breeding lines. NILs of OsHMA3-OsNramp5-OsNramp1 in different indica backgrounds (9311, H611 and H819) were developed by advanced-backcross and MAS. 9311 is an elite male parent of two-line middle-season hybrid rice, as well as a favourable indica variety in China. 9311 is the male parent of Liangyoupei9 and Y-liangyou1, which are representative varieties of the first- and second-stage super-hybrids in China. Liangyoupei9 was the hybrid rice variety with the largest yearly promotion area from 2002 to 2007, except in 2004, and Y-liangyou-1 had the largest yearly promotion area from 2010 to 2013 in China. However, 9311 and the hybrid rice varieties derived from 9311 have high Cd accumulation, especially in grains. H611 and H819 are the elite male parents of two-line early-season hybrid rice. Zhulinagyou819, the combination of Zhu1S and H819, is the first hybrid rice to be identified as a low Cd accumulation variety in Hanan province, China. The OsHMA3-OsNramp5-OsNramp1jap haplotype significantly decreased grain Cd concentration in a middle-season indica 9311 genetic background. The homozygotes of OsHMA3-OsNramp5-OsNramp1jap accumulated 24.7% less Cd in grains than did homozygotes of OsHMA3-OsNramp5-OsNramp1ind. These results indicate that the OsHMA3-OsNramp5-OsNramp1jap haplotype can be used in middle-season indica improvement for low Cd concentration in the grains. Among the 23 homozygous lines of OsHMA3-OsNramp5-OsNramp1jap, NIL 9311-58 showed the lowest Cd concentration in the grain, 40.7% less than 9311. Eight lines (improved 9311) were derived from NIL 9311-58 to evaluate whether the OsHMA3-OsNramp5-OsNramp1-Ghd7jap haplotype had negative effects on yield performance. The improved 9311 had significantly decreased grain Cd accumulation, by about 31.8%, and significantly increased yield, of about 4.6%. This improved 9311 line, a near isogenic line, is similar to 9311 variety in all agronomic aspects except low Cd accumulation in the grain. Therefore, the improved 9311 could replace 9311 for rice production in areas where Cd pollution is a potential threat. However, in contrast to the performance in middle-season indica background, the OsHMA3-OsNramp5-OsNramp1jap haplotype significantly increased grain Cd concentration in two early-season indica (H611 and H819) background. In addition, major effects were detected for HTD in NIL611 and NIL819, and the OsHMA3-OsNramp5-OsNramp1jap haplotype has significantly increased HTD. This observation might imply the existence of a linkage-drag or pleiotropic effect of the segment containing OsHMA3-OsNramp5-OsNramp1 for grain Cd concentration in the early-season indica background. Some previous works also reported the QTLs related to grain Cd concentration overlap or contained the QTLs/genes for DTH28,29. The pleiotropic effect of QTLs for grain Cd concentration and/or water management difference of the genotypes with different heading time in one trial plot is/are probably the main reason(s), which caused the experiment results above30,31.
Beside OsHMA3-OsNramp5-OsNramp1, an HTD gene, Ghd7, is located at a physical distance of about 181.5 kb from OsNramp1. Xue et al. (2008) reported that Ghd7-1 is a fully functional allele and Ghd7-2 is a weak functional allele. The Ghd7 locus was completely deleted in early-season rice (Ghd7-0). Our study showed that Ghd7 of japonica IRAT129, middle-season indica 9311, and early-season indica H611 and H819 was equivalent to the Ghd7-2, Ghd7-1 and Ghd7-0 type, respectively. All three NIL populations carried OsHMA3-OsNramp5-OsNramp1-Ghd7ind/jap. A minor HTD effect was identified in the NIL9311 population, possibly because of the effects of the Ghd7-1 and Ghd7-2 alleles on HTD. Linkage-drag with Ghd7 could be the reason for the OsHMA3-OsNramp5-OsNramp1jap haplotype to significantly increase HTD in early-season indica background. The linkage-drag with Ghd7 should be broken if the OsHMA3-OsNramp5-OsNramp1jap haplotype is to be used in early-season indica improvement. The improved early-season indica lines with OsHMA3-OsNramp5-OsNramp1-Ghd7jap showed late heading, and these cannot be cropped as early-season indica. Ghd7 is located in the centromeric region of chromosome 7, therefore the recombination rate is considerably low. Xue et al. (2008) reported that the recombination rate around Ghd7 region is about 37-fold lower than the genome average of approximately 200 kb/cM. A large segregating population (≥ 4077 plants, theoretically) should be developed to identify recombination events between OsNramp1 and Ghd7, if we are to use the OsHMA3-OsNramp5-OsNramp1jap haplotype to improve early-season indica for low Cd grain concentration. The target chromosome segment of OsHMA3-OsNramp5-OsNramp1 was selected using three intragenic markers for OsHMA3, OsNramp5 and OsNramp1 and none recombinant was used. To further investigate the effect of low Cd accumulation in grain is from one or several gene(s) of OsHMA3, OsNramp5 and OsNramp1, the recombinants should be identified from the large segregating population derived from heterozygotes of OsHMA3-OsNramp5-OsNramp1.
A novel allele of OsHMA3 was identified from a wide-compatibility japonica cultivar IRAT129 with low Cd-accumulating ability. Nucleotide variations were identified between IRAT129 and higher Cd-accumulating indica variety (9311, H611 and H819) promoter, but there were no significant differences in OsHMA3 expression between 9311 and NIL9311 tested in either the absence or the presence of Cd. Whether OsHMA3 allele from IRAT129 has stronger function on decreasing Cd-accumulating ability need be further studied. The expressions of OsNramp1 and OsNramp5 in roots all were lower in lower Cd-accumulating variety/line (IRAT129 and NIL9311-58) than that in higher Cd-accumulating variety (9311). The 400 bp deletion in the promoter region of OsNRAMP1 and sequence variations identified in the promoter region of OsNRAMP5 could lead to differences in the expression level of OsNramp1 and OsNramp5, as well as in Cd accumulation in the grain between 9311 and NIL9311-58. Our results provide evidence that segment covering OsHMA3-OsNramp5-OsNramp1 on chromosome 7 of a japonica cultivar IRAT129 can be used in indica varieties improvement with accumulating low levels of Cd. But, the linkage-drag between OsHMA3-OsNramp5-OsNramp1 and Ghd7 should be broken down by large segregating populations or gene editing in early-season indica improvement. The improved 9311 with introgression of OsHMA3-OsNramp5-OsNramp1IRAT129 showed significant decrease in grain Cd accumulation with no yield loss. However, the improved 9311 still cannot produce Cd-free grain (< 0.2 or 0.4 mg/kg) in mild Cd-polluted paddy field with intermittent irrigation management. Assembling large numbers of favourable low-Cd gene/QTL alleles or edit Cd accumulating genes (such as OsNramp5, etc.) could be further studied for improving rice with Cd-free grain.
Methods
Plant materials
A RIL (F7) population consisting of 133 lines was generated from an F1 hybrid between 9311 (an elite two-line middle-season indica hybrid restorer with high Cd-accumulating ability in China) and IRAT129 (a wide-compatibility japonica cultivar with low Cd-accumulating ability) by single seed descent.
Three sets of NILs including the genomic region covering the OsHMA3, OsNramp5 and OsNramp1 genes, with different genetic backgrounds, were also developed. IRAT129 was the paternal parent, and the IRTA129-type OsHMA3-OsNramp5- OsNramp1 haplotype (OsHMA3-OsNramp5- OsNramp1jap) donor was crossed with two elite two-line early-season indica hybrid restorers (H611 and H811) and an elite two-line middle-season indica hybrid restorer (9311) as maternal recipients. The resulting F1 plants were backcrossed with the recipients. Plants heterozygous for the target region in the progeny were selected using molecular markers and crossed to recipients. Finally, BC3F1 plants heterozygous for the target region were selected to produce BC3F2, which were genotyped using molecular markers, and japonica-type homozygotes (OsHMA3-OsNramp5-OsNramp1jap), indica-type homozygotes (OsHMA3-OsNramp5-OsNramp1ind) homozygotes and heterozygotes (OsHMA3-OsNramp5-OsNramp1jap/OsHMA3-OsNramp5-OsNramp1ind) were selected. The three sets of NILs were derived from the selected seeds and named NIL611, NIL819 and NIL9311 with H611, H819 and 9311 genetic backgrounds, respectively. Eight plants selected from the NIL9311-58 line were named improved 9311.
DNA preparation, molecular marker development, sequencing and expression analysis
Genomic DNA was extracted from 2-week-old seedlings using the CTAB method as described by Murray and Thomson32. Three newly developed intragenic markers were developed (Table 2) based on publicly available rice genome sequences23,33. Gene cloning and sequencing were done following the methods described in Yan et al.25 and Lu et al.34. Quantitative reverse transcription PCR (qRT-PCR) analysis was done following the methods described in Yan et al25. Primers for cloning, sequencing and qRT-PCR in this study were listed in Table S5.
Background genotyping and construction of a graphical genotype map
Genomic DNA was prepared using the CTAB method as described by Murray and Thomson31. A 1.2 K multiplex-PCR panel, which was based on GBTS platform from MolBreeding Biotechnol (http://www.molbreeding.com), was employed for background genotyping. A total of 48 SSR makers were also used in background genotyping, following the protocol of the identification of rice varieties by SSR marker method (NY/T 1433–2014).
Field experiments and phenotypic analysis
Field experiments were conducted in a Cd-polluted paddy field (Niangxiang, China) in the 2016, 2017 and 2018 rice-growing seasons. The soil Cd concentration was around 0.40 mg/kg with pH 6.4. The irrigation water Cd concentration was about 0.027 ug/L. Field trials of RILs were conducted in the 2016 and 2017 middle-season rice growing seasons. NILs of NIL611 and NIL819 were conducted in the 2017 early-season rice growing season, as well as NIL9311 in the 2017 middle-season rice growing season. The experiments were arranged in a randomised complete block design with three replicates, with 10 plants per line. Field management followed normal agricultural practices, except that intermittent irrigation was adopted to maximise the phenotypic differences. DTH was counted from seed sowing to the flowering of 50% of the plants of each line. At maturity, the middle eight plants of each line in each replication were harvested, and the Cd concentrations of brown rice were determined using NX-100FA.
The 9311 and improved 9311 lines were cropped in the 2018 middle-season rice growing season, arranged in a randomised complete block design with three replicates and 40 hills per replicate. Field management followed that of the 2016 and 2017 field experiments. Days to heading (DTH) was counted from seed sowing to the flowering of 50% of the plants of each line. The plant height (PH) of five normal plants in the middle of each plot was measured at the fully mature stage, and these five plants were harvested to evaluate various agronomic traits in the laboratory, including the number of grains per panicle (NGP), spikelet fertility (SF), 1000-grain weight (TGW) and yield per plant (YP).
Statistical analysis
The SAS (V8.01) procedure GLM was used to evaluate phenotypic variation among the different genotypic groups in the RILs and NILs35.
Supplementary Information
Acknowledgements
This work was supported by grants from Science and technology innovation program of Hunan (2019RS2054), National Key Technology Research and Development Program of China (2016YFD0101801), Major Project of Low-cadmium Early-season Rice Breeding Project of Hunan (Xiangnonglian [2018]70), State Key Laboratory of Hybrid Rice (Hunan Hybrid Rice Research Center) Open Project Fund (2018KF03), Special Project of National Independent Innovation Demonstration Zone (2018XK2005) and Science and Technology Innovation Program of Hunan (2018NK1020).
Author contributions
Y.Y. designed the experiments. Y.Y. and C.F. constructed the populations. T.Y., S.X., Y.X. and Y. Z. performed molecular marker development, sequencing and expression analysis. Q.Z., Z.W., X.Z. and Y.L. conducted the field trials. K.W., P.Q. and J.F. analyzed the data. K.W. and Y.Y. wrote the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets supporting the conclusions of this article are included within the article and its additional files. The seeds of RILs, NILs and the parents are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kai Wang, Tian-ze Yan and Shi-long Xu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85324-0.
References
- 1.Clemens S, et al. Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci. 2013;18(2):92–99. doi: 10.1016/j.tplants.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, et al. Genetic diversity, rather than cultivar type, determines relative grain Cd accumulation in hybrid rice. Front. Plant Sci. 2016;7:1407. doi: 10.3389/fpls.2016.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FAO & WHO. Codex Alimentarius Commission (2014). http://www.fao.org/news/story/en/item/238558/icode/
- 4.MOH. National Food Hygienic Standard of China (GB 2762-2012) (2012). http://www.nhc.gov.cn/ewebeditor/uploadfile/2013/01/20130128114248937.pdf.
- 5.Sebastian A, Prasad MNV. Cadmium minimization in rice. Agron. Sustain. Dev. 2014;34(1):155–173. doi: 10.1007/s13593-013-0152-y. [DOI] [Google Scholar]
- 6.Xie LH, et al. The cadmium and lead content of the grain produced by leading Chinese rice cultivars. Food Chem. 2017;217:217–224. doi: 10.1016/j.foodchem.2016.08.099. [DOI] [PubMed] [Google Scholar]
- 7.Bolan NS, et al. Cadmium contamination and its risk management in rice ecosystems. Adv. Agron. 2013;119:183–273. doi: 10.1016/B978-0-12-407247-3.00004-4. [DOI] [Google Scholar]
- 8.Arao T, Ae N. Genotypic variations in cadmium levels of rice grain. Soil Sci. Plant Nutr. 2003;49(4):473–479. doi: 10.1080/00380768.2003.10410035. [DOI] [Google Scholar]
- 9.He J, et al. Genotypic variation in grain cadmium concentration of lowland rice. J. Plant Nutr. Soil Sci. 2006;169(5):711–716. doi: 10.1002/jpln.200525101. [DOI] [Google Scholar]
- 10.Yan YF, et al. Genotypic variation of cadmium accumulation and distribution in rice. J. Crop Sci. Biotech. 2010;13(2):69–73. doi: 10.1007/s12892-010-0036-5. [DOI] [Google Scholar]
- 11.Nakanishi H, et al. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006;52(4):464–469. doi: 10.1111/j.1747-0765.2006.00055.x. [DOI] [Google Scholar]
- 12.Ueno D, et al. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad Sci. 2010;107(38):16500–16505. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyadate H, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol. 2011;189(1):190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi R, et al. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011;62(14):4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa S, et al. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. P. Natl. Acad. Sci. 2012;109(47):19166–19171. doi: 10.1073/pnas.1211132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki A, et al. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell. 2012;24(5):2155–2167. doi: 10.1105/tpc.112.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi R, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012;35(11):1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x. [DOI] [PubMed] [Google Scholar]
- 18.Shimo H, et al. Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J. Exp. Bot. 2011;62(15):5727–5734. doi: 10.1093/jxb/err300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uraguchi S, et al. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. 2011;108(52):20959–20964. doi: 10.1073/pnas.1116531109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu CL, et al. Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. 2020;62(3):314–329. doi: 10.1111/jipb.12794. [DOI] [PubMed] [Google Scholar]
- 21.Chang JD, et al. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020;43(10):2476–2491. doi: 10.1111/pce.13843. [DOI] [PubMed] [Google Scholar]
- 22.Liu C, et al. Characterization of a major QTL for manganese accumulation in rice grain. Sci. Rep. 2017;7(1):17704. doi: 10.1038/s41598-017-18090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawahara Y, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6(1):4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40(6):761. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, et al. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of Japonica rice cultivars. Plant Cell Environ. 2016;39(9):1941–1954. doi: 10.1111/pce.12747. [DOI] [PubMed] [Google Scholar]
- 26.MOA. Protocol for identification of rice varieties-SSR marker method (NY/T 1433-2014) (2014).
- 27.Cold Spring Harbor Laboratory, Oregon State University & EMBL-EBI. Gramene (2019). http://www.gramene.org/
- 28.Ishikawa S, et al. Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa) New Phytol. 2005;168(2):345–350. doi: 10.1111/j.1469-8137.2005.01516.x. [DOI] [PubMed] [Google Scholar]
- 29.Abe T, et al. Detection of QTLs to reduce cadmium content in rice grains using LAC23/Koshihikari chromosome segment substitution lines. Breeding Sci. 2013;63(3):284–291. doi: 10.1270/jsbbs.63.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honma T, et al. Optimal soil Eh, pH, and water management for simultaneously minimizing arsenic and cadmium concentrations in rice grains. Environ. Sci. Technol. 2016;50(8):4178–4185. doi: 10.1021/acs.est.5b05424. [DOI] [PubMed] [Google Scholar]
- 31.Arao T, et al. Effects of water management on cadmium and arsenic accumulation and dimethylarsinic acid concentrations in Japanese rice. Environ. Sci. Technol. 2009;43(24):9361–9367. doi: 10.1021/es9022738. [DOI] [PubMed] [Google Scholar]
- 32.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8(19):4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Rice Genome Sequencing Project. The rice annotation project database (2019). http://rapdb.dna.affrc.go.jp/
- 34.Lu L, et al. Evolution and association analysis of Ghd7 in rice. PLoS ONE. 2012;7(5):e34021. doi: 10.1371/journal.pone.0034021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.SAS Institute. SAS/STAT User’s Guide (1999) https://v8doc.sas.com/sashtml/stat/index.htm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files. The seeds of RILs, NILs and the parents are available from the corresponding author on reasonable request.