Abstract
Plant polyphenol gossypol has anticancer activities. This may increase cottonseed value by using gossypol as a health intervention agent. It is necessary to understand its molecular mechanisms before human consumption. The aim was to uncover the effects of gossypol on cell viability and gene expression in cancer cells. In this study, human colon cancer cells (COLO 225) were treated with gossypol. MTT assay showed significant inhibitory effect under high concentration and longtime treatment. We analyzed the expression of 55 genes at the mRNA level in the cells; many of them are regulated by gossypol or ZFP36/TTP in cancer cells. BCL2 mRNA was the most stable among the 55 mRNAs analyzed in human colon cancer cells. GAPDH and RPL32 mRNAs were not good qPCR references for the colon cancer cells. Gossypol decreased the mRNA levels of DGAT, GLUT, TTP, IL families and a number of previously reported genes. In particular, gossypol suppressed the expression of genes coding for CLAUDIN1, ELK1, FAS, GAPDH, IL2, IL8 and ZFAND5 mRNAs, but enhanced the expression of the gene coding for GLUT3 mRNA. The results showed that gossypol inhibited cell survival with decreased expression of a number of genes in the colon cancer cells.
Subject terms: Biochemistry, Cancer, Plant sciences, Molecular medicine
Introduction
Gossypol is a plant polyphenol with a highly colored yellow pigment (Fig. 1). It is found in the small intercellular pigment glands in the leaves, stems, roots, and seed of cotton plants (Gossypium hirsutum L.)1. Gossypol has traditionally been regarded as an anti-nutritional toxic compound. It has been known for a long time that consumption of gossypol-containing cottonseed oil contributes to its toxicity causing male infertility2. Therefore, gossypol is regarded as unsafe for most animal and human consumption. The residual gossypol in cottonseed meals limits its use primarily to feed ruminants, which have a relative high tolerance for the toxic compound2–6. Significant efforts have been conducted to reduce gossypol content in cottonseed by selecting glandless cotton varieties7–12 and genetic engineering of gossypol-free seeds of cotton plants13–15.
Figure 1.
Chemical structure of gossypol (molar mass: 518.56 g/mol) (Image was from public domain Gossypol—Wikipedia).
Gossypol was proposed recently to have potential biomedical applications. Gossypol and related compounds were reported to have anticancer activities associated with breast cancer16–18, colon cancer19, pancreatic cancer20,21 and prostate cancer22,23. It has antiobesity activities16,24, antiinflammatory activities25,26 and antifungal activities27,28. These new discoveries have generated intensive interest in biomedical field and enormous amounts of research have been directed at understanding the medical utilization of gossypol and related compounds.
Colon cancer is one of the deadliest diseases in the US and the World. American Cancer Society estimated that the lifetime risk of developing colorectal cancer is approximately 4.49% for men and 4.15% for women; which may cause 51,020 deaths during 2019 (https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html). Some studies have explored the effect of gossypol and its analogs on colon cancer cells. They bind to and inactivate BH3 domain-containing antiapoptotic proteins29. Gossypol analog Ch282-5 (2-aminoethanesulfonic acid sodium-gossypolone) exhibits anti-proliferative and pro-cell death activity against colon cancer cells both in vitro and in vivo, and the response of colon cancer cells to the drug correlates with Ch282-5′s inhibition of anti-apoptotic Bcl-2 proteins, induction of mitochondria-dependent apoptotic pathway, and disruption of mitophagy and mTOR pathway29. It was also shown that (-)-gossypol is a potential small molecule inhibitor of MSI1-RNA interaction, and may be incorporated into an effective anti-cancer strategy30. A novel water-soluble gossypol derivative increases chemotherapeutic sensitivity and promotes growth inhibition in colon cancer31. Gossypol sensitizes the antitumor activity of 5-FU through down-regulation of thymidylate synthase in human colon carcinoma cells32. These studies have provided a promising beginning to use gossypol in the prevention and treatment of colon cancer. However, not enough was done on the regulation of gene expression at the mRNA level by gossypol in colon cancer cells, especially related to cytokine expression regulated by the anti-inflammatory zinc finger protein 36/tristetraprolin (ZFP36/TTP). Much more information is required to develop an effective strategy to combat colon cancer.
It is our aim to uncover more profound effects of gossypol on regulating the cell viability and expression of a wide range of target genes involved in cancer development. In this study, we analyzed the cell viability and expression of 55 genes (Table 1) whose expression is regulated by gossypol in cancer cells20,33–39 and macrophages40 or regulated by ZFP36/TTP in tumor cells41–49 and macrophages50,51. Human colon cancer cells (COLO 225) were treated with multiple concentrations of gossypol followed by quantitative PCR analysis. Our results showed that gossypol inhibited cell viability and suppressed a number of gene expression in the human colon cancer cells.
Table 1.
Human qPCR primers.
| ID | mRNA | Name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Reference |
|---|---|---|---|---|---|
| H1 | Ahrr1 | Aryl hydrocarbon receptor repressor | AGGCTGCTGTTGGAGTCTCTTAA | CGATCGTTGCTGATGCATAAA | TTP45 |
| H2 | Bcl2 | B-cell lymphoma 2 | CAGCATGCGGCCTCTGTT | GGGCCAAACTGAGCAGAGTCT | Gossypol37 |
| H3 | Bcl2l1 | B-cell lymphoma 2 like 1 | GTGCGTGGAAAGCGTAGACA | ATTCAGGTAAGTGGCCATCCAA | TTP94 |
| H4 | Bnip3 | BCL2 protein-interacting protein 3 | GTCAAGTCGGCCGGAAAATA | TGCGCTTCGGGTGTTTAAAG | Gossypol20 |
| H5 | Cd36 | Cluster of differentiation 36/fatty acid translocase | CTCTTTCCTGCAGCCCAATG | TTGTCAGCCTCTGTTCCAACTG | TTP95 |
| H6 | Claudin1 | Maintain tissue integrity and water retention | GACAAAGTGAAGAAGGCCCGTAT | CAAGACCTGCCACGATGAAA | TTP49 |
| H7 | Cox1 | Cyclooxygenase 1 | CGCCCACGCCAGTGA | AGGCCGAAGCGGACACA | TTP96 |
| H8 | Cox2 | Cyclooxygenase 2 | CGATTGTACCCGGACAGGAT | TTGGAGTGGGTTTCAGAAATAATTT | TTP48 |
| H9 | Csnk2a1 | Casein kinase 2 alpha 1 | AGCGATGGGAACGCTTTG | AAGGCCTCAGGGCTGACAA | TTP46 |
| H10 | Ctsb | Cathepsin B | GACTTGTAGCTGCTGTCTCTCTTTGT | CAAGAGTCGCAAGAACATGCA | TTP97 |
| H11 | Cxcl1 | Chemokine (C-X-C motif) ligand 1 | GCCCAAACCGAAGTCATAGC | TGCAGGATTGAGGCAAGCT | TTP98 |
| H12 | Cyclind1 | Cyclin D1 | ACACGCGCAGACCTTCGT | CCATGGAGGGCGGATTG | Gossypol38 |
| H13 | Cyp19a1 | Cytochrome P450 family 19 subfamily A member 1 | GACATTGCAAGGACAGTGTGTTG | AGTCTCATCTGGGTGCAAGGA | Gossypol35 |
| H14 | Dgat1 | Diacylglycerol O-acyltransferase 1 | ACCTCATCTGGCTCATCTTCTTCTA | CCCGGTCTCCAAACTGCAT | DGAT99,100 |
| H15 | Dgat2a | Diacylglycerol O-acyltransferase 2a | CCCAGGCATACGGCCTTA | CAACACAGGCATTCGGAAGTT | DGAT100,101 |
| H16 | Dgat2b | Diacylglycerol O-acyltransferase 2b | ACTCTGGCCCTTCTCTGTTTTTTA | TCCACCTTGGTTGGGTGTGT | DGAT100,101 |
| H17 | E2f1 | E2F transcription factor 1 | CGGCGCATCTATGACATCAC | CAGCCACTGGATGTGGTTCTT | TTP44 |
| H18 | Elk1 | ETS transcription factor | CTCCTCCGCATCCCTCTTTAA | AGCGTCACAGATGGGTCCAT | TTP42 |
| H19 | Fas | Fas cell surface death receptor | GAACTCCTTGGCGGAAGAGA | AGGACCCCGTGGAATGTCA | Gossypol34 |
| H20 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | GGGTGTGAACCATGAGAAGTATGA | GGTGCAGGAGGCATTGCT | 83 |
| H21 | Glut1 | Glucose transporter 1 | TGCTCATGGGCTTCTCGAA | AAGCGGCCCAGGATCAG | GLUT51 |
| H22 | Glut2 | Glucose transporter 2 | GCATTTTTCAGACGGCTGGTA | GCGCCAACTCCAATGGTT | GLUT51 |
| H23 | Glut3 | Glucose transporter 3 | GAGGATATCACACGGGCCTTT | CCATGACGCCGTCCTTTC | GLUT51 |
| H24 | Glut4 | Glucose transporter 4 | CGTGGGCGGCATGATT | CCAGCATGGCCCTTTTCC | GLUT51 |
| H25 | Hif1a | Hypoxia inducible factor 1 subunit alpha | GGTGGATATGTCTGGGTTGAAAC | ATGCACTGTGGTTGAGAATTCTTG | TTP102 |
| H26 | Hmgr | 3-Hydroxy-3-methylglutaryl-CoA reductase | AAGTGAAAGCCTGGCTCGAA | CTAGTGCTGTCAAATGCCTCCTT | 103 |
| H27 | Hmox1 | Heme oxygenase 1 | CTTCTCCGATGGGTCCTTACACT | TCACATGGCATAAAGCCCTACA | TTP104 |
| H28 | Hua | Human antigen a | GATCCTCTGGCAGATGTTTGG | CGCGGATCACTTTCACATTG | Gossypol40 |
| H29 | Icam1 | Intercellular adhesion molecule 1/CD54 | GGAGCTTCGTGTCCTGTATGG | TTTCTGGCCACGTCCAGTTT | 105 |
| H30 | Inos | Inducible nitric oxide synthase | AGATCCGGTTCACAGTCTTGGT | GCCATGACCTTCCGCATTAG | 106 |
| H31 | Insr | Insulin receptor | CAACGGGCAGTTTGTCGAA | TGGTCGGGCAAACTTTCTG | 50 |
| H32 | Il2 | Interleukin 2 | TATGCAGATGAGACAGCAACCAT | TTGAGATGATGCTTTGACAAAAGG | TTP61 |
| H33 | IL6 | Interleukin 6 | CCCACACAGACAGCCACTCA | CCGTCGAGGATGTACCGAAT | TTP62 |
| H34 | IL8 | Interleukin 8 | CCATCTCACTGTGTGTAAACATGACTT | ATCAGGAAGGCTGCCAAGAG | TTP63 |
| H35 | Il10 | Interleukin 10 | GCCGTGGAGCAGGTGAAG | TGGCTTTGTAGATGCCTTTCTCT | TTP64 |
| H36 | Il12 | Interleukin 12 | TGCCTTCACCACTCCCAAA | TGTCTGGCCTTCTGGAGCAT | TTP65 |
| H37 | Il16 | Interleukin 16 | CAGGGCCTCACACGGTTT | GACAATCGTGACAGGTCCATCA | TTP47 |
| H38 | Il17 | Interleukin 17 | CCCAAAAGGTCCTCAGATTACTACA | TCATTGCGGTGGAGATTCC | TTP66 |
| H39 | Leptin | Body fat and obesity hormone | AGGGAGACCGAGCGCTTT | CACATCCCTCACCTCCTTCAAA | 107 |
| H40 | Map1lc3a | Microtubule-associated proteins 1 light chain 3A | GTGAACCAGCACAGCATGGT | CCTCGTCTTTCTCCTGCTCGTA | 108 |
| H41 | Map1lc3b | Microtubule-associated proteins 1 light chain 3B | AGGCGCTTACAGCTCAATGC | ACCATGCTGTGTCCGTTCAC | 108 |
| H42 | Nfkb | Nuclear factor kappa B | GGTGCCTCTAGTGAAAAGAACAAGA | GCTGGTCCCACATAGTTGCA | 109 |
| H43 | P53 | Tumor suppressor | CTTGCAATAGGTGTGCGTCAGA | GGAGCCCCGGGACAAA | Gossypol33 |
| H44 | Pim1 | Proto-oncogene serine/threonine-protein kinase | TGCTCCACCGCGACATC | TGAGCTCGCCGCGATT | TTP43 |
| H45 | Pparr | Peroxisome proliferator-activated receptor gamma | GAACGACCAAGTAACTCTCCTCAAA | CAAGGAGGCCAGCATTGTGT | Gossypol36 |
| H46 | Rab24 | Ras-related oncogene 24 | TCGGTCGGAGACGCACTT | TGGCCTCATAGCGCTCAGA | 110 |
| H47 | Rpl32 | Ribosomal protein L32 (60S ribosomal unit) | CCTCCAAGAACCGCAAAGC | GGTGACTCTGATGGCCAGTTG | 80 |
| H48 | Tnf | Tumor necrosis factor | GGAGAAGGGTGACCGACTCA | CAGACTCGGCAAAGTCGAGAT | TTP62 |
| H49 | Tnfsf10 | Tumor necrosis factor superfamily, member 10 | GCTCTGGGCCGCAAAAT | AGGAATGAATGCCCACTCCTT | Gossypol39 |
| H50 | Ulk2 | Unc-51 like autophagy activating kinase 2 | ACAGCTCCTTTCAAAATCCCTAAA | AGGCCCATGACGAGTAACCA | 111 |
| H51 | Vegf | Vascular endothelial growth factor | CCCACTGAGGAGTCCAACATC | GGCCTTGGTGAGGTTTGATC | TTP41 |
| H52 | Zfand5 | Zinc finger AN1-type containing 5 | AGGGTTTGACTGCCGATGTG | ACTGGATTCTCTTTTCTGATTTTTGC | TTP84 |
| H53 | Zfp36 | Zinc finger protein 36 | GGCGACTCCCCATCTTCAA | GACCGGGCAGTCACTTTGTC | TTP50 |
| H54 | Zfp36L1 | Zinc finger protein 36 like 1 | TCTGCCACCATCTTCGACTTG | TGGGAGCACTATAGTTGAGCATCT | TTP50 |
| H55 | Zfp36L2 | Zinc finger protein 36 like 2 | CCTTTCATACCATCGGCTTCTG | TCGTCCGCGTTGTGGAT | TTP50 |
Results
Effect of gossypol on colon cancer cell viability
Cell cytotoxicity of human colon cancer cells (COLO 225) was determined with MTT method after the cells were treated for 2, 4, 8 and 24 h with 0.1–100 µg/mL of gossypol. MTT assay showed that gossypol significantly reduced colon cancer cell survival under high concentrations or long time treatment (Fig. 2). The cell viability was reduced to 55% of the control by 100 µg/mL gossypol after being treated for 2 h. The cell viability was reduced to a quarter of the control by 100 µg/mL gossypol after being treated for 4 or 8 h. The cell viability was further reduced to only 19% of the control by 100 µg/mL gossypol treatment for 24 h. There were clear dosage effects on reducing colon cancer cell viability after 1 µg/mL of gossypol treatment (Fig. 2).
Figure 2.
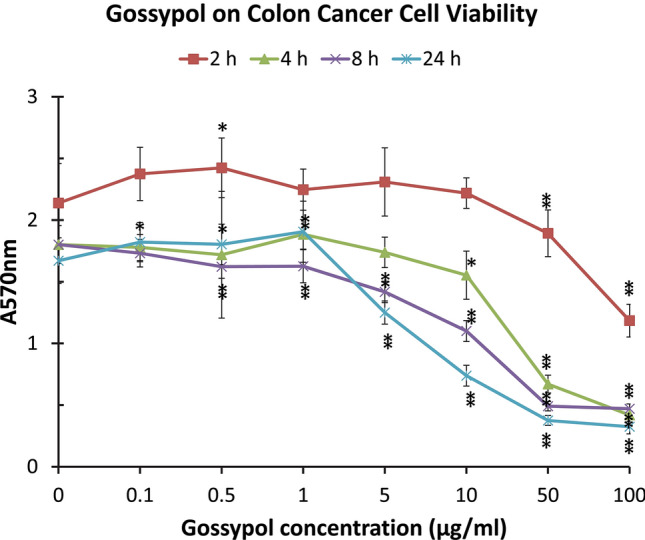
Effect of gossypol on human colon cancer cell viability. Human colon cancer cells (COLO 225) were treated with various concentrations of gossypol for 2, 4, 8 and 24 h. The cell media were added with MTT assay reagent, and incubated for 2 h before adding MTT solubilization solution, shaken at room temperature overnight. “0 µg/mL” treatment corresponding to 1% DMSO in the culture medium, the vehicle control for the experiment. The color density in the wells was recorded at A570. The data represent the mean and standard deviation of 12 independent samples. “*” and “**” near the data points represent significance between the treatment and the control at p < 0.05 and p < 0.01, respectively.
Basal expression level of selected genes in human colon cancer cells
To provide a basis for the comparison of gene expression regulation by gossypol in the colon cancer cells, we first measured the relative mRNA levels of 55 genes in the cells treated with 1% DMSO control for 8 h by SYBR Green qPCR assay using the specific primers (Table 1). The qPCR assay showed that the cycle of threshold (Cq) of BCL2 mRNA was one of the mRNAs with minimal variation among the 55 genes analyzed. The mean ± standard deviation of Cq for BCL2 mRNA was 29.39 ± 1.08 (n = 24) (Table 2, left column). The mRNA levels of GAPDH and RPL32 were the most abundant in the cells with 39 and 42 fold of BCL2 mRNA, respectively, whereas INOS mRNA was undetectable and AHRR1, COX1, CYCLIND1, GLUT4, ICAM1, IL10, IL12, RAB24, VEGF and ZFP36L2 mRNAs were minimally detected with less than 5% of BCL2 mRNA in the colon cancer cells (Table 2, left column).
Table 2.
Basal level, reference mRNA and gossypol effects on 55 gene expression.
| ID | mRNA | DMSO control | Gossypol | Gossypol/control | ||
|---|---|---|---|---|---|---|
| Mean ± SD | Fold of Bcl2 | Mean ± SD | Fold of Bcl2 | Fold of control | ||
| H1 | Ahrr1 | 33.88 ± 1.14 | 0.04 | 33.49 ± 1.09 | 0.03 | 0.65 |
| H2 | Bcl2 | 29.39 ± 1.08 | 1.00 | 28.37 ± 1.08 | 1.00 | 1.00 |
| H3 | Bcl2l1 | 27.64 ± 2.34 | 3.35 | 26.63 ± 1.81 | 3.34 | 1.00 |
| H4 | Bnip3 | 27.54 ± 1.22 | 3.60 | 26.47 ± 1.14 | 3.74 | 1.04 |
| H5 | Cd36 | 28.45 ± 1.23 | 1.91 | 27.55 ± 1.16 | 1.77 | 0.92 |
| H6 | Claudin1 | 28.03 ± 4.30 | 2.56 | 27.85 ± 3.73 | 1.44 | 0.56 |
| H7 | Cox1 | 34.92 ± 4.79 | 0.02 | 40.95 ± 5.37 | 0 | 0.01 |
| H8 | Cox2 | 30.65 ± 1.62 | 0.42 | 30.85 ± 2.80 | 0.18 | 0.43 |
| H9 | Csnk2a1 | 26.02 ± 1.95 | 10.33 | 25.22 ± 1.48 | 8.90 | 0.86 |
| H10 | Ctsb | 28.13 ± 2.73 | 2.38 | 27.34 ± 2.01 | 2.04 | 0.86 |
| H11 | Cxcl1 | 32.75 ± 2.60 | 0.10 | 31.46 ± 1.88 | 0.12 | 1.22 |
| H12 | Cyclind1 | 34.10 ± 5.42 | 0.04 | 33.59 ± 3.81 | 0.03 | 0.70 |
| H13 | Cyp19a1 | 32.40 ± 3.37 | 0.12 | 35.65 ± 2.03 | 0.01 | 0.05 |
| H14 | Dgat1 | 29.38 ± 1.86 | 1.00 | 28.55 ± 1.67 | 0.88 | 0.88 |
| H15 | Dgat2a | 31.80 ± 2.18 | 0.19 | 31.48 ± 2.30 | 0.12 | 0.62 |
| H16 | Dgat2b | 30.88 ± 1.79 | 0.35 | 30.37 ± 2.56 | 0.25 | 0.71 |
| H17 | E2f1 | 29.61 ± 1.08 | 0.85 | 29.72 ± 2.56 | 0.39 | 0.46 |
| H18 | Elk1 | 30.82 ± 2.84 | 0.37 | 31.22 ± 1.80 | 0.14 | 0.38 |
| H19 | Fas | 30.63 ± 4.60 | 0.42 | 28.97 ± 2.49 | 0.66 | 1.57 |
| H20 | Gapdh | 24.10 ± 4.01 | 39.09 | 22.66 ± 2.95 | 52.62 | 1.35 |
| H21 | Glut1 | 27.24 ± 2.54 | 4.41 | 26.80 ± 2.41 | 2.98 | 0.67 |
| H22 | Glut2 | 29.22 ± 1.81 | 1.12 | 27.72 ± 1.13 | 1.58 | 1.40 |
| H23 | Glut3 | 28.61 ± 1.31 | 1.71 | 27.00 ± 1.60 | 2.59 | 1.51 |
| H24 | Glut4 | 41.48 ± 4.81 | 0 | 41.17 ± 3.30 | 0 | 0.61 |
| H25 | Hif1a | 27.64 ± 2.33 | 3.36 | 26.82 ± 2.11 | 2.93 | 0.87 |
| H26 | Hmgr | 27.50 ± 1.91 | 3.70 | 26.46 ± 1.47 | 3.76 | 1.02 |
| H27 | Hmox1 | 29.68 ± 1.41 | 0.82 | 28.47 ± 1.37 | 0.94 | 1.15 |
| H28 | Hua | 32.25 ± 3.52 | 0.14 | 30.64 ± 2.34 | 0.21 | 1.51 |
| H29 | Icam1 | 33.93 ± 4.12 | 0.04 | 32.23 ± 1.50 | 0.07 | 1.61 |
| H30 | Inos | ud | ud | ud | ud | ud |
| H31 | Insr | 29.81 ± 3.12 | 0.74 | 29.58 ± 1.96 | 0.43 | 0.58 |
| H32 | Il2 | 31.27 ± 1.24 | 0.27 | 29.93 ± 1.19 | 0.34 | 1.25 |
| H33 | IL6 | 29.09 ± 1.39 | 1.23 | 27.29 ± 1.17 | 0.63 | 1.72 |
| H34 | IL8 | 29.14 ± 1.20 | 1.19 | 28.54 ± 1.52 | 0.89 | 0.75 |
| H35 | Il10 | 41.42 ± 5.26 | 0 | 39.31 ± 8.57 | 0 | 2.14 |
| H36 | Il12 | 37.55 ± 3.02 | 0 | 34.24 ± 4.17 | 0.02 | 4.94 |
| H37 | Il16 | 28.49 ± 1.17 | 1.87 | 28.57 ± 2.12 | 0.87 | 0.47 |
| H38 | Il17 | 29.56 ± 1.38 | 0.89 | 28.00 ± 1.30 | 1.29 | 1.46 |
| H39 | Leptin | 29.51 ± 1.22 | 0.92 | 28.34 ± 0.95 | 1.03 | 1.12 |
| H40 | Map1lc3a | 29.48 ± 1.87 | 0.94 | 27.84 ± 1.14 | 1.45 | 1.55 |
| H41 | Map1lc3b | 26.18 ± 1.73 | 9.25 | 25.25 ± 1.79 | 8.72 | 0.94 |
| H42 | Nfkb | 31.08 ± 2.97 | 0.31 | 30.21 ± 2.36 | 0.28 | 0.91 |
| H43 | P53 | 31.06 ± 2.58 | 0.31 | 30.22 ± 2.04 | 0.28 | 0.89 |
| H44 | Pim1 | 29.13 ± 1.09 | 1.20 | 28.25 ± 1.56 | 1.09 | 0.91 |
| H45 | Pparr | 29.63 ± 1.50 | 0.85 | 29.69 ± 1.36 | 0.40 | 0.47 |
| H46 | Rab24 | 42.25 ± 3.40 | 0 | 40.54 ± 3.54 | 0 | 1.62 |
| H47 | Rpl32 | 24.00 ± 3.68 | 41.78 | 22.94 ± 2.94 | 43.17 | 1.03 |
| H48 | Tnf | 30.88 ± 1.71 | 0.35 | 29.74 ± 1.21 | 0.39 | 1.10 |
| H49 | Tnfsf10 | 28.22 ± 1.48 | 2.24 | 27.72 ± 1.21 | 1.57 | 0.70 |
| H50 | Ulk2 | 29.20 ± 1.11 | 1.14 | 27.82 ± 1.08 | 1.46 | 1.28 |
| H51 | Vegf | 40.47 ± 4.57 | 0 | 38.68 ± 5.16 | 0 | 1.71 |
| H52 | Zfand5 | 27.20 ± 1.50 | 4.55 | 26.31 ± 1.49 | 4.19 | 0.92 |
| H53 | Zfp36 | 28.86 ± 1.85 | 1.44 | 27.37 ± 1.65 | 2.00 | 1.39 |
| H54 | Zfp36L1 | 29.29 ± 3.07 | 1.07 | 28.92 ± 2.44 | 0.69 | 0.64 |
| H55 | Zfp36L2 | 41.60 ± 3.61 | 0 | 39.81 ± 3.04 | 0 | 1.71 |
The data represent the mean and standard deviation of 24 independent samples. The fold was calculated using the mean data. ud: undetected. Bold italics: Genes with mRNA levels at least twofold of Bcl2, roughly interpreted as their expression more abundant than that of Bcl2. Italics: Genes with mRNA levels less than 50% of Bcl2, roughly interpreted as their expression less abundant than that of Bcl2.
Genes with mRNA levels at least twofold of BCL2 mRNA could be roughly interpreted as their expression more abundant than that of BCL2 mRNA. There were 13 more abundantly expressed genes than BCL2 mRNA including mRNAs of BCL2L1 (3.35 fold), BNIP3 (3.60 fold), CLUADIN1 (2.56 fold), CSNK2A1 (10.33 fold), CTSB (2.38 fold), GAPDH (39.09 fold), GLUT1 (4.41 fold), HIF1A (3.36 fold), HMGR (3.70 fold), MAP1LC3B (9.25 fold), RPL32 (41.78 fold), TNFSF10 (2.24 fold) and ZFAND5 (4.55 fold) (Table 2, left column). Genes with mRNA levels less than 50% of BCL2 mRNA could be roughly interpreted as their expression less abundant than that of BCL2 mRNA. There were 22 less abundantly expressed genes including mRNAs of AHRR1 (4%), COX1 (2%), COX2 (42%), CXCL1 (10%), CYCLIND1 (4%), CYP19A1 (12%), DGAT2A (19%), DGAT2B (35%), ELK1 (37%), FAS (42%), GLUT4 (< 1%), HUA (14%), ICAM1 (4%), IL2 (27%), IL10 (< 1%), IL12 (< 1%), NFKB (31%), P53 (31%), RAB24 (< 1%), TNF (35%), VEGF (< 1%) and ZFP36L2 (< 1%) (Table 2, left column). TaqMan qPCR assay showed similar trend as of SYBR Green qPCR (data not shown). SYBR Green qPCR assay was chosen to conduct gene expression analysis in the following experiments for cost saving and convenience.
Selection of reference gene for qPCR assays in human colon cancer cells
Reference gene for qPCR assays should be stably expressed without much variation by the experimental treatments. The less of standard deviation of Cq among the gossypol treatments might be indication of the more stable expression of the gene which could be served as an internal reference. To identify which gene could be served as an internal reference in the human colon cancer cells in our study, we pooled the data from triplicate each of the 8 concentrations (0, 0.1, 0.5, 1, 5, 10, 50 and 100 µg/mL of gossypol) and calculated the mean ± standard deviation from the 24 samples (Table 2, middle column). The Cq value of BCL2 mRNA was among the least varied in the cells with the mean ± standard deviation of 28.37 ± 1.08 (n = 24) (Table 2, middle column). The Cq value of other mRNAs with similar standard deviations were mRNAs of AHRR1 (1.09), BNIP3 (1.14), CD36 (1.16), GLUT2 (1.13), IL2 (1.19), IL6 (1.17), LEPTIN (0.95), MAP1LC3A (1.14) and ULK2 (1.08). GAPDH and RPL32 mRNAs were widely used as references for qPCR assays in mammalian cells. However, GAPDH and RPL32 genes were shown here with much larger standard deviations (2.95 and 2.94, respectively). Furthermore, GAPDH and RPL32 mRNAs were the most abundantly expressed in the cells with 53 and 43 fold of BCL2 mRNA, respectively, which were tens of fold higher than almost all of the other mRNAs analyzed in this study (Table 2, middle column). The large standard deviations and high expression levels of GAPDH and RPL32 mRNAs suggest that they were not good internal reference genes for qPCR assays in the human colon cancer cell. Since BCL2 was widely studied in colon cancer cells and was among the least regulated genes by gossypol, we therefore selected BCL2 mRNA as the internal reference for our qPCR analyses.
There were 13 genes with mRNA levels at least twofold of BCL2 mRNA in the 24 pooled samples including mRNAs of BCL2L1 (3.34 fold), BNIP3 (3.74 fold), CSNK2A1 (8.90 fold), CTSB (2.04 fold), GAPDH (52.62 fold), GLUT1 (2.98 fold), GLUT3 (2.59 fold), HIF1A (2.93 fold), HMGR (3.76 fold), MAP1LC3B (8.72 fold), RPL32 (43.17 fold), ZFAND5 (4.19 fold) and ZFP36 (twofold) (Table 2, middle column). These gene expression patterns from pooled gossypol samples were similar to those from DMSO control samples except that CLAUDIN1 and TNFSF10 were expressed more in DMSO samples but GLUT3 and ZFP36 were expressed more in the gossypol pooled samples (Table 2, left vs. middle columns). There were 24 genes with mRNA levels less than 50% of BCL2 mRNA including mRNAs of AHRR1 (3%), COX1 (< 1%), COX2 (18%), CXCL1 (12%), CLAUDIN1 (3%), CYP19A1 (1%), DGAT2A (12%), DGAT2B (25%), E2F1 (39%), ELK1 (14%), GLUT4 (< 1%), HUA (21%), ICAM1 (7%), INSR (43%), IL2 (34%), IL10 (< 1%), IL12 (2%), NFKB (28%), P53 (28%), PPARR (40%), RAB24 (< 1%), TNF (39%), VEGF (< 1%) and ZFP36L2 (< 1%) (Table 2, middle column). These gene expression patterns from gossypol pooled samples were similar to those from DMSO control samples except that FAS mRNA was expressed more in DMSO samples but E2F1, INSR and PPARR mRNAs were expressed more in the gossypol pooled samples (Table 2, left vs. middle columns).
Overall effect of gossypol on gene expression in human colon cancer cells
To provide a general idea how these genes were regulated by gossypol, we analyzed the pooled qPCR data using BCL2 mRNA as the internal reference and DMSO treatment as the sample control. As shown in the right column of Table 2, expression of a number of genes was affected by gossypol. Gossypol decreased the expression of six mRNAs with less than 50% of the control and only up-regulated the expression of two mRNAs with at least twofold of the control. The up-regulated mRNAs were IL10 (2.14 fold) and IL12 (4.94 fold) (Table 2, right column). The down-regulated mRNAs were COX1 (1%), COX2 (43%), CYP19A1 (5%), E2F1 (46%), ELK1 (38%) and PPARR (47%) (Table 2, right column).
Dosage effect of gossypol on gene expression in human colon cancer cells
Human colon cancer cells were treated with 8 concentrations of gossypol (0, 0.1, 0.5, 1, 5, 10, 50 and 100 µg/mL of gossypol). SYBR Green qPCR analyzed the expression of all 55 genes with BCL2 mRNA as an internal reference and 1% DMSO treatment as the sample control (Table 3). Generally, most of the gene expression was suppressed by higher concentrations of gossypol (Table 3). These genes include BCL2L1, CLAUDIN1, CSNK2A1, CTSB, CXCL1, DGAT1, DGAT2A, DGAT2B, ELK1, FAS, GAPDH, GLUT1, HMGR, HUA, ICAM1, MAP1LC3B, NFKB, P53, RPL32, VEGF, ZFAND5, ZFP36L1 and ZFP36L2. Some of the gene expression was increased by gossypol including mRNAs of COX1, COX2, GLUT3, GLUT4, PPARR and RAB24 (Table 3). Eight of the p values were less than 5% threshold as shown with a “*” in the right column of Table 3, suggesting their expression levels might be statistically significant. Among them, gossypol significantly regulated the expression of genes coding for mRNAs of CLAUDIN1, ELK1, FAS, GAPDH, GGLUT3, IL2, IL8 and ZFAND5; all down-regulated except GLUT3 mRNA (Table 3, highlighted row). TTP family mRNA levels (ZFP36, ZFP36L1 and ZFP36L2 mRNAs) were generally decreased and TTP-targeted cytokine mRNA levels (TNF, COX2, PPARR and RAB24 mRNAs) were relatively high in the colon cancer cells (Table 3). More specific analyses of gene expression under gossypol treatments are described below according to specific gene families.
Table 3.
Gossypol Dosages on Colon Cancer Cell Gene Expression.
| ID | mRNA | DMSO | 0.1 µg/mL | 0.5 µg/mL | 1 µg/mL | 5 µg/mL | 10 µg/mL | 50 µg/mL | 100 µg/mL | p value |
|---|---|---|---|---|---|---|---|---|---|---|
| Fold | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | |||
| H1 | Ahrr1 | 1.00 | 3.54 ± 2.16 | 2.91 ± 0.00 | 1.33 ± 0.51 | 1.41 ± 1.91 | 0.50 ± 0.17 | 1.51 ± 9.00 | ud | 0.226 |
| H2 | Bcl2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| H3 | Bcl2l1 | 1.00 | 1.29 ± 1.15 | 0.82 ± 0.28 | 0.82 ± 0.13 | 1.19 ± 1.24 | 0.33 ± 0.08 | 0.34 ± 0.06 | 0.25 ± 0.06 | 0.015 |
| H4 | Bnip3 | 1.00 | 0.91 ± 0.37 | 1.27 ± 0.72 | 0.80 ± 0.20 | 1.08 ± 0.45 | 0.55 ± 0.10 | 1.03 ± 0.14 | 0.55 ± 0.15 | 0.200 |
| H5 | Cd36 | 1.00 | 0.87 ± 0.11 | 1.12 ± 0.35 | 0.79 ± 0.42 | 1.36 ± 0.56 | 0.91 ± 0.20 | 1.09 ± 0.46 | 0.64 ± 0.12 | 0.304 |
| H6 | Claudin1 | 1.00 a | 1.72 ± 2.30 a | 0.64 ± 0.19 a | 0.77 ± 0.33 a | 0.77 ± 0.88 a | 0.20 ± 0.14 ab | 0.09 ± 0.11 ab | 0.005 ± 0.003 b | 0.03* |
| H7 | Cox1 | 1.00 | 2.08 ± 1.86 | 0.30 ± 0.44 | 1.15 ± 0.98 | 1.23 ± 2.08 | 1.35 ± 1.14 | 1.28 ± 1.11 | 6.60 ± 5.96 | 0.748 |
| H8 | Cox2 | 1.00 | 1.87 ± 2.90 | 2.35 ± 1.69 | 1.76 ± 2.13 | 3.69 ± 0.39 | 0.97 ± 1.22 | 3.61 ± 4.60 | 0.55 ± 0.47 | 0.370 |
| H9 | Csnk2a1 | 1.00 | 1.00 ± 0.68 | 0.70 ± 0.13 | 0.67 ± 0.18 | 0.87 ± 0.40 | 0.38 ± 0.14 | 0.33 ± 0.09 | 0.25 ± 0.07 | 0.032 |
| H10 | Ctsb | 1.00 | 1.47 ± 1.02 | 0.95 ± 0.26 | 0.84 ± 0.31 | 0.90 ± 1.09 | 0.24 ± 0.14 | 0.20 ± 0.06 | 0.14 ± 0.07 | 0.029 |
| H11 | Cxcl1 | 1.00 | 1.66 ± 0.74 | 1.45 ± 0.42 | 1.11 ± 0.55 | 1.53 ± 0.22 | 0.05 ± 0.08 | 0.03 ± 0.04 | 0.0001 ± 0.000 | 0.043 |
| H12 | Cyclind1 | 1.00 | ud | ud | ud | ud | ud | ud | ud | |
| H13 | Cyp19a1 | 1.00 | 0.77 ± 0.38 | 0.07 ± 0.07 | 19.49 ± 32.77 | 10.07 ± 14.2 | 0.44 ± 0.62 | 0.71 ± 1.15 | 3.17 ± 3.27 | 0.509 |
| H14 | Dgat1 | 1.00 | 0.78 ± 0.67 | 0.34 ± 0.08 | 0.53 ± 0.07 | 0.73 ± 0.49 | 0.31 ± 0.07 | 0.38 ± 0.16 | 0.19 ± 0.06 | 0.073 |
| H15 | Dgat2a | 1.00 | 1.37 ± 1.24 | 1.20 ± 0.62 | 0.06 ± 0.04 | 1.23 ± 0.82 | 0.40 ± 0.36 | 0.48 ± 0.09 | 0.11 ± 0.08 | 0.089 |
| H16 | Dgat2b | 1.00 | 0.08 ± 0.08 | 1.05 ± 0.53 | 0.43 ± 0.07 | 0.8 ± 0.06 | 0.57 ± 0.49 | 0.35 ± 0.33 | 0.28 ± 0.22 | 0.036 |
| H17 | E2f1 | 1.00 | 0.47 ± 0.44 | 0.39 ± 0.56 | 1.11 ± 0.65 | 1.64 ± 0.41 | 0.85 ± 0.75 | 1.44 ± 0.34 | 1.03 ± 0.10 | 0.075 |
| H18 | Elk1 | 1.00 a | 0.22 ± 0.11 b | 0.15 ± 0.06 b | 0.12 ± 0.07 b | 0.08 ± 0.07 b | 0.06 ± 0.03 b | 0.06 ± 0.05 b | 0.01 ± 0.00 b | 0.001* |
| H19 | Fas | 1.00 a | 1.25 ± 1.00 a | 0.44 ± 0.06 ab | 0.66 ± 0.25 ab | 0.50 ± 0.47 ab | 0.11 ± 0.07 b | 0.10 ± 0.13 b | 0.01 ± 0.01 c | 0.008* |
| H20 | Gapdh | 1.00 a | 1.09 ± 1.05 a | 0.48 ± 0.36 a | 0.69 ± 0.13 a | 0.63 ± 0.70 a | 0.26 ± 0.15 a | 0.17 ± 0.21 a | 0.03 ± 0.01 b | 0.046* |
| H21 | Glut1 | 1.00 | 0.94 ± 0.53 | 0.56 ± 0.22 | 0.74 ± 0.75 | 0.65 ± 1.04 | 0.98 ± 0.31 | 0.39 ± 0.31 | 0.11 ± 0.07 | 0.269 |
| H22 | Glut2 | 1.00 | 0.84 ± 0.23 | 1.14 ± 0.59 | 0.84 ± 0.27 | 1.37 ± 0.51 | 0.83 ± 0.09 | 1.02 ± 0.20 | 0.58 ± 0.14 | 0.180 |
| H23 | Glut3 | 1.00 b | 5.48 ± 2.75 ab | 5.75 ± 0.73 ab | 5.80 ± 2.90 ab | 3.22 ± 1.37 ab | 6.78 ± 0.46 a | 6.37 ± 1.95 a | 4.34 ± 2.10 ab | 0.023* |
| H24 | Glut4 | 1.00 | 2.77 ± 3.11 | 2.83 ± 1.04 | 31.14 ± 41.70 | 14.86 ± 17.21 | 5.75 ± 4.75 | 12.26 ± 16.28 | 3.70 ± 5.35 | 0.507 |
| H25 | Hif1a | 1.00 | 3.63 ± 4.39 | 1.89 ± 0.60 | 1.75 ± 0.59 | 1.09 ± 0.89 | 0.82 ± 0.29 | 0.64 ± 0.15 | 0.34 ± 0.14 | 0.039 |
| H26 | Hmgr | 1.00 | 0.93 ± 0.82 | 0.62 ± 0.31 | 0.58 ± 0.21 | 0.51 ± 0.26 | 0.32 ± 0.06 | 0.33 ± 0.14 | 0.38 ± 0.41 | 0.189 |
| H27 | Hmox1 | 1.00 | 0.66 ± 0.33 | 0.63 ± 0.11 | 1.15 ± 1.09 | 0.72 ± 0.11 | 0.50 ± 0.11 | 0.63 ± 0.16 | 0.35 ± 0.02 | 0.096 |
| H28 | Hua | 1.00 | 1.81 ± 1.45 | 0.98 ± 0.34 | 0.71 ± 0.31 | 0.63 ± 0.45 | 0.39 ± 0.17 | 0.30 ± 0.19 | 0.07 ± 0.06 | 0.032 |
| H29 | Icam1 | 1.00 | 0.70 ± 0.34 | 0.33 ± 0.14 | 0.54 ± 0.16 | 0.48 ± 0.44 | 0.13 ± 0.06 | 0.07 ± 0.07 | 0.06 ± 0.00 | 0.091 |
| H30 | Inos | ud | ud | ud | ud | ud | ud | ud | ud | |
| H31 | Insr | 1.00 | 2.51 ± 1.84 | 1.13 ± 0.48 | 1.38 ± 0.82 | 0.72 ± 0.75 | 0.34 ± 0.29 | 0.52 ± 0.42 | 0.05 ± 0.00 | 0.113 |
| H32 | Il2 | 1.00 ab | 0.95 ± 0.08 ab | 1.49 ± 0.29 a | 0.98 ± 0.13 ab | 1.040.49 ab | 0.83 ± 0.33 ab | 0.92 ± 0.18 ab | 0.60 ± 0.20 b | 0.04* |
| H33 | IL6 | 1.00 | 1.50 ± 0.43 | 1.79 ± 1.08 | 1.72 ± 1.00 | 1.81 ± 0.59 | 1.47 ± 0.48 | 1.49 ± 0.44 | 1.00 ± 0.37 | 0.623 |
| H34 | IL8 | 1.00 ab | 1.05 ± 0.44 a | 1.22 ± 0.19 a | 0.72 ± 0.17 ab | 1.23 ± 0.42 a | 0.88 ± 0.14 ab | 0.94 ± 0.08 ab | 0.36 ± 0.22 b | 0.013* |
| H35 | Il10 | 1.00 | ud | ud | ud | ud | ud | ud | ud | |
| H36 | Il12 | 1.00 | ud | ud | ud | ud | ud | ud | ud | |
| H37 | Il16 | 1.00 | 0.003 ± 0.001 | 0.003 ± 0.001 | 0.003 ± 0.001 | 0.001 ± 0.001 | 0.00 | 0.002 ± 0.001 | 0.001 ± 0.001 | |
| H38 | Il17 | 1.00 | 0.86 ± 0.49 | 1.87 ± 1.03 | 1.11 ± 0.33 | 1.91 ± 0.71 | 1.04 ± 0.53 | 1.54 ± 0.46 | 0.93 ± 0.37 | 0.179 |
| H39 | Leptin | 1.00 | 0.74 ± 0.21 | 1.15 ± 0.50 | 1.25 ± 0.10 | 1.18 ± 0.51 | 0.95 ± 0.25 | 1.08 ± 0.24 | 1.17 ± 0.35 | 0.604 |
| H40 | Map1lc3a | 1.00 | 0.94 ± 0.55 | 1.05 ± 0.47 | 1.01 ± 0.45 | 0.90 ± 0.31 | 0.57 ± 0.06 | 0.61 ± 0.05 | 0.60 ± 0.09 | 0.136 |
| H41 | Map1lc3b | 1.00 | 0.91 ± 0.98 | 0.57 ± 0.19 | 0.94 ± 0.68 | 0.68 ± 0.47 | 0.33 ± 0.03 | 0.31 ± 0.20 | 0.19 ± 0.09 | 0.132 |
| H42 | Nfkb | 1.00 | 0.96 ± 1.11 | 0.52 ± 0.18 | 0.47 ± 0.10 | 0.44 ± 0.51 | 0.20 ± 0.16 | 0.17 ± 0.21 | 0.02 ± 0.00 | 0.151 |
| H43 | P53 | 1.00 | 1.45 ± 1.30 | 0.29 ± 0.25 | 0.64 ± 0.29 | 0.73 ± 0.53 | 0.24 ± 0.20 | 0.25 ± 0.14 | 0.22 ± 0.06 | 0.128 |
| H44 | Pim1 | 1.00 | 0.69 ± 0.15 | 0.68 ± 0.33 | 0.65 ± 0.30 | 0.69 ± 0.63 | 0.69 ± 0.37 | 1.01 ± 0.10 | 0.62 ± 0.12 | 0.616 |
| H45 | Pparr | 1.00 | 1.39 ± 0.97 | 2.00 ± 0.91 | 2.72 ± 2.49 | 2.57 ± 1.34 | 1.61 ± 0.68 | 1.49 ± 1.13 | 9.39 ± 12.65 | 0.731 |
| H46 | Rab24 | 1.00 | 25.49 ± 31.18 | 10.76 ± 10.86 | 1.68 ± 0.00 | 1.36 ± 1.82 | 8.68 ± 12.21 | 1.69 ± 1.14 | 6.41 ± 6.64 | 0.630 |
| H47 | Rpl32 | 1.00 | 1.06 ± 0.90 | 0.68 ± 0.43 | 0.68 ± 0.22 | 0.46 ± 0.45 | 0.19 ± 0.11 | 0.15 ± 1.18 | 0.02 ± 0.01 | 0.029 |
| H48 | Tnf | 1.00 | 1.22 ± 0.57 | 1.24 ± 0.32 | 1.16 ± 0.52 | 1.47 ± 0.38 | 0.85 ± 0.29 | 1.29 ± 0.14 | 0.71 ± 0.23 | 0.241 |
| H49 | Tnfsf10 | 1.00 | 0.94 ± 0.30 | 1.23 ± 0.76 | 0.93 ± 0.39 | 1.26 ± 0.32 | 0.57 ± 0.57 | 1.01 ± 0.15 | 0.58 ± 0.14 | 0.342 |
| H50 | Ulk2 | 1.00 | 0.88 ± 0.12 | 1.61 ± 0.79 | 0.99 ± 0.37 | 1.22 ± 0.34 | 1.50 ± 0.56 | 1.19 ± 0.35 | 0.84 ± 0.15 | 0.273 |
| H51 | Vegf | 1.00 | 0.001 ± 0.001 | 0.01 ± 0.02 | 0.26 ± 0.35 | 0.001 ± 0.000 | 0.00002 ± 0.00002 | 0.0001 ± 0.0001 | 0.05 ± 0.00 | 0.107 |
| H52 | Zfand5 | 1.00 ab | 0.61 ± 0.47 a | 0.54 ± 0.21 ab | 0.53 ± 0.04 ab | 0.51 ± 0.17 ab | 0.35 ± 0.05 ab | 0.39 ± 0.12 ab | 0.20 ± 0.10 b | 0.048* |
| H53 | Zfp36 | 1.00 | 1.00 ± 0.75 | 0.65 ± 0.20 | 0.59 ± 0.08 | 0.72 ± 0.51 | 0.70 ± 0.11 | 0.81 ± 0.64 | 0.24 ± 0.08 | 0.243 |
| H54 | Zfp36L1 | 1.00 | 1.60 ± 1.48 | 0.12 ± 0.10 | 0.77 ± 0.07 | 0.83 ± 0.77 | 0.13 ± 0.09 | 0.07 ± 0.04 | 0.02 ± 0.00 | 0.050 |
| H55 | Zfp36L2 | 1.00 | 0.32 ± 0.42 | 0.90 ± 0.89 | 0.74 ± 0.97 | 0.08 ± 0.00 | 0.15 ± 0.15 | 0.18 ± 0.08 | 0.01 ± 0.00 | 0.112 |
The data represent the mean and standard deviation of three independent samples. Data with different lowcase letters represent significance between the treatments p < 0.05. ud: undetected. Bold italics: mRNA levels were statistically decreased by gossypol among the treatments with various concentrations. Italics: mRNA levels were statistically increased by gossypol among the treatments with various concentrations. "*" under P value column represents mRNA levels significantly affected by gossypol. Statistical analyses were conducted with Student–Newman–Keuls method for all pairwise multiple comparison. All pairwise multiple comparison was also performed with Tukey test yielding similar results (data not shown).
Gossypol decreased the expression of reference GAPDH and RPL32 genes in human colon cancer cells
The expression of the two well-known reference genes was analyzed in the colon cancer cell line under treatment with various concentration of gossypol using internal reference gene BCL2 selected in this study. The qPCR data showed that GAPDH and RPL32 mRNA levels were 39 and 42 fold of BCL2 in the controlled cells (Table 4). High concentrations of gossypol treatment resulted in a remarkable reduction of both GAPDH and RPL32 mRNA levels in the cells (Fig. 3A). Both GAPDH and RPL32 mRNA levels were reduced more than 80% by 40–100 µg/mL of gossypol treatment (Fig. 3A).
Table 4.
Relative mRNA levels of respective gene families in the human colon cancer cells.
| ID | mRNA | Mean ± SD (n = 24) | Fold |
|---|---|---|---|
| Reference gene | Mean ± SD | Fold of Bcl2 | |
| H2 | Bcl2 | 29.39 ± 1.08 | 1.00 |
| H20 | Gapdh | 24.10 ± 4.01 | 39.09 |
| H47 | Rpl32 | 24.00 ± 3.68 | 41.78 |
| ID | Reported gene | Mean ± SD | Fold of Bcl2 |
|---|---|---|---|
| H2 | Bcl2 | 29.39 ± 1.08 | 1.00 |
| H4 | Bnip3 | 27.54 ± 1.22 | 3.60 |
| H12 | Cyclind1 | 34.10 ± 5.42 | 0.04 |
| H13 | Cyp19a1 | 32.40 ± 3.37 | 0.12 |
| H19 | Fas | 30.63 ± 4.60 | 0.42 |
| H28 | Hua | 32.25 ± 3.52 | 0.14 |
| H43 | P53 | 31.06 ± 2.58 | 0.31 |
| H45 | Pparr | 29.63 ± 1.50 | 0.85 |
| H49 | Tnfsf10 | 28.22 ± 1.48 | 2.24 |
| ID | DGAT family | Mean ± SD | Fold of Dgat1 |
|---|---|---|---|
| H14 | Dgat1 | 29.38 ± 1.86 | 1.00 |
| H15 | Dgat2a | 31.80 ± 2.18 | 0.19 |
| H16 | Dgat2b | 30.88 ± 1.79 | 0.35 |
| ID | GLUT family | Mean ± SD | Fold of Glut1 |
|---|---|---|---|
| H21 | Glut1 | 27.24 ± 2.54 | 1.00 |
| H22 | Glut2 | 29.22 ± 1.81 | 0.25 |
| H23 | Glut3 | 28.61 ± 1.31 | 0.39 |
| H24 | Glut4 | 41.48 ± 4.81 | 0.00 |
| ID | TTP family | Mean ± SD | Fold of Zfp36 |
|---|---|---|---|
| H53 | Zfp36 | 28.86 ± 1.85 | 1.00 |
| H54 | Zfp36L1 | 29.29 ± 3.07 | 0.74 |
| H55 | Zfp36L2 | 41.60 ± 3.61 | 0.00 |
| ID | IL family | Mean ± SD | Fold of Zfp36 |
|---|---|---|---|
| H53 | Zfp36 | 28.86 ± 1.85 | 1.00 |
| H32 | Il2 | 31.27 ± 1.24 | 0.19 |
| H33 | IL6 | 29.09 ± 1.39 | 0.85 |
| H34 | IL8 | 29.14 ± 1.20 | 0.83 |
| H35 | Il10 | 41.42 ± 5.26 | 0.00 |
| H36 | Il12 | 37.55 ± 3.02 | 0.00 |
| H37 | Il16 | 28.49 ± 1.17 | 1.30 |
| H38 | Il17 | 29.56 ± 1.38 | 0.62 |
| ID | Proinflammatory gene | Mean ± SD | Fold of Zfp36 |
|---|---|---|---|
| H53 | Zfp36 | 28.86 ± 1.85 | 1.00 |
| H7 | Cox1 | 34.92 ± 4.79 | 0.01 |
| H8 | Cox2 | 30.65 ± 1.62 | 0.29 |
| H28 | Hua | 32.25 ± 3.52 | 0.10 |
| H39 | Leptin | 29.51 ± 1.22 | 0.64 |
| H48 | Tnf | 30.88 ± 1.71 | 0.24 |
| H49 | Tnfsf10 | 28.22 ± 1.48 | 1.56 |
| H51 | Vegf | 40.47 ± 4.57 | 0.00 |
| ID | TTP targeted other gene | Mean ± SD | Fold of Zfp36 |
|---|---|---|---|
| H53 | Zfp36 | 28.86 ± 1.85 | 1.00 |
| H1 | Ahrr1 | 33.88 ± 1.14 | 0.03 |
| H3 | Bcl2l1 | 27.64 ± 2.34 | 2.33 |
| H5 | Cd36 | 28.45 ± 1.23 | 1.33 |
| H6 | Claudin1 | 28.03 ± 4.30 | 1.78 |
| H9 | Csnk2a1 | 26.02 ± 1.95 | 7.17 |
| H10 | Ctsb | 28.13 ± 2.73 | 1.65 |
| H11 | Cxcl1 | 32.75 ± 2.60 | 0.07 |
| H17 | E2f1 | 29.61 ± 1.08 | 0.59 |
| H18 | Elk1 | 30.82 ± 2.84 | 0.26 |
| H25 | Hif1a | 27.64 ± 2.33 | 2.33 |
| H27 | Hmox1 | 29.68 ± 1.41 | 0.57 |
| H29 | Icam1 | 33.93 ± 4.12 | 0.03 |
| H44 | Pim1 | 29.13 ± 1.09 | 0.83 |
| H52 | Zfand5 | 27.20 ± 1.50 | 3.16 |
| ID | Other gene | Mean ± SD | Fold of Zfp36 |
|---|---|---|---|
| H53 | Zfp36 | 28.86 ± 1.85 | 1.00 |
| H26 | Hmgr | 27.50 ± 1.91 | 2.57 |
| H30 | Inos | ud | ud |
| H31 | Insr | 29.81 ± 3.12 | 0.51 |
| H40 | Map1lc3a | 29.48 ± 1.87 | 0.65 |
| H41 | Map1lc3b | 26.18 ± 1.73 | 6.42 |
| H42 | Nfkb | 31.08 ± 2.97 | 0.22 |
| H46 | Rab24 | 42.25 ± 3.40 | 0.00 |
| H50 | Ulk2 | 29.20 ± 1.11 | 0.79 |
The data represent the mean and standard deviation of 24 independent samples. ud: undetected. The relative mRNA levels were calculated using mean data of Bcl2 as the reference mRNA and the first mRNA in the respective gene family as the control.
Figure 3.
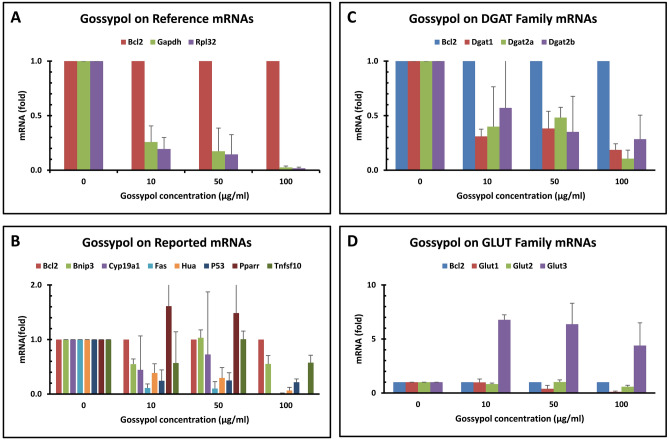
Gossypol regulated the expression of genes coded for qPCR reference mRNAs, genes reported to be regulated by gossypol, and genes coded for DGAT and GLUT mRNAs in human colon cancer cells. Human colon cancer cells (COLO 225) were treated with gossypol for 8 h. The data represent the mean and standard deviation of three independent samples. (A) genes coded for qPCR reference mRNAs, (B) genes reported to be regulated by gossypol, (C) genes coded for DGAT mRNAs, (D) genes coded for GLUT mRNAs.
Gossypol effect on reported gene expression in human colon cancer cells
The expression of a number of genes was shown previously to be regulated by gossypol in cancer cells20,33–39 and macrophages40. We analyzed the expression of BNIP3, CYP19A1, FAS, HUA, P53, PPARR and TNFSF10 genes under various concentrations of gossypol in the colon cancer cell line using BCL2 as the internal reference gene. In general, this group of genes was expressed lower than BCL2 control except BINP3 and TNFSF10 (Table 4). The expression of all these genes except PPARR gene was inhibited to a large extent by the highest concentration of gossypol tested at 100 µg/mL (Fig. 3B). It appears that PPARR gene expression was increased but the large standard deviation among the measurements prevented from making such a conclusion (Fig. 3B).
Gossypol effect on DGAT gene expression in human colon cancer cells
Diacylglycerol acyltransferases (DGATs) esterify sn-1,2-diacylglycerol with a long-chain fatty acyl-CoA and catalyze the rate-limiting step of triacylglycerol biosynthesis in eukaryotic organisms52. DGATs are divided into DGAT1 and DGAT2 subfamilies in animals and DGAT3 subfamily are present in plants52–55. DGAT2 mRNA was the major form of DGAT mRNAs in the mouse adipocytes and macrophages56,57. Gossypol was shown to be a strong stimulator of DGAT2 gene expression in mouse macrophages56. The qPCR data showed here that DGAT1 mRNA was the major form and the two variants of DGAT2 mRNA accounted for only half of the DGAT1 mRNA levels in the human colon cancer cells (Table 4). In contrast to mouse macrophages, we showed here that gossypol inhibited DGAT1 and DGAT2 expression in the human colon cancer cells (Fig. 3C).
Gossypol effect on GLUT gene expression in human colon cancer cells
Glucose transporter (GLUT) family proteins consist of four isoforms which are responsible for glucose uptake in mammalian cells. GLUT1 mRNA is the major form and GLUT2 mRNA is undetectable in macrophages by TaqMan qPCR51,58. In this study, GLUT1 mRNA was also shown to be the major form of GLUT mRNAs but GLUT4 mRNA was barely detected in the colon cancer cells (Table 4). Gossypol treatment at high concentrations significantly decreased GLUT1 mRNA levels but increased GLLUT3 mRNA levels (Fig. 3D).
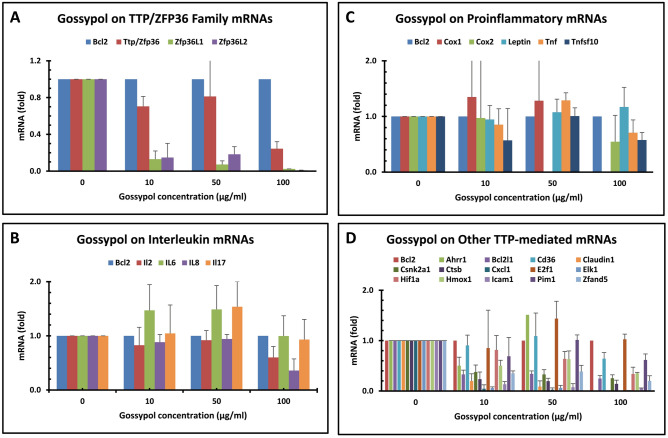
Gossypol effect on TTP gene expression in human colon cancer cells
Tristetraprolin (TTP/ZFP36/TIS11/G0S24/NUP475) family proteins are post-transcriptional factors controlling cytokine mRNA stability. TTP family proteins exhibit anti-inflammation effects with the potential for controlling inflammation-related diseases. TTP family proteins have three members in mammals (ZFP36 or TTP, ZFP36L1 or TIS11B, and ZFP36L2 or TIS11D) and fourth member in mouse and rat (ZFP36L3)59,60. SYBR Green qPCR showed that TTP/ZFP36 and ZFP36L1 were expressed in similar levels but ZFP36L2 mRNA was barely detectable in the colon cancer cells (Table 4). ZFP36, ZFP36L1 and ZFP36L2 mRNAs were all significantly reduced by high-dose of gossypol treatments (Fig. 4A).
Figure 4.
Gossypol regulated the expression of genes coded for TTP family, IL family, TTP-mediated proinflammatory cytokine and other mRNAs in human colon cancer cells. Human colon cancer cells (COLO 225) were treated with gossypol for 8 h. The data represent the mean and standard deviation of three independent samples. (A) genes coded for TTP family mRNAs. (B) genes coded for IL family mRNAs. (C) genes coded for TTP-mediated proinflammatory cytokine mRNAs. (D) genes coded for other TTP-mediated mRNAs.
Gossypol effect on IL gene expression in human colon cancer cells
Several members of the interleukins (ILs) are regulated by TTP family proteins which bind to AU-rich elements (ARE) in some cytokine mRNAs and destabilizes those transcripts. TTP-mediated ILs include IL261, IL662, IL863, IL1064, IL1265, IL1647 and IL1766. SYBR Green qPCR showed that IL10 and IL12 mRNAs were barely expressed and IL2 mRNA was low, whereas TTP and the other ILs were expressed in similar levels, which were several fold higher than IL2 mRNA in the human colon cancer cells (Table 4). The qPCR assays showed that gossypol decreased IL2 and IL8 mRNA levels but its effect on IL6 and IL17 was not apparent (Table 3 and Fig. 4B).
Gossypol effect on proinflammatory gene expression in human colon cancer cells
The major mRNAs destabilized by TTP family proteins are proinflammatory cytokine mRNA molecules. They down-regulate the expression of mRNAs encoding cytokines such as tumor necrosis factor-alpha (TNFα)67–70, granulocyte–macrophage colony-stimulating factor/colony-stimulating factor 2 (GM-CSF/CSF2)71,72 and cyclooxygenase 2/prostaglandin-endoperoxide synthase 2 (COX2/PTGS2)48. TNFα and GM-CSF mRNAs are stabilized in TTP knockout mice cells69,71. TTP knockout mice over-express these proinflammatory cytokines which cause a severe systemic inflammatory syndrome73,74. TTP over-expression reduces inflammatory responses75. These previous studies suggest that TTP is an anti-inflammatory protein. Except TNFSF10 mRNA, all the other proinflammatory mRNAs were expressed much lower than that of TTP and VEGF mRNA was almost undetectable in the colon cancer cells (Table 4). Gossypol did not exhibit significant effects on the mRNA levels of all these proinflammatory gene expression in the human colon cancer cells (Fig. 4C).
Gossypol effect on TTP-targeted other gene expression in human colon cancer cells
A number of other TTP-mediated mRNAs have been reported in the literature (Table 1). The basal levels of these mRNAs were either higher than that of TTP mRNA (BCL2L1, CsnK2A1, HIF1a and ZFAND5) or lower than that of TTP mRNA (Ahrr1, CXCL1, E2F1, ELK1, HMOX1 and ICAM1) (Table 4). Gossypol treatment resulted in a reduction of many of the mRNAs in the colon cancer cells (Fig. 4D).
Discussion
Cottonseed accounts for approximately 20% of the crop value. One way to increase the value of cottonseed is to isolate bioactive compounds aimed to improving nutrition and preventing diseases. The presence of toxic polyphenolic compound gossypol in the seeds limits its use as food and feed source for humans and non-ruminant animals2–6. On the other hand, recent studies have shown that gossypol has potential biomedical applications. This may significantly increase cottonseed value by using gossypol from the seeds as a health intervention agent. However, it is necessary to insure safety and effectiveness of gossypol as well as the underlining molecular mechanisms before human consumption. Therefore, we evaluated the effects of gossypol on toxicity and gene expression in human colon cancer cells.
In this study, we observed that gossypol significantly decreased the cell viability of human colon cancer cells (Fig. 2). Our previous study showed that gossypol inhibited breast and pancreas cancer cell viability76. The effect of gossypol on decreasing breast cancer cell (MCF-7) viability76 was in agreement with those using gossypol, gossypol derivative, and gossypol-enriched cottonseed oil16,17. Gossypol also decreased pancreatic cancer cell viability after short-term treatment76 which is in agreement with published reports20,77–79.
Before we examined the effect of gossypol on gene expression in human colon cancer cells, we evaluated the relative expression levels of 55 genes and selected the internal reference for qPCR analysis since it is important for normalization of gene expression levels80–83. Our study showed that BCL2 mRNA was the most stable among the mRNAs from 55 genes analyzed in human colon cancer cells treated with DMSO vehicle or various concentrations of gossypol (Table 2). This result is in consistent with a previous report showing that the effect of gossypol analog on BCL2 gene expression was minimal at the mRNA level31. Our results showed that GAPDH and RPL32 (60S ribosomal protein 32) mRNAs were not good qPCR assay references for the colon cancer cells since they were most abundant mRNAs with large variations under the cell culture conditions. This is in agreement with a previous report showing that GAPDH and 18s RNA mRNAs are not reliable references for qPCR assays in pharmacogenomics and toxicogenomics studies83. In contrast, RPL32 mRNA was shown to be a good reference for qPCR assays in mouse, rat and human post-infarction heart failure80 and mouse adipocytes and macrophages57,82. These results suggest that internal reference mRNA is probably different among the various tissues/cells tested.
Our study showed that most of the gene expression in human colon cancer cells was suppressed by high concentrations of gossypol (Table 3). Some of the p values were less than 5% threshold, suggesting their expression levels were statistically significant. By this standard, gossypol significantly decreased the expression of genes coding for mRNAs of CLAUDIN1, ELK1, FAS, GAPDH, IL2, IL8 and ZFAND5, but increased the expression of the gene coding for GLUT3 mRNA. These genes code for proteins involved in various biological processes and cancer development. CLAUDIN1 is a tight junction protein which is highly upregulated in colon cancer49. ELK1 is a transcription factor playing an important role in immunological response, which belongs to ETS protein family with the evolutionary conserved ETS domain stabilized by three key tryptophan residues interacting with DNA42. FAS is involved in the apoptotic system which is upregulated in human lung cancer cells (A549) by gossypol treatment for 12 h at 0.5 µmol/L (~ 0.26 µg/mL)34. GAPDH catalyzes the sixth step of glycolysis and serves to break down glucose for energy and carbon demands83. IL2 is an autocrine and paracrine growth factor involved in clonal T cell expansion, influences the magnitude and duration of an immune response, and contributes to the regulation of programmed cell death in T cells which is down-regulated by TTP through ARE-mediated mRNA decay61. IL8 induces chemotaxis in target cells, causes them to migrate toward the site of infection, and stimulates phagocytosis once they have arrived63. ZFAND5 enhances ARE-containing mRNA stability by competing with TTP for mRNA binding84. GLUT3 expressed specifically in neurons facilitates the transport of glucose across the plasma membranes of mammalian cells85.
Among the tested 55 genes, gossypol treatment inhibited the expression of well-known reference genes coding for GAPDH and RPL32 mRNAs (Fig. 3A). Among the genes reported to be regulated by gossypol in cancer cells20,33–39 and macrophages40, gossypol decreased the mRNA levels of BNIP3, CYP19A1, FAS, HUA, P53, PPARR and TNFSF10 genes in the human colon cancer cells (Fig. 3B). Gossypol decreased the mRNA levels of almost all of the DAGT, GLUT, TTP and IL gene families except GLUT3 in the cancer cells (Fig. 3C, D, 4A, B). The effect of gossypol on COX2, TNF and other proinflammatory cytokine mRNAs was not apparent (Fig. 4C), although it decreased the levels of a number of other TTP-regulated mRNAs coding for various functional proteins (Fig. 4D).
In our previous studies, gossypol significantly increased the expression of DGAT and HuR mRNAs in mouse RAW264.7 macrophages40,56. However, DGAT and HuR mRNAs were decreased by gossypol in the human colon cancer cells. Similarly, gossypol decreased FAS mRNA in colon cancer cells reported here but increased its expression in human lung cancer cells reported previously34. Finally, the mRNA levels of TTP family genes were shown here to be decreased in the colon cancer cells but reported to be increased in mouse macrophages86. These discrepancies might reflect the different responses of normal mouse macrophages and different human cancer cells to gossypol treatment.
This study provides evidence for the toxic effects of gossypol on cell viability and its effects on down-regulation of many gene expression at the mRNA level in the human colon cancer cells. However, there are a few limitations involved in the study which should be addressed in future study. First, the findings were derived from one colon cancer cell line (COLO225). It could be valuable to expand the scope of research with multiple cancer cell lines. Second, the general pattern of gossypol on decreasing mRNA levels of numerous genes was evident. However, the dosage effect of gossypol on mRNA levels was not strong and the standard deviations were large in many cases, probably due to extremely sensitive qPCR assays and many factors affecting the results from extracellular gossypol application, cell harvest, RNA extraction, cDNA synthesis to qPCR analysis. Third, it was a firm conclusion that gossypol negatively regulated many gene expression at the mRNA levels in the colon cancer cells. However, it could be great addition to confirm mRNA data at the protein level. Even though positive correlations between mRNA levels and protein levels are not guaranteed as shown in many publications, it is still valuable to perform experiments at the protein levels. Finally, this study provides evidence for gossypol’s toxic effects on cell viability and gene expression at the mRNA level in the human colon cancer cells. However, there is no functional analysis of gossypol affecting any intermediate steps between mRNA changes and cell viability. All these aspects are all worth of further investigation.
In conclusion, this study showed that gossypol significantly reduced the viability of human colon cancer cells. We further showed that BCL2 mRNA was the most stable among the 55 mRNAs in human colon cancer cells. Gossypol decreased the mRNA levels of DGAT, GLUT, TTP, ILs families and a number of previously reported genes. In particular, gossypol significantly suppressed the expression of the genes coding for CLAUDIN1, ELK1, FAS, GAPDH, IL2, IL8 and ZFAND5 mRNAs, but enhanced the expression of the gene coding for GLUT3 mRNA. This study provided evidence for potentially increasing the value of cottonseed by using cottonseed-derived gossypol as a health intervention agent.
Materials and methods
Colon cancer cell line
Human colon cancer cell line (COLO 205-ATCC CCL-222) was purchased from American Type Culture Collection (Manassas, VA) and kept under liquid nitrogen vapor in a Cryogenic Storage Vessel (Thermo Fisher Scientific, Waltham, MA). The cells were maintained at 37 °C in a humidified incubator with 5% CO2 in RPMI-1640 medium (Gibco, Life Technologies) supplemented with 10% (v:v) fetal bovine serum, 0.1 million units/L penicillin, 100 mg/L streptomycin, and 2 mmol/L L-glutamine.
Chemicals, reagents and equipment
The chemicals, reagents and equipment were described essentially as previously56. Gossypol (molar mass: 518.56 g/mol) was purified from cottonseed by HPLC and purchased from Sigma (St. Louis, MO). Gossypol stock was prepared in 100% DMSO at 10 mg/mL (approximately 19.2 mM). Cell cytotoxicity reagent (MTT based-In Vitro Toxicology Assay Kit) and DMSO were from Sigma. Tissue culture reagents (RPMI-1640, fetal bovine serum, penicillin, streptomycin, L-glutamine) were from Gibco BRL (Thermo Fisher). Tissue culture incubator was water jacket CO2 incubator, Forma Series II, Model 3100 Series (Thermo Fisher). Tissue culture workstation was Logic + A2 hood (Labconco, Kansas City, MO). Tissue culture plastic ware (flasks, plates, cell scraper) was from CytoOne (USA Scientific, Ocala, FL). Cell counting reagent (trypsin blue dye), slides (dual chamber), counter (TC20 Automatic Cell Counter) and microscope (Zoe Florescent Cell Imager) were from Bio-Rad (Hercules, CA). Microplate spectrophotometer (Epoch) was from BioTek Instruments (Winooski, VT).
Cell culture and chemical treatment
The basic cell culture protocol was following previous procedures51,68,87. Cancer cells were dissociated from the T-75 flask with 0.25% (w/v) trypsin-0.53 mM EDTA solution, stained with equal volume of 0.4% trypsin blue dye before counting the number of live cells with a TC20 Automatic Cell Counter. Cancer cells (0.5 mL) from trypsin-dissociated flasks were subcultured at approximately 1 × 105 cells/mL density in 24-well tissue culture plates. The cancer cells were routinely observed under a Zoe Florescent Cell Imager before and during treatment. Cancer cells were treated with 0, 0.1, 0.5, 1, 5, 10, 50 and 100 µg/mL (corresponding to 0, 0.19, 0.96, 1.92, 9.6, 19.2, 96 and 192 µM) of gossypol for 2, 4, 8 and 24 h (“0” treatment corresponding to 1% DMSO in the culture medium, the vehicle control for the experiment). These gossypol concentrations were in the range of previously published concentrations for gossypol (up to 100 µM)18,19,88, (-) gossypol (up to 100 µM)30, apogossypolone (up to 40 µM)89, and gossypol derivatives (IC50 concentrations of 6–28 µM)31.
Cell viability assay
Cell cytotoxicity was determined with the MTT based-In Vitro Toxicology Assay Kit76. Cancer cells in 96-well plates (12 wells/treatment) were treated with gossypol and incubated at 37 °C, 5% CO2 for 2, 4, 8 and 24 h. The cell media were added with 50 µL of MTT assay reagent (thiazolyl blue tetrazolium bromide) and incubated at 37 °C, 5% CO2 for 2 h before adding 500 µL MTT solubilization solution to each well, shaken at room temperature overnight. The color density in the wells was recorded by Epoch microplate spectrophotometer at A570.
qPCR primers and probes
A total of 55 genes were selected for qPCR analysis. The selection of genes was based on the literature showing those gene expression regulated by gossypol in cancer cells20,33–39 and macrophages40 or regulated by ZFP36/TTP in tumor cells41–49 and macrophages50,51 (Table 1). RNA sequences were obtained from the National Center for Biotechnology Information (NCBI)’s non-redundant protein sequence databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The qPCR primers were designed using Primer Express software (Applied Biosystems, Foster City, CA) and synthesized by Biosearch Technologies, Inc. (Navato, CA). The names of mRNAs and their nucleotide sequences (5′ to 3′) of the forward primers and reverse primers, and corresponding references are described in Table 1.
RNA isolation and cDNA synthesis
The methods for RNA isolation and cDNA synthesis were essentially as described56. Human colon cancer cells in 24-well plates (triplicate) treated with various concentrations of gossypol for 8 h, a treatment showing significant reduction on cell viability (Fig. 2). The dishes were washed twice with 1 mL 0.9% NaCl and lysed directly with 1 mL of TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). RNA was isolated according to the manufacturer’s instructions without DNase treatment and stored in -80 °C freezer. RNA concentrations were quantified with an Implen NanoPhotometer (Munchen, Germany). The cDNAs were synthesized from total RNA using SuperScript II reverse transcriptase. The cDNA synthesis mixture (20 μL) contained 5 μg total RNA, 2.4 μg oligo(dT)12–18 primer, 0.1 μg random primers, 500 μM dNTPs, 10 mM DTT, 40 u RNaseOUT and 200 u SuperScript II reverse transcriptase in 1X first-strand synthesis buffer (Life Technologies, Carlsbad, CA). The cDNA synthesis reaction was performed at 42 °C for 50 min. The cDNA was stored in − 80 °C freezer and diluted with water to 1 ng/µL before qPCR analyses.
Quantitative real-time PCR analysis
The qPCR assays followed the MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments90. The qPCR assays were described in details previously54,82,91,92. SYBR Green qPCR reaction mixture (12.5 μL) contained 5 ng of total RNA-derived cDNA, 200 nM each of the forward primer and reverse primer, and 1 × iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA). The reactions in 96-well clear plates sealed by adhesives were performed with CFX96 real-time system-C1000 Thermal Cycler (Bio-Rad Laboratories). The thermal cycle conditions were as follows: 3 min at 95 °C, followed by 40 cycles at 95 °C for 10 s, 65 °C for 30 s and 72 °C for 30 s. BCL2 mRNA was selected as the internal reference based on its minimal variation of gene expression among the 55 genes tested in the colon cancer cells (see “Results” for details). Ribosome protein 32 (RPL32) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs were widely used as the reference mRNAs in the qPCR analyses87 but they were not suitable for qPCR analysis for this cell type (see “Results” for details). TaqMan qPCR assay was used to confirm some SBYR Green qPCR assays using identical conditions as described previously82.
Data analysis and statistics
The 2−ΔCT and 2−ΔΔCT method of relative quantification was used to determine the fold change in expression93. This was done by first normalizing the resulting threshold cycle (CT) values of the target mRNAs to the CT values of the internal control BCL2 mRNA in the same cells (ΔCT = CTTarget − CTBcl2). Gossypol-treated qPCR ΔCT values was further normalized with DMSO qPCR ΔCT values in the same cells (ΔΔCT = ΔCTGossypol − ΔCTDMSO). The fold change in expression was then obtained (2−ΔΔCT). The data in the figures and tables represent the mean and standard deviation of various independent samples: Figure 2 (n = 12), Figs. 3 and 4 (n = 3), Tables 2 and 4 (n = 24), Table 3 (n = 3). They were analyzed by statistical analysis using ANOVA with SigmaStat 3.1 software (Systat Software). Multiple comparisons among the treatments with different concentrations of gossypol were performed with Student–Newman–Keuls method and Tukey test87.
Acknowledgements
We thank Dr. Thomas Klasson for suggesting statistical methods for data analysis and for general support of the research. This work was supported by the USDA-ARS Quality and Utilization of Agricultural Products National Program 306 through ARS Research Project 6054-41000-103-00-D. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Abbreviations
- DMSO
Dimethylsulfoxide
- LPS
Lipopolysaccharides
- MTT
3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide
- TTP
Tristetraprolin
- ZFP36
Zinc finger protein 36
Author contributions
H.C. designed the experiments. H.C. and K.S. performed the experiments. H.C., K.S. and F.C. analyzed the data, H.C. wrote the manuscript. H.C., F.C. and T.T.Y.W. revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kenar JA. Reaction chemistry of gossypol and its derivatives. J. Am. Oil Chem. Soc. 2006;83:269–302. doi: 10.1007/s11746-006-1203-1. [DOI] [Google Scholar]
- 2.Coutinho EM. Gossypol: a contraceptive for men. Contraception. 2002;65:259–263. doi: 10.1016/S0010-7824(02)00294-9. [DOI] [PubMed] [Google Scholar]
- 3.Camara AC, et al. Toxicity of gossypol from cottonseed cake to sheep ovarian follicles. PLoS ONE. 2015;10:e0143708. doi: 10.1371/journal.pone.0143708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadelha IC, Fonseca NB, Oloris SC, Melo MM, Soto-Blanco B. Gossypol toxicity from cottonseed products. Sci. World J. 2014;2014:231635. doi: 10.1155/2014/231635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randel RD, Chase CC, Jr, Wyse SJ. Effects of gossypol and cottonseed products on reproduction of mammals. J. Anim. Sci. 1992;70:1628–1638. doi: 10.2527/1992.7051628x. [DOI] [PubMed] [Google Scholar]
- 6.Zeng QF, et al. Effects of dietary gossypol concentration on growth performance, blood profiles, and hepatic histopathology in meat ducks. Poult. Sci. 2014;93:2000–2009. doi: 10.3382/ps.2013-03841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lusas EW, Jividen GM. Glandless cottonseed: a review of the first 25 years of processing and utilization research. J. Am. Oil Chem. Soc. 1987;64:839–854. doi: 10.1007/BF02641491. [DOI] [Google Scholar]
- 8.Alford BB, Liepa GU, Vanbeber AD. Cottonseed protein: what does the future hold? Plant Foods Hum. Nutr. 1996;49:1–11. doi: 10.1007/BF01092517. [DOI] [PubMed] [Google Scholar]
- 9.Cornu A, Delpeuch F, Favier JC. Utilization of gossypol-free cottonseed and its by-products as human food. Ann. Nutr. Aliment. 1977;31:349–364. [PubMed] [Google Scholar]
- 10.Sneed SM, Thomas MR, Alford BB. Effects of a glandless cottonseed protein diet on fasting plasma amino acid levels in college women. Am. J. Clin. Nutr. 1980;33:287–292. doi: 10.1093/ajcn/33.2.287. [DOI] [PubMed] [Google Scholar]
- 11.Thomas MR, Ashby J, Sneed SM, O'Rear LM. Minimum nitrogen requirement from glandless cottonseed protein for nitrogen balance in college women. J. Nutr. 1979;109:397–405. doi: 10.1093/jn/109.3.397. [DOI] [PubMed] [Google Scholar]
- 12.Hendricks JD, Sinnhuber RO, Loveland PM, Pawlowski NE, Nixon JE. Hepatocarcinogenicity of glandless cottonseeds and cottonseed oil to rainbow trout (Salmo gairdnerii) Science. 1980;208:309–311. doi: 10.1126/science.6892734. [DOI] [PubMed] [Google Scholar]
- 13.Palle SR, et al. RNAi-mediated ultra-low gossypol cottonseed trait: performance of transgenic lines under field conditions. Plant Biotechnol. J. 2013;11:296–304. doi: 10.1111/pbi.12013. [DOI] [PubMed] [Google Scholar]
- 14.Rathore KS, et al. Ultra-low gossypol cottonseed: generational stability of the seed-specific, RNAi-mediated phenotype and resumption of terpenoid profile following seed germination. Plant Biotechnol. J. 2012;10:174–183. doi: 10.1111/j.1467-7652.2011.00652.x. [DOI] [PubMed] [Google Scholar]
- 15.Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. USA. 2006;103:18054–18059. doi: 10.1073/pnas.0605389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong S, et al. (-)-Gossypol-enriched cottonseed oil inhibits proliferation and adipogenesis of human breast pre-adipocytes. Anticancer Res. 2013;33:949–955. [PubMed] [Google Scholar]
- 17.Liu S, et al. The (-)-enantiomer of gossypol possesses higher anticancer potency than racemic gossypol in human breast cancer. Anticancer Res. 2002;22:33–38. [PubMed] [Google Scholar]
- 18.Messeha SS, et al. Effects of gossypol on apoptosis-related gene expression in racially distinct triple-negative breast cancer cells. Oncol. Rep. 2019;42:467–478. doi: 10.3892/or.2019.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien CC, Ko CH, Shen SC, Yang LY, Chen YC. The role of COX-2/PGE2 in gossypol-induced apoptosis of colorectal carcinoma cells. J. Cell Physiol. 2012;227:3128–3137. doi: 10.1002/jcp.23067. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Y, et al. Gossypol and an HMT G9a inhibitor act in synergy to induce cell death in pancreatic cancer cells. Cell Death Dis. 2013;4:e690. doi: 10.1038/cddis.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakur A, et al. Pan-Bcl-2 inhibitor AT-101 enhances tumor cell killing by EGFR targeted T cells. PLoS ONE. 2012;7:e47520. doi: 10.1371/journal.pone.0047520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang X, et al. (-)-Gossypol suppresses the growth of human prostate cancer xenografts via modulating VEGF signaling-mediated angiogenesis. Mol. Cancer Ther. 2011;10:795–805. doi: 10.1158/1535-7163.MCT-10-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YW, Wang LS, Dowd MK, Wan PJ, Lin YC. (-)-Gossypol reduces invasiveness in metastatic prostate cancer cells. Anticancer Res. 2009;29:2179–2188. [PubMed] [Google Scholar]
- 24.Zhong S, et al. Aromatase expression in leptin-pretreated human breast pre-adipocytes is enhanced by zeranol and suppressed by (-)-gossypol. Anticancer Res. 2010;30:5077–5084. [PubMed] [Google Scholar]
- 25.Huo M, et al. Suppression of LPS-induced inflammatory responses by gossypol in RAW 264.7 cells and mouse models. Int. Immunopharmacol. 2013;15:442–449. doi: 10.1016/j.intimp.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Oskoueian E, Abdullah N, Hendra R, Karimi E. Bioactive compounds, antioxidant, xanthine oxidase inhibitory, tyrosinase inhibitory and anti-inflammatory activities of selected agro-industrial by-products. Int. J. Mol. Sci. 2011;12:8610–8625. doi: 10.3390/ijms12128610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellon JE, Zelaya CA, Dowd MK, Beltz SB, Klich MA. Inhibitory effects of gossypol, gossypolone, and apogossypolone on a collection of economically important filamentous fungi. J. Agric. Food Chem. 2012;60:2740–2745. doi: 10.1021/jf2044394. [DOI] [PubMed] [Google Scholar]
- 28.Puckhaber LS, Dowd MK, Stipanovic RD, Howell CR. Toxicity of (+)- and (-)-gossypol to the plant pathogen Rhizoctonia solani. J. Agric. Food Chem. 2002;50:7017–7021. doi: 10.1021/jf0207225. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, et al. A novel bioavailable BH3 mimetic efficiently inhibits colon cancer via cascade effects of mitochondria. Clin. Cancer Res. 2016;22:1445–1458. doi: 10.1158/1078-0432.CCR-15-0732. [DOI] [PubMed] [Google Scholar]
- 30.Lan L, et al. Natural product (-)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol. Oncol. 2015;9:1406–1420. doi: 10.1016/j.molonc.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan F, et al. A novel water-soluble gossypol derivative increases chemotherapeutic sensitivity and promotes growth inhibition in colon cancer. J. Med. Chem. 2010;53:5502–5510. doi: 10.1021/jm1001698. [DOI] [PubMed] [Google Scholar]
- 32.Yang D, et al. Gossypol sensitizes the antitumor activity of 5-FU through down-regulation of thymidylate synthase in human colon carcinoma cells. Cancer Chemother. Pharmacol. 2015;76:575–586. doi: 10.1007/s00280-015-2749-0. [DOI] [PubMed] [Google Scholar]
- 33.Barba-Barajas M, et al. Gossypol induced apoptosis of polymorphonuclear leukocytes and monocytes: involvement of mitochondrial pathway and reactive oxygen species. Immunopharmacol. Immunotoxicol. 2009;31:320–330. doi: 10.1080/08923970902718049. [DOI] [PubMed] [Google Scholar]
- 34.Chang JS, Hsu YL, Kuo PL, Chiang LC, Lin CC. Upregulation of Fas/Fas ligand-mediated apoptosis by gossypol in an immortalized human alveolar lung cancer cell line. Clin. Exp. Pharmacol. Physiol. 2004;31:716–722. doi: 10.1111/j.1440-1681.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 35.Dong Y, et al. Gossypol enantiomers potently inhibit human placental 3beta-hydroxysteroid dehydrogenase 1 and aromatase activities. Fitoterapia. 2015;109:132–137. doi: 10.1016/j.fitote.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Huang YW, et al. Molecular mechanisms of (-)-gossypol-induced apoptosis in human prostate cancer cells. Anticancer Res. 2006;26:1925–1933. [PubMed] [Google Scholar]
- 37.Kitada S, et al. Bcl-2 antagonist apogossypol (NSC736630) displays single-agent activity in Bcl-2-transgenic mice and has superior efficacy with less toxicity compared with gossypol (NSC19048) Blood. 2008;111:3211–3219. doi: 10.1182/blood-2007-09-113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ligueros M, et al. Gossypol inhibition of mitosis, cyclin D1 and Rb protein in human mammary cancer cells and cyclin-D1 transfected human fibrosarcoma cells. Br. J. Cancer. 1997;76:21–28. doi: 10.1038/bjc.1997.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeow WS, et al. Gossypol, a phytochemical with BH3-mimetic property, sensitizes cultured thoracic cancer cells to Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J. Thorac. Cardiovasc. Surg. 2006;132:1356–1362. doi: 10.1016/j.jtcvs.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 40.Cao H, Sethumadhavan K. Gossypol but not cottonseed extracts or lipopolysaccharides stimulates HuR gene expression in mouse cells. J. Funct. Foods. 2019;59:25–29. doi: 10.1016/j.jff.2019.05.022. [DOI] [Google Scholar]
- 41.Essafi-Benkhadir K, Onesto C, Stebe E, Moroni C, Pages G. Tristetraprolin inhibits ras-dependent tumor vascularization by inducing VEGF mRNA degradation. Mol. Biol. Cell. 2007;18:4648–4658. doi: 10.1091/mbc.e07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Florkowska M, et al. EGF activates TTP expression by activation of ELK-1 and EGR-1 transcription factors. BMC Mol. Biol. 2012;13:8. doi: 10.1186/1471-2199-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HK, et al. Expression of proviral integration site for Moloney murine leukemia virus 1 (Pim-1) is post-transcriptionally regulated by tristetraprolin in cancer cells. J. Biol. Chem. 2012;287:28770–28778. doi: 10.1074/jbc.M112.376483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HH, Lee SR, Leem SH. Tristetraprolin regulates prostate cancer cell growth through suppression of E2F1. J. Microbiol. Biotechnol. 2014;24:287–294. doi: 10.4014/jmb.1309.09070. [DOI] [PubMed] [Google Scholar]
- 45.Lee HH, et al. Tristetraprolin suppresses AHRR expression through mRNA destabilization. FEBS Lett. 2013;587:1518–1523. doi: 10.1016/j.febslet.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 46.Lee WH, et al. Casein kinase 2 regulates the mRNA-destabilizing activity of tristetraprolin. J. Biol. Chem. 2011;286:21577–21587. doi: 10.1074/jbc.M110.201137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milke L, et al. Depletion of tristetraprolin in breast cancer cells increases interleukin-16 expression and promotes tumor infiltration with monocytes/macrophages. Carcinogenesis. 2013;34:850–857. doi: 10.1093/carcin/bgs387. [DOI] [PubMed] [Google Scholar]
- 48.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3'-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 49.Sharma A, Bhat AA, Krishnan M, Singh AB, Dhawan P. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis. 2013;34:2610–2621. doi: 10.1093/carcin/bgt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao H, Polansky MM, Anderson RA. Cinnamon extract and polyphenols affect the expression of tristetraprolin, insulin receptor, and glucose transporter 4 in mouse 3T3-L1 adipocytes. Arch. Biochem. Biophys. 2007;459:214–222. doi: 10.1016/j.abb.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 51.Cao H, Urban JF, Jr, Anderson RA. Cinnamon polyphenol extract affects immune responses by regulating anti- and proinflammatory and glucose transporter gene expression in mouse macrophages. J. Nutr. 2008;138:833–840. doi: 10.1093/jn/138.5.833. [DOI] [PubMed] [Google Scholar]
- 52.Liu Q, Siloto RM, Lehner R, Stone SJ, Weselake RJ. Acyl-CoA:diacylglycerol acyltransferase: Molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012;51:350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Cao H. Structure-function analysis of diacylglycerol acyltransferase sequences from 70 organisms. BMC Res. Notes. 2011;4:249. doi: 10.1186/1756-0500-4-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao H, Shockey JM, Klasson KT, Mason CB, Scheffler BE. Developmental regulation of diacylglycerol acyltransferase family gene expression in tung tree tissues. PLoS ONE. 2013;8:e76946. doi: 10.1371/journal.pone.0076946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shockey JM, et al. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao H, Sethumadhavan K. Cottonseed extracts and gossypol regulate diacylglycerol acyltransferase gene expression in mouse macrophages. J. Agric. Food Chem. 2018;66:6022–6030. doi: 10.1021/acs.jafc.8b01240. [DOI] [PubMed] [Google Scholar]
- 57.Cao H. Identification of the major diacylglycerol acyltransferase mRNA in mouse adipocytes and macrophages. BMC. Biochem. 2018;19:1–11. doi: 10.1186/s12858-018-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fukuzumi M, Shinomiya H, Shimizu Y, Ohishi K, Utsumi S. Endotoxin-induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infect. Immun. 1996;64:108–112. doi: 10.1128/iai.64.1.108-112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 60.Blackshear PJ, et al. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the tristetraprolin family of CCCH tandem zinc finger proteins. Biol. Reprod. 2005;73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 61.Ogilvie RL, et al. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 62.Hochdorfer T, et al. LPS-induced production of TNF-alpha and IL-6 in mast cells is dependent on p38 but independent of TTP. Cell Signal. 2013;25:1339–1347. doi: 10.1016/j.cellsig.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balakathiresan NS, et al. Tristetraprolin regulates IL-8 mRNA stability in cystic fibrosis lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2009;296:L1012–L1018. doi: 10.1152/ajplung.90601.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaba A, et al. Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. J. Immunol. 2012;189:2089–2093. doi: 10.4049/jimmunol.1201126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu L, et al. Suppression of IL-12 production by tristetraprolin through blocking NF-kcyB nuclear translocation. J. Immunol. 2013;191:3922–3930. doi: 10.4049/jimmunol.1300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Datta S, et al. IL-17 regulates CXCL1 mRNA stability via an AUUUA/tristetraprolin-independent sequence. J. Immunol. 2010;184:1484–1491. doi: 10.4049/jimmunol.0902423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao H, Dzineku F, Blackshear PJ. Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases. Arch. Biochem. Biophys. 2003;412:106–120. doi: 10.1016/S0003-9861(03)00012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao H. Expression, purification, and biochemical characterization of the antiinflammatory tristetraprolin: a zinc-dependent mRNA binding protein affected by posttranslational modifications. Biochemistry. 2004;43:13724–13738. doi: 10.1021/bi049014y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 70.Lai WS, et al. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/MCB.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. doi: 10.1182/blood.V95.6.1891. [DOI] [PubMed] [Google Scholar]
- 72.Carballo E, et al. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc. Natl. Acad. Sci. USA. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/S1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 75.Sauer I, et al. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao H, Sethumadhavan K, Bland JM. Isolation of cottonseed extracts that affect human cancer cell growth. Sci. Rep. 2018;8:10458. doi: 10.1038/s41598-018-28773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banerjee S, et al. Preclinical studies of apogossypolone, a novel pan inhibitor of bcl-2 and mcl-1, synergistically potentiates cytotoxic effect of gemcitabine in pancreatic cancer cells. Pancreas. 2010;39:323–331. doi: 10.1097/MPA.0b013e3181bb95e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benz CC, et al. Biochemical correlates of the antitumor and antimitochondrial properties of gossypol enantiomers. Mol. Pharmacol. 1990;37:840–847. [PubMed] [Google Scholar]
- 79.Zubair H, et al. Mobilization of Intracellular Copper by Gossypol and Apogossypolone Leads to Reactive Oxygen Species-Mediated Cell Death: Putative Anticancer Mechanism. Int. J. Mol. Sci. 2016;17:973. doi: 10.3390/ijms17060973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brattelid, T. et al. Reference gene alternatives to Gapdh in rodent and human heart failure gene expression studies. BMC. Mol. Biol.11, 22 (2010). [DOI] [PMC free article] [PubMed]
- 81.Cao H, Shockey JM. Comparison of TaqMan and SYBR Green qPCR methods for quantitative gene expression in tung tree tissues. J. Agric. Food Chem. 2012;60:12296–12303. doi: 10.1021/jf304690e. [DOI] [PubMed] [Google Scholar]
- 82.Cao H, Cao F, Roussel AM, Anderson RA. Quantitative PCR for glucose transporter and tristetraprolin family gene expression in cultured mouse adipocytes and macrophages. Vitro Cell Dev. Biol. Anim. 2013;49:759–770. doi: 10.1007/s11626-013-9671-8. [DOI] [PubMed] [Google Scholar]
- 83.Chen D, Pan X, Xiao P, Farwell MA, Zhang B. Evaluation and identification of reliable reference genes for pharmacogenomics, toxicogenomics, and small RNA expression analysis. J. Cell Physiol. 2011;226:2469–2477. doi: 10.1002/jcp.22725. [DOI] [PubMed] [Google Scholar]
- 84.He G, Sun D, Ou Z, Ding A. The protein Zfand5 binds and stabilizes mRNAs with AU-rich elements in their 3'-untranslated regions. J. Biol. Chem. 2012;287:24967–24977. doi: 10.1074/jbc.M112.362020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao H, Graves DJ, Anderson RA. Cinnamon extract regulates glucose transporter and insulin-signaling gene expression in mouse adipocytes. Phytomedicine. 2010;17:1027–1032. doi: 10.1016/j.phymed.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 86.Cao, H. Proceedings of the 2019 Beltwide Cotton Conferences, New Orleans, Louisiana 559–571 (National Cotton Council of America, 2019).
- 87.Cao H, Anderson RA. Cinnamon polyphenol extract regulates tristetraprolin and related gene expression in mouse adipocytes. J. Agric. Food Chem. 2011;59:2739–2744. doi: 10.1021/jf103527x. [DOI] [PubMed] [Google Scholar]
- 88.Ko CH, Shen SC, Yang LY, Lin CW, Chen YC. Gossypol reduction of tumor growth through ROS-dependent mitochondria pathway in human colorectal carcinoma cells. Int. J. Cancer. 2007;121:1670–1679. doi: 10.1002/ijc.22910. [DOI] [PubMed] [Google Scholar]
- 89.Hu ZY, et al. Apogossypolone targets mitochondria and light enhances its anticancer activity by stimulating generation of singlet oxygen and reactive oxygen species. Chin. J. Cancer. 2011;30:41–53. doi: 10.5732/cjc.010.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 91.Cao H, Shockey JM. Comparison of TaqMan and SYBR Green qPCR methods for quantitative gene expression in tung tree tissues. J Agric. Food Chem. 2012;60:12296–12303. doi: 10.1021/jf304690e. [DOI] [PubMed] [Google Scholar]
- 92.Cao H, Cao F, Klasson KT. Characterization of reference gene expression in tung tree (Vernicia fordii) Ind. Crops Prod. 2013;50:248–255. doi: 10.1016/j.indcrop.2013.07.030. [DOI] [Google Scholar]
- 93.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 94.Frevel MA, et al. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell. Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu L, et al. Tristetraprolin induces cell cycle arrest in breast tumor cells through targeting AP-1/c-Jun and NF-kappaB pathway. Oncotarget. 2015;6:41679–41691. doi: 10.18632/oncotarget.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warzych E, Wolc A, Cieslak A, Lechniak-Cieslak D. 217 transcript abundance of cathepsin genes in cumulus cells as a marker of cattle oocyte quality. Reprod. Fertil. Dev. 2012;25:257. doi: 10.1071/RDv25n1Ab217. [DOI] [Google Scholar]
- 97.Fuhrmann DC, et al. Inactivation of tristetraprolin in chronic hypoxia provokes the expression of cathepsin B. Mol. Cell Biol. 2015;35:619–630. doi: 10.1128/MCB.01034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Datta S, et al. Tristetraprolin regulates CXCL1 (KC) mRNA stability. J. Immunol. 2008;180:2545–2552. doi: 10.4049/jimmunol.180.4.2545. [DOI] [PubMed] [Google Scholar]
- 99.Ludwig EH, et al. DGAT1 promoter polymorphism associated with alterations in body mass index, high density lipoprotein levels and blood pressure in Turkish women. Clin. Genet. 2002;62:68–73. doi: 10.1034/j.1399-0004.2002.620109.x. [DOI] [PubMed] [Google Scholar]
- 100.Cao H, Sethumadhavan K, Li K, Boue SM, Anderson RA. Cinnamon polyphenol extract and insulin regulate diacylglycerol acyltransferase gene expression in mouse adipocytes and macrophages. Plant Foods Hum. Nutr. 2019;74:115–121. doi: 10.1007/s11130-018-0709-7. [DOI] [PubMed] [Google Scholar]
- 101.Dey P, Chakraborty M, Kamdar MR, Maiti MK. Functional characterization of two structurally novel diacylglycerol acyltransferase2 isozymes responsible for the enhanced production of stearate-rich storage lipid in Candida tropicalis SY005. PLoS ONE. 2014;9:e94472. doi: 10.1371/journal.pone.0094472. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Fahling M, et al. Multilevel regulation of HIF-1 signaling by TTP. Mol. Biol. Cell. 2012;23:4129–4141. doi: 10.1091/mbc.e11-11-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kagami S, et al. HMG-CoA reductase inhibitor simvastatin inhibits proinflammatory cytokine production from murine mast cells. Int. Arch. Allergy Immunol. 2008;146(Suppl 1):61–66. doi: 10.1159/000126063. [DOI] [PubMed] [Google Scholar]
- 104.Jamal UM, et al. A functional link between heme oxygenase-1 and tristetraprolin in the anti-inflammatory effects of nicotine. Free Radic. Biol. Med. 2013;65C:1331–1339. doi: 10.1016/j.freeradbiomed.2013.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi JX, et al. CNOT7/hCAF1 is involved in ICAM-1 and IL-8 regulation by tristetraprolin. Cell Signal. 2014;26:2390–2396. doi: 10.1016/j.cellsig.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 106.Su NY, Tsai PS, Huang CJ. Clonidine-Induced Enhancement of iNOS Expression Involves NF-kappaB. J. Surg. Res. 2008;149:131–137. doi: 10.1016/j.jss.2007.11.725. [DOI] [PubMed] [Google Scholar]
- 107.Xu P, et al. Leptin and zeranol up-regulate cyclin D1 expression in primary cultured normal human breast pre-adipocytes. Mol. Med. Rep. 2010;3:983–990. doi: 10.3892/mmr.2010.372. [DOI] [PubMed] [Google Scholar]
- 108.Voss AK, Thomas T, Gruss P. Compensation for a gene trap mutation in the murine microtubule-associated protein 4 locus by alternative polyadenylation and alternative splicing. Dev. Dyn. 1998;212:258–266. doi: 10.1002/(SICI)1097-0177(199806)212:2<258::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 109.Jiang J, Slivova V, Jedinak A, Sliva D. Gossypol inhibits growth, invasiveness, and angiogenesis in human prostate cancer cells by modulating NF-kappaB/AP-1 dependent- and independent-signaling. Clin. Exp. Metastasis. 2012;29:165–178. doi: 10.1007/s10585-011-9439-z. [DOI] [PubMed] [Google Scholar]
- 110.Militello RD, et al. Rab24 is required for normal cell division. Traffic. 2013;14:502–518. doi: 10.1111/tra.12057. [DOI] [PubMed] [Google Scholar]
- 111.Gao W, Shen Z, Shang L, Wang X. Upregulation of human autophagy-initiation kinase ULK1 by tumor suppressor p53 contributes to DNA-damage-induced cell death. Cell Death. Differ. 2011;18:1598–1607. doi: 10.1038/cdd.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]