Summary
With the rapid advancement of single-cell RNA-sequencing (scRNA-seq) technology, many data-preprocessing methods have been proposed to address numerous systematic errors and technical variabilities inherent in this technology. While these methods have been demonstrated to be effective in recovering individual gene expression, the suitability to the inference of gene-gene associations and subsequent gene network reconstruction have not been systemically investigated. In this study, we benchmarked five representative scRNA-seq normalization/imputation methods on Human Cell Atlas bone marrow data with respect to their impacts on inferred gene-gene associations. Our results suggested that a considerable amount of spurious correlations was introduced during the data-preprocessing steps due to oversmoothing of the raw data. We proposed a model-agnostic noise-regularization method that can effectively eliminate the correlation artifacts. The noise-regularized gene-gene correlations were further used to reconstruct a gene co-expression network and successfully revealed several known immune cell modules.
Keywords: noise regularization, single-cell RNA-seq, gene-gene correlation, protein-protein interaction, gene network, single cell, data imputation
Highlights
-
•
scRNA-seq preprocessing methods were benchmarked on inferring gene-gene associations
-
•
Spurious correlations have been introduced during the data-preprocessing steps
-
•
A noise-regularization method was proposed to eliminate the correlation artifacts
-
•
Gene co-expression network can be constructed from the noise-regularized correlations
The bigger picture
In this study, we benchmarked five representative single-cell RNA-sequencing data-preprocessing methods with a focus on their influence in inferring gene-gene expression correlations. We found that substantial correlation artifacts have been introduced during the preprocessing steps due to data oversmoothing, raising the issue that correlation computed from these preprocessed data may not be reliable and should be treated with caution. We then proposed a noise-regularization method to penalize the oversmoothed data, which can effectively eliminate the artifacts while retaining the majority of the true correlations. The regularized correlations can be further applied to construct gene-gene correlation networks, which is helpful for obtaining mechanistic insights into the complex biological systems.
Reliable inference of gene-gene correlation from single-cell RNA-sequencing data can be valuable in reconstructing global gene networks and further uncovering biological insights. In our benchmarking study, we observed that a considerable amount of correlation artifacts was introduced during the data-preprocessing steps from various methods. We proposed a model-agnostic noise-regularization approach in the correlation calculation procedure that can effectively remove the spurious correlations and empower studies looking to dissect gene-gene association in scRNA-sequencing data.
Introduction
Gene co-expression network analysis is a common approach to gather biological information and uncover molecular mechanisms of biological processes. Microarray and RNA-sequencing (RNA-seq) data of bulk cells have been successfully used to infer gene-gene correlations and further reconstruct gene co-expression networks.1,2 However, these approaches are limited to measuring average gene expression across a pool of mixed cell types. Single-cell RNA-seq (scRNA-seq) technology makes it possible to profile gene expression at single-cell resolution, which allows for dissection of the heterogeneity within the superficially homogeneous cell populations and identification of hidden gene-gene correlations masked by bulk expression profiles.3,4
The rapid development of scRNA-seq technology provides the opportunity to gain new insights into complex biological systems. However, due to various factors in single-cell experiments, such as differences in cell lysis, reverse transcription efficiency, and molecular sampling during sequencing,5 scRNA-seq data are generally highly variable and noisy. To address these issues, numerous data-preprocessing methods have been proposed for scRNA-seq data analysis, which generally fall into two major categories: (1) transcript abundance normalization and (2) dropout imputation. The observed sequencing depth can vary dramatically from cell to cell. Data normalization is hence required to remove the technical noise while preserving true biological signals. scRNA-seq data are further complicated by high dropout rate,6,7 which refers to the phenomenon by which a large proportion of genes have a measured read count of zero due to the technical limitation in detecting the transcripts rather than true absence of the gene. Data imputation has been proposed to handle the dropouts and recover the undetected gene expressions.
scRNA-seq data-preprocessing methods have been benchmarked for various tasks, such as cell clustering, detection of differentially expressed genes, and trajectory analysis.8 The suitability of these methods for reverse engineering gene networks and, in general, for measuring gene-gene association, has not been systemically evaluated. Andrews and Hemberg tested several imputation methods on a small simulation dataset and found that dropout imputation would generate false-positive gene-gene correlations.9 However, the simulation dataset in that study represented the simplest case without technical confounders; thus, the effect of data preprocessing on real data remains unknown.
In this study, we benchmarked five normalization/imputation methods, which are representatives of their own methodology groups, in respect of their influence on gene-gene correlation inferences. The first method, global scaling normalization, normalizes a cell's gene expression levels (usually measured by the unique molecule index [UMI]) by its summed expression over all genes, e.g., total UMI. This method is usually followed by log transformation and Z-score scaling in the downstream analyses. Since the log transformation and Z-score scaling are monotonic (rank-preserved) functions, we only included total UMI normalization in our benchmarking (referred as NormUMI). The second normalization framework utilizes “Regularized Negative Binomial Regression” to normalize and stabilize variance of scRNA-seq data (referred as NBR). This method showed remarkable performance in removing the influence of technical noise while preserving biological heterogeneity.10 Three imputation methods were also included: (1) MAGIC, a data-smoothing approach that leverages the shared information across similar cells to denoise and fill in dropout values;11 (2) SAVER, a model-based approach that models the expression of each gene under a negative binomial distribution assumption and outputs the posterior distribution of the true expression;12 and (3) DCA, an adapted autoencoder framework that is able to capture the complexity and non-linearity in scRNA-seq data and infer gene expressions.13
To evaluate the influence of these preprocessing methods on gene-gene correlation inference, we applied them to bone marrow scRNA-seq data from the Human Cell Atlas (HCA) Project.14 We computed gene-gene correlation after the data preprocessing and compared results among the methods. With the exception of NormUMI, the normalization method with the least data manipulation, all other normalization/imputation methods presented a noticeable inflation of gene-gene correlation coefficients and introduced correlation artifacts for gene pairs that are not expected to be co-expressed. In addition, gene pairs with the highest correlations inferred from these methods had weak enrichments in protein-protein interactions from the STRING database,15 suggesting that many of these correlations may be the false signals introduced during the data preprocessing. Further data inspection using random and non-associated gene pairs as negative control indicated that the artifacts could be generated from data oversmoothing. In machine learning, adding noise under certain conditions has been previously shown to increase robustness of the results and reduce overfitting.16, 17, 18 To this end, we implemented a noise-regularization step to the preprocessed scRNA-seq data by adding noise drawing from uniform distribution that is scaled to the dynamic expression range of each individual gene. We found that this additional step efficiently reduced gene-gene correlation artifacts and improved overall evaluation metrics. We used the regularized expression data to reconstruct gene co-expression network and successfully revealed several known immune cell modules. The canonical cell-type marker genes were also rated higher in network topological properties, e.g., degree and PageRank, pinpointing their key roles in their respective cell clusters.
Results
Computing gene-gene correlation using scRNA-seq data
Previous benchmarking studies on scRNA-seq data-preprocessing methods were mostly based on simulated datasets with certain assumptions in the simulation process that might not be representative of real-world data. Depending on the simulation algorithm used, results might be biased toward certain methods. For instance, the method SAVER, which uses negative binomial distribution to model and impute the data, will stand out if the simulated dataset is also generated based on a negative binomial model. To avoid such biases, we employed real-world bone marrow scRNA-seq data from the HCA Preview Datasets as our benchmarking dataset14 for various data-preprocessing methods. The full dataset contains 378,000 bone marrow cells, which can be grouped into 21 cell clusters (Figure S1) covering all major immune cell types. We randomly sampled 50,000 cells from the original dataset and excluded genes expressing in fewer than 100 cells (0.2%) in this subset. The final benchmarking dataset contains 12,600 genes that could form over 79 million possible gene pairs.
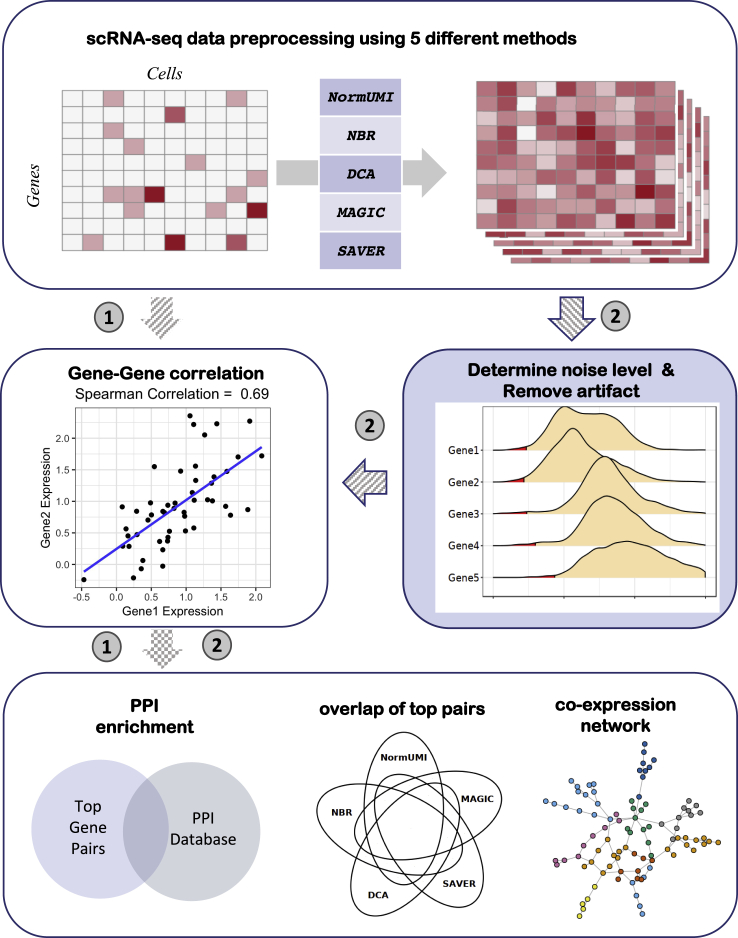
Five representative data-preprocessing methods were applied to the single-cell expression data matrix, including two normalization methods (NormUMI and NBR) and three imputation methods (DCA, MAGIC, and SAVER) (Figure 1). An important merit of scRNA-seq is its ability to unbiasedly capture the whole transcriptome of different cell types in a heterogeneous cell population. Expression of two genes could be highly correlated only in one specific cell type and therefore revealed cell-type-specific gene-gene associations. To capture the correlations across different cell types, we computed Spearman correlation of gene pairs within the ten largest clusters (>500 cells per cluster) in our benchmarking dataset, which included CD4 T cell, CD8 T cell, natural killer cell, B cell, Pre-B cell, CD14+ monocytes, FCGR3A+ monocytes, erythrocytes, granulocyte-macrophage progenitors, and hematopoietic stem cells (Figures 1 and S1). The highest correlation among these ten clusters was recorded as the final correlation for each gene pair (see Experimental procedures).
Figure 1.
Overview of the benchmarking framework
Five scRNA-seq data-preprocessing methods were applied to bone marrow single-cell expression data matrices. The gene-gene correlations were first calculated directly from the matrices after data preprocessing (denoted as route 1). We evaluated the methods by their derived gene-gene correlation enrichments in the STRING PPI database as well as the consistency between methods. The evaluation results indicated that the data-preprocessing procedure introduced artificial correlations. We then introduced a noise-regularization step (denoted as route 2): random noise generated based on gene expression level (regions in red) was applied to the expression matrices before proceeding to correlation calculation. This noise-regularization step effectively reduced the spurious correlations, and the refined gene-gene correlations could be used to construct gene co-expression networks.
Data preprocessing introduced spurious correlations
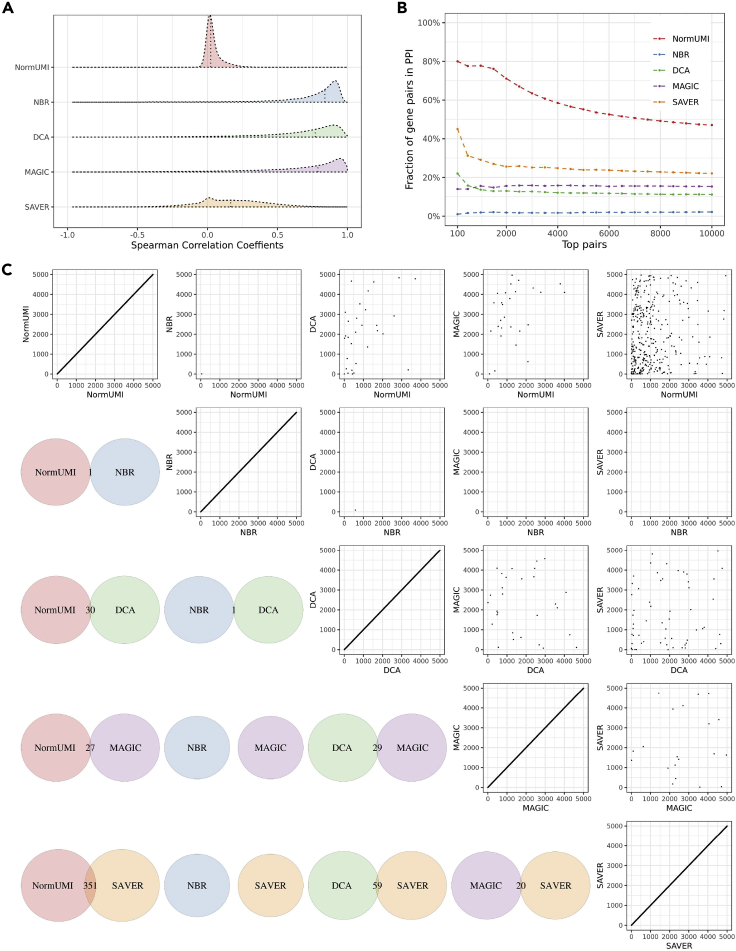
We first compared the distribution of the overall gene-gene correlations calculated from data matrices processed by the five methods. Since most of the gene pairs are not expected to have any association, we anticipated that the correlation distributions should peak around zero. However, with the exception of NormUMI, all other methods produced much higher median correlation values (NormUMI ρ = 0.023, NBR ρ = 0.839, MAGIC ρ = 0.789, DCA ρ = 0.770, SAVER ρ = 0.166) (Figure 2A). We proceeded to assess whether a higher correlation, after a specific data-preprocessing method, would reflect a higher chance of either functional or physical interaction between the two genes. Proteins encoded by a co-expressed gene pair are more frequently interacting with each other than a random pair. Therefore, if the resulting higher correlations are true positives, they should have relatively higher enrichment in the protein-protein interaction (PPI) database, while the spurious correlations would dilute the enrichment. We used the STRING database,15 which contains 5,772,157 interacting gene pairs, to evaluate the PPI enrichment of the top correlated gene pairs derived from each method. We selected top gene pairs (ranked by correlation coefficients) from each method and calculated the overlapping fraction of these pairs with the STRING database (Figure 2B). Our results showed that NormUMI had the highest PPI enrichments: 80% and 47% overlapped with STRING in the top 100 and 10,000 gene pairs, respectively. On the contrary, the top gene pairs from NBR had very low overlap with STRING (<2%), while MAGIC and DCA had similar PPI enrichments, ranging from 11% to 22%. SAVER yielded relatively better results, but the enrichments were merely half of those acquired by NormUMI. We also randomly sampled gene pairs and overlapped the random pairs with PPI to estimate the background enrichment level (Figure S2). The estimated background enrichment level was ∼3.6%, indicating that PPI enrichment of NBR was even lower than the background. Although this is a rather naive method that directly relates physical interactions with gene co-expression, the results here should still provide a fair comparison among the data-preprocessing methods given that the same assumption is made for all of them.
Figure 2.
Spurious gene-gene correlations are introduced during data preprocessing
(A) The distributions of the calculated correlations varied by preprocessing methods. NormUMI had a distribution centered close to zero, while NBR, DCA, and MAGIC all had apparently inflated correlation distributions. Vertical dotted lines indicate correlation medians.
(B) Enrichment curves of the top correlated gene pairs in PPI for each method. x axis indicates the top n gene pairs ranked by Spearman correlation coefficients; y axis indicates the fraction of the n gene pairs appearing in the STRING PPI database. NormUMI had the highest enrichment, followed by SAVER, MAGIC, DCA, and NBR.
(C) There was low consistency between the methods in inferring highly correlated gene pairs. Lower triangle indicates the overlapping of the top 5,000 gene pairs between the two denoted methods. The largest overlap was between NormUMI and SAVER, which has only 351 (∼7%) gene pairs ranked in the top 5,000 in both methods. Upper triangle compares the exact rank of the shared gene pairs between methods, which also shows low levels of agreement.
Bona fide gene-gene co-expression should be identified regardless of the data-preprocessing methods. To test this, we compared the consistency of highly correlated gene pairs derived from the five data-preprocessing procedures. We did a pairwise comparison of the top 5,000 gene pairs selected from each method and found that the overlapping gene pairs among methods were minimal. Only one gene pair was shared between NormUMI and NBR out of the top 5,000 pairs. The highest overlap was between NormUMI and SAVER, with only 351 pairs (∼7%) shared by the two methods (lower triangle in Figure 2C). We further compared the ranks of the shared pairs between the methods and found that there was also no clear trend in their top inference (upper triangle in Figure 2C). While this is not a fully quantitative assessment, it is clear that the high correlations derived from these data-preprocessing methods are likely to be artifacts.
Negative control
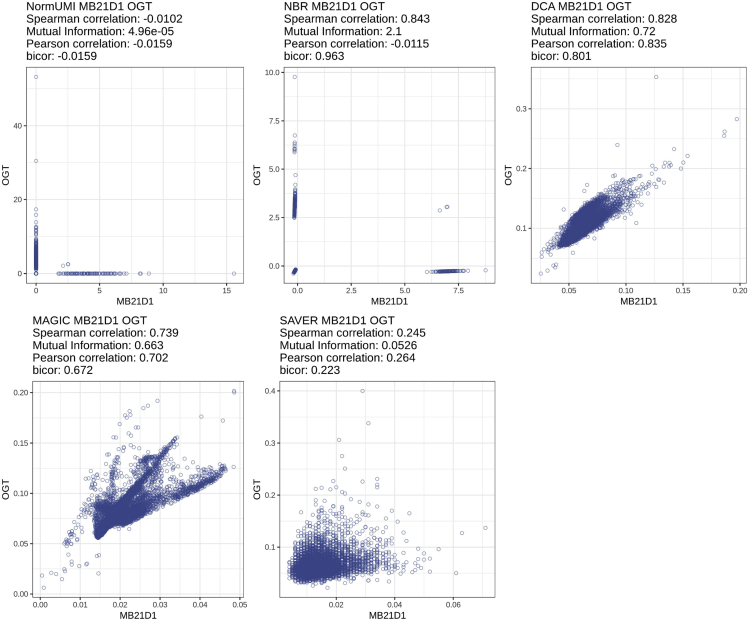
We next inspected several “negative control gene pairs” to obtain some insights into the potential cause of the spurious correlations. We defined a negative control pair using the following criteria: the two genes should not (1) appear as an interacting pair in the STRING database, (2) share any gene ontology term,19,20 and (3) be on the same chromosome. As an example, one of the negative control gene pairs, MB21D1 and OGT, had high correlation after data processing by NBR (ρ = 0.843), DCA (ρ = 0.828), and MAGIC (ρ = 0.739) in cell cluster #2. We also calculated the mutual information (MI) of the negative gene pairs, which can assess the strength of the association between two variables even when the relationship is highly non-linear.21 In this negative pair example, NBR (MI = 2.10 nat), DCA (MI = 0.72 nat), and MAGIC (MI = 0.663 nat) also showed much higher mutual information than the other two methods, NormUMI (MI = 5 × 10−5 nat) and SAVER (MI = 0.053 nat). Scatterplots of the gene pair expression values after data preprocessing are shown in Figure 3. Of the five methods, NormUMI was the only method that retained the zero counts from the raw data. From NormUMI, 6,110 cells out of 6,534 cells (93.5%) had zero values in both genes, 3 (0.04%) cells had non-zero values in both genes, while 1.3% and 5.2% cells had non-zero for MB21D1 and OGT, respectively. The other imputation methods intensely altered the zeros from the original expression matrix. We observed that after these procedures, the processed data all presented some degree of oversmoothing, especially in the double-zero regions in the original data, which created the correlation artifacts (Figure 3). Although NBR was not an imputation method and only shifted the zero values minimally, artificial rank correlations were introduced due to the difference in the adjusted magnitude per cell.
Figure 3.
Spurious gene-gene correlation caused by data oversmoothing
Scatterplot of expression values of non-associated gene pair, OGT and MB21D1, preprocessed by different methods. There is no existing evidence to indicate that these two genes are correlated, and only 3 out of 6,534 cells in cluster #2 had non-zero expression value in both genes in the original expression matrix. However, after preprocessing, NBR, DCA, and MAGIC all produced high correlations (0.843, 0.828, and 0.739) and high mutual information (2.1, 0.72, and 0.663 nat) between these two genes. The visualization suggested that this correlation artifact may be caused by data oversmoothing.
Noise regularization reduced spurious correlations
Regularization is a commonly used approach to prevent overfitting/oversmoothing in machine learning, and a previous work has demonstrated an equivalent form of regularization by introducing noise.16 Here, we proposed a method utilizing noise to penalize oversmoothed expression data and further reduce spurious correlations. To implement the method, we added random noise to every single feature in the expression matrix processed by the above preprocessing methods. Taking the expression value of gene i in cell j, denoted as V, as an example, we generated the noise by the following steps: (1) calculate the expression distribution of gene i after data-preprocessing procedure; (2) determine the 1 percentile of expression value of gene i, termed as M, to be used as the maximum of noise level (Figure 1); (3) generate a random value from a uniform distribution, ranging from 0 to M, and add this random value to V.
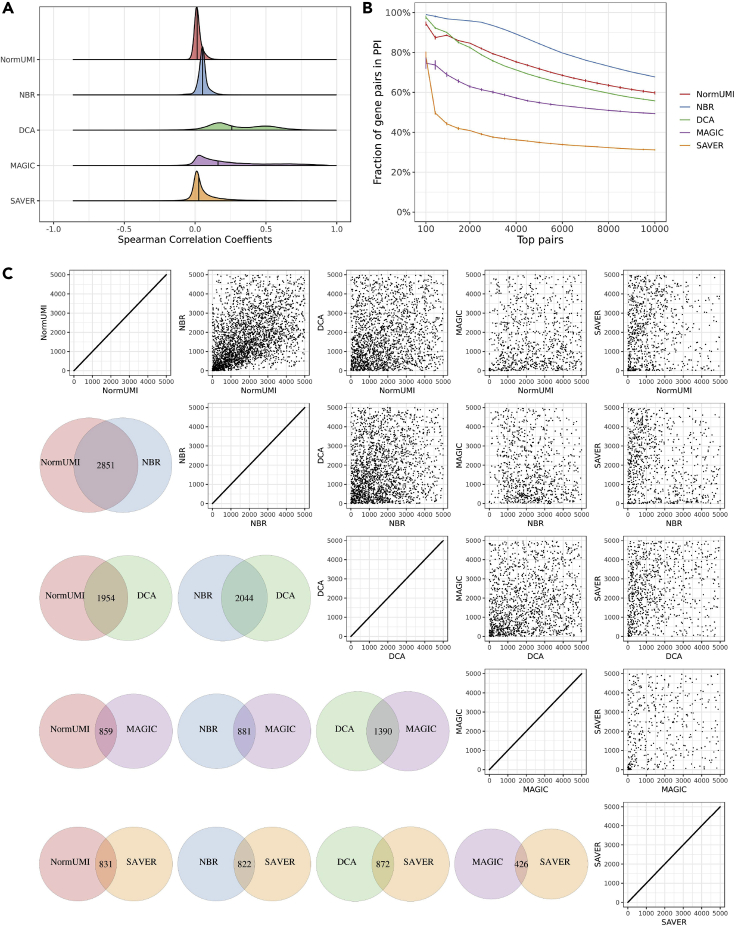
After applying noise regularization to the data matrices produced by each preprocessing method, we recomputed the gene-gene correlations. The correlation medians shifted toward zero for all five methods (Figure 4A), indicating a reduction in the correlation inflation. There were also substantial improvements in the PPI enrichment for all methods (Figure 4B). NBR, which previously had the lowest enrichment, yielded the highest PPI enrichment after noise regularization. In the top 100, 1,000, and 10,000 gene pairs in NBR, 99.0%, 96.8%, and 67.7% could be found in the PPI database, corresponding to 99.0-, 50.9-, and 31.6-fold improvement, respectively. DCA on average had ∼12% PPI enrichment in previous results. After noise regularization, it produced 97.6% enrichment in the top 100 pairs and 55.8% in the top 10,000 pairs, corresponding to a ∼5-fold improvement. NormUMI, which had the highest enrichment before noise regularization, also benefited from a ∼1.1- to 1.3-fold improvement. To test the robustness and reproducibility of the noise-regularization results, we repeated the procedure ten times with different random seeds to generate random noise and observed that the PPI enrichment performances were stable between repeats. The standard deviation of NBR in most points was less than 0.1% (error bar represents 99% confidence interval in Figure 4B).
Figure 4.
Noise regularization reduces spurious correlations
(A) After applying noise regularization, previously inflated correlation distributions from each method shifted toward zero. Vertical solid lines indicate correlation medians.
(B) There were substantial improvements of the PPI enrichment in the top correlated genes. Error bars indicate 99% confidence interval based on ten replicates, assuming error follows a Gaussian distribution.
(C) Compared with previous unregularized data (Figure 2C), there are higher levels of agreement among different methods. For example, more than 50% gene pairs were shared between NormUMI and NBR.
Different methods also showed higher agreements after applying noise regularization. Among the top 5,000 gene pairs, 2.851 (57%) overlapped between NormUMI and NBR (Figure 4C, lower triangle), and there was a significant correlation between the overlapped gene pairs (Spearman correlation, ρ = 0.50; Fisher's exact test, p = 1.77 × 10−181, Figure 4C, upper triangle). We also observed a higher degree of commonly identified gene-gene correlations between the other preprocessing methods, particularly between the top gene pairs.
Next, we compared the correlation coefficients of the top 5,000 gene pairs selected before and after noise regularization in each method (Figures S3 and S4). The most noticeable impact of the regularization was observed in NBR, where correlations of all the top gene pairs dropped dramatically after regularization. In DCA/MAGIC/SAVER, a wide range of correlations was observed after regularization, suggesting that not all gene pairs were equally affected. On the contrary, the top 5,000 gene pairs selected after regularization were also highly correlated before the regularization. We further selected several positive and negative control gene pairs to examine the effect of regularization on their gene expression and correlation (Figures S5–S7). In the negative control, the oversmoothed data points were randomized and the correlations were effectively diluted. In the positive controls (experimentally validated interactions), expressions of the gene pairs were not significantly changed, and the correlations remained relatively high after regularization. These results demonstrate that noise-regularization steps do not unvaryingly reduce correlation of all gene pairs, and the real signals are robust enough to tolerate the added noise.
Gene-gene correlation network inferred from scRNA-seq data
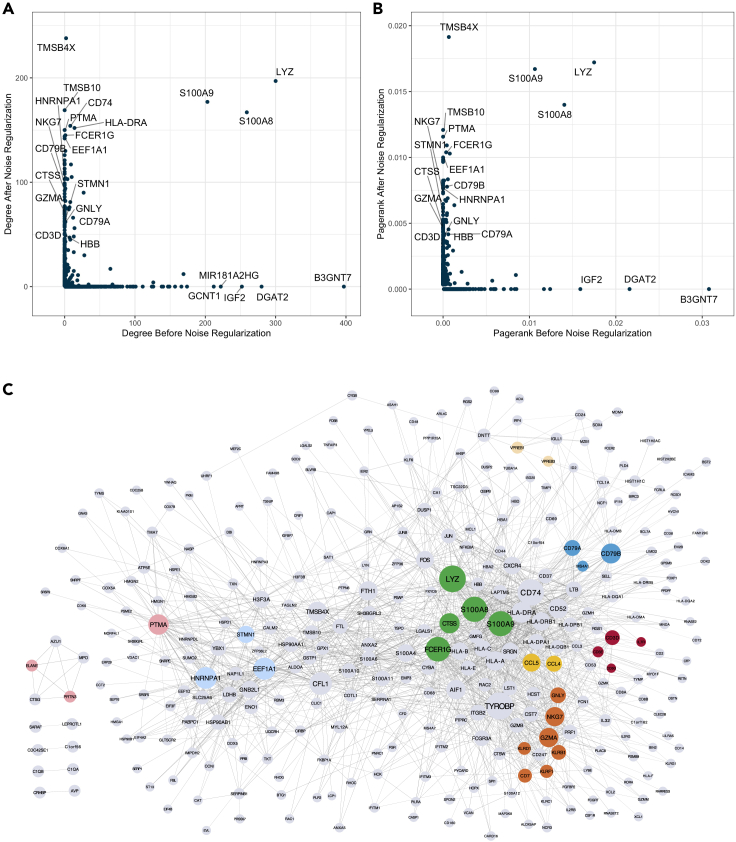
Co-expression networks can be used to identify gene modules with common biological functions, upstream regulators, and physically interacting proteins.22 With the gene expression measurement at single-cell resolution, scRNA-seq has fostered discoveries by improving our understanding of biological processes under different cell contexts. Therefore, gene-gene correlations revealed from single cells also have the potential to reconstruct more comprehensive networks uncovering cell-type-specific modules. Here, we used gene-gene correlations derived from NBR with noise regularization, since it yields the highest PPI enrichment among all the methods. To focus more on cell-type-specific interactions, we removed housekeeping genes that typically reflect the general cellular functions and are expected to express in all cells regardless of the cell types. There were 3,984 housekeeping genes removed from the original 12,600 genes. The 1,000 gene pairs with the highest correlations were then taken from each cluster (cluster #0 to cluster #9) to reconstruct the network. Degree and PageRank, two algorithms from graph theory, were used to measure the importance of each gene in the network. The degree of a gene in a network is simply the number of links (interactions) the gene has.23 Important genes tend to connect with many other genes and therefore should have relatively high degrees. In addition to the quantity of links, PageRank also takes into consideration quality of links to a gene and measures the overall “popularity” of a gene.24
We compared the gene co-expression networks reconstructed from pre- and postregularized data. Results showed that the latter network better represented the biological functions in the topological structure and had a higher degree or PageRank genes with more important functions in the immune system. For instance, LYZ, CD79B, and NKG7, the canonical marker genes for monocytes, B cells and natural killer cells, respectively, yielded higher PageRank and degree in the network with noise regularization. On the contrary, CD79B and NKG7 did not exist at all in the network without noise regularization (Figures 5A and 5B). We next overlaid existing PPI evidence to further refine the network by retaining only gene pairs from the STRING database.25,26 An algorithm providing efficient visualization of different network modules, EntOptLayout,27 was applied, and the network revealed several cell-type-related modules that can be associated with the known biology in our benchmarking dataset (Figure 5C). For instance, the upper right corner represents the B cell and pre-B cell module, with CD79A and CD79B having higher PageRank values that are proportional to the node size. Similarly, the natural killer cell module is represented in the lower right corner, and the middle right section represents T cell as well as a transit from cytotoxic CD8 T cell to natural killer cell (Figure 5C). These results demonstrate that, after implementing noise regularization, scRNA-seq data can be used to reconstruct gene co-expression networks that better reflect the underlying biology.
Figure 5.
Gene-gene correlation network inferred from scRNA-seq data
(A and B) Comparison of degree (A) and PageRank (B) of each gene in the correlation networks constructed before and after noise regularization. Genes present in one network but not in the other were assigned a zero value in the non-presenting one. Selected genes with high degree/PageRank before or after noise regularization were labeled. Cell-type marker genes such as NKG7, CD79B, and HBB had relatively higher degree and PageRank after noise regularization.
(C) Network construction with refined gene-gene correlations (NBR + noise regularization + removing links not in PPI), where the node size is proportional to its PageRank and the edge width is proportional to Spearman correlation between the two genes (nodes). Cell-type marker genes (colored nodes) such as CD79A, CD79B, NKG7, GNLY, LYZ, and STMN1 have high PageRank, indicating their importance in different cell types. Cell-type-related genes also formed cell-type-specific modules.
Discussion
scRNA-seq technology has been gaining increasingly more popularity over the past decade. Proper and efficient data preprocessing are crucial for downstream analyses such as cell clustering, differential gene expression detection, and novel cell-type discoveries.3,4 Here, we benchmarked five data-preprocessing methods for scRNA-seq with a focus on their influence in gene-gene correlation inference. Our results demonstrated that in a human bone marrow single-cell dataset, all the methods except NormUMI generated inflated gene-gene correlations. Furthermore, the highly correlated gene pairs had low enrichment in PPI, indicating that they were more likely to be artifacts introduced during the data-preprocessing procedure. Among these methods, NBR produced the lowest PPI enrichment, while NormUMI, the method with the least data manipulation, yielded much higher enrichment as compared with the other four sophisticated methods. Thus, our benchmarking results raise the issue that correlation computed directly from these preprocessed data may not be reliable and should be treated with caution.
Manual inspection of the negative control results suggested that major causes of the spurious correlations may come from overfitting or oversmoothing during data preprocessing. The preprocessing methods, especially those imputing dropout events, rely heavily on internal similarity information (either gene-gene similarity or cell-cell similarity) within the original dataset. For instance, MAGIC uses the data-diffusion algorithm to construct a more faithful neighborhood of cells and further imputes the missing values in one cell base on the expression pattern of the neighborhood. Indeed, this could be circular to measure the gene-gene correlation after applying these steps. Given that these methods rely on the similarity of gene expression to amend gene expression, it is not surprising that they produce augmented gene-gene correlations.
To resolve the correlation artifact issues, we proposed a model-agnostic noise-regularization method. False correlations from the overly smoothed data can be eliminated by the added noise while the true correlations should be robust enough to tolerate the noise. Since the dynamic range of expression varies gene by gene, magnitude of the added noise should also be set relative to an individual gene's expression level such that the true signal of genes with a lower expression range can be preserved. Thus, the level of random noise is determined as a percentile of a gene's dynamic range rather than a fixed value to be used for all genes. We further investigated the effect of different noise strengths (1, 5, 10, 20 percentile of the expression level), and found that use of the 1 percentile produced the optimal PPI enrichment (Figure S8). Finally, we generated random noise that ranged from 0 to 1 percentile of the gene expression level and applied them to the expression matrix. The noise-regularization step remarkably reduced the correlation artifacts and generated more reliable gene-gene association. However, it should be noted that the magnitude of the noise applied here was optimized to maximize the PPI enrichment, which may result in a higher true-positive rate. Since there is always a trade-off between sensitivity and specificity, whether this noise strength is optimal for revealing novel correlations likely requires further investigation.
Gene-gene correlations at the whole-transcriptome level for bulk cells have been established to reconstruct gene-gene interaction networks and further uncover gene functions and genetic modules.22,28,29 With the growing adoption of single-cell technology, the use of scRNA-seq to infer gene-gene correlations and reconstruct global gene network is also burgeoning. Pioneering work by Iacono et al. used single-cell data-derived correlation metrics to generate gene regulatory networks and found that the networks could detect latent regulatory changes.30 A deep-learning approach has also been developed to predict transcription factor targets from single-cell expression data.31 In this study, we used single-cell gene-gene correlations derived after noise regularization to reconstruct a gene network that produced clear immune cell-type-related modules. We also evaluated the importance of each gene in the network by applying well-established graph theory methods. We demonstrated that the canonical cell-type markers yield higher degree and PageRank, in general, indicating their critical roles in different cell types.
A limitation of this study is that these methods were mostly implemented using their default parameters, which may not be optimal for this dataset. Changing the parameters and hyperparameters could have noticeable impact on the results. Andrews and Hemberg tested different imputation methods on a simulation dataset and found that different parameters produced different degrees of false correlation.9 Unfortunately, the choice of parameters is often arbitrary and lacks clear guidelines. For instance, MAGIC applies data smoothing based on data diffusion between similar cells. Increasing the number of neighbors will lead to smoother data, in most cases resulting in inflated gene-gene correlations and more false positives in correlation-based analyses. In addition, the diffusion time (t) in the algorithm also strongly affects the data smoothness. By default, this parameter is determined according to the Procrustes disparity of the diffused data. However, default setting apparently generated oversmoothed data in our study. Using a different parameter value (e.g., decreased to a fixed number, 6), we found that the output can be visually improved, although a high amount of spurious correlations still exists. This challenge is further complicated when users need to consider combinations of several parameters. A similar issue is also noticed in the implementation of DCA that requires a series of parameters, including many routine deep-learning framework training parameters, such as learning rate and strength of L1/L2 regularization. The default architecture of DCA (three hidden layers with 64, 32, and 64 neurons) was originally optimized on a simulation dataset with only 200 genes. When it is applied to real datasets that contain over 10,000 genes, whether the default number of neurons can still capture the full picture and reconstruct reliable gene-gene networks becomes unclear. Furthermore, tuning the parameters could potentially help to reduce the correlation artifacts, but the tuned parameters may then be suboptimal for its original tasks such as cell clustering and differential gene expression analysis. In our framework, the noise regularization can serve as an additional step to infer reliable gene-gene correlations, and all other analyses can be performed directly on the data preprocessed using their optimized parameters and without noise regularization.
In summary, we compared five scRNA-seq data-preprocessing methods on a real single-cell dataset and found that several preprocessing procedures may have introduced a considerable amount of spurious gene-gene correlations. Therefore, single-cell analysis involving gene-gene correlations should be performed with caution. To address the issues, we proposed a model-agnostic method to regularize the preprocessed data, which can effectively remove the spurious correlations and empower studies looking to reconstruct co-expression networks from scRNA-seq data.
Experimental procedures
Resource availability
Lead contact
Wei Keat Lim: weikeat.lim@regeneron.com.
Materials availability
This study did not generate new single-cell RNA-seq data.
Data and code availability
The R code for analyses in this study is available at Github: https://github.com/RuoyuZhang/NoiseRegularization.
HCA scRNA-seq dataset
Bone marrow single-cell sequencing data were downloaded from the HCA Data Portal (https://data.humancellatlas.org). The dataset contains profiling of 378,000 immunocytes by the 10X Genomics chromium platform. Single-cell analysis was performed using the Seurat R package (Version 3.0).32 In the quality control step, low-quality cells were removed if they met one of the following criteria: (1) expressed less than 100 genes; (2) expressed more than 3,500 genes; (3) total UMI counts >10,000; (4) mitochondrial RNA percentage >10%. Remaining cells were clustered using k-nearest neighbor (KNN) graph-based clustering approach, with the first 30 principal components (PC) being used to construct the KNN graph. Clustering results were visualized with UMAP (Uniform Manifold Approximation and Projection), also using the first 30 PCs as inputs. In the subsequent correlation analysis, to reduce the computational burden we randomly sampled 50,000 cells from the original dataset. We further filtered out genes expressed in fewer than 100 cells (0.2%), which left 12,600 genes remaining in the final benchmarking dataset.
Normalization or imputation methods
NormUMI was performed using the Seurat R package (version 3.0) without log transformation.32 NBR, SAVER, and DCA were run with default parameters according to the software tutorials. Specifically, NBR was performed using sctransform R package (version 0.2.0).10 Poisson regression was performed for each gene under the negative binomial model. Regularized model parameters were used to transform observed UMI counts into Pearson residuals. DCA was performed with the dca python package:13 the deep-learning framework had three hidden layers with 64, 32, and 64 neurons. The learning rate used was 0.001 and batch size was set to 32. SAVER was run with the SAVER R package (version 1.1.1) without requiring additional parameters.12 MAGIC was run with MAGIC R implementation (version 1.5-9)11 with the following parameters: number of principal component npca = 30, power of the Markov affinity matrix t = 6, and number of nearest-neighbor k = 30.
Gene-gene correlation and mutual information calculation
Spearman correlation of each gene pair was calculated from cells in cluster 0 to cluster 9 (top ten clusters with the largest cell number, which range from 583 to 16,936 cells, Figure S1), respectively. A gene was considered present in a cluster if its expression was detected in more than 1% of the cells or 50 cells in that cluster, whichever is greater. The correlation of a gene pair in one cluster was considered an effective correlation if the two genes were both considered as expressed in that cluster. The highest effective correlation across the ten clusters was recorded as the final correlation for a given gene pair. MI of the selected gene pairs was measured using the infotheo R package (version 1.2.0), data were discretized using the equal frequencies binning algorithm, and the entropy was estimated with an empirical probability distribution.
Protein-protein interaction enrichment
Human protein-protein interaction data were retrieved from the STRING database (version 11) (http://string-db.org).15 The STRING database consists of comprehensively collected publicly available sources of protein-protein interaction information and is complemented with computational predictions. The final database includes both direct (physical) and indirect (functional) interactions. In this study, we used the Homo Sapiens version 11 (database9606.protein.links.full.v11.0) database, 5,772,157 PPIs involving both experimentally verified and computationally inferred pairs. After applying different data-preprocessing methods, gene pairs were ranked by their Spearman correlation coefficients. The top n gene pairs were then taken and overlapped with the STRING database. The fraction of the top n pairs appearing in the database was recorded as the PPI enrichment.
Noise regularization
Assuming that V is the expression value of gene i in cell j in the expression matrix processed by a specific method, a random noise value was generated and added to V by the following procedures. (1) Determine the expression distribution of gene i across all the cells. (2) Take 1 percentile of the gene i expression as the maximal noise level, denoted as M. If M equals zero, 0.1 will be used as the maximal noise level. (3) Generate a random number ranging from 0 to M under uniform distribution and add this random number to V to get the noise-regularized expression matrix. The noise regularization was applied to the expression data preprocessed by NormUMI/NBR/MAGIC/SAVER/DCA.
Network reconstruction
Within each cluster, we ranked the gene pairs by their Spearman correlation coefficients. In this study, since we were more interested in cell-type-specific gene interaction modules, we removed housekeeping genes from the network reconstruction. In general, housekeeping genes are required for basic cellular functions and are thus expected to express regardless of cell types. The housekeeping gene list used here was obtained from a previous publication,33 plus (1) typical housekeeping genes such as ACTB and B2M, (2) ribosomal, citrate cycle, and cytoskeleton genes from Reactome,34 and (3) mitochondrial DNA encoded genes. In total, 3,984 housekeeping genes were considered. After removing housekeeping genes, the top 1,000 gene pairs from each cluster were taken and put together to construct the draft network. The importance of each node in the network was measured by degree and PageRank using the igraph R package.35 We next refined the network by removing links that do not overlap with PPI in the STRING database. The final network was visualized using Cytoscape36 together with R package RCy3.37 The network layout was generated using the EntOptLayout Cytoscape plug-in.27
Acknowledgments
We thank The Human Cell Atlas for generating the “Census of Immune Cells” dataset and making it available to the research community. We thank Drs. Ian Setliff and Kaitlyn Gayvert for their helpful discussion and comments on the manuscript. This study was funded by Regeneron Pharmaceuticals.
Author contributions
R.Z. and W.K.L. conceived the study and drafted the manuscript. R.Z., G.S.A., and W.K.L. analyzed the data.
Declaration of interests
All authors are full-time employees of Regeneron Pharmaceuticals and receive options and stock as part of their compensation. All authors are named inventors on pending US Patent Application No. 17/032,848 and PCT Application No. PCT/US20/052787.
Published: February 15, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2021.100211.
Supplemental information
References
- 1.Freeman T.C., Goldovsky L., Brosch M., van Dongen S., Mazière P., Grocock R.J., Freilich S., Thornton J., Enright A.J. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput. Biol. 2007;3:2032–2042. doi: 10.1371/journal.pcbi.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballouz S., Verleyen W., Gillis J. Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics. 2015;31:2123–2130. doi: 10.1093/bioinformatics/btv118. [DOI] [PubMed] [Google Scholar]
- 3.Kolodziejczyk A.A., Kim J.K., Tsang J.C., Ilicic T., Henriksson J., Natarajan K.N., Tuck A.C., Gao X., Bühler M., Liu P. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18:35. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 5.Hicks S.C., Townes F.W., Teng M., Irizarry R.A. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics. 2017;19:562–578. doi: 10.1093/biostatistics/kxx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svensson V., Natarajan K.N., Ly L.H., Miragaia R.J., Labalette C., Macaulay I.C., Cvejic A., Teichmann S.A. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods. 2017;14:381. doi: 10.1038/nmeth.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Tian L., Dong X., Freytag S., Lê Cao K.A., Su S., JalalAbadi A., Amann-Zalcenstein D., Weber T.S., Seidi A., Jabbari J.S. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods. 2019;16:479–487. doi: 10.1038/s41592-019-0425-8. [DOI] [PubMed] [Google Scholar]
- 9.Andrews T., Hemberg M. False signals induced by single-cell imputation [version 1; peer review: 4 approved with reservations] F1000Res. 2018;7 doi: 10.12688/f1000research.16613.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Dijk D., Sharma R., Nainys J., Yim K., Kathail P., Carr A.J., Burdziak C., Moon K.R., Chaffer C.L., Pattabiraman D. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–727. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang M., Wang J., Torre E., Dueck H., Shaffer S., Bonasio R., Murray J.I., Raj A., Li M., Zhang N.R. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods. 2018;15:539–542. doi: 10.1038/s41592-018-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eraslan G., Simon L.M., Mircea M., Mueller N.S., Theis F.J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun. 2019;10:390. doi: 10.1038/s41467-018-07931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M. Science forum: The Human Cell Atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop C.M. Training with noise is equivalent to Tikhonov regularization. Neural Comput. 1995;7:108–116. [Google Scholar]
- 17.Neelakantan A., Vilnis L., Le Q.V., Sutskever I., Kaiser L., Kurach K., Martens J. Adding gradient noise improves learning for very deep networks. arXiv. 2015 1511.06807. [Google Scholar]
- 18.Smilkov D., Thorat N., Kim B., Viégas F., Wattenberg M. Smoothgrad: removing noise by adding noise. arXiv. 2017 1706.03825. [Google Scholar]
- 19.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2018;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinney J.B., Atwal G.S. Equitability, mutual information, and the maximal information coefficient. Proc. Natl. Acad. Sci. U S A. 2014;111:3354–3359. doi: 10.1073/pnas.1309933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 23.Bondy J.A., Murty U.S.R. Springer Publishing Company Inc; 2008. Graph Theory. [Google Scholar]
- 24.Page L., Brin S., Motwani R., Winograd T. Stanford InfoLab; 1999. The PageRank Citation Ranking: Bringing Order to the Web. [Google Scholar]
- 25.Cheng H., Jiang L., Wu M., Liu Q. Inferring transcriptional interactions by the optimal integration of ChIP-chip and knock-out data. Bioinform Biol. Insights. 2009;3:129–140. doi: 10.4137/bbi.s3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayyed-Ahmad A., Tuncay K., Ortoleva P.J. Transcriptional regulatory network refinement and quantification through kinetic modeling, gene expression microarray data and information theory. BMC Bioinformatics. 2007;8:20. doi: 10.1186/1471-2105-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ágg B., Császár A., Szalay-Beko M., Veres D.V., Mizsei R., Ferdinandy P., Csermely P., Kovács I.V. The EntOptLayout Cytoscape plug-in for the efficient visualization of major protein complexes in protein-protein interaction and signalling networks. Bioinformatics. 2019;35:4490–4492. doi: 10.1093/bioinformatics/btz257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iacono G., Massoni-Badosa R., Heyn H. Single-cell transcriptomics unveils gene regulatory network plasticity. Genome Biol. 2019;20:110. doi: 10.1186/s13059-019-1713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan Y., Bar-Joseph Z. Deep learning for inferring gene relationships from single-cell expression data. Proc. Natl. Acad. Sci. U S A. 2019;116:27151–27158. doi: 10.1073/pnas.1911536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. The reactome pathway knowledgebase. Nucleic Acids Res. 2017;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Csardi G., Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 36.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for Integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono K., Muetze T., Kolishovski G., Shannon P., Demchak B. CyREST: turbocharging Cytoscape access for external tools via a RESTful API. F1000Res. 2015;4:478. doi: 10.12688/f1000research.6767.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The R code for analyses in this study is available at Github: https://github.com/RuoyuZhang/NoiseRegularization.