Abstract
Objectives:
The 3rd generation epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR TKI) osimertinib has shown promising efficacy both in EGFR-mutant, T790M positive non-small cell lung cancer (NSCLC) patients who have become resistant to 1st or 2nd generation EGFR TKIs and patients with sensitizing EGFR mutations as the first line therapy. However, the degree and duration of response to osimertinib are heterogeneous. We hypothesized that the concurrent genomic landscape of these tumors could play a role in clinical outcomes and/or mechanisms of resistance.
Materials and methods:
We conducted a retrospective multicenter study of lung cancer patients who had developed resistance to osimertinib. Genomic profiling was done for all the patients by using targeted next-generation sequencing encompassing 59–1021 cancer-related genes.
Results and conclusion:
Known EGFR-dependent resistant mutations and activation of alternative pathways were identified in 44 % of all the patients with great heterogeneity. Gain-of-function mutations of CTNNB1 were highly enriched in our cohort. Some other putative resistance mechanisms to osimertinib, such as the recurrent EGFR V834 L mutation, were also identified. Moreover, pathogenic mutations of TP53 were negatively related to the efficacy of osimertinib. To sum up, heterogeneity of resistance to osimertinib was not only manifested by inter-individual differences, but also embodied in its intra-individual diversity.
Keywords: Next-generation sequencing, Osimertinib, Acquired resistance, TP53, Concurrent genomic landscape, Non-small cell lung cancer
1. Introduction
Osimertinib is a 3rd generation, highly mutant-selective, irreversible EGFR TKI targeting both EGFR sensitizing and T790M resistant mutations in preclinical models [1]. It is highly effective in patients bearing T790M mutation whose diseases have become resistant to prior treatment with EGFR TKIs. However, there is a great diversity of clinical outcomes to this therapy. We proposed that at least part of this diversity may be explained by co-occurring genetic alterations. Therefore, in this report we sought to probe the landscape of genomic changes in NSCLC patients at the time of developing resistance to osimertinib and correlated the co-mutation profile with time to treatment discontinuation.
2. Materials and methods
2.1. Patients and samples
In total, 100 lung cancer patients who were confirmed resistant to osimertinib were enrolled from multiple hospitals of China from August 2016 to August 2018. All patients received next-generation sequencing (NGS) testing in Geneplus-Beijing Institute with written informed consent. Samples were collected at the time of osimertinib discontinuation. Both tumor tissue and cell free DNA (cfDNA) from either plasma or pleural effusion were eligible for mutation profiling. The NGS testing covered ~1.4 Mbp genomic regions of 1021 cancer-related genes (or ~230 Kbp genomic regions of 59 genes for some patients) (Supplementary Tables 1 & 2).
2.2. Statistical analysis
As physicians almost always discontinue osimertinib only when obvious progression and clinical deterioration occurred in the real-world setting, time to treatment discontinuation (TTD) was used instead of progression-free survival (PFS) for analyzing the clinical outcome of osimertinib. TTD was defined here as the time from the start of treatment to the end of therapy. Patients discontinued osimertinib due to toxicity without disease progression were excluded from the study. TTD distribution was analyzed by the Kaplan-Meier method with a log rank test.
3. Results
3.1. Patient characteristics and mutation profiling
One hundred patients who had developed resistance to osimertinib were included. The time to treatment discontinuation (TTD) was known for 96 patients with a median duration of 8 months. The clinicopathological characteristics of all the patients are summarized in Supplementary Table 3. The median age was 57, and 61 % of the cohort were female. Ninety patients were of adenocarcinoma, and 94 were at stage IV. Almost all the patients had received 1st or 2nd generation EGFR TKIs prior to osimertinib, and 60 of them had received at least two lines of treatment before starting on osimertinib. Tumor tissue was available for genetic analysis in only 18 of these patients, so testing in the remaining 82 patients used cfDNA as an alternative. In total, a median of 4 (0–16) co-occurring mutations were detected in each patient. The most common mutated genes are EGFR, TP53, RB1, CTNNB1 and PIK3CA, which were detected in 85, 66, 12, 8 and 6 patients respectively (Supplementary Fig. 1).
3.2. Heterogeneity of resistance mechanisms to osimertinib
EGFR T790M was detected in 35 patients and absent in 65 patients, which was similar to a recent report [2]. C797S/G mutations were detected in 18 patients, with 2 having both C797S and C797 G mutations. All these 18 patients retained the T790M mutation, and 17 were in cis with the T790 M. The remaining patient had C797 G and T790 M in cis, and also C797S and T790 M in trans. Interestingly, 6 of the 18 patients had two DNA alterations each encoding for C797S/G mutation. Briefly, four had both c.2389 T > A and c.2390 G > C for C797S; one had c.2390 G > C for C797S and c.2389 T > G for C797 G; and one had c.2389 T > A for C797S and c.2389 T > G for C797 G. As these gene alterations were present on different sequencing reads in each person, we speculated that each mutation had developed independently in subclones of the tumor (Supplementary Fig. 2), suggesting convergent evolution, a novel finding. In addition to C797S/G, L718Q/V, L792H and G796S were detected in 4, 2 and 1 patients respectively, all of which have been reported to mediate resistance to osimertinib (Table 1) [3,4]. Interestingly, all these 7 patients were on the 3rd line osimertinib treatment (Table 1). All these resistant mutations coexisted with EGFR sensitizing mutations and all but one of those patients had a detectable T790M mutation. Notably, we identified four EGFR resistant mutations including C796S, L718Q and two DNA alterations encoding for C797S mutation with significantly different mutant allele frequency (MAF, 55.0 %, 10.2 %, 1.1 %, 0.2 % respectively) in one patient. This further demonstrated the heterogeneous evolution of tumor resistant subclones.
Table 1.
Known resistance mechanisms to osimertinib in all patients and patients with different lines of osimertinib.
| EGFR resistant mutations | Pts. (n = 100) | 2nd (n = 39) | 3rd (n = 41) | > 3rd (n = 19) |
|---|---|---|---|---|
| C797S / C797G | 17 / 3 | 7 / 1 | 8 / 2 | 2 / 0 |
| L718Q / L718V | 3 / 1 | 0 | 3 / 1 | 0 |
| L792H | 2 | 0 | 2 | 0 |
| G796S | 1 | 0 | 1 | 0 |
| Totala | 22 | 7 | 13 | 2 |
| Alternative pathways activation | ||||
| activating of PI3K-AKT-mTOR signaling | 10 | 5 | 3 | 2 |
| activating mutation of CTNNB1 | 8 | 3 | 4 | 1 |
| activating mutation of KRAS / NRAS | 4 / 1 | 2 / 1 | 2 / 0 | 0 |
| activating mutation of BRAF | 4 | 0 | 3 | 1 |
| activating mutation of ERBB2 | 3 | 1 | 2 | 0 |
| amplification of MET | 2 | 1 | 0 | 1 |
| RET fusion | 1 | 0 | 1 | 0 |
| Totalb | 26 | 11 | 11 | 4 |
Three patients have two or more concurrent EGFR resistant mutations.
Six patients have two or more concurrent gene alterations in alternative pathways.
Non-EGRR resistance mechanisms were identified in 26 patients with PI3K-AKT-mTOR signaling related genomic alterations occurring in 10 patients: 6 PIK3CA gain-of-function mutations, 3 PTEN and 1 NF1 loss-of-function mutations respectively. Moreover, activation of KRAS/NRAS and BRAF were detected in 5 and 4 patients respectively. Gain-of-function mutations in CTNNB1 (S37 F/C (5), D32 V (1), G34 V (1) and T41I (1)) were detected in 8 patients. These highly enriched mutations [5] were reported to mediate EGFR TKI drug persistence [6]. Gain-of-function mutations of ERBB2 were detected in 3 patients. No ERBB2 amplification was detected in this cohort. MET amplification was identified in 2 patients. Finally, one patient with CCDC6-RET fusion was identified in our cohort (Table 1). Remarkably, 6 of the 26 patients harbored at least 2 concomitant resistant mutations in alternative pathways, including KRAS/BRAF, KRAS/BRAF/CTNNB1, KRAS/PIK3CA, NRAS/PIK3CA mutations each in one patient and BRAF/CTNNB1 mutations in 2 patients. Besides, 4 patients were identified to have both EGFR resistant mutations and alternative pathway activation, which included EGFR/PIK3CA mutations in one patient and EGFR/CTNNB1 mutations in 3 patients.
3.3. Loss-of-function mutations in TP53 adversely impact the TTD of osimertinib
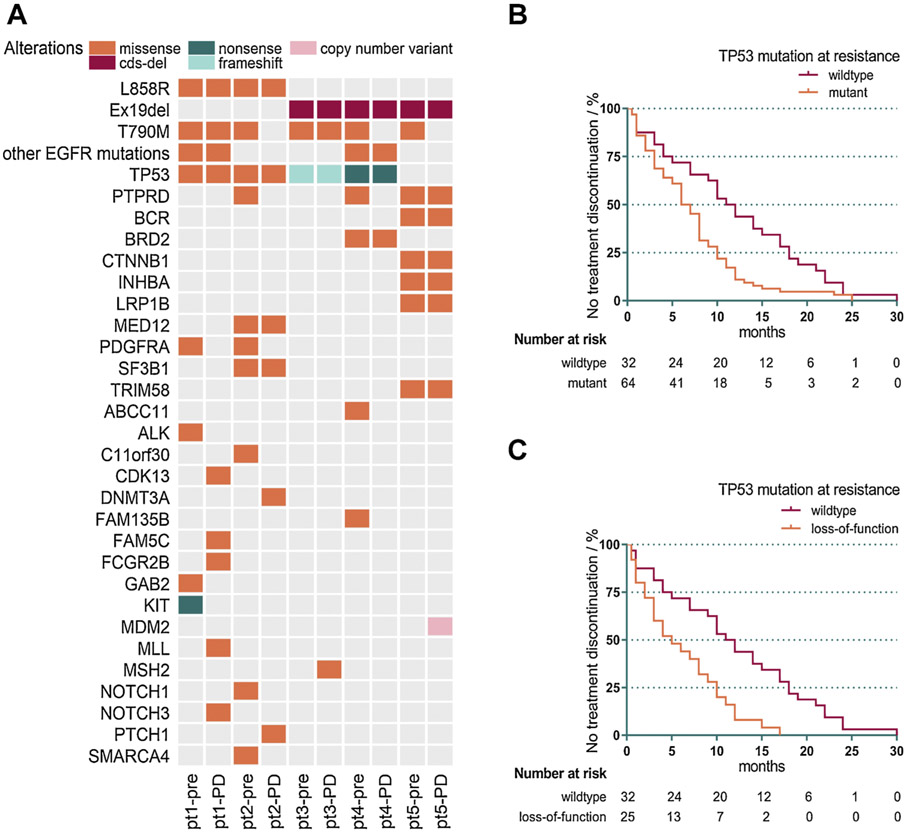
Five patients received NGS testing in the same institute right before starting on osimertinib (Fig. 1A). We noted that mutations of TP53 were detected in 4 of the 5 patients both pre- and post-osimertinib treatment, and none of the 4 patients had a TTD longer than 5 months. However, the remaining patient without TP53 mutation had a TTD of 9 months, the longest of the five. This prompted us to investigate whether mutations of TP53 could impact the efficacy of osimertinib. Our data showed that patients with TP53 mutations detected at the time of osimertinib discontinuation had significantly shorter TTD compared to patients with wildtype TP53 (median, 6.5 vs 11.5 months, p = 0.0029, Fig. 1B). When we focused on only pathogenic mutations of TP53, the difference was more significant (median, 5 vs 11.5 months, p = 0.0005, Fig. 1C).
Fig. 1. TP53 mutations adversely impact the TTD of osimertinib.
A. Pre- and post-osimertinib somatic mutation profiles of 5 patients. Patients are arranged on the x-axis while all genes detected are spread along the y-axis. We can see that co-occuring alterations in TP53 are common in these patients.
B. The Kaplan-Meier curve shows that patients positive for TP53 mutations at resistance have significantly shorter TTD compared with patients with wildtype TP53 (median, 6.5 vs 11.5 months, p = 0.0029).
C. The Kaplan-Meier curve shows that TTD of patients with pathogenic mutations of TP53 at resistance to osimertinib is markedly shorter compared with that of patients with wildtype TP53 (median, 5.0 vs 11.5 months, p = 0.0005).
4. Discussion
In this study, we reviewed a cohort of 100 patients with advanced NSCLC who became resistant to osimertinib and whose tumors demonstrated multiple resistance mechanisms to osimertinib at the time of development of resistance. In total, 44 patients were identified to have either tertiary EGFR mutations or activation of alternative pathways or both as resistance mechanisms.
Except for known EGFR resistance mutations mentioned above, some EGFR mutants with unknown significance were identified in this study (Supplementary Table 4). Most of those mutants occurred in only a single case, however, the EGFR V834 L mutation was detected in 3 patients. Recently it was predicted that V834 L might result in steric hindrance reducing binding ability of osimertinib and lead to resistance [7]. Primary resistance to osimertinib was noted in one patient harboring both L858R and V834 L mutations, further demonstrating the potential role of EGFR V834 L mutation in driving osimertinib resistance. Amplification of EGFR was noted in 5 patients, though its role in osimertinib resistance is still controversial. Histologic transformation of NSCLC to small-cell lung cancer (SCLC) has been reported in osimertinib-resistant patients. It was shown that SCLC transformed resistant cancers had many features of classical SCLC such as gene alterations in RB1 [8]. In our study, loss-of-function mutations of RB1 were detected in 12 patients, with 6 having none of the aforementioned resistance mechanisms. The FGFR3-TACC3 fusion was detected in 1 patient. Cell cycle related genomic alterations including amplification of CCND1 and loss of CDKN2A were detected each in 2 patients. Further studies on those mutations in osimertinib resistance are needed.
TP53 mutation has been shown to affect efficacy of 1st generation EGFR TKI when coexisting with EGFR sensitizing mutations. Here we demonstrated that inactivating mutations of TP53 were also associated with shorter TTD to osimertinib. Limited by the retrospective analysis of this real-world study, we did not have pre-treatment samples in all patients to analyze TP53 mutation before osimertinib treatment, but mutations of TP53 are more commonly present at baseline than to develop at progression in published studies of tumor evolution [9]. Thus, our data and those of others suggest that mutations in TP53 portend a shorter duration of efficacy of osimertinib.
It has been suggested that patients with indolent and small volume progression benefit from continuation of TKIs beyond progression [10]. Moreover, as there are limited attractive therapeutic options after osimertinib resistance, physicians tend to discontinue osimertinib only when obvious progression and clinical deterioration occur, thus this real world study used TTD instead of PFS to evaluate the efficacy of osimertinib. This study has certain limitations, the status of T790M mutation before osimertinib was uncertain in 66 patients. However, no difference in TTD was found between these 66 patients and patients with confirmed T790M mutation before the initiation of osimertinib (n = 34, p = 0.6924, Supplementary Fig. 3). Due to the nature of retrospective analysis, these results do not completely portray the evolution of genetic changes from diagnosis through progression and patient selection is not random. Additional larger cohorts will be needed to further confirm our findings.
In summary, we showed that resistance to osimertinib is highly heterogenous not only among different individuals but also within a single patient. The development of multiple different mutations within a single patient resulting in the same codon alteration suggest convergent evolution of independent clones developing the same mechanism of resistance. Mutations of TP53 seem to negatively affect patients’ duration of benefit to osimertinib.
Supplementary Material
Acknowledgements
This work was supported by National Key Research and Development Program of China (2016YFC0904900); Joint Funds for the Innovation of Science and Technology, Fujian province, China (2017Y9083); Fujian Provincial Health and Family Research Talent training program, Fujian province, China (2015-ZQN-ZD-9); Science and Technology Department guided projects, Fujian province, China (2018Y0016)
Abbreviations:
- EGFR
epidermal growth factor receptor
- NSCLC
non-small cell lung cancer
- TKI
tyrosine kinase inhibitor
- NGS
next-generation sequencing
- cfDNA
cell free DNA
- TTD
time to treatment discontinuation
- MAF
mutant allele frequency
Footnotes
Ethics
The study was approved by the institutional ethic review boards of Geneplus-Beijing. All patients in this study provided written informed consent for specimen collection, genetic testing, and use of this information for research purposes.
Declaration of Competing Interest
Huamin Xu, Rongrong Chen and Xuefeng Xia are employees of Geneplus-Beijing Ltd. All other authors declared no conflict of interest.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.lungcan.2019.10.021.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ, Hughes G, Rahi A, Jacobs VN, Red Brewer M, Ichihara E, Sun J, Jin H, Ballard P, Al-Kadhimi K, Rowlinson R, Klinowska T, Richmond GH, Cantarini M, Kim DW, Ranson MR, Pao W, AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer, Cancer Discov. 4 (2014) 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, Feeney N, Sholl LM, Dahlberg SE, Redig AJ, Kwiatkowski DJ, Rabin MS, Paweletz CP, Thress KS, Janne PA, Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-Positive lung cancer and acquired resistance to osimertinib, JAMA Oncol. 4 (2018) 1527–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ou SI, Cui J, Schrock AB, Goldberg ME, Zhu VW, Albacker L, Stephens PJ, Miller VA, Ali SM, Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib, Lung Cancer 108 (2017) 228–231. [DOI] [PubMed] [Google Scholar]
- [4].Callegari D, Ranaghan KE, Woods CJ, Minari R, Tiseo M, Mor M, Mulholland AJ, Lodola A, L718Q mutant EGFR escapes covalent inhibition by stabilizing a non-reactive conformation of the lung cancer drug osimertinib, Chem. Sci 9 (2018) 2740–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, McGranahan N, Wilson GA, Birkbak NJ, Olivas VR, Rotow J, Maynard A, Wang V, Gubens MA, Banks KC, Lanman RB, Caulin AF, St John J, Cordero AR, Giannikopoulos P, Simmons AD, Mack PC, Gandara DR, Husain H, Doebele RC, Riess JW, Diehn M, Swanton C, Bivona TG, Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers, Nat. Genet 49 (2017) 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Arasada RR, Shilo K, Yamada T, Zhang J, Yano S, Ghanem R, Wang W, Takeuchi S, Fukuda K, Katakami N, Tomii K, Ogushi F, Nishioka Y, Talabere T, Misra S, Duan W, Fadda P, Rahman MA, Nana-Sinkam P, Evans J, Amann J, Tchekneva EE, Dikov MM, Carbone DP, Notch3-dependent beta-catenin signaling mediates EGFR TKI drug persistence in EGFR mutant NSCLC, Nat. Commun 9 (2018) 3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Le X, Puri S, Negrao MV, Nilsson M, Robichaux JP, Boyle TA, Hicks JK, Lovinger K, Roarty EB, Rinsurongkawong W, Tang M, Sun H, Elamin YY, Lacerda L, Lewis J, Lee JJ, Roth JA, Swisher SG, Zhang J, William WN, Glisson BS, Papadimitrakopoulou VA, Gray JE, Heymach JV, Landscape of EGFR -dependent and -independent resistance mechanisms to osimertinib and continuation therapy post-progression in EGFR-mutant NSCLC, Clin. Cancer Res (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T, Howe E, Fulton LE, Mulvey HE, Bernardo LA, Mohamoud F, Miyoshi N, VanderLaan PA, Costa DB, Janne PA, Borger DR, Ramaswamy S, Shioda T, Iafrate AJ, Getz G, Rudin CM, Mino-Kenudson M, Engelman JA, RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer, Nat. Commun 6 (2015) 6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, Moore DA, Turajlic S, Xu H, Lee SM, Forster MD, Ahmad T, Hiley CT, Abbosh C, Falzon M, Borg E, Marafioti T, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Shah R, Joseph L, Quinn AM, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Dentro S, Taniere P, O’Sullivan B, Lowe HL, Hartley JA, Iles N, Bell H, Ngai Y, Shaw JA, Herrero J, Szallasi Z, Schwarz RF, Stewart A, Quezada SA, Le Quesne J, Van Loo P, Dive C, Hackshaw A, Swanton C, Consortium T.R, Tracking the evolution of non-small-cell lung cancer, N. Engl. J. Med 376 (2017) 2109–2121. [DOI] [PubMed] [Google Scholar]
- [10].Park K, Yu CJ, Kim SW, Lin MC, Sriuranpong V, Tsai CM, Lee JS, Kang JH, Chan KC, Perez-Moreno P, Button P, Ahn MJ, Mok T, First-line erlotinib therapy until and beyond response evaluation criteria in solid tumors progression in Asian patients with epidermal growth factor receptor mutation-positive non-small-cell lung cancer: the ASPIRATION study, JAMA Oncol. 2 (2016) 305–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.