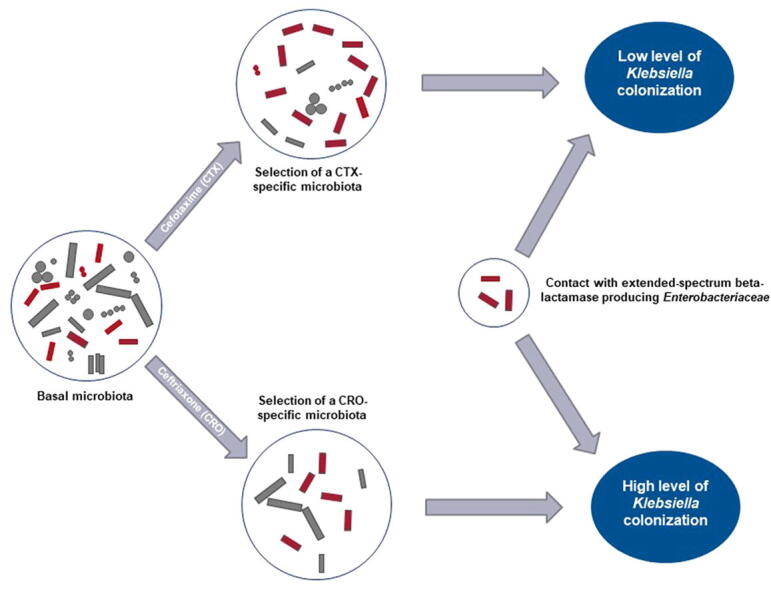
Graphical abstract
Keywords: Beta-lactamase, Gastrointestinal microbiome, Enterobacteriaceae, Extended-spectrum beta-lactamase
Abstract
Background
The globally increasing resistance due to extended-spectrum beta-lactamase producing Enterobacteriaceae is a major concern. The objective of this work was to develop a murine model to study the gut bacteria parameters during complex antibiotics like cefotaxime and ceftriaxone treatment and to compare the fecal carriage of ESBL-producing Enterobacteriaceae.
Methods
SWISS mice were treated either with ceftriaxone or with cefotaxime or with NaCl 0.9% as a control group from day 1 to day 5. We performed a gavage at day 4 with a Klebsiella pneumonia CTX-M9. We collected stools and performed pharmacological measurements, cultures and 16S rRNA gene amplification and sequencing during the 12 days of the stool collection.
Results
Mice treated with ceftriaxone were more colonized than mice treated with cefotaxime after gavage (p-value = 0.008; Kruskal-Wallis test). Ceftriaxone and cefotaxime were both excreted in large quantity in gut lumen but they drove architecture of the gut microbiota in different trajectories. Highest levels of colonization were associated with particular microbiota composition using principal coordinate analysis (PCoA) which were more often achieved in ceftriaxone-treated mice and which were preceded by highest fecal antibiotics concentrations in both cefotaxime or ceftriaxone groups. Using LEfSe, we found that twelve taxa were significantly different between cefotaxime and ceftriaxone-treated mice. Using SplinectomeR, we found that relative abundances of Klebsiella were significantly higher in CRO than in CTX-treated mice (p-value = 0.01).
Conclusion
Ceftriaxone selects a particular microbial community and its substitution for cefotaxime could prevent the selection of extended-spectrum beta-lactamase producing Enterobacteriaceae.
1. Introduction
The increasing global resistance of Enterobacteriaceae to beta-lactams is a major public health concern for the years to come and led the World Health Organization to classify the resistance of Enterobacteriaceae to third-generation cephalosporins (3GC) as a critical priority for research and development of new antibiotics in February 2017 [1]. The majority of 3GC-resistant Enterobacteriaceae produces extended-spectrum beta-lactamase (ESBL) which increases morbidity and mortality, length of stay and health costs [2], [3].
The microbiota would be composed of 800 to 1000 species comprising more than 7000 different strains [4]. Within this population of 1012 to 1013 individuals, bacteria are carrying genes for antibiotic resistance. Antibiotics conduct to a partial destruction of the gut bacteria (mainly anaerobic) susceptible to the given agent, thus leaving room and access to the necessary resources for organisms resistant to this treatment [5]. Knowing that gut microbiota is a reservoir for ESBL-producing Enterobacteriaceae, their multiplication can lead to excretion in the external environment and inter-individual transmission [6], [7]. These germs may be responsible for infections that will be difficult to treat [8]. One of the possibilities to limit the emergence of resistant mutants is to choose the antibiotics that have the lowest impact on the microbiota but knowledge that could drive this choice are still scarce.
Studies show that use of 3GC and exposure to 3GC favors ESBL-mediated resistance in Enterobacteriaceae [9], [10], [11]. Although they have similar antibacterial spectrum, ceftriaxone (CRO) and cefotaxime (CTX) may have different impact on ESBL-mediated resistance. Specifically, it is advocated that CRO has more impact on the gut microbiota than CTX, due to higher biliary elimination than CTX [12]. In a large multicentric study involving 701 health care facilities, use of CRO is positively associated with 3GC-resistant E. coli, whereas CTX use is not [13]. However, use of both 3GC is associated with 3GC-resistant Klebsiella. pneumonia (Kp) in the same study, and with 3GC-resistant E. cloacae in another one [12], [13]. The impact of replacing CRO by CTX on resistance to 3GC is reported in 2 hospital-level studies. In one center, it is associated with a slower growth of high-level cephalosporinase mediated resistance, mainly in E. cloacae, but has no impact on ESBL-mediated resistance [14]. More recently, Tan et al. show that switching from CRO to CTX is associated with a decreased incidence of hospital-acquired infection caused by ESBL-producing Enterobacteriaceae [15]. Lately, Burdet et al. do not found different effects on the microbiota in healthy volunteers treated 3 days with CRO or CTX [16]. Hence, the ecological advantage of CTX on CRO remains debated, and no experimental study has compared the effects of CRO and CTX on the fecal microbiota.
The main objective of this experimental work was to develop a murine model to study the gut bacteria parameters during complex antibiotics like CTX and CRO treatment and to compare the fecal carriage of ESBL-producing Enterobacteriaceae.
2. Materials and methods
2.1. Animals
SWISS non-consanguineous albino mice provided by the JANVIER laboratory (Le Genest-Saint-Isle, France) were used. These are robust and non-aggressive animals often used for pharmacological modeling and already used to study the carriage of multidrug-resistant Enterobacteriaceae [17]. We used females aged 6 weeks and weighing between 25 and 30 g, guaranteed without pathogens. The mice were housed at the Animal Research Center of the Institute of Health Research 2, University of Nantes, in a controlled environment (day/night cycle of 12 h, extinction 7:30 pm) with a controlled sterile diet. After a review of the literature on multidrug-resistant bacteria implantation in murine models, the number of mice per treatment group was set at 15 [18], [19], [20], [21]. In addition, mice were isolated in individual cages to prevent cross-contamination by coprophagia. Drinking water and food were provided ad libitum. Study was approved by the French Ministry of Higher Education and Research (APAFIS 8528, Site agreement A44279, ethics committee 006).
2.2. Antibiotics
Compared with CTX structure, CRO has a basic 2-(2–42,4–42)-2-(Z)-methoxyimino-acetyl side chain which led to a very long elimination half-life of 8 h in human, high beta-lactamase stability and extremely high chemotherapeutic efficacy against a broad spectrum of Gram-positive and Gram-negative pathogens. Therefore, in human, due to its extended half-life, CRO is used one or twice daily while CTX is used 3-time daily or in continuous infusion. Reconstitution of CRO and CTX was performed in physiological saline (NaCl 0.9%) to obtain initial concentrations of 100 g/L. Mice were randomized in three groups: Group 1 received CRO 250 mg/kg twice daily subcutaneously from day 1 to day 5, group 2 received CTX 500 mg/kg three times daily from day 1 to day 5 and the control group received NaCl 0.9% three times daily from day 1 to day 5. Taking into account the very short half-life of these molecules in mice and the concentrations usually found in humans, it was decided to use high dosages corresponding to 5 times the recommended dosage in humans when treating severe infections such as bacterial meningitis or infectious endocarditis [22], [23], [24]. Indeed, we carried out a preliminary pharmacokinetic study to choose the optimal dosage to be adopted based on the plasma and fecal concentrations usually found in patients treated for infections. Maximum plasma concentration and mean plasma half-life of CTX (after 250 mg/kg, n = 5) in our mice were 207 mg/L and 24 min, respectively and maximum plasma concentration and mean plasma half-life of CRO (after 500 mg/kg, n = 1) were 264 mg/L and 78 min, respectively. Plasma concentrations were similar to those usually observed in human and concentrations of CRO and CTX in the feces were they similar to those observed by previously published studies (279 µg/g and 167 µg/g respectively) [17], [25], [26]. We therefore chose to administer 500 mg/kg three times a day for CTX and 250 mg/kg twice a day for CRO. All subcutaneous injections were 200 μL.
2.3. Bacterial strain and gavage
The strain used for gavage was a clinical isolate of Kp, carrying the beta-lactamase CTX-M9 (MIC of CTX: 512 mg/L, MIC of CRO: 2046 mg/L). The plasmids harboring most blaCTX-M are transferable among bacterial cells, especially in the gut [27]. Inocula of 105 CFU were prepared immediately before gavage using a spectrophotometer. Gavage was done at day 4 of the antibiotic treatment.
Three runs of 5 mice in each group (CRO, CTX and NaCl) were performed.
2.4. Stool collection and culture
Stool sampling was performed at day 1, 4, 6, 8, 10 and 12. On the day of sampling, each mouse was placed in a clean cage for 1 h to collect his stool and was returned to his “accommodation” cage. Stools were immediately frozen at −80 °C.
Each stool was weighed and then crushed (Ultraturrax Tube Drive®, IKA™, Germany) with 10 μL saline per milligram of stool. Serial dilutions were performed in the fresh state, then manual inoculations of 100 μL on chromogenic media with mixture of antibiotics, including cefpodoxime (ChromID ESBL, BioMérieux™, Marcy L'Etoile, France) were carried out before incubation at 37 °C of 24 h.
The detection threshold was 2 log10 CFU/g stool. At least one colony per culture medium was identified using mass spectrometry (MALDI TOF, Vitek MS®, Biomérieux™, Marcy L'Etoile, France) to confirm genus and species.
2.5. 16S rRNA gene amplification and sequencing
Fecal samples were kept frozen at −80 °C until they were processed. After fecal DNA isolation using the PowerSoil® DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA fecal DNA kit) including an enzymatically cell lysis step, amplicons spanning the variable region 4 of bacterial 16S rRNA gene were generated and sequenced using Illumina Mi-seq platform at the University of Minnesota Genomic Center, Twin Cities, MN [28]. The 16S rRNA gene sequencing data from the Illumina runs were trimmed and filtered using SHI7 [29]. We then performed operational taxonomic units (OTUs) assignment using ‘NINJA-OPS’ against the Greengenes 13.8 database as a reference, by clustering the sequences with a threshold of 97% pairwise identity [30], [31]. Next, we used Quantitative Insights Into Microbial Ecology (QIIME) 1.9.1 for diversity analyses [32]. We presented beta diversity, based on Unweighted UniFrac distances, a β-diversity measure that uses phylogenetic information to compare samples, with principal coordinate analysis (PCoA), using the plugin beta_diversity_through_plots.py and a sampling depth of 17,236 [33]. We applied the PERMANOVA method on the previously obtained dissimilarity matrices. PERMANOVA was performed using 1000 permutations. We computed alpha diversity metrics using non-phylogeny metrics (Observed OTUs, Chao1 index, Shannon index) with the plugin alpha_rarefaction.py and a sampling depth of 17236. We also performed Random Forest (RF) classification with 500 trees and ten-fold cross-validation using the plugin supervised_learning.py and the OTU table [34]. To study longitudinal variation in our samples, we used SplinectomeR, that applied smoothing splines to summarize data for straightforward hypothesis testing in longitudinal studies [35].
To compare samples collected in CRO and CTX mice, we used the LEfSe tool on the OTU table collapsed at genus level. LEfSe (Linear discriminant analysis Effect Size) determines the features (here, taxa collapsed at genus level) most likely to explain differences between classes by coupling standard tests for statistical significance with additional tests encoding biological consistency and effect relevance [36]. To identify significant associations between microbial and phenotypic variables, we applied a linear multivariate regression model specifically adapted to microbiome data: MaAsLin, Multivariate microbial Association by Linear models [37]. MaAsLin constructs boosted, additive general linear models to associate metadata and transformed microbial taxonomic or functional relative abundances. Since microbial community profiles are typically high dimensional, boosting is used for feature selection over potential covariates to identify those most associated with each microbial feature. Selected metadata are then used in a general linear model with metadata as predictors and arcsin-square root transformed microbial relative abundances from the OTU table collapsed at genus level as the responses. The dataset generated and analyzed during the current study is available in the NCBI repository under the accession number PRJNA701545 (http://www.ncbi.nlm.nih.gov/bioproject/701545). The OTU table, collapsed at genus level is also provided as Table S1. Compositional biplot that simultaneously displays the sample clustering and phyla of the gut microbiomes of the fecal samples collected from CTX- and CRO-treated mice are represented in Fig. S1.
2.6. Pharmacokinetics
Stools collected were weighed, diluted in 0.9% NaCl (10 mg/100 μL) and homogenized by grinding in ball tubes and sonication. The ground material obtained was centrifuged (5 min, 13,000g, +4 °C).
For the CTX and desacetyl-CTX assay, 125 μL of the supernatant was mixed with 125 μL of a methanol/ZnSO4 3 M solution (80/20 v/v) and 250 μL of an acetonitrile solution containing the deuterated internal standard (13C2, 2H3-CTX). After centrifugation of the mixture (10 min, 13,000 g, +4 °C), 2 μL of the supernatant were injected into the chromatographic system (H-Class® Acquity UPLC system, Waters™, St Quentin en Yvelines, France). The system consisted of a Kinetex 2.6 μm C18 column in a thermostatically controlled oven at 50 °C, mobile phases with a binary gradient [(acetonitrile/formic acid 0.1% v/v) and (ultrapure water/formic acid 0.1% v/v)] at a flow rate of 0.8 mL/min and a mass spectrometer monitoring of the m/z ratios (456,460,414) for respectively CTX, 13C2, 2H3-CTX, and desacetyl-CTX for 4 min after each injection.
For the CRO assay, 250 μL of the supernatant was mixed with 250 μL of acetonitrile solution containing the deuterated internal standard (13C, 2H3-CRO). After centrifugation (5 min, 13,000g, +4 °C) of the mixture, 300 μL of the supernatant were mixed with 2 mL of dichloromethane. After centrifugation, 2 μL of the supernatant was injected into the H-Class® Acquity UPLC system. The chromatographic system consisted in a Kinetex 2.6 μm C18 column in a furnace thermostatically controlled at 50 °C, mobile phases with a binary gradient (acetonitrile and a 1 mM aqueous ammonium acetate solution) at a flow rate of 0.8 mL/min and a mass spectrometer monitoring of the m/z ratios (555,559) for the CRO and 13C, 2H3-CRO respectively for 4 min after each injection. For both methods, reproducibility and inaccuracy were less than 15%, the limit of quantification was 10 μg/g stool for CRO, CTX and desacetyl-CTX.
2.7. Quantification and statistical analysis
Statistical analyzes were performed using RStudio® (https://www.r-project.org/) and GraphPad Prism® 6.0 (GraphPad Software Inc. ™, La Jolla CA, USA). Areas under the curve (AUC) were calculated by trapezoidal method. Negative culture were fixed at 2 log10 CFU/g stool.
The Kruskal-Wallis test was used for unpaired data. The post hoc multiple comparison tests were performed using the Dunn method. Non-parametric Mann-Whitney tests were performed between the two-by-two groups with Bonferroni correction. All tests were defined with alpha risk determined a priori as significant if ≤ 0.05.
3. Results
3.1. CRO induced higher ESBL-Kp colonization than CTX
Results of growth culture are shown in Fig. 1A. The absence of pre-existing 3GC-resistant strains prior to gavage by ESBL-producing Kp is demonstrated by the negativity of stool cultures on the day of gavage. CRO-induced ESBL-producing Kp colonization was consistently higher than CTX-induced colonization from day 4 to day 12 (p-value = 0.008, Mann-Whitney test) (Fig. 1B).
Fig. 1.
Gut colonization by Klebsiella pneumoniae, transit modifications and fecal concentrations of CRO and CTX. (A) Quantification of fecal excretion of ESBL-producing Kp over time according to treatment administered (n = 15 per group). The bars represent the mean and the range. (B) Quantification of fecal excretion of ESBL-producing Kp over time according to treatment administered (n = 15 per group). Lines correspond to loess-smoothed conditional means and shading to SE. (C) Area under the curve (AUC) of ESBL-producing Kp excretion of control, CTX and CRO groups. A statistically significant difference in colonization between CRO and CTX was observed from day 4 to day 12 (p-value = 0.0078, Mann-Whitney test) and is symbolized by an asterisk. (D) CRO fecal concentrations. (E) CTX (squares) and desacetyl-CTX (stars) fecal concentrations. Values were indicated as mean ± SEM (n = 10 per group). Detection threshold was 10 μg/g.
3.2. CRO and CTX are both excreted in gut lumen
Detectable fecal concentrations for CRO were observed and were maintained until 10 days after the end of treatment (Fig. 1C). Detectable concentrations for CTX and its active metabolite desacetyl-CTX were observed until 10 days after stopping treatment (Fig. 1D). Fecal exposures to antibiotics were assessed using the area under the curve (AUC) of fecal concentrations over time. AUC of CTX and desacetyl-CTX were 1015 μg.g−1.day and 3962 μg.g−1.day respectively. AUC of CRO was 1859 μg.g−1.day.
3.3. CRO and CTX drive the architecture of the gut microbiota in different trajectories
At baseline (i.e., before antibiotic treatment was started), we did not find any significant difference between CTX, CRO-treated mice and control mice, in terms of alpha diversity (ANOVA, Shannon index: p-value = 0.556, observed OTUs: p-value = 0.143). Overall, we found a significant decrease in alpha diversity between antibiotic-treated mice and control mice (ANOVA, Shannon index: p-value = 0.001, observed OTUs: p-value = 0.001, chao1 index: p-value = 0.001). However, we did not find significant difference in alpha diversity between CTX and CRO-treated mice (ANOVA, Shannon index: p-value = 0.665, observed OTUs: p-value = 0.530, chao1 index: p-value = 0.855, Fig. S2). Importantly, alpha diversity decreased significantly with time in both CTX and CRO-treated mice (p-value = 0.01), but this decrease was not different between CTX-treated mice and CRO-treated mice (p-value = 0.50).
Using Unweighted UniFrac distances, at baseline (i.e., before antibiotic treatment was started), we did not find any significant difference between we did not find any significant difference between CTX, CRO-treated mice and control mice (PERMANOVA, r2 = 0.068, p-value = 0.492, Fig. S3A). Overall, we found that both CTX and CRO significantly alter the overall architecture of the gut microbiota when compared to control mice (PERMANOVA, r2 = 0.128, p-value = 0.001, principal component 1: ANOVA, p-value < 0.001, principal component 2: ANOVA, p-value < 0.001). Moreover, CRO and CTX drove the architecture of the gut microbiota in 2 different trajectories (PERMANOVA, r2 = 0.142, p-value = 0.001, principal component 1: ANOVA, p-value = 0.358, principal component 2: ANOVA, p-value = 0.002, Fig. S3A).
We observed that the time after the antibiotic treatment had a strong impact on the beta-diversity with higher Principal Coordinate (PC) 1 and PC2 scores observed at day 12 (Fig. 2A). Before treatment and until day 4, CRO- and CTX-treated mice were similar and differences appeared at day 8 and increased at day 12. A more important inter-individual variability was observed in CTX-treated than in CRO-treated mice at day 12. Beta-diversity trajectories by mouse are represented in Fig. S3B. Supervised learning using Random Forests, a machine learning method using OTUs as predictive features, accurately assigned samples to their source population (CTX or CRO-treated mice) based on taxonomic profiles at the OTU level (83.3% accuracy, 3 times better than the baseline error rate for random guessing). Thus, based on microbiome data only, we were able to predict if a mouse received CRO or CTX treatment.
Fig. 2.
CRO and CTX alter the overall architecture of gut microbiota and induce taxonomic changes. (A) Beta diversity comparisons of the gut microbiomes of the fecal samples collected from CTX- and CRO-treated mice. Beta diversity is represented by ellipse clustering according to the day after the start of antibiotic treatment. (B) Summary of the taxa that differentiate CTX from CRO-treated mice using Linear discriminant analysis Effect Size analysis (LEfSe). At genus level, 12 taxa were significantly different CTX from CRO-treated mice (absolute LDA log 10 score >2). (C) Longitudinal changes in Enterococcus compared between CTX and CRO-treated mice, using SplinectomeR with permuted spline test. (Left) Enterococcus relative abundance over time distinguishes CTX (group spline in blue) and CRO-treated mice (group spline in red; 999 permutations, p-value = 0.01). Permuted splines represented in grey. The permuted splines lie predominantly between the two observed curves, supporting the conclusion that this difference is larger than expected by chance. (Right) The plot output of the sliding spliner function shows the p-value at each specified interval derived from the distribution of points from individuals’ smoothed splines Dotted line indicates p-value = 0.05. (D) Longitudinal changes in Klebsiella compared between CTX and CRO-treated mice, using SplinectomeR with permuted spline test. (Left) Klebsiella relative abundance over time distinguishes CTX (group spline in blue) and CRO-treated mice (group spline in red; 999 permutations, p-value = 0.01). Permuted splines represented in grey. The permuted splines lie predominantly between the two observed curves, supporting the conclusion that this difference is larger than expected by chance (Right) The plot output of the sliding spliner function shows the p-value at each specified interval derived from the distribution of points from individuals’ smoothed splines. Dotted line indicates p-value = 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Using LEfSe on the OTU table collapsed at the genus level [36], we found that 12 taxa were significantly different between CTX and CRO-treated mice. Specifically, CRO-treated mice were associated with a significant gain in Lactobacillus, Klebsiella, unclassified Enterobacteriaceae, and Parabacteroides when compared to CTX-treated mice, that were, conversely, associated with increase in Enterococcus, unclassified Carnobacteriaceae, unclassified Planococcaceae, Granulicatella, unclassified Lactobacillales, unclassified Enterococcaceae, Vagococcus and Anaeoplasma (Fig. 2B). The most significant differentiating taxa between CTX and CRO-treated mice are represented in Fig. S4A and B. Significantly different taxa between CRO and control mice and CTX and control mice respectively are described in Fig. S4C and 5D. Comparison of antibiotic treated mice (all collected samples in CTX and CRO-treated mice) and control mice (all collected samples) is represented un Table S3.
To confirm these changes, we used SplinectomeR that enables group comparisons in longitudinal microbiome studies. First, we plotted the longitudinal changes in the three groups of mice, antibiotic-treated mice (CRO or CTX) and control mice. We found that in control mice, gut microbiota composition was relatively stable along time, based on the OTU table collapsed at genus level (Fig. S5A), whereas, gut microbiota composition was dramatically altered following CRO and CTX treatment (Fig. S5B and C). We confirm that Enterococcus was significantly higher in CTX than in CRO-treated mice along days of collection (p-value = 0.01, Fig. 2C) and that Klebsiella was significantly higher in CRO than in CTX-treated mice (p-value = 0.01, Fig. 2D). Unclassified Enterobacteriaceae, Parabacteroides and Lactobacillus were also higher in CRO than in CTX-treated mice (p-value = 0.04, p-value = 0.03 and p-value = 0.1, respectively).
3.4. Antibiotic fecal concentrations impact on OTU abundance
We investigated the relationship between CRO, CTX and desacetyl-CTX fecal concentrations and the gut microbiome by identifying significant multivariate linear associations using MaAsLin. The relative abundance of 34 OTUs was positively correlated with CRO fecal concentration levels, including members of Sphingomonadaceae, Microbacteriaceae, Sphingobacteriaceae and Enterobacteriaceae families. Two OTUs were positively correlated with CTX/desacetyl-CTX fecal concentration levels, corresponding to Lactobacillus and Leucobacter genera.
We observed that higher antibiotic concentration levels were not associated with highest PC1 and PC2 scores but preceded these metagenomics alterations leading to colonization by ESBL-producing Kp.
3.5. ESBL-producing Klebsiella pneumoniae carriage is associated to taxonomic changes
We observed that clustering of fecal samples was partly driven by the increased carriage of ESBL-producing Kp (r2 = 0.0641, p-value = 0.001, Fig. S6). Higher colonization levels being associated with higher PC1 and PC2 scores in the two groups, i.e. with the most altered microbiota. This beta-diversity pattern was mainly found in CRO-treated mice and especially at day 12 as described in Fig. S3B and suggested that CRO selects a microbial community which promotes colonization by ESBL-producing Kp. Conversely, highest inter-individual diversity observed at day 12 in CTX-treated mice was associated with in a lower level of colonization. We found that 8 taxa from the OTU table collapsed at genus level, were significantly different between mice displaying a high level of ESBL-producing Kp and those displaying a low level (Fig. S6B). Specifically, mice with a high level were associated with a significant gain in Klebsiella and unclassified Pseudomonadaceae when compared to low level mice, that were, conversely, associated with increase in unclassified Bacteroidales, unclassified Desulfovibrionaceae, Sutterella, unclassified Peptococcaceae, unclassified Clostridiaceae and Akkermansia.
These results were confirmed by a negative association found between intensity of ESBL-producing Kp carriage and OTUs from Bacteroidales and Clostridiaceae (Table S2).
4. Discussion
This experimental study shows three main results: (i) CRO appears to promote higher fecal carriage of ESBL-producing Kp than CTX; (ii) both antibacterial agents altered fecal microbiotas, but CRO had a more important impact than CTX; (iii) higher level of fecal colonization by ESBL-producing Kp was associated with more altered microbiota. These results suggest that the effect of CRO on Kp colonization could be mediated by its effect on the intestinal microbiota.
We observe that fecal Kp levels were significantly higher in CRO-treated mice than in 2 other groups (CTX and NaCl) but were equivalent in control and CTX-treated mice. We cannot explain these results neither by the fecal concentrations observed during the experience nor by the MIC of the strain. Indeed, concentrations of CRO and CTX in the feces were they similar to those observed by previously published studies [17], [25], [26]. Moreover, we cannot stated that MIC of CRO and CTX were different regarding the variability in measurement [38]. Therefore, we made 2 hypotheses: the first based on the modification of the architecture of the microbiota and the second based on the mechanism of action of these two 3GC.
Indeed, we found that CRO and CTX drove the architecture of the gut microbiota in two different trajectories. As expected, we found a significant decrease in alpha diversity between antibiotic-treated mice and control mice but we did not find significant difference in alpha diversity between CTX and CRO-treated mice [16]. If before the treatment and until day 4, it was impossible to tell the difference between CRO- and CTX-treated mice, differences appeared at day 8 and were accentuated at day 12. A more important inter-individual variability was observed in CTX-treated mice which do not reached all the maximum values of PC1 and PC2 at day 12 contrary to CRO-treated mice which reached high PC1 and PC2 values. Interestingly, higher Kp-colonization levels were associated with more altered microbiota in the two groups and higher antibiotic concentration levels were not associated with highest PC1 and PC2 scores but were preceded by these metagenomics alterations. Conversely, highest inter-individual diversity observed at day 12 in CTX-treated mice was associated with a lower level of colonization. These data suggested that CRO could select a microbial community which promotes colonization by ESBL-producing Kp more easily than CTX. These modifications have not been observed by Burdet et al. but their work focused on low dosages of CRO given over only three days and in healthy individuals which are not comparable to those potentially given in more serious infections [16]. The only two subjects with changes in the microbiota were those with detectable CRO in the stool but previous study that CRO is more frequently found in the stool in high concentrations and similar to that which we find [16], [26].
Our results challenge also the pharmacokinetic hypothesis considering that CRO only could selected more ESBL-producing Enterobacteriaceae because of a higher fecal elimination than CTX [12]. As explain before, we observed detectable concentration of CRO, CTX and its active metabolite desacetyl-CTX until 10 days after stopping treatment in faeces [39]. These results confirm those of Grall et al. who found digestive excretion of CTX in significant proportions [17]. We could also hypothesize that desacetyl-CTX plays a role in the prevention of carriage ESBL-producing Enterobacteriaceae. Indeed, if the bactericidal activity of desacetyl-CTX is generally considered to be lower than CTX (the MIC of desacetyl-CTX is 8-fold higher than that of CTX on susceptible strains of Kp), desacetyl-CTX could act as a beta-lactamase inhibitor and could potentiate the action of CTX as suggested by previous studies [40], [41]. To reinforce this hypothesis, Labia et al showed that desacetyl-CTX was much more stable to hydrolysis by beta-lactamase than CTX and could have a sustained bactericidal effect even in the presence of these enzymes [42].
These data are crucial for the comprehension of the parameters influencing the carriage of ESBL-producing Enterobacteriaceae, especially for patient displaying a high risk of colonization such as patient treated with antibiotics or travelers in endemic area.
Our study has several limitations. First, we used 16S rRNA sequencing that limits taxonomic identification at genus level. Therefore, we were not able to identify species or strains that differ between CRO and CTX treated. In addition, our work is limited to a single strain of ESBL-producing Enterobacteriaceae and deserves to be strengthened by testing other strains.
5. Conclusions
In mice, both CRO and CTX modify the microbiota but CRO promotes more the carriage of a strain of ESBL-producing Kp. If CRO appears to select a favorable specific microbial community for the installation of this bacteria in a concentration-dependent way, CTX and particularly its active metabolite desacetyl-CTX could prevent colonization because of a beta-lactamase inhibitor effect.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was funded by the association AGISMED TOXICOLOGIE.
Authors' contributions
M.G., E.M. and E.D. designed the experiments, M.G., F.B., R.B. conducted animal experiments, M.G., F.B., R.B., F.J. and P.B. conducted bacterial culture, MG., P.A., A.B. and J.G. conducted functional exploration experiments, Q.L. and E.M. performed metagenomics analysis, F.B., R.B. and E.D. conducted pharmacokinetics measurements, M.G., F.B., E.M, E.D., E.B, D.L. and M.N. wrote the manuscript and created figures.
Ethics approval and consent to participate
Ethics approval was not required for the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.02.019.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf [Internet]. [cited 2018 Aug 30]. Available from: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 2.Pitout J.D.D., Laupland K.B. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008 Mar;8(3):159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 3.de Kraker M.E.A., Wolkewitz M., Davey P.G., Koller W., Berger J., Nagler J. Burden of antimicrobial resistance in European hospitals: excess mortality and length of hospital stay associated with bloodstream infections due to Escherichia coli resistant to third-generation cephalosporins. J Antimicrob Chemother. 2011;66(2):398–407. doi: 10.1093/jac/dkq412. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Ruppé E., Andremont A. Causes, consequences, and perspectives in the variations of intestinal density of colonization of multidrug-resistant enterobacteria. Front Microbiol. 2013;4:129. doi: 10.3389/fmicb.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donskey C.J. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis Off Publ Infect Dis Soc Am. 2004;39(2):219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 7.Lerner A, Adler A, Abu-Hanna J, Cohen Percia S, Kazma Matalon M, Carmeli Y. Spread of KPC-producing carbapenem-resistant Enterobacteriaceae: the importance of super-spreaders and rectal KPC concentration. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2015 May;21(5):470.e1-7. [DOI] [PubMed]
- 8.Razazi K., Mekontso Dessap A., Carteaux G., Jansen C., Decousser J.-W., de Prost N. Frequency, associated factors and outcome of multi-drug-resistant intensive care unit-acquired pneumonia among patients colonized with extended-spectrum β-lactamase-producing Enterobacteriaceae. Ann Intensive Care. 2017;7(1):61. doi: 10.1186/s13613-017-0283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colodner R., Rock W., Chazan B., Keller N., Guy N., Sakran W. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2004;23(3):163–167. doi: 10.1007/s10096-003-1084-2. [DOI] [PubMed] [Google Scholar]
- 10.Kaier K., Frank U., Hagist C., Conrad A., Meyer E. The impact of antimicrobial drug consumption and alcohol-based hand rub use on the emergence and spread of extended-spectrum beta-lactamase-producing strains: a time-series analysis. J Antimicrob Chemother. 2009;63(3):609–614. doi: 10.1093/jac/dkn534. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Pai H., Kim Y.K., Kim N.H., Eun B.W., Kang H.J. Control of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a children’s hospital by changing antimicrobial agent usage policy. J Antimicrob Chemother. 2007;60(3):629–637. doi: 10.1093/jac/dkm225. [DOI] [PubMed] [Google Scholar]
- 12.Muller A., Lopez-Lozano J.M., Bertrand X., Talon D. Relationship between ceftriaxone use and resistance to third-generation cephalosporins among clinical strains of Enterobacter cloacae. J Antimicrob Chemother. 2004;54(1):173–177. doi: 10.1093/jac/dkh282. [DOI] [PubMed] [Google Scholar]
- 13.Gbaguidi-Haore H., Dumartin C., L’Hériteau F., Péfau M., Hocquet D., Rogues A.-M. Antibiotics involved in the occurrence of antibiotic-resistant bacteria: a nationwide multilevel study suggests differences within antibiotic classes. J Antimicrob Chemother. 2013;68(2):461–470. doi: 10.1093/jac/dks406. [DOI] [PubMed] [Google Scholar]
- 14.Grohs P., Kerneis S., Sabatier B., Lavollay M., Carbonnelle E., Rostane H. Fighting the spread of AmpC-hyperproducing Enterobacteriaceae: beneficial effect of replacing ceftriaxone with cefotaxime. J Antimicrob Chemother. 2014;69(3):786–789. doi: 10.1093/jac/dkt403. [DOI] [PubMed] [Google Scholar]
- 15.Tan B.K., Vivier E., Bouziad K.A., Zahar J.-R., Pommier C., Parmeland L. A hospital-wide intervention replacing ceftriaxone with cefotaxime to reduce rate of healthcare-associated infections caused by extended-spectrum β-lactamase-producing Enterobacteriaceae in the intensive care unit. Intensive Care Med. 2018;44(5):672–673. doi: 10.1007/s00134-018-5079-y. [DOI] [PubMed] [Google Scholar]
- 16.Burdet C., Grall N., Linard M., Bridier-Nahmias A., Benhayoun M., Bourabha K. Ceftriaxone and cefotaxime have similar effects on the intestinal microbiota in human volunteers treated by standard-dose regimens. Antimicrob Agents Chemother. 2019;63(6) doi: 10.1128/AAC.02244-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grall N., Massias L., Nguyen T.T., Sayah-Jeanne S., Ducrot N., Chachaty E. Oral DAV131, a charcoal-based adsorbent, inhibits intestinal colonization by β-lactam-resistant Klebsiella pneumoniae in cefotaxime-treated mice. Antimicrob Agents Chemother. 2013;57(11):5423–5425. doi: 10.1128/AAC.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyen C.K., Pultz N.J., Paterson D.L., Aron D.C., Donskey C.J. Effect of parenteral antibiotic administration on establishment of intestinal colonization in mice by Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2003;47(11):3610–3612. doi: 10.1128/AAC.47.11.3610-3612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiefel U., Pultz N.J., Donskey C.J. Effect of carbapenem administration on establishment of intestinal colonization by vancomycin-resistant enterococci and Klebsiella pneumoniae in mice. Antimicrob Agents Chemother. 2007;51(1):372–375. doi: 10.1128/AAC.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pultz M.J., Donskey C.J. Effects of imipenem-cilastatin, ertapenem, piperacillin-tazobactam, and ceftriaxone treatments on persistence of intestinal colonization by extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae strains in mice. Antimicrob Agents Chemother. 2007;51(8):3044–3045. doi: 10.1128/AAC.00194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertz F.B., Løbner-Olesen A., Frimodt-Møller N. Antibiotic selection of Escherichia coli sequence type 131 in a mouse intestinal colonization model. Antimicrob Agents Chemother. 2014;58(10):6139–6144. doi: 10.1128/AAC.03021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habib G, Lancellotti P, Antunes MJ, Bongiorni MG, Casalta J-P, Del Zotti F, et al. 2015 ESC Guidelines for the management of infective endocarditis: The Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM). Eur Heart J. 2015 Nov 21;36(44):3075–128. [DOI] [PubMed]
- 23.Tunkel A.R., Hasbun R., Bhimraj A., Byers K., Kaplan S.L., Scheld W.M. 2017 infectious diseases society of America’s clinical practice guidelines for healthcare-associated ventriculitis and meningitis. Clin Infect Dis. 2017;64(6):e34–e65. doi: 10.1093/cid/ciw861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Beek D., Cabellos C., Dzupova O., Esposito S., Klein M., Kloek A.T. ESCMID guideline: diagnosis and treatment of acute bacterial meningitis. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2016;22(Suppl 3):S37–S62. doi: 10.1016/j.cmi.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Léonard F., Andremont A., Leclerq B., Labia R., Tancrède C. Use of beta-lactamase-producing anaerobes to prevent ceftriaxone from degrading intestinal resistance to colonization. J Infect Dis. 1989;160(2):274–280. doi: 10.1093/infdis/160.2.274. [DOI] [PubMed] [Google Scholar]
- 26.Arvidsson A., Leijd B., Nord C.E., Angelin B. Interindividual variability in biliary excretion of ceftriaxone: effects on biliary lipid metabolism and on intestinal microflora. Eur J Clin Invest. 1988;18(3):261–266. doi: 10.1111/j.1365-2362.1988.tb01256.x. [DOI] [PubMed] [Google Scholar]
- 27.Ueda S., Ngan B.T.K., Huong B.T.M., Hirai I., Tuyen L.D., Yamamoto Y. Limited Transmission of blaCTX-M-9-Type-Positive Escherichia coli between Humans and Poultry in Vietnam. Antimicrob Agents Chemother. 2015;59(6):3574–3577. doi: 10.1128/AAC.00517-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gohl D.M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. 2016;34(9):942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 29.Al-Ghalith GA, Hillmann B, Ang K, Shields-Cutler R, Knights D. SHI7 Is a Self-Learning Pipeline for Multipurpose Short-Read DNA Quality Control. mSystems. 2018 Jun;3(3). [DOI] [PMC free article] [PubMed]
- 30.Al-Ghalith G.A., Montassier E., Ward H.N., Knights D. NINJA-OPS: fast accurate marker gene alignment using concatenated ribosomes. PLoS Comput Biol. 2016;12(1) doi: 10.1371/journal.pcbi.1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011 Feb;5(2):169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knights D., Costello E.K., Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35(2):343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 35.Shields-Cutler R.R., Al-Ghalith G.A., Yassour M., Knights D. SplinectomeR enables group comparisons in longitudinal microbiome studies. Front Microbiol. 2018;9:785. doi: 10.3389/fmicb.2018.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouton J.W., Meletiadis J., Voss A., Turnidge J. Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision. J Antimicrob Chemother. 2018;73(9):2374–2379. doi: 10.1093/jac/dky232. [DOI] [PubMed] [Google Scholar]
- 39.Limbert M., Seibert G., Schrinner E. The cooperation of cefotaxime and desacetyl-cefotaxime with respect to antibacterial activity and beta-lactamase stability. Infection. 1982;10(2):97–100. doi: 10.1007/BF01816732. [DOI] [PubMed] [Google Scholar]
- 40.Chin N.X., Neu H.C. Cefotaxime and desacetylcefotaxime: an example of advantageous antimicrobial metabolism. Diagn Microbiol Infect Dis. 1984;2(3 Suppl):21S–31S. [PubMed] [Google Scholar]
- 41.Marone P., Navarra A., Monzillo V., Traverso A. Antibacterial activity of combined cefotaxime and desacetyl-cefotaxime against aerobic and anaerobic gram-negative bacilli. Drugs Exp Clin Res. 1990;16(12):629–633. [PubMed] [Google Scholar]
- 42.Labia R., Morand A., Kazmierczak A. The action of beta-lactamases on desacetyl-cefotaxime and cefotaxime. J Antimicrob Chemother. 1984;(14 Suppl B):45–51. doi: 10.1093/jac/14.suppl_b.45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




