As part of JEM’s 125th anniversary celebration, Olivier Elemento discusses Peyton Rous’s discovery in 1911 that led to the identification of virus-inducing tumors, oncogene discovery, and the development of modern tumor biology.
Abstract
In 1911, more than a century ago, Peyton Rous described a curious observation, later explained by a virus named for him that led to the discovery of oncogenes, the modern era of cancer research, and the emergent field of precision medicine (1911. J. Exp. Med. https://doi.org/10.1084/jem.13.4.397).
Two years before his 1911 paper was published, Peyton Rous, a young physician-scientist at the Rockefeller Institute, had been given a Plymouth Rock hen with a large subcutaneous spindle-cell sarcoma in its breast. Inspired by reports of transmissible tumors in various animals, Rous injected small pieces of the tumor into hens from the same breed. Some of the pieces engrafted successfully, and the chickens developed tumors at the site of implantation (Rous, 1910). In 1911, Rous reported that after several rounds of implantation, the serially transplanted tumors became more invasive than the original and earlier tumors (Rous, 1911). Perhaps attempting to show that transplanted material did not form tumors unless it contained intact tumor cells, Rous then did something unusual and got a surprising result (Rous, 1911). He pulverized the tumor material; put the supernatant devoid of tumor fragments through a filter designed to remove any bacteria, remaining animal cells, or debris; and then injected the cell-free filtrate into other chickens. Unexpectedly, some of the animals developed sarcomas that formed at the site of injection and often metastasized widely (Rous, 1911).

Insights from Olivier Elemento.
What was the sarcomagenic agent in the filtrate? Rous initially speculated that it might have been either a “minute parasitic organism” or a “chemical stimulant, elaborated by the neoplastic cells” (Rous, 1911). Thanks to additional work by Rous and many others, we now know that it was a virus, now called Rous sarcoma virus (RSV), among the first of what are now widely known as retroviruses (Coffin et al., 1997). Several other cancer-causing animal viruses, many of them now known to be retroviruses, were subsequently discovered. In 1966, 55 years after his seminal paper, Rous was awarded the Nobel Prize in Physiology or Medicine “for his discovery of tumor-inducing viruses.”
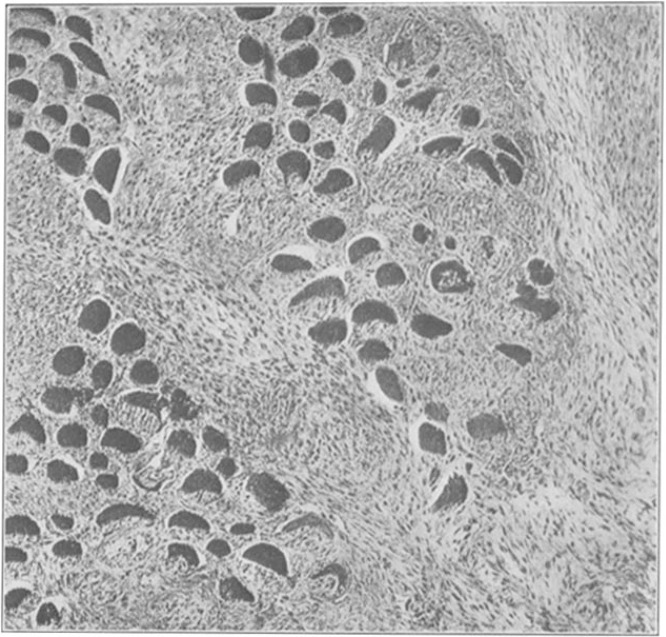
Filtrate from a chicken tumor was injected in the breast of another chicken resulting tumor growth. Image from Rous (1911).
A cascade of findings tracing their roots to Rous’s 1911 findings revolutionized our understanding of cancer. This started with the transforming agent encoded in the virus, the viral Src protein (v-Src), being identified (Brugge and Erikson, 1977) and shown to have tyrosine kinase capacity (Hunter and Sefton, 1980). Then, Varmus, Bishop, and colleagues found that a complementary DNA probe specific for the src nucleic acid sequence successfully hybridized to normal, nontransformed DNA from chicken cells (Stehelin et al., 1976a; Stehelin et al., 1976b), thus demonstrating that the viral Src oncogene had in fact a cellular origin. The cellular SRC gene, named c-Src, was found to have sequence homologues in other species including humans. Many other retroviruses found in a variety of vertebrates were subsequently found to carry host-derived oncogenes. Importantly, some of the corresponding cellular genes, e.g., RAS, ABL, and MYC, were found to be mutated and functionally carcinogenic in human cancers. Varmus and Bishop’s original and profound insight led to them being awarded the Nobel Prize in Physiology or Medicine in 1989 “for their discovery of the cellular origin of retroviral oncogenes” and set off an explosion of research into oncogenes.
Decades later, very large cancer genomic profiling projects like The Cancer Genome Atlas and the International Cancer Genome Consortium have now uncovered many more potential oncogenes that drive some of the common types of cancers. These studies confirmed that oncogenes are activated by a broad spectrum of mutations ranging from nucleotide substitutions to gene amplifications and translocations that fuse two genes together, e.g., BCR-ABL. These genes’ exact roles in cancer (and in normal cells) are being investigated in hundreds of laboratories worldwide.
Identification and study of a temperature-sensitive mutant viral strain showed that the function of v-Src (before its precise identification) was required for tumor maintenance and not just initiation of transformation (Martin, 1970). This phenomenon was demonstrated for many other oncogenes and was eventually defined as “oncogene addiction.” Enlightened by this critical finding, a vibrant biotechnology and pharmaceutical industry, frequently teaming up with academic scientists, developed numerous therapeutic strategies to block the protein product of oncogenes. One of the first targeted anti-oncogene therapies, imatinib, was designed to bind to the ABL protein (a tyrosine kinase in the same family as Src) and block its function. When used to treat cells that have mutations involving ABL and are addicted to them, such as chronic myelogenous leukemia cells, imatinib effectively kills these cells (O’Brien et al., 2003). Numerous such targeted agents have been designed to target a broad array of mutated proteins. In some cancers such as lung adenocarcinomas, mutations in up to seven different genes can now be targeted (Food and Drug Administration, 2020), highlighting the need for simultaneous and systematic DNA sequencing (now frequently done using next-generation sequencing) to assess which mutations are present in a patient’s tumor.
An important finding of large and systematic cancer genome projects has been the realization that certain oncogenic mutations can occur in a much wider variety of cancer types than initially thought, and that in some cases, molecules that target these oncogenes are effective in nearly all tumors that harbor the oncogenic mutation. This is the case for mutations in tropomyosin receptor kinases and tropomyosin receptor kinase inhibitors (Drilon et al., 2018). Thus, the rational design of oncogene-blocking molecules has culminated with the design of molecules that are effective in any tumor that has a mutation in a particular oncogene. Altogether, these relatively recent developments, whose roots trace back to Rous’s findings, have ushered in the era of precision medicine for cancer patients.
It is, however, important to note that despite all the recent progress, many oncogenes, including some of the earliest discovered ones such as KRAS, have resisted drugging (except for relatively rare mutations such as G12C amino acid changes in KRAS). Even Src, arguably the first oncogene, has resisted direct targeting except through inhibitors that target multiple tyrosine kinases, such as dasatinib. Some oncogenes such as those that act as transcription factors are hard to drug because they do not have a readily blockable enzymatic function like a kinase function. Thankfully, novel mechanisms such as targeted degradation, e.g., using proteolysis targeting chimera (Protacs; Sakamoto et al., 2001), may soon help drug many more oncogenes.
Finally, Rous’s findings offer an important lesson on the long-term importance of basic science research. Indeed his findings were ignored for decades because RSV and other oncogenic viruses occurred only in animals and were not transmissible to humans. 110 years after Rous’s pioneering discovery, numerous viruses causing human cancers have been discovered. We now know that up to 15% of human cancers are caused by viruses (zur Hausen, 1991); this number is likely up to 25% in developing countries. The first oncogenic human virus, Epstein–Barr virus, was observed in 1964 (Epstein et al., 1964) and causes Burkitt’s lymphoma. Kaposi’s sarcoma herpes virus was discovered in 1994 as the causal agent for Kaposi’s sarcoma (Chang et al., 1994), a disease frequently found in patients with HIV-associated AIDS. The human papillomavirus (HPV) was found in 1980 to be associated with cervical cancer (Dürst et al., 1983), a discovery that also led to a Nobel Prize in Physiology or Medicine awarded to Harald zur Hausen in 2008. Viruses that cause cancers without a clear direct oncogenic action have been discovered, such as hepatitis B and hepatitis C viruses (HBV and HCV, respectively), which cause liver cancer. Remarkably HPV-, HBV-, and HCV-associated cancers can be reliably prevented by targeting the cancer-causing viruses, either by vaccination (HBV, HPV) or antiviral treatment (HCV).
Altogether, it is hard not to be struck by the wide-ranging impact of Rous’s curious observation in chicken, which stretches from the discovery of oncogenes to human cancer–causing viruses and the success of precision medicine. The Rous legacy is truly formidable.
Acknowledgments
I am very grateful to Dr. Harold Varmus for his insightful comments on an earlier version of this article.
I am the founder of and hold equity in two companies, Volastra Therapeutics and OneThree Biotech, which use precision medicine technologies to develop anticancer therapies.
References
- Brugge, J.S., and Erikson R.L.. 1977. Nature. 10.1038/269346a0 [DOI] [Google Scholar]
- Chang, Y., et al. 1994. Science. 10.1126/science.7997879 [DOI] [Google Scholar]
- Coffin, J.M., et al. , editors. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Drilon, A., et al. 2018. N. Engl. J. Med. 10.1056/NEJMoa1714448 [DOI] [Google Scholar]
- Dürst, M., et al. 1983. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.80.12.3812 [DOI] [Google Scholar]
- Epstein, M.A., et al. 1964. Lancet. 10.1016/S0140-6736(64)91524-7 [DOI] [Google Scholar]
- Food and Drug Administration . 2020. https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling.
- Hunter, T., and Sefton B.M.. 1980. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.77.3.1311 [DOI] [Google Scholar]
- Martin, G.S. 1970. Nature. 10.1038/2271021a0 [DOI] [Google Scholar]
- O’Brien, et al. 2003. N. Engl. J. Med. 10.1056/NEJMoa022457 [DOI] [Google Scholar]
- Rous, P. 1910. J. Exp. Med. 10.1084/jem.12.5.696 [DOI] [Google Scholar]
- Rous, P. 1911. J. Exp. Med. 10.1084/jem.13.4.397 [DOI] [Google Scholar]
- Sakamoto, K.M., et al. 2001. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.141230798 [DOI] [Google Scholar]
- Stehelin, D., et al. 1976a. J. Mol. Biol. 10.1016/0022-2836(76)90152-2 [DOI] [Google Scholar]
- Stehelin, D., et al. 1976b. Nature. 10.1038/260170a0 [DOI] [Google Scholar]
- zur Hausen, H. 1991. Science. 10.1126/science.1659743 [DOI] [Google Scholar]