Abstract
Autophagy is mediated by membrane-bound organelles and it is an intrinsic catabolic and recycling process of the cell, which is very important for the health of organisms. The biogenesis of autophagic membranes is still incompletely understood. In vitro studies suggest that Atg2 protein transports lipids presumably from the ER to the expanding autophagic structures. Autophagy research has focused heavily on proteins and very little is known about the lipid composition of autophagic membranes. Here we describe a method for immunopurification of autophagic structures from Drosophila melanogaster (an excellent model to study autophagy in a complete organism) for subsequent lipidomic analysis. Western blots of several organelle markers indicate the high purity of the isolated autophagic vesicles, visualized by various microscopy techniques. Mass spectrometry results show that phosphatidylethanolamine (PE) is the dominant lipid class in wild type (control) membranes. We demonstrate that in Atg2 mutants (Atg2−), phosphatidylinositol (PI), negatively charged phosphatidylserine (PS), and phosphatidic acid (PA) with longer fatty acyl chains accumulate on stalled, negatively charged phagophores. Tandem mass spectrometry analysis of lipid species composing the lipid classes reveal the enrichment of unsaturated PE and phosphatidylcholine (PC) in controls versus PI, PS and PA species in Atg2−. Significant differences in the lipid profiles of control and Atg2− flies suggest that the lipid composition of autophagic membranes dynamically changes during their maturation. These lipidomic results also point to the in vivo lipid transport function of the Atg2 protein, pointing to its specific role in the transport of short fatty acyl chain PE species.
Keywords: autophagy, autophagic membranes, lipid composition, Atg2, lipidomics, Drosophila melanogaster
1. Introduction
Macroautophagy (autophagy hereafter) is a „self-eating” process of eukaryotic cells, during which damaged or obsolete cytoplasmic components (organelles, proteins, lipids, sugars etc.) are removed and degraded in lysosomes [1, 2]. Misregulation of autophagy contributes to neurodegenarative diseases, cancer, accelerated aging, etc [3–5]. This fundamental catabolic process relies on the biogenesis of unique, very dynamic membranes and membrane-bound autophagic vesicles. The pathway initiates with the nucleation of a double-membrane structure called phagophore (formerly also known as isolation membrane), which expands and engulfs a portion of the cytoplasm. After closure and sealing, it will form the autophagosome, a double membrane vesicle. In the last step, autophagosomes will fuse with lysosomes to form autolysosomes, where the sequestered cytoplasmic material will be degraded and recycled back to the cytosol [6].
Fluorescence and electron microscopy are widely used to study these specific organelles [7–9]. Autophagic structures can be isolated for example from human cell lines, yeast, mouse and rat tissues by applying subcellular fractionation [10], immunoprecipitation [11–13] or combination of these two methods [14]. Fruit flies are an excellent model organism to study autophagy in a complete animal as they can be genetically manipulated very easily, and about 75% of their genes show homology with disease associated human genes, so they can serve as models for various human diseases [2, 15, 16].
Autophagosomes are unique organelles regarding their lipid and protein composition, morphology and biogenesis. Autophagosome formation occurs very rapidly upon induction of autophagy, which requires a tremendous amount of membrane source(s). De novo synthesis of autophagic membranes is the most enigmatic field of autophagy; almost every compartment of the endomembrane system has been implicated in this process [17]. Interestingly, also the contact sites between organelles such as ER and mitochondria may play an important role in this biogenesis event [18]. During phagophore expansion, the synthesis or delivery of lipids must be distinctly controlled. Formation of these specific membranes relies on a collaborative work between proteins encoded by autophagy-related (Atg) genes and membrane lipids [19].
It has been shown recently that Atg2 protein is not only a potential tether between ER and phagophores, but it may also transport lipids from the ER to promote autophagosome biogenesis [20, 21]. Several in vitro studies on yeast (and human) Atg2 (Atg2A and B) showed the lipid transport activity of this special protein, which is able to transport several lipids at once using its hydrophobic cavity [21–24]. A very recent discovery is that the fast expansion of autophagic membranes after autophagy induction relies on localized “on-demand” de novo phospholipid synthesis, by the aid of Acyl-CoA synthetase Faa1 enzymes identified on nucleated phagophores in yeast, and the Atg2-Atg18 protein complex may also be involved in this process [13]. The P-element induced Drosophila mutant (Atg2EP3697), bearing the transposon insertion at the 5’ non-translated region of the Atg2 gene that resulted in a strong hypomorphic allele, showed an autophagy defect [25] similar to mammalian and C. elegans Atg2 mutants [26, 27]. Microscopy and biochemical investigations support that loss of Atg2 protein function in Drosophila, worms, and also in mammalian cells causes a sealing defect of phagophores, leading to accumulation of enlarged phagophore-like structures [9, 26–31]. Other key players of autophagy are Atg8 proteins together with their lipid conjugation (including the E3-like Atg5-Atg12-Atg16 complex) and deconjugation machinery. They have important roles in the biogenesis of autophagic membranes [32], and they are reversebly conjugated to the PE head groups present in the membranes of autophagic structures. The two distinct forms of Atg8 proteins: the non-lipidated Atg8 (Atg8-I) and lipidated Atg8 (Atg8-II) are the most widely used autophagic markers, including Atg8a in Drosophila [33, 34].
Lipids are largely unexplored players of the phagophore biogenesis machinery. The major structural lipids in eukaryotic membranes are glycerophospholipids, sphingolipids and sterols [35–37]. The most abundant glycerophospholipid classes of Drosophila are: phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylinositol (PI) and less abundant lipid classes are phosphatidylglycerol (PG) and phosphatidic acid (PA) [38–41]. PC accounts for more than 50% of the phospholipids in most eukaryotic membranes. Interestingly, in Drosophila, PE is the most abundant structural lipid and also a component of the lipoproteins (like Atg8), in contrast with more PC-centric lipidome of mammalian cells [38]. These main lipid classes differ in the chemical composition of their head groups and in length and saturation level of their fatty acyl chains [42]. This determines the shape and physicochemical behavior of these lipid molecules, such as phase transition properties and thickness of the membranes. All these properties influence membrane curvature and fusion events [35, 37, 43].
Lipids are not only structural components of the membranes but they also play a regulatory role in cellular processes, such as autophagy [44, 45]. Inactivation of Desat1 (desaturase coding gene, responsible for double bond generation) in Drosophila resulted in an autophagic defect: autolysosomes could not form properly [46]. Glycerolipids play important roles in the initiation of autophagy, elongation of phagophores, autophagosome maturation as well as in autophagosome-lysosome fusion. Different phosphorylated forms of PI including PI3P, PI4P, PI(4,5)P2, PI(3,5)P2 are also found in autophagic membranes, as different types of kinases present at these membranes are responsible for their generation. Although they are minor lipids (representing less than 1% of total lipids), together with PE they play important roles in autophagosome biogenesis by influencing the recruitment of specific proteins to the membrane [6, 47]. Interestingly, phosphatidic acid (PA) molecules may directly affect the physicochemical properties of lipid bilayers independently of protein effectors [44].
In this study, we established a method for isolating autophagic structures from adult Drosophila melanogaster, which we optimized for subsequent lipidomic investigations. Deciphering the lipid composition of autophagic membranes is crucial to fully understand the mechanism of autophagy. Using powerful Drosophila genetics and the advantage of high-throughput mass spectrometry, we point to the important in vivo lipid transport function of Atg2 protein.
2. Materials and methods
2.1. Fly work
Flies were kept on standard medium consisting of cornmeal, sucrose and yeast at 25 °C. The Atg2EP3697 EP element insertion line [25, 48] (FlyBase ID: FBal0123243) was obtained from the Bloomington Drosophila Stock Center (BDSC). The 3xmCherry-Atg8a autophagic reporter stock was published from our lab earlier [49]. For isolating autophagic membranes we used 4-hour starved homozygous 3xmCherry-Atg8a; Atg2EP3697 adult flies (Atg2− from here after), generated by standard genetic methods. We used as control a homozygous 3xmCherry-Atg8a transgenic line. Control flies were also starved (transferred to medium–free tubes) for 4 hours before autophagic membrane isolation. Starved flies were collected, snap frozen in liquid nitrogen and were stored at −80°C until further use.
2.2. Homogenization of adult flies
A mixture of male and female flies was cryogenically homogenized by liquid nitrogen, using mortar and pestle. To obtain enough amounts of membranes for lipidomic analysis we doubled the amount of control flies (400 mg) compared to those of Atg2− flies accumulating autophagic structures (200 mg). Homogenized fly powder was resuspended in buffer containing 250 mM sucrose, 20 mM HEPES, 1 mM EDTA, with freshly added protease inhibitors: 1 mM PMSF, 1 mM Aprotinin and 1 mM Leupeptin to a final volume of 1 ml (in case of control 2×1 ml). Homogenate was centrifuged at 3000 g for 15 min at 4 °C in order to get rid of unwanted material. The supernatant was used for the further experiments.
2.3. Direct Immunoprecipitation
First, we performed the pre-clearing of the lysate by using pre-blocked beads (binding-control magnetic agarose beads, Chromotek, bab-20). At each step of the immunprecipitation assay we collected 50 μl of supernatant for validation purposes (input, flow through/unbound fractions) and mixed with the same volume of 2X Laemmli buffer (Sigma, S3401–10VL), boiled at 100 °C for 6 min (these samples were used for SDS-PAGE separations). For pre-clearing, 1 ml of homogenate was added to 25 μl of pre-blocked equilibrated magnetic beads and rotated for 1 hr at 4 °C. For bead equilibration we used 3× 500 μl of ice-cold dilution buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA). Supernatant was obtained by using magnetic separator (DynaMag −2). Pre-cleared supernatant was transferred to 25 μl of equilibrated RFP-TRAP_AM beads (Chromotek, rtma-20) for binding, followed by 1 hr incubation at 4 °C with continuous rotation. After binding, the beads were washed 5 times with 500 μl of dilution buffer. We proceeded to elution by adding 50 μl of elution buffer (0.2 M glycine pH 2.5) and constantly pipetted up and down for 60 sec, followed by magnetic separation. The eluate fraction was transferred to a new tube and immediately neutralized with 5 μl of 1M Tris base (pH 10.4). To increase elution efficiency, this step was repeated once again. The first and the second eluted fractions were used either separately (for Western blot) or combined together (for Western blot, microscopy and mass spectrometry analysis).
2.4. Western blot
Proteins were separated by denaturing SDS-PAGE (10 or 15% gels) and transferred to Immobilon-P PVDF FL membranes. The membranes were incubated with the appropriate primary antibodies diluted in 10 ml Odyssey PBS blocking buffer, containing 0.1% Tween-20 (2h, RT). Membranes were washed 4×7 min with PBST, followed by incubation with Li-COR fluorescent secondary antibodies diluted in Odyssey buffer, containing 0.1% Tween-20 and 0.01% SDS (1h, RT). Finally, after 3×10 min washes with PBST, 1×10 min with PBS, 1×1 min with dH2O, 1×1 min with methanol, membranes were dried under a laminar air flow chamber for 20 min. The signal was detected with Odyssey fluorescent scanner, using Li-COR Image studio software. Blots were incubated with the following primary antibodies: rabbit monoclonal anti-GABARAP ab109364 (abcam) [1:2000], rabbit polyclonal anti-Atg5 A0856 (Sigma) [1:1000], mouse monoclonal anti-ATP5A ab14748 (abcam) [1:5000], mouse monoclonal anti-Cnx99A 6-2-1 (DSHB) [1:2000], rabbit polyclonal anti-Rab5 ab31261 (abcam) [1:1000], mouse monoclonal anti-Rab7 CG5915 (DSHB) [1:100], rabbit polyclonal anti-cathepsin L ab58991 (abcam) [1:1000] and mouse monoclonal anti-α-Tubulin AA4.3 (DSHB) [1:2000]. Species specific fluorescent secondary antibodies were used (Li-COR) [1:15000].
2.5. Fluorescent Microscopy
10 μl of the eluted fractions were pipetted onto a glass slide, mounted with a glass coverslip and immediately imaged in a Zeiss Axioimager M2 microscope equipped with Apotome 2 module and a Plan-Apochromat 100× 1.40 oil immersion objective. Raw images were processed in Zen 2 (Zeiss) software. Weak image deconvolution was performed to improve image sharpness from the ‘Apotome’ tab of Zen 2 and images were exported after adjusting brightness and contrast levels to remove noise. From these images, manual counting of the diameter of 3xmCherry-Atg8a positive structures (n=70) was performed in ImageJ (NIH) and statistics were performed in Graph Pad Prism. Statistical testing was performed by Mann-Whitney U test.
2.6. Atomic Force Microscopy (AFM) instrumentation and sample preparation
All experiments were carried out with an MFP-3D atomic force microscope and controller (Asylum Research, Santa Barbara, CA; driving software IgorPro 6.32A, Wavemetrics). Aluminium backside-coated silicon cantilevers (Olympus, AC240) with nominal spring constant of 2N/m, resonant frequency of 70 kHz in air, were used. All cantilevers were calibrated prior to experiments with the help of the driving software of the instrument using the Sader method [50]. Images were acquired using alternate-contact mode in closed loop in air medium at room temperature having 512 scan lines by 512 scan points. Background level correction and flattening was applied to each image to correct for eventual sample tilt.
5 μl of eluted fraction was dropped onto a freshly cleaved mica surface, and then let dry. Salt depositions from buffer were removed by gently washing with 200 μl of water, then dried again under high purity air flow (RH<5%).
2.7. Lipid extraction from isolated autophagic membranes
Total lipids were extracted from eluted fractions (stored at −80°C before isolation) of RFP-Trap immunoprecipitation experiments, according to Folch [51] and Bligh and Dyer [52] with small modifications. Eluted fractions were diluted with aqueous solution (final volume 1 ml) and 1 ml chloroform and 2 ml methanol were added. To protect our polyunsaturated lipids from oxidation 0.01% BHT (butylated hydroxytoluene) was added to the chloroform. After 30 min incubation period at 25 °C, 1 ml chloroform and 1 ml water were added. After phase separation happened, we collected the lower, lipid containing phase. Afterwards, combined lipid extraction was performed by adding 1 ml chloroform to the sample (this step was repeated three times). The combined lower (chloroform) layers were washed with 0.5 ml of 1 M KCl and then once with 0.5 ml of H2O. The solvent was evaporated under nitrogen gas using MD200 sample concentrator and the lipid extract was dissolved in 0.5 ml of chloroform/methanol (1:2), transferred to 2 ml clear glass vials with Teflon-lined screw cap and stored at −80 °C. Before shipping the samples to mass spectrometry analysis, the solvent was evaporated and the tube was filled with nitrogen.
2.8. Mass spectrometry analysis of lipid extracts
Lipidomic analysis of these total lipid extracts was performed at the Kansas Lipidomics Research Center Analytical Laboratory, using their tandem MS-based method [53, 54]. The lipid extract was prepared in 300 μl total volume. This was injected into the loop of the XEVO-TQS#WAA627 mass spectrometer for analysis. The background subtraction used the standard in front of each set of replicates for subtraction of contaminants. The amounts are reported as normalized signal/mg protein. This means that we are comparing the signals for the peaks in the sample to the signals for peaks of internal standards that were added in known amounts and we are reporting the data so that the signal that is represented as 1 = the same signal as 1 nmol of internal standard (usually with an adjustment for variation in response with m/z). Thus, the numbers that we report as normalized signal/mg protein for phospholipids can be considered to be equal to nmol/mg protein. To avoid many zeros, we report the data as picomol/mg protein. We also calculated the mol% of the lipids (relative amount of that compound compared to other compounds in the sample). The mol% of lipid species are averages of three (control) and four (mutant) independent biological replicates. Double bond indices of different lipid classes were calculated similarly as described in Falcone et al. [55], using the following equation [∑(% of normalized signal intensity/mg protein of lipid species × no. of double bonds)] / 100.
2.9. Protein content measurement
The protein content of the eluted immunoprecipitated fractions (ranging from 0.01683 mg to 0.0444 mg) prior to lipid extraction was measured with DeNovix DS-11FX spectrophotometer using the EasyApps software.
2.10. Statistics and reproducibility
Lipid statistical analyses were carried out using Microsoft Excel, OriginPro 8, and treated statistically by a t-test at significance level of at least p < 0.05, defined by Microsoft Excel. Significance levels in figures are designated as *p < 0.05, **p < 0.01, ***p < 0.005.
3. Results
3.1. Immunopurification of autophagic structures
To isolate autophagic structures we used the 3xmCherry-Atg8a transgene carrying Drosophila control animals. We starved the control flies for 4 hours before sample processing to enrich for autophagic structures. To isolate only phagophores, as well as to study the effect of Atg2 lipid transport protein loss on the lipid composition of initial autophagic membrane structures, we used Atg2− flies. To further enrich the autophagic structures in the mutant, the same starvation period was applied as in case of control flies. Autophagic organelles were isolated by immunopurification-assay and validated by biochemical approaches along with microscopy methods as follows.
3.1.1. Isolation of autophagic structures by RFP-Trap
Cytosolic fractions of the flies were obtained after cryohomogenization. Highly specific RFP-Trap was used for immunpurification of autophagic structures from the lysed flies. The RFP-Trap Magnetic Agarose consists of an anti-Red Fluorescent Protein (RFP) Nanobody, which is covalently bound to magnetic agarose beads and they can specifically recognize the mCherry-tag of our autophagic marker protein (3xmCherry-Atg8a). The lysate was pre-cleared before loading to the RFP-Trap, to avoid unspecific binding of other organelle membranes. For pre-clearing, we applied the “binding-control magnetic agarose beads” having the same magnetic agarose matrix as the RFP-Trap. The autophagic structures were eluted from the magnetic beads by a pH shift and these intact vesicles were used for characterization of their lipid composition (Fig. 1A).
Fig. 1. Immunopurification of autophagic structures from Drosophila control and Atg2− used for lipidomic analysis.
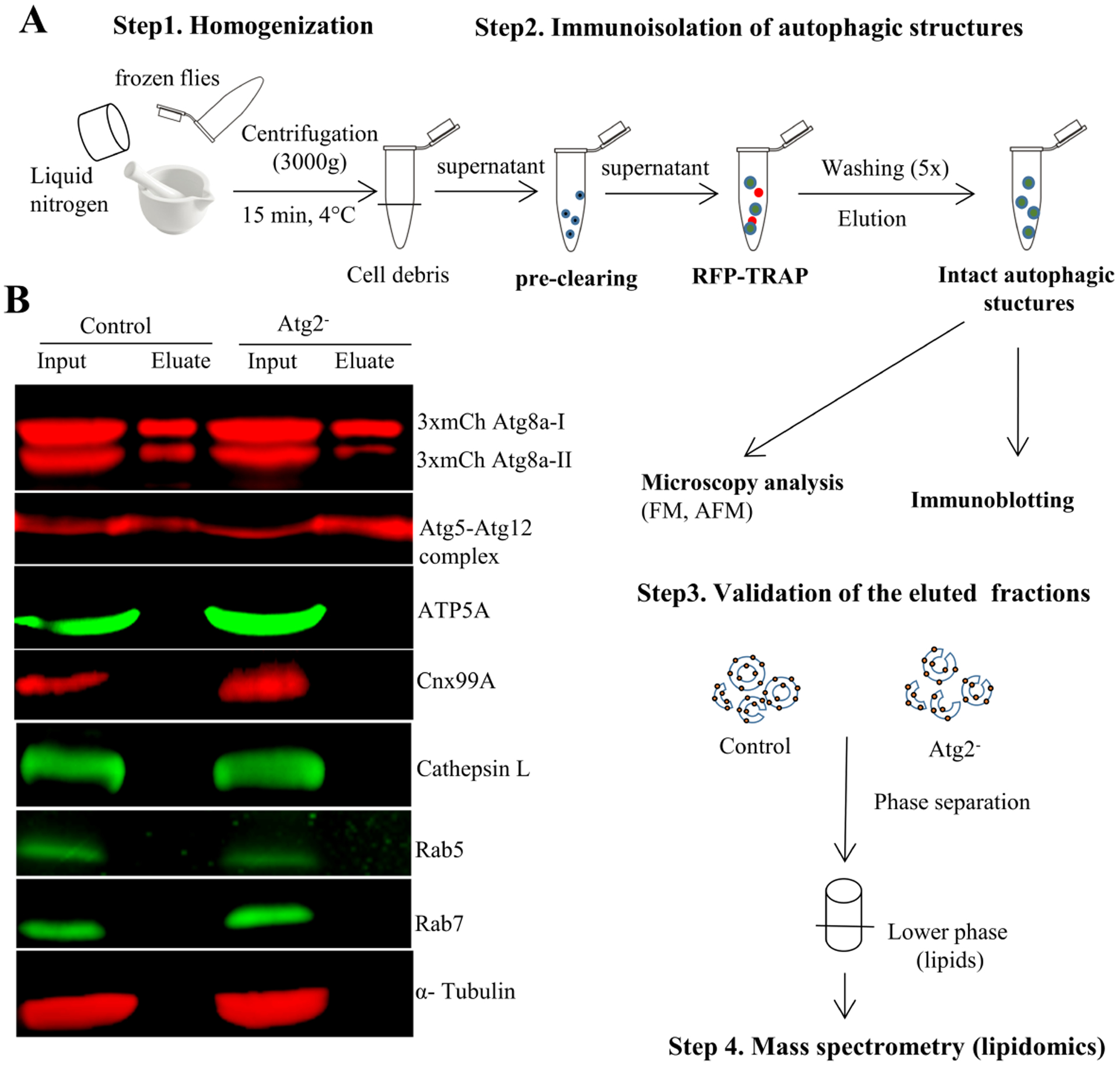
A) Schematic representation of the main steps of autophagic structures isolation procedure, as well as the experimental setup for validation of isolated fractions and for their lipid profiling. Frozen flies were homogenized gently (with mortar and pestle) using liquid nitrogen (until fine powder formed). Immunoisolation was carried out by using RFP-Trap, recognizing the 3xmCherry-tag of Atg8a autophagic marker protein. Prior to RFP-trap loading, the lysate was pre-cleared with “binding-control magnetic agarose beads” (which have the same magnetic agarose matrix as the RFP-Trap) to get rid of unspecific binding of other organelle membranes. Biochemical and microscopy methods were used for validation of the isolated autophagic structures. Lipids were isolated from these intact organelle membranes and analyzed by tandem mass spectrometry. B) Immunoblotting of pre-cleared lysate (input) and isolated autophagic structures (eluate) were used to validate the enrichment of isolated autophagic structures by using an anti-GABARAP antibody recognizing both non-lipidated, cytosolic (3xmCherry-Atg8a-I) and lipidated (3xmCherry-Atg8a-II) Atg8a, autophagic membrane-associated marker protein. Anti-Atg5 antibody was used to detect the presence of the Atg5-Atg12 conjugate, a marker of early autophagic structures. The high purity of isolated structures is shown by using antibodies against several organelle membrane specific marker proteins (ATP5A: mitochondria; Cnx99A: ER; Cathepsin-L: lysosome; Rab5: early endosome; Rab7: late endosome). The cytosolic α-Tubulin served as loading control.
3.1.2. Validation of isolated autophagic structures by Western blot
To verify the presence of autophagic structures (on the level of proteins) in our eluted fractions we performed Western blots of these fractions along with the pre-cleared lysate (Fig. 1B). Our autophagy marker proteins that served as the prey for the anti-RFP bait, 3xmCherry-Atg8a-I and -Atg8a-II, were enriched in the eluted fractions. The Atg5-Atg12 conjugate that is a common marker of early autophagic structures such as phagophores [56] was also enriched in the eluted fractions of control and Atg2− flies. To screen for the purity of autophagic structures we performed Western blots for a wide range of organelle-specific marker proteins including mitochondria (ATP5A), ER (Cnx99A), lysosomes (Cathepsin-L), early (Rab5) and late endosomes (Rab7). All of these organelle marker proteins were present in the lysate, but they were under the detection limit in the autophagic marker protein-enriched eluted fractions. Importance of pre-clearing of the lysate is illustrated by Fig S1. Western blots of immunprecipitation fractions without the pre-clearing step show the unspecific binding of some organelle membranes like mitochondria, as well as cytosolic proteins (like α-Tubulin). Thus, applying the pre-clearing step during the isolation protocol ensures the high purity of the isolated autophagic structures as well as the specificity of the method (Fig1B).
3.1.3. Validation of the isolated autophagic structures by microscopy
Since Western blot can only give information about proteins, to make sure that the identified autophagic marker protein Atg8a is associated with membranes, we applied fluorescence microscopy using the advantage of the native fluorescent mCherry tag of this marker protein. In eluted fractions from control and Atg2− flies we can observe a big variety of Atg8a-decorated autophagic vesicles with an average size of around 400–600 nm (Fig 2A and B): „semi-circle” like structures (initial autophagic structures: phagophores in early or middle stages of their expansion), phagophores which are ready to close, and sealed phagophores (autophagosomes). In the Atg2− flies we can detect enrichment of unsealed phagophores (Fig 2A) and their average sizes were smaller than in case of control flies (Fig. 2B). Resolution of this technique does not permit to differentiate clearly between the closed or ready to close structures, but according to prior literature we expect to have mostly unsealed vesicles in Atg2− flies [9, 26–28].
Fig. 2. Visualization of isolated mCherry-Atg8a structures.
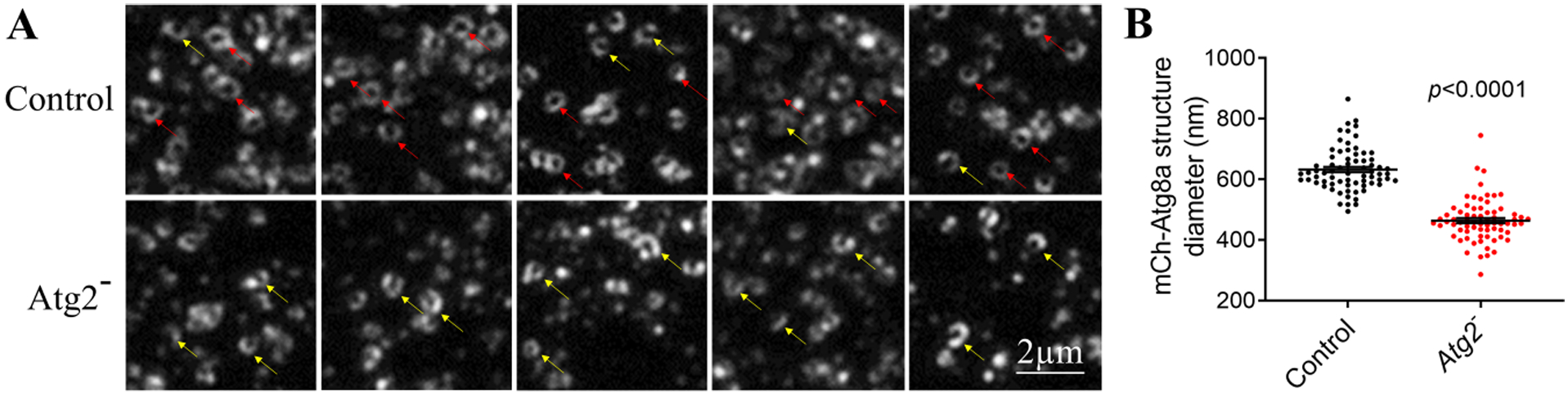
A) Fluorescence microscopy was used to image eluate fractions of Drosophila control and Atg2− flies. Red arrows indicate closed phagophores (autophagosomes), while yellow arrows indicate open ones. B) Diameter of mCherry-Atg8a structures (n=70) isolated from control and Atg2−. Statistical testing: Mann-Whitney U test.
We examined the eluted fractions with Atomic Force Microscopy (AFM), too (Fig. S2). AFM is a good complementary technique to monitor vesicle-like structures. As the instrument provides real three dimensional data, the size and volume characterization is easy and does not require difficult post processing. The studied and presented objects appear round shaped, with a size of a few hundred nanometers in diameter (Fig. S2A). The height of the flattened vesicles is slightly above 20 nm (Fig. S2B), which is roughly four fold the thickness of a lipid bilayer [57].
3.2. Lipid profiles of Drosophila autophagic membranes
After we elucidated the presence of autophagic marker proteins as well as membrane-surrounded autophagic vesicles in the eluted fractions, we isolated the lipids from these intact organelles. We applied a highly sensitive method, tandem mass spectrometry (lipidomics) to identify the lipid composition of control and Atg2− autophagic structures. This method allows us to detect lipids present even in the picomolar range. We identified a wide range of lipid species belonging to various lipid classes with shorter vs. longer, saturated vs. unsaturated (polyunsaturated) fatty acyl chain combinations. The amount of lipids were double normalized, first to the internal standards added in known amounts as well as to the protein content of the eluted fractions, which we used for lipid extraction. Our lipidomic data were filtered carefully, taking into consideration the known lipids and their biosynthetic pathways in Drosophila, excluding also any possible overlapping species.
3.2.1. Phospholipids, the building blocks of autophagic membranes
The applied lipid isolation method and lipidomic approach made possible the identification of several (around 70 species) amphipathic phospholipid molecules composing the lipid bilayer of autophagic membranes, in both control and Atg2− flies (Fig. 3). In control flies, the majority of the phospholipids belongs to PE (67 mol%), followed by PC (16 mol%) and PI (9 mol%) groups. PS (2 mol%) and PA (5 mol%) were less abundant. In contrast, the Atg2− structures exhibited much lower amounts of PE compared to the control flies, while PC levels remained unchanged. As a compensatory effect, we can observe a significant increase in the PI, PS and PA levels in the Atg2− flies (Fig. 3A). Total lipids were composed of various fatty acid containing species (pairs of fatty acids), ranging from 32 carbon atom containing species (32C) to 38C long species. Fig. 3B shows a clear shift in the fatty acyl chain lengths in autophagic structures of Atg2− flies. Longer fatty acyl chains are dominant in the mutant, while in the control the shorter fatty acid containing species are the most abundant ones.
Fig. 3. Phospholipid profiling of autophagic membranes isolated from Drosophila control and Atg2−.
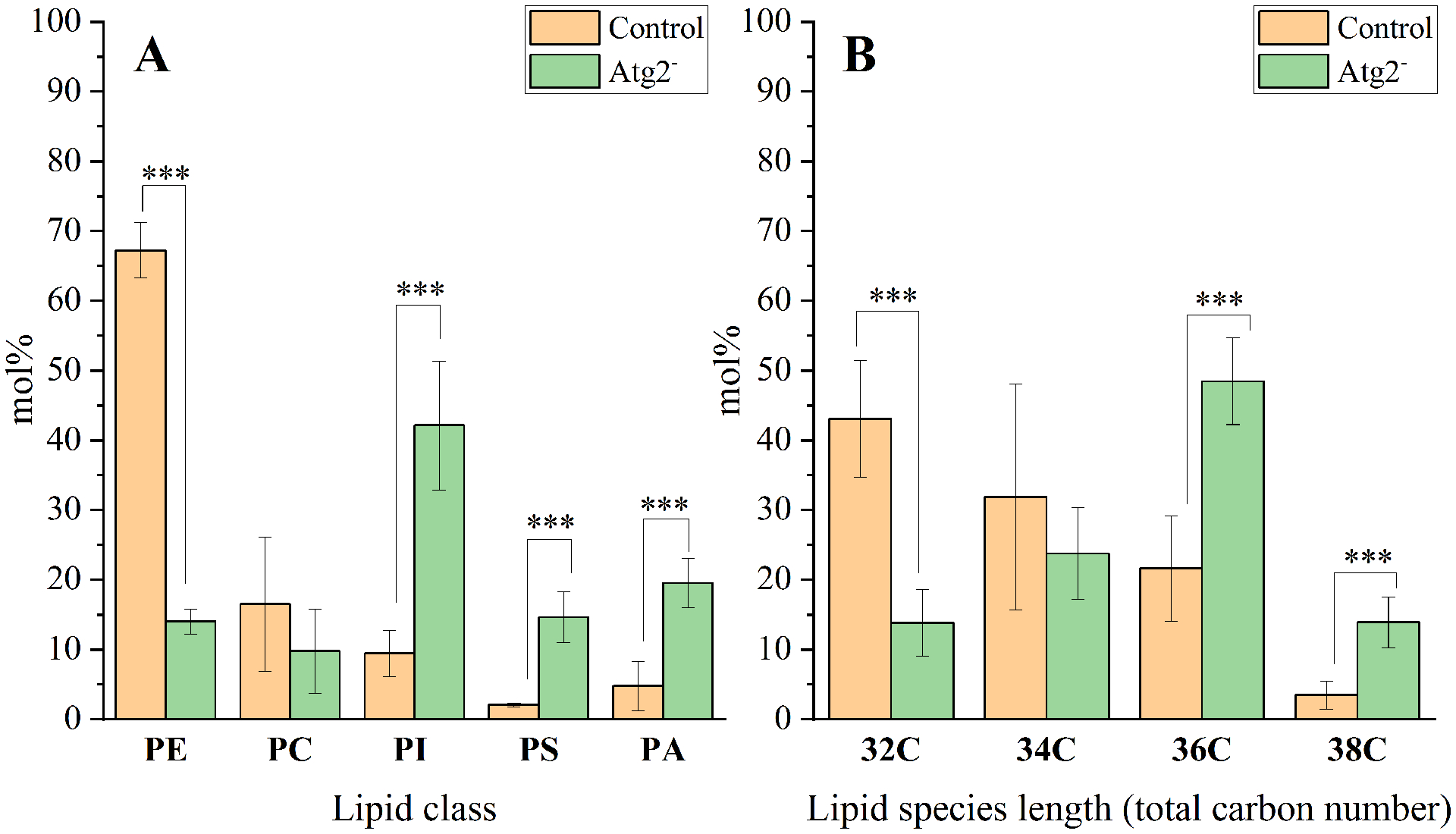
A) The main lipid classes composing autophagic membranes are PE (phosphatidyl-ethanolamine), PC (phosphatidylcholine), and PI (phosphatidylinositol). The less abundant lipids in controls are: PS (phosphatidylserine) and PA (phosphatidic acid). This ratio changes in Atg2 mutants, resulting in a significant increase of PI, PS and PA levels with a concomitant drop in PE. B) Fatty acid chain lengths (denoted as number of carbon atoms) of total lipid species belonging to various headgroups. Mol% distribution data are means ± SD (n=3 for control and n=4 for Atg2− flies). Statistically significant differences (based on t-tests) in figures are designated as *** for p < 0.005.
3.2.2. Specific lipid species composing the autophagic membranes
Fig. 4 summarizes the wide range of lipid species and their amounts belonging to individual lipid classes from control and Atg2− autophagic structures. Lipid species belonging to PE, PC, PS showed larger variety compared to PI and PA species. The level of monounsaturated PE (32:1, 34:1) and those of polysunsaturated PE (32:2, 36:4) species were significantly decreased in the Atg2− compared to the control (Fig. 4A). In case of PC we could not detect significant changes, but it is worth to mention the absence of PC (36:0, 38:4, 38:3, 38:2 38:1) species from the Atg2− (Fig. 4B). Changes in case of PI species belonging to Atg2− points to an opposite direction compared to those observed before with PE and PC species (Fig. 4C). Atg2− derived autophagic membranes exhibit large amounts of PI (36:3, 36:2, 36:1, 38: 2, 38:1), but some PI species like 34:2 are decreased or 34:1 is even undetectable in the mutant. The negatively charged PS (34:1, 36:5, 36:0) species are highly abundant in Atg2− (Fig.4D). Levels of species belonging to the lipid biosynthetic pathway intermediate, such as the PA (32:2, 32:1, 34:3, 34:1, 36:3) were significantly increased in Atg2− compared to the control (Figure 4E).
Fig. 4. Lipid species profiling of Drosophila control and Atg2− mutants.
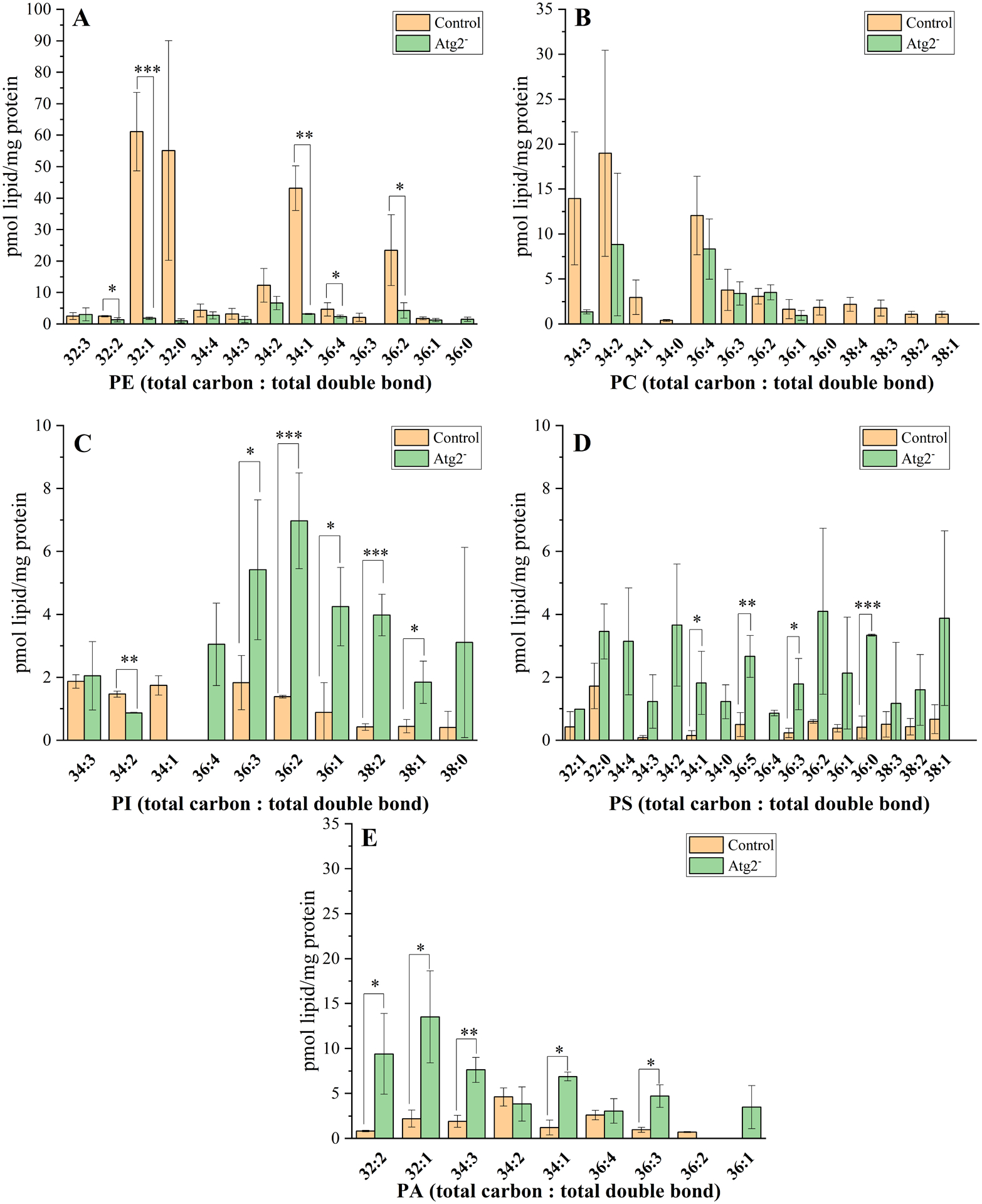
Amount of lipid species belonging to PE (panel A), PC (panel B), PI (panel C), PS (panel D) and PA (panel E) lipid classes. Data are means ± SD (n=3 for control and n=4 for Atg2− flies). Absolute lipid amounts in pmol are normalized to the protein content of the eluted autophagic membrane fractions. Significance levels (t-test) in figures are designated as * p < 0.05, ** p < 0.01, *** p < 0.005. Lipid species are denoted as total carbon number: total double bond. The identified lipid species are belonging to the following lipid classes: PE (phosphatidyl-ethanolamine); PC (phosphatidylcholine); PI (phosphatidylinositol); PS (phosphatidylserine); PA (phosphatidic acid).
3.2.3. Autophagic membranes are highly unsaturated
To get an overview of the unsaturation level in autophagic membranes of different stages, we calculated the mol% of lipid species belonging to the saturated (no double bonds) and unsaturated (one or more double bonds) categories (Fig. 5A). In case of control flies, almost a third of the species (63.4%) were unsaturated from the total lipids derived from autophagic structures, and in Atg2− flies, these exhibited even higher percentage of unsaturation (86%). The dominance of unsaturated lipids is indicated also by the unsaturated over saturated lipid ratios, which show greater numbers in case of membranes originating from Atg2− flies (Fig5B). To get a more detailed picture about the degree of unsaturation of the autophagic membranes, we determined the double bond indices of control and Atg2− flies on the basis of lipid species percentage and total double bonds identified by lipidomics. Table 1 compares the double bond indices of total lipids and those of the different lipid classes originating from autophagic membranes in control and Atg2− flies. There are no significant differences between the double bond indices of PC and total lipids. By contrast, important double bond index increases are seen for PS, PA and PI in Atg2− flies when compared to those of the control flies.
Fig. 5. Autophagic membranes from Drosophila control and Atg2− are highly unsaturated.
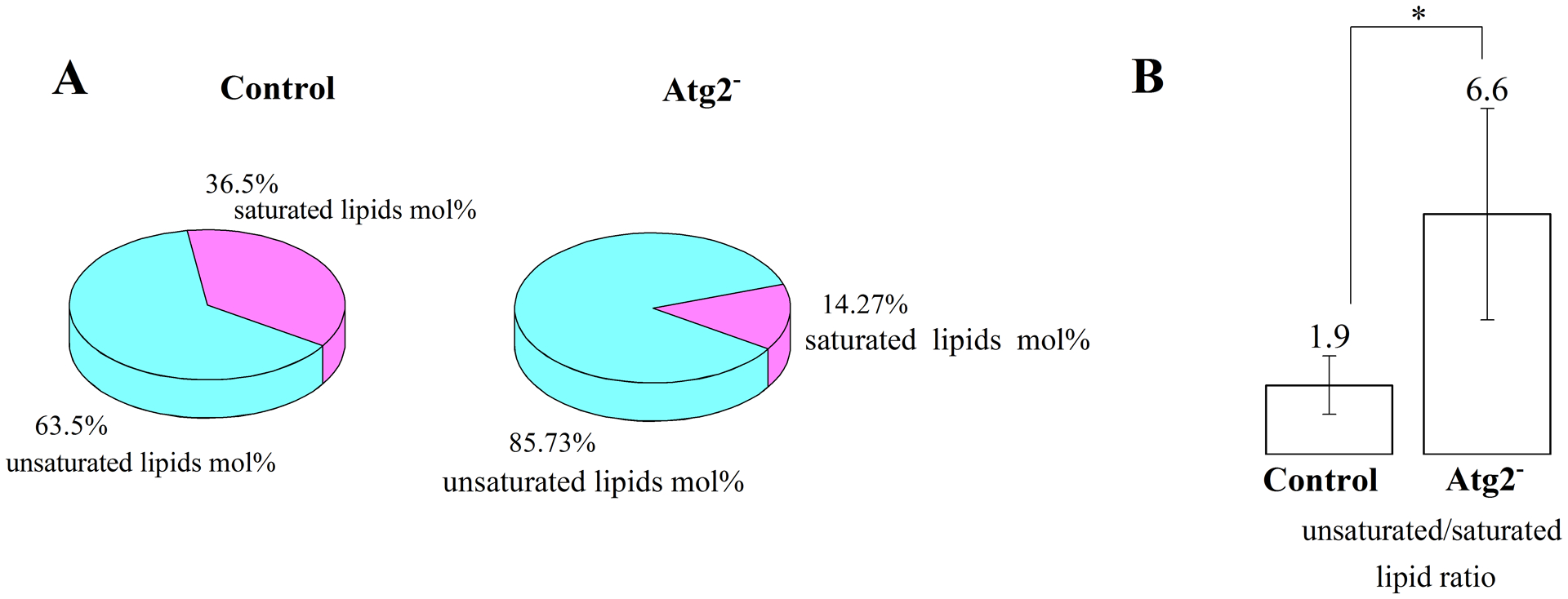
A) Mol% distribution of saturated and unsaturated total lipids in autophagic structures of control and Atg2− flies. Data are average of three (control) or four (Atg2−) biological replicates. B) Unsaturated versus saturated lipid ratios were calculated from the total identified lipid species. Data are mean ± SD (n=3 for control and n=4 for Atg2− flies). Significance level (t-test) is designated as * p < 0.05.
Table 1.
Double bond indices of various lipid classes, as well as total lipids, from autophagic membranes of control and Atg2− flies.
| Lipid class | Double bond index of lipid species | Statistical data | ||
|---|---|---|---|---|
| Control ±SD | Atg2− ±SD | p-value | Significance level (*) | |
| PC | 1.2 ±1.0 | 0.5 ±0.4 | 0.144 | NA |
| PI | 0.4 ±0.1 | 1.7 ±0.6 | 0.01 | * |
| PS | 0.1 ±0.0 | 0.6 ±0.2 | 0.002 | *** |
| PE | 1.7 ±0.7 | 0.6 ±0.1 | 0.01 | * |
| PA | 0.3 ±0.1 | 0.7 ±0.1 | 0.001 | *** |
| Total lipid | 3.7 ±1.5 | 4.2 ±1.0 | 0.305 | NA |
Double-bond indices were calculated as described in the Materials and Methods. ± SD values were calculated from 3 (control) or 4 (Atg2−) biological replicates. Significance levels (t-test) are designated as * p < 0.05, *** p < 0.005.
4. Discussion
Interestingly, autophagic membranes are composed mainly of lipids and contain very few proteins [1]. While autophagic proteins are intensively studied, we know much less about the lipids of the autophagic membranes. The exact lipid composition of autophagic structures at different stages - phagophores, autophagosomes and autolysosomes - are poorly characterized in any organism.
In this study, we propose a fast and easy biochemical method for the immunopurification of autophagic structures from a whole organism (Drosophila melanogaster) and characterize their lipid composition. Schematic Fig.6 illustrates our concept and methodology of isolation, as well as summarizes the lipids composing the autophagic membranes of control and Atg2– flies. It also highlights the consequences of the lack of Atg2 lipid transport protein on the lipid composition of phagophores and reveals some of the physical properties of these autophagic structures. Lipids of membranes are highly dynamic; they adapt very easily to environmental changes [37].
Fig. 6. Establishment of an easy and rapid biochemical protocol for isolation of various intact autophagic structures from Drosophila adult flies, optimized for lipidomic analysis.
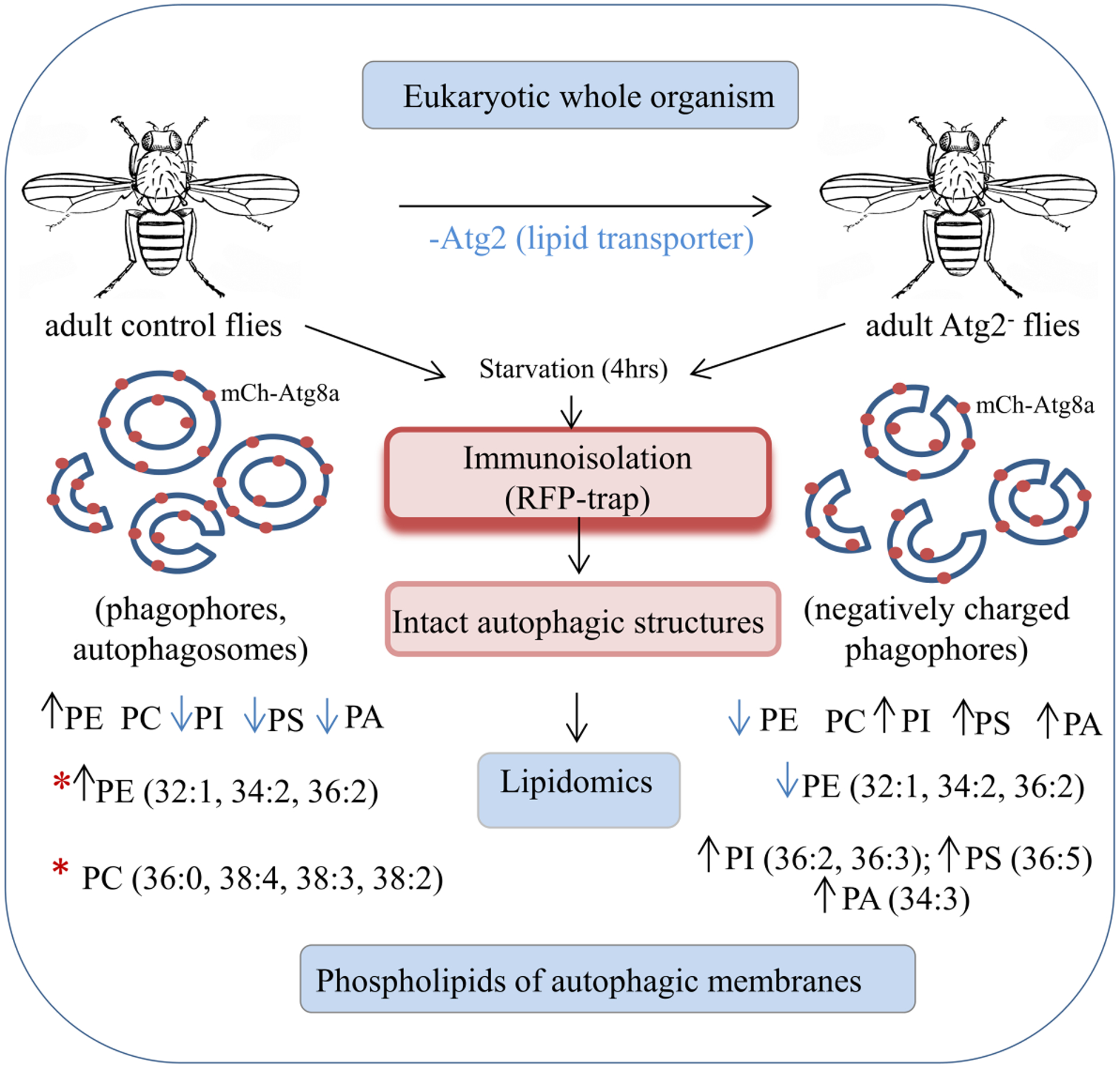
Up (↑) and down (↓) arrows indicate the directions of changes observed in the Atg2− flies compared to control lipid profile. Possible „favorite” lipid species of Atg2 lipid transporter are marked with red asterisks (*).
We show the successful purification of the autophagic structures by using the advantage of RFP-Traps, recognizing mCherry-tagged Atg8a, the autophagy specific marker protein in control and Atg2− flies. This protein co-purified with an early autophagic marker, the Atg5-Atg12 protein complex, confirming the autophagic nature of the isolated vesicles present in the eluted fractions. Autophagy is always occurring at a basal activity in cells, which can be enhanced by starvation. Enrichment of autophagic structures can be induced not only by starvation but also by applying chemical autophagy inhibitors. In this study, we tried to avoid the use of chemicals to tend toward more natural circumstances. The unique advantage of using a whole organism from the lipidomics point of view is that during the complex communication between different organs, lipids are subject to richer environmental stimuli than in cell cultures in vitro. This strongly influences the lipid composition of isolated membranes, because of the high rate of lipid remodeling events upon different environmental stimuli. We isolated a mixture of autophagic structures originating from all the different organs of the flies including brain, salivary gland, and fat body etc, which are favorably used in autophagy research. By using Atg2− flies we were able to enrich phagophores compared to the control flies where we have a mixture of phagophores and autophagosomes. Due to the autophagosome maturation process, Atg8 is deconjugated from the outer membranes of autophagosomes before fusing with endosomes or lysosomes [45]. This process prevents the extraction of amphisomes and autolysosomal structures from control animals, which serves as an advantage for us to avoid lysosomal or endosomal membrane lipids in our system.
We optimized our isolation protocol to be compatible with downstream lipidomic investigations. A great emphasis was placed on the degree of purity and integrity of the membranes (avoiding the use of detergent during immunopurification). Our immunoblot analysis of several organelle marker proteins suggests the high purity of the isolated autophagic organelles. We tested especially those organelles which can establish physical contact with autophagic structures (ER, lysosomes, endosomes, and mitochondria) [18].
Not only the purity, but also the visualization of these autophagic vesicles was an important issue for further lipidomic analysis. Our fluorescence microcopy approach, the most widely used and accepted technique to identify autophagy related organelles, confirmed the presence of vesicle-like structures (open or sealed) in our immunopurification eluted fractions. The average size of these vesicles (in the range of 400–600 nm) correspond well to the literature data [15]. AFM microscopy showed the three dimensional picture of autophagic structures and their membrane height verifies the double membrane nature of the isolated autophagosomes. The observed smaller sizes of the vesicles in the Atg2− is in correlation with the in vitro studies where they demonstrated that the size of the liposome is important in the lipid transport activity of Atg2−: smaller liposomes are preferred, indicating that Atg2 functions potentially at isolation membrane-ER exit contact sites [21].
Our work describes the phospholipid composition of phagophore and autophagosome membranes in the wild-type (control) flies (see a short summary in Fig.6). We used adult flies for the isolation to reduce the contaminants of triacylglycerols from the fat body tissues, which are overrepresented in larvae. The lipidome of Drosophila shows high variations from different developmental stages as well as from different organs [38, 40]. Our isolated autophagic structures represent the average lipid composition originating from different tissues of the body, which is dominated mainly by PE species. PE molecules are preferred by Atg8 type autophagic proteins. The covalent anchorage of Atg8 to PE can actively change the properties of the membranes, which is important for dynamic shape changes during autophagosome formation [47]. The fatty acid combinations of PE species bound to this protein is not yet known, but it would be interesting to explore in the future. Regarding the length of fatty acids esterified to the glycerol backbones; these are mainly in the 32C or 34C range (shorter fatty acids). More than half of the total lipids originating from the autophagic structures are unsaturated; mainly PE and PC contribute to this high degree of unsaturation level (see Table 1), as suggested earlier by other biochemical methods such as imidazole-buffered osmium tetroxide impregnation in mouse hepatocytes [58], as well as in case of yeast autophagic membranes [13].
We demonstrate that our immunoisolation protocol, as well as our Atg2− flies are suitable for characterization of specific autophagic membranes (like phagophores), as well as to follow the consequence of Atg2 lipid transporter loss on the phosopholipidome of these autophagic structures (see a short summary in Fig.6). Autophagic structures isolated from Atg2− flies exhibited substantially different lipid composition compared to control flies. According to our size measurements, control flies contained mainly autophagosomes in contrast with mostly phagophores isolated from Atg2 mutants, which may also contribute to the difference seen in lipid profiles. In Atg2− phagophore membranes the most dominat lipid class is PI (more than 40%), the precursor of PIP species identified earlier in autophagic membranes, which are the most studied lipids concerning their regulatory roles during autophagy. PI3P dynamics followed by the Atg18 family PI3P effectors WIPI1 and WIPI4 proteins can facilitate the potential ATG2-mediated transfer of lipids from the ER to the phagophores. PIPs provide negative charges to the membranes, which facilitates the lipid transfer activity of Atg2 and also the unidirectionality of lipid transfer from ER to negatively charged phagophores enriched in PI3P species [23]. Other phophoplipids including PS, PA, PE and PC contribute above 10% each to the total identified lipids. The high proportion of also negatively charged PS and PA in the mutant also contributes to the negative charges of the phagophores. It has been shown also in another in vitro study that presence of PS in the liposomes accelerated the lipid transport activity of human Atg2B protein [24].
Another impressive feature of Atg2− membranes is their high unsaturation levels, mainly caused by accumulation of polyunsaturated PS (36:5), PI (36:2, 36:3) and PA (34:3) species. Membrane fluidity increases with the degree of unsaturation, because double bonds generate kinks in the fatty acyl chains and result in lipids to be loosely packed [43]. These features help the phagophores to recruit Atg proteins as well as their fast reorganization to enclose cytosolic cargo. PA has been shown to play important roles in membrane curvature generation. The increased PA levels of Atg2− flies suggests the highly curved nature of phagophores, which are in agreement with earlier observations that Atg2 prefers to tether highly curved liposomes [21]. The presence of curvature-generating lipids are important for other Atg proteins, too: the curvature-sensing Atg3 involved in lipidation of Atg8a, as well as for Atg8-dependent membrane tethering and fusion events, which are curvature dependent and contribute to the in vivo biogenesis of autophagosomes [59].
Interestingly, there is a huge drop in the PE level of Atg2− flies suggesting that Atg2 might have a role in transporting of these PE (32:1, 34:2, 36:2) molecules from ER to the expanding phagophores. These results corroborate a recently published paper showing co-crystallization of yeast Atg2 with a PE molecule [21]. In vitro assays of purified human Atg2A protein confirmed the binding and transport capability of mainly PE, PC molecules, and less favored were the PI or PS molecules [22]. Although the overall PC level did not change significantly in the mutant, some PC species (like: 36:0, 38:4, 38:3 and 38:2) were not detectable in Atg2− flies compared to the control animals. These also suggest the possible role of the Atg2 proteins in their transport. A shift toward longer fatty acyl chain-containing lipid species in Atg2− phagophores may indicate their distinct biophysical properties. Since Atg2− flies manifested sealing and fusion defects we cannot exclude a minor effect of impaired autophagy on their phagophore lipid composition, too.
5. Conclusion
Our study reveals the phospholipid profile of autophagic structures from different stages (mixture of phagophores and autophagosomes versus mainly phagophores) isolated from Drosophila melanogaster control and Atg2− flies. We believe that the striking differences in the lipid pattern of autophagic structures from control and mutant flies primarily reflect the importance of the Atg2 lipid transporter, but we can not exclude the effect of dynamic changes during phagophore maturation and autophagosome closure that are also perturbed in the mutant. The unique properties of autophagic membranes largely depend on the interplay between proteins and lipids. Characterization of the lipid profile of autophagic membranes helps to understand this essential interplay between autophagy-specific lipids and autophagy proteins. Our Atg2− lipidomic results point to the in vivo function of the Atg2 lipid transport protein, especially its relevant contribution to de novo synthesis of initial autophagic structures, one of the most elusive steps of autophagy. Phagophores are formed even in the absence of Atg2 function, suggesting the existence of alternative lipid sources (other lipid transporters and close contact sites/vesicular routes with other organelles); but its contribution in shaping the lipid composition of autophagic membrane is likely critical, especially based on the level of PE species.
Supplementary Material
Supplementary Fig. S1. Unspecific bindings occur without pre-clearing of lysate, used for immunoisolation of autophagic structures from Drosophila control and Atg2−. Immunoblottings were performed from different fractions of immunopurification assay using RFP-Trap only: lysate (input), lysate after RFP-trap incubation (Flow-through), isolated autophagic structures (eluate). Without the pre-clearing step, mitochondrial marker proteins as well as cytosolic proteins (faint Tubulin signal) are also present in eluate (unspecific binding). Antibodies used are: anti-GABARAP (Atg8a-autophagic marker), anti-ATP5A (mitochondria), anti-Rab5 (early endosome), anti-Rab7 (late endosome), anti-Cathepsin-L (lysosome), anti-α-Tubulin (cytosolic marker and loading control).
Supplementary Fig. 2. Autophagic structures imaged with Atomic Force Microscope (AFM). A) Three dimensional images of eluate fractions from Drosophila control animals. B) Height of the vesicles along the red axis (see panel A) indicate the double membrane nature of the vesicles.
Acknowledgements
The authors are grateful to Gábor V. Horváth for his valuable discussions and technical help.
Funding: This work was supported by the National Research, Development and Innovation Office of Hungary NKFIH [PD128280 and KKP129797], as well as by Tempus Public Foundation TPF [SHE-19314-004/2019]. The lipid analyses described in this work were performed at the Kansas Lipidomics Research Center Analytical Laboratory. Instrument acquisition and lipidomics method development was supported by National Science Foundation (EPS 0236913, MCB 1413036, MCB 0920663, DBI 0521587, DBI 1228622), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of National Institute of Health (P20GM103418), and Kansas State University.
Abbreviations:
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- PA
phosphatidic acid
- PC
phosphatidylcholine
- Atg
autophagy-related
- Atg2−
Drosophila melanogaster homozygous viable adult flies of the genotype, 3xmCherry-Atg8a; Atg2EP3697
- Control
a homozygous 3xmCherry-Atg8a transgenic Drosophila melanogaster line
- AFM
Atomic Force Microscopy
- RFP
Red Fluorescent Protein
References
- [1].Juhasz G, Neufeld TP, Autophagy: a forty-year search for a missing membrane source, PLoS Biol, 4 (2006) e36, doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mulakkal NC, Nagy P, Takats S, Tusco R, Juhasz G, Nezis IP, Autophagy in Drosophila: from historical studies to current knowledge, Biomed Res Int, 2014 (2014) 273473, doi: 10.1155/2014/273473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bhattacharjee A, Szabo A, Csizmadia T, Laczko-Dobos H, Juhasz G, Understanding the importance of autophagy in human diseases using Drosophila, J Genet Genomics, 46 (2019) 157–169, doi: 10.1016/j.jgg.2019.03.007. [DOI] [PubMed] [Google Scholar]
- [4].Maruzs T, Simon-Vecsei Z, Kiss V, Csizmadia T, Juhasz G, On the Fly: Recent Progress on Autophagy and Aging in Drosophila, Front Cell Dev Biol, 7 (2019) 140, doi: 10.3389/fcell.2019.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Katheder NS, Khezri R, O’Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhasz G, Bilder D, Brech A, Stenmark H, Rusten TE, Microenvironmental autophagy promotes tumour growth, Nature, 541 (2017) 417–420, doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lorincz P, Juhasz G, Autophagosome-Lysosome Fusion, J Mol Biol, 432 (2020) 2462–2482, doi: 10.1016/j.jmb.2019.10.028. [DOI] [PubMed] [Google Scholar]
- [7].Jung M, Choi H, Mun JY, The autophagy research in electron microscopy, Applied Microscopy, 49 (2019), doi: 10.1186/s42649-019-0012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Takats S, Toth S, Szenci G, Juhasz G, Investigating Non-selective Autophagy in Drosophila, Methods Mol Biol, 1880 (2019) 589–600, doi: 10.1007/978-1-4939-8873-0_38. [DOI] [PubMed] [Google Scholar]
- [9].Nagy P, Varga A, Kovacs AL, Takats S, Juhasz G, How and why to study autophagy in Drosophila: it’s more than just a garbage chute, Methods, 75 (2015) 151–161, doi: 10.1016/j.ymeth.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Seglen PO, Brinchmann MF, Purification of autophagosomes from rat hepatocytes, Autophagy, 6 (2010) 542–547, doi: 10.4161/auto.6.4.11272. [DOI] [PubMed] [Google Scholar]
- [11].Yao J, Qiu Y, Jia L, Zacks DN, Autophagosome immunoisolation from GFP-LC3B mouse tissue, Autophagy, 15 (2019) 341–346, doi: 10.1080/15548627.2018.1539591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gao W, Kang JH, Liao Y, Ding WX, Gambotto AA, Watkins SC, Liu YJ, Stolz DB, Yin XM, Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment, J Biol Chem, 285 (2010) 1371–1383, doi: 10.1074/jbc.M109.054197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schutter M, Giavalisco P, Brodesser S, Graef M, Local Fatty Acid Channeling into Phospholipid Synthesis Drives Phagophore Expansion during Autophagy, Cell, 180 (2020) 135–149 e114, doi: 10.1016/j.cell.2019.12.005. [DOI] [PubMed] [Google Scholar]
- [14].Uematsu M, Nishimura T, Sakamaki Y, Yamamoto H, Mizushima N, Accumulation of undegraded autophagosomes by expression of dominant-negative STX17 (syntaxin 17) mutants, Autophagy, 13 (2017) 1452–1464, doi: 10.1080/15548627.2017.1327940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lorincz P, Mauvezin C, Juhasz G, Exploring Autophagy in Drosophila, Cells, 6 (2017), doi: 10.3390/cells6030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reiter LT, Potocki L, Chien S, Gribskov M, Bier E, A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster, Genome Res, 11 (2001) 1114–1125, doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lamb CA, Yoshimori T, Tooze SA, The autophagosome: origins unknown, biogenesis complex, Nat Rev Mol Cell Biol, 14 (2013) 759–774, doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- [18].Molino D, Nascimbeni AC, Giordano F, Codogno P, Morel E, ER-driven membrane contact sites: Evolutionary conserved machineries for stress response and autophagy regulation?, Commun Integr Biol, 10 (2017) e1401699, doi: 10.1080/19420889.2017.1401699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nishimura T, Tooze SA, Emerging roles of ATG proteins and membrane lipids in autophagosome formation, Cell Discov, 6 (2020) 32, doi: 10.1038/s41421-020-0161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Osawa T, Noda NN, Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis, Protein Sci, 28 (2019) 1005–1012, doi: 10.1002/pro.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, Ohsumi Y, Noda NN, Atg2 mediates direct lipid transfer between membranes for autophagosome formation, Nat Struct Mol Biol, 26 (2019) 281–288, doi: 10.1038/s41594-019-0203-4. [DOI] [PubMed] [Google Scholar]
- [22].Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ, ATG2 transports lipids to promote autophagosome biogenesis, J Cell Biol, 218 (2019) 1787–1798, doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Maeda S, Otomo C, Otomo T, The autophagic membrane tether ATG2A transfers lipids between membranes, Elife, 8 (2019), doi: 10.7554/eLife.45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Osawa T, Ishii Y, Noda NN, Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4, Genes Cells, 25 (2020) 65–70, doi: 10.1111/gtc.12733. [DOI] [PubMed] [Google Scholar]
- [25].Scott RC, Schuldiner O, Neufeld TP, Role and regulation of starvation-induced autophagy in the Drosophila fat body, Dev Cell, 7 (2004) 167–178, doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- [26].Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N, Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets, Mol Biol Cell, 23 (2012) 896–909, doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H, The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes, Dev Cell, 21 (2011) 343–357, doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- [28].Nagy P, Hegedus K, Pircs K, Varga A, Juhasz G, Different effects of Atg2 and Atg18 mutations on Atg8a and Atg9 trafficking during starvation in Drosophila, FEBS Lett, 588 (2014) 408–413, doi: 10.1016/j.febslet.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, Wang X, Kovacs AL, Yu L, Zhang H, C. elegans screen identifies autophagy genes specific to multicellular organisms, Cell, 141 (2010) 1042–1055, doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- [30].Tamura N, Nishimura T, Sakamaki Y, Koyama-Honda I, Yamamoto H, Mizushima N, Differential requirement for ATG2A domains for localization to autophagic membranes and lipid droplets, FEBS Lett, 591 (2017) 3819–3830, doi: 10.1002/1873-3468.12901. [DOI] [PubMed] [Google Scholar]
- [31].Tang Z, Takahashi Y, Chen C, Liu Y, He H, Tsotakos N, Serfass JM, Gebru MT, Chen H, Young MM, Wang HG, Atg2A/B deficiency switches cytoprotective autophagy to non-canonical caspase-8 activation and apoptosis, Cell Death Differ, 24 (2017) 2127–2138, doi: 10.1038/cdd.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y, Formation process of autophagosome is traced with Apg8/Aut7p in yeast, J Cell Biol, 147 (1999) 435–446, doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y, A ubiquitin-like system mediates protein lipidation, Nature, 408 (2000) 488–492, doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- [34].Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ, Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3, Nat Cell Biol, 16 (2014) 415–424, doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].de la Ballina LR, Munson MJ, Simonsen A, Lipids and Lipid-Binding Proteins in Selective Autophagy, J Mol Biol, 432 (2020) 135–159, doi: 10.1016/j.jmb.2019.05.051. [DOI] [PubMed] [Google Scholar]
- [36].Casares D, Escriba PV, Rossello CA, Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues, Int J Mol Sci, 20 (2019), doi: 10.3390/ijms20092167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].van Meer G, Voelker DR, Feigenson GW, Membrane lipids: where they are and how they behave, Nat Rev Mol Cell Biol, 9 (2008) 112–124, doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carvalho M, Sampaio JL, Palm W, Brankatschk M, Eaton S, Shevchenko A, Effects of diet and development on the Drosophila lipidome, Mol Syst Biol, 8 (2012) 600, doi: 10.1038/msb.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hammad LA, Cooper BS, Fisher NP, Montooth KL, Karty JA, Profiling and quantification of Drosophila melanogaster lipids using liquid chromatography/mass spectrometry, Rapid Commun Mass Spectrom, 25 (2011) 2959–2968, doi: 10.1002/rcm.5187. [DOI] [PubMed] [Google Scholar]
- [40].Guan XL, Cestra G, Shui G, Kuhrs A, Schittenhelm RB, Hafen E, van der Goot FG, Robinett CC, Gatti M, Gonzalez-Gaitan M, Wenk MR, Biochemical membrane lipidomics during Drosophila development, Dev Cell, 24 (2013) 98–111, doi: 10.1016/j.devcel.2012.11.012. [DOI] [PubMed] [Google Scholar]
- [41].Laurinyecz B, Peter M, Vedelek V, Kovacs AL, Juhasz G, Maroy P, Vigh L, Balogh G, Sinka R, Reduced expression of CDP-DAG synthase changes lipid composition and leads to male sterility in Drosophila, Open Biol, 6 (2016) 50169, doi: 10.1098/rsob.150169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Harayama T, Riezman H, Understanding the diversity of membrane lipid composition, Nat Rev Mol Cell Biol, 19 (2018) 281–296, doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- [43].Skowronska-Krawczyk D, Budin I, Aging membranes: Unexplored functions for lipids in the lifespan of the central nervous system, Exp Gerontol, 131 (2020) 110817, doi: 10.1016/j.exger.2019.110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dall’Armi C, Devereaux KA, Di Paolo G, The role of lipids in the control of autophagy, Curr Biol, 23 (2013) R33–45, doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ, Zou S, Chen Y, Zheng XL, Klionsky DJ, Liang Y, Xie Z, Dual roles of Atg8-PE deconjugation by Atg4 in autophagy, Autophagy, 8 (2012) 883–892, doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kohler K, Brunner E, Guan XL, Boucke K, Greber UF, Mohanty S, Barth JM, Wenk MR, Hafen E, A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila, Autophagy 5 (2009) 980–990, 10.4161/auto.5.7.9325. [DOI] [PubMed] [Google Scholar]
- [47].Knorr RL, Nakatogawa H, Ohsumi Y, Lipowsky R, Baumgart T, Dimova R, Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids, PLoS One, 9 (2014) e115357, doi: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G, Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila, PLoS One, 7 (2012) e44214, doi: 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hegedus K, Takats S, Boda A, Jipa A, Nagy P, Varga K, Kovacs AL, Juhasz G, The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy, Mol Biol Cell, 27 (2016) 3132–3142, doi: 10.1091/mbc.E16-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sader JE, Sanelli JA, Adamson BD, Monty JP, Wei X, Crawford SA, Friend JR, Marusic I, Mulvaney P, Bieske EJ, Spring constant calibration of atomic force microscope cantilevers of arbitrary shape, Rev Sci Instrum, 83 (2012) 103705, doi: 10.1063/1.4757398. [DOI] [PubMed] [Google Scholar]
- [51].Folch J, Lees M, Sloane Stanley GH, A simple method for the isolation and purification of total lipides from animal tissues, J Biol Chem, 226 (1957) 497–509. [PubMed] [Google Scholar]
- [52].Bligh EG, Dyer WJ, A rapid method of total lipid extraction and purification, Can J Biochem Physiol, 37 (1959) 911–917, doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- [53].Sparkes BL, Slone EE, Roth M, Welti R, Fleming SD, Intestinal lipid alterations occur prior to antibody-induced prostaglandin E2 production in a mouse model of ischemia/reperfusion, Biochim Biophys Acta, 1801 (2010) 517–525, doi: 10.1016/j.bbalip.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhou Z, Marepally SR, Nune DS, Pallakollu P, Ragan G, Roth MR, Wang L, Lushington GH, Visvanathan M, Welti R, LipidomeDB data calculation environment: online processing of direct-infusion mass spectral data for lipid profiles, Lipids, 46 (2011) 879–884, doi: 10.1007/s11745-011-3575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Falcone DL, Ogas JP, Somerville CR, Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition, BMC Plant Biol, 4 (2004) 17, doi: 10.1186/1471-2229-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mizushima N, The ATG conjugation systems in autophagy, Curr Opin Cell Biol, 63 (2020) 1–10, doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- [57].Vegh AG, Nagy K, Balint Z, Kerenyi A, Rakhely G, Varo G, Szegletes Z, Effect of antimicrobial peptide-amide: indolicidin on biological membranes, J Biomed Biotechnol, 2011 (2011) 670589, doi: 10.1155/2011/670589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Reunanen H, Punnonen EL, Hirsimaki P, Studies on vinblastine-induced autophagocytosis in mouse liver. V. A cytochemical study on the origin of membranes, Histochemistry, 83 (1985) 513–517, doi: 10.1007/BF00492453. [DOI] [PubMed] [Google Scholar]
- [59].Taniguchi S, Toyoshima M, Takamatsu T, Mima J, Curvature-sensitive trans-assembly of human Atg8-family proteins in autophagy-related membrane tethering, Protein Sci, 29 (2020) 1387–1400, doi: 10.1002/pro.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Unspecific bindings occur without pre-clearing of lysate, used for immunoisolation of autophagic structures from Drosophila control and Atg2−. Immunoblottings were performed from different fractions of immunopurification assay using RFP-Trap only: lysate (input), lysate after RFP-trap incubation (Flow-through), isolated autophagic structures (eluate). Without the pre-clearing step, mitochondrial marker proteins as well as cytosolic proteins (faint Tubulin signal) are also present in eluate (unspecific binding). Antibodies used are: anti-GABARAP (Atg8a-autophagic marker), anti-ATP5A (mitochondria), anti-Rab5 (early endosome), anti-Rab7 (late endosome), anti-Cathepsin-L (lysosome), anti-α-Tubulin (cytosolic marker and loading control).
Supplementary Fig. 2. Autophagic structures imaged with Atomic Force Microscope (AFM). A) Three dimensional images of eluate fractions from Drosophila control animals. B) Height of the vesicles along the red axis (see panel A) indicate the double membrane nature of the vesicles.


