Abstract
The extracellular matrix (ECM) provides an architectural meshwork that surrounds and supports cells. The dysregulation of heavily post-translationally modified ECM proteins directly contributes to various diseases. Mass spectrometry (MS)-based proteomics is an ideal tool to identify ECM proteins and characterize their post-translational modifications, but ECM proteomics remains extremely challenging owing to the extremely low solubility of the ECM. Herein, enabled by effective solubilization of ECM proteins using our recently developed photocleavable surfactant, Azo, we have developed a streamlined ECM proteomic strategy that allows fast tissue decellularization, efficient extraction and enrichment of ECM proteins, and rapid digestion prior to reversed-phase liquid chromatography (RPLC)-MS analysis. A total of 173 and 225 unique ECM proteins from mouse mammary tumors have been identified using 1D and 2D RPLC-MS/MS, respectively. Moreover, 87 (from 1DLC-MS/MS) and 229 (from 2DLC-MS/MS) post-translational modifications of ECM proteins, including glycosylation, phosphorylation, and hydroxylation, were identified and localized. This Azo-enabled ECM proteomics strategy will streamline the analysis of ECM proteins and promote the study of ECM biology.
Keywords: Extracellular matrix, matrisome, photocleavable surfactant, post-translational modification, glycoproteins
Graphical Abstract
The extracellular matrix (ECM) consists of approximately 300 core proteins including collagens, fibronectins, laminins, and proteoglycans and ~900 associated proteins, defined as the “matrisome”1–2, which forms an architectural meshwork and provides stability for the surrounding cells.1–5 Serving as a critical regulator of cell behaviors such as adhesion, migration, and proliferation, the ECM responds and communicates via biochemical cues to the intracellular milieu.3 ECM proteins are also known to be heavily post-translationally modified, most notably by glycosylation and hydroxylation.6,7 Dysregulation of ECM protein expression and post-translational modifications (PTMs) directly contribute to disease progression and regulate ECM protein structures, functions, and interactions contributing to pathogenesis.1–10 Particularly, the ECM is increasingly recognized as a major driver in tumor metastasis, which contributes to 90% of the cancer deaths.11–13 However, the biochemical characterization of ECM proteins remains a daunting task due to the extremely low solubility of ECM proteins.14–16
Advances in mass spectrometry (MS)-based proteomics make it an ideal tool to identify ECM proteins and characterize their PTMs occurring within the ECM microenvironment.2, 7, 17–28 Despite its immense potential, ECM proteomics remains extremely challenging mainly due to the dense network of cross-linked, fibrous proteins, which makes it exceedingly difficult to solubilize, digest, and analyze by MS.19 Moreover, the complexity and dynamic range of proteins present in the tissue lysates often result in underrepresentation of the important ECM sub-proteome in global proteomics studies.29 The current protocols for ECM proteomic analysis typically have lengthy and labor-intensive workflows that often include multiple digestion steps, some including chemical digestion using toxic chemicals such as cyanogen bromide (CNBr).20, 21, 30
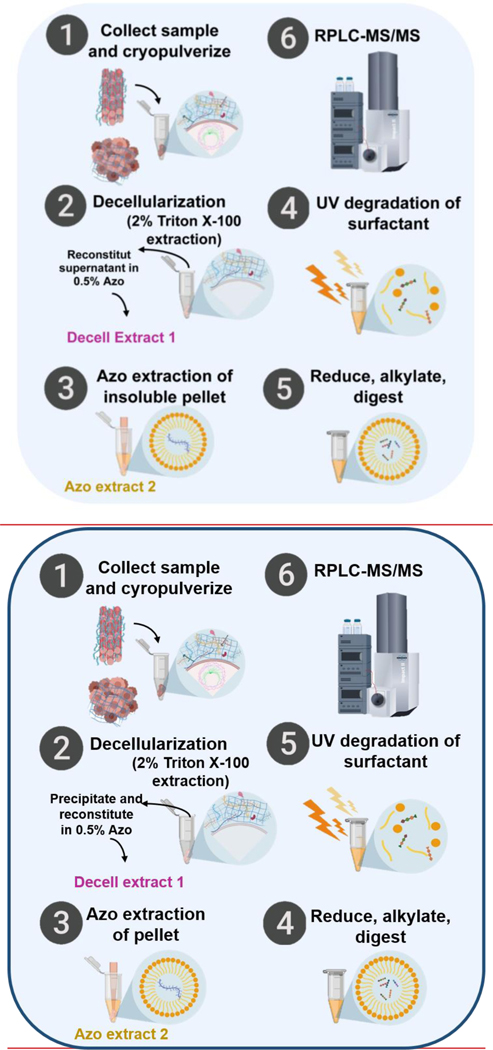
To address these challenges, here we developed a photocleavable surfactant-enabled ECM proteomics strategy to streamline the enrichment, extraction, and digestion steps for ECM proteomics (Figure 1). Specifically, we established a new decellularization/extraction method, enabled by our recently developed photocleavable anionic surfactant, Azo,31, 32 for the efficient enrichment of ECM proteins, which eliminates the need for multiple digestion steps and minimizes the sample clean-up prior to reversed phased liquid chromatography (RPLC)-MS analysis. Our sample preparation method takes approximately ~8 h (0.75 h for decellularization, 1.5 h for removal of Triton and work-up of the Decell extract, 1.5 h for extraction of the pellet, and 3.5 h for reduction, alkylation and trypsin digestion) compared to the conventional approaches that often take several days due to lengthy decellularization and multiple enzymatic digestions,20, 33 representing a significant improvement in the throughput of ECM proteomics. Specifically, we chose to analyze tumor tissues harvested from the transgenic mouse mammary tumor virus (MMTV) polyomavirus middle T (PyVT) mouse model as it represents a classic transgenic model for studying the microenvironment of metastatic human breast cancer34–37 and extensive collagen deposition is a pathological hallmark of many cancers.38, 39
Figure 1.
Schematic illustration of the Azo-enabled ECM proteomics strategy. (1) Tissues are collected, cryo-pulverized, and (2) undergo rapid decellularization in 2% Triton X-100. After centrifugation, the supernatants are collected, precipitated, and reconstituted in 0.5% Azo labeled as “Decell extract 1” for downstream analysis. (3) Azo was added to the remaining tissue pellet to extract the remaining proteins, resulting in “Azo extract 2”. (4) Both Decell Extract 1 and Azo extract 2 were reduced, alkylated, and digested with trypsin. (5) Azo was degraded by UV-irradiation and (6) the digested peptides were analyzed by RPLC-MS/MS.
RESULTS AND DISCUSSION
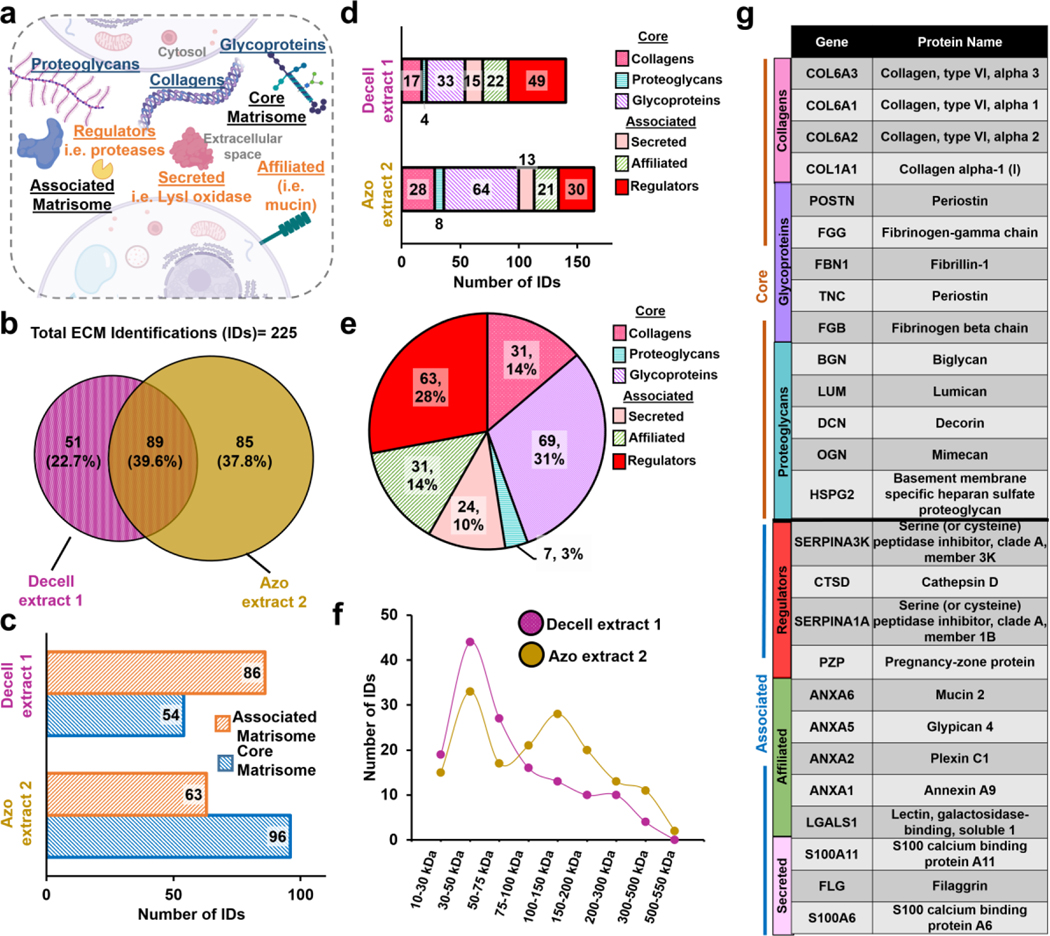
For characterization of the mouse tumor tissue, we considered both the core ECM domain-containing regions as well as regulatory and secreted ECM associated proteins, which constitute “the matrisome” as defined previously.1, 2 The core matrisome was classified into three subcategories, collagens, glycoproteins, and proteoglycans, whereas regulators, secreted factors, and affiliated proteins were classified as associated matrisome proteins (Figure 2a). Characterizing the ensemble of core and associated matrisome proteins is critical to achieving a comprehensive understanding of ECM biology.1, 2 However, core matrisome proteins, like collagens, are generally considered extremely challenging to be analyzed by proteomics since they are extremely difficult to be solubilized,40 and resistant to common extraction and enzymatic digestions.41
Figure 2.
ECM proteomics of mammary tumor tissue. a. Schematic of matrisome ECM composition and annotation.1, 2 b. Venn diagrams showing the overlap of ECM protein identifications found in the Decell extract 1 and Azo extract 2 identified using 2D RPLC MSMS. c. Division distribution of ECM identifications in each extract. d. Category distribution of ECM identifications in each extract. e. Pie chart illustrating the composition of all ECM identifications. f. The molecular weight (MW) distribution of all ECM identifications. g. Table of highest abundance ECM core matrisome proteins identified in Azo extract and associated matrisome proteins identified in Decell extract 1.
Typically, enrichment of the core matrisome proteins is obtained by decellularization,42 wherein the tissue is passively immersed in a buffer, such as SDS or Triton X-100, for several hours or days to remove soluble cellular material (proteins, lipids, metabolites, etc.), leaving an intact ECM.20, 21, 43 However, since an intact scaffold is not required for proteomics, we mechanically homogenized the tissue in Triton X-100 buffer44 to dramatically increase the throughput. The samples were centrifuged and the supernatant was collected and labeled as “Decell extract 1”, and a pellet remained (Figure S1). Commonly, proteins extracted during decellularization are discarded20 presumably because few ECM proteins are extracted during this step. To investigate whether the decellularization extraction contains soluble ECM factors, the proteins in the Decell extract 1 were precipitated and reconstituted in 0.5% Azo for downstream LC-MS analysis (Figure S1).
Next, the remaining tissue pellets were washed with ammonium bicarbonate (ABC) buffer (chosen for its MS-compatibility) to remove any remaining soluble proteins, surfactants, or salts (Figure S2). Azo was then used to extract the remaining proteins yielding “Azo extract 2” for further LC-MS analysis. We observed excellent reproducibility using SDS-PAGE analysis across technical replicates (Figure S3). Interestingly, we qualitatively observed that Decell extract 1 was rich in lower molecular weight proteins, whereas Azo extract 2 contained many higher molecular weight species (Figure S3).
Subsequently, Decell extract 1 and Azo extract 2 were digested in-solution with trypsin for 2 h followed by rapid degradation of Azo32 by UV light and subsequent analysis by RPLC-MS/MS. Recently we developed a high-throughput bottom-up proteomics method enabled by this photocleavable surfactant for robust protein extraction, rapid enzymatic digestion, and subsequent MS-analysis without additional sample clean-up following UV degradation.32 Identification and relative protein abundance including reproducibility across replicates were determined with MetaMorpheus45 and FlashLFQ46 software using intensity-based normalization. A full list of parameters can be found in Table S1 and a full list of ECM identifications can be found in Table S2. Transformed, normalized intensities of ECM peptide spectral matches (PSMs) in Azo extract 2 were plotted against each other, demonstrating a highly reproducible method with Pearson correlation coefficients from 0.91 to 0.93 (Figure S4).
Using the normalized peptide intensities, we analyzed the relative abundance of core and associated matrisome proteins in the Decell and Azo extracts (Figure S5a and Table S3). Significantly, we observed the Azo extract 2 generally contained a higher abundance of core matrisome proteins such as collagen alpha-1 (I) chain (COL1A1) and biglycan (BGN) (Figure S5b–c and Table S3). On the other hand, associated matrisome proteins, such as Galectin-1 (LGALS1) and Cathepsin D (CTSD), were enriched in the Decell extract 1 (Figure S5b–c) (Table S3). Additionally, we evaluated the cellular locations of proteins in both extractions to better understand their protein compositions (Figure S5a). Endoplasmic reticulum (ER), mitochondrial, cytoplasmic, secreted and membrane proteins were found in higher abundance in the Decell extract 1, whereas core matrisome and nuclear proteins were primarily present in Azo extract 2 (Figure S5c–d). Overall, the results demonstrated successful protein fractionation at the extraction level and highlighted Azo’s ability to solubilize and digest important ECM proteins such as type I collagen47 for a streamlined analysis.
To further assess Azo’s extraction efficacy, we directly compared its performance to 8 M urea, a common reagent used for ECM protein extraction.31,32,25 Pulverized tumor tissue was decellularized with Triton-X-100, washed, and extracted with 8 M urea in 25 mM ABC, 0.5 % Azo in 25 mM ABC, or 25 mM ABC (serving as a control). SDS-PAGE analysis demonstrated Azo extracted a unique protein profile compared to the other conditions (Figure S6a). Using RPLC-MS/MS analysis, we found the relative abundance of collagen species, a common benchmark for ECM enrichment,2, 14 was significantly increased in the Azo extract compared to the urea or ABC alone (Figure S6b and Table S3). Hence, this demonstrated the superior performance of Azo relative to urea, in solubilizing ECM proteins. In particular, fibrillar type I collagen chains, COL1A1 AND COL1A2, were solubilized more efficiently with Azo relative to urea extraction. Moreover, the use of photocleavable Azo eliminated the need for time-consuming desalting steps and greatly improved the throughput of ECM proteomics. Overall, we identified 173 ECM proteins using 1D RPLC-MS/MS. A total of 52 and 71 proteins were uniquely identified in the Decell extract 1 and Azo extract 2 samples, respectively, and 50 proteins were identified in both (Figure S7a). Next, we investigated whether the addition of protein fractionation by high-pH RPLC could increase the proteome coverage. This 2D high-pH low-pH RPLC approach contributed 52 new unique identifications (a 30% increase) compared to using 1D RPLC-MS/MS (Figure S7a–b). Although the increase in protein identification was moderate, we demonstrated our method could be easily combined with additional separation steps to increase proteome coverage.
Combined, we identified 225 unique ECM proteins from mouse mammary tumor tissue using this approach (Figure 2b and Table S2). Importantly, we observed both Decell extract 1 and Azo extract 2 contained many matrisome proteins. Of the ECM protein identifications, 89 ECM proteins were identified in both extracts, whereas 51 and 85 were uniquely identified in Decell extract 1 and Azo extract 2, respectively, which indicates that some proteins are preferentially solubilized and present in either Decell extract 1 or Azo extract 2 (Figure 2b).
We further investigated the protein compositions of each extract, observing that 54 core matrisome proteins and 86 associated proteins were identified in the Decell extract 1 whereas 96 core matrisome proteins and 63 associated proteins were identified in Azo extract 2 (Figure 2c). The results indicate enrichment of core matrisome proteins in the Azo extract 2, but also highlight the importance of analyzing the commonly disregarded decellularization fraction, Decell extract 1. Additionally, Decell extract 1 and Azo extract 2 contained similar numbers of proteoglycans, secreted proteins, and affiliated proteins (Figure 2d). On the other hand, ECM regulatory proteins were enriched in the Decell extract 1 whereas a large number of collagens and glycoproteins were identified in the Azo extract 2. All categories of ECM proteins were well represented, using our approach (Figure 2e). We observed that a larger number of higher molecular weight (>100 kDa) and core matrisome proteins were identified in Azo extract 2, whereas a larger number of lower molecular weight, associated matrisome species were identified in Decell extract 1 (Figure 2f). We reason that the higher molecular weight of core matrisome proteins makes them more difficult to be solubilized, thus require a strong surfactant such as Azo. A representative list of identified ECM proteins is shown in Figure 2g.
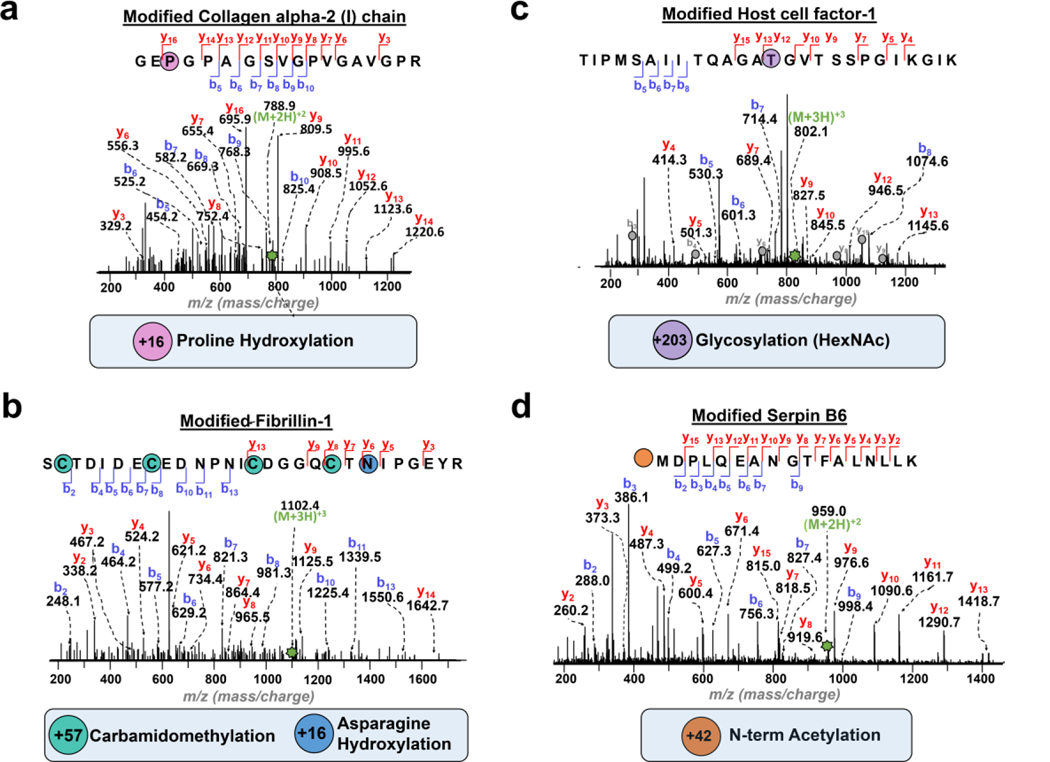
ECM proteins are known to be heavily post-translationally modified. Here, we used MetaMorpheus45 software to identify a diverse range of ECM PTMs including proline hydroxylation, asparagine hydroxylation, lysine hydroxylation, acetylation, phosphorylation and glycosylation (Figure S8). In total, 87 unique PTM sites were identified from mouse tumor tissue using 1D RPLC-MS/MS (Figure S8 and Table S4). Significantly, we observed a dramatic increase in PTM identifications, 229 total PTMs, with the 2DLC-MS/MS workflow (Figure S8 and Table S4), despite that the 2D approach provided only a moderate increase in the identifications of ECM proteins. Overall, proline hydroxylation, which is essential for the stability of collagen fibrils,47 accounted for 50% of all identified modifications from mouse mammary tumor tissue (Figure S8). Figure 3 shows some representative examples of unambiguously localized PTMs, such as proline hydroxylation of collagen alpha-2 (I) chain (COL1A2) and asparagine β hydroxylation, on proteins like fibrillin-1 (FBN1) (Figure 3a–b). N-acetylglucosamine (HexNAc) was localized to a threonine residue of Host cell-factor-1 (Figure 3c). Additionally, we have identified an N-terminal acetylation of Serpin B6 (Figure 3d).
Figure 3. Identification of PTMs in the ECM proteins.
Representative MS/MS spectra of identified ECM PTMs. Precursor ions are annotated with green stars. a. Precursor ion Collagen alpha-2 (I) chain peptide with proline (P) hydroxylation (+ 16 Da). [(M+2H)2+, 788.9 m/z, Expt’l: 1,575.8 Da, 0.8 ppm]. b. Fibrillin-1 peptide modified with cysteine (C) carbamidomethylation (+ 57 Da) and asparagine (N) hydroxylation (+16 Da). Precursor ion [(M+3H)3+, 1102.4 m/z, Expt’l: 3,304.3 Da, 1.4 ppm]. c. Host cell-factor-1 with (N)-linked acetylhexosamine (HexNAc) glycosylation (+203 Da). Precursor ion [(M+3H)3+, 802.1 m/z, Expt’l: 2,403.3 Da, 0.6 ppm]. HCFC1 co-eluted with a highly abundant peptide identified as splicing factor 3b subunit 2, where corresponding b and y ions are denoted with gray circles. d. Serpin B6 with N-terminal acetylation (+42 Da). Precursor ion [(M+2H)2+, 959.0 m/z, Expt’l: 1,916.0 Da, 0.2 ppm].
CONCLUSION
In summary, for the first time, we developed a streamlined ECM proteomics method enabled by a photocleavable surfactant, Azo, which addressed several challenges in the conventional ECM proteomics workflows. Most notably, Azo facilitated robust extraction of fibrous ECM proteins, aided in rapid trypsin digestion, and subsequently can be easily degraded by UV-irradiation before MS. This Azo-enabled ECM sample preparation including decellularization, ECM extraction, reduction, alkylation and digestion takes about 6 h compared to several days or a week using the conventional approaches. This sample preparation does not require chemical digestion, multiple enzymes, or deglycosylation. Moreover, we demonstrated the importance of analyzing the commonly discarded decellularization fraction to identify soluble ECM and ECM-associated proteins. Using both 1D and 2D RPLC-MS/MS, we have established an ECM protein catalog consisting of 225 ECM proteins from mouse tumor tissue and have further identified 229 total PTMs for ECM proteins including hydroxylation, phosphorylation, and glycosylation. We envision this Azo-enabled ECM proteomics strategy will streamline the analysis of ECM proteins and promote an understanding of ECM biology in various diseases such as tumor metastasis.
Supplementary Material
ACKNOWLEDGEMENT
The financial support is provided by NIH R01 GM117058 (to S.J. and Y. G.). Y.G. also would like to acknowledge GM125085, HL109810, HL096971, and the high-end instrument grant S10OD018475. S.K. is supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1747503 and the WiscAMP-BD program under Grant No. HRD-1612530. K.A.B. would like to acknowledge support from the Training Program in Translational Cardiovascular Science, T32 HL007936–19 and the University of Wisconsin Vascular Surgery Research Training Program, T32HL110853. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Graphics in main Figures 1, 2a and supplemental Figures 1, 5 were created with clipart from BioRender.com.
Footnotes
SUPPORTING INFORMATION
The Supplemental Materials and Methods are available free of charge on the ACS Publications website at
Chemicals and materials, Mouse mammary tumor tissue; ECM protein extraction; Digestion and 1D-RPLC-MS/MS; SDS-PAGE; Offline high-pH reverse-phase fractionation; Data analysis; Azo enabled two-step extraction for analysis of extracellular matrix (ECM) proteins (Figure S1); Pictures were taken after tissue decellularization and washing steps (Figure S2); 8% SDS-PAGE analysis of representative ECM extraction replicates of Decell extract 1 and Azo extract 2 (Figure S3); Scatter-plots of Log2 transformed relative ECM protein abundances in extraction replicates of Azo extract 2 from mammary mouse tumor tissue (Figure S4); Composition of Decell extract 1 and Azo extract 2 from mammary mouse tumor tissue (Figure S5); Comparison of Azo to the traditional ECM extraction buffers (Figure S6); Comparison of 1- and 2-dimensional LC separation strategies for ECM proteomics (Figure S7); Summary of PTM sites identified in ECM proteins from mammary mouse tumor tissue (Figure S8); Data analysis parameters (Table S1); Inventory of ECM protein identifications (Table S2); Summary of label-free quantitation of ECM proteins (Table S3); Summary of post-translational modifications of ECM proteins (Table S4).
REFERENCES
- 1.Shao X; Taha IN; Clauser KR; Gao Y; Naba A, MatrisomeDB: the ECM-protein knowledge database. Nucleic Acids Research 2020, 48 (D1), D1136–D1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naba A; Clauser KR; Hoersch S; Liu H; Carr SA; Hynes RO, The Matrisome:In SilicoDefinition andIn VivoCharacterization by Proteomics of Normal and Tumor Extracellular Matrices. Molecular & Cellular Proteomics 2012, 11 (4), M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hynes RO, The Extracellular Matrix: Not Just Pretty Fibrils. Science 2009, 326 (5957), 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouw JK; Ou G; Weaver VM, Extracellular matrix assembly: a multiscale deconstruction. Nature Reviews Molecular Cell Biology 2014, 15 (12), 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilkes DM; Semenza GL; Wirtz D, Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nature Reviews Cancer 2014, 14 (6), 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isra; Naba A, Exploring the extracellular matrix in health and disease using proteomics. Essays in Biochemistry 2019, 63 (3), 417–432. [DOI] [PubMed] [Google Scholar]
- 7.Yang C; Park AC; Davis NA; Russell JD; Kim B; Brand DD; Lawrence MJ; Ge Y; Westphall MS; Coon JJ; Greenspan DS, Comprehensive Mass Spectrometric Mapping of the Hydroxylated Amino Acid residues of the α1(V) Collagen Chain. Journal of Biological Chemistry 2012, 287 (48), 40598–40610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rienks M; Papageorgiou A-P; Frangogiannis NG; Heymans S, Myocardial Extracellular Matrix. Circulation Research 2014, 114 (5), 872–888. [DOI] [PubMed] [Google Scholar]
- 9.Lindsey ML; Iyer RP; Zamilpa R; Yabluchanskiy A; Deleon-Pennell KY; Hall ME; Kaplan A; Zouein FA; Bratton D; Flynn ER; Cannon PL; Tian Y; Jin Y-F; Lange RA; Tokmina-Roszyk D; Fields GB; De Castro Brás LE, A Novel Collagen Matricryptin Reduces Left Ventricular Dilation Post-Myocardial Infarction by Promoting Scar Formation and Angiogenesis. Journal of the American College of Cardiology 2015, 66 (12), 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walma DAC; Yamada KM, The extracellular matrix in development. Development 2020, 147 (10), dev175596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conklin MW; Eickhoff JC; Riching KM; Pehlke CA; Eliceiri KW; Provenzano PP; Friedl A; Keely PJ, Aligned Collagen Is a Prognostic Signature for Survival in Human Breast Carcinoma. The American Journal of Pathology 2011, 178 (3), 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henke E; Nandigama R; Ergün S, Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Frontiers in Molecular Biosciences 2020, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naba A; Clauser KR; Lamar JM; Carr SA; Hynes RO, Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett AS; Wither MJ; Hill RC; Dzieciatkowska M; D’Alessandro A; Reisz JA; Hansen KC, Hydroxylamine Chemical Digestion for Insoluble Extracellular Matrix Characterization. Journal of Proteome Research 2017, 16 (11), 4177–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyre DR; Paz MA; Gallop PM, Cross-Linking in Collagen and Elastin. Annual Review of Biochemistry 1984, 53 (1), 717–748. [DOI] [PubMed] [Google Scholar]
- 16.Frantz C; Stewart KM; Weaver VM, The extracellular matrix at a glance. Journal of Cell Science 2010, 123 (24), 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byron A; Humphries JD; Humphries MJ, Defining the extracellular matrix using proteomics. International Journal of Experimental Pathology 2013, 94 (2), 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merl-Pham J; Basak T; Knüppel L; Ramanujam D; Athanason M; Behr J; Engelhardt S; Eickelberg O; Hauck SM; Vanacore R; Staab-Weijnitz CA, Quantitative proteomic profiling of extracellular matrix and site-specific collagen post-translational modifications in an in vitro model of lung fibrosis. Matrix Biology Plus 2019, 1, 100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsey ML; Jung M; Hall ME; Deleon-Pennell KY, Proteomic analysis of the cardiac extracellular matrix: clinical research applications. Expert Review of Proteomics 2018, 15 (2), 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naba A; Pearce OMT; Del Rosario A; Ma D; Ding H; Rajeeve V; Cutillas PR; Balkwill FR; Hynes RO, Characterization of the Extracellular Matrix of Normal and Diseased Tissues Using Proteomics. Journal of Proteome Research 2017, 16 (8), 3083–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill RC; Calle EA; Dzieciatkowska M; Niklason LE; Hansen KC, Quantification of Extracellular Matrix Proteins from a Rat Lung Scaffold to Provide a Molecular Readout for Tissue Engineering. Molecular & Cellular Proteomics 2015, 14 (4), 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barallobre-Barreiro J; Didangelos A; Schoendube FA; Drozdov I; Yin X; Fernández-Caggiano M; Willeit P; Puntmann VO; Aldama-López G; Shah AM; Doménech N; Mayr M, Proteomics Analysis of Cardiac Extracellular Matrix Remodeling in a Porcine Model of Ischemia/Reperfusion Injury. Circulation 2012, 125 (6), 789–802. [DOI] [PubMed] [Google Scholar]
- 23.Raghunathan R; Sethi MK; Klein JA; Zaia J, Proteomics, Glycomics, and Glycoproteomics of Matrisome Molecules. Molecular & Cellular Proteomics 2019, 18 (11), 2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghunathan VK; Morgan JT; Chang Y-R; Weber D; Phinney B; Murphy CJ; Russell P, Transforming Growth Factor Beta 3 Modifies Mechanics and Composition of Extracellular Matrix Deposited by Human Trabecular Meshwork Cells. ACS Biomaterials Science & Engineering 2015, 1 (2), 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma F; Tremmel DM; Li Z; Lietz CB; Sackett SD; Odorico JS; Li L, In Depth Quantification of Extracellular Matrix Proteins from Human Pancreas. Journal of Proteome Research 2019, 18 (8), 3156–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomko LA; Hill RC; Barrett A; Szulczewski JM; Conklin MW; Eliceiri KW; Keely PJ; Hansen KC; Ponik SM, Targeted matrisome analysis identifies thrombospondin-2 and tenascin-C in aligned collagen stroma from invasive breast carcinoma. Scientific Reports 2018, 8 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langley SR; Willeit K; Didangelos A; Matic LP; Skroblin P; Barallobre-Barreiro J; Lengquist M; Rungger G; Kapustin A; Kedenko L; Molenaar C; Lu R; Barwari T; Suna G; Yin X; Iglseder B; Paulweber B; Willeit P; Shalhoub J; Pasterkamp G; Davies AH; Monaco C; Hedin U; Shanahan CM; Willeit J; Kiechl S; Mayr M, Extracellular matrix proteomics identifies molecular signature of symptomatic carotid plaques. Journal of Clinical Investigation 2017, 127 (4), 1546–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krasny L; Paul A; Wai P; Howard BA; Natrajan RC; Huang PH, Comparative proteomic assessment of matrisome enrichment methodologies. Biochem J 2016, 473 (21), 3979–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H; Sadygov RG; Yates JR, A Model for Random Sampling and Estimation of Relative Protein Abundance in Shotgun Proteomics. Analytical Chemistry 2004, 76 (14), 4193–4201. [DOI] [PubMed] [Google Scholar]
- 30.Goddard ET; Hill RC; Barrett A; Betts C; Guo Q; Maller O; Borges VF; Hansen KC; Schedin P, Quantitative extracellular matrix proteomics to study mammary and liver tissue microenvironments. The International Journal of Biochemistry & Cell Biology 2016, 81, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown KA; Chen B; Guardado-Alvarez TM; Lin Z; Hwang L; Ayaz-Guner S; Jin S; Ge Y, A photocleavable surfactant for top-down proteomics. Nature Methods 2019, 16 (5), 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown KA; Tucholski T; Eken C; Knott S; Zhu Y; Jin S; Ge Y, High-Throughput Proteomics Enabled by a Photocleavable Surfactant. Angewandte Chemie 2020, 132 (22), 8484–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Puig A; Mosquera JL; Jiménez-Delgado S; García-Pastor C; Jorba I; Navajas D; Canals F; Raya A, Proteomics Analysis of Extracellular Matrix Remodeling During Zebrafish Heart Regeneration. Molecular & Cellular Proteomics 2019, 18 (9), 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rennhack JP; To B; Swiatnicki M; Dulak C; Ogrodzinski MP; Zhang Y; Li C; Bylett E; Ross C; Szczepanek K; Hanrahan W; Jayatissa M; Lunt SY; Hunter K; Andrechek ER, Integrated analyses of murine breast cancer models reveal critical parallels with human disease. Nature Communications 2019, 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Mendoza MG; Inman DR; Ponik SM; Jeffery JJ; Sheerar DS; Van Doorn RR; Keely PJ, Neutrophils drive accelerated tumor progression in the collagen-dense mammary tumor microenvironment. Breast Cancer Research 2016, 18 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provenzano PP; Inman DR; Eliceiri KW; Beggs HE; Keely PJ, Mammary Epithelial-Specific Disruption of Focal Adhesion Kinase Retards Tumor Formation and Metastasis in a Transgenic Mouse Model of Human Breast Cancer. The American Journal of Pathology 2008, 173 (5), 1551–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provenzano PP; Inman DR; Eliceiri KW; Knittel JG; Yan L; Rueden CT; White JG; Keely PJ, Collagen density promotes mammary tumor initiation and progression. BMC Medicine 2008, 6 (1), 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu S; Xu H; Wang W; Li S; Li H; Li T; Zhang W; Yu X; Liu L, The role of collagen in cancer: from bench to bedside. Journal of Translational Medicine 2019, 17 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kai F; Drain AP; Weaver VM, The Extracellular Matrix Modulates the Metastatic Journey. Developmental Cell 2019, 49 (3), 332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey AJ, Collagen and elastin fibres. J Clin Pathol Suppl (R Coll Pathol) 1978, 12, 49–58. [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenblum G; Van Den Steen PE; Cohen SR; Bitler A; Brand DD; Opdenakker G; Sagi I, Direct Visualization of Protease Action on Collagen Triple Helical Structure. PLoS ONE 2010, 5 (6), e11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindsey ML; Hall ME; Harmancey R; Ma Y, Adapting extracellular matrix proteomics for clinical studies on cardiac remodeling post-myocardial infarction. Clinical Proteomics 2016, 13 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suna G; Wojakowski W; Lynch M; Barallobre-Barreiro J; Yin X; Mayr U; Baig F; Lu R; Fava M; Hayward R; Molenaar C; White SJ; Roleder T; Milewski KP; Gasior P; Buszman PP; Buszman P; Jahangiri M; Shanahan CM; Hill J; Mayr M, Extracellular Matrix Proteomics Reveals Interplay of Aggrecan and Aggrecanases in Vascular Remodeling of Stented Coronary Arteries. Circulation 2018, 137 (2), 166–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vavken P; Joshi S; Murray MM, TRITON-X is most effective among three decellularization agents for ACL tissue engineering. Journal of Orthopaedic Research 2009, 27 (12), 1612–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solntsev SK; Shortreed MR; Frey BL; Smith LM, Enhanced Global Post-translational Modification Discovery with MetaMorpheus. Journal of Proteome Research 2018, 17 (5), 1844–1851. [DOI] [PubMed] [Google Scholar]
- 46.Millikin RJ; Solntsev SK; Shortreed MR; Smith LM, Ultrafast Peptide Label-Free Quantification with FlashLFQ. Journal of Proteome Research 2018, 17 (1), 386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shoulders MD; Raines RT, Collagen Structure and Stability. Annual Review of Biochemistry 2009, 78 (1), 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.