Abstract
Haplotype-resolved de novo assembly is the ultimate solution to the study of sequence variations in a genome. However, existing algorithms either collapse heterozygous alleles into one consensus copy or fail to cleanly separate the haplotypes to produce high-quality phased assemblies. Here we describe hifiasm, a de novo assembler that takes advantage of long high-fidelity sequence reads to faithfully represent the haplotype information in a phased assembly graph. Unlike other graph-based assemblers that only aim to maintain the contiguity of one haplotype, hifiasm strives to preserve the contiguity of all haplotypes. This feature enables the development of a graph trio binning algorithm that greatly advances over standard trio binning. On three human and five non-human datasets, including California redwood with a ~30-gigabase hexaploid genome, we show that hifiasm frequently delivers better assemblies than existing tools and consistently outperforms others on haplotype-resolved assembly.
Introduction
De novo genome assembly is the most comprehensive method that provides unbiased insight to DNA sequences. With the rapid advances in long-read sequencing technologies such as Pacific Biosciences (PacBio) and Oxford Nanopore (ONT), many long-read assemblers have been developed to tackle this essential computational problem. Most of them1–9 collapse different homologous haplotypes into a consensus representation with heterozygous alleles frequently switching in the consensus. This approach works well for inbred samples that are nearly homozygous but necessarily misses half of the genetic information in a diploid genome. To solve this problem, Falcon-Unzip10 recovers heterozygous alleles by “unzipping” them in an initial collapsed assembly. It produces a pair of assemblies, one primary assembly representing a mosaic of homologous haplotypes, and one alternate assembly composed of short haplotype-specific contigs (haplotigs) for alleles absent from the primary assembly. The alternate assembly is often fragmented and does not represent a complete haplotype, making it less useful in practice. In addition, starting from a collapsed assembly, Falcon-Unzip may not recover highly heterozygous regions which are not properly collapsed in the initial assembly. Trio binning11 addresses these issues by globally partitioning long reads upfront with parental short reads and then performing two separate assemblies on the partitioned reads. This strategy works well for samples with high heterozygosity, but for a human sample sequenced with noisy long reads, it only produces fragmented assemblies with ~1.2 Mb contigs.
A great challenge to the assembly of heterozygous samples is caused by the 5–15% sequencing error rate of older long reads. With this high error rate, it is difficult to distinguish errors from heterozygotes occurring at a rate of <0.1% in humans. The recent availability of high-fidelity (HiFi) reads12 produced by PacBio has changed the equation. Generated from the consensus of multiple sequences of the same DNA molecule, HiFi reads have a much lower error rate of <1%. With HiFi, standard trio binning can produce contigs of 17 Mb12. Recent works relying on Hi-C or Strand-seq read binning13,14 can achieve better contiguity and phasing accuracy. These pre-binning algorithms all use short k-mers or short reads to partition HiFi reads15,16. They may not identify haplotype-specific markers in complex regions and result in wrong read partitions which will negatively affect the assembly as we will show later. In addition, both Hi-C and Strand-seq binning start with a collapsed assembly and have the same issues as Falcon-Unzip.
In 2012, we reasoned17 that a perfectly constructed unitig graph with read information is a lossless representation of single-end reads. Because this graph is lossless, we can compress input reads into a unitig graph and perform phasing later. This should maximize the power of long HiFi reads. Developed in parallel to our work, HiCanu18 follows a similar rationale and can produce Falcon-Unzip-style primary/alternate assemblies better than other assemblers especially around segmental duplications. However, HiCanu only tries to keep the contiguity of one parental haplotype and often breaks the contiguity of the other haplotype. When we separate parental haplotypes, these break points will lead to fragmented haplotype-resolved assemblies.
In this article we present hifiasm, an assembler for HiFi reads that generates a well-connected assembly graph and produces better assemblies in practice. We will first give an overview of the hifiasm algorithm, compare it to other assemblers for partially phased assemblies and then explain and evaluate the haplotype-resolved assembly algorithm used by hifiasm.
Results
Overview of the hifiasm algorithm
The first few steps of hifiasm broadly resemble the workflow of early long-read assemblers1,2 (Fig. 1). Hifiasm performs all-vs-all read overlap alignment and then corrects sequencing errors. Given a target read to be corrected, hifiasm inspects the alignment of reads overlapping with the target read. A position on the target read is said to be informative if there are two types of A/C/G/T bases (gaps ignored) at the position in the alignment and each type is supported by at least three reads. A read overlapping with the target read is inconsistent with the target if there are informative positions in the overlap and the read is not identical to the target read across all these positions; accordingly, the overlap between this and the target read is inconsistent. Inconsistent reads theoretically originate from a haplotype different from the target read. Hifiasm only uses consistent reads to correct the target read.
Figure 1: Outline of the hifiasm algorithm.
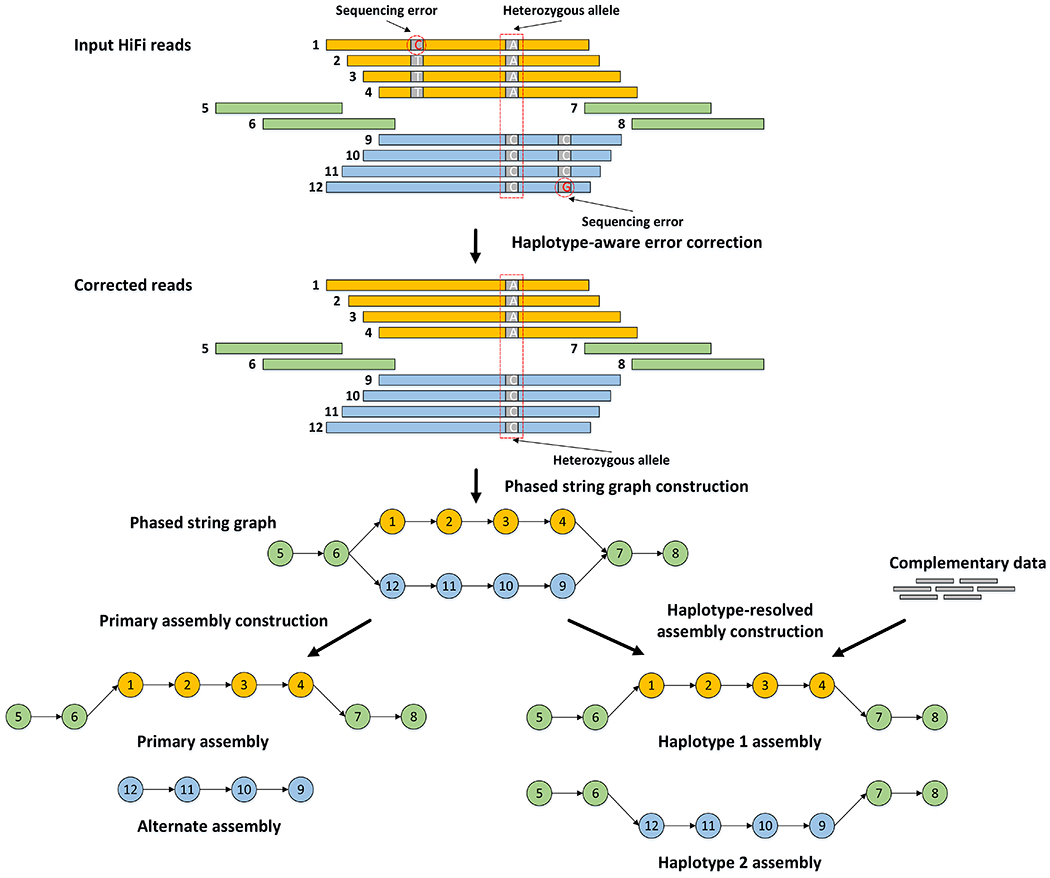
Orange and blue bars represent the reads with heterozygous alleles carrying local phasing information, while green bars come from the homozygous regions without any heterozygous alleles. In phased string graph, a vertex corresponds to the HiFi read with same ID, and an edge between two vertices indicates that their corresponding reads are overlapped with each other. Hifiasm first performs haplotype-aware error correction to correct sequence errors but keep heterozygous alleles, and then builds phased assembly graph with local phasing information from the corrected reads. Only the reads coming from the same haplotype are connected in the phased assembly graph. With complementary data providing global phasing information, hifiasm generates a completely phased assembly for each haplotype from the graph. Hifiasm also can generate unphased primary assembly only with HiFi reads. This unphased primary assembly represents phased blocks (regions) that are resolvable with HiFi reads, but does not preserve phasing information between two phased blocks.
Hifiasm performs three rounds of error correction by default. It then does overlap alignment again and builds a string graph19 where a vertex is an oriented read and an edge is a consistent overlap. After transitive reduction, a pair of heterozygous alleles will be represented by a “bubble” in the string graph (Fig. 1). No information is lost. If there are no additional data, hifiasm arbitrarily selects one side of each bubble and outputs a primary assembly similar to Falcon-Unzip and HiCanu. For a heterozygous genome, the primary assembly generated at this step may still contain haplotigs from more than one homologous haplotypes. HiCanu relies on third-party tools such as purge_dups20 to remove redundant haplotigs. Hifiasm natively implements a variant of the purge_dups algorithm. This simplifies the assembly pipeline.
If parents of the sample are also sequenced, hifiasm can use k-mer trio binning11 to label corrected reads in the string graph. In this case, hifiasm effectively discards the maternal unitigs to generate the paternal assembly, and vice versa. This graph-based trio binning may go through regions heterozygous in all three samples in the trio and is more robust to the mislabeling of reads. We will explain the advantage of hifiasm binning in a later section.
Assembling homozygous non-human genomes
We first evaluated hifiasm v0.12 along with Falcon-Unzip10 v1.8.1, Peregrine6 v0.1.6.1 and HiCanu18 v2.1 on two inbred samples23 including the C57/BL6J strain of M. musculus (mouse) and the B73 strain of Z. mays (maize). All assemblers produced long contigs for mouse (Table 1). To evaluate how often assemblers collapse paralogous regions and produce misassemblies, we mapped HiFi reads to each assembly, extracted apparently heterozygous SNPs at high coverage and clustered them into longer regions (Online Methods). These regions correspond to collapsed misassemblies. We identified 4 such misassemblies in the HiCanu assembly, 6 in hifiasm and more than 100 in both Falcon and Peregrine. HiCanu is the best at this metric although its contig N50 is the shortest.
Table 1.
Statistics of non-human assemblies
| Dataset | Assembler | Size (Gb) | N50 (Mb) | NG50 (Mb) | Alternate size (Gb) | Completeness (asmgene or BUSCO) |
|
|---|---|---|---|---|---|---|---|
| Complete (%) | Duplicated (%) | ||||||
| M. musculus (25×) | hifiasm | 2.610 | 21.1 | 20.6 | 0.044 | 99.73 | 0.23 |
| HiCanu | 2.594 | 16.0 | 14.8 | 0.077 | 99.68 | 0.22 | |
| Peregrine | 2.578 | 17.9 | 17.0 | 0.029 | 99.56 | 0.21 | |
| Falcon | 2.559 | 19.3 | 16.7 | 0.025 | 99.49 | 0.14 | |
| Z. mays (22×) | hifiasm | 2.190 | 37.5 | 37.5 | 0.095 | 99.85 | 0.17 |
| HiCanu | 2.145 | 27.1 | 24.1 | 0.040 | 99.84 | 0.13 | |
| Peregrine | 2.205 | 10.1 | 10.2 | 0.038 | 99.88 | 0.26 | |
| Falcon | 2.132 | 9.5 | 9.3 | 0.016 | 99.77 | 0.17 | |
| F. × ananassa (36×) | hifiasm (purge) | 0.829 | 17.6 | 17.6 | 0.458 | 98.45 | 93.43 |
| HiCanu | 1.044 | 8.4 | 9.8 | 0.295 | 98.08 | 92.94 | |
| HiCanu (purge) | 0.411 | 10.5 | 0.0 | 0.928 | 96.78 | 55.08 | |
| Peregrine | 0.930 | 5.5 | 6.7 | 0.260 | 98.33 | 91.70 | |
| Falcon | 0.971 | 5.4 | 7.3 | 0.213 | 98.27 | 92.81 | |
| R. muscosa (~29×) | hifiasm (purge) | 9.664 | 9.1 | 7.208 | 66.61 | 1.70 | |
| HiCanu | 9.645 | 5.2 | 6.361 | 65.54 | 3.92 | ||
| Peregrine | 9.415 | 0.9 | 2.936 | 66.84 | 1.72 | ||
| S. sempervirens (~33×) | hifiasm (purge) | 35.310 | 5.5 | 15.757 | 61.31 | 39.42 | |
| Peregrine | 35.662 | 0.8 | 63.20 | 35.93 | |||
HiCanu (purge) applies purge_dups to a HiCanu assembly. Hifiasm (purge) enables the built-in purge_dups equivalent strategy. The N50/NG50 of an assembly is defined as the sequence length of the shortest contig at 50% of the total assembly/genome size. To calculate the NG50, a genome size of 2730.9 Mb (AC:GCF_000001635.20), 2182.1 Mb (AC:GCA_902167145.1) and 813.4 Mb21 is used for M. musculus, Z. mays and F. × ananassa, respectively. The genome size is unknown for R. muscosa and S. sempervirens. “Alternate size” is the total length of the alternate assembly. The reference-based asmgene method22 is used to evaluate the gene completeness of M. musculus and Z. mays which have high-quality reference genomes. For these two samples, “Complete” gives the percentage of single-copy genes in the reference genome (one unique mapping at ≥97% identity) that are mapped at ≥97% identity to the assembly; “Duplicated” gives the percentage of reference single-copy genes that become multi-copy in the assembly. The BUSCO embryophyta dataset is used to evaluate the gene completeness of F. × ananassa and S. sempervirens; the tetrapoda dataset is used for R. muscosa. BUSCO scores of all samples can be found in Supplementary Table 7.
For the repeat-rich maize genome, hifiasm and HiCanu generated longer contigs and again produced much fewer collapsed misassemblies. There are 3 collapsed misassemblies in the hifiasm assembly and 9 in HiCanu, versus more than 100 in Falcon and Peregrine. Hifiasm and HiCanu perform better presumably because they can more effectively resolve repeats by requiring near perfect overlap18.
Assembling heterozygous non-human genomes
Since most natural samples are heterozygous, we next evaluated the assemblers on three heterozygous datasets from F. × ananassa (garden strawberry), R. muscosa (mountain yellow-legged frog) and S. sempervirens (California redwood). These samples are more challenging to assemble. F. × ananassa has an allopolyploid genome estimated to be 813.4 Mb in size21. All assemblers achieved a total assembly of ~1.2 Gb, including both primary and alternate contigs. However, they resolved the primary assembly differently. Hifiasm resulted in a primary assembly of similar size to the published genome. BUSCO24 regarded most single-copy genes to be duplicated, consistent with the previous observation21. HiCanu assigned most contigs to the primary. Applying purge_dups20 overcompressed the assembly and reduced the BUSCO completeness by 5%. Falcon-Unzip and Peregrine are somewhat between hifiasm and HiCanu. The varying primary assembly sizes highlight the difficulty in assembling polyploid genomes. On the other hand, all HiFi assemblies here have much longer contig N50 than the published assembly (>5 Mb vs 580 kb). HiFi enables better assembly.
R. muscosa is hard to assemble for its large genome size. We failed to run Falcon-Unzip for this sample using its released version. We did not apply purge_dups to the HiCanu assembly as it could not finish in 15 days. Without the purging step, the HiCanu assembly contains a higher rate of duplicated genes. The N50 of the hifiasm assembly is almost twice as long as the HiCanu assembly.
S. sempervirens poses an even greater challenge to assembly with a much larger hexaploid genome. Hifiasm took 875 Gb reads as input and produced a 35.6 Gb assembly in 3 days over 80 CPU threads using ~700 GB memory at the peak (Supplementary Table 10). The flow cytometric estimate of the full hexaploid genome is 62.8 Gb in size25. Our assembly is about half of that. Peregrine achieved a 35.6 Gb assembly as well. Its BUSCO score is 1.9% better than the hifiasm assembly. Peregrine took 15 days on a computer cluster. It ran slower and its assembly is more fragmented. Hifiasm overall performs better on large genomes.
Primary assembly of human genomes
We next evaluated hifiasm and other assemblers on three human datasets (Table 2). We introduced two new metrics, “multi-copy genes retained” and “resolved BACs” to evaluate how assemblers resolve difficult genomic regions such as long segmental duplications. If an assembler breaks contigs at such regions or misassemblies the regions, the resulting assembly will lose multi-copy genes and/or lead to fragmented BAC-to-contig alignment.
Table 2.
Statistics of human primary assemblies
| Dataset | Assembly | Size (Gb) | NG50 (Mb) | NGA50 (Mb) | QV | Multi-copy genes retained (%) | Resolved BACs (%) | Gene completeness (asmgene) |
|
|---|---|---|---|---|---|---|---|---|---|
| Complete (%) | Duplicated (%) | ||||||||
| CHM13 (HiFi 32×) | hifiasm | 3.052 | 88.9 | 86.7 | 54.2 | 99.7 | 98.8 | 99.97 | 0.05 |
| HiCanu | 3.037 | 69.7 | 67.9 | 54.1 | 98.9 | 97.6 | 99.97 | 0.04 | |
| Peregrine | 2.990 | 37.8 | 33.4 | 43.8 | 51.1 | 39.7 | 99.64 | 0.16 | |
| Falcon | 2.862 | 27.1 | 21.8 | 50.1 | 30.2 | 34.2 | 99.47 | 0.03 | |
| (ONT 120×) | Canu | 2.936 | 80.0 | 47.3 | 32.7 | 76.9 | 86.7 | 99.30 | 0.10 |
| Flye | 2.900 | 37.5 | 34.0 | 33.5 | 54.7 | 60.6 | 99.22 | 0.11 | |
| Shasta | 2.820 | 41.3 | 33.4 | 30.4 | 26.7 | 27.9 | 98.05 | 0.01 | |
| HG00733 (HiFi 33×) | hifiasm (purge) | 3.043 | 68.3 | 55.3 | 49.9 | 74.6 | 80.4 | 99.07 | 0.39 |
| HiCanu (purge) | 2.921 | 40.5 | 34.2 | 50.5 | 55.2 | 65.9 | 98.47 | 0.32 | |
| Peregrine | 3.035 | 30.1 | 30.1 | 40.5 | 37.2 | 38.5 | 98.70 | 0.31 | |
| Falcon | 2.861 | 24.4 | 23.2 | 46.3 | 33.6 | 38.0 | 96.51 | 0.15 | |
| (ONT 50×) | Canu | 2.923 | 41.1 | 36.6 | 29.5 | 54.6 | 69.3 | 98.32 | 0.66 |
| Flye | 2.890 | 26.7 | 25.4 | 29.9 | 34.2 | 44.7 | 97.88 | 0.20 | |
| Shasta | 2.805 | 21.2 | 20.8 | 30.0 | 17.0 | 22.9 | 97.19 | 0.05 | |
| HG002 (HiFi 36×) | hifiasm (purge) | 3.067 | 98.2 | 64.1 | 51.5 | 75.8 | 99.26 | 0.32 | |
| HiCanu (purge) | 2.953 | 48.3 | 39.4 | 52.1 | 59.7 | 98.71 | 0.18 | ||
| Peregrine | 3.081 | 33.4 | 32.5 | 41.3 | 42.5 | 99.14 | 0.36 | ||
| Falcon | 2.955 | 30.4 | 29.0 | 46.7 | 36.6 | 99.00 | 0.20 | ||
Polished ONT assemblies were generated by the Shasta developers8. HiCanu and hifiasm were run without duplication purging for the homozygous CHM13 cell line, and run with purging for the heterozygous HG00733 and HG002 cell lines. The NGA50 of an assembly is defined as the length of the correctly aligned block at 50% of the total reference genome size which is assumed to be 3.1 Gb. It was calculated based on the minigraph26 contig-to-reference alignment. The “QV” (quality value) equals the Phred-scaled contig base error rate measured by comparing 31-mers in contigs to 31-mers in short reads from the same sample. Percent “multi-copy genes retained” is reported by asmgene (Online Methods). It is the percentage of multi-copy genes in reference genome (multiple mapping positions at ≥99% sequence identity) that remain multi-copy in the assembly. A BAC is resolved if 99.5% of its bases can be mapped the assembly. There are 330 CHM13-specific BACs excluding those not resolved by the telomere-to-telomere (T2T) assembly, and there are 179 HG00733-specific BACs. HG002 does not have BAC data. Throughout the table, GRCh38 is used as the reference genome for HG00733 and HG002, and the T2T CHM13 assembly v0.9 is used as the reference for CHM13.
CHM13 is a homozygous cell line, similar to M. musculus and Z. mays. The telomere-to-telomere (T2T) consortium produced a near complete assembly for this sample with multiple data types and manual curation. Taking the T2T assembly as the ground truth, QUAST27 reported 349 misassemblies in the HiCanu assembly, 476 in the hifiasm assembly and more than 1500 misassemblies in others (Supplementary Table 1). HiCanu is the best in terms of the number of misassemblies. Nonetheless, it has shorter NG50, misses more multi-copy genes and resolves fewer BACs in comparison to hifiasm. Hifiasm and HiCanu are broadly comparable. Both of them are better than Peregrine, Falcon and ONT assemblies on all metrics by a large margin.
QUAST often takes structural variations (SVs) as misassemblies. Taking GRCh38 as the reference, it reported 23,541 misassemblies for the T2T assembly, greatly overestimating assembly errors. We thus did not apply QUAST to HG00733 and HG002 where the ground truth is missing. Instead, we inferred NGA50 based on minigraph alignment that can go through most SVs (Table 2). This more accurately measures large-scale misassemblies.
For these two heterozygous samples, HiCanu produced primary assemblies with several hundred megabases of heterozygous regions represented twice. We thus ran purge_dups20 to remove these falsely duplicated regions in the primary assembly. We tried a few purge_dups settings, including the default, and chose the one that gave the best primary assembly. Hifiasm can identify and remove falsely duplicated regions by inspecting inconsistent read overlaps between them. The other assemblers collapsed most heterozygous regions during assembly. They do not need additional tools like purge_dups, either.
For HG00733 and HG002, hifiasm and HiCanu consistently outperformed other assemblers. The hifiasm assembly was more complete and resolved more difficult regions than HiCanu. This difference probably has more to do with the duplicate purging algorithm than with the capability of the assembler. Nonetheless, this observation suggests it is easier to produce a high-quality primary assembly with hifiasm.
On running time, hifiasm took 7–9 wall-clock hours over 48 threads (Supplementary Table 10). The peak memory was below 150 GB. Peregrine was about twice as fast for human assembly but used more memory. HiCanu was about 7–8 times as slow as hifiasm using the same machine. Falcon was the slowest.
Improving haplotype-resolved assembly
A major issue with trio binning is that a fraction of heterozygous reads cannot be unambiguously partitioned to parental haplotypes: if both parents are heterozygous at a locus, a child read will harbor no informative k-mers and cannot be uniquely assigned to a parental haplotype; if, say, the father is heterozygous at a locus and the mother is homozygous, reads from the maternal haplotype cannot be partitioned, either. With standard trio binning, heterozygous reads that cannot be partitioned will be used in both parental assemblies. As a result, both alleles may be present in one haplotype assembly and lead to false duplications. Standard trio binning is unable to cleanly separate the two parental haplotypes.
Hifiasm draws power from HiFi read phasing in addition to trio binning. It does not partition reads upfront; it only labels reads in the string graph. In a long bubble representing a pair of heterozygous alleles, hifiasm may correctly phase it even if only a small fraction of reads are correctly labeled. This way hifiasm also rarely puts two alleles in one haplotype assembly.
Hi-C or Strand-seq based phasing13,14 can unambiguously phase most heterozygous reads and are naturally immune to false duplications. They however suffer from another issue shared by standard trio phasing: reads assigned to a wrong parental haplotype may break contigs (Fig. 2). By considering HiFi read phasing and the structure of the assembly graph, hifiasm may be able to identify and fix such binning errors.
Figure 2: Effect of false read binning.
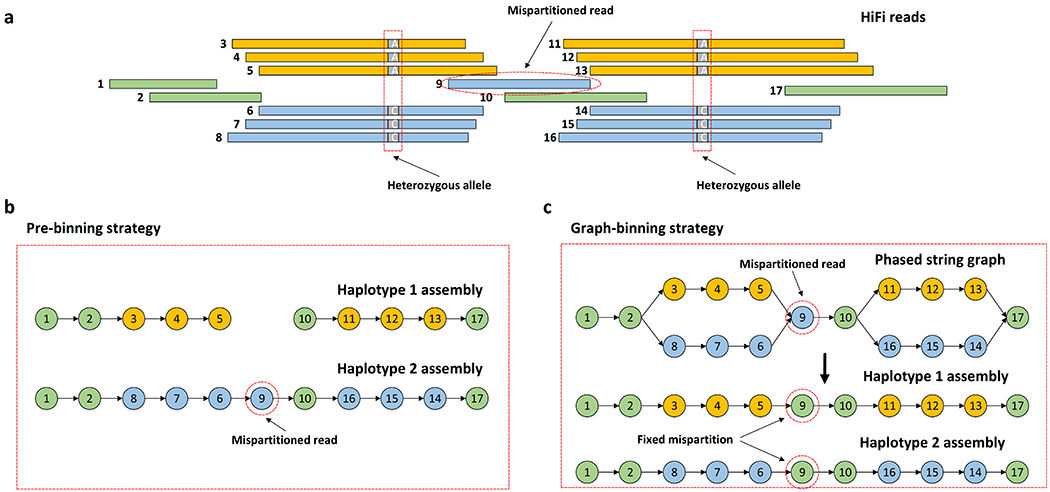
(a) A set of reads with global phasing information provided by the complementary data. Reads in orange and reads in blue are specifically partitioned into haplotype 1 and haplotype 2, respectively. The remaining reads in green are partitioned into both haplotypes. Read 9 without heterozygous alleles is mispartitioned into haplotype 2, instead of to both haplotypes. (b) Pre-binning assemblies produced by current methods which independently assemble two haplotypes. Haplotype 1 is broken into two contigs due to the mispartition of read 9. (c) Hifiasm fixes the mispartition by the local phasing information in the phased assembly graph. It is able to identify that read 9 does not have heterozygous alleles, so that read 9 should be partitioned into both haplotypes.
Haplotype-resolved assembly of heterozygous human genomes
To evaluate how well assemblers resolve both haplotypes, we applied trio binning assembly to HG00733 and HG002. Hifiasm performs graph trio binning that partitions a diploid assembly graph to generate the final assembly. HiCanu does standard trio binning11 that partitions HiFi reads upfront and assembles the two parental partitions separately. Peregrine does not natively support trio binning. We fed the HiCanu-partitioned reads to Peregrine for assembly. For comparison, we also acquired a Strand-seq HG00733 assembly14 and a Hi-C HG00733 assembly13 that use the same HiFi reads but are supplemented with additional data types for phasing.
On both datasets, trio hifiasm missed fewer variants and emitted longer contigs with higher QV and lower variant FDR than other assembly strategies (Table 3). The HiCanu contig NG50 was the shortest, which is probably caused by wrongly partitioned reads (Fig. 2) in combination with HiCanu’s strict requirement of exact overlapping. By collapsing inexact overlaps, Peregrine is more robust to partition errors in certain cases and can achieve longer contigs. However, this comes with the cost of fewer resolved BACs and increased FNR. The Strand-seq and Hi-C assemblies also use Peregrine and are affected by false read partitions in the same way. These two assemblies were not as good as hifiasm. It is not possible to get a good all-around assembly if we perform separate assemblies on pre-partitioned reads.
Table 3.
Statistics of haplotype-resolved human assemblies
| Dataset | Assembly | Size (Gb) | QV | NG50 (Mb) | Multi-copy genes retained (%) | Resolved BACs (%) | Switch error (%) | Hamming error (%) | FNR (%) | FDR (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| HG00733 | hifiasm (trio) | 6.071 | 49.9 | 34.9 | 84.0 | 95.5 | 0.08 | 0.22 | 2.43 | |
| HiCanu (trio) | 6.079 | 49.2 | 10.6 | 84.3 | 90.5 | 0.04 | 0.04 | 4.78 | ||
| Peregrine (trio) | 5.938 | 42.2 | 19.1 | 37.6 | 39.7 | 0.10 | 0.23 | 12.34 | ||
| Peregrine (Hi-C) | 5.867 | 41.6 | 26.1 | 33.2 | 35.2 | 0.12 | 0.67 | 3.31 | ||
| Peregrine (Strand-seq) | 5.805 | 45.8 | 26.6 | 33.0 | 46.9 | 0.18 | 0.72 | 3.99 | ||
| HG002 | hifiasm (trio) | 5.967 | 51.6 | 43.0 | 80.6 | 0.79 | 0.34 | 0.88 | 0.26 | |
| HiCanu (trio) | 6.003 | 50.4 | 12.1 | 80.4 | 0.75 | 0.19 | 1.57 | 0.32 | ||
| Peregrine (trio) | 5.888 | 42.7 | 25.8 | 38.7 | 0.70 | 0.18 | 4.42 | 4.18 | ||
Parental assemblies are merged together for computing QV, NG50 and BACs resolved. Calculating NG50 assumes a diploid human genome size of 6.2 Gb. Phased variants are called with dipcall28 for each pair of parental assemblies and are compared to HG002 truth variants from GIAB29 or HG00733 phased SNPs from HGSVC30. Phasing switch error rate: percent adjacent SNP pairs that are wrongly phased. Phasing hamming error rate: percent SNP sites that are wrongly phased. False negative rate (FNR): percent true variants that are missed in the assembly. False discovery rate (FDR): percent assembly-based variant calls that are not called in the truth data. RTG’s vcfeval31 is used for estimating variant FNR and FDR for HG002. For HG00733, FNR is estimated at heterozygous SNP sites only; FDR is not available because HGSVC does not provide confident regions. Percent “multi-copy genes retained” measures the percentage of multi-copy genes in GRCh38 (multiple mapping positions at ≥99% sequence identity) that remain multi-copy in the assembly, averaged between the two parental haplotypes. Gene completeness (asmgene) can be found in Supplementary Table 2.
The Human Leukocyte Antigen (HLA) region is highly heterozygous and enriched with complex variations. For HG002, we compared each assembly to the haplotype-resolved ground truth of the same sample32 (Supplementary Table 9). While hifiasm fully reconstructed both haplotypes, HiCanu failed to assemble paternal haplotype and peregrine likely introduced misassemblies in both haplotypes. These results are consistent with the BAC resolution of HG00733 assemblies (Table 3), showing that trio hifiasm can more effectively resolve hard regions.
Discussion
Hifiasm is a fast open-source de novo assembler specifically developed for HiFi reads. It mostly uses exact overlaps to construct the assembly graph and can separate different alleles or different copies of a segmental duplication involving a single segregating site. This greatly enhances its power for resolving near identical, but not exactly identical repeats and segmental duplications. In our evaluation, hifiasm consistently outperforms Falcon and Peregrine which do not take the advantage of exact overlaps.
In comparison to HiCanu which is developed in parallel to our work, hifiasm is able to generate a more complete assembly graph preserving all haplotypes more contiguously. This enables us to implement a graph trio binning algorithm that can produce a haplotype-resolved assembly tripling the contig N50 of a trio HiCanu assembly. Hifiasm can generate overall the best haplotype-resolved human assemblies so far.
Our graph binning algorithm can also work with reads labeled by Hi-C or Strand-seq binning that do not require parental data. However, because existing Hi-C or Strand-seq binning algorithms start with a collapsed assembly, they may not work well with highly heterozygous regions not represented well in the initial assembly. In our view, a better solution to pedigree-free phased assembly is to map Hi-C or Strand-seq data to the hifiasm assembly graph, group and order unitigs into chromosome-long scaffolds with the graph topology, and then phase heterozygous events along the scaffolds. We envision that haplotype-resolved assembly will become a common practice for both human and diploid non-human species, though haplotype-resolved assembly may remain challenging for polyploid plants in the near future.
Online Methods
Haplotype-aware error correction.
Hifiasm loads all reads into memory and performs all-vs-all pairwise alignment between them. For each read R, hifiasm effectively builds an approximate multi-sequence alignment from the pairwise alignment between R and each of its overlapping reads. Hifiasm then identifies positions on R at which there are two types of A/C/G/T bases in the alignment with each type supported by at least three overlapping reads. These positions inform base pair differences between haplotypes and are thus called informative positions. If R and its overlapping read Q are identical across all informative positions in the overlap, Q is regarded to come from the same haplotype as R. Hifiasm collects reads that are inferred to be on the same haplotype as R and use them to correct R with an algorithm similar to Falcon1, 10.
All-vs-all pairwise alignment is the major performance bottleneck in this step. Hifiasm uses a windowed version of Myers’ bit-vector algorithm33 to perform the base alignment. Instead of computing the alignment over the entire overlap, hifiasm splits read R into non-overlapping windows and does pairwise alignment in each window. This enables us to simultaneously align multiple windows using the SSE instructions34. In practice, one potential issue with windowing is that the alignment around window boundaries may be unreliable. To alleviate the issue, hifiasm realigns the subregion around the window boundary if it sees mismatches or gaps within 20bp around the boundary.
Constructing phased assembly graphs.
After haplotype-aware error correction, most sequencing errors have been removed while the marker positions are still kept. With nearly error-free reads, hifiasm is able to perform phasing accurately to determine if one overlap is among the reads coming from different haplotypes (i.e. inconsistent overlap). The next step is to build the assembly string graph3, 19. In this graph, nodes represent oriented reads and each edge between two nodes represents the overlap between the corresponding two reads. Note that only consistent overlaps are used to build the graph. Since hifiasm builds the graph on top of nearly error-free reads and highly accurate haplotype phasing, the produced assembly graph of hifiasm is simpler and cleaner than those of current assemblers for haploid genomes. However, for diploid genomes or polyploid genomes, its graph becomes more complicated as reads from different haplotypes are clearly separated out by phasing. Fig. 1 gives an example. Since there is a heterozygous allele on reads in orange and blue, hifiasm separates them into two groups in which all reads in the same color belong to one group. Only the reads from same group are overlapped with each other. For reads in green, they are overlapped with the reads in both groups because the overlaps among them are not long enough to cover at least one heterozygous allele. As a result, hifiasm generates a bubble in the assembly graph. A bubble is a subgraph consisting of a single source node v and a single sink node w with more than one path between v and w, and all nodes in this bubble except v and w do not connect to the rest of the whole graph. Most existing assemblers aim to produce one contiguous contig from the graph (i.e. single path in the graph) as much as possible. They tend to collapse bubbles when building the assembly graph. As a result, they will lose all but one allele in each bubble. In contrast, hifiasm is designed to retain all bubbles on the assembly graph. Owing to the fact that there are still a few errors at the corrected reads, hifiasm adopts a topological-aware graph cleaning strategy. It first identifies substructures embedding local phasing information like bubbles, and then only cuts too short overlaps outside these substructures. Hifiasm additionally records the inconsistent overlaps, which are helpful in the following assembly construction steps.
Constructing a primary assembly.
The construction of the primary assembly aims to produce contigs including one set of haplotypes but may switch subregions between haplotypes. In other words, each subregion in the primary assembly only comes from one haplotype, while the corresponding subregions of other haplotypes are removed as duplications. In this step, most existing assemblers follow the “best overlap graph” strategy or its variants35. Their key idea is to retain longer overlaps if there are multiple overlaps to a given read. In contrast, hifiasm produces a primary assembly mainly relied on the graph topological structures and the phasing relationship among different haplotypes. Ideally, the phased assembly graph of hifiasm should be a chain of bubbles for diploid genomes (Fig. 2c). It is easy and reliable to extract primary assembly from such chain of bubbles by bubble popping3. However, there are still tips (i.e. deadend contigs broken in single end) on the assembly graph caused by broken bubbles due to lack of coverage, phasing errors or unresolvable repeats. To fix this problem, hifiasm proposes a three-stage procedure (Supplementary Fig. 1). First, each bubble in the graph is reduced into a single path using bubble popping. This step removes most duplicated subregions on different haplotypes without hampering the contiguity of primary assembly. Second, given a tip unitig T that is broken in one end but connected to a unitig C in another end, hifiasm checks if there are other unitigs, which are also connected to C, coming from the different haplotypes of T. If such unitigs are identified, hifiasm removes tip T so that unitig C will become longer. The reason is that for T, its corresponding region from different haplotype has already been integrated into the new longer unitig C. Since hifiasm records overlaps between haplotypes (i.e. inconsistent overlaps), it can check if two unitigs come from different haplotypes. Last, hifiasm uses the “best overlap graph” strategy to deal with a few remaining unresolvable hard substructures on the assembly graph. In most cases, the graph topological information and the phasing information is more reliable than only keeping the longer overlaps. As a result, hifiasm is able to generate a better primary assembly than current assemblers which mainly rely on “best overlap graph” strategy.
Constructing a haplotype-resolved assembly.
The phased assembly graph in hifiasm embeds the local phasing information that is resolvable with HiFi reads. In this graph, the corresponding node of a homozygous read is at a single path connecting two bubbles, while the corresponding node of a heterozygous read is at a bubble (Fig. 2). Given parental short reads, hifiasm labels child HiFi reads with the existing k-mer based algorithm11. When generating a fully phased assembly for one haplotype, hifiasm drops reads of different haplotypes from the graph, while using the local phasing information in graph to correct the mispartition of global phasing. Hifiasm does not drop reads at a single path connecting two bubbles, since these are homozygous reads that must be contained in both haplotypes. For a bubble in which all reads are heterozygous, hifiasm applies bubble popping to select a single best path consisting of most reads with the expected haplotype label. If a few reads are assigned false labels by global phasing, they are likely to be corrected by the best path that traverses through them. In addition, instead of dropping any read with non-expected haplotype label, hifiasm drops a contig if the haplotype labels of most reads in it are non-expected.
Purging heterozygous duplications.
In the primary assembly construction step, accurately keeping one set of haplotypes is more challenging for haplotype-resolved assemblers. Although the bubble popping method and the tip removing method of hifiasm already purge large numbers of duplications from multiple haplotypes, some duplications still remain on the primary assembly, especially for subregions with a high heterozygosity rate. Existing assemblers postprocess the primary assembly using downstream tools like purge_dups20, which identify duplications by inexact all-vs-all contig alignment. If two contigs overlap with each other, the overlapped regions between them are duplications. However, inexact contig alignment might be not reliable on segmental duplications or repeats, leading to more duplications left or overpurged repetitive regions. To address this duplication challenge, hifiasm re-assembles the contigs by building a string graph regarding contigs as nodes, called a purge graph. Given contig A and contig B, we define A inconsistently overlaps B if there are enough reads of A that are inconsistently overlapped with the reads of B. Note that hifiasm records all inconsistent overlaps among reads in the initial phased assembly graph construction step by haplotype phasing. In the purge graph of hifiasm, each node is a contig, while an edge between two nodes is an inconsistent overlap between their corresponding contigs. Once the graph is built, hifiasm generates the non-redundant primary assembly by simple graph cleaning. As a result, the built-in purge duplication step of hifiasm is smoother and more reliable than existing downstream tools. This is because hifiasm identifies duplications from multiple haplotypes using accurate haplotype phasing of each read, while existing tools mainly rely on inexact contig alignment.
Evaluating collapsed misassemblies for inbred samples.
We mapped HiFi reads with minimap222 to each assembly and then called apparent heterozygous SNPs with htsbox, a fork of samtools. We selected biallelic SNPs such that each allele is supported by d reads where d is set to 75% of the average coverage of the sample. We then hierarchically cluster these apparent SNPs28 as follows: we merge two adjacent SNP clusters if (1) the minimum distance between them is within 10kb and (2) the density of SNPs in the merged cluster is at least 1 per 1kb. A cluster longer than 5kb and consisting of ≥10 SNPs is identified as a collapsed misassembly. Varying the thresholds changes the number of estimated misassemblies but does not alter the relative ranking between assemblers.
Evaluating gene completeness with asmgene.
BUSCO24 is a popular tool for evaluating gene completeness. It is very helpful for new species, but is underpowered for species with high-quality reference genomes. For example, BUSCO reports that the completeness of GRCh38 is only 94.8%, lower than the 95.2% percent completeness of the male HG002 hifiasm assembly (Supplementary Table 5).
In order to quantify gene completeness more accurately, we used the paftools script from the minimap2 package22 to calculate the asmgene scores. Unlike BUSCO, asmgene relies on a reference genome. It uses minimap2 to align EnsEMBL cDNAs (v99 for human and mouse and v47 for maize) to a reference genome or an assembly. For each transcript, asmgene records a hit if the transcript is mapped at ≥99% identity (≥97% for non-human species due to their higher diversity) over ≥99% of the transcript length. A transcript is considered to be single-copy (SC) if it has only one hit; otherwise it is considered to be multi-copy (MC). The asmgene script chooses the longest transcript to represent a gene. In Table 1 and 2, percent “Complete” equals |{SCorMCinASM}∩{SCinREF}|/|{SCinREF}|, where {SCinREF} denotes the set of genes single-copy in the reference genome and {SCorMCinASM} denotes the union sets of single- and multi-copy genes in the assembly. Similarly, percent “Duplicated” equals |{MCinASM}∩{SCinREF}|/|{SCinREF}|. In Table 2, percent “Multi-copy gene retained” is calculated by |{MCinASM}∩{MCinREF}|/|{MCinREF}|.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
This study was supported by US National Institutes of Health (grant R01HG010040, U01HG010971 and U41HG010972 to H.L.).
Footnotes
Competing interests
G.T.C. is an employee of Pacific Biosciences. H.L. is a consultant of Integrated DNA Technologies, Inc and on the Scientific Advisory Boards of Sentieon, Inc, BGI and OrigiMed.
Data availability
All HiFi data were obtained from NCBI Sequence Read Archive (SRA): SRR11606869 for Z. mays, SRR11606870 for M. musculus, SRR11606867 for F. × ananassa, SRR11606868 and SRR12048570 for R. muscosa, SRP251156 for S. sempervirens, SRR11292120 through SRR11292123 for CHM13, ERX3831682 for HG00733, and four runs (SRR10382244, SRR10382245, SRR10382248 and SRR10382249) for HG002. For trio binning and computing QV, short reads were also downloaded: SRR7782677 for HG00733, ERR3241754 for HG00731 (father), ERR3241755 for HG00732 (mother) and SRX1082031 for CHM13. GIAB’s “Homogeneity Run01” short-read runs were used for the HG002 trio. These HG002 reads were downsampled to 30-fold coverage. The BAC libraries of CHM13 and HG00733 can be found at https://www.ncbi.nlm.nih.gov/nuccore/?term=VMRC59+and+complete and https://www.ncbi.nlm.nih.gov/nuccore/?term=VMRC62+and+complete, respectively. The HG002 MHC reference sequences can be found at https://github.com/NCBI-Hackathons/TheHumanPangenome/tree/master/MHC/assembly/MHCv1.132. For BUSCO, the embryophyta dataset, the tetrapoda dataset and the mammalia dataset are available at https://busco-data.ezlab.org/v4/data/lineages/embryophyta_odb10.2020-09-10.tar.gz, http://busco.ezlab.org/v2/datasets/tetrapoda_odb9.tar.gz and http://busco.ezlab.org/v2/datasets/mammalia_odb9.tar.gz, respectively. The CHM13 reference (v0.9) generated by the T2T consortium can be found at https://s3.amazonaws.com/nanopore-human-wgs/chm13/assemblies/chm13.draft_v0.9.fasta.gz. The hifiasm assemblies produced in this work are available at ftp://ftp.dfci.harvard.edu/pub/hli/hifiasm/submission/.
Code availability
Hifiasm is available at https://github.com/chhylp123/hifiasm.
References
- 1.Chin C-S et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Berlin K et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol 33, 623–630 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Li H Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koren S et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolmogorov M, Yuan J, Lin Y & Pevzner PA Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Chin C-S & Khalak A Human Genome Assembly in 100 Minutes. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/705616v1 (2019). [Google Scholar]
- 7.Ruan J & Li H Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17, 155–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafin K et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol 38, 1044–1053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y et al. Fast and accurate assembly of Nanopore reads via progressive error correction and adaptive read selection. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.02.01.930107v1 (2020). [Google Scholar]
- 10.Chin C-S et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13,1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koren S et al. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol 36, 1174–1182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wenger AM et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garg S et al. Chromosome-scale, haplotype-resolved assembly of human genomes. Nat. Biotechnol (2020). 10.1038/s41587-020-0711-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porubsky D et al. Fully phased human genome assembly without parental data using single-cell strand sequencing and long reads. Nat. Biotechnol (2020). 10.1038/s41587-020-0719-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M et al. WhatsHap: fast and accurate read-based phasing. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/085050v2 (2016). [Google Scholar]
- 16.Edge P, Bafna V & Bansal V HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res 27, 801–812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28, 1838–1844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurk S et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res 30, 1291–1305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers EW The fragment assembly string graph. Bioinformatics 21, ii79–ii85 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Guan D et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896–2898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edger PP et al. Origin and evolution of the octoploid strawberry genome. Nat. Genet 51, 541–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hon T et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data 7, 399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV & Zdobnov EM BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Hizume M, Kondo T, Shibata F & Ishizuka R Flow Cytometric Determination of Genome Size in the Taxodiaceae, Cupressaceae sensu stricto and Sciadopityaceae. CYTOLOGIA 66, 307–311 (2001). [Google Scholar]
- 26.Li H, Feng X & Chu C The design and construction of reference pangenome graphs with minigraph. Genome Biol 21, 265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich A, Saveliev V, Vyahhi N & Tesler G QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat. Methods 15, 595–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zook JM et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol 37, 561–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleary JG et al. Comparing Variant Call Files for Performance Benchmarking of Next-Generation Sequencing Variant Calling Pipelines. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/023754v2 (2015). [Google Scholar]
- 32.Chin C-S et al. A Diploid Assembly-based Benchmark for Variants in the Major Histocompatibility Complex. Nat. Commun 11, 4794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers G A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM 46, 395–415 (1999). [Google Scholar]
- 34.Cheng H, Jiang H, Yang J, Xu Y & Shang Y BitMapper: an efficient all-mapper based on bit-vector computing. BMC bioinformatics 16, 192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller JR et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24, 2818–2824 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


