Abstract
The plant hormone indole-3-acetic acid (IAA) is one of the main signals playing a role in the communication between host and endophytes. Endophytes can synthesize IAA de novo to influence the IAA homeostasis in plants. Although much is known about IAA biosynthesis in microorganisms, there is still less known about the pathway by which IAA is synthesized in fungal endophytes. The aim of this study is to examine a possible IAA biosynthesis pathway in Cyanodermella asteris. In vitro cultures of C. asteris were incubated with the IAA precursors tryptophan (Trp) and indole, as well as possible intermediates, and they were additionally treated with IAA biosynthesis inhibitors (2-mercaptobenzimidazole and yucasin DF) to elucidate possible IAA biosynthesis pathways. It was shown that (a) C. asteris synthesized IAA without adding precursors; (b) indole-3-acetonitrile (IAN), indole-3-acetamide (IAM), and indole-3-acetaldehyde (IAD) increased IAA biosynthesis; and (c) C. asteris synthesized IAA also by a Trp-independent pathway. Together with the genome information of C. asteris, the possible IAA biosynthesis pathways found can improve the understanding of IAA biosynthesis in fungal endophytes. The uptake of fungal IAA into Arabidopsis thaliana is necessary for the induction of lateral roots and other fungus-related growth phenotypes, since the application of the influx inhibitor 2-naphthoxyacetic acid (NOA) but not the efflux inhibitor N-1-naphtylphthalamic acid (NPA) were altering these parameters. In addition, the root phenotype of the mutation in an influx carrier, aux1, was partially rescued by C. asteris.
Keywords: Arabidopsis thaliana, Cyanodermella asteris, endophyte, IAA biosynthesis, IAA uptake, Trp-dependent pathway, Trp-independent pathway
1. Introduction
The plant hormone indole-3-acetic acid (IAA) plays a role in the communication between host plant and microbes, including plant-associated microorganisms [1,2,3] and endophytes [3,4], but also plant pathogens [5,6,7]. IAA together with other phytohormones is responsible for plant growth and development. It is involved in nearly each plant process such as cell growth, root initiation, tropism, fruit ripening, or senescence [8]. If plant-associated microorganism can regulate the plant IAA levels, they are able to influence the plant metabolism to their benefit, either as pathogens or as beneficial microorganisms. Then, microbial IAA acts as a mediator between plants and microorganisms and plays a key role in the communication between both partners. Microorganisms alters plant auxin levels and can therefore influence the plant development.
The concentration of IAA in plants can be controlled via different mechanisms, mainly biosynthesis, transport, conjugation, and degradation [9]. In addition, the auxin level of plants can be influenced by the de novo biosynthesis of IAA by microorganisms. However, for IAA produced by microbes, they either need to be residing within a host plant or the IAA needs to be taken up usually through the roots. The influx of IAA is possible either by diffusion via the plasma membrane and/or by uptake carrier proteins. The auxin resistant1/like auxin resistant (AUX/LAX) family mediates these in Arabidopsis thaliana [10]. Auxin carriers might be involved in the interaction between plants and fungi; for example, a LAX family member (LAX3) was differentially regulated in A. thaliana roots inoculated with the growth-promoting fungus Serendipita indica [11], and Trichoderma virens needs IAA influx carriers to mediate its interaction with A. thaliana [1].
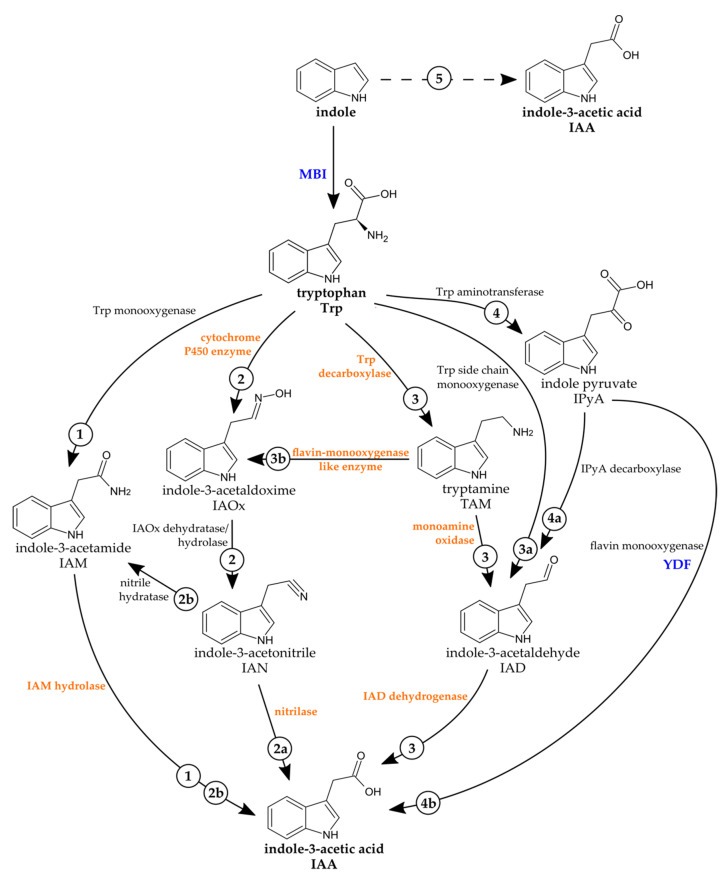
There are several well-known pathways to synthesize IAA in plants (reviewed in [12]). IAA can be synthesized by a tryptophan (Trp)-dependent and/or -independent pathway. The Trp-dependent pathway includes several different possible routes, named after the main intermediate: indole-3-pyruvic acid (IPyA), indole-3-acetamide (IAM), tryptamine (TAM), and indole-3-acetaldoxime/indole-3-acetonitrile (IAOx–IAN) pathways that may be interconnected (Figure 1). In several pathways, indole-3-acetaldehyde (IAD) is an intermediate. In the majority of plants, the major pathway is assumed to function via IPyA [13], but due to the existence of IAM [14] and also the IAOx-IAN pathway that is connected to indole glucosinolates in A. thaliana, the situation seems to be more complicated [15]. In the Trp-independent pathways, indole is most likely used as an IAA precursor [16,17,18]. Bacteria and fungi are also able to synthesize IAA [19], most likely using similar ways as found in plants, but there is not as much known. The genes and enzymes of the IAA biosynthesis steps are not completely resolved [20,21]. Fungi interacting with plants can produce IAA to increase root growth as endophytes in the rhizosphere or in the host plant itself. Pathogenic fungi can use IAA to cause disease symptoms or to interfere with the plant defense system [22]. Therefore, the focus was often on plant pathogens, especially for fungi [23]. For example, the smut fungus Ustilago maydis is causing galls on host tissues and can synthesize its own IAA [24]. Genes encoding proteins to synthesize IAA via the IAD pathway but also via TAM have been identified. Double knockout mutants had lower IAA levels, indicating the role of this pathway for fungal IAA production. Similarly, Ustilago esculenta converted IAD to IAA, but in addition IPyA and indole-3-lactic acid [25]. An early study on different Taphrina species, which also cause hyperplastic diseases in plants, indicated a pathway via IPyA and IAD, but the fungi could also convert IAN to IAA [26]. In four different plant-associated Fusarium species, among them corn, rice, and tomato pathogens, genes for the IAM pathway were identified [27]. F. graminearum could use TAM and IAN as intermediates rather than IAM [28]. Colletotrichum gloeosporioides causing anthracnose disease synthesized IAA in culture through IAM [29], similar to the rust fungus Puccinia graminis from which the respective genes were identified [30]. Sclerotinia sclerotiorum seemed to use the IAOx–IAN pathway due to an enzymatic activity for an IAOx dehydratase that was isolated and characterized [31].
Figure 1.
Indole-3-acetic acid (IAA) biosynthesis pathways in bacteria, fungi, and plants with the known enzymes (modified after KEGG [36,37,38]). IAA can be synthesized from tryptophan (Trp-dependent pathways, 1–4) and directly from indole (Trp-independent pathways, 5). Plants synthesize IAA over the indole-3-acetamide (IAM) (1), the indole-3-acetaldoxime/indole-3-acetonitrile (IAOx–IAN) (2-2a), the tryptamine (TAM) (3-3b-2-2a), and the indole-3-pyruvic acid (IPyA) pathway (4-4a-3 or 4-4b). Bacteria and fungi prefer the IAM (1), the IAOx–IAN (2-2a or 2-2b), the TAM (3), or the IPyA (4-4a-3) pathway. The IAA pathway with the direct conversion of Trp to indole-3-acetaldehyde (IAD, without IPyA as intermediate, 3a-3) is only known in Pseudomonas species [39,40,41,42]. The reaction site of the IAA biosynthesis inhibitors 2-mercaptobenzimidazole (MBI) (Trp synthase) and yucasin DF (YDF) (YUC enzymes) is shown in blue. Enzymes for which candidate genes were found in C. asteris are highlighted in orange (see also Table 1).
The mechanisms, especially how endophytes synthesize IAA, are important to understand the communication between them and their host [32,33]. However, for endophytic fungi, not so many studies have been performed. T. virens was capable of interacting with A. thaliana by its own IAA synthesis [1]. Since the fungus produced IAD and indole-3-ethanol in addition to IAA, it can be assumed that at least the IAD pathway is active in this endophyte. The arbuscular mycorrhizal fungus Rhizophagus irregularis was analyzed for the presence of IAA production, but no biosynthetic pathway was elucidated so far [34]. In the beneficial endophytic basidiomycete S. indica, IAA production was shown also, again without describing a specific pathway [35].
Cyanodermella asteris is an endophyte from the inflorescence axis of Aster tataricus [43], which is a perennial plant from the Northern part of Asia. Roots and rhizomes of A. tataricus are well known in the traditional Chinese medicine due to their antibacterial, antiviral, antiulcer, and diuretic activities [44,45,46]. The antitumor compounds astins, originally found in A. tataricus [47,48], can be synthesized by the endophytic fungus C. asteris, but some bioactive variants are only found in the plant [49]. Therefore, understanding the communication between both partners could shed light on possible mechanisms of how the synthesis of the antitumor compounds astins might be regulated. Since A. tataricus grows too large for laboratory studies, A. thaliana was chosen instead for co-cultivation experiments with C. asteris under sterile conditions on Petri dishes.
Co-cultivation assays with A. thaliana showed a root phenotype well known to be induced by IAA-producing microorganisms in plant-beneficial interactions [50,51,52]: shorter main root, increased lateral root growth, enhanced root biomass, and an auxin response detected in auxin-inducible promoter–reporter plants specifically in root tips and lateral root primordia.
A further question was whether C. asteris is capable of synthesizing IAA de novo and whether the IAA can also be secreted into the medium, and if so, by which pathway(s). In answering these questions, another piece would be added to the IAA biosynthesis puzzle in microorganisms and might contribute to resolving some of the loose ends. Genome analysis of genes possibly involved in IAA biosynthesis and feeding experiments with C. asteris using the IAA precursors and intermediates (Trp, TAM, IAN, IAM, IAD IPyA, indole) and the IAA biosynthesis inhibitors 2-mercaptobenzimidazole (MBI) [53] and yucasin DF (YDF) [54,55] will show us possible IAA biosynthetic routes.
In addition, the possible contribution of auxin uptake and efflux was analyzed for the co-cultivation between C. asteris and A. thaliana on plates containing IAA influx (2-naphthoxyacetic acid, NOA) [56] and efflux (N-1-naphtylphthalamic acid, NPA) inhibitors [57]. This could show if the root phenotype might be caused by fungal IAA secreted into the medium.
2. Results
2.1. The Endophyte C. asteris Induces a Root Phenotype on the Non-Host Plant A. thaliana that Is Reminiscent of High Auxin
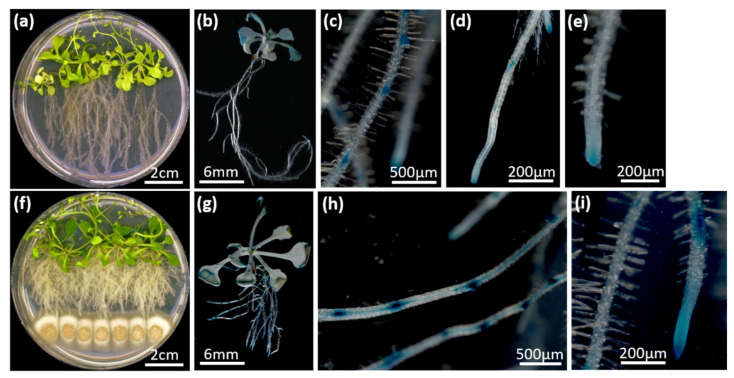
C. asteris induced a root phenotype with a massive lateral root and a reduced main root growth on the non-host plant A. thaliana that is reminiscent of auxin application (Figure 2a,f). The induction of the auxin response in root tips and lateral root primordia was also detected in a DR5::GUS line of A. thaliana (Figure 2b–e,g–i): Co-cultivated plants showed an increased amount of lateral root primordia that were additionally more intensely stained compared to controls.
Figure 2.
Phenotype of A. thaliana ecotype Columbia (Col-0) with C. asteris. A. thaliana and C. asteris were cultivated on ½ MS/MEAlow under long-day conditions. (a) Phenotype of A. thaliana Col-0 after 35 days of cultivation. (b–e) β-glucuronidase (GUS) staining of DR5::GUS lines of A. thaliana after 14 days of cultivation. (f) Phenotype of A. thaliana Col-0 co-cultivated with C. asteris after 35 days of cultivation. (g–i) GUS staining of roots of DR5::GUS lines of A. thaliana co-cultivated with C. asteris after 14 days of cultivation.
Since the co-cultivation studies of C. asteris with A. thaliana showed an increased lateral root growth indicating a change in the plant IAA level, the possibility of an IAA biosynthesis by C. asteris was analyzed.
2.2. The Fungal Genome of C. asteris Reveals Several Putative Genes Involved in IAA Biosynthesis
Different candidate genes for IAA biosynthesis enzymes could be identified in the automated annotated genome sequence of C. asteris [58] (Table 1). Following the candidate genes, a TAM pathway would be possible, since all necessary genes were found in the genome of C. asteris. Three genes of the IAOx–IAN pathway were found, but two genes were missing for the IAOx dehydratase/hydrolase (IAOx into IAN) and the nitrile hydratase (IAN into IAM). Only one gene was found in the annotation for the IAM pathway, where the gene for the Trp monooxygenase in the first step (Trp into IAM) was missing. Genes for the conversion of Trp into IPyA were not found in the genome annotation of C. asteris, indicating that this pathway might not be used for IAA biosynthesis. In addition to the Trp-dependent pathways, there were also putative genes for the direct conversion of indole into IAA. However, since this is information from the genome and not transcriptome, it cannot be concluded that the genes detected would be indeed expressed.
Table 1.
Candidate genes of C. asteris involved in IAA biosynthesis. Shown are the candidate genes in C. asteris to the corresponding enzyme reaction, but certainly this is no evidence that they are expressed in C. asteris. The putative IAA biosynthesis pathways in C. asteris are highlighted in orange in Figure 1.
| Reaction | Enzyme | Candidate Genes (Gene ID) |
|---|---|---|
| Trp → TAM | Trp decarboxylase | 1234 |
| TAM → IAOx | flavin monooxygenase enzyme * | 17, 2090, 4995, 5792, 7832 2451, 3001, 6023, 9811 |
| TAM → IAD | monoamine oxidase | 3466, 3467, 8359 |
| IAD → IAA | IAD dehydrogenase | 10233, 6228, 7132, 8151, 9123 |
| Trp → IAOx | cytochrome P450 | 17, 2090, 4995, 5792, 7832 |
| IAOx → IAN | IAOx dehydratase/hydrolase | - |
| IAN → IAA | nitrilase | 1223, 5809 |
| IAN → IAM | nitrile hydratase | - |
| Trp → IAM | Trp monooxygenase | - |
| IAM → IAA | IAM hydrolase | 1862, 6419 |
| Trp → IPyA | Trp aminotransferase | - |
| IPyA → IAD | IPyA decarboxylase | - |
| indole → IAA | - | 2451, 3001, 6023, 9811 |
* This reaction involves two different enzymes (TAM → N-hydroxy-TAM → IAOx).
2.3. C. asteris Secretes the Synthesized IAA into the Liquid Medium
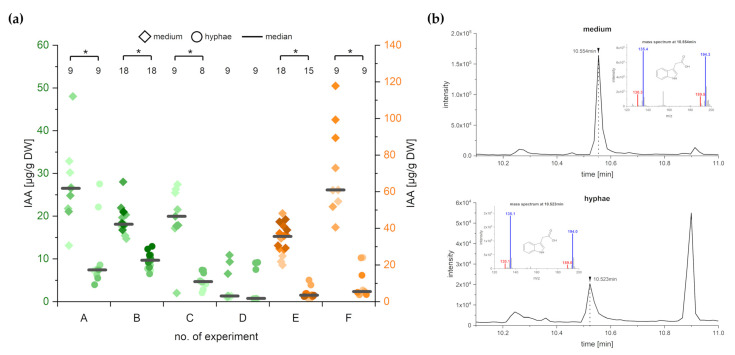
Several candidate gene of C. asteris indicated a de novo biosynthesis of IAA, including different Trp-dependent pathways such as via TAM or IAOx-IAN but also a Trp-independent pathway with indole as the IAA precursor. Therefore, fungal in vitro cultures of C. asteris were analyzed for their IAA production after 10 days of cultivation (Figure 3). The specific mass spectrum of methylated IAA (Figure 3b), including the internal standard D5-IAA, was found in medium and hyphae of C. asteris. IAA was detected in all six experiments with higher IAA amounts in the medium compared to hyphae (Figure 3a, p < 0.05). C. asteris secreted most of the synthesized IAA into the medium (64% to 92%), since only 8% to 36% of the IAA was retained in the hyphae.
Figure 3.
Amount of IAA in medium and hyphae of C. asteris in in vitro cultures. C. asteris was cultivated in MEAlow medium at 23 °C and 130 rpm in the dark for 10 days. (a) IAA levels in medium and hyphae of C. asteris in different experiments (A–F). Experiments A to D are shown with the left y-axis, experiments E and F are shown with the right y-axis. Data were analyzed using an unpaired t-test with Welch correction (test statistics can be found in Table S1). Significant differences between medium and hyphae are labeled with an asterisk. Sample size n is given in the upper part below the asterisks. (b) Extracted ion chromatogram of IAA (m/z 130) from medium and hyphae samples of C. asteris, including mass spectra (MS) at the specific retention times (red: MS data of IAA; blue: MS data of the internal standard D5-IAA).
2.4. IAA Biosynthesis in C. asteris
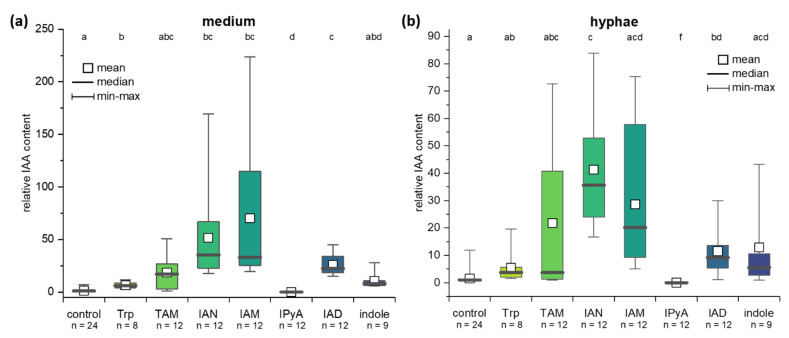
To elucidate the contribution of different possible pathways, C. asteris was incubated with different precursors and intermediates of IAA pathways (Figure 1) to evaluate their possible contribution to the biosynthesis. C. asteris was cultivated with 1 mM Trp, 1 mM TAM, 100 µM IAN, 100 µM IAM, 100 µM IPyA, 100 µM IAD, and 1 mM indole to induce the IAA biosynthesis (Figure 4). All precursors, except IPyA, increased the levels of IAA in C. asteris. IPyA is very unstable compared to other intermediates and was converted non-enzymatically to IAA (data not shown). In general, the direct precursors of IAA (IAN, IAM, and IAD) enhanced the biosynthesis very strongly in medium and hyphae (p < 0.05), whereas an intermediate earlier in the IAA biosynthesis, TAM, increased less strong and only in the hyphae. The early IAA biosynthesis precursors Trp and indole showed a small induction.
Figure 4.
IAA levels in medium and hyphae of C. asteris after feeding with different IAA precursors. C. asteris was cultivated in MEAlow for seven days, before IAA precursors (Trp, TAM, IAN, IAM, IPyA, IAD, or indole) were added to the culture. IAA levels were detected after 10 days of cultivation (3 days with precursors). The IAA level in (a) medium and (b) hyphae of C. asteris is correlated to the respective control (without any IAA precursor), which is set to 1. Small letters above the box plots show the statistical significance; test statistics can be found in Table S2. Sample size n is given below the x-axis.
2.5. The IAA Biosynthesis Inhibitor MBI Reveals a Trp-Independent Biosynthesis Pathway in C. asteris
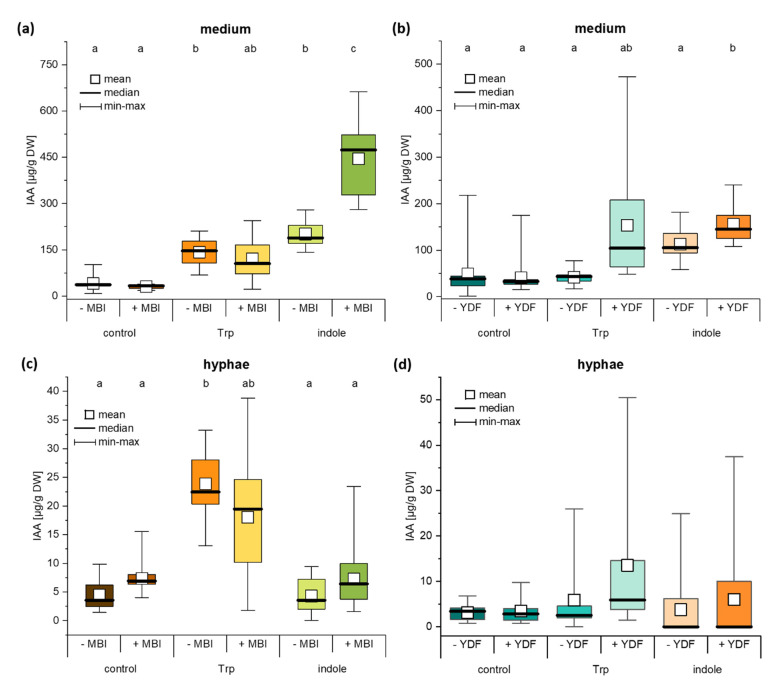
In addition to the Trp-dependent IAA biosynthesis, we were also interested in the possibility of a Trp-independent biosynthetic pathway. Here, two different IAA biosynthesis inhibitors MBI and YDF were used to disrupt the pathways and the two main IAA precursors Trp and indole were added to enhance possible effects (Figure 5).
Figure 5.
IAA content in medium and hyphae of C. asteris containing IAA precursors and IAA biosynthesis inhibitors. C. asteris was cultivated in MEAlow containing IAA biosynthesis inhibitors MBI or YDF. IAA precursors Trp and indole were added separately after 7 days of cultivation. IAA was detected after 10 days of cultivation. IAA content in (a) medium and (c) hyphae of C. asteris containing MBI as IAA biosynthesis inhibitor. IAA content in (b) medium and (d) hyphae of C. asteris containing YDF as IAA biosynthesis inhibitor. Small letters indicate statistically significant differences (Welch-ANOVA, post-hoc Games–Howell; see Table S3 for test statistics).
The IAA biosynthesis inhibitor MBI increased the IAA level 4.2-fold in C. asteris cultures with indole (Figure 5a, p < 0.001). This was interpreted as the possibility to change to an alternative pathway from indole when the Trp-dependent one was blocked. The controls without any IAA precursor and with Trp did not change their IAA levels in the presence of MBI. Without any biosynthesis inhibitor, similar IAA levels were detected for Trp and indole as substrate in the cultures, and they were increased to the 4.4- to 5.5-fold compared to controls. These differences were only seen in the medium, not in hyphae (Figure 5c). Only Trp increased the IAA content in the hyphae, no matter whether MBI was present or not.
The addition of YDF, a second IAA biosynthesis inhibitor, did not change the IAA pattern as MBI did (Figure 5b). Controls without any IAA precursor and the cultures incubated with indole showed no significant differences between cultures with and without YDF. Only the cultures where Trp was added increased threefold their IAA levels, when YDF was present. Similar to MBI, the hyphae did not change their IAA levels when YDF was present (Figure 5d). This experiment showed evidence for a Trp-independent pathway to synthesize IAA from the precursor indole in C. asteris.
2.6. Auxin Synthesis by C. asteris and Uptake into the Host Plant Are Important for the Root Phenotype
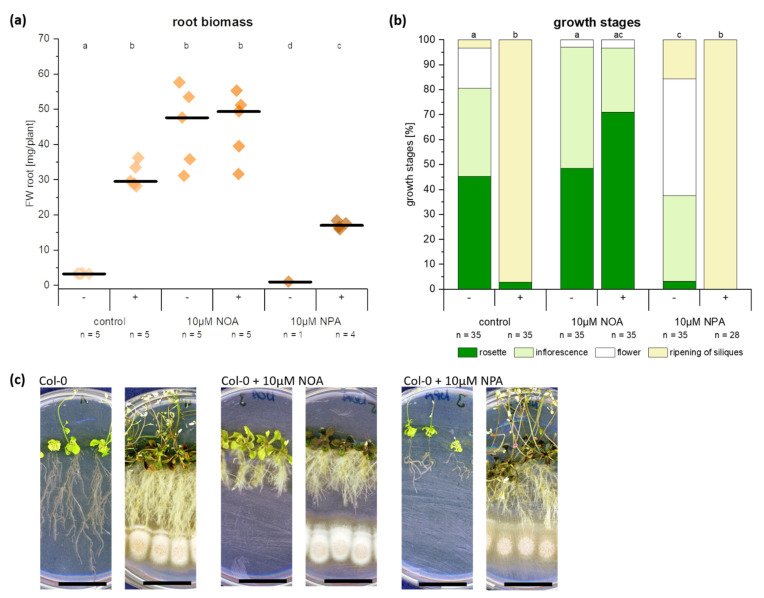
The fungus C. asteris was co-cultivated with A. thaliana on medium containing two different IAA transport inhibitors: NOA and NPA (Figure 6). A. thaliana control plants developed a very strong phenotype in co-cultivation with C. asteris, including reduced main root length, massive lateral root growth, increased root and leaf biomass (Figure 2 and Figure S1). The addition of 10 µM NOA to the medium changed the phenotype of the A. thaliana plants: shorter main roots (Figure S1b), agravitropic root growth, and a higher root biomass (Figure 6a), resulting from the small, compact phenotype (Figure 6c) and a delayed plant development compared to the controls (Figure 6b). The co-cultivation with C. asteris changed the root phenotype of the NOA-treated plants only slightly: the root biomass (Figure 6a) and root length (Figure S1b) were similar; only the reduction of the root growth decreased slightly (Figure 6c). The plants co-cultivated with C. asteris developed similar to the plants without C. asteris on NOA (Figure 6b). NPA-treated plants had shorter roots (Figure S1b) and smaller rosettes (Figure S1a) compared to control plants; but they developed similar to controls with similar biomasses in leaves (Figure S1c) and roots (Figure 6a). Co-cultivation with C. asteris led in NPA-treated plants to longer roots (Figure S1b) and larger rosettes (Figure S1c) as seen already in control plants.
Figure 6.
Co-cultivation of A. thaliana and C. asteris with IAA transport inhibitors 2-naphthoxyacetic acid (NOA) and N-1-naphtylphthalamic acid (NPA). A. thaliana and C. asteris were co-cultivated on ½ MS/MEAlow including either 10 µM NOA or 10 µM NPA under long day conditions for 35 days. (a) Root biomass and (b) growth stages of A. thaliana plants. Small letters indicate significant differences. –median. (c) Growth of A. thaliana and C. asteris on NOA or NPA containing medium. The bar indicates 2 cm. Statistical analyses can be found in Table S4.
Increasing NOA and NPA concentrations in the medium (Figure S2) led to stronger effects: NOA-treated plants developed shorter roots and were delayed in plant development, which was not affected by C. asteris; NPA-treated plants showed still an enhanced lateral root growth and a faster plant development in the presence of C. asteris, but the main root growth was not reduced.
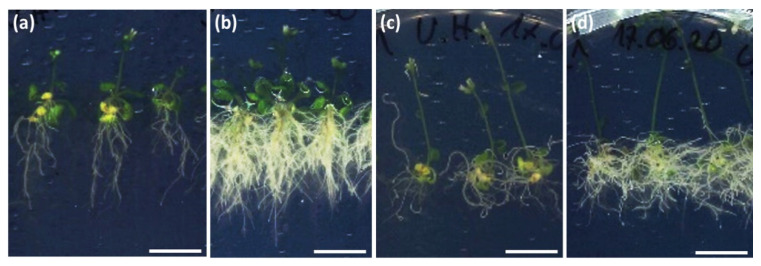
aux1 mutants of A. thaliana (background: Landsberg erecta Ler-0) that lack the auxin resistant 1 (AUX1) influx carrier showed still an increased lateral roots growth (Figure 7c,d) in the presence of C. asteris. The agravitropic root growth, as seen in the aux1 controls, was still present, but it was not as strong as in the aux1 controls. The ecotype Ler-0 showed a similar root phenotype in the presence of C. asteris as the Columbia (Col-0) ecotype (Figure 2 and Figure 7a,b).
Figure 7.
Root phenotype of ecotype Landsberg erecta (Ler-0) and auxin resistant 1 (aux1)-mutants of A. thaliana when incubated with C. asteris for 21 days. A. thaliana and C. asteris were co-cultivated on ½ MS/MEAlow under long day conditions. (a) Ecotype Ler-0 control, (b) ecotype Ler-0 co-cultivated with C. asteris. (c) aux1-mutant of A. thaliana, (d) aux1-mutant of A. thaliana co-cultivated with C. asteris. The bar indicates 1 cm.
The mutant tir1, lacking the transport inhibitor response 1 (TIR1) receptor, showed a similar phenotype as already known from control plants with an increase in lateral roots and a reduction of main root growth, but no difference to Col-0 control roots (Figure S3). Since the TIR receptor family consists of several proteins, it is possible that others are responsible for IAA perception in the tir1 mutant.
Summarizing these experiments, the IAA influx carrier inhibitor NOA reduced the effects caused by C. asteris, whereas NPA did not influence the phenotype of A. thaliana caused by C. asteris. The absence of the AUX1 influx carrier had a slight effect on the root phenotype, namely the agravitropic response was rescued by C. asteris to some extent.
3. Discussion
It was shown that the endophytic fungus C. asteris synthesized IAA via different pathways. In addition to Trp-dependent pathways, C. asteris uses also a Trp-independent pathway using indole as precursor to synthesize IAA. C. asteris secreted most of the IAA into the culture medium and retained only small amounts in its hyphae (Figure 3, Figure 4 and Figure 5). The root phenotype of A. thaliana co-cultivated with C. asteris was reminiscent of high auxin concentrations, and inoculated auxin-responsive promoter reporter lines (DR5-GUS) showed an increased staining in areas where lateral root primordia are formed compared to control roots (Figure 2). Similarly, it was reported for the interaction of T. virens and A. thaliana that in a DR5::GUS line, an increased activity in the root tip for auxin responses was found [1]. Unfortunately, the pictures did not reveal any changes in other parts of the roots, so it is not possible to compare these with the results found in the interaction of C. asteris and A. thaliana in this study (Figure 2).
Most of the IAA was secreted into the medium, indicating that fungal IAA most likely acts in the host plant and not in the fungal endophyte. In the natural environment inside the plant, secreted fungal IAA would influence the plant IAA homeostasis and thus plant AUX1 growth and development. IAA was synthesized using Trp-dependent and -independent pathways in C. asteris (for the different pathways, see Figure 1). The direct precursor of IAA in the Trp-dependent pathways—IAM, IAN and IAD—led to an increase in the IAA content, and this is corroborated by the existence of the candidate genes involved in the different pathways that were found in the genome of C. asteris (Table 1). Despite the fact that expression of the genes has not been determined so far, the increase of IAA production after incubation with the respective precursors indicate that these gene products could be involved in the auxin production.
The existence of the IPyA pathway could not be clearly verified, since IPyA did not increase the de novo synthesized IAA level in C. asteris. However, since IPyA was rapidly degraded in the fungal medium (data not shown), its possible role as an intermediate cannot completely be ruled out. In addition, candidate genes were not found in the genome, indicating that IAA might not be synthesized via the IPyA pathway. The IPyA pathway is widely spread in bacteria and fungi: Azospirillum brasilense [59,60], Pseudomonas stutzeri [61], and Ustilago maydis [24] are just a few examples. Pseudomonas species convert Trp directly into IAD without synthesizing IPyA, which is catalyzed by a Trp side chain monooxygenase [39,40,41,42]. Such a pathway cannot be excluded in C. asteris, but a candidate gene for a Trp monooxygenase in the genome was not found.
The IAM pathway would also be possible, but here again, a Trp monooxygenase would be needed. The first IAM pathway was described for Pseudomonas savastanoi [40], but it is found by now also in other plant-associated bacteria [62] and fungi. The fungal pathogens Fusarium proliferatum [27], Colletotrichum fruticola [63], and C. gloeosporioides f.sp. aeschynomene [64] synthesize IAA also over the IAM pathway. The latter fungus uses the IAM and IPyA pathway, preferring IAM as the main biosynthetic way [64]. The IAM pathway is usually used by pathogens to induce an uncontrolled plant cell growth [65]. However, different Streptomyces species indicate more than one IAA biosynthetic pathway, including the IAM pathway as the dominant one [66].
IAM is not only an intermediate in the IAM pathway, but it can also be an intermediate in the IAOx–IAN pathway. Candidate genes were found for the Trp conversion into IAOx in C. asteris, but no gene was found for the IAOx conversion into IAN. IAN can be converted either directly into IAA using nitrilases or via IAM into IAA, but the genome was lacking candidate genes for the nitrile hydratase (conversion of IAN into IAM) in C. asteris. Nitrilases are a large family in bacteria, fungi, plants, and animals [67]. IAN-specific nitrilases were found among others in Pseudomonas species [68,69] and Alcaligenes faecalis JM3 [70,71]. A nitrile hydratase could be isolated from the plant pathogen Agrobacterium tumefaciens and the leguminous Rhizobium spp. [72], which catalyzes the transformation of IAN into IAM.
The most possible IAA biosynthesis pathway in C. asteris might be the TAM pathway: here, candidate genes could be identified for each enzymatic reaction, and an induced biosynthesis with exogenous IAD could be shown (Figure 4). The TAM pathway is known in different bacteria and fungi. Several microorganisms are described to possess a Trp decarboxylase activity, which is responsible for the Trp decarboxylation of Trp to TAM (Taphrina deformans [73], Metarhizium robertsii [74]). Other microorganisms increase their IAA level when they were supplemented with exogenous TAM (Azospirillium [75], Ustilago maydis [76], Leptosphaeria maculans [6]). In addition to the TAM pathway, which might be the main pathway for IAA biosynthesis in C. asteris, there could exist also other pathways for IAA biosynthesis with partial enzymatic coverage, where the substrates for missing steps could be provided by the host plant.
In addition to the different Trp-dependent IAA biosynthesis pathways, there is a Trp-independent pathway. There is evidence that Trp-independent pathways exist in bacteria [60] and fungi [77], but it is not clear whether IAA is synthesized using indole or indole-3-glycerol phosphate (IGP) as a precursor. The IAA biosynthesis inhibitor MBI can inhibit the Trp-dependent biosynthesis of IAA [53,78]. C. asteris synthesizes high levels of IAA in the presence of indole (Figure 5), if the Trp-dependent biosynthesis was inhibited by MBI.
Different plant mutants suggest that IAA can be synthesized from indole and IGP. Orp mutants of maize, which show a defect in the last step of Trp biosynthesis [16,17], synthesized IAA from 15N1-anthranilate (indole precursor), but not from D5-Trp. The mutants accumulated up to 50 times more IAA than controls [79]. trp2 mutants of A. thaliana also synthesized IAA from 15N1-anthranilate and not from D5-Trp. trp2 and trp3 mutants accumulated very high levels of IAA (38-fold and 19-fold more than controls) [80]. A. thaliana plants with an antisense gene copy of IGP synthase as well as trp2-1 and trp3-1 mutants produced different amounts of IAA: whereas the trp mutants increased their IAA level, IGP antisense mutants decreased it [18]. Whether these results from plants can be transferred to bacteria and fungi has to be shown by further research, but there is a strong evidence for the existence of a Trp-independent IAA biosynthesis starting with indole and/or IGP also in these organisms.
The second IAA biosynthesis inhibitor YDF used in our study is a more stable derivative of yucasin and blocks the conversion of Trp into IPyA by the YUCCA enzymes in plants [54,55]. The application of YDF to C. asteris changed the IAA levels in Trp-treated cultures but not dramatically. The IPyA pathway is only one out of many different IAA biosynthesis pathways, which could compensate the possible reduction of IAA. Furthermore, possible genes involved in the IPyA pathway were not found in the genome, which does not exclude them since it is only an automatic annotation. The cultures supplemented with indole showed an increased IAA level indicating alternative IAA biosynthesis pathways in C. asteris beside over IPyA. In consequence, the IAA level increased only very slightly (not significant) in these cultures in the presence of YDF (Figure 5).
In the co-cultivation experiment with A. thaliana and C. asteris on medium containing the IAA transport inhibitors NOA or NPA, the IAA efflux carrier inhibitor NPA did not influence the root phenotype remodeling associated with C. asteris. Only the IAA influx carrier inhibitor NOA inhibited the alterations in the root architecture of A. thaliana in most instances. The roots developed, compared to the non-co-cultivated plants on NOA, only slightly elongated lateral roots without an increase in root biomass. IAA is transported into the cell passively by diffusion and actively by AUX1/LAX transporters [81]. These transporters are blocked by NOA [56], reducing the intracellular IAA level. Diffusion of IAA still takes place and explains why NOA-treated plants slightly increased their root length in the presence of C. asteris. NPA is targeting the IAA efflux carriers ABCB1 and ABCB19 [82,83,84], which blocks the polar and thus basipetal auxin transport [10]. There is disagreement regarding whether NPA directly inhibits the PIN efflux transporters [57]. pin1 loss-of-function mutants of A. thaliana show the same phenotype as NPA-treated plants [85,86,87]. NPA-treated plants showed the typical phenotype, described in the literature [88]: agravitropic root growth with reduced lateral roots. The influx carrier proteins are not affected by NPA [89], so IAA from the fungus can still be transported into the cell and influence the IAA homeostasis of the plant.
A. thaliana plants without any NOA developed a root phenotype (Figure 2 and Figure 6), which is known to be induced by other plant-beneficial microorganisms. Treating plants with IAA transport inhibitor reduces the effect of microorganisms on root architecture [90,91,92,93]. For example, the plant-growth promoting fungus Aspergillus ustus induced lateral root growth and thus root biomass [94]. The bacterium Bacillus megaterium STM196 reduced primary root growth, but it induced lateral root growth and root biomass [51]. Several microorganisms are known to induce these changes in root architecture (reviewed in [95]). The induction of lateral root growth was lost in aux-mutants of A. thaliana when inoculated with different microorganisms [91,96]. The increased root system, induced by plant-beneficial microorganisms, can improve nutrition uptake, which leads to a better tolerance against different environmental stresses [97,98,99].
Further experiments should investigate the potential contribution of the different candidate genes involved in IAA biosynthesis that were identified. Additional feeding experiments with heavy-isotope labeled Trp and identification of the intermediates will also improve our knowledge about which Trp-dependent pathway is preferred by C. asteris.
4. Materials and Methods
4.1. Plant and Fungal Material
All A. thaliana ecotypes (Col-0 and Ler-0) and mutants (aux1 and tir1) used in this study were obtained from Nottingham Arabidopsis Stock Centre (NASC, Loughborough, UK). C. asteris was derived from our own culture (deposited under the accession number DSM 100826 at DSMZ, Braunschweig, Germany).
4.2. Co-Cultivation of A. thaliana with C. asteris
A. thaliana plants of the ecotypes Col-0, Ler-0, and auxin mutants aux1 and tir1 were co-cultivated with C. asteris on Petri dishes, sealed with Parafilm (Bemis, Neenah, WI, USA). A. thaliana seeds were sterilized with 70% ethanol containing 0.1% Triton-X 100 for 1 min, followed by 1.2% sodium hypochlorite containing 0.1% Triton-X 100 and three to four times washing with autoclaved water. Seven sterilized seeds were sown in the upper part of Petri dishes with ½ MS/MEAlow (1.1 g/L MS incl. vitamins, 5 g/L malt extract, 5 g/L D-glucose, 0.5 g/L peptone, 0.5 mL/L Hutner’s trace elements [100]; pH 6.1) and cultivated under long day conditions (16 h with 23°C with light intensities between 90 and 120 µmol s−1 m−2; 8 h with 18 °C in the dark). After one week, C. asteris conidia were applied to the lower part of the Petri dish. Conidia were harvested from a one-week-old C. asteris culture in MEAlow by filtering (gauze, 20 µm mesh size) and centrifugation (5000 rcf, 4 °C, 10 min). The pellet with conidia was dissolved in MEAlow and used for inoculation. Each plant was inoculated with 105 conidia in a distance of 4.5 cm to the seeds (seven spots per Petri dish). The phenotype of A. thaliana plants was recorded weekly (root length, growth stage). The plants were harvested after 35 days of cultivation for fresh and dry weight determination. For treatment with auxin transport inhibitors, A. thaliana seeds were sterilized and sown as above. After one week, A. thaliana seedlings were transferred to Petri dishes containing ½ MS/MEAlow including NOA (10 µM and 30 µM; dissolved in water and filter sterilized) and NPA (10 µM and 20 µM; dissolved in DMSO), and co-cultivated with C. asteris conidia as described above.
4.3. GUS Staining of A. thaliana DR5::GUS
Harvested plants of A. thaliana DR5::GUS were incubated in 5-bromo-4-chloro-1H-indol-3-yl β-D-glucopyranosiduronic acid (X-Gluc) incubation buffer (0.1 M phosphate buffer, 10 mM Na2EDTA, 0.5 M K3[Fe(CN6)], 0.5 M K4[Fe(CN6)], 0.5% Triton X-100; filter sterilized and 1.2 µM X-Gluc added) for 2 h at 37 °C in the dark. Reaction was stopped by transferring plants into 0.1 M phosphate buffer. Green tissues were destained in ethanol (ascending order of 30%–50%–70%–90%–100%). The plants were screened with an Axio Zoom.V16 (Carl Zeiss Microscopy, Jena, Germany).
4.4. Incubation of C. asteris with IAA Precursors and/or Biosynthesis Inhibitors
C. asteris was cultivated in liquid MEAlow on a shaking incubator (23 °C, 130 rpm) for 10 days. After centrifugation (5000 rcf, 10 °C, 10 min) to separate medium and hyphae, hyphae were freeze-dried and dry weight was determined. IAA was extracted and analyzed from medium and freeze-dried hyphae.
To determine an IAA biosynthesis pathway in C. asteris, liquid cultures of C. asteris were fed with different IAA precursors (1 mM Trp dissolved in water, sterilized by autocaving, and 1 mM indole dissolved in methanol) and intermediates (1 mM TAM; 100 µM of IAN/ IAM/IAD/IPyA; all dissolved in methanol) after seven days of cultivation. The cultures were harvested (see above) after three additional days for IAA extraction and analysis (see Section 4.4).
In a third experiment, C. asteris was cultivated in liquid MEAlow including the IAA biosynthesis inhibitors MBI (60 µM; dissolved in isopropanol) or YDF (100 µM; dissolved in isopropanol). IAA precursors Trp or indole (each 1 mM) were added after seven days. After an additional three days, the fungal culture was harvested (see above) to extract and analyze the IAA content.
4.5. Extraction and Detection of IAA
For IAA extraction, each medium sample was divided into three technical replicates, which was set up with 1 µg internal standard D5-IAA (Euriso-top SAS, Saint-Aubin, France). Then, the medium was shaken for 2 h in the dark at 4 to 8 °C to guarantee a uniformly distribution of the internal standard. After this, the pH value was set to three, and the medium was extracted twice with the same volume of ethyl acetate (HPLC grade, Carl Roth, Karlsruhe, Germany) (vortexing for 10 s, centrifugation for 5 min, 4 °C, 5000 rcf). The organic phases were collected and evaporated at a Multivapor P-12 (Büchi Labortechnik GmbH, Essen, Germany). The dried sediments were resuspended twice in 500 µL methanol (HPLC grade, Carl Roth, Karlsruhe, Germany) and again evaporated under gaseous nitrogen. In the next step, the 200 µL of methanol resolved IAA was methylated with the same volume of (trimethylsilyl)diazomethane (1:100 dilution in diethylether) at room temperature for 30 min, before the methanol was evaporated under gaseous nitrogen. The IAA sample was finally resuspended in 50 µL of ethyl acetate and measured by gas chromatography-mass spectrometry (GC-MS).
Freeze-dried hyphae samples were ground and mixed with 80% methanol, including 5% acetic acid. The samples were divided into three technical replicates, and 1 µg internal standard D5-IAA was added to each replicate. After shaking for 2 h in the dark at 4 to 8 °C, the samples were centrifuged to remove the pellet (10,000 rcf, 4 °C, 10 min). The methanol of the supernatant was evaporated under gaseous nitrogen until the water phase was left. The pH of the water phase was set to three and extracted twice with the same volume of ethyl acetate. The combined organic phases of ethyl acetate were evaporated under gaseous nitrogen before the methylation of IAA was performed as described above. The sample was treated as described for medium, and the IAA contents were measured by GC-MS.
Samples were separated by GC 3900 on an Agilent HP-5 column (30 m/Ø 0.250 mm/0.25 µm film) with 1 mL/min helium (10 psi). Sample (1 µL) was injected splitless at 250 °C and separated by the following method: 2 min at 70 °C, then a temperature increase about 20 °C/min until 280 °C, and held until 17.5 min. The separated sample was analyzed with an ion trap (Saturn 2100 MS; Varian, Palo Alto, CA, USA) and ionized by electron ionization (70 eV) at 200 °C.
The IAA content of each sample was calculated according to the isotope dilution equation [101] with the corresponding m/z (methylated IAA: m/z 130; methylated D5-IAA: m/z 135). The background of the IAA precursors and intermediates at m/z 130 were subtracted to get only the de novo synthesized IAA levels of C. asteris. To compare the different experiments with the different IAA precursors/intermediates, the IAA levels in each experiment were related to the respective control without any supplementation.
4.6. Data Analysis
Statistical analyses were carried out using OriginPro 2021 version 9.8.0.200 (OriginLab Corporation, Northampton, MA, USA).
Data of the interaction study between A. thaliana and C. asteris on NOA- or NPA-containing medium were analyzed with different statistical analyses: (a) A Welch-ANOVA (due to an unequal sample size and dissimilar variances) was used for biomarkers with unequal samples size and dissimilar variances, with IAA transport inhibitor included as a six-level between-subjects factor (control, NOA, NPA without and with C. asteris). Post-hoc tests were performed using a Games–Howell test. (b) A Kruskal–Wallis test was used for biomarkers with equal sample size but unsimilar variances or for growth stages, with IAA transport inhibitor included as a three-level (control, NOA, NPA) and C. asteris included as two-level (without or with C. asteris) between-subjects factor. Post-hoc tests were performed using Dunn’s test. Effect sizes were calculated [102,103].
For determination of biosynthetic pathways, data of the controls (without any precursor or intermediate) were analyzed using an unpaired t-test on the medians of the three technical replicates, assuming unequal variances (Welch correction).
IAA levels of C. asteris cultures, which were supplemented with different IAA precursors/intermediates and/or IAA biosynthesis inhibitors, were statistically analyzed by performing a Welch-ANOVA on the medians of the three technical replicates, with IAA precursors as an eight-level (control, Trp, TAM, IAN, IAM, IPyA, IAD, indole) or IAA biosynthesis inhibitor as a six level (control, MBI, and YDF without and with C. asteris) between-subjects factor. A post-hoc test was performed (Games–Howell test) and effect sizes were calculated [102,103]. The respective test used is given in the results and/or figure legends.
5. Conclusions
This study revealed that the endophytic fungus C. asteris produces the plant hormone IAA via different biosynthetic pathways: Trp-dependent and Trp-independent. The most likely Trp-dependent way is the TAM pathway, which does not exclude other pathways such as via IPyA or IAM. The Trp-independent pathway starts with indole as the substrate. By synthesizing and secreting IAA, C. asteris influences the root growth of A. thaliana that results in more lateral but shorter main roots. The phenotype was reduced after the addition of an auxin transport inhibitor for influx but not efflux. Further experiments should lead to the identification of responsible genes in the endophyte for the biosynthesis and secretion of IAA. This study needs to be investigated further in a future study to see how the growth of the roots behave and change through different stages of the plant growth ending to the greenhouse stage.
Acknowledgments
We thank Jerry D. Cohen, University of Minnesota, USA, for the gift of yucasin DF.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/5/2651/s1, Figure S1: Co-cultivation of A. thaliana and C. asteris on IAA transport inhibitor containing medium, Figure S2: Co-cultivation of A. thaliana and C. asteris in the presence of IAA transport inhibitors, Figure S3: Growth of tir1 mutant of A. thaliana with C. asteris after 35 days, Table S1: Test statistics on IAA levels in medium and hyphae of C. asteris, using an unpaired t-test. Table S2: Test statistics on IAA levels in medium and hyphae of C. asteris in presence of IAA precursors, Table S3: Test statistics on IAA levels in medium and hyphae of C. asteris in the presence of MBI or YDF, Table S4: Test statistics on co-cultivation of A. thaliana and C. asteris on 10 µM NOA or 10 µM NPA, Table S5: Test statistics on co-cultivation of A. thaliana and C. asteris on 30 µM NOA or 20 µM NPA. Table S6: Raw data.
Author Contributions
Conceptualization, L.J. and J.L.-M.; methodology, L.J.; software, L.J.; validation, L.J. and J.L.-M.; formal analysis, L.J.; investigation, L.J. and U.H.; resources, J.L.-M.; data curation, L.J.; writing—original draft preparation, L.J.; writing—review and editing, J.L.-M.; visualization, L.J.; supervision, J.L.-M.; project administration, J.L.-M.; funding acquisition, J.L.-M. All authors have read and agreed to the published version of the manuscript.
Funding
Open Access Funding by the Publication Fund of the TU Dresden.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting this study are available in this paper and in its supplementary data published online.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Contreras-Cornejo H.A., Macías-Rodríguez L., Cortés-Penagos C., López-Bucio J. Trichoderma virens, a Plant Beneficial Fungus, Enhances Biomass Production and Promotes Lateral Root Growth Through an Auxin-dependent Mechanism in Arabidopsis. Plant. Physiol. 2009;149:1579–1592. doi: 10.1104/pp.108.130369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz-Castro R., Díaz-Pérez C., Martínez-Trujillo M., del Río R.E., Campos-García J., López-Bucio J. Transkingdom Signaling Based on Bacterial Cyclodipeptides with Auxin Activity in Plants. Proc. Natl. Acad. Sci. USA. 2011;108:7253–7258. doi: 10.1073/pnas.1006740108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rai M., Varma A. Arbuscular Mycorrhiza-like Biotechnological Potential of Piriformospora indica, Which Promotes the Growth of Adhatoda Vasica Nees. Electron. J. Biotechnol. 2005;8:1–6. doi: 10.2225/vol8-issue1-fulltext-5. [DOI] [Google Scholar]
- 4.Suebrasri T., Harada H., Jogloy S., Ekprasert J., Boonlue S. Auxin-producing Fungal Endophytes Promote Growth of Sunchoke. Rhizosphere. 2020;16:100271. doi: 10.1016/j.rhisph.2020.100271. [DOI] [Google Scholar]
- 5.Jahn L., Mucha S., Bergmann S., Horn C., Staswick P., Steffens B., Siemens J., Ludwig-Müller J. The Clubroot Pathogen (Plasmodiophora brassicae) Influences Auxin Signaling to Regulate Auxin Homeostasis in Arabidopsis. Plants. 2013;2:726–749. doi: 10.3390/plants2040726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leontovycová H., Trdá L., Dobrev P.I., Šašek V., Gay E., Balesdent M.-H., Burketová L. Auxin Biosynthesis in the Phytopathogenic Fungus Leptosphaeria maculans Is Associated with Enhanced Transcription of Indole-3-pyruvate Decarboxylase LmIPDC2 and Tryptophan Aminotransferase LmTAM1. Res. Microbiol. 2020;171:174–184. doi: 10.1016/j.resmic.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Vandeputte O., Oden S., Mol A., Vereecke D., Goethals K., El Jaziri M., Prinsen E. Biosynthesis of Auxin by the Gram-positive Phytopathogen Rhodococcus fascians Is Controlled by Compounds Specific to Infected Plant Tissues. Appl. Environ. Microbiol. 2005;71:1169–1177. doi: 10.1128/AEM.71.3.1169-1177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies P.J. The Plant Hormones: Their Nature, Occurrence, and Functions. In: Davies P.J., editor. Plant Hormones: Biosynthesis, Signal Transduction, Action! Springer; Dordrecht, The Netherlands: 2010. pp. 1–15. [Google Scholar]
- 9.Ludwig-Müller J. Auxin Conjugates: Their Role for Plant Development and in the Evolution of Land Plants. J. Exp. Bot. 2011;62:1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- 10.Parry G., Delbarre A., Marchant A., Swarup R., Napier R., Perrot-Rechenmann C., Bennett M.J. Novel Auxin Transport Inhibitors Phenocopy Auxin Influx Carrier Mutation aux1. Plant. J. 2001;25:399–406. doi: 10.1046/j.1365-313x.2001.00970.x. [DOI] [PubMed] [Google Scholar]
- 11.Pérez-Alonso M.-M., Guerrero-Galán C., Scholz S.S., Kiba T., Sakakibara H., Ludwig-Müller J., Krapp A., Oelmüller R., Vicente-Carbajosa J., Pollmann S. Harnessing Symbiotic Plant–Fungus Interactions Unleash Hidden Forces from Extreme Plant Ecosystems. J. Exp. Bot. 2020;71:3865–3877. doi: 10.1093/jxb/eraa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y. Auxin Biosynthesis. Arab. Book Am. Soc. Plant Biol. 2014:12. doi: 10.1199/tab.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H., et al. The Main Auxin Biosynthesis Pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann T., Hoffmann M., Hentrich M., Pollmann S. Indole-3-Acetamide-Dependent Auxin Biosynthesis: A Widely Distributed Way of Indole-3-Acetic Acid Production? Eur. J. Cell Biol. 2010;89:895–905. doi: 10.1016/j.ejcb.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Kriechbaumer V., Glawischnig E. Auxin Biosynthesis within the Network of Tryptophan Metabolism. JNBT. 2005;2:55–58. [Google Scholar]
- 16.Wright A.D., Moehlenkamp C.A., Perrot G.H., Neuffer M.G., Cone K.C. The Maize Auxotrophic Mutant Orange Pericarp Is Defective in Duplicate Genes for Tryptophan Synthase β. Plant Cell. 1992;4:711–719. doi: 10.2307/3869529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright A.D., Neuffer M.G. Orange Pericarp in Maize: Filial Expression in a Maternal Tissue. J. Hered. 1989;80:229–233. doi: 10.1093/oxfordjournals.jhered.a110840. [DOI] [Google Scholar]
- 18.Ouyang J., Shao X., Li J. Indole-3-glycerol Phosphate, a Branchpoint of Indole-3-acetic Acid Biosynthesis from the Tryptophan Biosynthetic Pathway in Arabidopsis thaliana. Plant J. 2000;24:327–334. doi: 10.1046/j.1365-313x.2000.00883.x. [DOI] [PubMed] [Google Scholar]
- 19.Patten C.L., Glick B.R. Bacterial Biosynthesis of Indole-3-acetic Acid. Can. J. Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 20.Hull A.K., Vij R., Celenza J.L. Arabidopsis Cytochrome P450s That Catalyze the First Step of Tryptophan-dependent Indole-3-acetic Acid Biosynthesis. Proc. Natl. Acad. Sci. USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duca D., Lorv J., Patten C.L., Rose D., Glick B.R. Indole-3-acetic Acid in Plant–microbe Interactions. Antonie Van Leeuwenhoek. 2014;106:85–125. doi: 10.1007/s10482-013-0095-y. [DOI] [PubMed] [Google Scholar]
- 22.Fu S.-F., Wei J.-Y., Chen H.-W., Liu Y.-Y., Lu H.-Y., Chou J.-Y. Indole-3-Acetic Acid: A Widespread Physiological Code in Interactions of Fungi with Other Organisms. Plant Signal. Behav. 2015;10:e1048052. doi: 10.1080/15592324.2015.1048052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanclud E., Morel J.-B. Plant Hormones: A Fungal Point of View. Mol. Plant Pathol. 2016;17:1289–1297. doi: 10.1111/mpp.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reineke G., Heinze B., Schirawski J., Buettner H., Kahmann R., Basse C.W. Indole-3-acetic Acid (IAA) Biosynthesis in the Smut Fungus Ustilago maydis and Its Relevance for Increased IAA Levels in Infected Tissue and Host Tumour Formation. Mol. Plant Pathol. 2008;9:339–355. doi: 10.1111/j.1364-3703.2008.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung K.R., Tzeng D.D. Biosynthesis of Indole-3-Acetic Acid by the Gall-Inducing Fungus Ustilago esculenta. J. Biol. Sci. 2004;4:744–750. [Google Scholar]
- 26.Yamada T., Tsukamoto H., Shiraishi T., Nomura T., Oku H. Detection of Indoleacetic Acid Biosynthesis in Some Species of Taphrina Causing Hyperplastic Diseases in Plants. Jpn. J. Phytopathol. 1990;56:532–540. doi: 10.3186/jjphytopath.56.532. [DOI] [Google Scholar]
- 27.Tsavkelova E., Oeser B., Oren-Young L., Israeli M., Sasson Y., Tudzynski B., Sharon A. Identification and Functional Characterization of Indole-3-acetamide-mediated IAA Biosynthesis in Plant-associated Fusarium Species. Fungal Genet. Biol. 2012;49:48–57. doi: 10.1016/j.fgb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Luo K., Rocheleau H., Qi P.-F., Zheng Y.-L., Zhao H.-Y., Ouellet T. Indole-3-Acetic Acid in Fusarium graminearum: Identification of Biosynthetic Pathways and Characterization of Physiological Effects. Fungal Biol. 2016;120:1135–1145. doi: 10.1016/j.funbio.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Maor R., Haskin S., Levi-Kedmi H., Sharon A. In Planta Production of Indole-3-Acetic Acid by Colletotrichum gloeosporioides f.sp. aeschynomene. Appl. Environ. Microbiol. 2004;70:1852–1854. doi: 10.1128/AEM.70.3.1852-1854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin C., Park J.-J., Gang D.R., Hulbert S.H. Characterization of a Tryptophan 2-Monooxygenase Gene from Puccinia graminis f.sp. tritici Involved in Auxin Biosynthesis and Rust Pathogenicity. Mol. Plant Microbe Interact. 2014;27:227–235. doi: 10.1094/MPMI-09-13-0289-FI. [DOI] [PubMed] [Google Scholar]
- 31.Pedras M.S.C., Minic Z., Thongbam P.D., Bhaskar V., Montaut S. Indolyl-3-Acetaldoxime Dehydratase from the Phytopathogenic Fungus Sclerotinia sclerotiorum: Purification, Characterization, and Substrate Specificity. Phytochemistry. 2010;71:1952–1962. doi: 10.1016/j.phytochem.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Mehmood A., Hussain A., Irshad M., Hamayun M., Iqbal A., Khan N. In Vitro Production of IAA by Endophytic Fungus Aspergillus awamori and Its Growth Promoting Activities in Zea mays. Symbiosis. 2019;77:225–235. doi: 10.1007/s13199-018-0583-y. [DOI] [Google Scholar]
- 33.Mehmood A., Hussain A., Irshad M., Hamayun M., Iqbal A., Tawab A., Khan N. Yucasin and Cinnamic Acid Inhibit IAA and Flavonoids Biosynthesis Minimizing Interaction Between Maize and Endophyte Aspergillus nomius. Symbiosis. 2020;81:149–160. doi: 10.1007/s13199-020-00690-z. [DOI] [Google Scholar]
- 34.Pons S., Fournier S., Chervin C., Bécard G., Rochange S., Frey N.F.D., Pagès V.P. Phytohormone Production by the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis. PLoS ONE. 2020;15:e0240886. doi: 10.1371/journal.pone.0240886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vadassery J., Ritter C., Venus Y., Camehl I., Varma A., Shahollari B., Novák O., Strnad M., Ludwig-Müller J., Oelmüller R. The Role of Auxins andCytokinins in the Mutualistic Interaction of Arabidopsis and Piriformospora indica. Mol. Plant Microbe Interact. 2008;21:1371–1383. doi: 10.1094/MPMI-21-10-1371. [DOI] [PubMed] [Google Scholar]
- 36.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. A Publ. Protein Soc. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oberhänsli T., Défago G., Haas D. Indole-3-acetic Acid (IAA) Synthesis in the Biocontrol Strain CHA0 of Pseudomonas fluorescens: Role of Tryptophan Side Chain Oxidase. Microbiology. 1991;137:2273–2279. doi: 10.1099/00221287-137-10-2273. [DOI] [PubMed] [Google Scholar]
- 40.Magie A.R., Wilson E.E., Kosuge T. Indoleacetamide as an Intermediate in the Synthesis of Indoleacetic Acid in Pseudomonas savastanoi. Science. 1963;141:1281–1282. doi: 10.1126/science.141.3587.1281. [DOI] [PubMed] [Google Scholar]
- 41.Kosuge T., Heskett M.G., Wilson E.E. Microbial Synthesis and Degradation of Indole-3-acetic Acid I.The Conversion of L-tryptophan to Indole-3-acetamide by an Enzyme System from Pseudomonas savastoi. J. Biol. Chem. 1966;241:3738–3744. doi: 10.1016/S0021-9258(18)99834-0. [DOI] [PubMed] [Google Scholar]
- 42.Hutzinger O., Kosuge T. Microbial Synthesis and Degradation of Indole-3-acetic Acid II. The Source of Oxygen in the Conversion of L-tryptophan to Indole-3-acetamide. Biochim. Biophys. Acta (BBA) Gen. Subj. 1967;136:389–391. doi: 10.1016/0304-4165(67)90085-2. [DOI] [PubMed] [Google Scholar]
- 43.Jahn L., Schafhauser T., Pan S., Weber T., Wohlleben W., Fewer D., Sivonen K., Flor L., van Pée K.-H., Caradec T., et al. Cyanodermella Asteris Sp. Nov. (Ostropales) from the Inflorescence Axis of Aster Tataricus. Mycotaxon. 2017;132:107–123. doi: 10.5248/132.107. [DOI] [Google Scholar]
- 44.Shao Y., Ho C.-T., Chin C.-K., Rosen R.T., Hu B., Qin G.-W. Triterpenoid Saponins from Aster auriculatus. Phytochemistry. 1997;44:337–340. doi: 10.1016/S0031-9422(96)00551-1. [DOI] [PubMed] [Google Scholar]
- 45.Shirota O., Morita H., Takeya K., Itokawa H., Iitaka Y. Cytotoxic Triterpenes from Aster tataricus. Nat. Med. 1997;51:170–172. [Google Scholar]
- 46.Shao Y., Ho C.T., Chin C.K., Poobrasert O., Yang S.W., Cordell G. Asterlingulatosides C and D, Cytotoxic Triterpenoid Saponins from Aster lingulatus. J. Nat. Prod. 1997;60:743–746. doi: 10.1021/np970080t. [DOI] [PubMed] [Google Scholar]
- 47.Xu H.-M., Zeng G.-Z., Zhou W.-B., He W.-J., Tan N.-H. Astins K–P, Six New Chlorinated Cyclopentapeptides from Aster tataricus. Tetrahedron. 2013;69:7964–7969. doi: 10.1016/j.tet.2013.07.006. [DOI] [Google Scholar]
- 48.Morita H., Nagashima S., Uchiumi Y., Kuroki O., Takeda K., Itokawa H. Cyclic Peptides from Higher Plants. XXVIII. Antitumor Activity and Hepatic Microsomal Biotransformation of Cyclic Pentapeptides, Astins, from Aster tataricus. Chem. Pharm. Bull. 1996 doi: 10.1248/cpb.44.1026. [DOI] [PubMed] [Google Scholar]
- 49.Schafhauser T., Jahn L., Kirchner N., Kulik A., Flor L., Lang A., Caradec T., Fewer D.P., Sivonen K., Berkel W.J.H., et al. Antitumor Astins Originate from the Fungal Endophyte Cyanodermella asteris Living Within the Medicinal Plant Aster tataricus. Proc. Natl. Acad. Sci. USA. 2019;116:26909–26917. doi: 10.1073/pnas.1910527116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi C.-L., Park H.-B., Lee J.S., Ryu S., Ryu C.-M. Inhibition of Primary Roots and Stimulation of Lateral Root Development in Arabidopsis thaliana by the Rhizobacterium Serratia marcescens 90-166 Is Through Both Auxin-dependent and -independent Signaling Pathways. Mol. Cells. 2010;29:251–258. doi: 10.1007/s10059-010-0032-0. [DOI] [PubMed] [Google Scholar]
- 51.López-Bucio J., Campos-Cuevas J.C., Hernández-Calderón E., Velásquez-Becerra C., Farías-Rodríguez R., Macías-Rodríguez L.I., Valencia-Cantero E. Bacillus megaterium Rhizobacteria Promote Growth and Alter Root-system Architecture Through an Auxin- and Ethylene-independent Signaling Mechanism in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2007;20:207–217. doi: 10.1094/MPMI-20-2-0207. [DOI] [PubMed] [Google Scholar]
- 52.Splivallo R., Fischer U., Göbel C., Feussner I., Karlovsky P. Truffles Regulate Plant Root Morphogenesis via the Production of Auxin and Ethylene. Plant Physiol. 2009;150:2018–2029. doi: 10.1104/pp.109.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilic N., Östin A., Cohen J.D. Differential Inhibition of Indole-3-acetic Acid and Tryptophan Biosynthesis by Indole Analogues. I. Tryptophan Dependent IAA Biosynthesis. Plant Growth Regul. 1999;27:57–62. doi: 10.1023/A:1006103902155. [DOI] [Google Scholar]
- 54.Tsugafune S., Mashiguchi K., Fukui K., Takebayashi Y., Nishimura T., Sakai T., Shimada Y., Kasahara H., Koshiba T., Hayashi K. Yucasin DF, a Potent and Persistent Inhibitor of Auxin Biosynthesis in Plants. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-14332-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishimura T., Hayashi K., Suzuki H., Gyohda A., Takaoka C., Sakaguchi Y., Matsumoto S., Kasahara H., Sakai T., Kato J., et al. Yucasin Is a Potent Inhibitor of YUCCA, a Key Enzyme in Auxin Biosynthesis. Plant J. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 56.Lanková M., Smith R.S., Pešek B., Kubeš M., Zažímalová E., Petrášek J., Hoyerová K. Auxin Influx Inhibitors 1-NOA, 2-NOA, and CHPAA Interfere with Membrane Dynamics in Tobacco Cells. J. Exp. Bot. 2010;61:3589–3598. doi: 10.1093/jxb/erq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teale W., Palme K. Naphthylphthalamic Acid and the Mechanism of Polar Auxin Transport. J. Exp. Bot. 2017;69:303–312. doi: 10.1093/jxb/erx323. [DOI] [PubMed] [Google Scholar]
- 58.Jahn L., Schafhauser T., Wibberg D., Rückert C., Winkler A., Kulik A., Weber T., Flor L., van Pée K.-H., Kalinowski J., et al. Linking Secondary Metabolites to Biosynthesis Genes in the Fungal Endophyte Cyanodermella asteris: The Anti-cancer Bisanthraquinone Skyrin. J. Biotechnol. 2017;257:233–239. doi: 10.1016/j.jbiotec.2017.06.410. [DOI] [PubMed] [Google Scholar]
- 59.Costacurta A., Keijers V., Vanderleyden J. Molecular Cloning and Sequence Analysis of an Azospirilium brasilense Indole-3-pyruvate Decarboxylase Gene. Mol. Gen. Genet. 1994;243:463–472. doi: 10.1007/BF00280477. [DOI] [PubMed] [Google Scholar]
- 60.Prinsen E., Costacurta A., Michiels K., Vanderleyden J., Van Onckelen H. Azospirillum brasilense Indole-3-acetic Acid Biosynthesis: Evidence for a Non-tryptophan Dependent Pathway. Mol. Plant Microbe Interact. 1993;6:609. doi: 10.1094/MPMI-6-609. [DOI] [Google Scholar]
- 61.Pedraza R.O., Ramírez-Mata A., Xiqui M.L., Baca B.E. Aromatic Amino Acid Aminotransferase Activity and Indole-3-acetic Acid Production by Associative Nitrogen-fixing Bacteria. Fems Microbiol. Lett. 2004;233:15–21. doi: 10.1016/j.femsle.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 62.Pollmann S., Neu D., Weiler E.W. Molecular Cloning and Characterization of an Amidase from Arabidopsis thaliana Capable of Converting Indole-3-Acetamide into the Plant Growth Hormone, Indole-3-Acetic Acid. Phytochemistry. 2003;62:293–300. doi: 10.1016/S0031-9422(02)00563-0. [DOI] [PubMed] [Google Scholar]
- 63.Numponsak T., Kumla J., Suwannarach N., Matsui K., Lumyong S. Biosynthetic Pathway and Optimal Conditions for the Production of Indole-3-acetic Acid by an Endophytic Fungus, Colletotrichum fructicola CMU-A109. PLoS ONE. 2018;13:e0205070. doi: 10.1371/journal.pone.0205070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson M., Riov J., Sharon A. Indole-3-acetic Acid Biosynthesis in Colletotrichum gloeosporioides f. sp. aeschynomene. Appl. Environ. Microbiol. 1998;64:5030–5032. doi: 10.1128/AEM.64.12.5030-5032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Follin A., Inzé D., Budar F., Genetello C., Van Montagu M., Schell J. Genetic Evidence That the Tryptophan 2-mono-oxygenase Gene of Pseudomonas savastonoi Is Functionally Equivalent to One of the T-DNA Genes Involved in Plant Tumour Formation by Agrobacterium tumefaciens. Mol. Gen. Genet. 1985;201:178–185. doi: 10.1007/BF00425657. [DOI] [Google Scholar]
- 66.Manulis S., Shafrir H., Epstein E., Lichter A., Barash I. Biosynthesis of Indole-3-acetic Acid via the Indole-3-acetamide Pathway in Streptomyces spp. Microbiology. 1994;140:1045–1050. doi: 10.1099/13500872-140-5-1045. [DOI] [PubMed] [Google Scholar]
- 67.Pace H.C., Brenner C. The Nitrilase Superfamily: Classification, Structure and Function. Genome Biol. 2001;2:reviews0001.1. doi: 10.1186/gb-2001-2-1-reviews0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feil H., Feil W.S., Chain P., Larimer F., DiBartolo G., Copeland A., Lykidis A., Trong S., Nolan M., Goltsman E., et al. Comparison of the Complete Genome Sequences of Pseudomonas syringae pv. syringae B728a and Pv. Tomato DC3000. Proc. Natl. Acad. Sci. USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiziak C., Conradt D., Stolz A., Mattes R., Klein J. Nitrilase from Pseudomonas fluorescens EBC191: Cloning and Heterologous Expression of the Gene and Biochemical Characterization of the Recombinant Enzyme. Microbiology. 2005;151:3639–3648. doi: 10.1099/mic.0.28246-0. [DOI] [PubMed] [Google Scholar]
- 70.Kobayashi M., Izui H., Nagasawa T., Yamada H. Nitrilase in Biosynthesis of the Plant Hormone Indole-3-acetic Acid from Indole-3-acetonitrile: Cloning of the Alcaligenes Gene and Site-directed Mutagenesis of Cysteine Residues. Proc. Natl. Acad. Sci. USA. 1993;90:247–251. doi: 10.1073/pnas.90.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagasawa T., Mauger J., Yamada H. A Novel Nitrilase, Arylacetonitrilase, of Alcaligenes faecalis JM3. Eur. J. Biochem. 1990;194:765–772. doi: 10.1111/j.1432-1033.1990.tb19467.x. [DOI] [PubMed] [Google Scholar]
- 72.Kobayashi M., Suzuki T., Fujita T., Masuda M., Shimizu S. Occurrence of Enzymes Involved in Biosynthesis of Indole-3-acetic Acid from Indole-3-acetonitrile in Plant-associated Bacteria, Agrobacterium and Rhizobium. Proc. Natl. Acad. Sci. USA. 1995;92:714–718. doi: 10.1073/pnas.92.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perley J.E., Stowe B.B. On the Ability of Taphrina deformans to Produce Indoleacetic Acid from Tryptophan by Way of Tryptamine. Plant Physiol. 1966;41:234–237. doi: 10.1104/pp.41.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao X., Lovett B., Fang W., St Leger R.J. Metarhizium robertsii Produces Indole-3-acetic Acid, Which Promotes Root Growth in Arabidopsis and Enhances Virulence to Insects. Microbiology. 2017;163:980–991. doi: 10.1099/mic.0.000494. [DOI] [PubMed] [Google Scholar]
- 75.Hartmann A., Singh M., Klingmüller W. Isolation and Characterization of Azospirillum Mutants Excreting High Amounts of Indoleacetic Acid. Can. J. Microbiol. 1983;29:916–923. doi: 10.1139/m83-147. [DOI] [Google Scholar]
- 76.Basse C.W., Lottspeich F., Steglich W., Kahmann R. Two Potential Indole-3-acetaldehyde Dehydrogenases in the Phytopathogenic Fungus Ustilago maydis. Eur. J. Biochem. 1996;242:648–656. doi: 10.1111/j.1432-1033.1996.0648r.x. [DOI] [PubMed] [Google Scholar]
- 77.Rao R.P., Hunter A., Kashpur O., Normanly J. Aberrant Synthesis of Indole-3-acetic Acid in Saccharomyces cerevisiae Triggers Morphogenic Transition, a Virulence Trait of Pathogenic Fungi. Genetics. 2010;185:211–220. doi: 10.1534/genetics.109.112854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ludwig-Müller J., Denk K., Cohen J.D., Quint M. An Inhibitor of Tryptophan-dependent Biosynthesis of Indole-3-acetic Acid Alters Seedling Development in Arabidopsis. J. Plant Growth Regul. 2010;29:242–248. doi: 10.1007/s00344-009-9128-1. [DOI] [Google Scholar]
- 79.Wright A.D., Sampson M.B., Neuffer M.G., Michalczuk L., Slovin J.P., Cohen J.D. Indole-3-acetic Acid Biosynthesis in the Mutant Maize Orange Pericarp, a Tryptophan Auxotroph. Science. 1991;254:998–1000. doi: 10.1126/science.254.5034.998. [DOI] [PubMed] [Google Scholar]
- 80.Normanly J., Cohen J.D., Fink G.R. Arabidopsis thaliana Auxotrophs Reveal a Tryptophan-independent Biosynthetic Pathway for Indole-3-acetic Acid. Proc. Natl. Acad. Sci. USA. 1993;90:10355–10359. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldsmith M.H.M. The Polar Transport of Auxin. Ann. Rev. Plant Physiol. 1977;28:439–478. doi: 10.1146/annurev.pp.28.060177.002255. [DOI] [Google Scholar]
- 82.Geisler M., Blakeslee J.J., Bouchard R., Lee O.R., Vincenzetti V., Bandyopadhyay A., Titapiwatanakun B., Peer W.A., Bailly A., Richards E.L., et al. Cellular Efflux of Auxin Catalyzed by the Arabidopsis MDR/PGP Transporter AtPGP1. Plant J. 2005;44:179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 83.Rojas-Pierce M., Titapiwatanakun B., Sohn E.J., Fang F., Larive C.K., Blakeslee J., Cheng Y., Cuttler S., Peer W.A., Murphy A.S., et al. Arabidopsis P-Glycoprotein 19 Participates in the Inhibition of Gravitropism by Gravacin. Chem. Biol. 2007;14:1366–1376. doi: 10.1016/j.chembiol.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 84.Nagashima A., Uehara Y., Sakai T. The ABC Subfamily B Auxin Transporter AtABCB19 Is Involved in the Inhibitory Effects of N-1-naphthyphthalamic Acid on the Phototropic and Gravitropic Responses of Arabidopsis Hypocotyls. Plant Cell Physiol. 2008;49:1250–1255. doi: 10.1093/pcp/pcn092. [DOI] [PubMed] [Google Scholar]
- 85.Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell. 1991;3:677–684. doi: 10.2307/3869249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reinhardt D., Mandel T., Kuhlemeier C. Auxin Regulates the Initiation and Radial Position of Plant Lateral Organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. The PIN Auxin Efflux Facilitator Network Controls Growth and Patterning in Arabidopsis Roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 88.Mattsson J., Sung Z.R., Berleth T. Responses of Plant Vascular Systems to Auxin Transport Inhibition. Development. 1999;126:2979–2991. doi: 10.1242/dev.126.13.2979. [DOI] [PubMed] [Google Scholar]
- 89.Bailly A., Sovero V., Vincenzetti V., Santelia D., Bartnik D., Koenig B.W., Mancuso S., Martinoia E., Geisler M. Modulation of P-Glycoproteins by Auxin Transport Inhibitors Is Mediated by Interaction with Immunophilins. J. Biol. Chem. 2008;283:21817–21826. doi: 10.1074/jbc.M709655200. [DOI] [PubMed] [Google Scholar]
- 90.Rincón A., Priha O., Sotta B., Bonnet M., Le Tacon F. Comparative Effects of Auxin Transport Inhibitors on Rhizogenesis and Mycorrhizal Establishment of Spruce Seedlings Inoculated with Laccaria bicolor. Tree Physiol. 2003;23:785–791. doi: 10.1093/treephys/23.11.785. [DOI] [PubMed] [Google Scholar]
- 91.Felten J., Kohler A., Morin E., Bhalerao R.P., Palme K., Martin F., Ditengou F.A., Legué V. The Ectomycorrhizal Fungus Laccaria bicolor Stimulates Lateral Root Formation in Poplar and Arabidopsis Through Auxin Transport and Signaling. Plant Physiol. 2009;151:1991–2005. doi: 10.1104/pp.109.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Niemi K., Vuorinen T., Ernstsen A. Ectomycorrhizal Fungi and Exogenous Auxins Influence Root and Mycorrhiza Formation of Scots Pine Hypocotyl Cuttings In Vitro. Tree Physiol. 2002;22:1231–1239. doi: 10.1093/treephys/22.17.1231. [DOI] [PubMed] [Google Scholar]
- 93.Fitze D., Wiepning A., Kaldorf M., Ludwig-Müller J. Auxins in the Development of an Arbuscular Mycorrhizal Symbiosis in Maize. J. Plant Physiol. 2005;162:1210–1219. doi: 10.1016/j.jplph.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 94.Salas-Marina M.A., Silva-Flores M.A., Cervantes-Badillo M.G., Rosales-Saavedra M.T., Islas-Osuna M.A., Casas-Flores S. The Plant Growth-promoting Fungus Aspergillus ustus Promotes Growth and Induces Resistance Against Different Lifestyle Pathogens in Arabidopsis thaliana. J. Microbiol. Biotechnol. 2011;21:686–696. doi: 10.4014/jmb.1101.01012. [DOI] [PubMed] [Google Scholar]
- 95.Sukumar P., Legué V., Vayssières A., Martin F., Tuskan G.A., Kalluri U.C. Involvement of Auxin Pathways in Modulating Root Architecture During Beneficial Plant–microorganism Interactions. Plant Cell Environ. 2013;36:909–919. doi: 10.1111/pce.12036. [DOI] [PubMed] [Google Scholar]
- 96.Contesto C., Milesi S., Mantelin S., Zancarini A., Desbrosses G., Varoquaux F., Bellini C., Kowalczyk M., Touraine B. The Auxin-signaling Pathway Is Required for the Lateral Root Response of Arabidopsis to the Rhizobacterium Phyllobacterium brassicacearum. Planta. 2010;232:1455–1470. doi: 10.1007/s00425-010-1264-0. [DOI] [PubMed] [Google Scholar]
- 97.Glick B.R., Cheng Z., Czarny J., Duan J. Promotion of Plant Growth by ACC Deaminase-producing Soil Bacteria. Eur. J. Plant Pathol. 2007;119:329–339. doi: 10.1007/s10658-007-9162-4. [DOI] [Google Scholar]
- 98.Marschner H., Dell B. Nutrient Uptake in Mycorrhizal Symbiosis. Plant Soil. 1994;159:89–102. doi: 10.1007/BF00000098. [DOI] [Google Scholar]
- 99.Slankis V. Soil Factors Influencing Formation of Mycorrhizae. Annu Rev. Phytopathol. 1974;12:437–457. doi: 10.1146/annurev.py.12.090174.002253. [DOI] [Google Scholar]
- 100.Hutner S.H., Provasoli L., Schatz A., Haskins C.P. Some Approaches to the Study of the Role of Metals in the Metabolism of Microorganisms. PAPHS. 1950;94:152–170. [Google Scholar]
- 101.Ilic N., Normanly J., Cohen J.D. Quantification of Free Plus Conjugated Indoleacetic Acid in Arabidopsis Requires Correction for the Nonenzymatic Conversion of Indolic Nitriles. Plant Physiol. 1996;111:781–788. doi: 10.1104/pp.111.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lenhard W., Lenhard A. Calculation of Effect Sizes. Dettelbach (Germany): Psychometrica. [(accessed on 12 January 2021)];2016 Available online: https://www.psychometrica.de/effect_size.html.
- 103.Lakens D. Calculating and Reporting Effect Sizes to Facilitate Cumulative Science: A Practical Primer for T-Tests and ANOVAs. Front. Psychol. 2013;4 doi: 10.3389/fpsyg.2013.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this study are available in this paper and in its supplementary data published online.